Abstract
Objective
Aconitum plants (Ranunculaceae) exhibit toxicity, and accidental ingestion of the plants has been reported in Japan. Identifying the cause of poisoning is important for emergency medical treatment, and a rapid and simple detection technique is required for the identification of poisoning cause. In the present study, we developed a rapid and simple method for detecting Aconitum plant DNA using a loop-mediated isothermal amplification (LAMP) assay.
Results
Specific LAMP primers for Aconitum plants were designed based on the trnL–trnF intergenic spacer region. Using the LAMP primers, the LAMP assay included an initiation reaction of 10 min followed by amplification for 20 min at the isothermal reaction temperature of 65 °C. The LAMP reaction was demonstrated to be specific and highly sensitive to Aconitum plants, given that the assay can be used for 1 pg of purified DNA. Using raw extracted DNA as template, the entire detection procedure from DNA extraction to final detection required only 30 min. Moreover, the protocol identified samples containing approximately 5 mg of Aconitum plants cooked and digested with artificial gastric juice. The currently proposed protocol exhibits good potential as a screening method of Aconitum plant poisoning for emergency medical care.
Electronic supplementary material
The online version of this article (10.1186/s13104-019-4463-1) contains supplementary material, which is available to authorized users.
Keywords: Aconitum, Isothermal amplification, LAMP, Accidental ingestion, Toxic plants
Introduction
In the case of accidental ingestion of toxic plants, rapid identification is important for emergency medical treatment. Aconitum plants, which belong to the family Ranunculaceae, comprise over 200 species worldwide, and approximately 20 species grow naturally in Japan [1–5]. The leaves of Aconitum plants are similar to those of edible plants, such as Anemone flaccida (Ranunculaceae) and Parasenecio delphiniifolius (Compositae), and often grow in the same habitat; therefore, accidental ingestion of Aconitum plants could occur [6]. Aconitum plants contain alkaloids, such as aconitine, which causes systemic paralysis, nausea, and vomiting, followed by dizziness, palpitation, hypotension, arrhythmia, shock, and coma [7, 8]. Some cases of poisoning lead to fatal accidents because of respiratory failure [6, 9–11]. In Japan, a total of 14 patients were reported to be poisoned by Aconitum plants from 2008 to 2017, out of which three patients died because of toxicity [12]. Toxic plants can be identified by morphological analysis or chemical analysis using high-performance liquid chromatography (HPLC) or liquid chromatography-mass spectrometry (LC/MS) [6, 9–11, 13]. Recently, DNA-based analytical methods, such as real time polymerase chain reaction (PCR), have been proposed for the detection of toxic plants, such as Aconitum plants and Veratrum album [14, 15]. However, these analytical methods require long processing time and a well-equipped laboratory. Therefore, a rapid and simple detection method is desirable to facilitate emergency medical treatment of poisoning. A loop-mediated isothermal amplification (LAMP) assay is a DNA amplification method performed under isothermal conditions at 60 °C to 65 °C; the amplification efficiency of the assay is 100- to 1000-fold higher than that of PCR [16]. The LAMP method can be performed within a short time using a simple device, such as a block incubator, and has consequently been widely used for nucleic acid detection of viruses, bacteria, and plants [17–19]. In the present study, we developed a rapid and simple LAMP-based method for the identification of Aconitum plants. Our protocol could be useful as a screening method for poisoning cases that are suspected to be caused by Aconitum plants in emergency medical care.
