Abstract
Endogenous compounds have been reported to be the regulators of UDP-glucuronosyltransferases (UGTs) isoforms. This study aims to investigate the regulatory effects of the activity of UGT isoforms by two important lipid components phosphatidylcholine (PC) and lysophosphatidylcholines (LPC) using in vitro incubation system.
UGTs supersomes-catalyzed 4-methylumbelliferone (4-MU) glucuronidation was used as the probe reaction to evaluate the inhibition of compounds towards UGT isoforms except UGT1A4, and UGT1A4-catalyzed trifluoperazine (TFP) glucuronidation reaction was utilized to phenotype the activity of UGT1A4.
About 50 μM of LPC15:0, LPC16:0, LPC17:0, LPC18:0, LPC18:1 and PC16:0, 2:0 exhibited inhibition towards more than 90% activity of UGT isoforms, and other LPC and PC components showed negligible inhibitory potential towards all the UGT isoforms. UGT1A6 and UGT1A8 were identified to be the most sensitive UGT isoforms susceptible for the inhibition by LPC15:0, LPC16:0, LPC17:0, LPC18:0, LPC18:1 and PC16:0, 2:0, indicating the strong influence of these LPC and PC components towards UGT1A6 and UGT1A8-catalyzed metabolic reaction when the concentrations of these components increased.
Keywords: Allosteric activation, lysophosphatidylcholines, phosphatidylcholine, UDP-glucuronosyltransferases
Introduction
Lipids, defined as fat-soluble molecules based on their solubility in non-polar solvents, exhibit complex structural diversity. Besides their structural functions, lipids have been demonstrated to play a key role in important biological functions, including signaling pathways (Wenk, 2005). Phospholipids are a class of lipids playing important structural functions through forming lipid bilayers which are the major structural basis of biological membrane (Lagace & Ridgway, 2013). Phosphatidylcholines (PCs, Figure 1) are one main component of phospholipids, and lysophosphatidylcholines (LPCs, Figure 1) are derived from phosphatidylcholines through the removal of one of the fatty acid groups from PC. PC and LPC play the important biological functions in the body. For example, LPC has been demonstrated to exhibit key roles in modulating immune response through affecting the function of immune-regulatory cells, which have been speculated to be main contributor towards some chronic inflammatory diseases, such as atherosclerosis (Matsumoto et al., 2007).
Figure 1.
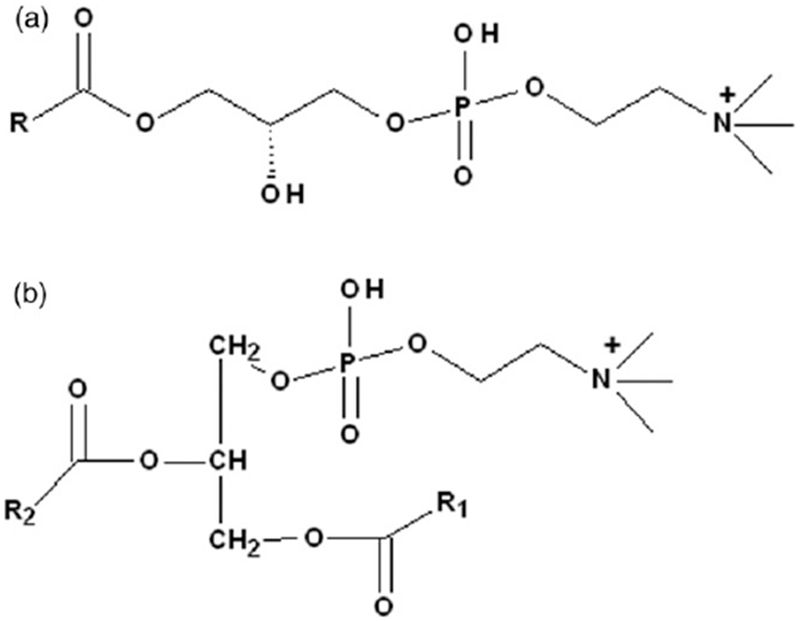
The structures of Lysophosphatidylcholines (LPC) (A) and Phosphatidylcholine (PC) (B).
PC and LPC have been previously demonstrated to be good substrates and modulators of drug-metabolizing enzymes. LPC 18:1 has been reported to be a good substrate of cytochrome P450 2W1 (Xiao & Guengerich, 2012). The activity of CYPs also strongly depends on PC and LPC components. The experiment performed by Diatlovitskaia et al. (1977) showed that LPC can inactivate the CYPs through enhancing the conversion of the active form of CYPs into the inactivate form cytochrome P-420. In addition, PC can reactivate CYP inactivated by LPC (Diatlovitskaia et al., 1982).
Human UDP-glucuronosyltransferases (UGTs) are important membrane proteins located in endoplasmic reticulum, and play important roles in metabolism of a variety of endogenous and exogenous compounds (Fang et al., 2013a). Altered activity of UGT isoforms will result in the disrupted elimination of endogenous and exogenous compounds. For example, indinavir, an HIV therapeutic drug, can significantly induce the elevation of unconjugated bilirubin in serum through inhibition of UGT1A1-mediated bilirubin glucuronidation (Zucker et al., 2001). Inhibition of UGT1A1 by sorafenib has been speculated to be main reason sorafenib-induced elevation of serum bilirubin (Meza-Junco et al., 2009).
The influence of UGT isoforms’ activity by fatty acids has been widely investigated, and the results showed that unsaturated fatty acids were potent inhibitors of renal UGT isoforms (Tsoutsikos et al., 2004). As is known, the structures of PC and LPC components contained fatty acid chains. Therefore, the inhibition potential of PC and LPC components towards UGTs activity was speculated in this study. As previously described (Fang et al., 2013b), recombinant human UGTs-catalyzed 4-MU glucuronidation reaction was used to evaluate the inhibitory effects of PC and LPC components towards UGT isoforms except UGT1A4, and where trifluoperazine (TFP) glucuronidation was performed as the standard reaction to evaluate the inhibition potential of PC and LPC components toward UGT1A4 activity.
