Abstract
Actinosynnema is a small but well-known genus of actinomycetes for production of ansamitocin, the payload component of antibody-drug conjugates against cancers. However, the secondary metabolite production profile of Actinosynnema pretiosum ATCC 31565, the most famous producer of ansamitocin, has never been fully explored. Our antiSMASH analysis of the genomic DNA of Actinosynnema pretiosum ATCC 31565 revealed a NRPS–PKS gene cluster for polyene macrolactam. The gene cluster is very similar to gene clusters for mirilactam and salinilactam, two 26-membered polyene macrolactams from Actinosynnema mirum and Salinispora tropica, respectively. Guided by this bioinformatics prediction, we characterized a novel 26-membered polyene macrolactam from Actinosynnema pretiosum ATCC 31565 and designated it pretilactam. The structure of pretilactam was elucidated by a comprehensive analysis of HRMS, 1D and 2D-NMR, with absolute configuration of chiral carbons predicted bioinformatically. Pretilactam features a dihydroxy tetrahydropyran moiety, and has a hexaene unit and a diene unit as its polyene system. A preliminary antibacterial assay indicated that pretilactam is inactive against Bacillus subtilis and Candida albicans.
Keywords: Actinosynnema pretiosum ATCC 31565, polyene macrolactam, pretilactam, antiSMASH
1. Introduction
Genome-guided discovery of secondary metabolites is a powerful strategy for rational identification of novel compounds from microorganisms [1,2]. Recent advances in DNA sequencing technologies allow rapid and low-cost sequencing of microbial genomic DNA, and bioinformatics tools (antiSMASH, ClusterMine360, and NORINE, etc.) are also developed for detection of various gene clusters for secondary metabolites (even their structures) in microbial genomes, which greatly facilitate the de-replication of known metabolites and targeted mining of new ones in the chemistry study of secondary metabolites produced by microorganisms [3,4,5]. The gene clusters predicted by the comprehensive antiSMASH may provide valuable information about secondary metabolite production profiles of microbial strains analyzed.
Polyene macrolactams are a growing family of microbial secondary metabolites biosynthesized by NRPS–PKS (nonribosomal peptide synthase–polyketide synthase) complexes. Quite a few polyene macrolactams, together with their (putative) gene clusters, have been identified from actinomycetes in the past decade, such as auroramycin from Streptomyces, micromonolactam and lobosamide from Micromonospora, mirilactam from Actinosynnema mirum, and salinilactam from Salinispora tropica [6,7,8,9,10,11,12,13,14]. It is worthy to note that some of these polyene macrolactams display antibacterial, antiprotozoal, or cytotoxic activities [9,10,11].
Actinosynnema is a small but well-known genus of actinomycetes for its production of ansamitocin, the payload component of antibody-drug conjugates against cancers. Actinosynnema pretiosum ATCC 31565 (hereafter in brief ATCC 31565) is the most famous ansamitocin producer [14,15,16,17]. However, the secondary metabolite production profile of ATCC 31565 has never been fully explored, although it was reported to produce secondary metabolites dnacin (a naphthyridinomycin-type antitumor antibiotic) and actinosynneptide (a cytotoxic hexapeptide) [18,19,20].
To get a deep insight into the secondary metabolite biosynthetic potential of ATCC 31565, we sequenced its genomic DNA to direct our chemical investigation of secondary metabolites for new compound(s). Herein, the genome-guided identification of a novel 26-membered polyene macrolactam from ATCC 31565 is described as below.