Main text
Materials and methods
Plant materials and DNA extraction
Aconitum japonicum Thunb. subsp. subcuneatum (Nakai) Kadota, Aconitum sachalinense F. Schmidt subsp. yezoense (Nakai) Kadota, Aconitum chinense Siebold ex Paxton, Anemone flaccida, and Parasenecio delphiniifolius were obtained from the medicinal botanical gardens of Kanazawa University and Josai University. A total of 24 plant samples used for specificity test were obtained from the medicinal botanical garden of Kanazawa University and are provided in Additional file 1. Total genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Germany) or NucleoSpin Plant II (Takara, Japan) and quantified using a NanoDrop 1000 instrument (ThermoFisher Scientific, USA). Extracted DNA was diluted to 5.0 ng/µL and used as DNA template. Optimization of the LAMP assay was performed using 5.0 ng/µL Ac. japonicum subsp. subcuneatum DNA as positive control. LAMP sensitivity was evaluated using 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, and 100 fg of purified Ac. japonicum subsp. subcuneatum genomic DNA as template. Samples were prepared by frying with vegetable oil for 1, 3, and 10 min or boiling in water for 5 and 10 min. Afterwards, samples were treated with or without artificial gastric juice for 2 h [15]. Artificial gastric juice 1 L contained 2.0 g of sodium chloride, 3.2 g of pepsin, and 7.0 mL of 35% HCl and diluted with water to a final volume of 1 L. Template DNA was prepared with extracted DNA using an Easy DNA Extraction Kit version 2 (Kaneka, Japan). Approximately 5 mg of leaf was digested with 100 µL of alkaline lysis solution (Solution A) and heated at 98 °C for 8 min. The heat-treated sample was mixed with 14 µL of neutralization solution (Solution B), and the tenfold-diluted lysate was used as DNA template.
Sequencing and LAMP primer design
PCR targeting the trnL–trnF intergenic spacer region was conducted at the final volume of 25 µL with the following components: 2.5 µL of 1× PCR buffer (Takara), 0.2 mM each dNTP, 0.5 µM each of forward primer (5ʹ-3ʹ; CGA AAT CGG TAG ACG CTA CG) and reverse primer (5ʹ-3ʹ; ATT TGA ACT GGT GAC ACG AG), 0.625 U of Ex Taq DNA polymerase (TaKaRa), and 1 µL of genomic plant DNA. The PCR products were purified and sequenced using BigDye Terminator v1.1 (Applied Biosystems, USA) on a 3130xl DNA genetic analyzer or 310 DNA genetic analyzer (Applied Biosystems) following the manufacturer’s protocol. Obtained sequences of Ac. japonicum subsp. subcuneatum, Anemone flaccida, and Parasenecio delphiniifolius were aligned, and primers specific to Ac. japonicum subsp. subcuneatum were designed using PrimerExplore v4 (https://primerexplorer.jp). A loop primer was designed manually to improve reaction efficiency [20]. The LAMP primers consisted of FIP (5ʹ-3ʹ; TGG GGG TAA AGC GAA CTT TTT ATG ACT TTT TAA ATC GTG AGG GT), BIP (5ʹ-3ʹ; TAA TCC TTT TTT CAG CGG TTC CAA TCC GAT CCA TTT GTG AGA), F3 (5ʹ-3ʹ; TTA TAG TAA GAG GAA AAT CCG TC), B3 (5ʹ-3ʹ; AAA CTT GTG ATA AAA GAG AAA CC), and a loop primer (5ʹ-3ʹ; TGG GGA TAG AGG GAC TTG A).
LAMP assay
The LAMP assay was carried out using an Isothermal amplification kit (Nippon Gene, Japan) with the total volume of 20 µL containing 10 µL of 2× reaction mix, 0.2 µM each F3 and B3 primers, 1.6 µM each FIP and BIP primers, 0.8 µM loop primer, and 1 µL of genomic DNA as template. Fluorescence data of the LAMP reaction were obtained using Smart Cycler (Cepheid, USA) or Step One Plus Real-Time PCR System (ThermoFisher Scientific, USA). The LAMP reaction was carried out for 20 min at the constant temperature of 65 °C. Fluorescence signals were collected at 15-s intervals. The cycle threshold value was determined and converted to the amplification time. The reactions were carried out in triplicates. Results were expressed as standard deviation (min ± SD).