Materials and methods
Chemicals and reagents
4-Methylumbelliferone (4-MU), 4-methylumbelliferone-β-D-glucuronide (4-MUG), Tris-HCl, 7-hydroxycoumarin, trifluoperazine (TFP, purity≥99%) and uridine-5′-diphosphoglucuronic acid trisodium salt (UDPGA) were purchased from Sigma-Aldrich (St. Louis, MO). UGT supersomes (UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A10, UGT2B4 and UGT2B7) expressed in baculovirus-infected insect cells were obtained from BD Gentest Corp. (Woburn, MA). All the PC and LPC (1-acyl) components were purchased from Avanti Polar Lipids (Alabaster, AL). The purity of these compounds was above 95%. All other reagents were of HPLC grade or of the highest grade commercially available.
Investigation of the inhibition of UGTs’ activity by LPC and PC components
The in vitro incubation system to evaluate the inhibition of UGTs’ activity by LPC and PC components has been described in the previous literatures (Fang et al., 2013a,b). For UGT supersomes (except UGT1A4) catalyzed in 4-MU glucuronidation probe reactions, the incubation system (total volume = 200 μL) contained UGT supersomes, 5mM UDPGA, 5mM MgCl2, 50mM Tris-HCl (pH = 7.4) and 4-MU in the absence or presence of different concentrations of various PC and LPC components. The used incubation time and protein concentration were previously determined to ensure the reaction rate within the linear range. The 4-MU concentration was equal to known Km or S50 values for each UGT form. The concentrations of 4-MU and UGT supersomes, and incubation time were given in Supplemental Table S1. The incubation reaction was initiated through addition of UDPGA to the mixture after a 5 min pre-incubation at 37 °C. The reactions were quenched by adding 100 μL acetonitrile with 7-hydroxycoumarin (100 μM) as internal standard. The mixture was centrifuged at 20 000 × g for 10 min, and an aliquot of supernatant was transferred to an auto-injector vial for HPLC analysis. The HPLC system (Shimadzu, Kyoto, Japan) contained a SCL-10A system controller, two LC-10AT pumps, a SIL-10A auto injector, a SPD-10AVP UV detector. Chromatographic separation was carried out using a C18 column (4.6 × 200 mm, 5 μm, Kromasil) at a flow rate of 1 mL/min and UV detector at 316 nm. The mobile phase consisted of acetonitrile (A) and H2O containing 0.5% (v/v) formic acid (B). The following gradient condition was used: 0–15 min, 95–40% B; 15–20 min, 10% B; 20–30 min, 95% B. The calculation curve was generated by peak area ratio (4-MUG/internal standard) over the concentration range of 4-MUG 0.1–100 mM. The curve was linear over this concentration range, with an r2 value > 0.99. The limits of detection and quantification were determined at signal-to-noise ratios of 3 and 10, respectively. The accuracy and precision for each concentration were more than 95%. For UGT1A4-catalyzed TFP glucuronidation reaction, 40 μM (close to its Km value) of TFP was incubated with human recombinant UGT1A4 (0.1 mg/mL) at 37 °C for 20 min in the absence or presence of LPC and PC components as previously described (Uchaipichat et al., 2006).
Determination of inhibition kinetic type and parameters of some representative components
The inhibition kinetic behaviors (including kinetic type and parameters) were determined for some representative PC or LPC components. The reaction velocity was determined at different concentrations of substrates and inhibitors, and the incubation time is the same as that in the initial screening study. Two most common plot methods (Dixon and Lineweaver-Burk plots) to determine the inhibition type were employed in this study. The inhibition parameters (Ki) were calculated through correlating the slopes from the Lineweaver-Burk plots versus the concentrations of inhibitors.
Molecular docking
Till now, none of the integrated 3D-structure of UGT family members has yet been crystallized. For this reason, the knowledge-based comparative modeling method, the package of Advanced Protein Modeling (APM) distributed within SYBYL, was employed to establish the 3D structure of UGT1A6. The ORCHESTRAR suite within APM package was designed for comparative protein modeling, with which the all-atom model can be created based on a target protein sequence and one or more structural homologs. With ORCHESTRAR, reliable models can be built only if the similarity between the target and template sequence is higher than 10%. As known, the recognition of homology between protein sequences provides invaluable information toward understanding the biological behavior and biochemical function of uncharacterized sequences. Here, the target protein sequence of UGT1A6 was obtained from NCBI database (accession number: NP_476446.2, rattus) with a length of 530 aa. This sequence was used to search the similarity within the Protein Data Bank with the Blast tool, which was re-aligned with the Fugue module distributed within APM. We tried to specify out a template sequence with relatively high identity and a ligand crystallized together with the protein. Finally, the crystal structure of macrolide glycosyltransferases (PDB code: 2IYF) was selected as the homology template, with which was the backbone of the structurally conserved regions (SCR) modeled, then the structurally variable regions were built through Loop Search and side chains adding. Finally, the 3D-model of UGT1A6 was obtained after the optimization with staged energy minimization.
With the comparative modeled 3D-structure of UGT1A6, the interaction between the protein and the LPC inhibitors were evaluated. Here, in the molecular docking method, Surflex-Dock was used to model the binding affinity and interaction mode of LPCs with UGT1A6. Surflex-Dock is a knowledge-based molecular docking method which is guided by the protomol, an idealized representation of a ligand that makes every potential interaction with the binding site. With the “AMBER7 FF99” partial charges was calculated for the protein, a ligand-based mode was adopted to recognize the active site that may bind with LPC ligands with the setup of float radius as 1. Here, an empirical scoring function, Chemscore, was used to evaluate the reasonableness of the binding conformation of the ligand, which can properly describe the binding affinities. Before performing Docking, the LPC ligands were optimized by energy minimization and conformational search using Tripos force field, also was the partial atomic charges were calculated with the Gasteiger-Huckel charge method.
