Abstract
While most melanomas can be distinguished from nevi by histopathology, the histology is ambiguous for some melanocytic tumors, contributing to diagnostic uncertainty. Therefore molecular assays, including FISH or SNP array, and more recently a gene expression test (myPath, Myriad Genetics) have been proposed to aid in the work-up of ambiguous tumors. Two hundred and sixty-eight prospectively submitted cases were gathered, with the goal of comparing the myPath assay to morphologic diagnosis in (1) morphologically unequivocal cases (198), and to morphologic diagnosis and FISH in (2) morphologically ambiguous cases (70). Melanoma FISH was performed using probes for 6p25, 6q23, 11q13, Cep6, 9p21, and Cep9 and scored according to established criteria. The myPath assay was scored by the manufacturer as benign, indeterminate, or malignant. In the unequivocal group, myPath assay showed 75% agreement with morphologic diagnosis, with 67% sensitivity and 81% specificity. In the ambiguous group, FISH and myPath showed 69% inter-test agreement. For these cases agreement with histopathologic interpretation was 84% for FISH and 74% for myPath. Sensitivity and specificity of FISH was 61 and 100%, 50 and 93% for myPath, respectively. Cases from both groups in which myPath was discordant with either morphologic diagnosis and/or FISH (81/268 cases), were submitted for evaluation by two experienced dermatopathologist and also by SNP-array. SNP-array results correlated better than FISH, which correlated better than myPath, with the morphologic interpretation. Our findings document that molecular diagnostics show good correlation with consensus diagnoses, but discordant results occur, and vary in level of correlation with consensus interpretations. Studies with long-term outcomes data within specific ambiguous lesion subsets are required to establish the accuracy of this test, as each molecular diagnostic technique has limitations based on both lack of clinical outcomes data in ambiguous melanocytic tumors and in terms of their sensitivity and specificity in melanocytic lesion subtypes.
Introduction
Histopathologic evaluation of melanocytic neoplasms allows for accurate classification of the majority of cases as either benign or malignant. However, the biologic behavior of a subset of melanocytic tumors remains notoriously challenging to predict based on morphologic evaluation alone due to conflicting features, leading to both under and over diagnosis of melanoma [1–3]. Given the critical need for accurate diagnosis of melanocytic tumors to ensure proper clinical management, ancillary molecular diagnostic tools have been recently proposed to aid in the classification of lesions with morphologic features that show conflicting findings of both benign and malignant melanocytic lesions (reviewed in the ref. [4]).
Melanomas typically contain multiple chromosomal aberrations, in contrast to benign nevi [5, 6], and the harnessing of this genomic information (in particular utilizing knowledge of recurrent aberrations found across the different genetic melanoma subtypes) has led to the development of the melanoma fluorescence in situ (FISH) test as well as array comparative genomic hybridization (aCGH) as ancillary tools to aid in melanoma diagnosis, both of which assess for chromosomal gains and/or losses. aCGH has the advantage of querying the entire genome for chromosomal aberrations however it is currently limited in its applicability due to its higher tumor tissue requirements, lower resolution (in comparison to FISH), limited published data on its application to morphologically ambiguous melanocytic tumors, expense and lack of coverage by many insurers, and the unknown significance of isolated genetic aberrations detected by aCGH [4, 7, 8].
Melanoma FISH is a commercially available test utilizing 3 probes to chromosomes 6 (6p25, 6q23, and CEP6), and 1 on chromosome 11 (11q13), and more recently 2 additional probes on chromosome 9 (9p21 and CEP9), with or without an additional probe on chromosome 8 (8q24) [9–16]. The test is considered positive if any one of 6 criteria are met in a subset percentage of tumor cells above a previously established threshold, including an absolute or relative gain of 6p25 or relative loss of 6q23 (with CEP6 as a control), gain of 11q13, homozygous deletion of 9p21 (using CEP9 as a control), or gain of 8q24. The reported sensitivity of FISH in morphologically unequivocal melanomas and benign nevi using the original 4 probes (6p25, 6q23, CEP6, and 11q13) ranged from 75 to 100% [9, 10, 13, 17–22] and a specificity of approximately 95%, with lowest sensitivity in spitzoid melanomas and melanomas from intermittently sun-damaged skin. Cep 9 and 9p21 (a region showing homozygous deletion in some spitzoid melanomas) were added to the 4 probe assay to improve the sensitivity in spitzoid tumors [14, 16]. As the majority of published literature on the test is from morphologically unequivocal cases, however, the diagnostic utility of the FISH assay in truly histopathologically ambiguous melanocytic remains controversial, due to the limited amount of published data on the test’s sensitivity and specificity in ambiguous melanocytic tumors with long-term clinical outcomes data [9, 13, 21–23]. Based on the limited data to date on this latter case cohort, it appears that 9p21 homozygous deletion and to a somewhat lesser extent, gains in RREB1 or CCND1, tend to show a higher correlation with clinically aggressive behavior in ambiguous melanocytic tumors with long-term outcomes data, at least in ambiguous spitzoid melanocytic tumors [16, 24].
Recently, a diagnostic gene expression-based test (myPath, Myriad Genetics) has been marketed as an ancillary diagnostic tool to aid in the classification of challenging melanocytic tumors [25–28]. The test analyzes the expression of 23 genes (including 14 genes implicated in melanoma pathogenesis and 9 housekeeping genes) via quantitative RT-PCR, and calculates a gene expression score by utilizing a proprietary algorithm to assess the relative levels of gene expression into a negative (benign), indeterminate, or positive (malignant) score. It has been clinically validated in a retrospective study authored by Myriad, containing 437 morphologically unequivocal melanocytic lesions (211 melanomas and 216 nevi) [25]. There is as yet relatively limited clinical experience with the test, including on unequivocally benign and malignant melanocytic tumors, with a reported sensitivity and specificity (including indeterminate cases which comprise between 10 and 16% of reported cases), ranging from 53–90 to 90–100%, respectively and of 62–94 to 90–100%, respectively excluding indeterminate cases, in classifying unequivocally benign from malignant lesions [25–29]. Two studies comparing the test to other ancillary molecular diagnostic tests have been performed, however the number of cases in these studies was limited [27, 29]. A single study assessed the results of the myPath assay on 78 ambiguous melanocytic tumors [29]; however, there is no information on the test’s performance on ambiguous melanocytic tumors with long-term clinical outcomes data.
