Abstract
Background and Objectives: Several studies inspected the impact of P2X7 polymorphisms on individual susceptibility to tuberculosis (TB), but the findings are still controversial and inconclusive. To achieve a more precise estimation, we conducted a meta-analysis of all eligible studies on the association between P2X7 polymorphisms and TB risk. Materials and Methods: Relevant studies were identified by searching the PubMed, Web of Science, Scopus and Google scholar databases up to November 2018. Twenty-four full-text articles were included in our meta-analysis. The strength of association between P2X7 polymorphisms and TB risk was evaluated by odds ratios (ORs) and 95% confidence intervals (95% CIs) under five genetic models. Results: The findings of this meta-analysis revealed that the rs3751143 variant significantly increased the risk of TB in heterozygous codominant (OR = 1.44, 95%CI = 1.17–1.78, p = 0.0006, AC vs. AA), homozygous codominant (OR = 1.87, 95% CI = 1.40–2.49, p = 0.0004, CC vs. AA), dominant (OR = 1.50, 95% CI = 1.22–1.85, p = 0.0002, AC + CC vs. AA), recessive (OR = 1.61, 95% CI = 1.25–2.07, p = 0.001, CC vs. AC + AA), and allele (OR = 1.41, 95% CI = 1.19–1.67, p < 0.0001, C vs. A) genetic models. Stratified analysis showed that rs3751143 increased the risk of pulmonary tuberculosis (PTB) and extrapulmonary tuberculosis (EPTB) in all genetic models. Furthermore, the rs3751143 increased risk of TB in the Asian population. The findings did not support an association between the rs2393799, rs1718119, rs208294, rs7958311, and rs2230911 polymorphisms of P2X7 and TB risk. Conclusions: The findings of this meta-analysis suggest that P2X7 rs3751143 polymorphism may play a role in susceptibility to TB in the Asian population. More well-designed studies are required to elucidate the exact role of P2X7 polymorphisms on TB development.
Keywords: P2X7, polymorphism, tuberculosis, meta-analysis
1. Introduction
Tuberculosis (TB) is a chronic infectious disease caused by the bacillus Mycobacterium tuberculosis (MTB). It remains a serious public global health problem. According to the World Health Organization (WHO) report, there were an estimated 10.4 million new cases of TB worldwide and approximately 1.3 million deaths in 2016 [1]. Approximately one-third of the general population is currently infected with Mtb, and nearly 5–10% of these infected individuals will progress to active TB [2,3]. Mounting evidence has proposed that host genetic factors play an important role in determining inter-individual difference in susceptibility to TB [4,5,6].
The human P2X7 gene is mapped to chromosome 12 (12q24.31). It encodes cell-surface nucleotide receptors called P2X7 receptor (P2X7R) [7]. The P2X7R, a ligand-gated cation channel, is highly expressed on macrophages and other immune cells [8]. Activation of P2X7R by extracellular adenosine triphosphate (eATP) causes immediate opening of a cation selective channel, permitting the influx of Ca2+ and Na+ and the efflux of K+ [9]. In M. tuberculosis infection, activation of the P2X7R promotes a range of signaling cascades leading to the apoptosis of MTB-infected macrophages [10,11].
The P2X7R gene is indeed polymorphic. The precise correlation between the P2X7 polymorphisms and susceptibility to TB is not completely documented. Several single nucleotide polymorphisms (SNPs) have been revealed that affect the function of this receptor which cause P2X7R loss-of-function (LOF) or gain-of-function (GOF) [8]. Many studies have inspected the association between P2X7 polymorphisms and risk of tuberculosis in various populations, but the findings were inconsistent [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. So we conducted an updated meta-analysis of all available eligible case-control studies published to date, focusing on the association between P2X7 polymorphisms and tuberculosis risk.
2. Methods
2.1. Literature Search
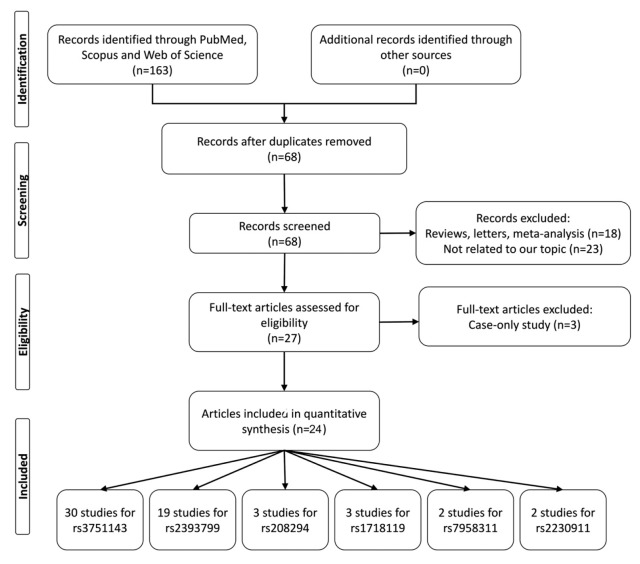
The PubMed, Web of Science, Scopus, and Google scholar databases for all potentially eligible research articles up to November 2018 on the relationship between P2X7 polymorphisms and TB risk were searched. The search key words used were “P2X7 or P2X7R” and “tuberculosis” and “polymorphism or variant”. Figure 1 shows the process of recognizing eligible studies. The inclusion criteria were as follows: case-control studies focusing on the association between P2X7 polymorphisms and TB risk; the frequencies distribution of alleles and genotypes in patients and controls can be extracted. The exclusion criteria were studies that are not associated with P2X7 polymorphisms and TB risk; overlapping data, conference papers, reviews, meta-analyses; no sufficient data reported.
Figure 1.
Flow chart illustrates the detailed study selection process of this meta-analysis.
2.2. Data Extraction
Two investigators independently inspected and evaluated the articles for eligibility according to inclusion and exclusion criteria. The following data were recorded from the selected studies such as the first author’s name, publication year, ethnicity, genotyping methods, genotype and allelic profile, as well as the source of controls.
2.3. Statistical Analysis
The chi-square test was used to check whether genotypes within the controls conformed to the Hardy-Weinberg equilibrium (HWE). We calculated the pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs) to assess the association between the P2X7 polymorphisms and TB susceptibility. The significance of the pooled OR was determined by the Z-test, and a p-value less than 0.05 was considered significant. Heterogeneity between the studies was estimated by Q statistic and the I2 test. p < 0.10 designated significant heterogeneity. If heterogeneity did not exist, a fixed-effects model was used to calculate the pooled ORs; otherwise, a random-effects model was utilized.
Publication bias was inspected with the funnel plot and an asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was further measured using Egger’s linear regression test. p value < 0.05 was considered a significant publication bias. Sensitivity analysis was done by neglecting each study in turn to assess the quality and consistency of the results. All statistical analyses were executed using STATA v14.1 software (College Station, TX, USA).