Results
Inhibition profiles of PC and LPC components towards various UGT isoforms
About 50 μM of compounds were firstly utilized to evaluate the inhibitory potential of various PC and LPC components towards various UGT isoforms, and the residual activity [% residual activity = (the activity at 100 μM of compounds/the control activity at 0 μM compounds) × 100%] was given in Tables 1 and 2. For the inhibition of LPCs towards UGT isoforms, LPC 6:0 to LPC 14:0 showed negligible inhibition towards all the UGT isoforms at 50 μM. About 50 μM of LPC 15:0 to LPC 18:0 showed strong inhibition towards UGT1A6 and UGT1A8. 50 μM of LPC18:1 showed strong inhibition towards UGT1A6, UGT1A8 and UGT2B7, with the inhibition percentage to be 97.5, 98.7 and 87.3%, respectively. As shown in Table 2, among the tested PC components, PC 8:0, PC 9:0, PC 10:0, PC 19:0 and PC 16:0, 2:0 exhibited inhibition potential towards some UGT isoforms. PC 8:0 inhibited the activity of UGT1A6 by 74.4%. PC 9:0 inhibited the activity of UGT1A6 and UGT1A9 by 78.6 and 89.3%, respectively. The activity of UGT1A6 was inhibited by 76.8% by PC 10:0. 50 μM of PC 19:0 inhibited the activity of UGT2B7 by 76.9%. About 50 μM of PC 16:0, 2:0 exhibited the inhibition towards UGT1A4, UGT1A6 and UGT1A8 with the inhibition percentage to be 94.5, 94.2 and 99.3%, respectively.
Table 1.
Initial screening of LPC components (50 μM) towards various recombinant human UGT forms.
| 1A1 | 1A3 | 1A4 | 1A6 | 1A7 | 1A8 | 1A9 | 2B4 | 2B7 | |
|---|---|---|---|---|---|---|---|---|---|
| LPC 6:0 | 176.7 | 84.4 | 107.3 | 72.7 | 130.6 | 108.0 | 95.9 | 183.6 | 70.0 |
| LPC 7:0 | 174.9 | 80.4 | 104.2 | 71.0 | 138.3 | 101.1 | 92.9 | 183.5 | 73.8 |
| LPC 8:0 | 184.9 | 80.3 | 100.5 | 74.2 | 133.3 | 102.7 | 99.6 | 185.0 | 70.1 |
| LPC 9:0 | 179.6 | 79.6 | 78.0 | 67.2 | 134.9 | 101.2 | 94.5 | 179.1 | 76.2 |
| LPC 10:0 | 170.2 | 75.9 | 107.5 | 65.7 | 139.8 | 100.7 | 99.2 | 208.4 | 77.3 |
| LPC 11:0 | 167.3 | 75.9 | 60.3 | 65.7 | 149.9 | 124.0 | 104.6 | 217.6 | 78.0 |
| LPC 12:0 | 165.9 | 74.8 | 63.0 | 80.2 | 138.6 | 89.2 | 89.2 | 255.5 | 72.8 |
| LPC 13:0 | 160.6 | 88.1 | 57.3 | 83.8 | 120.1 | 101.6 | 99.4 | 239.4 | 79.9 |
| LPC 14:0 | 171.3 | 74.2 | 52.8 | 33.3 | 153.6 | 19.2 | 97.5 | 217.1 | 90.5 |
| LPC 15:0 | 162.9 | 52.8 | 53.4 | 2.9 | 156.2 | 1.6 | 82.8 | 152.6 | 43.9 |
| LPC 16:0 | 162.4 | 59.1 | 55.0 | 1.2 | 144.7 | 1.3 | 84.5 | 153.3 | 77.1 |
| LPC 17:0 | 167.6 | 119.0 | 46.7 | 8.5 | 164.3 | 2.2 | 87.5 | 333.4 | 81.5 |
| LPC 18:0 | 160.5 | 72.4 | 24.4 | 4.3 | 133.4 | 1.2 | 64.4 | 280.4 | 39.3 |
| LPC 18:1 | 175.3 | 54.8 | 44.8 | 2.5 | 146.8 | 1.3 | 80.8 | 262.8 | 12.7 |
The values shown are the residual activity, which was calculated using the following equation: % residual activity = (the activity at 50 μM LPC components/the control activity at 0 μM LPC components) × 100%. The values were given as mean value of triplicate experiments.
Table 2.
Initial screening of PC components (50 μM) towards various recombinant human UGT forms.
| 1A1 | 1A3 | 1A4 | 1A6 | 1A7 | 1A8 | 1A9 | 2B4 | 2B7 | |
|---|---|---|---|---|---|---|---|---|---|
| PC 6:0, 6:0 | 187.8 | 105.1 | 99.5 | 67.3 | 166.0 | 116.2 | 96.1 | 178.7 | 70.2 |
| PC 7:0, 7:0 | 176.8 | 109.9 | 102.2 | 50.7 | 150.2 | 106.1 | 137.7 | 186.9 | 64.5 |
| PC 8:0, 8:0 | 155.6 | 119.1 | 52.5 | 25.6 | 114.8 | 78.3 | 180.5 | 173.1 | 71 |
| PC 9:0, 9:0 | 162.3 | 117.7 | 60.5 | 21.4 | 137 | 33.2 | 10.7 | 156.7 | 79.6 |
| PC 10:0, 10:0 | 197.9 | 107.4 | 52.6 | 23.2 | 163.2 | 59.3 | 158 | 168.4 | 86.2 |
| PC 11:0, 11:0 | 232.3 | 110.7 | 49.3 | 35.0 | 175.5 | 116.8 | 97.8 | 181.0 | 100.5 |
| PC 12:0, 12:0 | 208.8 | 93.0 | 50.6 | 40.8 | 171.4 | 114.1 | 84.1 | 182.7 | 85.8 |
| PC 13:0, 13:0 | 201.1 | 110.7 | 57.7 | 56.5 | 160.6 | 114.3 | 99.2 | 193.8 | 81.4 |
| PC 14:0, 14:0 | 208.9 | 97.3 | 84.5 | 54.3 | 162.3 | 110.2 | 86.3 | 179.0 | 66.1 |
| PC 15:0, 15:0 | 210.1 | 103.4 | 55.2 | 55.0 | 172.7 | 105.9 | 64.7 | 191.3 | 51.7 |
| PC 16:0, 16:0 | 192.6 | 92.6 | 60.1 | 53.7 | 155.2 | 107.6 | 67.5 | 185.1 | 60.3 |
| PC 17:0, 17:0 | 192.3 | 81.4 | 55.7 | 49.1 | 138.9 | 92.1 | 65.7 | 175.4 | 35.5 |
| PC 18:0, 18:0 | 208.2 | 83.7 | 80.9 | 50.7 | 139.9 | 84.1 | 67.7 | 93.4 | 36.0 |
| PC 19:0, 19:0 | 184.9 | 75.9 | 66.5 | 49.1 | 135.5 | 60.6 | 42.3 | 83.4 | 23.1 |
| PC 20:0, 20:0 | 202.3 | 90.8 | 117.6 | 88.2 | 125.5 | 94.9 | 65.6 | 79.5 | 88.9 |
| PC 21:0, 21:0 | 185.2 | 88.2 | 136.6 | 85.4 | 122.0 | 36.9 | 38.1 | 80.3 | 26.2 |
| PC 22:0, 22:0 | 170.3 | 92.1 | 53.8 | 60.2 | 86.2 | 38.0 | 43.9 | 60.0 | 33.0 |
| PC 23:0, 23:0 | 133.7 | 80.4 | 86.3 | 98.9 | 118.8 | 93.5 | 77.3 | 168.3 | 69.5 |
| PC 16:0, 2:0 | 181.1 | 42.6 | 5.5 | 5.8 | 92.1 | 0.7 | 91.0 | 166.8 | 36.2 |
| PC 16:0, 14:0 | 209.5 | 84.4 | 63.8 | 87.3 | 150.0 | 90.0 | 91.7 | 158.1 | 80.1 |
| PC 16:0, 18:0 | 195.0 | 78.7 | 90.0 | 93.8 | 148.1 | 86.4 | 109.8 | 114.4 | 154.7 |
| PC 16:0, 18:1 | 207.2 | 90.9 | 51.8 | 85.6 | 146.4 | 105.5 | 120.8 | 177.6 | 45.7 |
| PC 18:0, 18:2 | 212.0 | 109.2 | 54.3 | 91.2 | 159.8 | 101.8 | 107.9 | 170.1 | 46.8 |
| PC 18:0, 20:4 | 203.5 | 90.0 | 52.1 | 94.2 | 144.7 | 107.5 | 85.8 | 186.9 | 56.5 |
| PC 18:0, 22:6 | 186.6 | 90.0 | 59.7 | 88.8 | 167.9 | 116.1 | 94.4 | 183.2 | 73.6 |
