Abstract
The southern African Miombo and Mopane ecoregions constitute a unique repository of plant diversity whose diversification and evolutionary history is still understudied. In this work, we assessed the diversity, distribution, and conservation status of Miombo and Mopane tree legumes within the Zambezian phytoregion. Data were retrieved from several plant and gene databases and phylogenetic analyses were performed based on genetic barcodes. Seventy-eight species (74 from Miombo and 23 from Mopane, 19 common to both ecoregions) have been scored. Species diversity was high within both ecoregions, but information about the actual conservation status is scarce and available only for ca. 15% of the species. Results of phylogenetic analyses were consistent with current legume classification but did not allow us to draw any conclusion regarding the evolutionary history of Miombo and Mopane tree legumes. Future studies are proposed to dissect the diversity and structure of key species in order to consolidate the network of conservation areas.
Keywords: diversity, Miombo, Mopane, tree legumes, Zambezian phytoregion
1. Introduction
Tropical dry forests and woodlands constitute a large portion of the world’s vegetation, covering one-sixth of the earth’s surface and more than half of the African continent [1,2]. Among them, the Miombo–Mopane woodlands are the most predominant type of vegetation in Southern Africa, and together with Amazonia, Congo, New Guinea and the North American deserts, are considered wilderness areas of global conservation significance [3]. The woodlands play a crucial role in formal and informal economies, supporting the livelihoods of millions of rural and urban people, by providing important resources such as timber, food, medicines, biofertilizers, housing and energy [4,5,6,7,8]. The Miombo and Mopane woodlands also play an important role in the ecosystem dynamics, particularly with respect to biodiversity, water, carbon and energy balance [9,10,11,12,13].
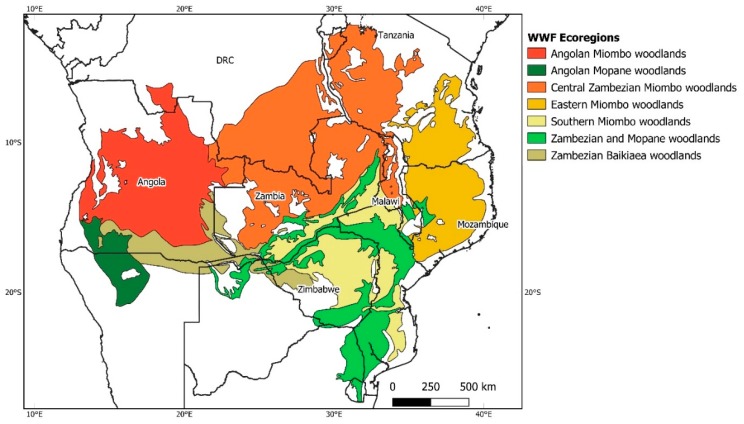
The plant diversity of these ecosystems comprises a wealthy repository of biodiversity, with a high proportion of native species, which makes it biologically unique [14,15]. According to Olson et al. [16], five sub-regions have been delineated through the Miombo woodlands (i.e., Angolan Miombo woodlands, Central Zambezian Miombo woodlands, Zambezian Baikiaea woodlands, Eastern Miombo woodlands and Southern Miombo woodlands) that cover about 3,000,000 km2 across the Zambezian region of Angola, Democratic Republic of Congo, Malawi, Mozambique, Tanzania, Zambia and Zimbabwe [17,18]. The Mopane woodlands represent the second most significant type of vegetation in the Zambezian phytoregion, covering approximately 600,000 km2. This region includes two sub-areas (i.e., Zambezian and Mopane woodlands, and Angolan Mopane woodlands), and is distributed over northern Namibia, southern Angola, Zimbabwe, Botswana, Zambia, Malawi, southern Mozambique and northern South Africa [17,19].
The Miombo and Mopane woodlands are dominated by species belonging to the Leguminosae [2], which is considered the second most economically important plant family [20,21,22,23]. This family includes over 19,500 species spanning about 770 genera and six subfamilies, namely Caesalpinioideae, Cercidoideae, Detarioideae, Dialioideae, Duparquetioideae and Papilionoideae, many of which establish root-nodule symbiosis with N2 fixing rhizobia bacteria [23]. The Miombo woodlands are dominated by trees of the genera Brachystegia, Julbernardia and Isoberlinia, while the Mopane woodlands are dominated by Colosphosperum mopane (Benth.) Leonard [9,17,24]. Most of these trees are under severe ecological pressure, due to logging and charcoal production [25,26], as well as fires related to animal, human and climate factors [6,10,11,13], which have contributed to the massive degradation of these woodlands and raised the need for their conservation [4,24,27,28].
African forests are among the most understudied regions in the world with respect to phylogenetic diversity [29]. Characterization of a region’s phylogenetic diversity, which is based on the evolutionary relationships between species from a given geographical area, can be a powerful tool to analyze the phylogenetic structure of natural communities and can assist the study of processes governing the assembly of species in ecological communities [30,31]. The identification of areas with more or less phylogenetic diversity based on species richness is another important element for conservation studies [32]. Although the African woodlands remain of enormous evolutionary interest, assessing species diversification within Miombo and Mopane plant communities across West and East African woodlands is still understudied. Since the dominant plant lineages in Miombo and Mopane are tree legumes, this provides an excellent case study to understand the diversity and the evolutionary history of these two predominant types of vegetation in Southern Africa. In this context, the objectives of this study were: (i) to characterize the diversity and distribution of Miombo and Mopane tree legumes within the Zambezian phytoregion (Figure 1); (ii) to provide a unique view of their phylogenetic diversity and how these lineages have evolved across West and East African woodlands; and (iii) to provide new data to assist the implementation of conservation strategies and the sustainable management of the Mopane and Miombo woodlands.
Figure 1.
Geographical distribution of the Miombo–Mopane woodlands in the Zambezian phytoregion.
2. Results
2.1. Tree Legumes Diversity and Conservation Status
Our study categorized 78 Leguminosae trees, of which 74 were representative of the Miombo woodlands and 23 of the Mopane woodlands (19 species common to both habitats) (Table 1). Overall, the largest number of species was found in Zambia, with 71 out of the 78 species (91%), while the Democratic Republic of Congo (DRC) had the lowest number (51 species or 65%) (Table 1). Five of the six recognized subfamilies were found in the Miombo woodlands: Caesalpinioideae, the most frequent (27 species), followed by Papilionoideae (22 species), Detarioideae (21 species), and Cercidoideae and Dialioideae with only three and one species, respectively. Among the 34 genera, Brachystegia and Acacia were the most diverse with 12 and 11 species, respectively. The number of species per country ranged from 50 in the DRC to 67 in Zambia. The Mopane woodlands harbored four subfamilies of legume trees, 14 species belonging to the Caesalpinioideae subfamily, followed by Papilionoideae with six species. Only two species from Detarioideae and one from Cercidoideae were retrieved. The number of genera was lower in Mopane than in Miombo (15 versus 34). The highest number of species was recorded in Zimbabwe (23) and the lowest in the DRC (11).
Table 1.