The goal of this study was to independently compare the myPath assay to morphologic diagnosis in a large series of prospectively submitted (1) morphologically unequivocal cases, and to expert consensus morphologic diagnosis, FISH, and SNP array analysis in (2) morphologically ambiguous cases and gene score discordant cases, collected from a large community-based practice.
Materials and methods
Patients
This study was approved by the Institutional Review Board at Inform Diagnostics. Patients with a diagnosis of a borderline (histologically ambiguous) melanocytic tumor diagnosed at Inform Diagnostics between November 2013 and April 2015 for which material was available, were prospectively analyzed. Patients from the same time period with a morphologically unequivocal diagnosis of melanoma, common nevus, dysplastic nevus, Spitz nevus, deep penetrating nevus, and blue nevus served as controls. Re-excision specimens were excluded from the study. Clinical and histopathological information regarding the patient’s age, gender, lesion type, involved anatomic site was obtained. All cases had either previously stained hematoxylin and eosin slides, and/or sufficient tissue available in paraffin embedded blocks for subsequent evaluation. Overall the control portion of the data set included 117 benign nevi (19 common nevus, 73 dysplastic nevus [16 mild, 36 moderate, 21 severe), 4 blue nevus, 15 Spitz nevus [1 junctional, 14 compound/intradermal], 5 deep penetrating nevus, 1 Bapoma), and 81 melanomas (61 invasive melanoma, 20 melanoma in situ). The second data set was comprised of 70 morphologically ambiguous cases, which were deemed to be histopathologically challenging by two independent experienced dermatopathologists and also by an additional pathologist (JR or SS) and to therefore warrant additional molecular analysis.
FISH
Melanoma FISH was performed at Inform Diagnostics on FFPE tissue using probes RREB1 (Ras-Responsive Element Binding Protein-1) on chromosome 6p25 (Empire Genomics, Buffalo, NY, USA, MYB (myeloblastosis) on chromosome 6p23 (Abbott Molecular Inc, Abbott Park, IL, USA), CCND1 (cyclin D1) on chromosome 11q13 (Abbott Molecular Inc, Abbott Park, IL, USA), CDKN2A on 9p21 (Abbott Molecular Inc, Abbott Park, IL, USA), CEP6 (Abbott Molecular Inc, Abbott Park, IL, USA), and CEP9 (Abbott Molecular Inc, Abbott Park, IL, USA), according to manufacturer’s instructions. Two cytotechnologists trained in the melanoma FISH assay each enumerate 50 nuclei, scoring independently (for a total of 100 nuclei). These results are compared and if they are not concordant, a third scorer is brought in to score, to ensure maximal accuracy. Cases were interpreted as positive if greater than 55% of nuclei contained more 6p25 (RREB1) than CEP6 signals, greater than 29% of nuclei contained greater than 2 6p25 (RREB1) signals, greater than 40% of nuclei showed less 6q23 (MYB) than CEP6 signals, greater than 38% of nuclei contained greater than 2 11q13 (CCND1) signals, or greater than 33% of nuclei showed homozygous loss of 9p21, according to previously published criteria[30]. All cases were assessed and controlled for the presence of a significant tetraploid population.
Gene expression score
Cases were submitted to Myriad Genetics (Salt Lake City, UT, USA) for analysis with the myPath melanoma gene expression score, and were classified as either benign, indeterminate, or malignant based on previously demarcated thresholds by the manufacturer [25].
Histopathologic review of discordant cases
All cases for which the gene expression score was discordant with either the histopathologic final diagnosis and/or the melanoma FISH result, were additionally submitted for histopathologic review by two experienced dermatopathologists (KB and EV). This is in addition to the initial review by three other experienced dermatopathologists which was performed on all the cases in the morphologically challenging cohort at the time of initial diagnosis. Each case was anonymized and was reviewed independently by EV and KB, and included the slide material on which the initial diagnosis was made (including H&E slides, levels when necessary, and immunostains). The available clinical information, including patient age, anatomic location, and sex, was provided. They were blinded to the sign out diagnosis as well as to the gene expression score, FISH and SNP array results. Cases were categorized as favor benign or favor malignant only if there was full concordance on expert review; all the rest were categorized as ambiguous.
SNP array analysis
Cases in which the myPath gene expression score was discordant with either the morphologic evaluation and/or the FISH result, for which sufficient material was available, were submitted for a more comprehensive cytogenetic analysis at Memorial Sloan Kettering Cancer Center by SNP array using the Affymetrix OncoScan assay (Affymetrix, Santa Clara, CA) as previously described [7].
Statistical analysis
The Fisher’s exact test was utilized to assess differences between variables and was performed with VassarStats. As it is recommended to avoid exclusion of indeterminate results in statistical analysis in a test that can result in equivocal results (as this can lead to bias in reporting of diagnostic test performance, including falsely elevated measurements of sensitivity and specificity) [31, 32], the indeterminate results (−2 to −0.1), were grouped as negative results in the calculation of sensitivity and specificity.