3. Results
3.1. Study Characteristics
Through a comprehensive literature search and selection based on the inclusion criteria, 59 relevant case-control studies from 24 selected articles [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35] were included in the pooled analysis. There were 30 studies with 5247 cases and 7614 controls on rs3751143 (1513A > C), 19 studies with 3235 cases and 4685 controls on rs2393799 (−762 C > T), 3 studies with 2185 cases and 2107 controls on rs1718119 (Thr348Ala), 3 studies with 1994 cases and 2037 controls on rs208294 (His155Tyr), 2 studies with 2000 cases and 2006 controls on rs7958311, 2 studies with 1853 cases and 1797 controls on rs2230911 included into meta-analysis. The main characteristics of included studies are shown in Table 1.
Table 1.
Characteristics of all studies included in the meta-analysis.
| First Author | Year | Country | Ethnicity | TB | Source of Control | Genotyping Method | Case/Control | Cases | Controls | HWE (p) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3751143 A > C | AA | AC | CC | A | C | AA | AC | CC | A | C | ||||||||
| Amiri | 2018 | Iran | Asian | PTB | PB | PCR-RFLP | 100/100 | 76 | 21 | 3 | 173 | 27 | 40 | 58 | 2 | 138 | 62 | <0.001 |
| Ben-Selma | 2011 | Tunisia | African | PTB | HB | PCR-RFLP | 168/150 | 130 | 34 | 4 | 294 | 42 | 104 | 40 | 6 | 248 | 52 | 0.395 |
| Ben-Selma | 2011 | Tunisia | African | EPTB | HB | PCR-RFLP | 55/150 | 19 | 23 | 13 | 61 | 49 | 104 | 40 | 6 | 248 | 52 | 0.395 |
| Chaudhary | 2018 | India | Asian | PTB | PB | ARMS-PCR | 145/247 | 63 | 73 | 9 | 199 | 91 | 141 | 95 | 11 | 377 | 117 | 0.315 |
| Chaudhary | 2018 | India | Asian | EPTB | PB | ARMS-PCR | 100/247 | 42 | 42 | 16 | 126 | 74 | 141 | 95 | 11 | 377 | 117 | 0.315 |
| De | 2017 | India | Asian | PTB | PB | PCR-RFLP | 56/60 | 26 | 18 | 12 | 70 | 42 | 36 | 21 | 3 | 93 | 27 | 0.978 |
| Fernando | 2007 | Southeast Asia | Asian | PTB | PB | TaqMan | 56/167 | 34 | 17 | 5 | 85 | 27 | 105 | 55 | 7 | 265 | 69 | 0.952 |
| Fernando | 2007 | Southeast Asia | Asian | EPTB | PB | TaqMan | 30/167 | 9 | 17 | 4 | 35 | 25 | 105 | 55 | 7 | 265 | 69 | 0.952 |
| Fernando | 2007 | Australia | Caucasian | PTB | PB | TaqMan | 49/102 | 28 | 21 | 0 | 77 | 21 | 64 | 34 | 4 | 162 | 42 | 0.845 |
| Fernando | 2007 | Australia | Caucasian | EPTB | PB | TaqMan | 50/102 | 18 | 28 | 4 | 64 | 36 | 64 | 34 | 4 | 162 | 42 | 0.845 |
| Li | 2002 | Gambia | African | PTB | HB | PCR-RFLP | 325/297 | 261 | 58 | 6 | 580 | 70 | 256 | 37 | 4 | 549 | 45 | 0.057 |
| Mokrousov | 2008 | Russia | Caucasian | PTB | HB | PCR-RFLP | 188/126 | 120 | 59 | 9 | 299 | 77 | 96 | 27 | 3 | 219 | 33 | 0.511 |
| Nino-Moreno | 2007 | México | Mixed | PTB | HB | PCR-RFLP | 94/110 | 53 | 33 | 8 | 139 | 49 | 70 | 38 | 2 | 178 | 42 | 0.215 |
| Ozdemir | 2014 | Turkey | Asian | PTB | PB | PCR-RFLP | 71/160 | 44 | 18 | 9 | 106 | 36 | 76 | 63 | 21 | 215 | 105 | 0.176 |
| Ozdemir | 2014 | Turkey | Asian | EPTB | PB | PCR-RFLP | 89/160 | 47 | 34 | 8 | 128 | 50 | 76 | 63 | 21 | 215 | 105 | 0.176 |
| Sambasivan | 2010 | India | Asian | PTB | HB | PCR-RFLP | 156/100 | 89 | 55 | 12 | 233 | 79 | 71 | 21 | 8 | 163 | 37 | 0.002 |
| Shamsi | 2016 | Iran | Asian | PTB | HB | PCR-RFLP | 100/100 | 33 | 66 | 1 | 132 | 68 | 83 | 16 | 1 | 182 | 18 | 0.817 |
| Sharma | 2010 | India | Asian | PTB | PB | T-ARMS-PCR | 181/177 | 102 | 75 | 4 | 279 | 83 | 126 | 48 | 3 | 300 | 54 | 0.515 |
| Sharma | 2010 | India | Asian | EPTB | PB | T-ARMS-PCR | 23/177 | 8 | 13 | 2 | 29 | 17 | 126 | 48 | 3 | 300 | 54 | 0.515 |
| Singla | 2012 | India | Asian | PTB | PB | PCR-RFLP | 286/392 | 162 | 112 | 12 | 436 | 136 | 258 | 123 | 11 | 639 | 145 | 0.420 |
| Singla | 2012 | India | Asian | EPTB | PB | PCR-RFLP | 71/392 | 45 | 22 | 4 | 112 | 30 | 258 | 123 | 11 | 639 | 145 | 0.420 |
| Souza de Lima | 2016 | Brazil | South American | PTB | HB | TaqMan | 288/287 | 170 | 95 | 23 | 435 | 141 | 184 | 89 | 14 | 457 | 117 | 0.450 |
| Taype | 2010 | Peru | Caucasian | PTB | HB | PCR-RFLP | 498/513 | 352 | 130 | 16 | 834 | 162 | 347 | 149 | 17 | 843 | 183 | 0.838 |
| Taype | 2010 | Peru | Caucasian | EPTB | HB | PCR-RFLP | 121/513 | 82 | 37 | 2 | 201 | 41 | 347 | 149 | 17 | 843 | 183 | 0.838 |
| Tekin | 2010 | Turkey | Caucasian | EPTB | HB | PCR-RFLP | 74/192 | 39 | 28 | 7 | 106 | 42 | 141 | 46 | 5 | 328 | 56 | 0.595 |
| Velayati | 2013 | Iran | Asian | PTB | HB | PCR- RFLP | 79/50 | 42 | 35 | 2 | 119 | 39 | 37 | 12 | 1 | 86 | 14 | 0.981 |
| Wu | 2015 | China | Asian | PTB | PB | PCR-RFLP | 103/87 | 33 | 49 | 21 | 115 | 91 | 51 | 27 | 9 | 129 | 45 | 0.075 |
| Xiao | 2009 | China | Asian | PTB | HB | PCR-RFLP | 41/384 | 21 | 18 | 2 | 60 | 22 | 221 | 119 | 44 | 561 | 207 | <0.001 |
| Xiao | 2009 | China | Asian | EPTB | HB | PCR-RFLP | 55/384 | 30 | 19 | 6 | 79 | 31 | 221 | 119 | 44 | 561 | 207 | <0.001 |