2. Results and Discussion
2.1. ATCC 31565 Contains a Gene Cluster for Polyene Macrolactam
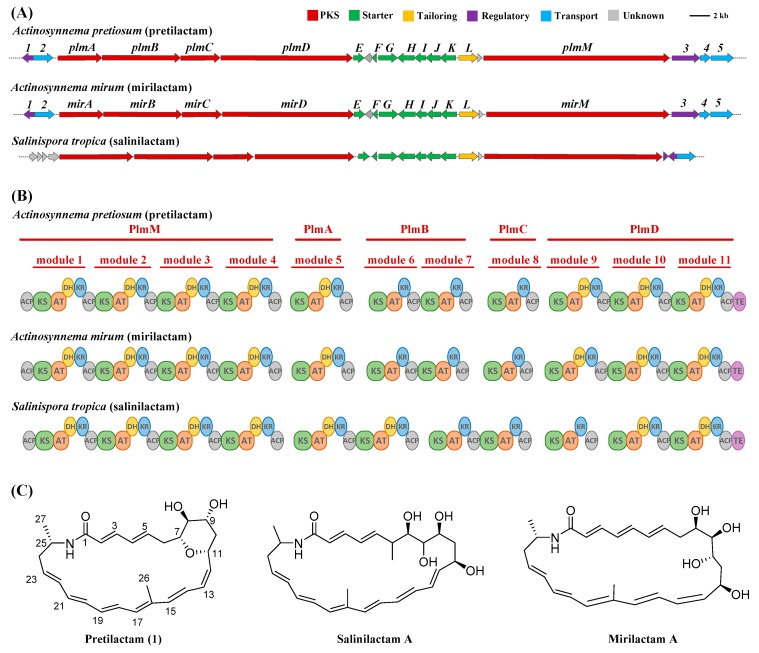
Genome sequencing and assembly resulted in a linear chromosome DNA of 8,131,271 bp for ATCC 31565. AntiSMASH analysis of the chromosome DNA revealed over 20 gene clusters for various secondary metabolites, including the expected gene cluster for ansamitocin biosynthesis. Among them, an 80 kb gene cluster for NRPS–PKS showed very high percentages of similar genes to mirilactam and salinilactam gene clusters. Specifically, the gene cluster contained 5 genes for type I modular polyketide synthases (PKS), 1 gene for cytochrome P450 oxidase, and a set of 7 genes for 3-aminobutyrate (the starter of some polyene macrolactams) biosynthesis. The enzymes/proteins encoded by these genes showed ≥96% amino acid sequence identities to those for mirilactam biosynthesis. In particular, the 11 modules of the five PKSs revealed the same or very similar domain organization to PKSs for mirilactam or salinilactam biosynthesis (Figure 1A,B, Table 1). Thus, the gene cluster should be responsible for the biosynthesis of polyene macrolactam(s) very similar or identical to mirilactam or salinilactam. The gene cluster was designated as plm, and deposited in GenBank with the accession number MK341065.
Figure 1.
(A) The plm gene cluster for a putative polyene macrolactam (pretilactam, 1) from Actinosynnema pretiosum ATCC 31565 and its comparison with gene clusters for mirilactam from Actinosynnema mirum and salinilactam from Salinispora tropica. (B) Alignment of PKS domains for biosynthesis of the putative polyene macrolactam (pretilactam, 1), mirilactam and salinilactam. ACP, acyl carrier protein; AT, acyltransferase; DH, dehydrotase; KR, ketoreductase; KS, ketosynthase. (C) Structures of pretilactam (1), salinilactam A, and mirilactam A.
Table 1.
Predicted function of ORFs from plm gene cluster for a putative polyene macrolactam (pretilactam, 1) in ATCC 31565.
| ORF | Size (aa) | Homologue [Accession No./Strain] | Identity/Positive | Predicted Function |
|---|---|---|---|---|
| plm1 | 210 | TetR/AcrR family [ACU36614.1/Actinosynnema mirum DSM 43827] | 92/96 | transcriptional regulator |
| plm2 | 613 | major facilitator superfamily MFS_1 [ACU36615.1/Actinosynnema mirum DSM 43827] | 95/96 | MFS transporter |
| plmA | 1816 | short-chain dehydrogenase/reductase SDR [ACU36616.1/Actinosynnema mirum DSM 43827] | 97/98 | type I modular PKS |
| plmB | 3092 | short-chain dehydrogenase/reductase SDR [ACU36617.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmC | 1606 | short-chain dehydrogenase/reductase SDR [ACU36618.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmD | 5400 | short-chain dehydrogenase/reductase SDR [ACU36619.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmE | 296 | proline-specific peptidase [ACU36620.1/Actinosynnema mirum DSM 43827] | 96/97 | peptidase |
| plmF | 82 | phosphopantetheine-binding protein [ACU36622.1/Actinosynnema mirum DSM 43827] | 100/100 | phosphopantetheine-binding protein |