Results
MyPath test results in unequivocal cases
To assess the performance of the myPath assay on histopathologically unequivocal cases, we submitted 81 malignant (61 invasive melanoma [1 acral, 48 superficial spreading melanomas, 2 nodular melanomas, 5 unclassified melanomas, 4 lentigo maligna melanomas, 1 malignant blue MM arising in a blue nevus] and 20 melanoma in situ [12 lentigo maligna and 8 superficial spreading], and 117 benign melanocytic neoplasms (19 common nevi, 73 dysplastic nevi [16 mild, 36 moderate, 21 severe], 4 blue nevi, 15 Spitz nevi [1 junctional, 14 compound/intradermal], 5 deep penetrating nevi, 1 Bapoma) for analysis. The myPath gene expression score had a 75% overall agreement with histopathologic diagnosis (67% sensitivity and 89% specificity). In the malignant group, 11 of the invasive melanomas (18%) (10 superficial spreading melanomas, 1 malignant melanoma arising in a blue nevus), and 5 melanoma in situ cases (25%) were negative on the myPath assay (Fig. 1). Six invasive melanomas (10%) (1 nodular melanoma, 5 superficial spreading melanomas) and 4 melanoma in situ cases (20%) were scored as indeterminate (Fig. 1). Two invasive melanomas (3%) yielded no result due to insufficient RNA. In the benign group, 12 nevi (10%) (3 mildly dysplastic, 5 moderately dysplastic, 3 severely dysplastic, and 1 bapoma) were scored as malignant with the myPath assay (Fig. 2), 8 nevi (7%) (1 mildly dysplastic, 3 moderately dysplastic, 3 severely dysplastic, 1 common compound nevus) were scored as indeterminate, and 9 nevi (8%) (2 blue nevi, 2 moderately dysplastic, 2 severely dysplastic, 1 common compound nevus, 2 spitz nevi) yielded no result due to insufficient RNA. The diagnoses in the myPath discordant cases from the morphologically unequivocal group were additionally confirmed as either benign or malignant on the expert review (see below), with the exception of 3 invasive melanomas arising in nevi, 4 dysplastic nevi, and 2 melanoma in situ, all of which were categorized as ambiguous as an unequivocal expert consensus diagnosis was not reached. These were therefore removed from this cohort of cases, yielding a final cohort of 189 unequivocal cases. In this final cohort, the myPath gene expression score had a 79% overall agreement with histo-pathologic diagnosis (72% sensitivity and 94% specificity) (Table 1). Excluding indeterminate cases, the sensitivity and specificity of the myPath assay in unequivocal cases was 83% and 89%, respectively. Although all dyplastic nevi (including all dysplastic nevi categorized as dysplastic nevus with severe atypia) in the final cohort of unequivocal cases in our study were verified as benign on expert review, as definitive classification of severely dysplastic nevi is a notoriously challenging diagnostic area even amongst experts in dermatopathology, we additionally calculated the myPath assay sensitivity and specificity and overall agreement in the unequivocal case cohort excluding all nevi with severe atypia. The sensitivity, specificity, and overall agreement excluding all dysplastic nevi with severe atypia were essentially the same as with including the severely dysplastic nevi (72%, 91%, and 80%, respectively).
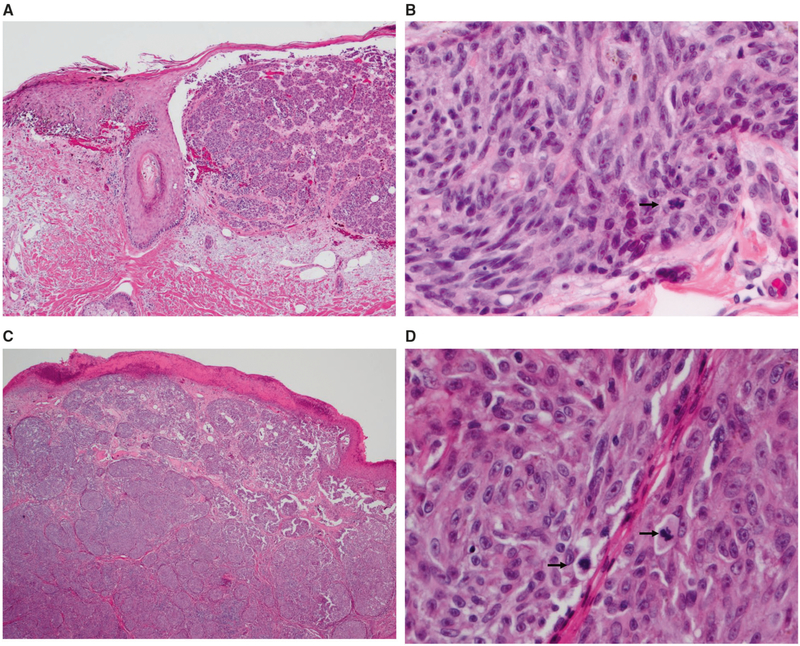
Fig. 1.