| Zheng | 2017 | China | Asian | PTB | PB | TaqMan | 1595/1521 | 972 | 551 | 72 | 2495 | 695 | 900 | 544 | 77 | 2344 | 698 | 0.655 |
| rs2393799 C > T |
CC | CT | TT | C | T | CC | CT | TT | C | T | ||||||||
| Amiri | 2018 | Iran | Asian | PTB | PB | PCR-RFLP | 100/100 | 8 | 88 | 4 | 104 | 96 | 4 | 95 | 1 | 103 | 97 | <0.001 |
| Bahari | 2013 | Iran | Asian | PTB | PB | ARMS-PCR | 150/150 | 71 | 54 | 25 | 196 | 104 | 104 | 40 | 6 | 248 | 52 | 0.395 |
| Ben-Selma | 2011 | Tunisia | African | PTB | HB | ARMS-PCR | 168/150 | 16 | 57 | 95 | 89 | 247 | 14 | 51 | 85 | 79 | 221 | 0.130 |
| Ben-Selma | 2011 | Tunisia | African | EPTB | HB | ARMS-PCR | 55/150 | 4 | 15 | 36 | 23 | 87 | 14 | 51 | 85 | 79 | 221 | 0.130 |
| Chaudhary | 2018 | India | Asian | PTB | PB | ARMS-PCR | 145/247 | 62 | 67 | 16 | 191 | 99 | 101 | 111 | 35 | 313 | 181 | 0.614 |
| Chaudhary | 2018 | India | Asian | EPTB | PB | ARMS-PCR | 100/247 | 44 | 48 | 8 | 136 | 64 | 101 | 111 | 35 | 313 | 181 | 0.614 |
| Li | 2002 | Gambia | African | PTB | HB | PCR-RFLP | 323/347 | 23 | 118 | 182 | 164 | 482 | 44 | 140 | 163 | 228 | 466 | 0.111 |
| Mokrousov | 2008 | Russia | Caucasian | PTB | HB | ARMS | 190/127 | 86 | 87 | 17 | 259 | 121 | 65 | 46 | 16 | 176 | 78 | 0.093 |
| Nino-Moreno | 2007 | México | Mixed | PTB | HB | ARMS | 92/110 | 8 | 32 | 52 | 48 | 136 | 15 | 44 | 51 | 74 | 146 | 0.275 |
| Sambasivan | 2010 | India | Asian | PTB | HB | PCR-RFLP | 156/100 | 38 | 88 | 30 | 164 | 148 | 15 | 49 | 36 | 79 | 121 | 0.801 |
| Shamsi | 2016 | Iran | Asian | PTB | HB | PCR-RFLP | 100/100 | 1 | 99 | 0 | 101 | 99 | 6 | 93 | 1 | 105 | 95 | <0.001 |
| Singla | 2012 | India | Asian | PTB | PB | ARMS | 286/392 | 143 | 115 | 28 | 401 | 171 | 231 | 143 | 18 | 605 | 179 | 0.485 |
| Singla | 2012 | India | Asian | EPTB | PB | ARMS | 71/392 | 40 | 25 | 6 | 105 | 37 | 231 | 143 | 18 | 605 | 179 | 0.485 |
| Songane | 2012 | Indonesia | Asian | PTB | PB | MassARRAY | 842/844 | 181 | 413 | 248 | 775 | 909 | 177 | 412 | 255 | 766 | 922 | <0.001 |
| Velayati | 2013 | Iran | Asian | PTB | HB | ARMS | 79/50 | 10 | 67 | 2 | 87 | 71 | 3 | 47 | 0 | 53 | 47 | <0.001 |
| Wu | 2015 | China | Asian | PTB | PB | PCR-RFLP | 103/87 | 35 | 47 | 21 | 117 | 89 | 9 | 30 | 48 | 48 | 126 | 0.202 |
| Xiao | 2009 | China | Asian | PTB | HB | ARMS | 38/384 | 23 | 11 | 4 | 57 | 19 | 208 | 135 | 41 | 551 | 217 | 0.009 |
| Xiao | 2009 | China | Asian | EPTB | HB | ARMS | 58/384 | 40 | 12 | 6 | 92 | 24 | 208 | 135 | 41 | 551 | 217 | 0.009 |
| Zhou | 2018 | China | Asain | EPTB | HB | Mass Spectrometry | 179/324 | 81 | 77 | 21 | 239 | 119 | 122 | 143 | 59 | 387 | 261 | 0.137 |
| rs1718119 G > A |
GG | AG | AA | G | A | GG | AG | AA | G | A | ||||||||
| Bahari | 2013 | Iran | Asian | PTB | PB | T-ARMS-PCR | 150/150 | 63 | 72 | 15 | 198 | 102 | 66 | 69 | 15 | 201 | 99 | 0.622 |
| Zheng | 2017 | China | Asian | PTB | PB | TaqMan | 1568/1454 | 1090 | 440 | 38 | 2620 | 516 | 978 | 417 | 59 | 2373 | 535 | 0.087 |
| Zhu | 2016 | China | Asian | PTB | HB | MassARRAY | 467/503 | 372 | 91 | 4 | 835 | 99 | 412 | 89 | 2 | 913 | 93 | 0.222 |
| rs208294 G > A |
GG | AG | AA | G | A | GG | AG | AA | G | A | ||||||||
| Chaudhary | 2018 | India | Asian | Mixed | PB | PCR-RFLP | 245/246 | 56 | 147 | 42 | 259 | 231 | 49 | 143 | 54 | 241 | 251 | 0.011 |
| Zheng | 2017 | China | Asian | PTB | PB | TaqMan | 1570/1467 | 597 | 732 | 241 | 1926 | 1214 | 578 | 679 | 210 | 1835 | 1099 | 0.642 |
| Zhou | 2018 | China | Asian | EPTB | HB | Mass Spectrometry |
179/324 | 22 | 80 | 77 | 124 | 234 | 70 | 145 | 109 | 285 | 363 | 0.099 |
| rs7958311 G > A |
GG | AG | AA | G | A | GG | AG | AA | G | A | ||||||||
| Zheng | 2017 | China | Asian | PTB | PB | TaqMan | 1533/1503 | 402 | 797 | 334 | 1601 | 1465 | 396 | 775 | 332 | 1567 | 1439 | 0.199 |
| Zhu | 2016 | China | Asian | PTB | HB | MassARRAY | 467/503 | 114 | 215 | 138 | 443 | 491 | 137 | 262 | 104 | 536 | 470 | 0.300 |
| rs2230911 C > G |
CC | CG | GG | C | G | CC | CG | GG | C | G | ||||||||
| Souza de Lima | 2016 | Brazil | South American | PTB | HB | TaqMan | 288/288 | 170 | 95 | 23 | 435 | 141 | 193 | 89 | 6 | 475 | 101 | 0.245 |
| Zheng | 2017 | China | Asian | PTB | PB | TaqMan | 1565/1509 | 1029 | 482 | 54 | 2540 | 590 | 997 | 467 | 45 | 2461 | 557 | 0.274 |
List of abbreviations: PTB: Pulmonary Tuberculosis; TB: Tuberculosis; EPTB: Extrapulmonary Tuberculosis; PCR-RFLP: PCR-Restriction fragment length polymorphism; ARMS-PCR: Amplification-refractory mutation system-PCR; TaqMan: probes used in quantitative PCR; T-ARMS-PCR: Multiplex Tetra-Primer Amplification Refractory Mutation System-PCR; MassARRAY: Non-fluorescent detection platform utilizing mass spectrometry to accurately measure PCR-derived amplicons.