| PC 18:1, 14:0 | 201.8 | 75.3 | 49.8 | 100.8 | 136.7 | 111.2 | 89.8 | 176.0 | 60.8 |
| PC 18:1, 16:0 | 212.1 | 80.0 | 52.6 | 94.1 | 154.1 | 108.7 | 156.5 | 179.9 | 71.4 |
| PC 18:1, 18:0 | 191.0 | 79.1 | 60.4 | 102.6 | 164.6 | 110.0 | 96.4 | 214.8 | 81.2 |
The values shown are the residual activity, which was calculated using the following equation: % residual activity = (the activity at 50 μM PC components/the control activity at 0 μM PC components) × 100%. The values were given as mean value of triplicate experiments.
PC 16:0, 2:0, 1-palmitoyl-2-acetyl-sn-glycero-3-phosphocholine; PC 16:0, 14:0, 1-palmitoyl-2-myristoyl-sn-glycero-3-phosphocholine; PC 16:0, 18:0, 1-palmitoyl-2-stearoyl-sn-glycero-3-phosphocholine; PC 16:0, 18:1, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; PC 18:0, 18:2, 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphocholine; PC 18:0, 20:4, 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphocholine; PC 18:0, 22:6, 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine; PC 18:1, 14:0, 1-oleoyl-2-myristoyl-sn-glycero-3-phosphocholine; PC 18:1, 16:0, 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine; PC 18:1, 18:0, 1-oleoyl-2-stearoyl-sn-glycero-3-phosphocholine.
Molecular docking to understand the inhibition of UGT isoforms by lipid components
To more deeply understand the interaction between the UGTs and lipid components, we used the molecular docking method to elucidate the interaction between LPC 12:0-LPC 18:0 and LPC 18:1 and UGT1A6. Typically, the inhibitory potency shall be highly attributed to the binding affinity of the bio-macromolecule to its ligands. However, there has not been an available crystallized structure of UGT1A6 for the investigation. Therefore, we have employed the comparative modeling to construct a 3D model of UGT1A6. It should be mentioned that there has not been any animal UGT family members been crystallized completely, and we had to refer the plant or enkaryocyte 3D structure as the homology template. When submitting the protein sequence of 1A6 to search the similar homologue within the PDB database with Blast tool, we found the identity between the target and homologue sequences mainly distributed between 20–30%. Considering the factors like query cover, E-score, identity and bounded ligands, we selected the crystal structure of macrolide glycosyltransferases (PDB code: 2IYF) as the homologue template which shared an identity of 27% with the target sequence. The homology re-alignment by Fugue was shown as Supplemental Table S1. Followed by the procedure of constructing the structurally conserved regions (SCR), modeling the structurally variable regions, evaluating the model as well as minimizing staged energy, the 3D-model of UGT1A6 was obtained. With Surflex–Dock, an active cavity was recognized for the binding of the ligand in adjacent to the UDP binding area. The active packet was constituted by Asn95 Phe93 Gly94 Leu83 Asp34 Gln33 Phe147 Asp149 Phe170 Tyr168 Ser176 Arg238 Lys237 Met180 Ser189 HIS179 Ser186 Phe194 Try195, etc. Then LPC inhibitors were docked into the active site to evaluate the interaction mode and binding affinity. Nearly for all the LPCs, Leu236 and Ser241 play significant role to form hydrogen bonds with phosphonyl group, which predominate the location of the LPC head of quaternary ammonium salt, but the situation of the tail varied according to the length of the alkyl chain. Ranking by Chemscore, we select the most suitable binding conformation among the top five poses. The interaction modes of LPCs can be categorized into two types: (1) the longer LPCs like (LPC16:0, 17:0, 18:0 and 18:1) tend to occupy the whole cavity by situating the head in the inner area while the tail of alkane towards the entrance of the pocket that keeps off the access to UDP (Figure 2). (2) The shorter LPCs adopt conformations get tightly close to the wall of the cavity also by placing the head of the quaternary ammonium salt group in the inner area but the tail of alkane towards the edge area of the pocket which still leaves enough space for the access to UDP for other ligands (Figure 3). Among these LPCs, LPC15:0 was an exception. It was located crookedly at the entrance of the cavity with the head of quaternary ammonium salt group towards the UDP. Such a conformation can ultimately block the binding of other ligands with UGT1A6. Through analyzing the chemscore value of the docked LPCs combinationally, we found chemscores of the longer alkyl chain (LPC16:0-LPC18:0 and LPC18:1) are remarkably higher than the shorter ones (LPC12:0-LPC14:0), as shown in Table 3. Without considering LPC15:0, there could be an approximately increasing trend between the residual activity and the chemscore value, which is a main factor that can explain why the longer LPCs show much higher inhibitory potency. As for LPC15:0, even its chemscore value is not very high, the special interaction conformation contributed greatly to its higher inhibition activity. Therefore, we can draw a conclusion that the inhibition potency of LPCs rely combinational both on their binding affinities and the interaction poses.