Leguminosae tree species from Miombo and Mopane ecosystems: main habitat, distribution in the Zambezian area, conservation status (International Union for Conservation of Nature - IUCN) and habit.
| Taxon | Subfamily | Habitat | Distribution 1 | Conservation Status 2 | Habit | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AO | DRC | ZM | ZW | TZ | MW | MZ | |||||
| Acacia arenaria Schinz | Caesalpinioideae | Miombo and Mopane | X | X | X | NE | Shrub or small tree 2–9 m | ||||
| Acacia erubescens Welw. ex Oliv. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | X | NE | Shrub or tree 2–10 m |
| Acacia galpinii Burtt Davy | Caesalpinioideae | Miombo | X | X | X | X | X | X | NE | Large tree 8–36 m | |
| Acacia gerrardii Benth. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | NE | Shrub or tree 5–15 m | |
| Acacia hebeclada DC. | Caesalpinioideae | Mopane | X | X | X | NE | Shrub or small tree to 3 m | ||||
| Acacia kirkii Oliv. | Caesalpinioideae | Mopane | X | X | X | X | X | X | X | NE | Tree 2.5–18 m |
| Acacia mellifera (Vahl) Benth. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | NE | Tree 4–9 | ||
| Acacia nigrescens Oliv. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | NE | Tree 4–30 m | |
| Acacia nilotica (L.) Willd. ex Delile | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | LC | Tree 3–15 m | |
| Acacia polyacantha | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Large tree 3.5–20 m |
| Acacia robusta Burch. | Caesalpinioideae | Miombo | X | X | X | X | NE | Tree 5-30 m | |||
| Acacia sieberiana DC. | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 3–25 m |
| Acacia welwitschii Oliv. | Caesalpinioideae | Miombo | X | X | X | NE | Large tree 3–20 m | ||||
| Afzelia quanzensis Welw. | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 1.5–35 m |
| Albizia adianthifolia (Schum.) W.Wight | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 2.5–40 m |
| Albizia anthelmintica Brongn. | Caesalpinioideae | Miombo | X | X | X | X | X | X | NE | Shrub or tree 2–12 m | |
| Albizia antunesiana Harms | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 6–18 m |
| Albizia glaberrima (Schum. & Thonn.) Benth. | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 9–25 m |
| Albizia harveyi E.Fourn. | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 1.5–20 m |
| Albizia versicolor Oliv. | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 3–20 m |
| Amblygonocarpus andongensis (Oliv.) Exell & Torre | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 6–25 m |
| Baikiaea plurijuga Harms | Detarioideae | Miombo and Mopane | X | X | X | LR/NT | Tree 6–25 m | ||||
| Baphia bequaertii De Wild. | Papilionoideae | Miombo | X | X | X | NE | Shrub or tree 3–10 m | ||||
| Baphia massaiensis Taub. | Papilionoideae | Miombo | X | X | X | X | X | X | X | LC | Shrub or tree to 8–10 m |
| Bauhinia petersiana Bolle | Cercidoideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 3–10 m |
| Bauhinia thonningii Schum. | Cercidoideae | Miombo and Mopane | X | X | X | X | X | X | NE | Shrub or tree to 2–20 m | |
| Bauhinia tomentosa L. | Cercidoideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 1–8 m |
| Bobgunnia madagascariensis (Desv.) J.H.Kirkbr. & Wiersema | Papilionoideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 2–15 m |
| Brachystegia allenii Hutch. & Burtt Davy | Detarioideae | Miombo | X | X | X | X | X | X | NE | Tree 3–20 m | |
| Brachystegia bakeriana Burtt Davy & Hutch. | Detarioideae | Miombo | X | X | X | VU B1+2c | Shrub or tree to 10 m | ||||
| Brachystegia boehmii Taub | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 2.5–25 m |
| Brachystegia floribunda Benth. | Detarioideae | Miombo | X | X | X | X | X | X | NE | Tree 4–15 m | |
| Brachystegia gossweileri Burtt Davy & Hutch. | Detarioideae | Miombo | X | X | X | NE | Tree 6–24 m | ||||
| Brachystegia longifolia Benth. | Detarioideae | Miombo | X | X | X | X | X | X | NE | Tree 2–30 m | |
| Brachystegia puberula Burtt Davy & Hutch. | Detarioideae | Miombo | X | X | X | LC | Tree 6–12 m | ||||
| Brachystegia spiciformis Benth | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 5–40 m |
| Brachystegia tamarindoides Benth. | Detarioideae | Miombo | X | X | X | X | X | NE | Tree 4–30 m | ||
| Brachystegia taxifolia Harms | Detarioideae | Miombo | X | X | X | X | NE | Shrub or tree 2–16 m | |||
| Brachystegia utilis Hutch. & Burtt Davy | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 6–20 m |
| Brachystegia wangermeeana De Wild. | Detarioideae | Miombo | X | X | X | X | X | NE | Tree 1.5–20 m | ||
| Burkea africana Hook. | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 4–20 m |
| Cassia abbreviata Oliv. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | NE | Shrub or tree 3–15 m | |
| Colophospermum mopane (Benth.) Leonard | Detarioideae | Mopane | X | X | X | X | X | NE | Tree 4–18 m tall | ||
| Cordyla africana Lour. | Papilionoideae | Miombo | X | X | X | X | X | NE | Tree 9–40 m | ||
| Craibia zimmermannii (Harms) Dunn | Papilionoideae | Miombo | X | X | NE | Tree 4–5 m | |||||
| Dalbergia arbutifolia Baker | Papilionoideae | Miombo | X | X | X | X | X | X | NE | Shrub or tree 3–18 m | |
| Dalbergia boehmii Taub. | Papilionoideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 4.5–21 m |
| Dalbergia melanoxylon Guill. & Perr. | Papilionoideae | Miombo and Mopane | X | X | X | X | X | X | X | LR/NT | Spiny shrub or tree 1–30 m |
| Dalbergia nitidula Baker | Papilionoideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 2–12 m |
| Dialium englerianum Henriq. | Dialioideae | Miombo | X | X | X | X | NE | Tree 6–23 m | |||
| Dichrostachys cinerea (L.) Wight & Arn. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | X | LC | Shrub or tree 1–12 m |
| Elephantorrhiza goetzei (Harms) Harms | Caesalpinioideae | Miombo | X | X | X | X | X | X | X | NE | Shrub or tree 1–7 m |
| Entada abyssinica Steud. ex A. Rich. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | X | NE | Tree 2.7–15 m |
| Erythrophleum africanum (Benth.) Harms | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | X | NE | Tree 4–18 m |
| Faidherbia albida (Delile) A.Chev. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | X | NE | Tree 6–30 m |
| Guibourtia coleosperma (Benth.) J. Léonard | Detarioideae | Miombo | X | X | X | X | NE | Tree 12–30 m | |||
| Isoberlinia angolensis (Benth.) Hoyle & Brenan | Detarioideae | Miombo | X | X | X | X | X | NE | Shrub or tree 1–20 m | ||
| Isoberlinia scheffleri (Harms) Greenway | Detarioideae | Miombo | X | X | VU B1+2b | Tree 30–46 m | |||||
| Isoberlinia tomentosa (Harms) Craib & Stapf | Detarioideae | Miombo | X | X | X | X | X | NE | Tree 3–18 m | ||
| Julbernardia globiflora (Benth.) Troupin | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 5–15 m |
| Julbernardia paniculata (Benth.) Troupin | Detarioideae | Miombo | X | X | X | X | X | X | X | NE | Tree 2–20 m |
| Millettia stuhlmannii Taub. | Papilionoideae | Miombo | X | X | X | NE | Tree 6–25 m | ||||
| Millettia usaramensis Taub. | Papilionoideae | Miombo | X | X | X | X | NE | Shrub or tree 2–10 m | |||
| Mundulea sericea (Willd.) A. Chev. | Papilionoideae | Miombo | X | X | X | X | X | X | NE | Large shrub or tree 1–8 m | |
| Ormocarpum trichocarpum (Taub.) Engl. | Papilionoideae | Miombo and Mopane | X | X | X | X | NE | Shrub or small tree 1–6 m | |||
| Peltophorum africanum Sond. | Caesalpinioideae | Miombo and Mopane | X | X | X | X | X | X | NE | Tree 3–12 m | |
| Pericopsis angolensis (Baker) Meeuwen | Papilionoideae | Miombo | X | X | X | X | X | X | X | NE | Tree 3–20 m |
| Philenoptera bussei (Harms) Shrire | Papilionoideae | Miombo | X | X | X | X | X | NE | Tree 3–15 m | ||
| Philenoptera nelsii (Schinz) Schrire | Papilionoideae | Mopane | X | X | X | NE | Shrub or tree 4–12 m | ||||
| Philenoptera violacea (Klotze) Shrire | Papilionoideae | Miombo | X | X | X | X | X | X | X | NE | Tree 4.5–27 m |
| Pterocarpus angolensis DC. | Papilionoideae | Miombo | X | X | X | X | X | X | X | LR/NT | Tree 5–30 m |
| Pterocarpus brenanii Barbosa & Torre | Papilionoideae | Miombo | X | X | X | X | X | X | X | LC | Tree 4–12 m |
| Pterocarpus lucens Guill. & Perr. | Papilionoideae | Miombo and Mopane | X | X | X | X | X | LC | Tree 7.5–18 m | ||
| Pterocarpus rotundifolius (Sond.) Druce | Papilionoideae | Miombo and Mopane | X | X | X | X | X | X | X | NE | Tree 3–25 m |
| Senna petersiana (Bolle) Lock | Caesalpinioideae | Miombo | X | X | X | X | X | X | NE | Shrub or small tree 2–6 m | |
| *Tamarindus indica L. | Detarioideae | Miombo | X | X | X | X | X | X | X | LC | Tree to 25 m |
| Xanthocercis zambesiaca (Baker) Dumaz-le-Grand | Papilionoideae | Miombo | X | X | X | X | NE | Tree 7–30 m | |||
| Xeroderris stuhlmannii (Taub.) Mendonça & E.P. Sousa | Papilionoideae | Miombo and Mopane | X | X | X | X | X | NE | Tree up to 20 m | ||
| Total numbers | - | ||||||||||
| Number of species (Miombo) | - | - | 58 | 50 | 67 | 62 | 64 | 60 | 62 | - | - |
| Number of species (Mopane) | - | - | 19 | 11 | 21 | 23 | 17 | 18 | 17 | - | - |
| Number of species (Miombo and/or Mopane) | - | - | 62 | 51 | 71 | 66 | 65 | 62 | 64 | - | - |
* The origin of Tamarindus indica is uncertain and some sources (Africa Plant Database) suggest that it is native only in Madagascar. 1 Distribution: AO—Angola; DRC—Democratic Republic of Congo; ZM—Zambia; ZW—Zimbabwe; TZ—United Republic of Tanzania; MW—Malawi; MZ—Mozambique. 2 Conservation status according to IUCN Red List: LC—least concern; NT—near threatened; VU—vulnerable; NE—not evaluated.