| plmG | 529 | AMP-dependent synthetase and ligase [ACU36623.1/Actinosynnema mirum DSM 43827] | 96/97 | synthetase and ligase |
| plmH | 507 | AMP-dependent synthetase and ligase [ACU36624.1/Actinosynnema mirum DSM 43827] | 98/98 | synthetase and ligase |
| plmI | 308 | S-malonyltransferase-like protein [ ACU36625.1/Actinosynnema mirum DSM 43827] | 98/98 | ACP S-malonyl transferase |
| plmJ | 419 | pyridoxal phosphate-dependent aminotransferase [ACU36626.1/Actinosynnema mirum DSM 43827] | 97/98 | aminotransferase |
| plmK | 450 | l-lysine 2,3-aminomutase [ACU36627.1/Actinosynnema mirum DSM 43827] | 99/99 | glutamate 2,3-aminomutase |
| plmL | 395 | cytochrome P450 [ACU36628.1/Actinosynnema mirum DSM 43827] | 98/99 | cytochrome P450 oxidase |
| plmM | 7110 | short-chain dehydrogenase/reductase SDR [ACU36630.1/Actinosynnema mirum DSM 43827] | 97/98 | type I modular PKS |
| plm3 | 931 | transcriptional regulator, LuxR family [ACU36631.1/Actinosynnema mirum DSM 43827] | 98/98 | transcriptional regulator |
| plm4 | 279 | ABC transporter related [ACU36632.1/Actinosynnema mirum DSM 43827] | 99/100 | ATP-binding protein |
| plm5 | 855 | FtsX-like permease family protein [ACU36633.1/Actinosynnema mirum DSM 43827] | 98/99 | FtsX-like permease family |
2.2. The Putative Polyene Macrolactam(s) was Detected by LC-MS
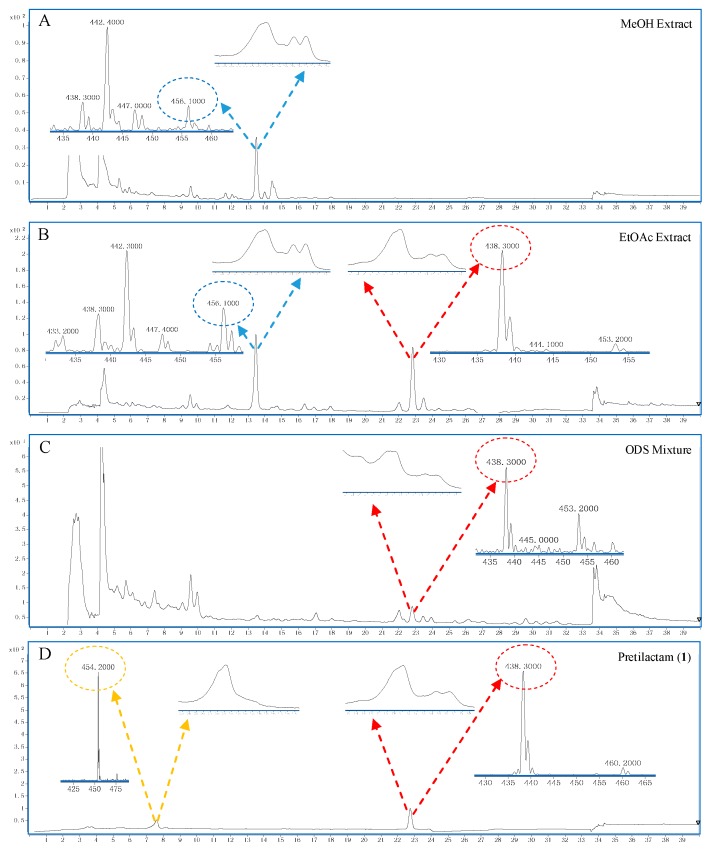
The fermentation culture of ATCC 31565 was extracted with methanol or ethyl acetate, and the extracts were analyzed by LC-MS. The LC traces revealed peaks with UV absorption profiles very similar to mirilactam A (Figure 2). The hyphenated MS spectra of these peaks displayed molecular ions m/z 456 (the same as that of mirilactam A) and/or 438 [M + H]+. Thus, putative polyene macrolactams were found in ATCC 31565.
Figure 2.
LC-MS for putative polyene macrolactam and the purified polyene macrolactam pretilactam (1). (A) A molecular ion m/z 456 [M + H]+ appeared at 13.5 min in the MeOH extract of ATCC 31565. (B) Two molecular ions m/z 456 [M + H]+ and 438 [M + H]+ appeared at 13.5 and 22.7 min, respectively, in the EtOAc extract of ATCC 31565. (C) Only the molecular ion m/z 438 [M + H]+ appeared at 22.7 min in the ODS mixture before column fractionation. (D) A molecular ion m/z 438 [M + H]+ appeared at 22.7 min for the purified pretilactam (1). The molecular ion m/z 454 [M + H]+ at 7.6 min should be a degradation molecule due to incubation of the purified pretilactam (1) at 4 °C for 24 h before analysis.
2.3. Structure of the Polyene Macrolactam (1, Pretilactam) was Elucidated by NMR
Compound 1 is a white amorphous solid. Its molecular formula was established as C27H35O4N by HRMS (Figure S1), indicating 11 degrees of unsaturation. The 13C-NMR, DEPT, and HSQC spectra (Figures S2–S8) of 1 differentiated 27 carbon atoms, including 1 carbonyl group (δC 166.2), 1 nonprotonated olefinic carbon (δC 135.7), 20 methine groups (including 15 olefinic groups δC 124.7–139.7, 4 oxymethine groups δC 66.8–71.4, and 1 nitrogen-substituted methine group δC 44.7), 3 methylene groups δC 34.8–42.2, and 2 methyl groups δC 13.1–21.3. The presence of many olefinic groups indicated that 1 is a polyene.
