Abstract
Background
It is important to identify biomarkers associated with BRCA mutation in women with early breast cancer (BC) to improve early identification of mutation carriers. Thus, in this study, we examined the protein expression of claudin (CLDN) 3, CLDN4, CLDN7, and E-cadherin. Moreover, we analyzed additional histopathological variables and their associations in familial BC.
Methods
Immunohistochemical analysis for CLDNs and E-cadherin was performed on 237 BC cases of three different subsets of BC tumors: 62 from BRCA1 mutation carriers, 59 from BRCA2 mutation carriers, and 116 tumors from patients with BRCA wild type (WT) as controls. Histopathological data were also analyzed in the different subgroups. Logistic regression and receiver operation characteristic (ROC) curve were conducted to investigate factors associated with BRCA tumors.
Results
Expression of CLDN3 positively correlated with BRCA-mutated BC. CLDN3 was expressed in 58% of BRCA1-mutated tumors compared to only 7% in BRCA2-mutated tumors (p < 0.001) and 1% in WT tumors (p < 0.001). CK5 and CK14 expression were also more likely to arise in BRCA1 tumors (44 and 16%, respectively) than in the control group (8 and 4%) (p < 0.001, p = 0.012, respectively). We also found a significantly higher proportion of CK5+ among BRCA1 tumors (44%) in comparison with BRCA2-related BC (8%) (p < 0.001). In addition, there was a significant difference between both groups regarding CK14: positive expression in 16 and 5%, respectively (p = 0.030). CK5 and CK14 did not differ between the BRCA2 group and the WT tumors significantly. In a multivariate regression model, expression of CK5 (Odds ratio (OR): 6.46; 95% confidence interval (CI): 1.52–27.43; p = 0.011), and CLDN3 (OR: 200.48; 95% CI: 21.52–1867.61; p < 0.001) were associated with BRCA1 mutation status.
Conclusions
Our data suggests that CLDN3, CK5, and CK14 in combination with ER, PR and HER2 are associated with BRCA1 mutation status.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5908-6) contains supplementary material, which is available to authorized users.
Keywords: Familial breast cancer, BRCA1, BRCA2, Tissue microarray, Immunohistochemistry, Claudin
Background
Breast cancer (BC) is the leading cancer type among women in the world [1]. Familial BC, representing 5–7% of all BC, are hereditary and are associated with inherited gene mutations [2]. Approximately 25% of familial BC are due to germline mutations in the BRCA1 and BRCA2 genes, which are located on chromosome 17 and 13, respectively [2–4]. The average cumulative BC risk in BRCA1 mutation carriers by age 70 is 57–65%, whereas the cumulative BC risk in patients with BRCA2 mutation is 45–49% [5, 6].
BRCA1-associated tumors show a more aggressive phenotype, the majority of these tumors are invasive ductal adenocarcionomas (74%) and are poorly differentiated (high histological grade) [7–13]. More than 75% of BRCA1-mutated tumors are triple-negative, have a basal-like phenotype, or both [2, 7, 10, 14–20]. Triple-negative BC is characterized by lack of expression of hormone receptors (ie. estrogen receptor (ER) and progesterone receptor (PR)), and human epidermal growth factor receptor 2 (HER2) [21]. Basal-like BC, a subtype of triple-negative BC, can be characterized by the expression of basal cytokeratins (CK) (such as CK5/6, CK14) and epidermal growth factor receptor (EGFR), among others [14, 16, 22–28].
Claudin-low is another subtype of triple-negative BC, and can be characterized by low expression of claudin (CLDN) 3, CLDN4, CLDN7, and E-cadherin. The majority of claudin-low tumors have a poor prognosis [29–31]. CLDNs are structural and functional components of tight junctions which provide cell-cell adhesion in epithelial to endothelial cells [32]. There are at least 24 different CLDNs existing in humans, the expression of each seems to be tissue specific [33]. E-cadherin is one of the most important molecules in cell-cell adhesion in epithelial tissues [34]. Loss of intercellular adhesion by E-cadherin correlates with increased invasiveness and metastasis of tumors [34–39].
On the contrary, BRCA2-mutated tumors are more heterogeneous. The immunophenotype of BRCA2-associated tumors is very similar to sporadic BC. They are frequently characterized by low/intermediate histological grade. They often show no or low expression of HER2 and are often positive for ER and PR than in BRCA1-related tumors. Furthermore, BRCA2-mutated tumors do not express CK5, CK6 and CK14 [2, 7, 10, 13, 17, 40–44].
In this study, we analyzed clinicohistopathological features which are already associated with BRCA1/2 tumors (ER, PR, HER2, CK5 and 14, EGFR, among others). In addition, we selected three important CLDNs in BC, ie. CLDN3/4/7, and E-cadherin, which are used for characterization of the claudin-low subtype. We aim to define the expression profiles of these biomarkers in BRCA1 BC and compare these with BRCA2 and BRCA WT patients to improve early identification of mutation carriers.
We presented the study as an abstract at the 15th St. Gallen International Breast Cancer Conference in Vienna, Austria. [Danzinger S et al.: Intratumoral Cytokeratin 5 and Claudin 3 protein expression predicts for the presence of BRCA1 germline mutation in women with early breast cancer. The Breast 2017, 32 (Suppl 1):S22–77.]
Methods
Study population
A total of 242 BC tissue microarrays (TMA) were obtained from the Kathleen Cuningham Foundation Consortium for research into Familial Breast cancer (kConFab) [http://www.kconfab.org]. We evaluated one case per patient, and one core per case. The core diameter was 0.6 mm. Three BC cases were excluded due to TP53 (n = 1) and PALP2 (n = 2) mutation status. We also eliminated two cases of ductal carcinoma in situ. Our analysis was therefore based on 237 BC tumors where 62 tumors originated from BRCA1 carriers, 59 tumors were from BRCA2 carriers, and we obtained the remaining 116 from BRCA WT patients. The BRCA WT subgroup served as controls in our study. The control group consists of tumors from non BRCA1/2 mutation carriers. We used these tumors from consecutive BC patients with familial history. BRCA testing and analysis are described in the Supplemental (Additional file 1). Clinicopathological information collected included age at diagnosis, tumor size, tumor morphology, tumor grade, ER, PR, HER2, CK5 and 14, and EGFR.
