Abstract
The attempt to describe complex diseases by solely genetic determination has not been successful. There is increasing recognition that the development of disease is often a consequence of interactions between multiple genetic and environmental factors. To date, much of the research on environmental determinants of disease has focused on single exposures generally measured at a single time point. In order to address this limitation, the concept of the exposome has been introduced as a comprehensive approach, studying the full complement of environmental exposures from conception onwards. However, exposures are vast, dynamic, and diverse, and only a small proportion can be reasonably measured due to limitations in technology and feasibility. In addition, the interplay between genes and exposure as well as between different exposures is complicated and multifaceted, which leads to difficulties in linking disease or health outcomes with exposures. The large numbers of collected samples require well-designed logistics. Furthermore, the immense data sets generated from exposome studies require a significant computational investment for both data analysis and data storage. This report summarizes discussions during an international exposome symposium held at Gunma University in Japan regarding the concept of the exposome, challenges in exposome research, and future perspectives in the field.
Keywords: exposome, metabolomics, exposure, epigenetics, environment
1. Introduction
Since the human genome was sequenced, an extreme effort has been placed into mapping the role of genes in the onset of disease. It was expected that we would be able to explain the cause of disease, as well as understand the genetic basis of health. However, we have found that while the genetic contribution to different diseases varies, nongenetic factors have much greater attributable risks, often in the range of 80 to 90% for complex diseases [1]. This dominance of nongenetic components emphasizes the role of the environment in the risk of the development of chronic disease. In 2010, there were >50 million global deaths, of which ~50% could be attributed to a small set of exposures consisting primarily of particulate air pollution, smoking, and diet [2]. This suggests that if we can identify relevant exposures, we can potentially intervene to reduce the burden of these chronic diseases.
We now know that adverse gene–environment interactions probably influence most chronic diseases, including respiratory disease, allergy, and cancer [3]. However, the tools for quantitative assessment of exposures, based on measurements of chemicals in air, water, food, and the human body, are still limited in their capacity to detect multiple exposures simultaneously. This lack of comprehensive profiling methods to assess exposure has resulted in a need to rely upon self-reported information supplemented with population level data from multiple sources (e.g., demographics, census data, green space surveys, and GIS approaches) to categorize exposures. These self-reports, with the general exceptions of alcohol consumption and smoking, are unreliable predictors of long-term exposure levels and are poorly suited for assessing exposome interactions. While the extensive investment in genomics has successfully illuminated the genetic determinants of diseases, we are still in the dark ages when it comes to quantifying exposures.
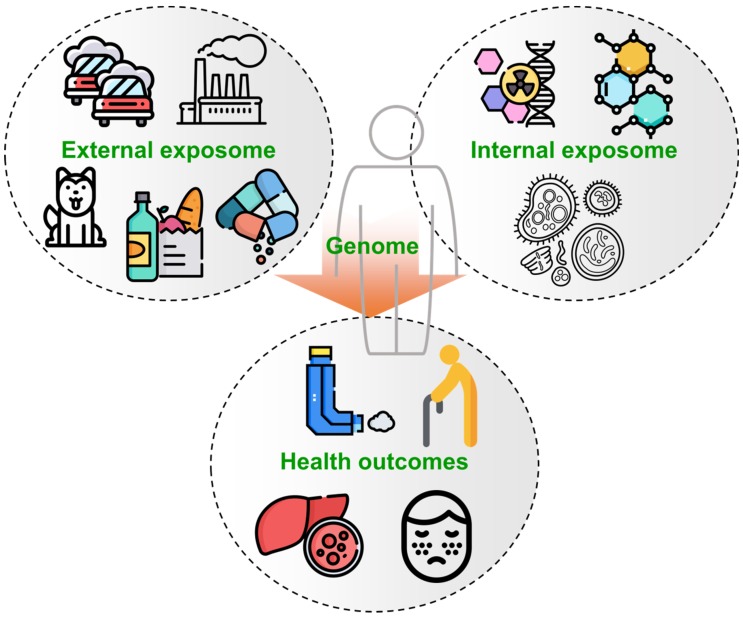
In recognition of this disparity in current knowledge on the health associations of the interactions between genes and environmental exposures, the term exposome has been proposed to represent all environmental exposures (including those from diet, behavior, and endogenous sources) from conception onward [4,5] (Figure 1). The exposome encompasses all nongenetic influences, and has become of critical relevance to understanding disease etiology [4], with particular interest in examining the pregnancy exposome and birth cohorts in disease presentation [6]. A few initiatives have begun, including EXPOsOMICs [7,8], HEALS (Health and Environment-wide Associations based on Large population Surveys) [9], and HELIX (the Human Early-life Exposome) [10] supported by the EU; CHEAR (the Children’s Health Exposure Analysis Resource) [11] and HERCULES (The Emory Health and Exposome Research Center: Understanding Lifetime Exposures) funded by the National Institute of Environmental Health Sciences (NIEHS) in the US; JECS (the Japan Environment and Children’s Study) launched in Japan [12]; and the I3CARE International Exposome Center collaboration between the Chinese University of Hong Kong, the University of Utrecht and the University of Toronto. However, extensive efforts are still needed to develop the methodologies to capture the exposome. An international symposium focusing on the exposome was held at Gunma University, Japan, on 23 October 2018. All of the authors on this report attended the meeting and a list of the talks is provided in Figure S1. Based upon the current status of the exposome field, this report summarizes opinions and perspectives with respect to study design, exposure measurement, sample collection and storage and future outlooks from discussions during the symposium.