The COSY and TOCSY spectra of 1 revealed two spin systems. The first spin system (C-2 to C-15) contained 4 oxymethines, 2 methylenes, and two diene fragments (C-2 to C-5, C-12 to C-15). The COSY correlations between H-8 (δH 3.13) and an active hydrogen (δH 4.58, 8-OH), and between H-9 (δH 3.90) and another active hydrogen (δH 4.53, 10-OH), established two hydroxymethines: C-8 (δC 71.4) and C-9 (δC 67.0). The chemical shifts of δC 71.0 (δH 3.62) and δC 66.8 (δH 4.58) disclosed the other two oxymethines at C-7 and C-11, respectively. The HMBC correlation from H-7 to C-11 indicated that the two oxymethines must share an oxygen (as a bridge to join C-7 and C-11), and this was also supported by the molecular formula requirement of 1, i.e., two oxygens in hydroxys, one oxygen in carbonyl, thus only one oxygen left for two oxymethines. Therefore, a dihydroxy tetrahydropyran moiety was determined in 1. The HMBC correlations from H-2 and H-3 to the carbonyl at δC 166.2 and NOESY correlation between H-2 and NH proved that the diene fragment of C-2 to C-5 conjugated with an amide group. The HMBC correlations from H-14 (δH 6.70) to the nonprotonated olefinic carbon δC 135.7 (C-16), H3-26 (δH 1.70) to the olefinic carbon C-17 (δC 130.9), and NOESY correlation between H-14 and C-26 methyl group (δC 13.1, δH 1.70 s) further extended the diene fragment of C-12 to C15 to a triene fragment of C-12 to C-17.
The second spin system (C-27, C-25 to C-22) contained a methyl, a methine, a methylene, and two sp2 methines. The COSY correlation between the methine (δH 3.82, H-25) and an active hydrogen (δH 7.35, -NH-), together with the chemical shifts of the methine (δH 3.82, δC 44.7), demonstrated that C-25 connected to the nitrogen atom. The HMBC correlation from NH group (δC 7.35) to C-1 (δC 166.2), as well as only one nitrogen atom in the molecular formula of 1, determined the connection of the carbonyl C-1 with C-25 via NH.
Four sp2-hybridized methine carbons (δC 128.7, 131.0, 132.1, and 139.0) could not be assigned due to the imperfectness of 1D- and 2D-NMR signals, with their corresponding proton signals overlapped except the δH 6.19 for δC 128.7. However, based on the unsaturation degrees of 1 and UV absorption profile (maximal absorption at 301, 343, and 364 nm suggested a hexaene unit; Figure 2), the four sp2-hybridized methines must form a diene fragment that connected the above two spin systems at C-17 and C-22 to form a hexaene unit, thereby completing the 26-membered polyene macrolactam of 1 (Figure 3).
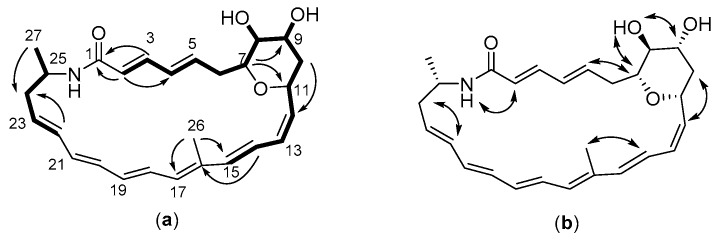
Figure 3.
(a) 1H-1H COSY and key HMBC correlations of pretilactam (1). (b) Key NOESY correlations of pretilactam (1).
The geometries of five C-C double bonds in 1 were assigned to be 2E, 12Z, 14Z, 16E, and 22E based on their vicinal coupling constants (JH2-H3 = 14.4 Hz, JH12-H13 = 11.2 Hz, and JH14-H15 = 15.2 Hz, JH18-H19 = 14.4 Hz, JH22-H23 = 14.4 Hz) and the NOESY correlations of H-14/H-26. Owing to the overlaps of proton signals and NOESY correlations, the geometries of C-C double bonds at 4(5), 18(19), and 20(21) in 1 could not be determined. Thus, the planar structure of 1 was determined by NMR interpretation. Compound 1 was designated as pretilactam. Its NMR data were assigned in Table 2.
Table 2.
NMR data of pretilactam (1).