Immunohistochemical analysis for ER, PR, HER2, CK5 and 14, and EGFR.
Immunohistochemical staining of the samples was performed as described in our previous study [45]. The following antibodies were used: clone SP1 against ER (prediluted, 790–4296, Ventana Medical Systems Inc., Tucson, AZ, USA), clone 1E2 against PR (790–4325, Ventana Medical Systems Inc.), clone 4B5 against HER2 (prediluted, 800–2996, Ventana Medical Systems Inc.), clone EP1601Y against CK5 (305R-16, Cell Marque Corporation, Rocklin, CA, USA), clone LL002 against CK14 (LL002-L-CE, Leica, Novocastra, Newcastle upon Tyne, United Kingdom), and clone 31G7 against EGFR (28–0005, Zymed, South San Francisco, CA, USA).
ER and PR were considered positive if there were ≥ 1% tumor nuclei stained according to the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) Guideline [46]. HER2-positivity was defined by staining of > 10% of tumor cells as proposed by the update of the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) clinical practice guideline. Staining of HER2 should also be strong and circumferentially membranous [47]. CK5, CK14, and EGFR were regarded as positive if any cytoplasmic and/or membranous staining was seen in the tumor cells [33].
Immunohistochemical analysis for CLDN3/4/7 and E-cadherin.
Immunohistochemical staining for CLDN3, CLDN4, CLDN7, and E-cadherin was performed according to the protocol of our previous study [45]. For immunohistochemistry of paraffin-embedded sections (5 μm), we used the ultraView Universal DAB Detection Kit (5269806001, 760–500, Ventana Medical Systems Inc., Tucson, AZ, USA), and an automated immunostainer system (Ventana Benchmark, Ventana Medical Systems Inc.). Tissues were processed with high-temperature technique for 30 min (CLDN3 and CLDN7) or 60 min (CLDN4, E-cadherin) and incubated with antibodies.
We used the following antibodies for staining the tissue sections: Rabbbit anti-Claudin-3 against CLDN3 (32 min; 34–1700, Invitrogen, Thermo Fisher Scientific, Rockford, IL, USA), Mouse anti-Claudin-4 (monoclonal) against CLDN4 (64 min; 32–9400, Invitrogen), Rabbit anti-Claudin-7 against CLDN7 (32 min; 34–9100, Invitrogen), and anti-E-Cadherin (36) Mouse Monoclonal against E-cadherin (16 min; 790–4497, Ventana Medical Systems Inc.). The antibody concentrations for CLDN3, CLDN4, and CLDN7 were in the range of 2–3 μg/ml, the antibody concentration for E-cadherin was 0.314 μg/ml.
We used hydrogen peroxide and 3,3’-diaminobenzidine-tetrahydrochloride for visualization of the reaction. The slides were counterstained with haemalaun and exposed to a bluing reagent for different times. We used colon (CLDN3, E-cadherin) and breast tissue (CLDN7) as positive controls. CLDN4 was overexpressed in ovarian cancer.
Immunohistochemical staining was centrally reviewed by an experienced pathologist (MR) to ensure comparable results. The stained sections were visualized on an Olympus BX50 microscope (Olympus Corporation, Tokyo, Japan). The imaging software cell^P (Olympus Corporation) was used for taking pictures of the slides. The tumor samples were evaluated by differentiation between positive and negative staining. Only membranous staining was classified as positive. There is no standard for assessing the CLDN expression [48]. Positive expression of CLDN3, CLDN4, and CLDN7 was defined by any detectable staining in the membrane of the tumor cell. E-cadherin was regarded positive if any staining was observed. Thus, complete absence of any membranous E-cadherin immunoreactivity was considered as E-cadherin-negative [49].
Statistical methods
Descriptive statistics were performed to determine the characteristics of our study sample, which comprise three groups: BRCA1, BRCA2, and BRCA WT. ANOVA test and student’s t-test were used to compare the mean age at diagnosis between two groups. Chi-square and Fisher’s Exact (for smaller sample size) tests were used to compare the proportions of clinicohistopathological (categorical) parameters. To further determine the relationship between clinicohistopathological parameters with mutation status, Spearman’s correlation analysis was performed. Logistic regression was conducted to identify independent factors associated with BRCA1 (and BRCA2) mutation status. Associations were summarized using the odds ratio (OR) and corresponding 95% confidence interval (CI) derived from the model estimates. The receiver operation characteristic (ROC) curves were constructed for the prediction of BRCA1 mutation status. The predictive ability of each model was summarized by the area under the curve (AUC), and optimal models were classified as those that yielded the highest AUC in the ROC analysis. We excluded all unknowns/undetermined values from analysis. Statistical significance was considered as p < 0.05 (2-tailed). We performed all statistical analyses using SPSS (v. 23.0 (SPSS Inc., Chicago, IL, USA)).
Results
The characteristics of the patients are summarized in Table 1. Immunohistochemical staining of CLDN3, CLDN4, and CLDN7, and E-cadherin is shown in Fig. 1 and Fig. 2. The tumor size of most of the tumors in all of the three groups (45% of BRCA1, 53% of BRCA2, and 63% of WT tumors) was < 2 cm. The histological type is dominated by invasive ductal carcinoma in all groups (72–90%). Grade 3 was the most common tumor grade in BRCA1 tumors (65%), followed by BRCA2 (44%) tumors, compared to only 27% of BRCA WT tumors (p < 0.001, p = 0.006, respectively).
Table 1.