3.2. Main Analysis Results
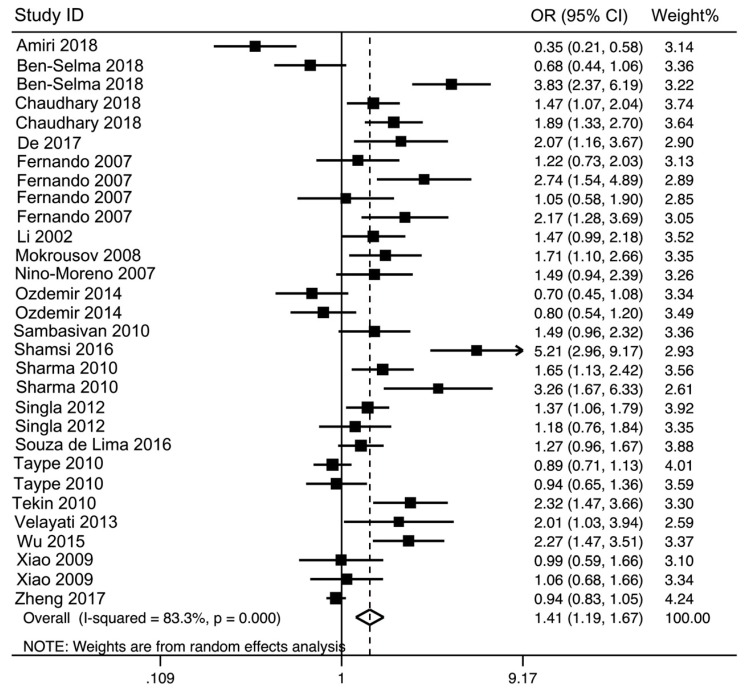
The Forest plots were applied to show meta-analysis results for each genetic model. Overall, the rs3751143 variant significantly increased the risk of TB in heterozygous codominant (OR = 1.44, 95% CI = 1.17–1.78, p = 0.0006, AC vs. AA), homozygous codominant (OR = 1.87, 95% CI = 1.40–2.49, p = 0.0004, CC vs. AA), dominant (OR = 1.50, 95% CI = 1.22–1.85, p = 0.0002, AC + CC vs. AA), recessive (OR = 1.61, 95% CI = 1.25–2.07, p = 0.001, CC vs. AC + AA), and allele (OR = 1.41, 95% CI = 1.19–1.67, p < 0.0001, C vs. A) genetic models (Table 2 and Figure 2).
Table 2.
The pooled ORs and 95% CIs for the association between P2X7 polymorphisms and tuberculosis susceptibility.
| Polymorphism | No. | Genetic Model | Association Test | Heterogeneity | Publication Bias Tests | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | Z | P | χ2 | I2 (%) | P | Egger’s Test p-Value |
Begg’s Test p-Value |
|||
| rs3751143 | 30 | AC vs. AA | 1.44 (1.17–1.78) | 3.42 | 0.0006 | 158.86 | 81.7 | 0.000 | 0.016 | 0.016 |
| CC vs. AA | 1.87 (1.40–2.49) | 4.26 | 0.0004 | 61.79 | 54.7 | 0.000 | 0.002 | 0.047 | ||
| AC + CC vs. AA | 1.50 (1.22–1.85) | 3.78 | 0.0002 | 178.85 | 83.8 | 0.000 | 0.007 | 0.018 | ||
| CC vs. AC + AA | 1.61 (1.25–2.07) | 3.65 | 0.001 | 50.79 | 44.9 | 0.005 | 0.006 | 0.051 | ||
| C vs. A | 1.41 (1.19–1.67) | 3.97 | <0.0001 | 173.41 | 83.3 | 0.000 | 0.006 | 0.066 | ||
| rs2393799 | 19 | CT v CC | 1.00 (0.83–1.20) | 0.01 | 0.989 | 32.34 | 44.3 | 0.020 | 0.460 | 0.753 |
| TT vs. CC | 0.99 (0.68–1.44) | 0.04 | 0.965 | 73.55 | 75.5 | 0.000 | 0.935 | 0.510 | ||
| CT + TT vs. CC | 0.97 (0.77–1.23) | 0.22 | 0.825 | 58.41 | 69.2 | 0.000 | 0.557 | 0.649 | ||
| TT vs. CT + CC | 0.99 (0.74–1.32) | 0.08 | 0.938 | 71.34 | 74.8 | 0.000 | 0.962 | 0.680 | ||
| T vs. C | 0.98 (0.83–1.17) | 0.20 | 0.844 | 95.15 | 81.1 | 0.000 | 0.657 | 0.763 | ||
| rs1718119 | 3 | AG vs. GG | 0.99 (0.86–1.13) | 0.16 | 0.88 | 1.13 | 0 | 0.57 | 0.308 | 0.602 |
| AA vs. GG | 0.70 (0.49–0.99) | 1.99 | 0.05 | 3.56 | 44 | 0.17 | 0.136 | 0.117 | ||
| AG + AA vs. GG | 0.96 (0.84–1.09) | 0.68 | 0.50 | 2.22 | 10 | 0.33 | 0.312 | 0.602 | ||
| AA vs. AG + GG | 0.70 (0.49–1.00) | 1.98 | 0.05 | 3.24 | 38 | 0.20 | 0.141 | 0.117 | ||
| G vs. A | 0.93 (0.83–1.04) | 1.24 | 0.21 | 3.49 | 43 | 0.17 | 0.242 | 0.602 | ||
| rs208294 | 3 | AG vs. GG | 1.03 (0.93–1.23) | 0.89 | 0.37 | 3.77 | 47 | 0.15 | 0.694 | 0.602 |
| AA vs. GG | 1.18 (0.69–2.02) | 0.61 | 0.54 | 8.99 | 78 | 0.010 | 0.900 | 0.602 | ||
| AG + AA vs. GG | 1.16 (0.80–1.68) | 0.76 | 0.45 | 6.50 | 69 | 0.04 | 0.751 | 0.602 | ||
| AA vs. AG + GG | 1.08 (0.78–1.49) | 0.47 | 0.64 | 5.60 | 64 | 0.06 | 0.904 | 0.602 | ||
| A vs. G | 1.09 (0.85–1.40) | 0.70 | 0.49 | 8.82 | 77 | 0.01 | 0.860 | 0.602 | ||
| rs7958311 | 2 | AG vs. GG | 1.01 (0.87–1.17) | 0.09 | 0.93 | 0.02 | 0 | 0.88 | ||
| AA vs. GG | 1.23 (0.77–1.95) | 0.87 | 0.38 | 5.15 | 81 | 0.02 | ||||
| AG + AA vs. GG | 0.81 (0.50–1.31) | 0.87 | 0.38 | 8.09 | 88 | 0.004 | ||||
| AA vs. AG + GG | 1.24 (0.76–2.01) | 0.87 | 0.38 | 8.09 | 88 | 0.004 | ||||
| A vs. G | 1.11 (0.88–1.40) | 0.87 | 0.38 | 5.18 | 81 | 0.02 | ||||
| rs2230911 | 2 | CG vs. CC | 1.03 (0.90–1.19) | 0.42 | 0.67 | 0.95 | 0 | 0.33 | ||
| GG vs. CC | 2.10 (0.58–7.66) | 1.13 | 0.26 | 6.65 | 85 | 0.010 | ||||
| CG + GG vs. CC | 1.01 (0.88–1.16) | 0.13 | 0.89 | 0.28 | 0 | 0.60 | ||||
| GG vs. CG + CC | 2.03 (0.60–6.94) | 1.13 | 0.26 | 6.12 | 84 | 0.01 | ||||
| G vs. C | 1.22 (0.83–1.80) | 1.03 | 0.30 | 6.09 | 84 | 0.01 | ||||
List of Abbreviations: OR: Odds Ratio; CI: Confidence interval Z: Z-score; P: Probability; χ2: χ2 test; I2: I2 value.