Figure 1.
The concept of the exposome. The human exposome is often divided into the external exposome (e.g., air, diet, and social factors) and internal exposome (biological responses to exposure). Interactions between exposure and genetic factors can lead to disease or adverse health outcomes (e.g., aging, allergy, asthma, and cancer). Icons are designed by Freepik.
2. What is the Exposome?
The exposome can be defined as the total sum of all exposures from conception onwards [13,14]. The concept of the exposome was introduced as the environmental counterpart to the genome in order to establish the role of environmental exposures on a given genetic background. An exposure comes from the external environment (e.g., environment, lifestyle, stress, diet, and noise) and includes our internal biological processes (e.g., metabolism). The exposome is therefore often recognized as being comprised of the external and internal exposome (Figure 1) [15]. The external exposome refers to the full set of external exposures and experiences potentially contributing to disease or health outcomes (see Table 1 for some examples). The internal exposome includes biological responses to the external environment, interactions with exogenous compounds (e.g., interactions between diet and the gut microbiome [16,17]) and exogenous compounds entering the internal environment via diet, smoking, water, air, etc.
Table 1.
Relevant exposures for inclusion in exposome studies.
| Exposure Group | Exposure |
|---|---|
| External | |
| - Meteorology | Climate change, temperature, humidity, wind, atmospheric pressure |
| - Outdoor exposures | NO2, SO2, CO, O3, VOCs, PM, radiation, UV, traffic, pollen |
| - Built environment | Population density, building density, facilities, green space, walkability, neighborhood safety, accessibility to resources (e.g., hospitals, bus stations), noise |
| - Home environment | VOCs, PM, NO2, CO, aldehydes, metals, plasticizers, dust, pets, pests, allergen (e.g., house dust mites), mold, fungi, microbes, endotoxin |
| - Personal behavior | Diets, physical activity, tobacco smoke, alcohol, drugs, sleep, sex, cosmetics |
| - Social economic factors | Social factors, education, economy, psychological and mental stress |
| - Food and water contaminants | Fertilizers, metals, pesticides, plasticizers, DBPs, PCBs, flame retardants, PFASs |
| - Medications | Medicines, surgeries |
| - Occupational exposures | Chemicals, dust, metals, virus, animal proteins, plants, heat/cold stress |
| Internal | |
| Primary external exposures and associated metabolites, epigenetic (e.g., methylations, histone modifications), microbiome/metabolome/proteome/transcriptome/genome changes, etc. | |
Abbreviations: VOCs: volatile organic compounds; PM: particulate matter; PCB: polychlorinated biphenyl; NO2: nitrogen dioxide; UV: ultraviolet; CO: carbon oxide; O3: ozone; SO2: sulfur dioxide; DBPs: (water) disinfection by-products; PFASs: per- and polyfluoroalkyl substances.
3. How to Perform an Exposome Study?
Implementing the exposome concept to explain the causalities of chronic disease poses multiple challenges [18]. Herein, we discuss four basic questions: (1) Which variables should be included in the study design? (2) How to develop methods to comprehensively assess all exposures from both external and internal sources? (3) Which sample types can be collected and how to effectively manage large sample numbers? (4) How to dissect the associations between exposome data and health end points?
3.1. Study Design
As the field of exposome science continues to develop, a fundamental question is how to perform an exposome study. There are no clear methodologies and only a few examples in the literature. To date, three overall strategies have been applied. The first one is called “bottom-up”, in which external exposures are comprehensively measured and then linked to disease or health outcomes [6,19]. The second strategy is “top-down”, in which omics technologies (e.g., metabolomics, adductomics, and epigenomics) are applied to identify biomarkers of exposures and new causes of or associations with disease [20,21]. The last approach is “meet-in-the-middle”, in which the association between exposures and disease or biomarkers of exposures are initially investigated and then the relationship between disease and biomarkers is assessed. No matter which strategy is applied, an exposome study should be assumption-directed. For example, if the study is designed to explore the role of environmental factors on asthma, information on all exposures that could potentially affect airway inflammation should be collected [22]. However, this will come at the cost of not identifying rare exposures that are not yet known to correlate with a given disease; whereas, if the study aims to discover effects of exposures on unexpected health outcomes, it is desirable to include all measurable exposures in a given time period. However, this approach is not feasible and therefore some basic assumptions must be made in the initial experimental design in terms of which exposure variables are most relevant to the study at hand. The epidemiology field is well versed in designing these complex studies, with multiple options depending upon the study design (e.g., cross-sectional, longitudinal, cohort, and case–control). The different study designs can also complement one another, and there may be advantages in integrating them to gain an improved understanding of the effects of exposure.