Characteristics of tumors (WT = BRCA wild type breast cancer, BRCA1, BRCA2 = breast cancer in BRCA1/BRCA2 mutation carriers, ER = estrogen receptor, PR = progesterone receptor, HER2 = human epidermal growth factor receptor 2, CK = cytokeratin, EGFR = epidermal growth factor receptor, CLDN = claudin)
| Characteristics | WT (N = 116) | BRCA1 (N = 62) | BRCA2 (N = 59) | WT v BRCA1 | p-value (Chi = square or Fisher’s test (if cells < 5))* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Col % | N | Col % | N | Col % | WT v BRCA2 | BRCA1 v BRCA2 | WT v BRCA1/2 | |||
| Age at diagnosis | <50y | 79 | 68 | 47 | 76 | 38 | 64 | 0.282 | 0.623 | 0.170 | 0.721 |
| > = 50y | 37 | 32 | 15 | 24 | 21 | 36 | |||||
| Tumor grade | 1 | 29 | 25 | 1 | 2 | 4 | 7 | <.001 | 0.006 | 0.021 | <.001 |
| 2 | 37 | 32 | 12 | 19 | 22 | 37 | |||||
| 3 | 31 | 27 | 40 | 65 | 26 | 44 | |||||
| Unknown | 19 | 16 | 9 | 15 | 9 | 15 | |||||
| Tumor size | < 2 cm | 73 | 63 | 28 | 45 | 31 | 53 | 0.152 | 0.272 | 0.411 | 0.147 |
| 2-5 cm | 35 | 30 | 26 | 42 | 20 | 34 | |||||
| > 5 cm | 2 | 2 | 1 | 2 | 3 | 5 | |||||
| Unknown | 6 | 5 | 7 | 11 | 7 | 12 | |||||
| Tumor morphology | Invasive ductal carcinoma | 83 | 72 | 56 | 90 | 47 | 80 | 0.027 | 0.327 | 0.072 | 0.068 |
| Invasive lobular carcinoma | 8 | 7 | 0 | 0 | 5 | 8 | |||||
| Invasive ductal+lobular carcinoma | 6 | 5 | 0 | 0 | 2 | 3 | |||||
| Carcinoma – undefined | 4 | 3 | 2 | 3 | 3 | 5 | |||||
| Other | 15 | 13 | 4 | 6 | 2 | 3 | |||||
| ER | Negative | 28 | 24 | 47 | 76 | 13 | 22 | <.001 | 0.448 | <.001 | <.001 |
| Positive | 78 | 67 | 14 | 23 | 45 | 76 | |||||
| Unknown | 10 | 9 | 1 | 2 | 3 | 5 | |||||
| PR | Negative | 42 | 36 | 49 | 79 | 23 | 39 | <.001 | 0.89 | <.001 | 0.001 |
| Positive | 68 | 59 | 11 | 18 | 34 | 58 | |||||
| Unknown | 6 | 5 | 2 | 3 | 4 | 7 | |||||
| HER2 | Negative | 77 | 66 | 43 | 69 | 22 | 37 | 0.887 | <.001 | <.001 | 0.003 |
| Positive | 22 | 19 | 13 | 21 | 32 | 54 | |||||
| Unknown | 17 | 15 | 6 | 10 | 7 | 12 | |||||
| CK5 | Negative | 78 | 67 | 28 | 45 | 44 | 75 | <.001 | 0.702 | <.001 | 0.001 |
| Positive | 9 | 8 | 27 | 44 | 5 | 8 | |||||
| Unknown | 29 | 25 | 7 | 11 | 12 | 20 | |||||
| CK14 | Negative | 89 | 77 | 45 | 73 | 49 | 83 | 0.012 | 0.999 | 0.030 | 0.129 |
| Positive | 5 | 4 | 10 | 16 | 3 | 5 | |||||
| Unknown | 22 | 19 | 7 | 11 | 9 | 15 | |||||
| EGFR | Negative | 76 | 66 | 52 | 84 | 52 | 88 | 0.585 | 0.052 | 0.209 | 0.122 |
| Positive | 10 | 9 | 5 | 8 | 1 | 2 | |||||
| Unknown | 30 | 26 | 5 | 8 | 8 | 14 | |||||
| CLDN3 | Negative | 107 | 92 | 24 | 39 | 50 | 85 | <.001 | 0.039 | <.001 | <.001 |
| Positive | 1 | 1 | 36 | 58 | 4 | 7 | |||||
| Unknown | 8 | 7 | 2 | 3 | 7 | 12 | |||||
| CLDN4 | Negative | 6 | 5 | 6 | 10 | 0 | 0 | 0.423 | 0.094 | 0.999 | 0.059 |
| Positive | 100 | 86 | 56 | 90 | 58 | 98 | |||||
| Unknown | 10 | 9 | 5 | 8 | 3 | 5 | |||||
| CLDN7 | Negative | 109 | 94 | 62 | 100 | 57 | 97 | n/a | 0.339 | 0.475 | 0.999 |
| Positive | 0 | 0 | 0 | 0 | 1 | 2 | |||||
| Unknown | 7 | 6 | 0 | 0 | 3 | 5 | |||||
| E-cadherin | Negative | 10 | 9 | 9 | 15 | 16 | 27 | 0.296 | 0.002 | 0.081 | 0.016 |
| Positive | 89 | 77 | 48 | 77 | 40 | 68 | |||||
| Unknown | 17 | 15 | 5 | 8 | 5 | 8 | |||||
*unknown/undetermined values are excluded from analysis
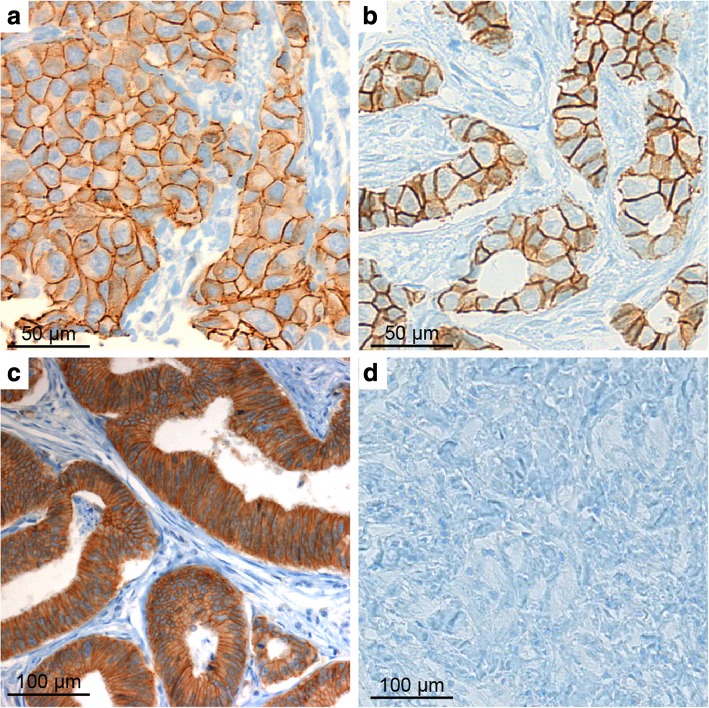
Fig. 1.