Figure 2.
The forest plot for association between P2X7 rs3751143 polymorphism and tuberculosis risk under allelic genetic model (C vs. A).
No significant association was found between P2X7 rs2393799, rs1718119, rs208294, rs7958311, and rs2230911 polymorphisms and TB risk (Table 2).
3.3. Subgroup Analysis Results
Stratified analysis was achieved (Table 3). The findings proposed that rs3751143 polymorphism increased the risk of pulmonary tuberculosis (PTB) and extrapulmonary tuberculosis (EPTB) in all genetic models. Besides, this polymorphism only contributes to the risk of TB in the Asian population, but not in the Caucasian population (Table 3). The rs2393799 polymorphism was not associated with the risk of TB in the Asian population (Table 3).
Table 3.
Stratified analysis of P2X7 polymorphisms and tuberculosis risk.
| Parameters | No. | AC vs. AA | CC vs. AA | AC + CC vs. AA | CC vs. AC + AA | C vs. A | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | ||
| rs3751143 | |||||||||||
| Tuberculosis | |||||||||||
| PTB | 21 | 1.35 (1.05–1.74) | 0.020 | 1.50 (1.10–2.04) | 0.010 | 1.39 (1.09–1.78) | 0.009 | 1.34 (1.04–1.73) | 0.020 | 1.31 (1.09–1.58) | 0.004 |
| EPTB | 9 | 1.68 (1.17–2.42) | 0.005 | 2.62 (1.19–5.78) | 0.020 | 1.84 (1.21–2.79) | 0.004 | 2.05 (1.07–3.93) | 0.030 | 1.67 (1.16–2.42) | 0.006 |
| Ethnicities | |||||||||||
| Asian | 19 | 1.48 (1.09–2.00) | 0.010 | 1.70 (1.17–2.48) | 0.006 | 1.53 (1.13–2.06) | 0.006 | 1.47 (1.07–2.00) | 0.020 | 1.57 (1.22–2.02) | 0.0005 |
| Caucasian | 6 | 1.47 (0.99–2.17) | 0.05 | 1.56 (0.70 − 3.51) | 0.28 | 1.49 (0.98–2.26) | 0.06 | 1.36 (0.70–2.66) | 0.37 | 1.37 (0.96–1.97) | 0.09 |
| African | 3 | 1.45 (0.66–3.19) | 0.36 | 2.16 (0.33–13.95) | 0.42 | 1.60 (0.62–4.13) | 0.33 | 1.89 (0.41–8.81) | 0.42 | 1.56 (0.62–3.94) | 0.35 |
| rs2393799 | CT vs. CC | TT vs. CC | CT + TT vs. CC | TT vs. CT + CC | C vs. T | ||||||
| Asian | 14 | 0.92 (0.74–1.14) | 0.44 | 0.87 (0.54–1.41) | 0.58 | 0.86 (0.66–1.14) | 0.30 | 0.92 (0.61–1.40) | 0.70 | 0.98 (0.83–1.17) | 0.84 |
3.4. Heterogeneity and Publication Bias
In our study, relatively obvious heterogeneities existed under all five genetic models for rs3751143 and rs2393799 (Table 2). For rs1718119, heterogeneities were not observed under all genetic models. For rs208294, heterogeneities were not observed under heterozygous codominant and recessive genetic models. For rs7958311, heterogeneities were not observed under heterozygous codominant and for rs2230911 variant, heterogeneities were not observed under heterozygous codominant and dominant models.
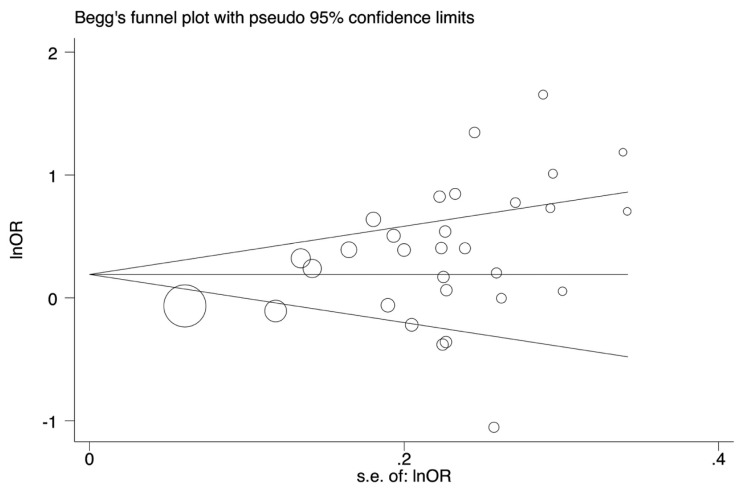
Begg’s tests were done with funnel plot to assess publication bias. Publication bias was found for rs3751143 under five genetic models (Table 2 and Figure 3).
Figure 3.
The funnel plot for the test of publication bias. The funnel plot for rs3751143 polymorphism under allele genetic model (C vs. A).