Figure 2.
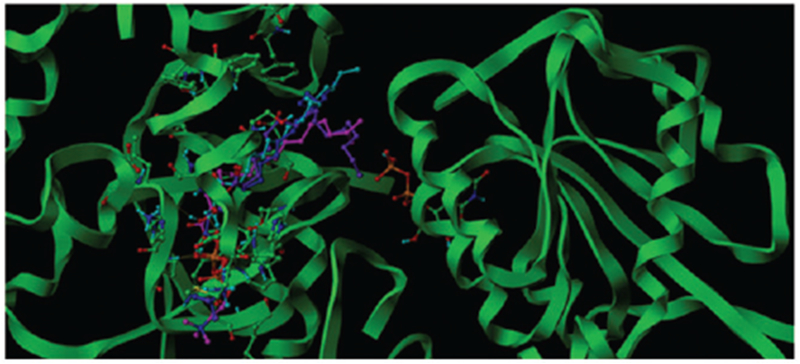
The binding poses of LPC16:0, 17:0, 18:0 and 18:1 in the active cavity in adjacent to UDP (the molecule in the right domain) of UGT1A6 of UGT1A6, which was colored in blue, Cyan, Magenta and Purple color respectively.
Figure 3.
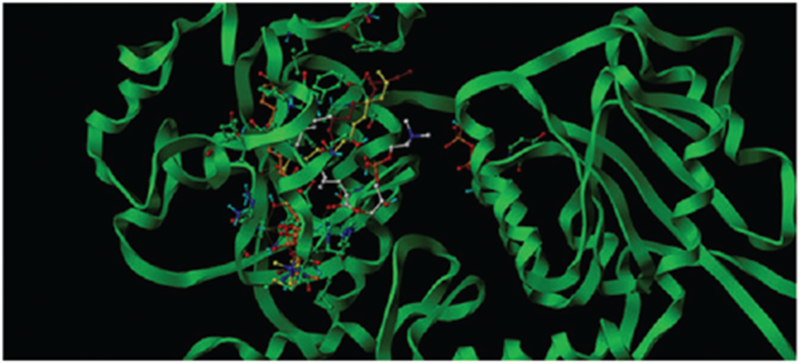
The binding poses of LPC12:0, 13:0, 14:0 and 15:0 in the active cavity in adjacent to UDP (the molecule in the right domain) of UGT1A6, which was denoted in orange, yellow, dark red and atom-based color respectively.
Table 3.
Chemscore values for the interacted poses with UGT1A6 of LPCs.
| LPCs | 12:0 | 13:0 | 14:0 | 15:0 | 16:0 | 17:0 | 18:0 | 18:1 |
|---|---|---|---|---|---|---|---|---|
| Chemoscore | −25.31 | −33.79 | −36.64 | −31.72 | −41.26 | −40.22 | −39.92 | −41.23 |
Inhibitory kinetic type and parameters of representative PC and LPC components
Some representative LPC and PC components were selected to determine the inhibition kinetic type and parameters (Ki). LPC 15:0 and LPC 18:0 were chosen as the representative LPC component, and PC 16:0, 2:0 was selected as the representative PC component. The inhibition type was determined through drawing the plot using the reaction velocity at different concentrations of substrates and inhibitors versus the concentrations of substrates and inhibitors with Dixon fitting equation and Lineweaver-Burk fitting equation. For the inhibition of LPC 15:0 towards UGT1A6, LPC 18:0 towards UGT1A8, PC 16:0, 2:0 towards UGT1A6, the intersection point was located in the second quadrant in Dixon plot (Figures 4A, 6A and 7A) and in the vertical axis in Lineweaver-Burk plot (Figures 4B, 6B and 7B), indicating the competitive inhibition. For the inhibition of LPC 18:0 towards UGT1A6, and PC 16:0, 2:0 towards UGT1A8, the intersection point was located in the horizontal axis in both Dixon plot (Figures 5A and 8A) and Lineweaver-Burk plot (Figures 5B and 8B), demonstrating that LPC 18:0 and PC16:0, 2:0 non-competitively inhibited the activity of UGT1A6 and UGT1A8, respectively.
Figure 4.

Evaluation of the inhibition of LPC 15:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. (A) Dixon plot of the inhibition of LPC 15:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (B) Lineweaver–Burk plot of the inhibition of LPC 15:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (C) Second plot of LPC 15:0’s inhibition towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. The data point represents the mean of the duplicate experiments.
Figure 6.