Claudin (CLDN) 3. Positive expression in BRCA1 breast cancer (BC) (a) and negative control (isotypic antibody) (b). Positive CLDN4 expression in BRCA1-mutated BC (c), negative control of CLDN4 (isotypic antibody) (d). Positive CLDN7 in BRCA2 BC (e), negative CLDN7 in BRCA1-related tumor (f), and negative control of CLDN7 (isotypic antibody) (g). Positive CK5 staining in BRCA1 BC (h)
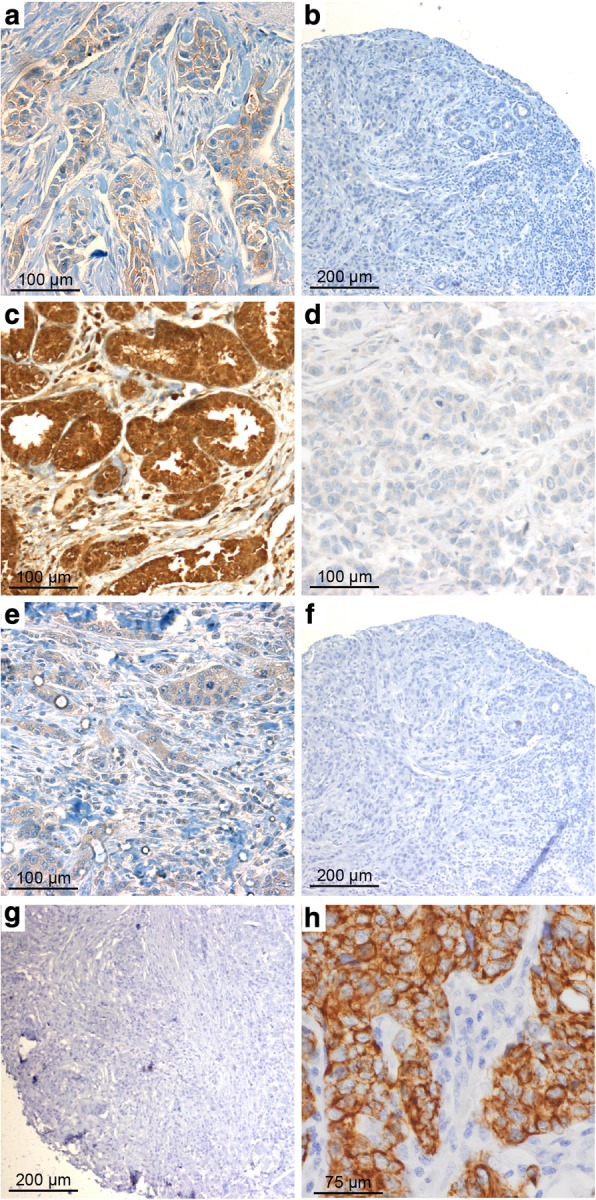
Fig. 2.
E-cadherin. Positive staining in BRCA1-mutated (a) and BRCA wild type breast cancer (b). Positive benign epithelium (colon) as positive control (c). Negative staining in the wild type group (d)
Negative expression of ER and PR was significantly more common among BRCA1 mutation carriers (76 and 79%, respetively) compared to the BRCA2 group (22 and 39%, respectively) (both p < 0.001) and in comparison with the WT subgroup (24 and 36%, respectively) (both p < 0.001). HER2 was positive in 54% of BRCA2 versus 19% of WT tumors (p < 0.001) and versus 21% of BRCA1-mutated tumors (p < 0.001). Positive HER2 was found to be associated with a BRCA2 mutation compared to the WT.
Tumors with expression of CK5 and CK14 were more likely to arise in BRCA1 (44 and 16%, respectively) than in the control group (8 and 4%) (p < 0.001, p = 0.012, respectively). We also found a significantly higher proportion of positive CK5 expression among BRCA1 tumors (44%) in comparison with BRCA2-related BC (8%) (p < 0.001). CK14 was positive in 16% of BRCA1 versus 5% of BRCA2 tumors (p = 0.030). Furthermore, EGFR negatively correlated with the BRCA2 mutation status in comparison with BRCA WT with statistical significance.
Thirty-six of 41 CLDN3-positive cases had a BRCA1 mutation, and 40 of 41 such cases have a mutation in either BRCA1 or BRCA2. CLDN3 was positively correlated with BRCA1-mutated BC. Positive CLDN3 was found in 58% of BRCA1-mutated tumors compared to only 7% of BRCA2-mutated tumors (p < 0.001) and 1% of WT tumors (p < 0.001). Positive CLDN3 was also significantly more frequent in the BRCA2 in comparison with the WT group (p = 0.039) (Table 1). A positive correlation was observed between CLDN3 with tumor grade and CK5. In contrast, CLDN3 negatively correlated with ER and PR.
E-cadherin expression was significantly different in proportion between tumors of BRCA WT and BRCA2 mutation (77% versus 68%, respectively, p = 0.002). With regard to CLDN4 and CLDN7, there were no significant differences observed among the groups. Positive CLDN4 was very common (86–98%), but CLDN7 was rarely positive (0–2%).
In an univariate analysis of clinicopathological factors associated with BRCA1-mutated BC versus the WT subtype, tumor grade, ER, PR, and expression of CK5, CK14, and CLDN3 were found to be independent parameters. However, when these features were put into a multivariate regression model and adjusted with age, only CK5+ (Odds ratio (OR): 6.46; 95% Confidence interval (CI): 1.52–27.43; p = 0.011), and CLDN3+ (OR: 200.48; 95% CI: 21.52–1867.61; p < 0.001) they revealed to be associated with BRCA1 mutation status (Table 2).
Table 2.