One key challenge is to identify cause and effect in exposome association studies. While it is not possible to fully assign cause and effect in the design of an exposome study, some concepts can aid in interpreting findings. Due to the observational and cross-sectional study design of most studies conducted to date, there is often a temporal discordance between exposure and observed effect. It is therefore difficult to understand the effect of potential exposures from single spot samples. Exposome-based studies therefore benefit from longitudinal sampling, particular during critical life stages [23]. Accordingly, study design needs to incorporate multiple longitudinal time periods as well as temporal variability [24]. Many exposome studies employ birth cohorts, which enables studies to monitor the health trajectory of a population over time (including critical periods early in life) and investigation of the effects of multiple exposures [6].
The potential scale of exposome work can be massive by definition and is often beyond the ability of a single research group, requiring collaborations. It is not necessary to initiate an exposome study from scratch, and finished or ongoing associated projects can be leveraged and re-analyzed to suit exposome research aims. For example, HELIX combines six established and ongoing longitudinal population-based birth cohort studies in six European countries, serving as a good example of integrating existing resources [19].
Nonetheless, there remain challenges of study design and exposure reconstruction. First, for rare and low frequency disorders, especially those with long latency periods, establishing prospective studies is not always feasible as it would require very large samples sizes and many years of follow-up to obtain sufficient cases. For example, amyotrophic lateral sclerosis (ALS) is a disease that occurs with a frequency of 2 per 100,000 and is most often diagnosed in the fifth decade of life [25]. To study the early life environmental determinants of this neurodegenerative disorder, a sample size in excess of a million participants who are studied over decades would be needed in a prospective cohort design. A second major challenge is to retrospectively measure fetal exposure, which is a critical period of development. Many extant studies do not have biobanked samples of umbilical cord blood. An alternative to this approach is the use of naturally shed deciduous teeth or permanent adult teeth. Deciduous teeth commence development prenatally and grow in an incremental pattern (akin to growth rings in trees). Using beam-based methods, such as laser ablation, it is now possible to reconstruct past exposures (e.g., metals and organic chemicals) at fine temporal resolution, including over the fetal developmental period [26,27]. However, this approach requires significant expertise in dental tissue biology and has relatively complex sample preparation methods. For the external environment, there exist methods to reconstruct particulate air pollution, ambient temperature, green-space, light-at-night, and other measures using satellite-based monitoring [28,29,30]. These tools allow fine temporal scale reconstruction of historical exposure that may be added to existing studies.
An exposome study can also be fulfilled without extensively measuring exposures in large-scale biological samples. For example, efforts have been made to dissect the relationship between the genome and environment in large data streams such as electronic health records and epidemiological cohorts using tools of bioinformatics [31,32]. In a recent study [33], a large health insurance dataset was analyzed to assess the genetic and environmental contributions of 560 disease-related phenotypes in 56,396 twin pairs and 724,513 sibling pairs. The contribution of specific environmental risk factors like socioeconomic status and air pollution in phenotype were also estimated. Notably, this type of study requires massive data sets, vast experience in data analysis as well as comprehensive knowledge in genetics and epidemiology.
3.2. Which Exposures to Measure
Due to the comprehensive nature of the exposome, it is essentially not possible to measure the full complement of exposures. It then becomes a question of which exposures are the most informative as well as most realistic to obtain. A nonexclusive list of potential exposome categories and examples is shown in Table 1. External exposures such as climate and air quality can be acquired from local meteorological stations and environment databases (e.g., Global Environmental Database, Japan). The use of GIS (Geographical Intelligent Software) is also particularly useful for acquiring this type of data. Personal life style and behavior as well as social factors are typically obtained by questionnaires or self-reports. Smartphone-based personal wearable devices can monitor basic functions including physical activity, heart rate, sleep quality, food nutrition, etc. Silicone bands that are capable of capturing airborne or skin contact exposures including pesticides and flame retardants are now available (MyExposome, Inc., Corvallis, OR, USA) [34,35]. More professional devices have also been invented, for example a modified MicroPEM™ (RTI International, Research Triangle Park, NC, USA), which includes an infrared laser nephelometer, a PTFE (polytetrafluoroethylene) or PES (polyethersulfone) filter, and a cartridge filled with zeolite adsorbent beads that can capture hundreds of airborne biological and chemical exposures [36].
Current exposome studies mostly focus on measurable environmental exposures (e.g., air pollution, smoke, and occupational exposure) due to knowledge and technology limitations. However, there is a need to have as comprehensive of a measurement as possible. Measurable internal biological changes can serve as good indicators of external exposures, for example, telomere length is a master integrator of stressors that result from a variety of lifestyle and behavioral factors [37]. More comprehensively, omics technologies are recognized as promising approaches for obtaining biomarkers of exposure, biomarkers of effect, and biomarkers of susceptibility. In particular, it would be useful to identify biomarkers of susceptibility in order to identify at-risk or sensitive populations for certain environmental exposures. While top-down measurement of exposome signals through measurement of the global biological response using omics technologies addresses the issue to some extent, the current analytical technology is not sufficiently sensitive or flexible to capture and identify the components of the exposome in a single method [38,39]. Accordingly, current approaches will have to focus on specific chemical classes and integrate multi-platform data to generate more informative datasets.