The Begg’s tests indicated no evidence of publication bias for rs2393799, rs1718119, and rs208294 (Table 2) under all genetic models.
3.5. Sensitivity Analysis
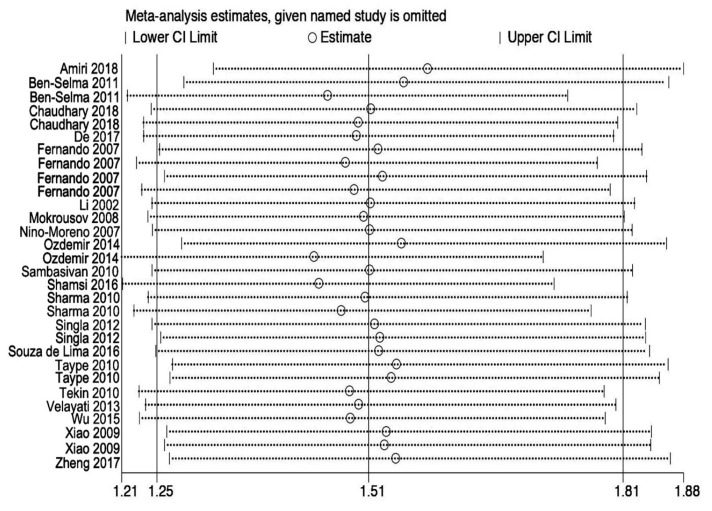
To better inspect the impact of individual study on the pooled OR, we performed sensitivity analysis through deleting each study one by one. Outcomes indicated that ORs were not statistically influenced in all genetic models for rs3751143 (Figure 4), as well as for rs2393799, showing that our results are stable and reliable.
Figure 4.
Sensitivity analyses for studies on P2X7 rs3751143 polymorphism and the risk of tuberculosis for C vs. A.
4. Discussion
Mounting evidence proposed that host genetic factors are implicated in tuberculosis susceptibility [4,6]. The P2X7R is highly expressed on macrophages and other immune cells [8]. It is a key molecule in the clearance of MTB in macrophages by adenosine triphosphate (ATP)-induced apoptosis of macrophage [8,10]. P2X7 is polymorphic and several studies investigated the impact of P2X7 polymorphisms on predisposition to TB [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. But these studies failed to reach a consistent conclusion. Therefore, to provide a comprehensive and reliable conclusion, we conducted the present meta- analysis to increase the statistical power of the association. Our findings suggest the P2X7 rs3751143 (Glu498Ala) polymorphism significantly increased the risk of overall TB. Stratified analysis of this polymorphism significantly increased the risk of PTB and EPTB. Also, the rs3751143 polymorphism increased the risk of TB in the Asian population. Findings did not support an association between rs2393799 (−762 C > T), rs1718119 (Thr348Ala), rs208294 (His155Tyr), rs7958311 (Arg270His), and rs2230911(Thr357Ser) polymorphisms and TB risk.
Ge et al. [36] performed a meta-analysis (n = 10) on the association between P2X7 rs3751143 polymorphism and PTB risk and found that this variant significantly increased the risk of PTB. Another meta-analysis (n = 11) performed by Alshammari et al. [37] showed no significant association between rs3751143 polymorphism and risk of TB. Stratified analysis revealed an association between this variant and the risk of TB in the Asian population. The results of a meta-analysis of 8 studies indicated that rs3751143 polymorphism significantly increased the risk of EPTB [38]. Another meta-analysis of 9 studies conducted by Wu et al. [39] revealed that rs3751143 significantly increased the risk of TB. A meta-analysis published by Yi et al. [40] on the association between rs2393799 (−762 C > T) polymorphism and TB susceptibility indicated that this variant is associated with TB risk. Our meta-analysis has more advantages than previous meta-analyses. We included a higher number of relevant published studies. Besides, we evaluated 6 polymorphisms in this meta-analysis.
Several polymorphisms have been described that cause P2X7R loss-of-function (LOF) or gain-of-function (GOF) [8]. The common polymorphism of P2X7 is rs3751143 (A1513C; Glu498Ala) polymorphism located in exon 13, accountable for LOF. This polymorphism affects the sensitivity of P2X7R to ATP and may contribute to increased susceptibility to MTB infection in humans [13,14,41]. The findings of the present meta-analysis support an association between rs3751143 polymorphism and the risk of TB. Another LOF polymorphism is rs2393799 (−762 C > T), which is located in the promoter of P2X7 and decrease the expression of P2X7R. The relationship between the rs2393799 polymorphism and susceptibility to TB is still debated [12,13,14,17,18,19,21,23,27,28,29,31,32,33,34], and pooled analysis of all available data did not support an association between this variant and susceptibility to TB. The rs208294 (489 C > T; His155Tyr) is GOF polymorphism. This polymorphism increases the affinity of P2X7R to ATP [42]. Limited studies investigated the impact of this polymorphism on TB susceptibility [14,30,34]. Pooled analysis revealed no evidence of association between this variant and TB risk.
Up until now, only 3 studies investigated the association between rs1718119 (1068 G > A; Thr348Ala) polymorphism and TB risk [30,31,34]. Our findings did not support an association between this polymorphism and TB risk.
Porphyromonas gingivalis, a bacterial carcinogen, plays a key role in cancer development by inhibiting apoptosis through several mechanisms. It has been shown that this bacterium secretes an anti-apoptotic enzyme nucleoside diphosphate kinase (NDK) which cleaves ATP and prevents proapoptotic P2X7 receptor activation, consequently modulating ATP/P2X7-signaling pathway [43]. It has been proposed that MTB secrete NDK, which act as a Rho-GTPase-activating protein (Rho-GAP), and covert Guanosine triphosphate (GTP)-bound active form to guanosine diphosphate (GDP)-bound inactive form, eventually facilitating its pathogenesis [44].
Some limitations of our meta-analysis should be acknowledged. Firstly, heterogeneity between studies was evident, which might distort the conclusion of this meta-analysis. Heterogeneity may be partly arising in the differences of ethnicities. Secondly, the sample sizes for some polymorphisms were small. Therefore, the results of this meta-analysis should be interpreted with caution.
Despite these limitations, however, there are still some advantages to having done this meta-analysis. First, this meta-analysis involved more studies than the previous meta-analyses, so the statistical power of our study is higher than the published meta-analysis. Second, we evaluated six polymorphisms of P2X7.
5. Conclusions
Overall, our meta-analysis proposed that P2X7 rs3751143 polymorphism may serve as a risk factor for TB in the Asian population. However, further well-designed studies with large sample sizes are necessary to confirm our findings.