Logistic regression for BRCA1 versus WT and BRCA2 versus WT: univariate and multivariate analysis
| Characteristics | Univariate LR OR (95%CI) |
p-value | Multivariate LRa OR (95%CI) |
p-value | Univariate LR OR (95%CI) |
p-value | Multivariate LRa OR (95%CI) |
p-value | |
|---|---|---|---|---|---|---|---|---|---|
| BRCA1 vs WT | BRCA2 vs WT | ||||||||
| Age at diagnosis (years) | <50y | 1.0 | 1.0 | ||||||
| ≥50y | 0.68 (0.34–1.37) | 0.283 | 1.18 (0.61–2.28) | 0.624 | |||||
| Tumor grade | 1 + 2 | 1.0 | 1.0 | 1.0 | |||||
| 3 | 6.55 (3.07–13.97) | <.001 | 2.13 (1.06–4.29) | 0.034 | 2.41 (1.02–5.69) | 0.045 | |||
| Tumor size | < 2 cm | 1.0 | 1.0 | – | |||||
| > = 2 cm | 1.90 (.98–3.68) | 0.056 | 1.51 (0.77–2.96) | 0.227 | – | ||||
| ER | Negative | 1.0 | 1.0 | – | |||||
| Positive | .11 (.05–.22) | <.001 | 0.46 (0.09–2.66) | 0.400 | 1.35 (0.62–2.91) | 0.449 | – | ||
| PR | Negative | 1.0 | 1.0 | – | |||||
| Positive | 0.14 (0.07–0.30) | <.001 | 0.35 (0.07–1.87) | 0.354 | 0.96 (0.49–1.85) | 0.890 | – | ||
| HER2 | Negative | 1.0 | 1.0 | 1.0 | |||||
| Positive | 1.06 (.49–2.31) | 0.887 | 1.03 (0.24–4.48) | 0.970 | 5.33 (2.58–11.03) | <.001 | 5.21 (2.18–12.45) | <.001 | |
| CK5 | Negative | 1.0 | 1.0 | – | |||||
| Positive | 8.36 (3.50–19.93) | <.001 | 6.46 (1.52–27.43) | 0.011 | 0.79 (0.23–2.71) | 0.705 | – | ||
| CK14 | Negative | 1.0 | 1.0 | – | |||||
| Positive | 3.96 (1.28–12.27) | 0.017 | 0.73 (0.14–3.89) | 0.709 | – | ||||
| EGFR | Negative | 1.0 | 1.0 | ||||||
| Positive | 0.73 (.24–2.26) | 0.586 | 0.15 (0.02–1.20) | 0.074 | |||||
| CLDN3 | Negative | 1.0 | 1.0 | ||||||
| Positive | 160.50 (20.96–1229.06) | <.001 | 200.48 (21.52–1867.61) | <.001 | 8.92 (0.97–81.90) | 0.053 | |||
| CLDN4 | Negative | 1.0 | – | – | – | ||||
| Positive | 3.36 (0.39–28.62) | 0.267 | – | – | – | ||||
| CLDN7 | Negative | – | – | – | – | ||||
| Positive | – | – | – | – | |||||
| E-cadherin | Negative | 1.0 | – | 1.0 | 1.0 | ||||
| Positive | 0.60 (0.23–1.58) | 0.299 | – | 0.27 (0.11–0.64) | 0.003 | 0.33 (0.11–0.98) | 0.046 | ||
aadjusted for age at diagnosis
For BRCA2 BC versus WT, univariate analysis showed that tumor grade, the expression of HER2, and E-cadherin were independent markers of BRCA2 status. In a multivariate regression model, the expression of HER2 (OR: 5.21; 95% CI: 2.18–12.45; p < 0.001) showed an association with BRCA2 mutation status (Table 2).
For BRCA2 versus BRCA1 tumors, univariate analysis showed that tumor grade, ER, PR, HER2, CK5, CK14, and CLDN3 were independent parameters of BRCA2 status. However, in a multivariate model, all parameters were not significantly associated with the mutation status, except for CLDN3. Positive CLDN3 in BRCA2-related tumors has an OR of 0.05 (95% CI: 0.01–0.26; p < 0.001) with reference to BRCA1 tumors. Thus, negative CLDN3 was found in 85% of BRCA2-related tumors versus 39% of BRCA1 BC (p < 0.001).
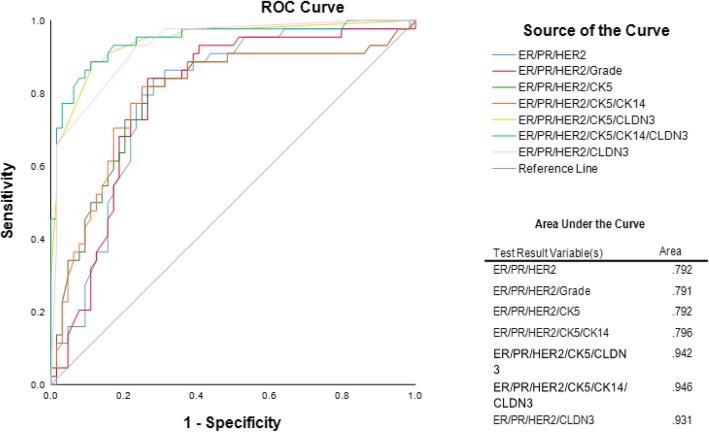
Our receiver operation characteristic (ROC) analysis showed that the base model that correlates with BRCA1 status, which consisted of only ER, PR and HER2, yielded an area under the curve (AUC) of 0.792. The addition of CLDN3 to this model resulted in an AUC of 0.931. When CK5 was added to this model, the model yielded an AUC of 0.942. Further addition of CK14 resulted in an AUC of 0.946 in the ROC curve which is shown in Fig. 3.
Fig. 3.
ROC curve for the discrimination between BRCA1-mutated tumors and BRCA WT tumors
Discussion
Taken together, results from our study showed that CLDN3, CK5, and CK14 are associated with BRCA1 mutation status when used in a model where ER, PR and HER2 status are already known. Our research shows that staining of CLDN3, CK5, and CK14 in combination with ER, PR and HER2 indicate an association with BRCA1 mutation status.