Results
Sequencing and primer design
First, we sequenced the intergenic spacer regions between the trnL–trnF of Ac. japonicum subsp. subcuneatum, Ac. sachalinense subsp. yezoense, Ac. chinense, Anemone flaccida, and Parasenecio delphiniifolius. In Japan, Ac. japonicum subsp. subcuneatum and Ac. sachalinense subsp. yezoense are is predominant, and Ac. chinense is cultivated as an ornamental plant. Sequencing analyses showed that the three Aconitum plants had the same sequences of the trnL–trnF regions. The sequence alignment of the trnL–trnF regions of Ac. japonicum subsp. subcuneatum (DDBJ/EMBL/GenBank database accession No. LC435033), Anemone flaccida (accession No. LC435034), and Parasenecio delphiniifolius (accession No. LC435035) were performed and we designed the Ac. japonicum subsp. subcuneatum-specific LAMP primer set (FIP, BIP, F3, and B3) and the loop primer to shorten the reaction time (Additional file 2).
LAMP reaction
Using the LAMP primer set, the LAMP assay was performed using Ac. japonicum subsp. subcuneatum DNA to optimize the LAMP conditions (Fig. 1). The LAMP reaction using the loop primer started at 7.2 ± 0.1 min, which was shorter than the reaction time without the loop primer (10.1 ± 0.39 min) (Fig. 1a). Next, the LAMP reaction was carried out at 61, 63, 65, and 67 °C. DNA amplification during the LAMP reaction at the incubation temperatures of 61, 63, and 65 °C was detected at approximately 7 min (7.5 ± 0.1, 7.1 ± 0.1, and 7.0 ± 0.2 min, respectively), whereas DNA amplification at 67 °C (9.5 ± 0.3 min) was slightly delayed (Fig. 1b). To evaluate the sensitivity of the LAMP assay, a serial tenfold dilution of purified DNA (10 ng, 1 ng, 100 pg, 10 pg, 1 pg, and 100 fg) was used as DNA template (Table 1). The DNA templates at 10 ng, 1 ng, 100 pg, 10 pg, and 1 pg showed successful amplification (6.8 ± 0.2, 8.0 ± 0.3, 8.9 ± 0.2, 10.3 ± 0.4, and 11.9 ± 0.3 min); however, amplification was not detected using 100 fg of DNA template. The sensitivity of the LAMP assay was higher than the previously reported sensitivity of real-time PCR (100 pg) [14]. We also confirmed that amplification of no-template controls did not occur for 20 min. To evaluate the specificity of the LAMP assay for Aconitum plants, the assay was conducted using three Aconitum species (Ac. japonicum subsp. subcuneatum, Ac. sachalinense subsp. yezoense, and Ac. chinense), Anemone flaccida, and Parasenecio delphiniifolius. Ac. japonicum subsp. subcuneatum, Ac. sachalinense subsp. yezoense, and Ac. chinense showed positive results from the LAMP assay (7.1 ± 0.1, 7.3 ± 0.0, and 7.2 ± 0.2 min, respectively), whereas Anemone flaccida and Parasenecio delphiniifolius tested negative. We further evaluated the results using 24 other randomly selected plant species to determine any false positive reaction (Additional file 1). No amplification was detected in the LAMP reactions with other plant species, except for Coptis japonica, which belongs to Ranunculaceae. However, amplification of C. japonica was observed in 2 of 5 replicates and the initiation time of amplification was approximately 18 min (17.8 min and 19.3 min), which was markedly longer than that of Aconitum plants (approximately 7 min) (Additional file 1). When the mixture containing 50 pg Ac. japonicum subsp. subcuneatum DNA and 50 ng Anemone flaccida DNA was used as template, amplification was observed at 9.8 ± 0.3 min. Therefore, we concluded that the LAMP primer set is specific to Aconitum plants.
Fig. 1.