Images from 2 representative morphologically unequivocally malignant cases with negative or indeterminate gene expression scores. a, b Case 98: 79-year-old man with a pigmented lesion on the back, diagnosed as melanoma (1.1 mm, no ulceration), diagnosis confirmed on expert review. Negative myPath score. SNP array was positive for multiple chromosomal abnormalities (gain 4q21.23-qter; gain of 6p; gain 7p22.1-qter; del(21q)). a There is expansile nodular dermal growth and lentiginous epidermal growth of melanocytes with adnexal extension. b The dermal melanocytes are spindled, with mitotic activity (arrow). c, d Case 96: 34-year-old male with a pigmented lesion on the back, diagnosed as melanoma (5.1 mm, ulcerated), diagnosis confirmed on expert review. Indeterminate gene expression score. SNP array was positive for multiple chromosomal abnormalities (del 1p13.1–p13.3; multiple segmental gains (at different levels) across 6p23.3–p24.3, gain at 6p21.2; del 11q12.1–q13.1; multiple segmental gains (at different levels) across 16p12.3–p13.3, gain at 16p12.1; multiple segmental gains (at different levels) and losses across 22q). c There is dense nodular dermal growth of melanocytes without maturation and over-lying epidermal ulceration. d The dermal melanocytes are epithelioid with prominent nucleoli and frequent mitoses (arrows)
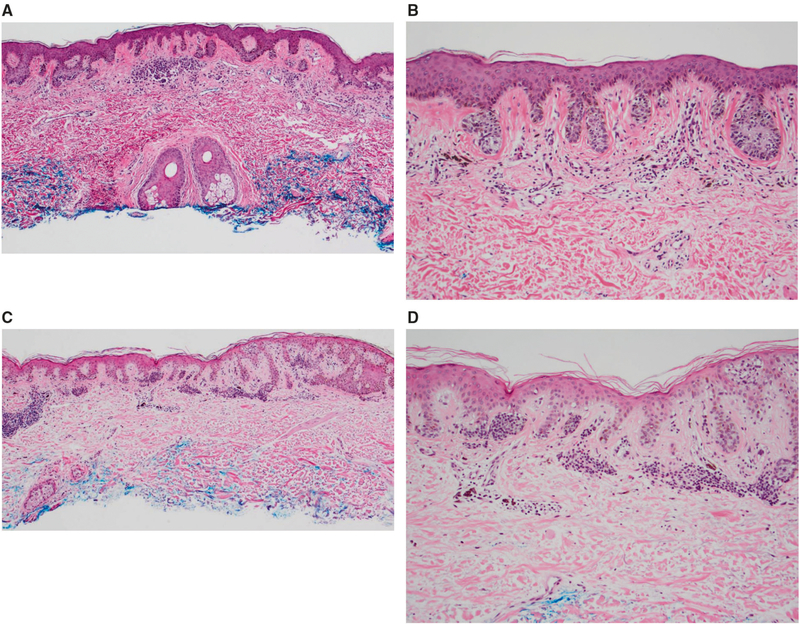
Fig. 2.
Images from 2 representative morphologically unequivocally benign cases with positive gene expression scores. a, b Case 45: 46 male with a pigmented lesion on the right chest, diagnosed as a compound dysplastic nevus with mild atypia, diagnosis confirmed on expert review. Positive gene expression score. Tissue was insufficient for SNP array analysis. a There is a bland dermal component with overlying lentiginous and nested growth of melanocytes, with mild nuclear enlargement (b). c, d Case 40: 81 year old male with a pigmented lesion on the left upper arm, diagnosed as a compound dysplastic nevus with mild atypia, diagnosis confirmed on expert review. Positive gene expression score. Tissue was insufficient for SNP array analysis. c Junctional melanocytes show slight nuclear enlargement with a lentiginous and nested growth along the dermoepidermal junction, with nests of bland melanocytes in the dermis (d)
Table 1.
Correlation of myPath results with histopathologic interpretation in unequivocal cases
| Unequivocal cases | Histologically malignant | Histologically benign | Total | ||
|---|---|---|---|---|---|
| myPath | 189 | ||||
| Malignant | 53 | 11 | Sensitivity | 72% | |
| Benign | 11 | 88 | Specificity | 94% | |
| Indeterminate | 10 | 5 | Agreement | 79% | |
| No result | 2 | 9 |
Results of MyPath and FISH tests in morphologically challenging cases
We next compared the results of the myPath assay to the melanoma FISH assay in 70 prospectively submitted morphologically challenging cases. Melanoma FISH and the myPath score showed a 69% intertest agreement (Table 2). The myPath assay showed a 74% overall agreement with the final interpretation (50% sensitivity, 96% specificity), excluding 17 cases which remained in the histopathologically ambiguous category despite expert review (Table 3). Melanoma FISH showed an 84% agreement with final interpretation (61% sensitivity, 100% specificity) (Table 3), again excluding the 17 morphologically ambiguous cases. Excluding indeterminate cases, the myPath sensitivity and specificity was 67% and 96%, respectively. Table 4 lists the breakdown of morphologically challenging cases by specific histopathologic subcategories: spitzoid (45), lesions in which the differential diagnosis was between melanoma and dysplastic or mitotically active atypical nevus (nevoid category [17 cases]), and 4 deep penetrating nevus-like cases). The sensitivity and specificity of the melanoma FISH and myPath assays were similar in the spitzoid category (60% and 100% respectively for FISH and 67% and 95% for myPath, respectively) as was the overall agreement with morphologic interpretation (87%); however, the sensitivity of the myPath assay was lower than FISH in the nevoid cases (63% for FISH and 50% for myPath, respectively). The overall agreement with morphologic interpretation was also lower for the myPath assay as compared to FISH in the nevoid cases (54% and 75%, respectively). Both FISH and the myPath assay showed similar results in the DPN-like category, although the small size of the case cohort precluded assessment of sensitivity and specificity.
Table 2.
Correlation of myPath with FISH in morphologically ambiguous case cohort
| FISH positive |
FISH negative |
No FISH result | Total cases |
Intertest agreement | |
|---|---|---|---|---|---|
| MyPath | 70 | 69% | |||
| 14 Malignant | 9 | 5 | 0 | ||
| 43 Benign | 7 | 31 | 5 | ||
| 8 Indeterminate | 2 | 4 | 2 | ||
| 5 No result | 0 | 4 | 1 |
Table 3.
Comparison of FISH and myPath test results with expert histologic evaluation in morphologically ambiguous case cohort
| Favor malignant | Favor benign | Ambiguous | No. cases | ||
|---|---|---|---|---|---|
| Challenging cases | 70 | ||||
| FISH | |||||
| Positive | 11 | 0 | 7 | Sensitivity | 61% |
| Negative | 7 | 27 | 10 | Specificity | 100% |
| No result | 4 | 4 | 0 | Agreement | 84% |
| MyPath | |||||
| Malignant | 10 | 1 | 3 | Sensitivity | 50% |
| Benign | 5 | 25 | 13 | Specificity | 96% |
| Indeterminate | 5 | 1 | 1 | Agreement | 74% |
| No result | 2 | 4 | 0 | ||
Table 4.