Our findings show that tumors with expression of CK5 and CK14 were more likely to arise in BRCA1 than in the control group. This has been reported previously by Lakhani et al. where they investigated immunohistochemical staining for basal markers. They found that CK5/6, CK14, and EGFR were more frequent in BRCA1 tumors compared to non-mutation BC (58% versus 7, 61% versus 12, 67% versus 21%, p < 0.0001 in each case, respectively) [42]. This supports that most of BRCA1-related cancers belong to the basal-like subtype [2]. Foulkes and colleagues showed that the expression of CK5/6 was statistically significantly associated with BRCA1-related BC [18, 50]. Furthermore, an association between positive CK5/6 and BRCA mutation status was shown by Murria Estal et al. [51] However, Mohanty et al. could not show a statistically significant difference of CK14 expression between BRCA-mutated and sporadic BC [52]. In addition, CK5/6, CK14, and E-cadherin were not associated with BRCA1 status in a study from Hassanein et al. [53] Eerola et al. reported that CK14 was significantly associated with BRCA1 tumors in univariate analysis [54]. In our study, the model which consisted of ER, PR, HER2, positive expression of CLDN3, CK5, and CK14 yielded an AUC of 0.946 in the ROC analysis to indicate an association with the BRCA1 mutation status. We also found a significantly higher proportion of CK5 expression in BRCA1 than BRCA2 BC. EGFR, an additional biomarker of the basal-like subtype, negatively correlated with the BRCA2 mutation status in comparison with BRCA WT in our study. Expression of CK5/6 and CK14 in BRCA2-related BC is rare and does not differ from sporadic tumors [7, 40–42, 55]. Results from our study showed similar findings. Otherwise, Eerola et al. showed a positive expression of CK5/6 and CK14 in 7.7 and 26.9%, respectively, in BRCA2 tumors. Only regarding CK14, there was a significant difference between these tumors and the sporadic ones [54].
CLDN3 and CLDN4 are commonly expressed in BC [32, 56, 57]. Madaras et al. examined the expression of CLDN3, 4, and 7, among other variables, in BRCA-mutated and BRCA WT tumor tissues. In BRCA-mutated tumors, CLDN3, 4, and 7 were expressed at higher level compared to BRCA WT tumors [58]. Higher overexpression rates for CLDN3, 4, and 7 were found in BRCA1-related BC compared to sporadic BC [48]. We showed that CLDN3 was positively correlated with BRCA1-mutated BC. Positive CLDN3 was also significantly more frequent in the BRCA2 in comparison with the WT group. However, in our study there was no association between CLDN4 or CLDN7 and BRCA-mutated BC found. Positive CLDN4 was very common, but CLDN7 was rarely positive in all groups (BRCA1, BRCA2, WT).
In the analysis of CIMBA (Consortium of Investigators of Modifiers of BRCA1/2), Mavaddat et al. showed that 78% of BRCA1-related tumors were ER-negative, but in patients with BRCA2 mutation status ER was negative only in 23% of the tumors. The authors could demonstrate that in the BRCA1 group, 79% were PR-negative and 90% HER2-negative compared to 36 and 87% in BC patients with BRCA2 mutation status, respectively. These results allow to conclude that ER-positive tumors occur more likely in BRCA2 than in BRCA1 cases [44]. Several studies could also demonstrate that negative ER and PR were significantly more common among BRCA1-mutated tumors compared to sporadic cases, among others [11–13, 20, 48, 53]. Therefore, these results are similar to our findings. A low prevalence of positive HER2 (6.8%) expression was shown among patients with BRCA2-related BC by Evans et al., too [59]. BRCA2 tumors often show no or low expression of HER2 [7, 13, 17, 40] However, Armes et al. showed a strong expression of HER2 in 44% (4 of 9) of BRCA2 tumors [60]. Accordingly in our study, positive HER2 was found to be associated with a BRCA2 mutation compared to the WT.
The limitations of this study are a small sample size resulting in a large OR as observed in the results for CLDN3, and should consequently be interpreted with caution. A larger sample size for future studies would be necessary to confirm these findings. Moreover, we did not analyze different molecular subtypes in this study. Future studies should therefore investigate the relationship between the molecular subtypes and the BRCA mutation status. Additionally, the problem of selection should be mentioned as a further limitation of our study. We selected patients with familial breast cancer based on the BRCA1/2 mutation status. Thus, the controls consist of predominantly luminal subtypes.
The dataset of this study should be considered as a training dataset, the analysis is hypothesis generating. This aspect is one of the limitations of our study which is exploratory. We do not have a sufficiently high number of independently collected samples to test the parameters in an independent dataset, too. Thus, further studies are necessary regarding this point.
The examination of TMA should be mentioned as a further limitation of this study. However, the staining was fairly homogenous across the tumor. There were no areas with stronger or weaker intensity observed. TMA is an acceptable and sufficient device, which has been developed and used in research settings [61–65].
In conclusion, findings from our study showed that CLDN3, CK5, and CK14 in combination with ER, PR and HER2 are associated with BRCA1 mutation status.
Conclusions
Our data suggests that CLDN3, CK5, and CK14 in combination with ER, PR and HER2 indicate an association with BRCA1 mutation status.
Additional files
BRCA testing and analysis (DOCX 16 kb)
Data (XLSX 40 kb)
Data dictionary (DOCX 21 kb)
Acknowledgements
This work was supported by the Austrian Society of Endogrinological Oncology (Österreichische Gesellschaft für endokrinologische Onkologie, OEGEO). We want to thank the society for supporting this project.
We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
Abbreviations
- AUC
Area under the curve
- BC
Breast cancer
- CI
Confidence interval
- CK14
Cytokeratin 14
- CK5
Cytokeratin 5
- CLDN
Claudin
- EGFR
Epidermal growth factor receptor
- ER
Estrogen receptor
- HER2
Human epidermal growth factor receptor 2
- OR
Odds ratio
- PR
Progesterone receptor
- ROC
Receiver operation characteristic
- TMA
Tissue microarray
- WT
Wild type
Authors’ contributions
SD, DM, YYT, and CFS contributed to study conception and design. SD and CFS contributed to provision of study materials and patients. SD, YYT, and CFS contributed to collection and assembly of data. MK contributed to immunohistochemical staining of the tissue sections. SD, YYT, MR, SW, and CFS contributed to data analysis and interpretation. SD, YYT, and CFS contributed to writing the manuscript. All authors contributed to the review and approval of the manuscript.