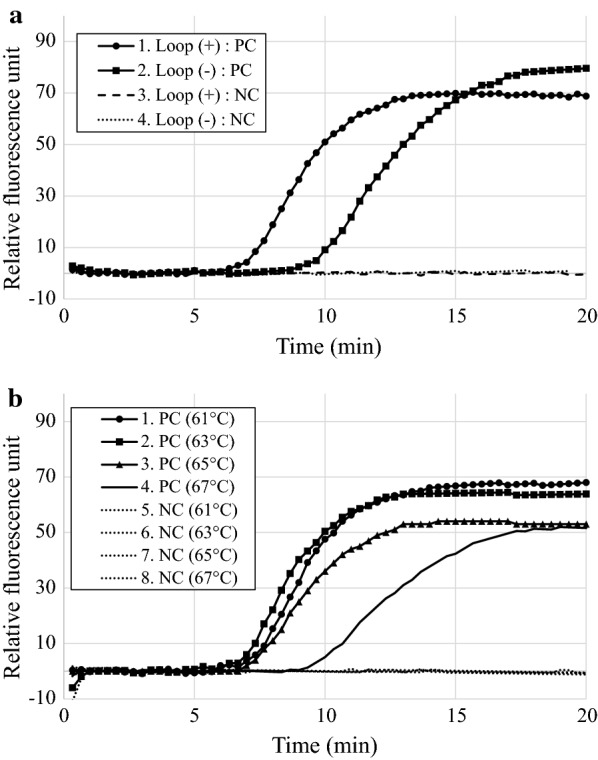
Optimization of the LAMP assay. a The LAMP assay was performed using the LAMP primer set with or without a loop primer. b The LAMP reaction was performed at 61, 63, 65, and 67 °C. Amplification profiles were obtained from positive control (PC) samples using Aconitum. japonicum subsp. subcuneatum DNA (5 ng) and negative control (NC) samples using Anemone flaccida DNA (5 ng)
Table 1.
Results of LAMP detection assay
| Sample | DNA template | Reaction timea | |
|---|---|---|---|
| Sensitivity | Ac. japonicum subsp. subcuneatum | 10 ng | 6.8 ± 0.2 min |
| 1 ng | 8.0 ± 0.3 min | ||
| 100 pg | 8.9 ± 0.2 min | ||
| 10 pg | 10.3 ± 0.4 min | ||
| 1 pg | 11.9 ± 0.3 min | ||
| 100 fg | nd | ||
| (No template) | nd | ||
| Specificity | Ac. japonicum subsp. subcuneatum | 5 ng | 7.1 ± 0.1 min |
| Ac. sachalinense subsp. yezoense | 5 ng | 7.3 ± 0.0 min | |
| Ac. chinense | 5 ng | 7.2 ± 0.2 min | |
| Anemone flaccida | 5 ng | nd | |
| Parasenecio delphiniifolius | 5 ng | nd | |
| Mixture | Ac. japonicum subsp. subcuneatum | 50 pg | 10.0 ± 0.2 min |
| Ac. japonicum subsp. subcuneatum + Anemone flaccida | 50 pg + 50 ng | 9.8 ± 0.3 min | |
| Anemone flaccida | 50 ng | nd |
nd not detected
aThe reactions were carried out in triplacates
Simple protocol of LAMP and reactivity for processed samples
To simplify Aconitum plant detection, the LAMP assay was conducted using raw extracted DNA, which has been previously reported [19]. Processed samples were tested using the proposed protocol. Considering that Anemone flaccida and Parasenecio delphiniifolius are generally eaten after boiling or frying, Ac. japonicum subsp. subcuneatum leaves were boiled for 5 and 10 min or fried for 1, 3, and 10 min. Next, the processed samples were treated with artificial gastric juice at 2 h. Regardless of whether or not the samples were treated with artificial gastric juice, positive reactions were observed in the LAMP assay using non-processed samples, samples boiled for 5 and 10 min, and samples fried for 1 min and 3 min (Table 2). No amplification was observed using fried samples for 10 min probably because the DNA was degraded by excessive heating.
Table 2.