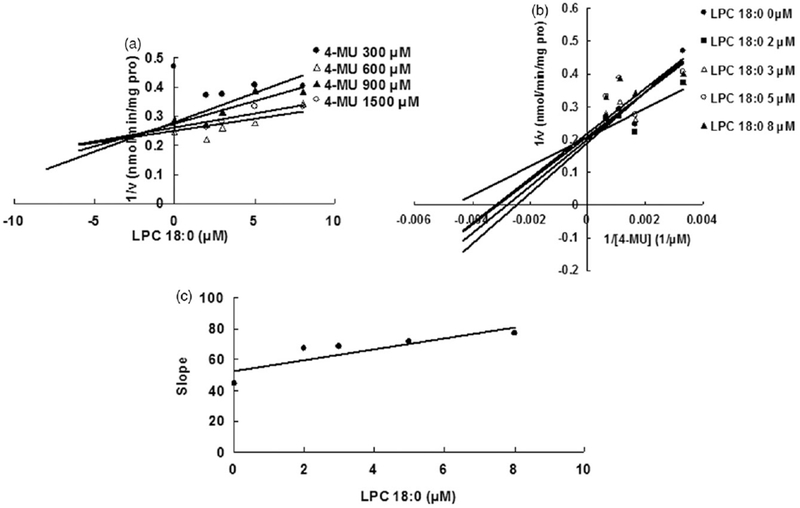
Evaluation of the inhibition of LPC 18:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation. (A) Dixon plot of the inhibition of LPC 18:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation; (B) Lineweaver–Burk plot of the inhibition of LPC 18:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation; (C) Second plot of LPC 18:0’s inhibition towards recombinant UGT1A8-catalyzed 4-MU glucuronidation. The data point represents the mean of the duplicate experiments.
Figure 7.
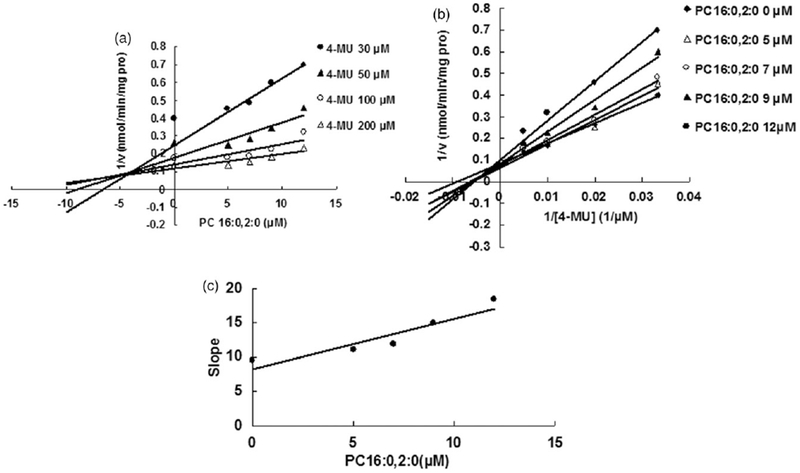
Evaluation of the inhibition of PC 16:0,2:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. (A) Dixon plot of the inhibition of PC 16:0,2:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (B) Lineweaver–Burk plot of the inhibition of PC 16:0,2:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (C) Second plot of PC 16:0,2:0’s inhibition towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. The data point represents the mean of the duplicate experiments.
Figure 5.

Evaluation of the inhibition of LPC 18:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. (A) Dixon plot of the inhibition of LPC 18:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (B) Lineweaver–Burk plot of the inhibition of LPC 18:0 towards recombinant UGT1A6-catalyzed 4-MU glucuronidation; (C) Second plot of LPC 18:0’s inhibition towards recombinant UGT1A6-catalyzed 4-MU glucuronidation. The data point represents the mean of the duplicate experiments.
Figure 8.
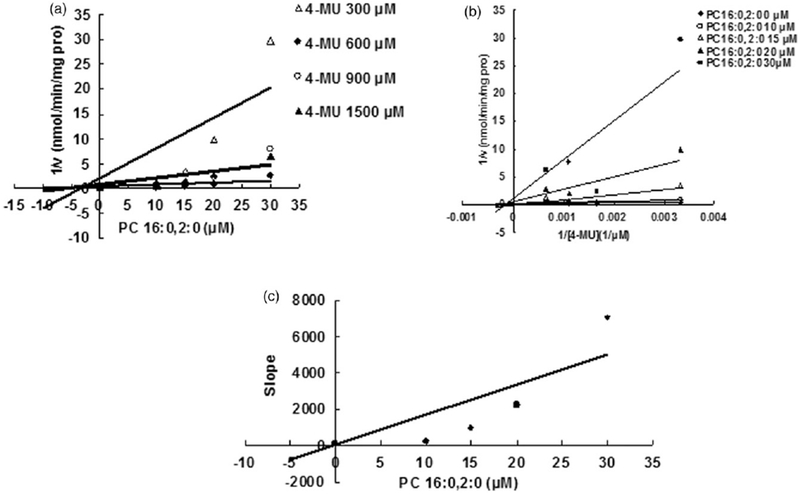
Evaluation of the inhibition of PC 16:0,2:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation. (A) Dixon plot of the inhibition of PC 16:0,2:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation; (B) Lineweaver-Burk plot of the inhibition of PC 16:0,2:0 towards recombinant UGT1A8-catalyzed 4-MU glucuronidation; (C) Second plot of PC 16:0,2:0’s inhibition towards recombinant UGT1A8-catalyzed 4-MU glucuronidation. The data point represents the mean of the duplicate experiments.
Discussion
Endogenous substances as the inhibitors of UGT isoforms have been increasingly reported, and this inhibitory effect has some physiological or pathological role. For example, ATP has been reported to be an endogenous inhibitor of UDP-glucuronosyltransferases (UGTs), and the inhibition capability depends on the ratio of ATP/ADP/AMP. Therefore, the dynamical alteration for the concentration of ATP and AMP can switch UGTs-catalyzed glucuronidation reaction on/off (Ishii et al., 2012). Our recent study showed that bile acid component taurolithocholic acid (TLCA) exhibited strong inhibitory effects towards multiple UGT isoforms, including UGT1A1, UGT1A3, UGT1A4, UGT1A7, UGT1A10, UGT2B7 and UGT2B15 (Fang et al., 2013b), indicating the possible disruption of metabolic disorders induced by the elevated level of bile acids components in some diseases such as cholestasis.