3.3. Sample Collection and Management
The most common and readily available biological samples include blood and urine. Additional matrices of interest include feces, hair, sputum [40], and breast milk. Another approach that has been particularly successful is the use of teeth [41,42,43]. As mentioned earlier, the incremental and archival nature of teeth is an attractive option for obtaining detailed information on exposure timing. For example, a tooth-matrix biomarker method identified that metal toxicant uptake and essential element deficiency increased the risk of autism spectrum disorder in children [44].
Exposome studies generally involve large cohorts in order to have sufficient statistical power to detect meaningful effect sizes. For example, the JECS study is only halfway through its targeted completion, and has already amassed >5,000,000 individual sample tubes collected from >250,000 participants (>100,000 expecting mothers, >100,000 children, and >50,000 fathers). Exposure to environmental factors are being assessed by chemical analyses of biospecimens (e.g., blood, cord blood, urine, breast milk, and hair), household environment measurements, and computational simulations using monitoring data (e.g., ambient air quality monitoring), as well as questionnaires. The logistics of managing this many samples should not be underestimated and protocols with respect to sample collection, pretreatment, transportation, aliquoting, labeling, and storage should be decided early in the study design [12]. Biological samples should be aliquoted into small volumes to minimize potential effects from repeated freeze–thaw cycles [45]. Another important component is the question of sample integrity during storage. Some cohort studies may include samples that are decades old and there is little information on the stability of materials overtime. It would be particularly valuable to have molecular markers of sample viability, and there have been some initial efforts in this area [46,47].
3.4. Exposome Data Analysis
The basic concept of the exposome is to determine how environmental factors affect human health. Therefore, when it comes to data analysis, it is vital to link exposures to disease or health [48]. However, prior to data analysis, obtained exposures (variants) need to be preprocessed or cleaned (e.g., address missing values, classify the exposures into categories, calculate the correlations between and within categories, narrow down the variant dimensions by excluding co-occurring exposures, and data transformation). These efforts are nontrivial, and require a clear data analysis plan prior to study initiation.
While GWAS has been extensively used to identify genetic variants associated with disease, the concept of exposome-wide association study (EWAS) is still relatively new [9,49]. EWAS borrows the methods of GWAS to identify environmental factors that are associated with specific phenotypes. The logical next step in these analyses is the concept of genome-wide by environment interaction studies (GWEIS) in large-scale settings [50]. GWEIS approaches have been applied in various studies to investigate if there are specific variants associated with disease risk following exposure, or to identify relevant disease mechanisms. For example, novel loci for childhood asthma in relation to traffic-related air pollution exposure have been identified in recent GWEIS [51], as well as loci for respiratory symptoms in relation to occupational exposures [52] and for serum lipids in relation to smoking [53]. However, appropriate statistical models to conduct genome-wide studies in a true exposome setting have yet to be developed and well-powered studies to be performed. Likewise, epigenome-wide studies have been conducted for single environmental exposures linking, for example, maternal tobacco smoke during pregnancy [54] and prenatal air pollution exposure [55] to newborn DNA methylation changes, but no comprehensive exposome–epigenetics study has been published to date. Yet, these epigenetics studies highlight the complex interplay between environmental exposures and our genetic setup.
An interesting method to present exposure interactions is the exposome interaction network, which combines human- and environment-related exposures. Data analysis was designed to examine the intersection of interacting ecosystems (e.g., human-related bacteria, fungi, and arthropods). An exposome cloud was generated by querying species-interaction databases based on all measured biotic exposures. The number of nodes/edges/average interactions in the cloud provides an overview of personal lifestyles and work–home routines [36]. This method represents a realistic approach to interrogate and analyze this large body of data in a meaningful fashion that is accessible for the interested reader.
4. Future Perspectives
There is a need to define the composition of the exposome in order to assist in experimental design and to move the field forward. A number of efforts have already been made to develop exposome databases [56,57,58]. For example, metabolomics databases have begun to focus on exogenous chemicals and METLIN has expanded its library to include >700,000 chemicals [59]. In addition, there are decades of studies performed in the environmental field on a compound-by-compound basis. Accordingly, a vast, scattered exposure database already exists, representing an opportunity to collect a large body of information on the effects of exposure. There are also significant data available from workplace and occupational exposures [60]. In addition, multiple groups have performed targeted environmental exposures [61,62,63] as well as work performed in model organisms [64]. However, in order to make effective use of these resources, there is a need for extensive data sharing and for platforms capable of processing and analyzing large-scale data.