| Position | δH (mult., J in Hz) | δC | 1H-1H COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 1 | 166.2 | ||||
| 2 | 5.84 (d, 14.4) | 124.7 | H-3 | C-1, C-4 | NH |
| 3 | 6.52 (dd, 14.4, 10.4) | 139.7 | H-2, H-4 | C-1 | |
| 4 | 6.11 (ovl a) | 127.8 | H-5 | ||
| 5 | 5.54 (m) | 138.3 | H-4, H-6 | H-7 | |
| 6 | 2.55 (m), 2.10 (m) | 34.8 | H-5, H-7 | C-5, C-7 | |
| 7 | 3.62 (t, 8.8) | 71.0 | H-6, H-8 | C-6, C-8, C-9, C-11 | 8-OH |
| 8 | 3.13 (br.t, 6.4) | 71.4 | H-7, H-9, OH-8 | ||
| 9 | 3.90 (brs) | 67.0 | H-8, H-10, OH-9 | 8-OH | |
| 10 | 1.81 (brs) | 35.5 | H-9, H-11 | C-11, C-12 | H-12 |
| 11 | 4.58 (ovl a) | 66.8 | H-10, H-12 | ||
| 12 | 5.85 (m) | 133.1 | H-11, H-13 | C-11 | |
| 13 | 6.16 (t, 11.2) | 132.9 | H-12, H-14 | ||
| 14 | 6.70 (dd, 14.4, 12.0) | 126.0 | H-13, H-15 | C-16 | H3-26 |
| 15 | 6.24 (d, 15.2) | 136.6 | H-14 | C-26 | |
| 16 | 135.7 | ||||
| 17 | 6.10 (ovl a) | 130.9 a | |||
| 18 | 6.19 (dd, 14.4, 12.8) | 128.7 | |||
| 19 | 6.10 (ovl a) | 131.0 a | |||
| 20 | 6.10 (ovl a) | 132.1 a | |||
| 21 | 6.10 (ovl a) | 139.0 | |||
| 22 | 5.90 (dd, 14.4, 11.2) | 134.8 | H-23 | C-24 | H-24 |
| 23 | 5.46 (m) | 130.7 | H-22, H-24 | ||
| 24 | 2.33 (m), 1.69 (ovl a) | 42.2 | H-23, H-25 | C-25, C-23 | |
| 25 | 3.82 (brs) | 44.7 | H-24, H-27, NH | ||
| 26 | 1.70 (s) | 13.1 | C-16, C-17 | ||
| 27 | 1.12 (d, 6.4) | 21.3 | H-25 | C-24, C-25 | |
| NH | 7.35 (d, 9.6) | H-25 | C-1, C-25 | H-2 | |
| 8-OH | 4.58 (d, 6.4) | H-8 | H-7, H-9 | ||
| 9-OH | 4.53 (d, 2.4) | H-9 |
a Overlapped with other peaks.
The absolute configuration of chiral carbons in pretilactam may be predicted bioinformatically. The chiral C-9 and C-11 should take the same S configuration because substrate predictions for KR stereochemistry in module 8 and module 7 (Figure 1B) were both A1 by antiSMASH [3,21]. Chiral C-25 should take the same S configuration as in mirilactam A because the seven genes for 3-aminobutyrate biosynthesis from ATCC 31565 could be regarded as the same with those from Actinosynnema mirum (based on ≥96% amino acid sequence identities for each enzyme or protein encoded by the 7 genes) [6]. Besides, the S configuration of C-25 was also supported by the stereochemistry of mirilactams C-E [12]. The H-7, OH-8, and H-9 were strongly suggested to locate on the same surface of the tetrahydropyran ring by NOESY correlations of H-7/OH-8 and OH-8/H-9. Thus, chiral C-7 and C-8 should take the same R configuration (Figure 1C).
The structure of pretilactam (1) elucidated by NMR matches rather well with the structure predicted by its gene cluster, except the following three minor points. First, the formation of C-C double bond 12(13) requires an active DH domain in module 6. Second, the formation of C-6(hydroxy),7 single bond demands an inactive DH domain in module 9. Third, the substrate predictions of acyltransferase domains AT2, AT3, AT4, and AT8 are not accurate. Similar disagreements have occurred many times in structure prediction by other type I modular PKSs [6,22,23,24,25].
2.4. The Dihydroxy Tetrahydropyran Ring Might Come from Dehydration of a Tetrahydroxy Pentane Unit in Pretilactam Biosynthesis
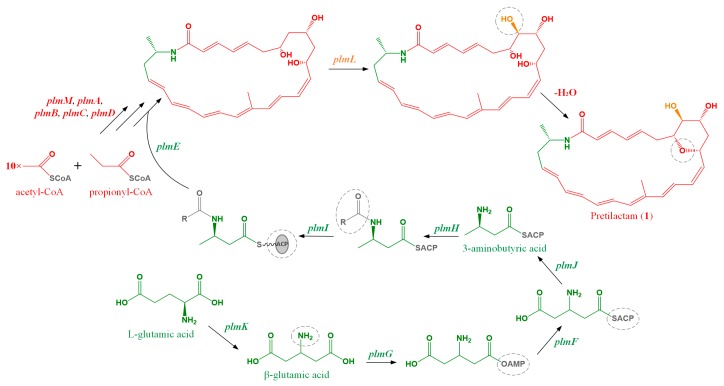
Pretilactam features a dihydroxy tetrahydropyran ring, which might be formed by spontaneous dehydration of a tetrahydroxy pentane unit in the final stage of pretilactam biosynthesis. Based on present understanding of microbial polyene macrolactams biosynthesis, we proposed a plausible biosynthetic pathway for pretilactam (Figure 4).
Figure 4.
The plausible biosynthetic pathway of pretilactam (1).
2.5. Pretilactam was Inactive Against Bacillus Subtilis and Candida Albicans
A preliminary biological activity assay indicated that pretilactam showed no growth inhibition against Bacillus subtilis CMCC 63501 and Candida albicans ATCC 10231 at an amount of ca. 25 μg/paper disk. Due to unstable property of pretilactam (and insufficient amount of pretilactam), other biological activity assays of pretilatam were not conducted.