Information on the conservation status of the Miombo and Mopane trees is still scarce as most of the species (66 species or 85%) have not yet been assessed globally, according to the categories and criteria of the International Union for Conservation of Nature (IUCN) Red List (Table 1). Therefore, only 12 species could be evaluated, of which seven were classified with least concern (Acacia nilotica (L.) Willd. ex Delile; Baphia massaiensis Taub.; Brachystegia puberula Burtt Davy & Hutch.; Dichrostachys cinerea (L.) Wight & Arn.; Pterocarpus brenanii Barbosa & Torre; Pterocarpus lucens Guill. & Perr.; Tamarindus indica L.), three as near threatened (Dalbergia melanoxylon Guill. & Perr.; Pterocarpus angolensis DC.; Baikiaea plurijuga Harms), and two as vulnerable species (Isoberlinia scheffleri (Harms) Greenway; Brachystegia bakeriana Burtt Davy & Hutch) (Figure 2).
Figure 2.
Leguminosae species from Miombo and Mopane woodlands and their conservation status according to the IUCN Red List. (A) Baikiaea plurijuga occurs in Miombo and Mopane woodlands and is classified as near threatened; (B) Brachystegia bakeriana occurs in Miombo and is classified as vulnerable; (C) Dichrostachys cinerea occurs in both ecoregions and is classified as least concern; (D) Pterocarpus angolensis occurs in Miombo woodlands and is classified as near threatened.
2.2. Molecular Phylogenetic Analysis
As DNA sequences were not available for all the selected taxa, the analyzed set was reduced to a total of 67 species from the original set of 78 (Table 2). A total of eight datasets were created: two for each single locus (internal transcribed spacer (ITS), matK and rbcL), and two combined (ITS + matK + rbcL). The ITS matrix had the lower number of taxa and haplotype diversity (n = 38, Hd = 0.9986), while rbcL had the highest of the three (n = 53, Hd = 1.0000), followed by matK (n = 69, Hd = 0.9987). The combined set of ITS+matK+rbcL had a total of 70 sequences, with a single shared haplotype (Hd = 0.9996). A summary of the processed molecular data of this study is available in Table S1 of the Supplementary Materials.
Table 2.
Leguminosae tree species from Miombo and Mopane ecosystems used in the phylogenetic analyses. GenBank accession numbers for the corresponding internal transcribed spacer (ITS) and cpDNA (rbcL and matK) sequences.
| Taxon | Subfamily | rbcL | matK | ITS |
|---|---|---|---|---|
| Acacia arenaria Schinz | Caesalpinioideae | JX572181 | JX517408 | - |
| Acacia erubescens Welw. ex Oliv. | Caesalpinioideae | JF265248 | JF270605 | JQ265878 |
| Acacia galpinii Burtt Davy | Caesalpinioideae | JX572194 | JX518092 | JQ265866 |
| Acacia gerrardii Benth. | Caesalpinioideae | JF265250 | JF270607 | JQ265879 |
| Acacia hebeclada DC. | Caesalpinioideae | JX572199 | JX517672 | JQ265920 |
| Acacia kirkii Oliv. | Caesalpinioideae | JX572204 | JX517387 | JQ265829 |
| Acacia mellifera (Vahl) Benth. | Caesalpinioideae | JX572211 | JX517310 | - |
| Acacia nigrescens Oliv. | Caesalpinioideae | EU213440 | EU214210 | KY688811 |
| Acacia nilotica (L.) Willd. ex Delile | Caesalpinioideae | JF265255 | JF270612 | JX139101 |
| Acacia polyacantha | Caesalpinioideae | - | - | JQ265902 |
| Acacia robusta Burch. | Caesalpinioideae | JX572222 | JX517547 | - |
| Acacia sieberiana DC. | Caesalpinioideae | JF265259 | JX517353 | JQ265854 |
| Acacia welwitschii Oliv. | Caesalpinioideae | JX572234 | JX518159 | - |
| Afzelia quanzensis Welw. | Detarioideae | JX572247 | JX518045 | KY306488 |
| Albizia adianthifolia (Schum.) W.Wight | Caesalpinioideae | JQ025020 | JQ024935 | - |
| Albizia anthelmintica Brongn. | Caesalpinioideae | JX572254 | JX517977 | - |
| Albizia antunesiana Harms | Caesalpinioideae | - | - | - |
| Albizia glaberrima (Schum. & Thonn.) Benth. | Caesalpinioideae | JX572256 | JX518104 | - |
| Albizia harveyi E.Fourn. | Caesalpinioideae | JX572257 | JX518176 | - |
| Albizia versicolor Oliv. | Caesalpinioideae | JX572260 | AF274210 | - |
| Amblygonocarpus andongensis (Oliv.) Exell & Torre | Caesalpinioideae | JX572301 | AF521812 | - |
| Baikiaea plurijuga Harms | Detarioideae | JX572322 | JX517704 | KY306501 |
| Baphia bequaertii De Wild. | Papilionoideae | - | - | - |
| Baphia massaiensis Taub. | Papilionoideae | JF265298 | JF270652 | - |
| Bauhinia petersiana Bolle | Cercidoideae | JX572327 | JX517937 | - |
| Bauhinia thonningii Schum. | Cercidoideae | KU568124 | KT461985 | - |
| Bauhinia tomentosa L. | Cercidoideae | JX572328 | JX517621 | KX057838 |
| Bobgunnia madagascariensis (Desv.) J.H.Kirkbr. & Wiersema | Papilionoideae | JX572335 | AY386940 | EF560800 |
| Brachystegia allenii Hutch. & Burtt Davy | Detarioideae | KU568100 | KX146320 | - |
| Brachystegia bakeriana Burtt Davy & Hutch. | Detarioideae | - | - | - |
| Brachystegia boehmii Taub | Detarioideae | JX572347 | EU361886 | KY306513 |
| Brachystegia floribunda Benth. | Detarioideae | KU568148 | KX146363 | KY306515 |
| Brachystegia gossweileri Burtt Davy & Hutch. | Detarioideae | - | - | - |
| Brachystegia longifolia Benth. | Detarioideae | KU568078 | KX146300 | AF513687 |
| Brachystegia puberula Burtt Davy & Hutch. | Detarioideae | - | - | - |
| Brachystegia spiciformis Benth | Detarioideae | - | EU361888 | KY306518 |
| Brachystegia tamarindoides Benth. | Detarioideae | - | - | - |
| Brachystegia taxifolia Harms | Detarioideae | - | - | - |
| Brachystegia utilis Hutch. & Burtt Davy | Detarioideae | - | - | - |
| Brachystegia wangermeeana De Wild. | Detarioideae | - | - | - |