Funding
There is no funding of the study.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
[Additional supporting files: “Data” (Additional file 2), “Data dictionary” (Additional file 3)]
Ethics approval and consent to participate
The study has been approved by the ethics committee of the Medical University of Vienna.
All procedures performed in our study were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Written informed consent was obtained from all individual participants included in the study.
Consent for publication
We have obtained written consent to publish from the participants.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sabine Danzinger, Phone: +43 (0)1 40400 28010, Email: sabine.danzinger@meduniwien.ac.at.
Yen Yen Tan, Email: yen.tan@meduniwien.ac.at.
Margaretha Rudas, Email: margaretha.rudas@meduniwien.ac.at.
Marie-Theres Kastner, Email: marie-theres.kastner@meduniwien.ac.at.
Sigrid Weingartshofer, Email: sigrid.weingartshofer@meduniwien.ac.at.
Daniela Muhr, Email: daniela.muhr@meduniwien.ac.at.
Christian F. Singer, Email: christian.singer@meduniwien.ac.at
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Melchor L, Benitez J. The complex genetic landscape of familial breast cancer. Hum Genet. 2013;132(8):845–863. doi: 10.1007/s00439-013-1299-y. [DOI] [PubMed] [Google Scholar]
- 3.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 4.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Groep P, van der Wall E, van Diest PJ. Pathology of hereditary breast cancer. Cell Oncol (Dordr) 2011;34(2):71–88. doi: 10.1007/s13402-011-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stratton Michael R. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. The Lancet. 1997;349(9064):1505–1510. [PubMed] [Google Scholar]
- 9.Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, Farid LM, Venter D, Antoniou A, Storfer-Isser A, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90(15):1138–1145. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 10.Honrado E, Benitez J, Palacios J. The molecular pathology of hereditary breast cancer: genetic testing and therapeutic implications. Mod Pathol. 2005;18(10):1305–1320. doi: 10.1038/modpathol.3800453. [DOI] [PubMed] [Google Scholar]
- 11.Eerola H, Heikkila P, Tamminen A, Aittomaki K, Blomqvist C, Nevanlinna H. Histopathological features of breast tumours in BRCA1, BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2005;7(1):R93–100. doi: 10.1186/bcr953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappuis PO, Nethercot V, Foulkes WD. Clinico-pathological characteristics of BRCA1- and BRCA2-related breast cancer. Semin Surg Oncol. 2000;18(4):287–295. doi: 10.1002/(sici)1098-2388(200006)18:4<287::aid-ssu3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, et al. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin Cancer Res. 2003;9(10 Pt 1):3606–3614. [PubMed] [Google Scholar]
- 14.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26(15):2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 15.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 16.Lachapelle J, Foulkes WD. Triple-negative and basal-like breast cancer: implications for oncologists. Curr Oncol. 2011;18(4):161–164. doi: 10.3747/co.v18i4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Moreno-Bueno G, Hardisson D, Calero F, Benitez J, Palacios J. Vimentin and laminin expression is associated with basal-like phenotype in both sporadic and BRCA1-associated breast carcinomas. J Clin Pathol. 2007;60(9):1006–1012. doi: 10.1136/jcp.2006.042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triantafyllidou O, Vlachos IS, Apostolou P, Konstantopoulou I, Grivas A, Panopoulos C, Dimitrakakis C, Kassanos D, Loghis C, Bramis I, et al. Epidemiological and clinicopathological characteristics of BRCA-positive and BRCA-negative breast cancer patients in Greece. J BUON. 2015;20(4):978–984. [PubMed] [Google Scholar]
- 21.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 23.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14(5):1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina breast Cancer study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 27.Tang P, Skinner KA, Hicks DG. Molecular classification of breast carcinomas by immunohistochemical analysis: are we ready? Diagn Mol Pathol. 2009;18(3):125–132. doi: 10.1097/PDM.0b013e31818d107b. [DOI] [PubMed] [Google Scholar]
- 28.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24(2):157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 29.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 32.Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human malignancy: a clinical perspective. Cancer Manag Res. 2013;5:367–375. doi: 10.2147/CMAR.S38294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, Singh K, Mangray S, Tavares R, Noble L, Resnick MB, Yakirevich E. Claudin expression in high-grade invasive ductal carcinoma of the breast: correlation with the molecular subtype. Mod Pathol. 2013;26(4):485–495. doi: 10.1038/modpathol.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65(23):3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strumane K, Berx G, Van Roy F. Cadherins in cancer. Handb Exp Pharmacol. 2004;165:69–103. doi: 10.1007/978-3-540-68170-0_4. [DOI] [PubMed] [Google Scholar]
- 36.David JM, Rajasekaran AK. Dishonorable discharge: the oncogenic roles of cleaved E-cadherin fragments. Cancer Res. 2012;72(12):2917–2923. doi: 10.1158/0008-5472.CAN-11-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113(1):173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66(1):107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 39.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392(6672):190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 40.Bane AL, Beck JC, Bleiweiss I, Buys SS, Catalano E, Daly MB, Giles G, Godwin AK, Hibshoosh H, Hopper JL, et al. BRCA2 mutation-associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am J Surg Pathol. 2007;31(1):121–128. doi: 10.1097/01.pas.0000213351.49767.0f. [DOI] [PubMed] [Google Scholar]