Processed samples detected using the proposed protocol
| Non-processed sample (leaf) | Boiled sample | Fried sample | ||||
|---|---|---|---|---|---|---|
| 5 min | 10 min | 1 min | 3 min | 10 min | ||
| Ac. japonicum subsp. subcuneatum | + (+) | + (+) | + (+) | + (+) | + (+) | − (−) |
| Anemone flaccida | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) |
Dried leaves were processed by boiling for 5 or 10 min or frying for 1, 3, and 10 min. The symbol ‘+’ indicates successful amplification within 20 min. No amplification within 20 min was denoted by the symbol ‘−’. The results of processed samples that were treated with artificial gastric juice for 2 h are shown in parentheses
Discussion
In the present study, we developed a LAMP-based method for detecting Aconitum plant DNA. The LAMP primer set was designed based on the trnL–trnF intergenic spacer region of chloroplast DNA. The sequences of many Aconitum species are provided in the DDBJ/EMBL/GenBank databases. Out of 39 Aconitum species, 24 species had fully matched sequences with the LAMP primer binding sites, whereas the sequences of 15 species did not completely match the binding sites. Considering the diversity of Aconitum plants, which include over 200 Aconitum species worldwide, LAMP reactivity for each Aconitum species should be carefully evaluated. Using raw extracted DNA, the whole detection procedure from DNA extraction to detection was completed within approximately 30 min. Given that the protocol successfully identified most of the processed samples treated with artificial gastric juice, it can be used to screen unknown samples that are not identifiable by morphological analysis. It could test vomit samples from a patient, samples of a suspicious food that a patient ate, and samples from a suspicious plant. In the present study, we performed the LAMP assay on a real-time PCR device to monitor the amplification signals. However, the LAMP assay can be performed on a block incubator, and amplification can be checked by visual inspection (Additional file 3). Therefore, the protocol is expected to be useful as a screening method for poisoning cases that are suspected to be caused by Aconitum plants in emergency medical care. In summary, we developed a rapid and simple method for the detection of Aconitum plants based on a LAMP assay, a nucleic acid detection method. Our method showed high sensitivity, and the whole detection procedure can be completed within 30 min. Therefore, the proposed LAMP method could be useful as a screening method for poisoning in emergency medical treatment.
Limitations
Considering that Aconitum plants comprise more than 200 species and several Aconitum plants have varieties and subspecies, LAMP reactivity for each Aconitum species should be evaluated in a careful manner.
Additional files
Additional file 1. Plant information and LAMP Specificity for 24 randomly selected samples.
Additional file 2. Primer design for the LAMP assay.
Additional file 3. Sample tubes after LAMP reaction.
Acknowledgements
Not applicable.
Abbreviations
- bp
base pair
- DNA
deoxyribonucleic acid
- dNTP
deoxyribonucleotide triphosphate
- LAMP
loop-mediated isothermal amplification
- PCR
polymerase chain reaction
Authors’ contributions
MK conceived and designed the study protocol. MK, AK, TY, HA, and YohS performed sample collection, data collection, and data analysis. MK performed data presentation and drafted the manuscript. HA, YosS, RS, and YohS critically reviewed the manuscript. Planning, coordination, and supervision of the project was conducted by HA, YohS, RS, and YosS. All authors read and approved the final manuscript.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP17H00685. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed in the current study are described in the manuscript. Any additional details will be made available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Masashi Kitamura, Phone: +81-49-271-8089, Email: kitamura@josai.ac.jp.
Akira Kazato, Email: yk15019@josai.ac.jp.
Tadashi Yamamuro, Email: yamamuro@nrips.go.jp.
Hirokazu Ando, Email: ando@p.kanazawa-u.ac.jp.
Yohei Sasaki, Email: sasaki@p.kanazawa-u.ac.jp.
Ryuichiro Suzuki, Email: ryu_suzu@josai.ac.jp.
Yoshiaki Shirataki, Email: shiratak@josai.ac.jp.