| Burkea africana Hook. | Caesalpinioideae | JQ025025 | JQ024940 | KX057840 |
| Cassia abbreviata Oliv. | Caesalpinioideae | JX572384 | JF270682 | - |
| Colophospermum mopane (Benth.) Leonard | Detarioideae | JF265343 | JF270696 | AY955788 |
| Cordyla africana Lour. | Papilionoideae | JF265371 | KP177913 | - |
| Craibia zimmermannii (Harms) Dunn | Papilionoideae | JX572478 | JX518072 | - |
| Dalbergia arbutifolia Baker | Papilionoideae | JX572499 | JX517956 | AB828608 |
| Dalbergia boehmii Taub. | Papilionoideae | JX572501 | JX517962 | AB828617 |
| Dalbergia melanoxylon Guill. & Perr. | Papilionoideae | KU748232 | KY484235 | KM276150 |
| Dalbergia nitidula Baker | Papilionoideae | - | JX970899 | - |
| Dialium englerianum Henriq. | Dialioideae | - | - | - |
| Dichrostachys cinerea (L.) Wight & Arn. | Caesalpinioideae | JQ025041 | KX302328 | AF458820 |
| Elephantorrhiza goetzei (Harms) Harms | Caesalpinioideae | JX572549 | JX517358 | - |
| Entada abyssinica Steud. ex A. Rich. | Caesalpinioideae | JX572556 | AF521829 | KX057869 |
| Erythrophleum africanum (Benth.) Harms | Caesalpinioideae | JX572568 | JX517525 | - |
| Faidherbia albida (Delile) A.Chev. | Caesalpinioideae | KX119293 | KX119382 | KX057872 |
| Guibourtia coleosperma (Benth.) J. Léonard | Detarioideae | JX572650 | JX518076 | - |
| Isoberlinia angolensis (Benth.) Hoyle & Brenan | Detarioideae | KU568126 | KX146343 | HM041837 |
| Isoberlinia scheffleri (Harms) Greenway | Detarioideae | AM234240 | EU361983 | HM041838 |
| Isoberlinia tomentosa (Harms) Craib & Stapf | Detarioideae | KX119306 | KX162205 | KX057885 |
| Julbernardia globiflora (Benth.) Troupin | Detarioideae | JX572701 | JX517829 | - |
| Julbernardia paniculata (Benth.) Troupin | Detarioideae | KU568145 | KX146360 | - |
| Millettia stuhlmannii Taub. | Papilionoideae | JX572773 | JX517411 | - |
| Millettia usaramensis Taub. | Papilionoideae | JX905971 | JX905956 | - |
| Mundulea sericea (Willd.) A. Chev. | Papilionoideae | JQ025063 | JQ024975 | AF467482 |
| Ormocarpum trichocarpum (Taub.) Engl. | Papilionoideae | JX572810 | JX517885 | AF068158 |
| Peltophorum africanum Sond. | Caesalpinioideae | JX572846 | KX302342 | - |
| Pericopsis angolensis (Baker) Meeuwen | Papilionoideae | KU568030 | KX584412 | KX584402 |
| Philenoptera bussei (Harms) Shrire | Papilionoideae | JX572848 | JX518116 | - |
| Philenoptera nelsii (Schinz) Schrire | Papilionoideae | - | - | - |
| Philenoptera violacea (Klotze) Shrire | Papilionoideae | JF265547 | JF270890 | JX506439 |
| Pterocarpus angolensis DC. | Papilionoideae | KY829237 | KY829168 | KY829139 |
| Pterocarpus brenanii Barbosa & Torre | Papilionoideae | JX572903 | JN083540 | JN083475 |
| Pterocarpus lucens Guill. & Perr. | Papilionoideae | KU568062 | KX146285 | JN083486 |
| Pterocarpus rotundifolius (Sond.) Druce | Papilionoideae | JF265565 | JF270907 | JN083509 |
| Senna petersiana (Bolle) Lock | Caesalpinioideae | JF265596 | JX517765 | - |
| Tamarindus indica L. | Detarioideae | AB378732 | JQ587877 | KY306654 |
| Xanthocercis zambesiaca (Baker) Dumaz-le-Grand | Papilionoideae | JX573092 | JX517427 | - |
| Xeroderris stuhlmannii (Taub.) Mendonça & E.P. Sousa | Papilionoideae | JX573093 | JX517470 | AF467485 |
For each data matrix, two independent phylogenies were constructed, one using maximum likelihood (ML), and another using Bayesian inference (BI). In both methods, tree rooting was performed using Polygalaceae species as outgroups, namely Monnina xalapensis, Rhinotropis acanthoclada and Xanthophyllum hainanense. Overall, the BI phylogenies achieved higher support values, but were in turn less resolved, with more polytomies than ML trees. Topologies were highly similar within the same paired datasets, and to a lesser extent between locus, showing similar clustering patterns between species.
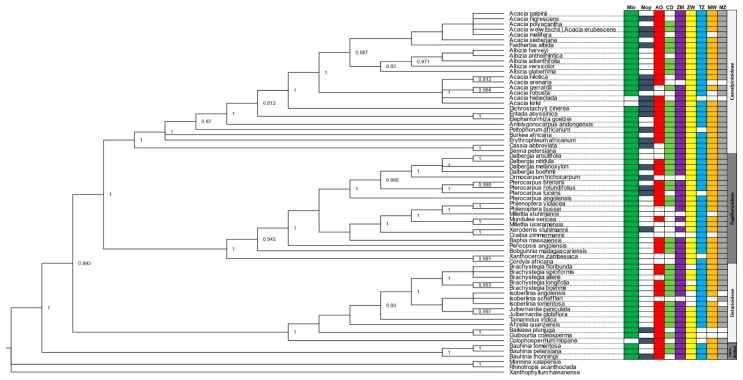
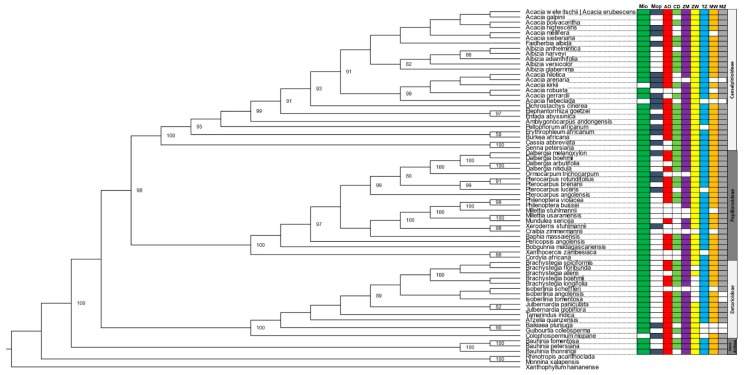
The phylogenetic trees obtained using the matK matrix provided the best resolution and support from the singular gene analysis, showing a clear split between subfamilies and clustering between closely resembling species (Figure S1). ITS and rbcL phylogenies were generally less supported in both ML and BI analysis (Figures S2 and S3), and some taxa were also poorly positioned (i.e., found outside of the subfamily clade). The concatenated set of the three selected genes (ITS + matK + rbcL) achieved the best overall result both using BI (Figure 3) and ML (Figure 4), with a lower number of unresolved nodes, as well as higher values of support, following the clustering pattern obtained by The Legume Phylogeny Working Group [23].
Figure 3.
Phylogenetic tree of the Leguminosae tree species from Miombo and Mopane ecosystems. The phylogeny was constructed with three selected genes (ITS + matK + rbcL) using the Bayesian inference (BI) method. Acronyms for the countries/regions: Mio—Miombo; Mop—Mopane; AO—Angola; DRC—Democratic Republic of the Congo; ZM—Zambia; ZW—Zimbabwe; TZ—United Republic of Tanzania; MW—Malawi; MZ—Mozambique.
Figure 4.
Phylogenetic tree of the Leguminosae tree species from Miombo and Mopane ecosystems. The phylogeny was constructed with three selected genes (ITS + matK + rbcL) and the maximum likelihood (ML) method. Acronyms for the countries/regions: Mio—Miombo; Mop—Mopane; AO—Angola; DRC—Democratic Republic of the Congo; ZM—Zambia; ZW—Zimbabwe; TZ—United Republic of Tanzania; MW—Malawi; MZ—Mozambique.