3. Discussion
Pretilactam is a novel 26-membered polyene macrolactam. Its discovery indicates that ATCC 31565, the famous ansamitocin producer, is a talent strain for various secondary metabolites.
It is rather challenging to determine the absolute configuration of many polyene macrolactams because of their unstable property and unassigned E/Z geometries of some C-C double bonds. We found that pretilactam was sensitive to light, air, and acidic pH, and changed slowly in organic solvents such as methanol, acetonitrile, and DMSO. In addition, pretilactam has three C-C double bonds unassigned of their E/Z geometries. It is, therefore, very difficult to determine the absolute configuration of chiral carbons in pretilactam by chemical analysis.
The plm is believed to be responsible for pretilactam biosynthesis because it is the only candidate gene cluster for polyene macrolactam biosynthesis by antiSMASH of the genomic DNA of ATCC 31565. Still, genetic manipulation of plm should be carried out to confirm its biological function experimentally. Besides, the biological activity of pretilactam needs to be further explored in the future.
4. Materials and Methods
4.1. Cultivation of ATCC 31565
Frozen stock spores of ATCC 31565 were inoculated onto ISP2 (glucose 4.0 g/L, malt extract 10.0 g/L, yeast extract 4.0 g/L, agar 15.0 g/L) plates and incubated at 28 °C for 5–7 days to develop mycelia with fresh spores. The fresh spores were collected and spread onto fermentation plates (dextrin 50.0 g/L, maltose 20.0 g/L, Pharmamedia 25.0 g/L, corn steep liquor 1.5 g/L, K2HPO4, 0.5 g/L, NaCl 2.0 g/L, agar 20.0 g/L, pH 7.0), then incubated at 28 °C for 8–10 days to produce mycelia lawns.
4.2. Genomic DNA Sequence Analysis
The genomic DNA of ATCC 31565 was extracted from mycelia using DNeasy blood and tissue kit from QIAGEN (Hilden, Germany). The DNA was sequenced on Illumina NGS and PacBio RSII with P4/C6 chemistry using the SMRTbell template prep kit v1.0 (Pacific Biosciences, CA, USA). Sequence data were processed and assembled using HGAP3, and sequence assembly was polished with the Quiver algorithm implemented in the PacBio SMRT analysis suite v2.3.0 (Pacific Biosciences, CA, USA).
4.3. LC-MS for Polyene Macrolactam
Fermentation culture of ATCC 31565 was extracted with an equal volume of MeOH or EtOAc. The MeOH or EtOAc extract (50 mL) was vacuum-dried then re-dissolved in 1.0 mL MeOH. An amount of 10 μL MeOH solution was used for LC-MS.
The above EtOAc extract of ATCC 31565 was mixed with ODS (12 nm, S-50 μm, AAG12S50, YMC) at a proportion of 1:1.5, then vacuum-dried at room temperature. The dried mixture was re-extracted with MeOH, and the MeOH solution was used for LC-MS.
A purified sample of pretilactam (1), dissolved in MeCN and stored at 4 °C for 24 h, was used for LC-MS.
LC-MS parameters: For LC analysis, an Agilent 1100 was used, with a Diamonsil C18 (4.6 × 250 mm, 5 μm); mobile phase CH3CN-H2O, 1.0 mL/min, 30–50% in 30 min; wavelength 300 nm; 25 °C. For MS analysis, an Agilent 6410 QQQ was used, with MS2 scan type; fragmentor 60 V; positive polarity.
4.4. Isolation of Polyene Macrolactam (Pretilactam, 1)
A large-scale cultivation of ATCC 31565 was performed to obtain enough material for isolation and characterization of the putative polyene macrolactam(s). The fermentation cultures (50 L; from ca. 1000 plates, 50 mL/plate) were pooled and extracted with EtOAc. The organic layer was collected and evaporated under reduced pressure, which gave 175 g of oily brown residue. The residue was loaded on an ODS column (12 nm, S-50 μm, AAG12S50, YMC; 460 × 49 mm), and fractionated by a step-wise gradient (20%, 30%, 40%, 50%, and 100%) of ethanol-H2O. Fraction 4 from 50% ethanol-H2O elution was evaporated under reduced pressure, which afforded 257.2 mg crude preparation for the putative polyene macrolactam(s). The crude preparation was subjected to preparative HPLC (YMC-Pack ODS-A, 250 × 20 mm, S-5 μm, 12 nm; 50–80% CH3CN-H2O, 25 min), which gave 11.2 mg refined preparation of 1 after evaporation. The refined preparation was subjected to semi-preparative HPLC (YMC-Pack ODS-A, 250 × 10 mm, S-5 μm, 12 nm; 78% CH3OH-H2O) for a final polishing, which yielded 4.1 mg pure preparation of 1 (with purity at 98% by HPLC analysis). The isolation process was conducted under dark, and dried samples in the purification process were stored in glass vials filled with N2 gas, as polyene macrolactams are sensitive to light and O2.