Challenging case broken down by category, comparing myPath and FISH test results with expert histologic evaluation
| Favor malignant | Favor benign | Ambiguous | No. cases | |||
|---|---|---|---|---|---|---|
| Challenging Spitzoid cases | 45 | |||||
| FISH | ||||||
| Positive | 6 | 0 | 6 | Sensitivity | 60% | |
| Negative | 4 | 21 | 6 | Specificity | 100% | |
| No result | 0 | 2 | 0 | Agreement | 87% | |
| MyPath | ||||||
| Malignant | 6 | 1 | 2 | Sensitivity | 67% | |
| Benign | 2 | 20 | 9 | Specificity | 95% | |
| Indeterminate | 1 | 0 | 1 | Agreement | 87% | |
| No result | 1 | 2 | 0 | |||
| Challenging nevoid cases | 17 | |||||
| FISH | ||||||
| Positive | 5 | 0 | 0 | Sensitivity | 63% | |
| Negative | 3 | 4 | 3 | Specificity | 100% | |
| No result | 1 | 1 | 0 | Agreement | 75% | |
| MyPath | ||||||
| Malignant | 4 | 0 | 1 | Sensitivity | 50% | |
| Benign | 1 | 2 | 2 | Specificity | 100% | |
| Indeterminate | 3 | 1 | 0 | Agreement | 54% | |
| No result | 1 | 2 | 0 | |||
| Challenging DPN-like cases | 4 | |||||
| FISH | ||||||
| Positive | 0 | 0 | 1 | |||
| Negative | 0 | 2 | 1 | |||
| MyPath | ||||||
| Malignant | 0 | 0 | 0 | |||
| Benign | 0 | 2 | 2 | |||
| Indeterminate | 0 | 0 | 0 | |||
DPN deep penetrating nevus
Results of expert histopathologic review and SNP array on myPath discordant cases
All cases from both the histologically unequivocable group as well as the histologically ambiguous group in which the myPath result was discordant with either the morphologic final diagnosis (unequivocable group) or the consensus final signout and/or the FISH result in the ambiguous group (81/268 cases), were submitted for independent evaluation by 2 additional experienced dermatopathologist and also for SNP array (if sufficient material was available). As mentioned previously, the diagnoses in the myPath discordant cases from the morphologically unequivocal group were confirmed as either benign or malignant on the expert review, with the exception of 3 invasive melanomas arising in nevi, 4 dysplastic nevi, and 2 melanoma in situ, all of which were categorized as ambiguous as an expert consensus diagnosis was not reached.
The myPath result showed a 14% concordance rate with the concordant expert consensus diagnosis (excluding cases in which there was a lack of consensus between the expert reviewers and the final sign out diagnosis) (Table 5). Of these discordant cases, false positives (52%; 11/21 cases) occurred at approximately the same rate as false negatives (48%; 14/29 (Table 5; Fig. 3). Only 31 of the myPath discordant cases had sufficient material for SNP array. The concordance of SNP array with consensus diagnosis was 71%, which was higher than FISH (54%) (Table 5). Interestingly, the one case categorized as benign on expert review that was positive on SNP array was also scored as malignant on the myPath assay (FISH was not performed). This lesion was diagnosed as a dysplastic nevus with re-excision recommended, and was also categorized as dysplastic nevus in the expert review. The lesion was completely excised.
Table 5.
Discordant cases: comparison of myPath, FISH, and aCGH test results with expert diagnostic review
| Benign consensus diagnosis | Malignant consensus diagnosis | No consensus | Agreement with consensus diagnosis | No. cases | |
|---|---|---|---|---|---|
| MyPath | 14% | 81 | |||
| Malignant | 11 | 3 | 4 | ||
| Benign | 4 | 14 | 20 | ||
| Indeterminate | 6 | 12 | 7 | ||
| FISH | 54% | 32 | |||
| Positive | 0 | 2 | 8 | ||
| Negative | 5 | 6 | 11 | ||
| SNP array | 71% | 31 | |||
| Positive | 1 | 9 | 10 | ||
| Negative | 3 | 4 | 4 |
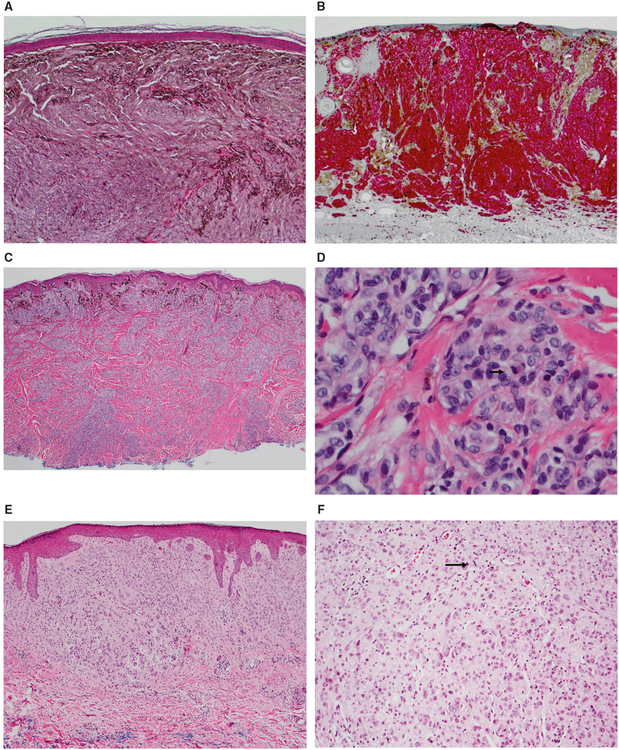
Fig. 3.