The standard approach to an exposome study generally involves large numbers of individuals in order to obtain reliable exposure-phenotype interactions. However, it is also possible to focus on fewer participants for a longer period (i.e., longitudinal study). In a recent study, 15 individuals were followed for up to 890 days to discover the diversity and dynamics of the exposome and its potential impact on human health [36]. Apart from revealing the casualties of disease, the exposome might also be useful for providing individual feedback (e.g., health status, local environment and quality), heritability prediction, and identifying individuals at risk. The exposome will most likely become an important component of personalized health care, together with genetic and other omics data, in order to establish which exposures may result in adverse health effects for individuals. The combination of genetic determinants with exposure data may be able to identify susceptible subpopulations that have higher risk levels of disease development in association with specific exposures. It is likely that the near future of health care will incorporate a combination of personal, home, and work space monitoring to measure exposure profiles, which will be correlated with individual susceptibility to disease and ultimately assist in maintaining health.
5. Concluding Remarks
There is a strong link between environmental exposure and disease onset; however, few efforts have been made to determine the total human exposure from our environment. The advent of the exposome is an attempt to understand the important, but complex nature of the interactions between environmental exposures, genetics, and health. An exposome study must incorporate multiple potential confounders including population vs. individual variation, age effects, seasonal effects, spatial effects, etc. In the future, it will be important to systematically expand the depth and breadth of our exposure knowledge. More importantly, the development of exposome science will require extraordinary efforts in many disciplines (environmental science, analytical chemistry, bioinformatics, genetics, systems biology, and epidemiology). Scientists from these fields, policy-makers, and funding agencies need to harmonize their approach in order to tackle the complexity of the exposome, which offers nothing less than the potential to understand the etiology of heterogeneous diseases.
Acknowledgments
We thank Komugi Kanazawa and Kyoko Ogura for extensive support for the planning of the symposium.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-1989/9/6/106/s1, Figure S1: Program of the 5th International Symposium of the Gunma University Initiative for Advanced Research (GIAR).
Author Contributions
P.Z. and C.E.W. planned and drafted the manuscript; M.A., R.C., T.I., M.J., I.M., E.M., M.P., F.T., and M.R.W. assisted in editing and reviewing.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant (JP18H06121, JP19K21239, JP19K17662) and the Japanese Environment Research and Technology Development Fund (No. 5-1752). We acknowledge the Gunma University Initiative for Advanced Research (GIAR), the STINT Foundation, the Swedish Heart Lung Foundation (HLF 20170734, HLF 20180290), and the Swedish Research Council (2016-02798). M.A. was supported by NIH (DP2ES025453). M.J. was supported by NIH (R01ES027595). I.M. was supported by a JSPS postdoctoral fellowship (P17774). E.M. was supported by grants from the European Research Council (757919), the Swedish Research Council, and the Strategic Research Area Epidemiology at KI. M.P. was supported by NIH (P30 ES09089).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Willett W.C. Balancing life-style and genomics research for disease prevention. Science. 2002;296:695–698. doi: 10.1126/science.1071055. [DOI] [PubMed] [Google Scholar]
- 2.Lim S.S., Vos T., Flaxman A.D., Danaei G., Shibuya K., Adair-Rohani H., Amann M., Anderson H.R., Andrews K.G., Aryee M., et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappaport S.M. Genetic Factors Are Not the Major Causes of Chronic Diseases. PLoS ONE. 2016;11:e0154387. doi: 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappaport S.M., Barupal D.K., Wishart D., Vineis P., Scalbert A. The blood exposome and its role in discovering causes of disease. Environ. Health Perspect. 2014;122:769–774. doi: 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wild C.P. The exposome: From concept to utility. Int. J. Epidemiol. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 6.Robinson O., Basagana X., Agier L., de Castro M., Hernandez-Ferrer C., Gonzalez J.R., Grimalt J.O., Nieuwenhuijsen M., Sunyer J., Slama R., et al. The Pregnancy Exposome: Multiple Environmental Exposures in the INMA-Sabadell Birth Cohort. Environ. Sci. Technol. 2015;49:10632–10641. doi: 10.1021/acs.est.5b01782. [DOI] [PubMed] [Google Scholar]