- 41.Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, et al. Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat. 2005;90(1):5–14. doi: 10.1007/s10549-004-1536-0. [DOI] [PubMed] [Google Scholar]
- 42.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11(14):5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 43.Larsen MJ, Thomassen M, Gerdes AM, Kruse TA. Hereditary breast cancer: clinical, pathological and molecular characteristics. Breast Cancer (Auckl) 2014;8:145–155. doi: 10.4137/BCBCR.S18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Spurdle A, Robson M, Sherman M, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the consortium of investigators of modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomark Prev. 2012;21(1):134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danzinger S, Tan YY, Rudas M, Kastner MT, Weingartshofer S, Muhr D, Singer CF. kConFab I: differential Claudin 3 and EGFR expression predicts BRCA1 mutation in triple-negative breast Cancer. Cancer Investig. 2018;36(7):378–388. doi: 10.1080/07357907.2018.1499934. [DOI] [PubMed] [Google Scholar]
- 46.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heerma van Voss MR, van Diest PJ, Smolders YH, Bart J, van der Wall E, van der Groep P. Distinct claudin expression characterizes BRCA1-related breast cancer. Histopathology. 2014;65(6):814–827. doi: 10.1111/his.12490. [DOI] [PubMed] [Google Scholar]
- 49.Christgen M, Noskowicz M, Schipper E, Christgen H, Heil C, Krech T, Langer F, Kreipe H, Lehmann U. Oncogenic PIK3CA mutations in lobular breast cancer progression. Genes Chromosomes Cancer. 2013;52(1):69–80. doi: 10.1002/gcc.22007. [DOI] [PubMed] [Google Scholar]
- 50.Palacios J, Honrado E, Osorio A, Diez O, Rivas C, Benitez J. Re: Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2004;96(9):712–714. doi: 10.1093/jnci/djh115. [DOI] [PubMed] [Google Scholar]
- 51.Murria Estal R, Palanca Suela S, de Juan JI, Alenda Gonzalez C, Egoavil Rojas C, Garcia-Casado Z, Lopez Guerrero JA, Juan Fita MJ, Sanchez Heras AB, Segura Huerta A, et al. Relationship of immunohistochemistry, copy number aberrations and epigenetic disorders with BRCAness pattern in hereditary and sporadic breast cancer. Familial Cancer. 2016;15(2):193–200. doi: 10.1007/s10689-015-9864-2. [DOI] [PubMed] [Google Scholar]
- 52.Mohanty SK, Lai JP, Gordon OK, Pradhan D, Bose S, Dadmanesh F. BRCA-mutated invasive breast carcinomas: Immunohistochemical analysis of insulin-like growth factor II mRNA-binding protein (IMP3), cytokeratin 8/18, and cytokeratin 14. Breast J. 2015;21(6):596–603. doi: 10.1111/tbj.12494. [DOI] [PubMed] [Google Scholar]
- 53.Hassanein M, Huiart L, Bourdon V, Rabayrol L, Geneix J, Nogues C, Peyrat JP, Gesta P, Meynard P, Dreyfus H, et al. Prediction of BRCA1 germ-line mutation status in patients with breast cancer using histoprognosis grade, MS110, Lys27H3, vimentin, and KI67. Pathobiology. 2013;80(5):219–227. doi: 10.1159/000339432. [DOI] [PubMed] [Google Scholar]
- 54.Eerola H, Heinonen M, Heikkila P, Kilpivaara O, Tamminen A, Aittomaki K, Blomqvist C, Ristimaki A, Nevanlinna H. Basal cytokeratins in breast tumours among BRCA1, BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2008;10(1):R17. doi: 10.1186/bcr1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honrado E, Osorio A, Palacios J, Milne RL, Sanchez L, Diez O, Cazorla A, Syrjakoski K, Huntsman D, Heikkila P, et al. Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J Clin Oncol. 2005;23(30):7503–7511. doi: 10.1200/JCO.2005.01.3698. [DOI] [PubMed] [Google Scholar]
- 56.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P, Sukumar S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164(5):1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madaras L, Balint N, Gyorffy B, Tokes AM, Barshack I, Yosepovich A, Friedman E, Paluch-Shimon S, Zippel D, Baghy K, et al. BRCA mutation-related and Claudin-low breast Cancer: blood relatives or stepsisters. Pathobiology. 2016;83(1):1–12. doi: 10.1159/000439135. [DOI] [PubMed] [Google Scholar]
- 59.Evans DG, Lalloo F, Howell S, Verhoef S, Woodward ER, Howell A. Low prevalence of HER2 positivity amongst BRCA1 and BRCA2 mutation carriers and in primary BRCA screens. Breast Cancer Res Treat. 2016;155(3):597–601. doi: 10.1007/s10549-016-3697-z. [DOI] [PubMed] [Google Scholar]
- 60.Armes JE, Trute L, White D, Southey MC, Hammet F, Tesoriero A, Hutchins AM, Dite GS, McCredie MR, Giles GG, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59(8):2011–2017. [PubMed] [Google Scholar]
- 61.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 62.Kyndi M, Sorensen FB, Knudsen H, Overgaard M, Nielsen HM, Andersen J, Overgaard J. Tissue microarrays compared with whole sections and biochemical analyses. A subgroup analysis of DBCG 82 b&c. Acta Oncol. 2008;47(4):591–599. doi: 10.1080/02841860701851871. [DOI] [PubMed] [Google Scholar]
- 63.Rossing HH, Talman ML, Laenkholm AV, Wielenga VT. Implementation of TMA and digitalization in routine diagnostics of breast pathology. APMIS. 2012;120(4):341–347. doi: 10.1111/j.1600-0463.2011.02871.x. [DOI] [PubMed] [Google Scholar]
- 64.Dekker TJ, ter Borg S, Hooijer GK, Meijer SL, Wesseling J, Boers JE, Schuuring E, Bart J, van Gorp J, Bult P, et al. Quality assessment of estrogen receptor and progesterone receptor testing in breast cancer using a tissue microarray-based approach. Breast Cancer Res Treat. 2015;152(2):247–252. doi: 10.1007/s10549-015-3444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dekker TJ, ter Borg S, Hooijer GK, Meijer SL, Wesseling J, Boers JE, Schuuring E, Bart J, van Gorp J, Bult P, et al. Erratum to: quality assessment of estrogen receptor and progesterone receptor testing in breast cancer using a tissue microarray-based approach. Breast Cancer Res Treat. 2015;152(2):253. doi: 10.1007/s10549-015-3478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BRCA testing and analysis (DOCX 16 kb)
Data (XLSX 40 kb)
Data dictionary (DOCX 21 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
[Additional supporting files: “Data” (Additional file 2), “Data dictionary” (Additional file 3)]