The distribution of species among ecoregions (Miombo and Mopane) was as predictable and in general evenly distributed across the seven countries. The smaller subfamily, Cercidoideae, was well represented in Miombo (3 out of 3 species), holding a single taxon (Bauhinia thonningii), which was found in Mopane as well. The Detarioideae subfamily included 15 species, one common to both Miombo and Mopane (Baikiaea plurijuga), one exclusively from Mopane (Colophospermum mopane) and 13 exclusively from Miombo (Brachystegia spp., Isoberlinea spp., Julbernardia spp., Tamarindus indica and Guibourtia coleosperma). One of these species, I. scheffleri, was restricted to Tanzania and Mozambique, while B. plurijuga and G. coleosperma were not present in Tanzania, Malawi, Mozambique and the DRC. The 21 species belonging to the Papilionoideae were all present in Miombo, five of them (Dalbergia melanoxylon, Ormocarpum trichocarpum, Pterocarpus rotundifolius, Pterocarpus lucens, and Xeroderris stuhlmannii) being also present in Mopane. Three species, O. trichocarpum, Millettia stuhlmannii and M. usaramensis were present only in the eastern countries. The largest subfamily Caesalpinioideae, included 25 species, mostly Acacia spp. and Albizia spp. homogeneously distributed by country and ecoregion (Figure 3 and Figure 4).
3. Discussion
As expected, the Miombo woodlands presented a higher diversity of species (74 overall taxa, 55 Miombo-exclusive) than Mopane (23 taxa, 4 Mopane-exclusive) [17,33]. This is likely related to the larger area and therefore to a rather diverse edaphic (e.g., drainage, soil depth and texture) and climate (warm to hot climate, 710 to 1365 mm mean annual precipitation and 18 to 23 °C mean annual temperature) conditions in the Miombo ecoregion [9,34]. Additionally, Mopane is usually characterized by clayed soils [33] with a discontinuous tree cover and a continuous C4 grass layer [35,36]. Thus, environmental determinants might have underlined a slight deviation on the evolutionary history of Miombo and Mopane tree legumes [37]. Also expected was the exclusive presence of the typically dominant genera of Miombo (Brachystegia, Isoberlinia and Julbernardia) and Mopane (Colophospermum mopane) in either ecosystem [17,19,24].
When comparing the results with the combined areas of Miombo and Mopane woodlands there was not a logical correlation between area size and species number in some countries. This was particularly evident in Angola, which holds the largest woodland area of Miombo and Mopane but houses only 62 out of the 78 species. Other large countries such as Zambia and Mozambique however, did not follow this trend, as the identified species were well represented. These results could be explained by three hypotheses: i) the origin point of dispersal for the Leguminosae family was Zambia and/or Zimbabwe with a more recent expansion to Angola, compared to the other neighboring countries; ii) the knowledge on species diversity in Angola is incipient; or iii) a combination of the two. Among the seven countries included in this study, the highest species diversity was found in Zambia (i.e., 71 out of the 78 scored species, and 16 out of 17 typical Miombo species), likely related to the fact that Zambia is the center of endemism for Brachystegia [9]. Except for Zambia, the number of taxa was nearly the same for countries with dry Miombo (Malawi, Mozambique and Zimbabwe) and wet Miombo (Angola, DRC, Tanzania and Malawi). However, it is important to consider that the scored diversity in this study does not reflect species abundance and frequency, which could explain this similarity despite the fact that wet Miombo is often a floristically richer region [9,38].
The rarity of a certain species or ecosystem is frequently the first and most important feature when deciding its need of protection, as is the higher risk of extinction and loss of possible unique lineages [39,40,41]. This feature is particularly significant when information on the threat status of a species is insufficient [42] as is the case of Miombo and Mopane tree legumes, whose conservation status is available for a minority of species (Table 1) [43]. Such a trend was also found for other tree lineages endemic from Africa such as the nitrogen-fixing actinorhizal trees and shrubs [44]. This issue is of upmost importance within the context of the Bonn Challenge under which many countries have pledged to restore millions of degraded and deforested woodlands and forests [45]. Thus, more efforts are needed to investigate the vegetation dynamics, anthropogenic and environmental drivers as well as the different conservation management strategies across Miombo and Mopane countries. Examples of such efforts include the recent work of, (i) Chiteculo and Surovy [46] and Chiteculo et al. [47], that characterized the vegetation composition and structure and deforestation patterns of the Miombo woodlands in the Huambo province, Angola, respectively; (ii) Ribeiro et al. [6] that conducted a 12-year analysis of the spatio-temporal patterns of fire to refine the fire management strategy in one of the most pristine areas of Miombo, the Niassa National Reserve, Mozambique; (iii) Mugasha et al. [48] that provided a pioneer study on modeling tree growth in the Miombo woodlands from Tanzania based on long-term monitoring data; and (iii) Chidumayo [49] that performed a long-term study (1982–2018) across the Miombo woodlands in Zambia to investigate the woodland drivers and contribute to the design of management plans.
The use of phylogenetic diversity as an effective complementary mean of conservation has often been the object of debate. Since the proposal of this metric by Vane-Wright [50], several studies have been able to test and evaluate this hypothesis with various results and opinions [51,52]. Regarding molecular data, the results obtained in this study corroborate previous molecular findings showing that the use of taxonomic barcodes or DNA barcoding have great applicability on the identification, phylogeny reconstruction and evolutionary analysis for forest dwelling flora [53,54,55], in our case tree legumes from the Miombo and Mopane ecoregions. The results are in line with a recent study by The Legume Phylogeny Working Group [23], clustering together four out of the six new subfamilies: Caesalpinioideae, Cercidoideae, Detarioideae and Papilionoideae (Figure 3 and Figure 4).
Functional diversity is another useful parameter to assess phylogenetic diversity and the evolutionary potential (i.e., the ability of a species to adapt to environmental changes) [56,57,58,59]. With few exceptions, the distribution of the 67 lineages and subfamilies across the seven countries was quite uniform and therefore, not informative regarding their evolutionary history across the Mopane and Miombo axis. This was particularly the case of the typically dominant genera of Miombo (i.e., Brachystegia, Isoberlinia and Julbernardia) and Mopane (C. mopane). Thus, further studies are needed in order to assess the genetic diversity and population structure of key species from Miombo and Mopane. Such complementary studies will be essential to provide better and more well-founded areas of protection for the Miombo and Mopane woodlands. In some countries where protected areas are still scarce, such as Angola, this might support the establishment of a network of protected areas spanning different sub-regions.
In conclusion, the Miombo and Mopane woodlands hold a differential phylogenetic diversity: the Miombo covers a larger area and holds a higher number of legume species; while the Mopane spans a smaller land mass that houses several unique and rare lineages. Both ecoregions hold a high value of biodiversity, even with a somewhat dissimilar composition, and as such future studies should take in account their exclusive characteristics alongside their shared ones when proposing new conserved species and areas.
4. Materials and Methods
4.1. Study Area and Spatial Analyses
The study area included the Mopane and Miombo ecoregions from the Zambezian phytoregion (i.e., Angola, Democratic Republic of Congo (DRC), Zambia, Zimbabwe, Tanzania, Malawi, and Mozambique) (Figure 1). Only the southern provinces of DRC (i.e., Tanganyika, Haut-Lomami, Lualaba, and Haut-Katanga (former province of Katanga, extinguished in 2009)), were included in this study, as only these four provinces are part of the Zambezian phytoregion [60]. The map of the terrestrial World Wildlife Fund (WWF) ecoregions [61] and the five Miombo and two Mopane sub-regions were analyzed and overlapped with the digital maps of the Zambezian region.