4.5. NMR Assay
1H and 13C-NMR spectra data were obtained at 800 and 200 MHz, respectively, on a BRUKER AVANCE III 800 spectrometer, and measured in DMSO-d6 at room temperature.
4.6. Assay of Pretilactam (1) Against Bacillus Subtilis CMCC 63501 and Candida Albicans ATCC 10231
A stock suspension (5–10 μL) of Candida albicans ATCC 10231 (or Bacillus cereus CMCC 63501) was inoculated into LB broth (100 mL in a 500 mL flask) for shaking at 37 °C overnight. The fresh culture of Candida albicans ATCC 10,231 (or Bacillus cereus CMCC 63501) was added into LB broth (with agar at 1.5%, 45 °C) at a proportion of 1.0% and mixed well, then poured into plates (12.0 mL for each plate, with a diameter of 8.5 cm; 106−7 CFU/mL). After solidification, the LB agar plates were ready for bioassay uses.
Filter paper disks (diameter 6.0 mm) loaded with ca. 6.25, 12.5, and 25 μg pretilactam (1) per paper disk were overlaid on the LB agar plates, then incubated at 37 °C for 12~15 h for microbial growth and inhibition zone formation.
Pretilactam (1): amorphous white powder; UV (MeOH) λmax (log ε) = 298 (4.54), 345 (3.22), 362 (3.09). IR (MeOH, cm−1): 3344.9, 2919.8, 2854.2, 1730.2, 1643.9, 1608.8, 1430.6, 1373.3, 1318.1, 1258.7, 1163.0, 1110.3, 1059.4, 898.0, 801.2, 662.9, 615.3. 1H-NMR and 13C-NMR data, Table 2; HRESIMS m/z 438.2646 [M + H]+ (calculated for C27H36O4N, 438.2639).
Acknowledgments
The NMR assays were performed at Nuclear Magnetic Resonance Center of School of Pharmaceutical Sciences, Capital Medical University, China.
Supplementary Materials
The supplementary materials are available online.
Author Contributions
L.W. conceived the study. J.W. and G.S. performed culture and fermentation; J.W., H.L., and L.Y. performed DNA extraction and genome assembly; J.W., L.B., and L.W. performed genome analysis; J.W., X.H., L.L. and S.L. performed chemical analysis and compound isolation; X.H. and B.J. performed NMR structure determination. J.W. performed biological assay; J.W., B.J., and L.W. wrote the manuscript.
Funding
This work was supported by Drug Innovation Major Project (2018ZX09711-001), CAMS Initiative for Innovative Medicine (CAMS-I2M-3-012), and National Infrastructure of Microbial Resources (No. NIMR-2018-3).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Nikolouli K., Mossialos D. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol. Lett. 2012;34:W1393–W1403. doi: 10.1007/s10529-012-0919-2. [DOI] [PubMed] [Google Scholar]
- 2.Amoutzias G.D., Chaliotis A., Mossialos D. Discovery strategies of bioactive compounds synthesized by nonribosomal peptide synthetases and type-I polyketide synthases derived from marine microbiomes. Mar. Drugs. 2016;14:80. doi: 10.3390/md14040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber T., Blin K., Duddela S., Krug D., Kim HU., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conway K.R., Boddy C.N. ClusterMine360: A database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013;41:D402–D407. doi: 10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caboche S., Pupin M., Leclere V., Fontaine A., Jacques P., Kucherov G. NORINE: A database of nonribosomal peptides. Nucleic Acids Res. 2008;36:D326–D331. doi: 10.1093/nar/gkm792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulze C.J., Donia M.S., Siqueira-Neto J.L., Ray D., Raskatov J.A., Green R.E., McKerrow J.H., Fischbach M.A., Linington R.G. Genome-directed lead discovery: Biosynthesis, structure elucidation, and biological evaluation of two families of polyene macrolactams against Trypanosoma brucei. Acs Chem. Biol. 2015;10:2373–23781. doi: 10.1021/acschembio.5b00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawahara T., Fujiwara T., Kagaya N., Shin-Ya K. JBIR-150, a novel 20-membered polyene macrolactam from marine-derived Streptomyces sp. OPMA00071. J. Antibiot. (Tokyo) 2018;71:390–392. doi: 10.1038/s41429-017-0010-2. [DOI] [PubMed] [Google Scholar]
- 8.Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W., Jensen P.R., Moore B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim Y.H., Wong F.T., Yeo W.L., Ching K.C., Lim Y.W., Heng E., Chen S., Tsai D.J., Lauderdale T.L., Shia K.S., et al. Auroramycin: A potent antibiotic from Streptomyces roseosporus by CRISPR-Cas9 activation. Chembiochemistry. 2018;19:1716–1719. doi: 10.1002/cbic.201800266. [DOI] [PubMed] [Google Scholar]