Author Contributions
M.T., designed the study. H.S. and A.M.-R. searched the literatures and extracted the data. M.H. performed the statistical analyses. M.T. and M.H. wrote the manuscript. All authors read and approved the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.WHO . Global Tuberculosis Report. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 2.Trebucq A., Schwoebel V. Numbers of tuberculosis cases: Dreams and reality. Int. J. Tuberc. Lung Dis. 2016;20:1288–1292. doi: 10.5588/ijtld.15.0873. [DOI] [PubMed] [Google Scholar]
- 3.Zumla A., Raviglione M., Hafner R., von Reyn C.F. Tuberculosis. N. Engl. J. Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 4.Naderi M., Hashemi M., Ansari H. Macrophage migration inhibitory factor −173 G > C polymorphism and risk of tuberculosis: A meta-analysis. EXCLI J. 2017;16:313–320. doi: 10.17179/excli2016-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naderi M., Hashemi M., Mirshekari H., Bahari G., Taheri M. Toll-like Receptor 1 Polymorphisms Increased the Risk of Pulmonary Tuberculosis in an Iranian Population Sample. Biomed. Environ. Sci. 2016;29:825–828. doi: 10.3967/bes2016.110. [DOI] [PubMed] [Google Scholar]
- 6.Qiu Y., Cao S., Gou C., Yue Y., Jiang S., Ma T., Xue X. Associations of tumor necrosis factor-alpha polymorphisms with the risk of tuberculosis: A meta-analysis. Scand. J. Immunol. 2018:e12719. doi: 10.1111/sji.12719. [DOI] [PubMed] [Google Scholar]
- 7.Ralevic V., Burnstock G. Receptors for purines and pyrimidines. Pharmacol. Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- 8.Miller C.M., Boulter N.R., Fuller S.J., Zakrzewski A.M., Lees M.P., Saunders B.M., Wiley J.S., Smith N.C. The role of the P2X7 receptor in infectious diseases. PLoS Pathog. 2011;7:e1002212. doi: 10.1371/journal.ppat.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khakh B.S., North R.A. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442:527–532. doi: 10.1038/nature04886. [DOI] [PubMed] [Google Scholar]
- 10.Placido R., Auricchio G., Falzoni S., Battistini L., Colizzi V., Brunetti E., Di Virgilio F., Mancino G. P2X7 purinergic receptors and extracellular ATP mediate apoptosis of human monocytes/macrophages infected with Mycobacterium tuberculosis reducing the intracellular bacterial viability. Cell. Immunol. 2006;244:10–18. doi: 10.1016/j.cellimm.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Fairbairn I.P., Stober C.B., Kumararatne D.S., Lammas D.A. ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X7-dependent process inducing bacterial death by phagosome-lysosome fusion. J. Immunol. 2001;167:3300–3307. doi: 10.4049/jimmunol.167.6.3300. [DOI] [PubMed] [Google Scholar]
- 12.Amiri A., Sabooteh T., Ahmadi S.A.Y., Azargoon A., Shahsavar F. Association of P2X7 gene common polymorphisms with pulmonary tuberculosis in Lur population of Iran. Egypt. J. Med Hum. Genet. 2018;19:231–234. doi: 10.1016/j.ejmhg.2017.12.002. [DOI] [Google Scholar]
- 13.Ben-Selma W., Ben-Kahla I., Boukadida J., Harizi H. Contribution of the P2X7 1513A/C loss-of-function polymorphism to extrapulmonary tuberculosis susceptibility in Tunisian populations. FEMS Immunol. Med. Microbiol. 2011;63:65–72. doi: 10.1111/j.1574-695X.2011.00824.x. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhary A., Singh J.P., Sehajpal P.K., Sarin B.C. P2X7 receptor polymorphisms and susceptibility to tuberculosis in a North Indian Punjabi population. Int. J. Tuberc. Lung Dis. 2018;22:884–889. doi: 10.5588/ijtld.18.0023. [DOI] [PubMed] [Google Scholar]
- 15.De R., Kundu J.K. Tuberculosis risk in P2X7 1513A/C polymorphism of the tribes of Jhargram, West Bengal. Int. J. Zool. Stud. 2017;2:189–193. [Google Scholar]
- 16.Fernando S.L., Saunders B.M., Sluyter R., Skarratt K.K., Goldberg H., Marks G.B., Wiley J.S., Britton W.J. A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2007;175:360–366. doi: 10.1164/rccm.200607-970OC. [DOI] [PubMed] [Google Scholar]
- 17.Li C.M., Campbell S.J., Kumararatne D.S., Bellamy R., Ruwende C., McAdam K.P., Hill A.V., Lammas D.A. Association of a polymorphism in the P2X7 gene with tuberculosis in a Gambian population. J. Infect. Dis. 2002;186:1458–1462. doi: 10.1086/344351. [DOI] [PubMed] [Google Scholar]
- 18.Mokrousov I., Sapozhnikova N., Narvskaya O. Mycobacterium tuberculosis co-existence with humans: Making an imprint on the macrophage P2X7 receptor gene? J. Med. Microbiol. 2008;57:581–584. doi: 10.1099/jmm.0.47455-0. [DOI] [PubMed] [Google Scholar]
- 19.Nino-Moreno P., Portales-Perez D., Hernandez-Castro B., Portales-Cervantes L., Flores-Meraz V., Baranda L., Gómez-Gómez A., Acuña-Alonzo V., Granados J., González-Amaro R. P2X7 and NRAMP1/SLC11 A1 gene polymorphisms in Mexican mestizo patients with pulmonary tuberculosis. Clin. Exp. Immunol. 2007;148:469–477. doi: 10.1111/j.1365-2249.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozdemir F.A., Erol D., Konar V., Yuce H., Kara Senli E., Bulut F., Deveci F. Lack of association of 1513 A/C polymorphism in P2X7 gene with susceptibility to pulmonary and extrapulmonary tuberculosis. Tuberkuloz ve toraks. 2014;62:7–11. doi: 10.5578/tt.4740. [DOI] [PubMed] [Google Scholar]
- 21.Sambasivan V., Murthy K.J., Reddy R., Vijayalakshimi V., Hasan Q. P2X7 gene polymorphisms and risk assessment for pulmonary tuberculosis in Asian Indians. Dis. Markers. 2010;28:43–48. doi: 10.1155/2010/843729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S., Kumar V., Khosla R., Kajal N., Sarin B., Sehajpal P. Association of P2X7 receptor +1513 (A-->C) polymorphism with tuberculosis in a Punjabi population. Int. J. Tuberc. Lung Dis. 2010;14:1159–1163. [PubMed] [Google Scholar]
- 23.Singla N., Gupta D., Joshi A., Batra N., Singh J. Genetic polymorphisms in the P2X7 gene and its association with susceptibility to tuberculosis. Int. J. Tuberc. Lung Dis. 2012;16:224–229. doi: 10.5588/ijtld.11.0076. [DOI] [PubMed] [Google Scholar]