4.2. Database of Legume Trees
A list of the native tree legume species (Leguminosae family) from the Mopane and Miombo woodlands was created through an extensive research in scientific publications [6,12,17,33,62] and references therein and online databases, such as the African Plant Database [63], Plants of the World Online [64], Flora of Mozambique [65], Flora of Zambia [66], Flora of Malawi [67] and Flora of Zimbabwe [68]. Scientific names were updated according to The Plant List [69], while subfamilies were compiled using the organization defined by the The Legume Phylogeny Working Group (LPWG) [23]. The main distribution in the Zambezian phytoregion, species’ habit, conservation status and molecular data were also searched and compiled. Distribution data was attained in several bibliographic sources [70,71,72,73,74] and online databases, namely the Global Biodiversity Information Facility (GBIF) platform [75], and the African Plant Database [63]. The conservation status of each species was consulted in the International Union for Conservation of Nature—Red List [40].
4.3. Sequence Data and Phylogenetic Analysis
To perform the molecular analysis, three markers were selected: two chloroplast genes often used in plant barcoding, rbcL and matK [76,77]; and the internal transcribed spacer (ITS) region, which has been shown to be a useful complementary barcode [78].
The software Geneious Prime 2019.0.3 [79] was used to retrieve the selected DNA sequences from the National Center for Biotechnology Information (NCBI) GenBank database, and each locus dataset was subsequently aligned using the multiple sequence alignment tool MAFFT [80], available online [81]. The haplotype diversity of the data was verified using DNASP6 v.6.12.01 [82], and then processed with trimAl v.1.3 [83], through the Phylemon2 framework [84], to remove poorly aligned regions and improve the quality of the alignments. A concatenated set using the three loci was created using Concatenator [85] to assess the collective phylogeny of the selected taxa. PartitionFinder2 v.2.1.1 [86,87,88] was used to find the adequate partitioning and models of evolution for the four groups (three single and one combined), using the corrected Akaike Information Criterion (AICc) as the model, and partitioning by gene and codon position.
Phylogeny reconstructions were made using two different methodologies for the individual and combined locus datasets, namely maximum likelihood (ML) and Bayesian inference (BI). In both cases, species from the Polygalaceae family, a sister group of Leguminoseae, were selected as outgroups (Monnina xalapensis, Rhinotropis acanthoclada and Xanthophyllum hainanense, the latter functioning as the most exterior outgroup) [89]. ML analyses were performed using RAxML v.8.2.10 [90], through RaxmlGUI v.1.5b [91], using a ML + rapid bootstrap search and an autoMRE bootstrap. BI analyses were made using MrBayes v.3.2.6 [92], with 1.5 × 107 generations and a sample every 100 steps, with default chains and temperature. Convergence on all parameters was verified using Tracer v.1.6 [93] across all runs. The BI analyses were performed using the Cipres Gateway services [94]. Finally, we summarized, annotated and later exported the resulting phylogenetic trees using FigTree v.1.4.3 [95].
Acknowledgments
The authors are thankful to Luis Catarino (Faculty of Science, University of Lisbon) for providing the photographs for Figure 2.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/6/182/s1, Figure S1: The phylogenetic trees obtained using the matK matrix with ML (A) and BI (B). Figure S2: The phylogenetic trees obtained using the rcbL matrix with ML(A) and BI (B). Figure S3: The phylogenetic trees obtained using the ITS matrix with ML (A) and BI (B). Table S1: Summary of the analyzed molecular data.
Author Contributions
Conceptualization, M.M.R. and A.I.R.-B.; methodology, I.M., S.C., A.R.P.; software, D.B., A.R.P.; validation, I.M., S.C., N.R., M.M.R., A.I.R.-B.; formal analysis, I.M., S.C.; investigation, I.M., S.C.; resources, D.B., A.R.P.; data curation, M.M.R., A.I.R.-B., N.S.R.; writing—original draft preparation, I.M., S.C.; writing—review and editing, I.M., S.C., M.M.R., N.S.R., A.I.R.-B.; visualization, S.C., A.R.P.; supervision, N.S.R., M.M.R., A.I.R.-B.; project administration and funding acquisition, M.M.R., N.S.R., A.I.R.-B.
Funding
This research was funded by Fundo para a Investigação Aplicada e Multissectorial (Mozambique and Italy) and Portuguese national funds from Camões, Instituto da Cooperação e da Língua and Fundação para a Ciência e a Tecnologia (research units UID/GEO/04035/2013-GeoBioTec and UID/AGR/04129/2013-LEAF; and the grants SFRH/BD/113951/2015 to IM and SFRH/BD/120054/2016 to SC).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Murphy P.G., Lugo A.E. Ecology of tropical dry forest. Annu. Rev. Ecol. Syst. 1986;17:67–88. doi: 10.1146/annurev.es.17.110186.000435. [DOI] [Google Scholar]
- 2.Grace J., José J.S., Meir P., Miranda H.S., Montes R.A. Productivity and carbon fluxes of tropical savannas. J. Biogeogr. 2006;33:387–400. doi: 10.1111/j.1365-2699.2005.01448.x. [DOI] [Google Scholar]
- 3.Mittermeier R.A., Mittermeier C.G., Brooks T.M., Pilgrim J.D., Konstant W.R., da Fonseca G.A.B., Kormos C. Wilderness and biodiversity conservation. Proc. Natl. Acad. Sci. USA. 2003;100:10309–10313. doi: 10.1073/pnas.1732458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syampungani S., Chirwa P.W., Akinnifesi F.K., Sileshi G., Ajayi O.C. The miombo woodlands at the cross roads: Potential threats, sustainable livelihoods, policy gaps and challenges. Nat. Resour. Forum. 2009;33:150–159. doi: 10.1111/j.1477-8947.2009.01218.x. [DOI] [Google Scholar]
- 5.Djoudi H., Vergles E., Blackie R.R., Koame C.K., Gautier D. Dry forests, livelihoods and poverty alleviation: Understanding current trends. Int. For. Rev. 2015;17:54–69. doi: 10.1505/146554815815834868. [DOI] [Google Scholar]
- 6.Moura I., Maquia I., Rija A.A., Ribeiro N., Ribeiro-Barros A.I. Biodiversity studies in key species from the African mopane and miombo woodlands. In: Bitz L., editor. Genetic Diversity. IntechOpen; London, UK: 2017. pp. 91–109. [Google Scholar]
- 7.Moura I., Duvane J.A., Silva M.J.E., Ribeiro N., Ribeiro-Barros I. Woody species from the Mozambican Miombo woodlands: A review on their ethnomedicinal uses and pharmacological potential. J. Med. Plants Res. 2018;12:15–31. doi: 10.5897/JMPR2017.6540. [DOI] [Google Scholar]
- 8.Ribeiro-Barros A.I., Silva M.J., Moura I., Ramalho J.C., Máguas-Hanson C., Ribeiro N.S. The Potential of Tree and Shrub Legumes in Agroforestry Systems. In: Amanullah K., Fahad S., editors. Nitrogen in Agriculture-Updates. IntechOpen; London, UK: 2017. pp. 223–239. [Google Scholar]
- 9.Frost P. The Ecology of Miombo Woodlands. In: Campbell B., editor. The Miombo in Transition: Woodlands and Welfare in Africa. Center for International Forestry Research (CIFOR); Bogor, Indonesia: 1996. pp. 11–57. [Google Scholar]
- 10.Ribeiro N.S., Saatchi S.S., Shugart H.H., Washington-Alen R.A. Aboveground biomass and leaf area index (LAI) mapping for Niassa Reserve, northern Mozambique. J. Geophys. Res. 2008;113:G02S02. doi: 10.1029/2007JG000550. [DOI] [Google Scholar]
- 11.Ribeiro N.S., Shugart H.H., Washington-Allen R. The effects of fire and elephants on species composition and structure of the Niassa Reserve, northern Mozambique. J. For. Ecol. Manag. 2008;255:1626–1636. doi: 10.1016/j.foreco.2007.11.033. [DOI] [Google Scholar]