- 7.Turner M.C., Vineis P., Seleiro E., Dijmarescu M., Balshaw D., Bertollini R., Chadeau-Hyam M., Gant T., Gulliver J., Jeong A., et al. EXPOsOMICS: Final policy workshop and stakeholder consultation. BMC Public Health. 2018;18:260. doi: 10.1186/s12889-018-5160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vineis P., Chadeau-Hyam M., Gmuender H., Gulliver J., Herceg Z., Kleinjans J., Kogevinas M., Kyrtopoulos S., Nieuwenhuijsen M., Phillips D.H., et al. The exposome in practice: Design of the EXPOsOMICS project. Int. J. Hyg. Environ. Health. 2017;220:142–151. doi: 10.1016/j.ijheh.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steckling N., Gotti A., Bose-O’Reilly S., Chapizanis D., Costopoulou D., De Vocht F., Gari M., Grimalt J.O., Heath E., Hiscock R., et al. Biomarkers of exposure in environment-wide association studies—Opportunities to decode the exposome using human biomonitoring data. Environ. Res. 2018;164:597–624. doi: 10.1016/j.envres.2018.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Vrijheid M., Slama R., Robinson O., Chatzi L., Coen M., van den Hazel P., Thomsen C., Wright J., Athersuch T.J., Avellana N., et al. The human early-life exposome (HELIX): Project rationale and design. Environ. Health Perspect. 2014;122:535–544. doi: 10.1289/ehp.1307204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balshaw D.M., Collman G.W., Gray K.A., Thompson C.L. The Children’s Health Exposure Analysis Resource: Enabling research into the environmental influences on children’s health outcomes. Curr. Opin. Pediatr. 2017;29:385–389. doi: 10.1097/MOP.0000000000000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamoto T., Nitta H., Murata K., Toda E., Tsukamoto N., Hasegawa M., Yamagata Z., Kayama F., Kishi R., Ohya Y., et al. Rationale and study design of the Japan environment and children’s study (JECS) BMC Public Health. 2014;14:25. doi: 10.1186/1471-2458-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rappaport S.M. Implications of the exposome for exposure science. J. Expos. Sci. Environ. Epidemiol. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- 14.Wild C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 15.Vrijheid M. The exposome: A new paradigm to study the impact of environment on health. Thorax. 2014;69:876–878. doi: 10.1136/thoraxjnl-2013-204949. [DOI] [PubMed] [Google Scholar]
- 16.Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S., Deng L., Bry L., Gordon J.I., Kahn C.R. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A., Zapata R.C., Pezeshki A., Workentine M.L., Chelikani P.K. Host genetics and diet composition interact to modulate gut microbiota and predisposition to metabolic syndrome in spontaneously hypertensive stroke-prone rats. FASEB J. 2019 doi: 10.1096/fj.201801627RRR. [DOI] [PubMed] [Google Scholar]
- 18.Wild C.P., Scalbert A., Herceg Z. Measuring the exposome: A powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagenesis. 2013;54:480–499. doi: 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- 19.Maitre L., de Bont J., Casas M., Robinson O., Aasvang G.M., Agier L., Andrusaityte S., Ballester F., Basagana X., Borras E., et al. Human Early Life Exposome (HELIX) study: A European population-based exposome cohort. BMJ Open. 2018;8:e021311. doi: 10.1136/bmjopen-2017-021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooke M.S., Hu C.W., Chang Y.J., Chao M.R. Urinary DNA adductomics—A novel approach for exposomics. Environ. Int. 2018;121:1033–1038. doi: 10.1016/j.envint.2018.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu S.S., Grigoryan H., Edmands W.M., Hu W., Iavarone A.T., Hubbard A., Rothman N., Vermeulen R., Lan Q., Rappaport S.M. Profiling the Serum Albumin Cys34 Adductome of Solid Fuel Users in Xuanwei and Fuyuan, China. Environ. Sci. Technol. 2017;51:46–57. doi: 10.1021/acs.est.6b03955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A., Khatri S.B. The Exposome and Asthma. Clin. Chest Med. 2019;40:107–123. doi: 10.1016/j.ccm.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer R.M., Smith M.N., Faustman E.M. Developing the Regulatory Utility of the Exposome: Mapping Exposures for Risk Assessment through Lifestage Exposome Snapshots (LEnS) Environ. Health Perspect. 2017;125:085003. doi: 10.1289/EHP1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D.P. Sequencing the exposome: A call to action. Toxicol. Rep. 2016;3:29–45. doi: 10.1016/j.toxrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chio A., Logroscino G., Traynor B.J., Collins J., Simeone J.C., Goldstein L.A., White L.A. Global epidemiology of amyotrophic lateral sclerosis: A systematic review of the published literature. Neuroepidemiology. 2013;41:118–130. doi: 10.1159/000351153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora M., Austin C., Sarrafpour B., Hernandez-Avila M., Hu H., Wright R.O., Tellez-Rojo M.M. Determining prenatal, early childhood and cumulative long-term lead exposure using micro-spatial deciduous dentine levels. PLoS ONE. 2014;9:e97805. doi: 10.1371/journal.pone.0097805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velthorst E., Smith L., Bello G., Austin C., Gennings C., Modabbernia A., Franke N., Frangou S., Wright R., de Haan L., et al. New Research Strategy for Measuring Pre- and Postnatal Metal Dysregulation in Psychotic Disorders. Schizophr. Bull. 2017;43:1153–1157. doi: 10.1093/schbul/sbx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorek-Hamer M., Just A.C., Kloog I. Satellite remote sensing in epidemiological studies. Curr. Opin. Pediatr. 