- 24.De Lima D.S., Ogusku M.M., Sadahiro A., Pontillo A. Inflammasome genetics contributes to the development and control of active pulmonary tuberculosis. Infect. Genet. Evol. 2016;41:240–244. doi: 10.1016/j.meegid.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Taype C.A., Shamsuzzaman S., Accinelli R.A., Espinoza J.R., Shaw M.A. Genetic susceptibility to different clinical forms of tuberculosis in the Peruvian population. Infect. Genet. Evol. 2010;10:495–504. doi: 10.1016/j.meegid.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Tekin D., Kayaalti Z., Dalgic N., Cakir E., Soylemezoglu T., Kutlubay B.I., Kilic B.A. Polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis in Turkish children. Pediatr. Infect. Dis. J. 2010;29:779–782. doi: 10.1097/INF.0b013e3181d9932e. [DOI] [PubMed] [Google Scholar]
- 27.Velayati A.A., Farnia P., Farahbod A.M., Karahrudi M.A., Derakhshaninezhad Z., Kazampour M., Sheikhghomi S., Saeif S. Association of receptors, purinergic P2X7 and tumor necrosis factor-alpha gene polymorphisms in susceptibility to tuberculosis among Iranian patients. Arch. Clin. Infect. Dis. 2013 doi: 10.5812/archcid.16087. [DOI] [Google Scholar]
- 28.Wu J., Lu L., Zhang L., Ding Y., Wu F., Zuo W., Zhang W. Single Nucleotide Polymorphisms in P2X7 Gene Are Associated with Serum Immunoglobulin G Responses to Mycobacterium tuberculosis in Tuberculosis Patients. Dis. Markers. 2015;2015:671272. doi: 10.1155/2015/671272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao J., Sun L., Jiao W., Li Z., Zhao S., Li H., Jin J., Jiao A., Guo Y., Jiang Z., et al. Lack of association between polymorphisms in the P2X7 gene and tuberculosis in a Chinese Han population. FEMS Immunol. Med. Microbiol. 2009;55:107–111. doi: 10.1111/j.1574-695X.2008.00508.x. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X., Li T., Chen Y., Pan H., Zhang Z., Dai Y., Wang J. Genetic polymorphisms of the P2X7 gene associated with susceptibility to and prognosis of pulmonary tuberculosis. Infect. Genet. Evol. 2017;53:24–29. doi: 10.1016/j.meegid.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Bahari G., Hashemi M., Taheri M., Naderi M., Moazeni-Roodi A., Kouhpayeh H.R., Eskandari-Nasab E. Association of P2X7 gene polymorphisms with susceptibility to pulmonary tuberculosis in Zahedan, Southeast Iran. Genet. Mol. Res. GMR. 2013;12:160–166. doi: 10.4238/2013.January.24.8. [DOI] [PubMed] [Google Scholar]
- 32.Shamsi M., Zolfaghari M.R., Farnia P. Association of IFN-gamma and P2X7 Receptor Gene Polymorphisms in Susceptibility to Tuberculosis among Iranian Patients. Acta Microbiol. Immunol. Hung. 2016;63:93–101. doi: 10.1556/030.63.2016.1.7. [DOI] [PubMed] [Google Scholar]
- 33.Songane M., Kleinnijenhuis J., Alisjahbana B., Sahiratmadja E., Parwati I., Oosting M., Plantinga T.S., Joosten L.A., Netea M.G., Ottenhoff T.H., et al. Polymorphisms in autophagy genes and susceptibility to tuberculosis. PLoS ONE. 2012;7:e41618. doi: 10.1371/journal.pone.0041618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Tan C.Y., Mo Z.J., Gao Q.L., He D., Li J., Huang R.F., Li Y.B., Guo C.F., Guo Q., et al. P2X7 receptor in spinal tuberculosis: Gene polymorphisms and protein levels in Chinese Han population. Infect. Genet. Evol. 2018;57:138–144. doi: 10.1016/j.meegid.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X., Guo W., Ren G., He X., Hu Q., Zhang Y., Kang L., Yuan D., Jin T. P2X7R Gene Polymorphisms are Associated with Increased Risk of Pulmonary Tuberculosis in the Tibetan Chinese Population. Am. J. Trop. Med. Hyg. 2016;95:1016–1020. doi: 10.4269/ajtmh.16-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge H.B., Chen S. A meta-analysis of P2X7 gene-1513A/C polymorphism and pulmonary tuberculosis susceptibility. Hum. Immunol. 2016;77:126–130. doi: 10.1016/j.humimm.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Alshammari E.M., Mandal R.K., Wahid M., Dar S.A., Jawed A., Areeshi M.Y., Khan S., Khan M.E.A., Panda A.K., Haque S. Genetic association study of P2X7 A1513C (rs 3751143) polymorphism and susceptibility to pulmonary tuberculosis: A meta-analysis based on the findings of 11 case-control studies. Asian Pac. J. Trop. Med. 2016;9:1150–1157. doi: 10.1016/j.apjtm.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Areeshi M.Y., Mandal R.K., Dar S., Wahid M., Khan M.E., Panda A.K., Jawed A., Haque S. P2X7 1513 A>C Polymorphism Confers Increased Risk of Extrapulmonary Tuberculosis: A Meta-analysis of Case-Control Studies. Curr. Genom. 2015;17:450–458. doi: 10.2174/1389202917666160513104737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu G., Zhao M., Gu X., Yao Y., Liu H., Song Y. The effect of P2X7 receptor 1513 polymorphism on susceptibility to tuberculosis: A meta-analysis. Infect. Genet. Evol. 2014;24:82–91. doi: 10.1016/j.meegid.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Yi L., Cheng D., Shi H., Huo X., Zhang K., Zhen G. A meta-analysis of P2X7 gene-762T/C polymorphism and pulmonary tuberculosis susceptibility. PLoS ONE. 2014;9:e96359. doi: 10.1371/journal.pone.0096359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wesselius A., Bours M.J., Arts I.C., Theunisz E.H., Geusens P., Dagnelie P.C. The P2X7 loss-of-function Glu496Ala polymorphism affects ex vivo cytokine release and protects against the cytotoxic effects of high ATP-levels. BMC Immunol. 2012;13:64. doi: 10.1186/1471-2172-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cabrini G., Falzoni S., Forchap S.L., Pellegatti P., Balboni A., Agostini P., Cuneo A., Castoldi G., Baricordi O.R., Di Virgilio F. A His-155 to Tyr polymorphism confers gain-of-function to the human P2X7 receptor of human leukemic lymphocytes. J. Immunol. 2005;175:82–89. doi: 10.4049/jimmunol.175.1.82. [DOI] [PubMed] [Google Scholar]
- 43.Karpinski T.M. Role of Oral Microbiota in Cancer Development. Microorganisms. 2019;7:20. doi: 10.3390/microorganisms7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chopra P., Koduri H., Singh R., Koul A., Ghildiyal M., Sharma K., Tyagi A.K., Singh Y. Nucleoside diphosphate kinase of Mycobacterium tuberculosis acts as GTPase-activating protein for Rho-GTPases. FEBS Lett. 2004;571:212–216. doi: 10.1016/j.febslet.2004.06.073. [DOI] [PubMed] [Google Scholar]