- 12.Marunda C., Chidumayo E. Dry forests and woodlands in sub-Saharan Africa: Context and challenges. In: Chidumayo E.N., Gumbo D.J., editors. The Dry Forests and Woodlands of Africa. Earthscan; London, UK: 2010. pp. 14–22. [Google Scholar]
- 13.Ribeiro N.S., Syampungani S., Nangoma D., Ribeiro-Barros A. Miombo Woodlands Research Towards the Sustainable use of Ecosystem Services in Southern Africa. In: Lo Y., Blanco J.A., Roy S., editors. Biodiversity in Ecosystems-Linking Structure and Function. IntechOpen; London, UK: 2015. pp. 475–491. [Google Scholar]
- 14.Linder H.P. Plant diversity and endemism in sub-Saharan tropical Africa. J. Biogeogr. 2001;28:169–182. doi: 10.1046/j.1365-2699.2001.00527.x. [DOI] [Google Scholar]
- 15.Linder H.P. The evolution of African plant diversity. Front. Ecol. Evol. 2014;2:38:1–38:14. doi: 10.3389/fevo.2014.00038. [DOI] [Google Scholar]
- 16.Olson D.M., Dinerstein E., Wikramanayake E.D., Burgess N.D., Powell G.V.N., Underwood E.C., D’amico J.A., Itoua I., Strand H.E., Morrison J.C., et al. Terrestrial Ecoregions of the World: A New Map of Life on Earth: A new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioScience. 2001;51:933–938. doi: 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2. [DOI] [Google Scholar]
- 17.Burgess N., Hales J.A., Underwood E., Dinerstein E., Olson D., Itoua I., Schipper J., Ricketts T., Newman K. Terrestrial Ecoregions of Africa and Madagascar: A Conservation Assessment. Island Press; Washington, DC, USA: 2004. [Google Scholar]
- 18.Dewees P.A., Campbell B.M., Katerere Y., Sitoe A., Cunningham A.B., Angelsen A., Wunder S. Managing the miombo woodlands of southern Africa: Policies, incentives and options for the rural poor. J. Nat. Resour. Policy Res. 2010;2:57–73. doi: 10.1080/19390450903350846. [DOI] [Google Scholar]
- 19.Makhado R.A., Mapaure I., Potgieter M.J., Luus-Powell W.J., Saidi A.T. Factors influencing the adaptation and distribution of Colophospermum mopane in southern Africa’s mopane savannas—A review. Bothalia-Afr. Biodivers. Conserv. 2014;44:1–9. doi: 10.4102/abc.v44i1.152. [DOI] [Google Scholar]
- 20.Lewis G.P., Schrire B.D., Mackinder B.A., Rico L., Clark R. A 2013 linear sequence of legume genera set in a phylogenetic context—A tool for collections management and taxon sampling. S. Afr. J. Bot. 2013;89:76–84. doi: 10.1016/j.sajb.2013.06.005. [DOI] [Google Scholar]
- 21.LPWG Towards a new classification system for legumes: Progress report from the 6th International Legume Conference. S. Afr. J. Bot. 2013;89:3–9. doi: 10.1016/j.sajb.2013.07.022. [DOI] [Google Scholar]
- 22.LPWG Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich clades. Taxon. 2013;62:217–248. doi: 10.12705/622.8. [DOI] [Google Scholar]
- 23.LPWG A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon. 2017;66:44–77. doi: 10.12705/661.3. [DOI] [Google Scholar]
- 24.Jew E.K., Dougill A.J., Sallu S.M., O’Connell J., Benton T.G. Miombo woodland under threat: Consequences for tree diversity and carbon storage. For. Ecol. Manag. 2016;361:144–153. doi: 10.1016/j.foreco.2015.11.011. [DOI] [Google Scholar]
- 25.Graham P.H., Vance C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catarino S., Duarte M.C., Costa E., Carrero P.G., Romeiras M.M. Conservation and sustainable use of the medicinal Leguminosae plants from Angola. PeerJ. 2019;7:e6736. doi: 10.7717/peerj.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chirwa P.W., Syampungani S., Geldenhuys C.J. Managing southern African woodlands for biomass production: The potential challenges and opportunities. In: Seifert T., editor. Bioenergy from Wood. Springer; Dordrecht, The Netherlands: 2014. pp. 67–87. [Google Scholar]
- 28.Romeiras M.M., Figueira R., Duarte M.C., Beja P., Darbyshire I. Documenting biogeographical patterns of African timber species using herbarium records: A conservation perspective based on native trees from Angola. PLoS ONE. 2014;9:e103403. doi: 10.1371/journal.pone.0103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y., Chen S., Hu G., Mwachala G., Yan X., Wang Q. Species richness and phylogenetic diversity of seed plants across vegetation zones of Mount Kenya, East Africa. Ecol. Evol. 2018;8:8930–8939. doi: 10.1002/ece3.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy O.J., Senterre B. Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. J. Ecol. 2007;95:493–506. doi: 10.1111/j.1365-2745.2007.01222.x. [DOI] [Google Scholar]
- 31.Winter M., Devictor V., Schweiger O. Phylogenetic diversity and nature conservation: Where are we? Trends Ecol. Evol. 2013;28:199–204. doi: 10.1016/j.tree.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Forest F., Grenyer R., Rouget M., Davies T.J., Cowling R.M., Faith D.P., Balmford A., Manning J.C., Procheş S., van der Bank M., et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- 33.Timberlake J., Chidumayo E., Sawadogo L. Distribution and Characteristics of African Dry Forests and Woodlands. In: Chidumayo E.N., Gumbo D., editors. The Dry Forests and Woodlands of Africa: Managing for Products and Services. Earthscan; London, UK: 2010. pp. 11–41. [Google Scholar]
- 34.Meerts P. An annotated checklist to the trees and shrubs of the Upper Katanga (DR Congo) Phytotaxa. 2016;258:201–250. doi: 10.11646/phytotaxa.258.3.1. [DOI] [Google Scholar]
- 35.Scholes R.J., Archer S.R. Tree-grass interactions in savannas. Annu. Rev. Ecol. Syst. 1997;28:517–544. doi: 10.1146/annurev.ecolsys.28.1.517. [DOI] [Google Scholar]
- 36.Ratnam J., Bond W.J., Fensham R.J., Hoffmann W.A., Archibald S., Lehmann C.E.R., Anderson M.T., Higgins S.I., Sankaran M. When is a ‘forest’ a savanna, and why does it matter? Glob. Ecol. Biogeogr. 2011;20:653–660. doi: 10.1111/j.1466-8238.2010.00634.x. [DOI] [Google Scholar]
- 37.Daru B.H., Van der Bank M., Davies T.J. Unravelling the evolutionary origins of biogeographic assemblages. Divers. Distrib. 2018;24:313–324. doi: 10.1111/ddi.12679. [DOI] [Google Scholar]
- 38.White F. The Vegetation of Africa. Unesco; Paris, France: 1983. [Google Scholar]
- 39.Gauthier P., Debussche M., Thompson J.D. Regional priority setting for rare species based on a method combining three criteria. Biol. Conserv. 2010;143:1501–1509. doi: 10.1016/j.biocon.2010.03.032. [DOI] [Google Scholar]
- 40.IUCN Guidelines for Using the IUCN Red List Categories and Criteria, Version 13. Standards and Petitions Subcommittee. [(accessed on 14 February 2019)]; Available online: http://www.iucnredlist.org/documents/RedListGuidelines.pdf.
- 41.Romeiras M.M., Catarino S., Gomes I., Fernandes C., Costa J.C., Caujapé-Castells J., Duarte M.C. IUCN Red List assessment of the Cape Verde endemic flora: Towards a global strategy for plant conservation in Macaronesia. Bot. J. Linn. Soc. 2016;180:413–425. doi: 10.1111/boj.12370. [DOI] [Google Scholar]
- 42.Isaac N.J., Redding D.W., Meredith H.M., Safi K. Phylogenetically-Informed Priorities for Amphibian Conservation. PLoS ONE. 2012;7:e43912. doi: 10.1371/journal.pone.0043912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.IUCN The IUCN Red List of Threatened Species. Version 2017-3. [(accessed on 3 June 2018)]; Available online: http://www.iucnredlist.org.
- 44.Ribeiro-Barros A.I., Catarino S., Moura I., Ramalho J.C., Romeiras M.M., Ghodhbane-Gtari F. Actinorhizal trees and shrubs from Africa: Distribution, conservation and uses. Antonie Leeuwenhoek. 2019;112:31–46. doi: 10.1007/s10482-018-1174-x. [DOI] [PubMed] [Google Scholar]
- 45.The Bonn Challenge. [(accessed on 19 June 2019)]; Available online: http://www.bonnchallenge.org.