References
- 1.Singhuber J, Zhu M, Prinz S, Kopp B. Aconitum in traditional Chinese medicine: a valuable drug or an unpredictable risk? J Ethnopharmacol. 2009;126(1):18–30. doi: 10.1016/j.jep.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 2.Jabbour F, Renner SS. A phylogeny of Delphinieae (Ranunculaceae) shows that Aconitum is nested within Delphinium and that Late Miocene transitions to long life cycles in the Himalayas and Southwest China coincide with bursts in diversification. Mol Phylogenet Evol. 2012;62(3):928–942. doi: 10.1016/j.ympev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Meng J, Li X, Li H, Yang J, Wang H, He J. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 2018;23(5):E1015. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makino T. Makino’s new illustrated flora of Japan. Tokyo: The Hokuryukan Co. Ltd.; 1989. pp. 135–141. [Google Scholar]
- 5.Kakiuchi N, Atsumi T, Higuchi M, Kamikawa S, Miyako H, Wakita Y, Ohtsuka I, Hayashi S, Hishida A, Kawahara N, Nishizawa M, Yamagishi T, Kadota Y. Diversity in aconitine alkaloid profile of Aconitum plants in Hokkaido contrasts with their genetic similarity. J Nat Med. 2015;69(1):154–164. doi: 10.1007/s11418-014-0867-3. [DOI] [PubMed] [Google Scholar]
- 6.Kazuma K, Satake M, Konno K. Case of fatal aconite poisoning, and its background. Shokuhin Eiseigaku Zasshi. 2013;54(6):419–425. doi: 10.3358/shokueishi.54.419. [DOI] [PubMed] [Google Scholar]
- 7.Ameri A. The effects of Aconitum alkaloids on the central nervous system. Prog Neurobiol. 1998;56(2):211–235. doi: 10.1016/S0301-0082(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 8.Shaw D. Toxicological risks of Chinese herbs. Planta Med. 2010;76(17):2012–2018. doi: 10.1055/s-0030-1250533. [DOI] [PubMed] [Google Scholar]
- 9.Pullela R, Young L, Gallagher B, Avis SP, Randell EW. A case of fatal aconitine poisoning by Monkshood ingestion. J Forensic Sci. 2008;53(2):491–494. doi: 10.1111/j.1556-4029.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Zhuo L, Liu L, Zhu S, Sunnassee A, Liang M, Zhou L, Liu Y. Seven cases of fatal aconite poisoning: forensic experience in China. Forensic Sci Int. 2011;212(1–3):e5–e9. doi: 10.1016/j.forsciint.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Elliott SP. A case of fatal poisoning with the aconite plant: quantitative analysis in biological fluid. Sci Justice. 2002;42(2):111–115. doi: 10.1016/S1355-0306(02)71807-8. [DOI] [PubMed] [Google Scholar]
- 12.Ministry of Health, Labour and Welfare: Shizendoku no risk profile. https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/kenkou_iryou/shokuhin/syokuchu/poison/index.html. Accessed 15 Dec 2018.
- 13.Usui K, Hayashizaki Y, Hashiyada M, Nakano A, Funayama M. Simultaneous determination of 11 aconitum alkaloids in human serum and urine using liquid chromatography-tandem mass spectrometry. Leg Med (Tokyo). 2012;14(3):126–133. doi: 10.1016/j.legalmed.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Matsuyama S, Nishi K. Genus identification of toxic plant by real-time PCR. Int J Legal Med. 2011;125(2):211–217. doi: 10.1007/s00414-010-0487-8. [DOI] [PubMed] [Google Scholar]
- 15.Kikkawa HS, Tsuge K, Kubota S, Aragane M, Ohta H, Sugita R. Species identification of white false hellebore (Veratrum album subsp. oxysepalum) using real-time PCR. Forensic Sci Int. 2017;275:160–166. doi: 10.1016/j.forsciint.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosaki Y, Takada A, Ebihara H, Grolla A, Kamo N, Feldmann H, Kawaoka Y, Yasuda J. Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplification. J Virol Methods. 2007;141(1):78–83. doi: 10.1016/j.jviromet.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 18.Ravan H, Amandadi M, Sanadgol N. A highly specific and sensitive loop-mediated isothermal amplification method for the detection of Escherichia coli O157:H7. Microb Pathog. 2016;91:161–165. doi: 10.1016/j.micpath.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura M, Aragane M, Nakamura K, Adachi T, Watanabe K, Sasaki Y. Improved on-site protocol for the DNA-based species identification of Cannabis sativa by loop-mediated isothermal amplification. Biol Pharm Bull. 2018;41(8):1303–1306. doi: 10.1248/bpb.b18-00272. [DOI] [PubMed] [Google Scholar]
- 20.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Plant information and LAMP Specificity for 24 randomly selected samples.
Additional file 2. Primer design for the LAMP assay.
Additional file 3. Sample tubes after LAMP reaction.
Data Availability Statement
The datasets used and/or analyzed in the current study are described in the manuscript. Any additional details will be made available from the corresponding author upon reasonable request.