2016;28:228–234. doi: 10.1097/MOP.0000000000000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld A., Dorman M., Schwartz J., Novack V., Just A.C., Kloog I. Estimating daily minimum, maximum, and mean near surface air temperature using hybrid satellite models across Israel. Environ. Res. 2017;159:297–312. doi: 10.1016/j.envres.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Bose S., Ross K.R., Rosa M.J., Chiu Y.M., Just A., Kloog I., Wilson A., Thompson J., Svensson K., Rojo M.M.T., et al. Prenatal particulate air pollution exposure and sleep disruption in preschoolers: Windows of susceptibility. Environ. Int. 2019;124:329–335. doi: 10.1016/j.envint.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manrai A.K., Ioannidis J.P.A., Patel C.J. Signals Among Signals: Prioritizing Non-genetic Associations in Massive Datasets. Am. J. Epidemiol. 2019 doi: 10.1093/aje/kwz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Goallec A., Patel C.J. Age-dependent co-dependency structure of biomarkers in the general population of the United States. Aging. 2019;11:1404–1426. doi: 10.18632/aging.101842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakhani C.M., Tierney B.T., Manrai A.K., Yang J., Visscher P.M., Patel C.J. Repurposing large health insurance claims data to estimate genetic and environmental contributions in 560 phenotypes. Nat. Genet. 2019;51:327–334. doi: 10.1038/s41588-018-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Connell S.G., Kincl L.D., Anderson K.A. Silicone wristbands as personal passive samplers. Environ. Sci. Technol. 2014;48:3327–3335. doi: 10.1021/es405022f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon H.M., Scott R.P., Holmes D., Calero L., Kincl L.D., Waters K.M., Camann D.E., Calafat A.M., Herbstman J.B., Anderson K.A. Silicone wristbands compared with traditional polycyclic aromatic hydrocarbon exposure assessment methods. Anal. Bioanal. Chem. 2018;410:3059–3071. doi: 10.1007/s00216-018-0992-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang C., Wang X., Li X., Inlora J., Wang T., Liu Q., Snyder M. Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring. Cell. 2018;175:277–291.e31. doi: 10.1016/j.cell.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epel E.S., Blackburn E.H., Lin J., Dhabhar F.S., Adler N.E., Morrow J.D., Cawthon R.M. Accelerated telomere shortening in response to life stress. Proc. Natl. Acad. Sci. USA. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappaport S.M. Biomarkers intersect with the exposome. Biomarkers. 2012;17:483–489. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis K.K., Marder E., Balshaw D.M., Cui Y., Lynes M.A., Patti G.J., Rappaport S.M., Shaughnessy D.T., Vrijheid M., Barr D.B. Biomonitoring in the Era of the Exposome. Environ. Health Perspect. 2017;125:502–510. doi: 10.1289/EHP474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bessonneau V., Pawliszyn J., Rappaport S.M. The Saliva Exposome for Monitoring of Individuals’ Health Trajectories. Environ. Health Perspect. 2017;125:077014. doi: 10.1289/EHP1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andra S.S., Austin C., Arora M. The tooth exposome in children’s health research. Curr. Opin. Pediatr. 2016;28:221–227. doi: 10.1097/MOP.0000000000000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andra S.S., Austin C., Wright R.O., Arora M. Reconstructing pre-natal and early childhood exposure to multi-class organic chemicals using teeth: Towards a retrospective temporal exposome. Environ. Int. 2015;83:137–145. doi: 10.1016/j.envint.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora M., Austin C. Teeth as a biomarker of past chemical exposure. Curr. Opin. Pediatr. 2013;25:261–267. doi: 10.1097/MOP.0b013e32835e9084. [DOI] [PubMed] [Google Scholar]
- 44.Arora M., Reichenberg A., Willfors C., Austin C., Gennings C., Berggren S., Lichtenstein P., Anckarsater H., Tammimies K., Bolte S. Fetal and postnatal metal dysregulation in autism. Nat. Commun. 2017;8:15493. doi: 10.1038/ncomms15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuhadar S., Koseoglu M., Atay A., Dirican A. The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem. Med. 2013;23:70–77. doi: 10.11613/BM.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheelock A.M., Paulson L., Litton J.E., EuPA Biobank Initiative Group The EuPA Biobank Initiative: Meeting the future challenges of biobanking in proteomics & systems medicine. J. Proteom. 2015;127:414–416. doi: 10.1016/j.jprot.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Liu X., Hoene M., Yin P., Fritsche L., Plomgaard P., Hansen J.S., Nakas C.T., Niess A.M., Hudemann J., Haap M., et al. Quality Control of Serum and Plasma by Quantification of (4E,14Z)-Sphingadienine-C18-1-Phosphate Uncovers Common Preanalytical Errors During Handling of Whole Blood. Clin. Chem. 2018;64:810–819. doi: 10.1373/clinchem.2017.277905. [DOI] [PubMed] [Google Scholar]