- 46.Chiteculo V., Surovy P. Dynamic Patterns of Trees Species in Miombo Forest and Management Perspectives for Sustainable Production—Case Study in Huambo Province, Angola. Forests. 2018;9:321. doi: 10.3390/f9060321. [DOI] [Google Scholar]
- 47.Chiteculo V., Abdollahnejad A., Panagiotidis D., Surový P., Sharma R. Defining Deforestation Patterns Using Satellite Images from 2000 and 2017: Assessment of Forest Management in Miombo Forests—A Case Study of Huambo Province in Angola. Sustainability. 2019;11:98. doi: 10.3390/su11010098. [DOI] [Google Scholar]
- 48.Mugasha W.A., Eid T., Bollandsås O.M., Mbwambo L. Modelling diameter growth, mortality and recruitment of trees in miombo woodlands of Tanzania. South. For. A J. For. Sci. 2017;9:51–64. doi: 10.2989/20702620.2016.1233755. [DOI] [Google Scholar]
- 49.Chidumayo E.N. Management implications of tree growth patterns in miombo woodlands of Zambia. For. Ecol. Manag. 2019;436:105–116. doi: 10.1016/j.foreco.2019.01.018. [DOI] [Google Scholar]
- 50.Vane-Wright R.I., Humphries C.J., Williams P.H. What to protect?—Systematics and the agony of choice. Biol. Conserv. 1991;55:235–254. doi: 10.1016/0006-3207(91)90030-D. [DOI] [Google Scholar]
- 51.Pollock L.J., Rosauer D.F., Thornhill A.H., Kujala H., Crisp M.D., Miller J.T., McCarthy M.A. Phylogenetic diversity meets conservation policy: Small areas are key to preserving eucalypt lineages. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140007. doi: 10.1098/rstb.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazel F., Pennell M.W., Cadotte M.W., Diaz S., Dalla Riva G.V., Grenyer R., Leprieur F., Mooers A.O., Mouillot D., Tucker C.M., et al. Prioritizing phylogenetic diversity captures functional diversity unreliably. Nat. Commun. 2018;9:2888:1–2888:9. doi: 10.1038/s41467-018-05126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kress W.J., Erickson D.L., Swenson N.G., Thompson J., Uriarte M., Zimmerman J.K. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS ONE. 2010;5:e15409. doi: 10.1371/journal.pone.0015409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang Y., Deng Z., Zang R., Long W. DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests. Sci. Rep. 2017;7:12564. doi: 10.1038/s41598-017-13057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Liu J., Shan Y.X., Ge X.J., Burgess K.S. The use of DNA barcodes to estimate phylogenetic diversity in forest communities of southern China. Ecol. Evol. 2019;9:5372–5379. doi: 10.1002/ece3.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krajewski C. Phylogeny and Diversity. Science. 1991;254:918. doi: 10.1126/science.254.5034.918-a. [DOI] [Google Scholar]
- 57.Losos J.B. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 58.Srivastava D.S., Cadotte M.W., MacDonald A.A.M., Marushia R.G., Mirotchnick N. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 2012;15:637–648. doi: 10.1111/j.1461-0248.2012.01795.x. [DOI] [PubMed] [Google Scholar]
- 59.Mouquet N., Devictor V., Meynard C.N., Munoz F., Bersier L.F., Chave J., Couteron P., Dalecky A., Fontaine C., Gravel D., et al. Ecophylogenetics: Advances and perspectives. Biol. Rev. 2012;87:769–785. doi: 10.1111/j.1469-185X.2012.00224.x. [DOI] [PubMed] [Google Scholar]
- 60.Linder H.P., De Klerk H.M., Born J., Burgess N.D., Fjeldså J., Rahbek C. The partitioning of Africa: Statistically defined biogeographical regions in sub-Saharan Africa. J. Biogeogr. 2012;39:1189–1205. doi: 10.1111/j.1365-2699.2012.02728.x. [DOI] [Google Scholar]
- 61.World Wildlife Fund. [(accessed on 19 June 2019)]; Available online: https://www.worldwildlife.org/publications/terrestrial-ecoregions-of-the-world.
- 62.Figueiredo E., Smith G. Plants of Angola/Plantas de Angola. South African National Biodiversity Institute (SANBI Publishing); Pretoria, South Africa: 2008. [Google Scholar]
- 63.African Plant Database. [(accessed on 19 June 2019)]; Available online: http://www.ville-ge.ch/musinfo/bd/cjb/africa/index.php.
- 64.Plants of the World Online. [(accessed on 19 June 2019)]; Available online: http://powo.science.kew.org.
- 65.Flora of Mozambique. [(accessed on 19 June 2019)]; Available online: http://www.mozambiqueflora.com.
- 66.Flora of Zambia. [(accessed on 19 June 2019)]; Available online: http://www.zambiaflora.com.
- 67.Flora of Malawi. [(accessed on 19 June 2019)]; Available online: http://www.malawiflora.com.
- 68.Flora of Zimbabwe. [(accessed on 19 June 2019)]; Available online: http://www.zimbabweflora.co.zw.
- 69.The Plant List. [(accessed on 19 June 2019)]; Available online: http://www.theplantlist.org.
- 70.Brenan J.P.M. Flora Zambesiaca: Mozambique, Malawi, Zambia, Zimbabwe, Botswana. Volume Three: Part One. Crown Agents for Oversea Governments and Administration; London, UK: 1970. [Google Scholar]
- 71.Pope G.V. Flora Zambesiaca: Mozambique, Malawi, Zambia, Zimbabwe, Botswana. Royal Botanic Gardens; Kew, UK: 2000. Volume Three: Part Six. [Google Scholar]
- 72.Pope G.V., PolHill R.M. Flora Zambesiaca: Mozambique, Malawi, Zambia, Zimbabwe, Botswana. Royal Botanic Gardens; Kew, UK: 2001. Volume Nine: Part Five. [Google Scholar]
- 73.Sanfilippo M. Trinta Árvores e Arbustos do Miombo Angolano. Guia de Campo para a Identificação. COSPE; Firenze, Italy: 2013. [Google Scholar]
- 74.Urso V. Vinte Árvores e Arbustos do Mopane Angolano. Guia de Campo para a Identificação. COSPE; Firenze, Italy: 2013. [Google Scholar]
- 75.Global Biodiversity Information Facility (GBIF) [(accessed on 19 June 2019)]; Available online: http://www.gbif.org.
- 76.CBOL Plant Working Group. Hollingsworth P.M., Forrest L.L., Spouge J.L., Hajibabaei M., Ratnasingham S., Fazekas A.J. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Vere N., Rich T.C., Trinder S.A., Long C. DNA barcoding for plants. In: Batley J., editor. Plant Genotyping Methods in Molecular Biology (Methods and Protocols) Humana Press; New York, NY, USA: 2015. pp. 101–118. [DOI] [PubMed] [Google Scholar]
- 78.Tripathi A.M., Tyagi A., Kumar A., Singh A., Singh S., Chaudhary L.B., Roy S. The internal transcribed spacer (ITS) region and trnhH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE. 2013;8:e57934. doi: 10.1371/journal.pone.0057934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017:bbx108. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 83.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T., et al. Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pina-Martins F., Paulo O.S. Concatenator: Sequence data matrices handling made easy. Mol. Ecol. Resour. 2008;8:1254–1255. doi: 10.1111/j.1755-0998.2008.02164.x. [DOI] [PubMed] [Google Scholar]
- 86.Lanfear R., Calcott B., Ho S.Y., Guindon S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 87.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 88.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 89.Soltis D.E., Soltis P.S., Chase M.W., Mort M.E., Albach D.C., Zanis M., Savolainen V., Hahn W.H., Hoot S.B., Fay M.F., et al. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 2000;133:381–461. doi: 10.1111/j.1095-8339.2000.tb01588.x. [DOI] [Google Scholar]
- 90.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Silvestro D., Michalak I. RaxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 92.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 93.Rambaut A., Drummond A.J. Tracer v1.6. [(accessed on 1 March 2019)]; Available online: http://tree.bio.ed.ac.uk/software/tracer/
- 94.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 95.Rambaut A. FigTree, a Graphical Viewer of Phylogenetic Trees. Institute of Evolutionary Biology University of Edinburgh. [(accessed on 1 March 2019)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.