PC and LPC components are important constituents of biological membranes, and UGTs are located in membrane of endoplasmic reticulum (ER). Therefore, the UGTs’ activity can be regulated by the compositions of PC and LPCs, and some preliminary results in the previous literatures have reported some relationship between PC and LPC components and UGTs’ activity. For example, the experiment carried out by Dragacci et al. (1987) has shown that LPC components can regulate the activity of UGTs, and oleoyl LPC (LPC 18:1) exhibited maximal regulation capability (approximately 4-fold increase for UGTs’ activity). The experiment performed by Zakim & Eibl (1992) used purified UGT isoform from pig liver demonstrated the influence of charge and the distribution of charge in the polar region of phospholipids towards the activity of UGTs. This study aims to clarify the different regulation role of LPC and PC components towards different UGT isoforms using UGT supersomes. We used the equal quantity of UGT supersomes in which the lipid components were supposed to be equal, and different quantity of LPC and PC components were added to investigate the influence of PC and LPC components towards UGTs’ activity. Among the tested LPC components (as shown in Table 1), LPC 6:0–LPC 14:0 exhibited negligible inhibition effect towards all the UGT isoforms, and LPC 15:0–LPC 18:0 and LPC 18:1 exhibited strong inhibitory behavior towards UGT1A6 and UGT1A8 (the residual activity less than 10% at 50 μM of these compounds), indicating that UGT1A6 and UGT1A8 are the most sensitive UGT isoforms susceptible for the inhibition by LPC components. The similar sensitivity of UGT1A6 and UGT1A8 towards PC components was also observed. About 50 μM of PC 16:0, 2:0 exhibited the strongest inhibition potential towards UGT1A6 and UGT1A8. The reasons for the sensitivity of UG1A6 and UGT1A8 towards the LPCs and PCs with medium and long chain fatty acid remain unclear for us according to the recent papers. The possible reasons might be that the longer fatty acid chains might block the binding site of UDP binding sites. More efforts should be performed for elucidating the possible reasons in the future. In addition, UGT1A4 activity was also strongly inhibited by the supplement of 50 μM of PC 16:0, 2:0 (as shown in Table 2). It should be noted that PC16:0, 2:0 reduced UGT1A6 activity, whereas the same is not true for other related PC such as PC16:0, 14:0 and PC16:0, 18:0, which might indicate an interesting structure-inhibitory potential, and needs further study. The inhibition kinetic determination was also determined to predict the possible binding sites in UGT isoforms, and the binding sites will depend on not only the species of LPC and PC but also the UGTs isoforms, indicating that the inhibition effect of LPCs and PCs towards UGT isoforms through different regulatory mode. It should be also noted that the reasons for the inhibition phenomena might be different. LPCs might directly inhibit the activity of UGTs. For PCs components, PCs might form liposomes which might also interact with the membrane for UGT supersomes. Therefore, besides the direct inhibition towards the activity of UGTs, PCs might also exhibit their inhibition through the interaction with membrane structures. In addition, different from the inhibitions of fatty acids towards UGT isoforms, some LPCs and PCs components showed the activation role towards UGTs activity. The detailed mechanism should be also investigated in the future.
We tried to explain the detailed molecular mechanism why the one-carbon distinction from LPC14:0 to LPC15:0 can significantly affect the inhibition potential towards the activity of UGT1A6. Due to no crystal structure of UGT1A6, homology modeling was firstly employed to obtain the 3D-model of UGT1A6. Through comparison of the binding results of LPC 12:0–LPC18:0 and LPC18:1 towards UGT1A6, we found that the longer LPCs like (LPC16:0, 17:0, 18:0 and 18:1) tend to occupy the whole cavity by situating the head inside the inner area while the tail of alkane towards the entrance of the pocket that keeps off the access to UDP, and the shorter LPCs adopt conformations get tightly close to the wall of the cavity also by placing the head of quaternary ammonium salt group inside the inner area but the tail of alkane towards the edge area of the pocket which still leaves enough space for the access to UDP for other ligands.
The disturbance of LPC and PC homeostasis has been frequently reported in many diseases and toxicity response towards xenobiotics. For example, in the toxicity response of mice towards trichloroethylene (TCE)-induced liver injury, the level of LPC 18:0 and LPC 18:1 in serum significantly increased (Fang et al., 2013c). Increased LPC content has also been regarded to be an effector of fatty acid-induced insulin resistance (Han et al., 2011). All these alteration might result in the influence towards the activity of UGT isoforms, especially for UGT1A6 and UGT1A8. UGT1A6 has been widely considered to be an important UGT isoform involved in the metabolism of xenobiotic and exogenous substances. For example, UGT1A6 has been reported to be the main enzyme involved in the metabolism of serotonin which is an important neurotransmitter (Bock & Kohle, 2005). In addition, UGT1A6 can detoxify the carcinogenic arylamines and aryl hydrocarbons, which plays a key role in preventing the carcinogenesis of these compounds (Bock & Kohle, 2005). UGT1A8 is a key UGT isoform located in gastrointestinal tract, and also plays an important in elimination of some important clinical drugs, including raloxifene (Kokawa et al., 2013) and mycophenolic acid (Mackenzie, 2000). In addition, low activity of UGT1A8 has also reported to have some relationship with increased risk of cancers (Dura et al., 2012). Therefore, when the level of LPC and PC components increased in some diseases or toxicity response, especially for LPC15:0, LPC16:0, LPC17:0, LPC18:0, LPC18:1 and PC 16:0, 2:0, the metabolic process of UGT1A6 and UGT1A8-mediated metabolism of endogenous and exogenous substances might be needed to monitored.