- 10.Skellam E.J., Stewart A.K., Strangman W.K., Wright J.L. Identification of micromonolactam, a new polyene macrocyclic lactam from two marine Micromonospora strains using chemical and molecular methods: Clarification of the biosynthetic pathway from a glutamate starter unit. J. Antibiot. (Tokyo) 2013;66:431–441. doi: 10.1038/ja.2013.34. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino S., Okada M., Awakawa T., Asamizu S., Onaka H., Abe I. Mycolic acid containing bacterium stimulates tandem cyclization of polyene macrolactam in a lake sediment derived rare actinomycete. Org. Lett. 2017;19:4992–4995. doi: 10.1021/acs.orglett.7b02508. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino S., Ozeki M., Wong C.P., Zhang H., Hayashi F., Awakawa T., Morita H., Onaka H., Abe I. Mirilactams C-E, novel polycyclic macrolactams isolated from combined-culture of Actinosynnema mirum NBRC 14064 and mycolic acid-containing bacterium. Chem. Pharm. Bull. (Tokyo) 2018;66:660–667. doi: 10.1248/cpb.c18-00143. [DOI] [PubMed] [Google Scholar]
- 13.Aigle B., Lautru S., Spiteller D., Dickschat J.S., Challis G.L., Leblond P., Pernodet J.L. Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 2014;41:251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- 14.Tanida S., Izawa M., Hasegawa T. Ansamitocin analogs from a mutant strain of Nocardia. I. Isolation of the mutant, fermentation and antimicrobial properties. J. Antibiot. (Tokyo) 1981;34:489–495. doi: 10.7164/antibiotics.34.489. [DOI] [PubMed] [Google Scholar]
- 15.Hatano K., Higashide E., Akiyama S.I., Yoneda M. Selective accumulation of ansamitocins P-2, P-3 and P-4, and biosynthetic origins of their acyl moieties. Agric. Biol. Chem. 1984;48:1721–1729. doi: 10.1080/00021369.1984.10866408. [DOI] [Google Scholar]
- 16.Du Z.Q., Zhong J.J. Rational approach to improve ansamitocin P-3 production by integrating pathway engineering and substrate feeding in Actinosynnema pretiosum. Biotechnol. Bioeng. 2018;115:2456–2466. doi: 10.1002/bit.26775. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Zhao P., Kang Q., Ma J., Bai L., Deng Z. Dual carbamoylations on the polyketide and glycosyl moiety by asm21 result in extended ansamitocin biosynthesis. Chem. Biol. 2011;18:1571–1580. doi: 10.1016/j.chembiol.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Hida T., Muroi M., Tanida S., Harada S. Structures of dnacin A1 and B1, new naphthyridinomycin-type antitumor antibiotics. J. Antibiot. (Tokyo) 1994;47:917–921. doi: 10.7164/antibiotics.47.917. [DOI] [PubMed] [Google Scholar]
- 19.Tanida S., Hasegawa T., Muroi M., Higashide E. Dnacins, new antibiotics. I. Producing organism, fermentation, and antimicrobial activities. J. Antibiot. (Tokyo) 1980;33:1443–1448. doi: 10.7164/antibiotics.33.1443. [DOI] [PubMed] [Google Scholar]
- 20.Lu C., Xie F., Shan C., Shen Y. Two novel cyclic hexapeptides from the genetically engineered Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2017;101:2273–2279. doi: 10.1007/s00253-016-8017-3. [DOI] [PubMed] [Google Scholar]
- 21.Keatinge-Clay A.T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Kudo F., Kawamura K., Uchino A., Miyanaga A., Numakura M., Takayanagi R., Eguchi T. Genome mining of the hitachimycin biosynthetic gene cluster: Involvement of a phenylalanine-2,3-aminomutase in biosynthesis. Chembiochemistry. 2015;16:909–914. doi: 10.1002/cbic.201500040. [DOI] [PubMed] [Google Scholar]
- 23.Low Z.J., Pang L.M., Ding Y., Cheang Q.W., Le Mai Hoang K., Thi Tran H., Li J., Liu X.W., Kanagasundaram Y., Yang L., et al. Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci Rep. 2018;8:1594. doi: 10.1038/s41598-018-20018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y., Wang M., Wu C., Tan Y., Li J., Hao X., Duan Y., Guan Y., Shang X., Wang Y., et al. Identification and proposed relative and absolute configurations of niphimycins C-E from the marine-derived Streptomyces sp. IMB7-145 by genomic analysis. J. Nat. Prod. 2018;81:178–187. doi: 10.1021/acs.jnatprod.7b00859. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Chen X., Li Z., Xu W., Tao W., Wu J., Yang J., Deng Z., Sun Y. Functional analysis of cytochrome P450s involved in streptovaricin biosynthesis and generation of anti-MRSA analogues. ACS Chem. Biol. 2017;12:2589–2597. doi: 10.1021/acschembio.7b00467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.