- 48.Patel C.J. Analytic Complexity and Challenges in Identifying Mixtures of Exposures Associated with Phenotypes in the Exposome Era. Curr. Epidemiol. Rep. 2017;4:22–30. doi: 10.1007/s40471-017-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel C.J., Bhattacharya J., Butte A.J. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunn E.C., Wiste A., Radmanesh F., Almli L.M., Gogarten S.M., Sofer T., Faul J.D., Kardia S.L., Smith J.A., Weir D.R., et al. Genome-Wide Association Study (Gwas) and Genome-Wide by Environment Interaction Study (Gweis) of Depressive Symptoms in African American and Hispanic/Latina Women. Depress. Anxiety. 2016;33:265–280. doi: 10.1002/da.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gref A., Merid S.K., Gruzieva O., Ballereau S., Becker A., Bellander T., Bergstrom A., Bosse Y., Bottai M., Chan-Yeung M., et al. Genome-Wide Interaction Analysis of Air Pollution Exposure and Childhood Asthma with Functional Follow-up. Am. J. Respir. Crit. Care Med. 2017;195:1373–1383. doi: 10.1164/rccm.201605-1026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng X., Vonk J.M., van der Plaat D.A., Faiz A., Pare P.D., Joubert P., Nickle D., Brandsma C.A., Kromhout H., Vermeulen R., et al. Genome-wide interaction study of gene-by-occupational exposures on respiratory symptoms. Environ. Int. 2019;122:263–269. doi: 10.1016/j.envint.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Bentley A.R., Sung Y.J., Brown M.R., Winkler T.W., Kraja A.T., Ntalla I., Schwander K., Chasman D.I., Lim E., Deng X., et al. Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet. 2019;51:636–648. doi: 10.1038/s41588-019-0378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joubert B.R., Felix J.F., Yousefi P., Bakulski K.M., Just A.C., Breton C., Reese S.E., Markunas C.A., Richmond R.C., Xu C.J., et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruzieva O., Xu C.J., Breton C.V., Annesi-Maesano I., Anto J.M., Auffray C., Ballereau S., Bellander T., Bousquet J., Bustamante M., et al. Epigenome-Wide Meta-Analysis of Methylation in Children Related to Prenatal NO2 Air Pollution Exposure. Environ. Health Perspect. 2017;125:104–110. doi: 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neveu V., Moussy A., Rouaix H., Wedekind R., Pon A., Knox C., Wishart D.S., Scalbert A. Exposome-Explorer: A manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017;45:D979–D984. doi: 10.1093/nar/gkw980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faisandier L., Bonneterre V., De Gaudemaris R., Bicout D.J. Occupational exposome: A network-based approach for characterizing Occupational Health Problems. J. Biomed. Inform. 2011;44:545–552. doi: 10.1016/j.jbi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Wishart D., Arndt D., Pon A., Sajed T., Guo A.C., Djoumbou Y., Knox C., Wilson M., Liang Y., Grant J., et al. T3DB: The toxic exposome database. Nucleic Acids Res. 2015;43:D928–D934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warth B., Spangler S., Fang M., Johnson C.H., Forsberg E.M., Granados A., Martin R.L., Domingo-Almenara X., Huan T., Rinehart D., et al. Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Anal. Chem. 2017;89:11505–11513. doi: 10.1021/acs.analchem.7b02759. [DOI] [PubMed] [Google Scholar]
- 60.Walker D.I., Uppal K., Zhang L., Vermeulen R., Smith M., Hu W., Purdue M.P., Tang X., Reiss B., Kim S., et al. High-resolution metabolomics of occupational exposure to trichloroethylene. Int. J. Epidemiol. 2016;45:1517–1527. doi: 10.1093/ije/dyw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carlsten C., Blomberg A., Pui M., Sandstrom T., Wong S.W., Alexis N., Hirota J. Diesel exhaust augments allergen-induced lower airway inflammation in allergic individuals: A controlled human exposure study. Thorax. 2016;71:35–44. doi: 10.1136/thoraxjnl-2015-207399. [DOI] [PubMed] [Google Scholar]
- 62.Mookherjee N., Piyadasa H., Ryu M.H., Rider C.F., Ezzati P., Spicer V., Carlsten C. Inhaled diesel exhaust alters the allergen-induced bronchial secretome in humans. Eur. Respir. J. 2018;51 doi: 10.1183/13993003.01385-2017. [DOI] [PubMed] [Google Scholar]
- 63.Brook J.R., Setton E.M., Seed E., Shooshtari M., Doiron D., CANUE—The Canadian Urban Environmental Health Research Consortium The Canadian Urban Environmental Health Research Consortium—A protocol for building a national environmental exposure data platform for integrated analyses of urban form and health. BMC Public Health. 2018;18:114. doi: 10.1186/s12889-017-5001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.David A., Lange A., Abdul-Sada A., Tyler C.R., Hill E.M. Disruption of the Prostaglandin Metabolome and Characterization of the Pharmaceutical Exposome in Fish Exposed to Wastewater Treatment Works Effluent As Revealed by Nanoflow-Nanospray Mass Spectrometry-Based Metabolomics. Environ. Sci. Technol. 2017;51:616–624. doi: 10.1021/acs.est.6b04365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.