Images from 3 morphologically ambiguous cases. a, b Case 148: 28 year old female with a pigmented lesion on the left upper back, diagnosed as favor malignant (2 mm, no ulceration), on expert review. Negative gene expression score, negative FISH, SNP array positive for CN-LOH at 17q12-qter. a There is a densely cellular proliferation of melanocytes in the dermis without maturation and with areas of heavy pigmentation. b A MART-1/Ki-67 double stain shows an elevated proliferative index in the tumor (Mart-1 red, Ki-67 brown). c, d Case 143: 36-year-old male with a pigmented lesion on the chest, diagnosed as a severely atypical intradermal melanocytic tumor of uncertain malignant potential on expert review. Negative gene expression score, negative FISH, SNP array positive for multiple chromosomal aberrations (del 1p12-pter; amp 1p11.2–p12, gain 1q; del 7q34-qter; gain 11q22.1–q22.3, gain 11q23.3–q24.3; del 17p11.2–p13.2, amp at 17p11.2, del 17p11.2-qter; del at 20p13, gain 20p12.3–p13, del 20p11.2–p12.3, gain 20q). c There is compound proliferation of melanocytes with heavy pigmentation superficially and dense aggregates of melanocytes in the dermis in a plexiform fashion without maturation. d The melanocytes are epithelioid with variably prominent nucleoli and mitotic activity (including deep mitoses, arrow). e, f Case 6: 44-year-old female with a pigmented lesion on the back, diagnosed as spitzoid melanoma on expert review (1.6 mm, no ulceration). Positive gene expression score, negative FISH, SNP array positive for monosomy 9. e There is a cellular compound proliferation of epithelioid melanocytes with prominent pleomorphism and no maturation. f Mitotic activity is easily identified (arrow)
Discussion
For a screening test to be clinically relevant, it should have a satisfactory sensitivity and specificity, be cost effective, and provide useful clinically actionable information to guide patient care, based on robust and sufficiently powered independent and ideally prospective studies. Outside of studies co-authored by the employees of and/or consultants for the manufacturer (Myriad), limited data has been published on the myPath test’s performance in accurately categorizing unequivocally benign or malignant melanocytic tumors or compared it’s performance in categorizing ambiguous melanocytic tumors against the current gold standard, histopathologic evaluation, although it is available as a lab developed test and is being utilized to influence diagnostic decision-making [33, 34]. Additionally, comparison of myPath to the melanoma FISH test in the published literature prior to this study is limited to only 15 unequivocally malignant, 24 benign, and 78 morphologically challenging cases [29]. Comparison of myPath to a comprehensive cytogenetic analysis, such as SNP array has been performed in 9 published malignant cases to date [27]. In this study, we sought to independently evaluate the performance of the myPath test in a larger prospectively submitted set of unequivocally benign and unequivocally malignant melanocytic tumors, as well as compare the test’s performance to that of melanoma FISH and SNP array in the categorization of challenging melanocytic lesions (for which the test was created), using expert histopathologic evaluation as the gold standard. The myPath test was assessed on a cohort of 189 morphologically unequivocal cases (76 malignant and 113 benign melanocytic neoplasms), and was found to have a 79% overall agreement with histopathologic diagnosis, with a sensitivity of 72% and specificity of 94%. This is in contrast to the manufacturer’s reported sensitivity and specificity of 90% and 91%, respectively, although it is relatively similar (62% and 97%, respectively) to that reported in Minca et al. [29].
In contrast to the manufacturer, our results include the indeterminate cases, as for a test that can result in equivocal results the recommendation is to avoid exclusion of the indeterminate result in statistical analysis, as this can lead to bias in reporting of diagnostic test performance, including falsely elevated measurements of sensitivity and specificity [31, 32], which is why samples with indeterminate test scores (−2.0 to −0.1) were not excluded in our study, but instead were grouped as negative. The presence of an indeterminate category within the myPath assay also remains a limitation of the assay itself, as this equivocal result is difficult to know how to interpret. Given the fear of underdiagnosis of malignancy within our litigious medical atmosphere in the United States in particular, this equivocal category may lead to overclassification/overtreatment of benign lesions with atypical morphologic features and an indeterminate result as malignant.
Our cohort of unequivocally malignant cases included a majority of superficial spreading melanomas (69%). The superficial spreading histological pattern is the most common melanoma subtype, which is likely why the majority of the invasive melanomas were this subtype in the prospectively and consecutively submitted unequivocal malignant cases sent in. The malignant cohorts in the initial training and validation performed by the manufacturer also contained a majority of superficial spreading type (63% of all melanomas) [25]. While melanomas are still conventionally separated into 4 histologic subtypes (superficial spreading, nodular, lentigo maligna, and acral) the histologic subtype does not always correlate directly with genetic subtype, as the genetic drivers within these subtypes are diverse (for example while the majority of superficial spreading melanomas have BRAF mutations, others harbor NRAS, or KIT mutations, or are triple wildtype). Despite this genetic heterogeneity however, the predominance of superficial spreading type melanoma is a limitation to our study as well as to the initial validation of the assay, as the sensitivity and specificity of the assay may be different in different melanoma subtypes.
Within the 70 additional ambiguous melanocytic tumors submitted, the myPath assay showed a 74% overall agreement with the final expert histopathologic interpretation (50% sensitivity, 96% specificity; excluding 17 cases which remained in the histopathologically ambiguous category despite expert review, and a 69% intertest agreement with melanoma FISH. Similarly to the previously published data comparing myPath to melanoma FISH [29], the sensitivity and specificity of the melanoma FISH assay was higher than the myPath test (61% and 100%, respectively) as was the agreement with final expert histopathologic interpretation (84%). The 100% specificity of the melanoma FISH test in this cohort was unexpected, since we have seen a number of false-positive FISH test results even after careful adjustments for tetraploidy. Based on experience with a much larger set of cases in our practice, the specificity of the melanoma FISH test is around 95%.