It should be noted that some LPC and PC components can up-regulate the activity of some UGT isoforms in our present in vitro incubation systems. For example, many LPC components can increase the activity of UGT2B4 nearly 1- to 2-fold, indicating the allosteric activation of some LPC and PC components towards UGT2B4. This phenomenon has been observed in our previous study in which cholic acid (CA) and glycocholic acid (GCA) were found to exhibit allosteric activation towards the activity of UGT1A3 (Fang et al., 2014b). Therefore, the complex regulation effects of LPC and PC components towards UGT isoforms’ activity will complicate the explanation of final results due to the disruption of homeostasis of LPC and PC components. In addition, in most situations, the alteration of LPC and PC components will be accompanied with the alteration of other exogenous substances, such as bile acids components, which might further complicate the final results on the regulation towards UGT isoforms.
In conclusion, the relatively complete the regulatory profiles of LPC and PC components towards various UGT isoforms were given. The obvious inhibition of UGT isoforms by LPC 15:0, LPC 16:0, LPC 17:0, LPC 18:0, LPC 18:1 and PC16:0, 2:0 was observed, and UGT1A6 and UGT1A8 were most sensitive UGT isoforms susceptible to these LPC species. Given that the allosteric activation of some LPC species towards UGTs and the possibly accompanied alteration of other exogenous substances (e.g. bile acids, etc.), the translation of these in vitro data towards in vivo situation should be paid more caution.
Supplementary Material
Acknowledgments
Declaration of interest
The authors have declared no conflict of interest. This work was supported by the National Natural Science Foundation of China (No. 81202586).
Footnotes
Supplementary material available online.
References
- Bock KW, Kohle C. (2005). UDP-glucuronosyltransferase 1A6: structural, functional, and regulatory aspects. Methods Enzymol 400: 57–75. [DOI] [PubMed] [Google Scholar]
- Diatlovitskaia EV, Lemenovskaia AF, Archakov AI, et al. (1977). Alteration of the lipid composition of rat liver microsomes on reconstitution of the cytochrom P-450 system. Inactivation of the enzyme by lysolecithin. Biokhimiia 42:139–43. [PubMed] [Google Scholar]
- Diatlovitskaia EV, Petkova DKH, Bergel’son LD. (1982). Lipid dependence of the activity of cytochrome P-450 from microsomes of rat liver by the phosphatidylcholine transfer protein from bovine liver. Biokhimiia 47:1366–9. [PubMed] [Google Scholar]
- Dragacci S, Thomassin J, Magdalou J, et al. (1987). Properties of human hepatic UDP-glucuronosyltransferases. Relationship to other inducible enzymes in patients with cholestasis. Eur J Clin Pharmacol 32: 485–91. [DOI] [PubMed] [Google Scholar]
- Dura P, Salomon J, Te Morsche RH, et al. (2012). High enzyme activity UGT1A1 or low activity UGT1A8 and UGT2B4 genotypes increase esophageal cancer risk. Int J Oncol 40:1789–96. [DOI] [PubMed] [Google Scholar]
- Fang ZZ, Cao YF, Hu CM, et al. (2013a). Structure-inhibition relationship of ginsenosides towards UDP-glucuronosyltransferases (UGTs). Toxicol Appl Pharmacol 267:149–54. [DOI] [PubMed] [Google Scholar]
- Fang ZZ, He RR, Cao YF, et al. (2013b). A model of in vitro UDP-glucuronosyltransferase inhibition by bile acids predicts possible metabolic disorders. J Lipid Res 54:3334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZZ, Krausz KW, Tanaka N, et al. (2013c). Metabolomics reveals trichloroacetate as a major contributor to trichloroethylene-induced metabolic alterations in mouse urine and serum. Arch Toxicol 87: 1975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MS, Lim YM, Quan W, et al. (2011). Lysophosphatidylcholine as an effector of fatty acid-induced insulin resistance. J Lipid Res 52: 1234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, An K, Nishimura Y, Yamada H. (2012). ATP serves as an endogenous inhibitor of UDP-glucuronosyltransferase (UGT): a new insight into the latency of UGT. Drug Metab Dispos 40:2081–9. [DOI] [PubMed] [Google Scholar]
- Kokawa Y, Kishi N, Jinno H, et al. (2013). Effect of UDP-glucuronosyltransferase 1A8 polymorphism on raloxifene glucuronidation. Eur J Pharm Sci 49:199–205. [DOI] [PubMed] [Google Scholar]
- Lagace TA, Ridgway ND. (2013). The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta 1833:2499–510. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI. (2000). Identification of uridine diphosphate glucuronosyl transferases involved in the metabolism and clearance of mycophenolic acid. Ther Drug Monit 22:10–3. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Kobayashi T, Kamata K. (2007). Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr Med Chem 14: 3209–20. [DOI] [PubMed] [Google Scholar]
- Meza-Junco J, Chu QS, Christensen O, et al. (2009). UGT1A1 polymorphism and hyperbilirubinemia in a patient who received sorafenib. Cancer Chemother Pharmacol 65:1–4. [DOI] [PubMed] [Google Scholar]
- Tsoutsikos P, Miners JO, Stapleton A, et al. (2004). Evidence that unsaturated fatty acids are potent inhibitors of renal UDP-glucuronosyltransferases (UGT): kinetic studies using human kidney cortical microsomes and recombinant UGT1A9 and UGT2B7. Biochem Pharmacol 67:191–9. [DOI] [PubMed] [Google Scholar]
- Uchaipichat V, Mackenzie PI, Elliot DJ, Miners JO. (2006). Selectivity of substrate (trifl uoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfi npyrazone) “probes” for human UDP-glucuronosyltransferases. Drug Metab Dispos 34:449–56. [DOI] [PubMed] [Google Scholar]
- Wenk MR. (2005). The emerging field of lipidomics. Nat Rev Drug Discov 4:594–610. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Guengerich FP. (2012). Metabolomic analysis and identification of a role for the orphan human cytochrome P450 2W1 in selective oxidation of lysophospholipids. J Lipid Res 53:1610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakim D, Eibl H. (1992). The influence of charge and the distribution of charge in the polar region of phospholipids on the activity of UDP-glucuronosyltransferase. J Biol Chem 267:13166–70. [PubMed] [Google Scholar]
- Zucker SD, Qin X, Rouster SD, et al. (2001). Mechanism of indinavir-induced hyperbilirubinemia. Proc Natl Acad Sci USA 98:12671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