Within the 31 cases with sufficient tissue for evaluation by SNP array, SNP array showed the highest agreement with expert consensus diagnosis (71%), followed by FISH (54%), and then myPath (14%), with the caveat that this population of cases was by definition enriched for cases in the myPath score was discordant with the initial sign out diagnosis and/or the FISH result, as it was this cohort of gene score-discordant cases which were specifically submitted for additional expert review and for SNP array. MyPath had a lower sensitivity in borderline melanocytic tumors with a nevoid morphology (50%) than it did in the spitzoid category of melanocytic tumors (67%). This may be due to an enrichment for lesions representing malignant transformation within a benign precursor within the ambiguous nevoid category in general, and consequent intermingling of benign melanocytes with malignant ones in the RNA extracted from these lesions for the myPath assay.
Within the unequivocally benign category of lesions, the false positive gene expression scores were seen exclusively within dysplastic nevi (9/73) and BAPomas (1/1).
Predicting the clinical behavior of histopathologically borderline melanocytic tumors remains one of the most challenging areas of dermatopathology, and hence the need for additional diagnostic tools other than conventional light microscopic analysis is pressing. Fear of under diagnosis of melanoma often leads to over diagnosis of malignancy and consequent overtreatment, particularly in borderline melanocytic lesions. Molecular tools have enlightened our understanding of the genetic pathways involved in the pathogenesis of many nevic and melanoma genetic subtypes [4], and have highlighted the genetic heterogeneity of melanoma and melanocytic tumors. Given this genomic heterogeneity within cutaneous melanocytic neoplasia, it is not surprising that assays (including immunohistochemistry, melanoma FISH, and the myPath gene expression assay) relying on a narrow subset of biomarkers, show significant variability in sensitivity and specificity in accurately classifying melanocytic tumors. Interestingly, with regards to gene expression assay development, although gene expression profiling of melanomas in the literature has in the past identified genes whose expression was associated with either good or bad prognosis [35–38], no significant overlap of genes was seen between these studies. A molecular approach that is able to query the entire genome at high resolution (such as next generation sequencing) will likely yield the highest sensitivity and specificity. Until this type of analysis becomes sufficiently cost effective and there is data on the application of this technology as well as others (including the myPath gene expression assay, FISH, and SNP array) to ambiguous melanocytic tumors with long-term clinical outcomes data, histopathologic evaluation remains the standard of care in diagnostic evaluation of melanocytic tumors. If an ancillary diagnostic tool is to be utilized, the choice of assay is best guided by knowledge of the genetic aberrations specifically associated with disease progression within specific melanocytic tumor subtypes, existence of studies on application of the assay to borderline melanocytic tumors with outcomes data, as well as cost benefit analysis to the patient given the high deductibles and lack of coverage for many of these assays with insufficient peer reviewed evidence to validate their coverage.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Cerroni L, Barnhill R, Elder D, et al. Melanocytic tumors of uncertain malignant potential: results of a tutorial held at the XXIX Symposium of the International Society of Dermato-pathology in Graz, October 2008. Am J Surg Pathol. 2010;34:314–26. [DOI] [PubMed] [Google Scholar]
- 2.Scolyer RA, Shaw HM, Thompson JF, et al. Interobserver reproducibility of histopathologic prognostic variables in primary cutaneous melanomas. Am J Surg Pathol. 2003;27:1571–6. [DOI] [PubMed] [Google Scholar]
- 3.Troxel DB. Medicolegal aspects of error in pathology. Arch Pathol Lab Med. 2006;130:617–9. [DOI] [PubMed] [Google Scholar]
- 4.Lang UE, Yeh I, McCalmont TH. Molecular melanoma diagnosis update: gene fusion, genomic hybridization, and massively parallel short-read sequencing. Clin Lab Med. 2017;37:473–84. [DOI] [PubMed] [Google Scholar]
- 5.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. [DOI] [PubMed] [Google Scholar]
- 6.Blokx WA, van Dijk MC, Ruiter DJ. Molecular cytogenetics of cutaneous melanocytic lesions—diagnostic, prognostic and therapeutic aspects. Histopathology. 2010;56:121–32. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Rao M, Fang Y, et al. A genome-wide high-resolution array-CGH analysis of cutaneous melanoma and comparison of array-CGH to FISH in diagnostic evaluation. J Mol Diagn. 2013;15:581–91. [DOI] [PubMed] [Google Scholar]
- 8.Ali L, Helm T, Cheney R, et al. Correlating array comparative genomic hybridization findings with histology and outcome in spitzoid melanocytic neoplasms. Int J Clin Exp Pathol. 2010;3:593–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009; 33:1146–56. [DOI] [PubMed] [Google Scholar]
- 10.Morey AL, Murali R, McCarthy SW, Mann GJ, Scolyer RA. Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology. 2009;41:383–7. [DOI] [PubMed] [Google Scholar]
- 11.Gerami P, Zembowicz A. Update on fluorescence in situ hybridization in melanoma: state of the art. Arch Pathol Lab Med. 2011;135:830–7. [DOI] [PubMed] [Google Scholar]
- 12.Pouryazdanparast P, Haghighat Z, Beilfuss BA, Guitart J, Gerami P. Melanocytic nevi with an atypical epithelioid cell component: clinical, histopathologic, and fluorescence in situ hybridization findings. Am J Surg Pathol. 2011;35:1405–12. [DOI] [PubMed] [Google Scholar]
- 13.Vergier B, Prochazkova-Carlotti M, de la Fouchardiere A, et al. Fluorescence in situ hybridization, a diagnostic aid in ambiguous melanocytic tumors: European study of 113 cases. Mod Pathol. 2011;24:613–23. [DOI] [PubMed] [Google Scholar]
- 14.Gammon B, Beilfuss B, Guitart J, Gerami P. Enhanced detection of spitzoid melanomas using fluorescence in situ hybridization with 9p21 as an adjunctive probe. Am J Surg Pathol. 2012; 36:81–8. [DOI] [PubMed] [Google Scholar]
- 15.Requena C, Rubio L, Traves V, et al. Fluorescence in situ hybridization for the differential diagnosis between Spitz naevus and spitzoid melanoma. Histopathology. 2012;61:899–909. [DOI] [PubMed] [Google Scholar]
- 16.Gerami P, Scolyer RA, Xu X, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol. 2013; 37:676–84. [DOI] [PubMed] [Google Scholar]
- 17.Gerami P, Mafee M, Lurtsbarapa T, Guitart J, Haghighat Z, Newman M. Sensitivity of fluorescence in situ hybridization for melanoma diagnosis using RREB1, MYB, Cep6, and 11q13 probes in melanoma subtypes. Arch Dermatol. 2010;146:273–8. [DOI] [PubMed] [Google Scholar]
- 18.Dalton SR, Gerami P, Kolaitis NA, et al. Use of fluorescence in situ hybridization (FISH) to distinguish intranodal nevus from metastatic melanoma. Am J Surg Pathol. 2010;34:231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busam KJ, Fang Y, Jhanwar SC, Pulitzer MP, Marr B, Abramson DH. Distinction of conjunctival melanocytic nevi from melanomas by fluorescence in situ hybridization. J Cutan Pathol. 2010;37:196–203. [DOI] [PubMed] [Google Scholar]
- 20.Gammon B, Beilfuss B, Guitart J, Busam KJ, Gerami P. Fluorescence in situ hybridization for distinguishing cellular blue nevi from blue nevus-like melanoma. J Cutan Pathol. 2011;38:335–41. [DOI] [PubMed] [Google Scholar]
- 21.Massi D, Cesinaro AM, Tomasini C, et al. Atypical Spitzoid melanocytic tumors: a morphological, mutational, and FISH analysis. J Am Acad Dermatol. 2011;64:919–35. [DOI] [PubMed] [Google Scholar]
- 22.Gaiser T, Kutzner H, Palmedo G, et al. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol. 2010;23:413–9. [DOI] [PubMed] [Google Scholar]
- 23.Tetzlaff MT, Wang WL, Milless TL, et al. Ambiguous melanocytic tumors in a tertiary referral center: the contribution of fluorescence in situ hybridization (FISH) to conventional histopathologic and immunophenotypic analyses. Am J Surg Pathol. 2013;37:1783–96. [DOI] [PubMed] [Google Scholar]
- 24.Wiesner T, Kutzner H, Cerroni L, Mihm MC Jr., Busam, Murali R Genomic aberrations in spitzoid melanocytic tumours and their implications for diagnosis, prognosis and therapy. Pathology. 2016;48:113–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke LE, Warf MB, Flake DD 2nd, et al. Clinical validation of a gene expression signature that differentiates benign nevi from malignant melanoma. J Cutan Pathol. 2015;42:244–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke LE, Flake DD 2nd, Busam K, et al. An independent validation of a gene expression signature to differentiate malignant melanoma from benign melanocytic nevi. Cancer. 2017;123:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke LE, Pimentel JD, Zalaznick H, Wang L, Busam KJ. Gene expression signature as an ancillary method in the diagnosis of desmoplastic melanoma. Hum Pathol. 2017;70:113–20. [DOI] [PubMed] [Google Scholar]
- 28.Ko JS, Matharoo-Ball B, Billings SD, et al. Diagnostic distinction of malignant melanoma and benign nevi by a gene expression signature and correlation to clinical outcomes. Cancer Epidemiol Biomark Prev. 2017;26:1107–13. [DOI] [PubMed] [Google Scholar]
- 29.Minca EC, Al-Rohil RN, Wang M, et al. Comparison between melanoma gene expression score and fluorescence in situ hybridization for the classification of melanocytic lesions. Mod Pathol. 2016;29:832–43. [DOI] [PubMed] [Google Scholar]
- 30.Gerami P, Li G, Pouryazdanparast P, et al. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol. 2012;36:808–17. [DOI] [PubMed] [Google Scholar]
- 31.Services USDoHaH, Administration FaD, Health CfDaR, Branch DD, Biostatistics Do, Biometrics OoSa. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. U.S. Food and Drug Administration; 2007. [Google Scholar]
- 32.Shinkins B, Thompson M, Mallett S, Perera R. Diagnostic accuracy studies: how to report and analyse inconclusive test results. BMJ. 2013;346:f2778. [DOI] [PubMed] [Google Scholar]
- 33.Cockerell C, Tschen J, Billings SD, et al. The influence of a gene-expression signature on the treatment of diagnostically challenging melanocytic lesions. Per Med. 2017;14:123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cockerell CJ, Tschen J, Evans B, et al. The influence of a gene expression signature on the diagnosis and recommended treatment of melanocytic tumors by dermatopathologists. Medicine. 2016;95:e4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jewell R, Elliott F, Laye J, et al. The clinicopathological and gene expression patterns associated with ulceration of primary melanoma. Pigment Cell Melanoma Res. 2015;28:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nsengimana J, Laye J, Filia A, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6:11683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunner G, Reitz M, Heinecke A, et al. A nine-gene signature predicting clinical outcome in cutaneous melanoma. J Cancer Res Clin Oncol. 2013;139:249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–82. [DOI] [PubMed] [Google Scholar]