Abstract
Background
Adaptive responses to stress are essential for cell and organismal survival. In metazoans, little is known about the impact of environmental stress on RNA homeostasis.
Results
By studying the regulation of a cadmium-induced gene named numr-1 in Caenorhabditis elegans, we discovered that disruption of RNA processing acts as a signal for environmental stress. We find that NUMR-1 contains motifs common to RNA splicing factors and influences RNA splicing in vivo. A genome-wide screen reveals that numr-1 is strongly and specifically induced by silencing of genes that function in basal RNA metabolism including subunits of the metazoan integrator complex. Human integrator processes snRNAs for functioning with splicing factors, and we find that silencing of C. elegans integrator subunits disrupts snRNA processing, causes aberrant pre-mRNA splicing, and induces the heat shock response. Cadmium, which also strongly induces numr-1, has similar effects on RNA and the heat shock response. Lastly, we find that heat shock factor-1 is required for full numr-1 induction by cadmium.
Conclusion
Our results are consistent with a model in which disruption of integrator processing of RNA acts as a molecular damage signal initiating an adaptive stress response mediated by heat shock factor-1. When numr-1 is induced via this pathway in C. elegans, its function in RNA metabolism may allow it to mitigate further damage and thereby promote tolerance to cadmium.
Electronic supplementary material
The online version of this article (10.1186/s12915-019-0675-z) contains supplementary material, which is available to authorized users.
Keywords: Caenorhabditis elegans, Cadmium, snRNA, RNA splicing, Stress response, Surveillance mechanism
Introduction
For most eukaryotic genes, introns must be spliced out of precursor mRNA (pre-mRNA) to produce mature mRNA transcripts ready for translation [1], a process that is essential to organismal development and homeostasis [2, 3]. Splicing is predominantly conducted in the nucleus by the spliceosome and associated RNA-binding proteins that form a dynamic, extremely complicated, and poorly understood macromolecular complex of small nuclear RNAs (snRNAs) and up to 300 distinct proteins [4]. In the last decade, an additional metazoan-specific complex of at least 14 proteins, named integrator, was discovered that indirectly promotes splicing by processing snRNAs into their functional forms [5–7].
Although mechanisms for regulating protein homeostasis during environmental stress have been studied in great detail [8], little is known about the regulation of RNA homeostasis during stress. Disruption of pre-mRNA processing has been observed during environmental stress [9, 10], and recent studies have revealed associations and causative relationships between splicing fidelity and aging, cancer, and myotonic dystrophy [11–13]. Splicing is also being explored as a therapeutic target [13]. A molecular understanding of how environmental stress affects RNA processing and adaptive cellular responses would provide novel insights into this core eukaryotic cell process with broad relevance to cell homeostasis and disease.
Responses to environmental stress almost always include transcriptional activation of genes encoding proteins that mitigate damage. However, translation of these stress response genes can be delayed if they contain introns and splicing is disrupted. In eukaryotes of all kingdoms, genes that are rapidly induced during stress contain few introns as a strategy to bypass splicing [14]. Genes encoding canonical heat shock protein (HSP) chaperone genes exemplify this strategy [15]. Coincidentally, HSPs have been shown to help restore RNA splicing during stress, presumably by promoting proper folding of splicing factor proteins [10, 15–17]. Other stress-responsive and intronless genes are poorly studied.
Using the genetic model nematode Caenorhabditis elegans, a previous study reported that a small intronless gene named nuclear localized metal responsive-1 (numr-1) is highly induced by cadmium, a heavy metal and environmental contaminant [18–20]. Biochemical functions of NUMR-1 are unknown, but the protein was shown to localize to the nucleus and to promote C. elegans longevity and adult survival in cadmium [18]. NUMR-1 protein sequence has no resemblance to canonical metal-responsive metallothioneins that function as metal ion chelators [21].
We find that NUMR-1 contains motifs common in RNA-binding proteins and influences RNA splicing. We conducted a genome-wide RNA interference (RNAi) screen for regulators of numr-1 and find that silencing of core splicing factors or integrator complex subunits strongly induces numr-1 in the absence of cadmium. Cadmium or silencing of integrator complex subunits disrupts RNA splicing and processing of snRNAs suggesting a common molecular damage surveillance mechanism. Lastly, heat shock factor-1 (HSF-1), the guardian of the proteome, is activated by cadmium or integrator complex silencing and is required for full numr-1 induction. Our results are consistent with a model where cadmium disrupts the processing of snRNAs by the integrator complex leading to HSF-1 activation of numr-1 and HSPs as mechanisms to restore homeostasis.
Results
numr-1 encodes an SR-like protein
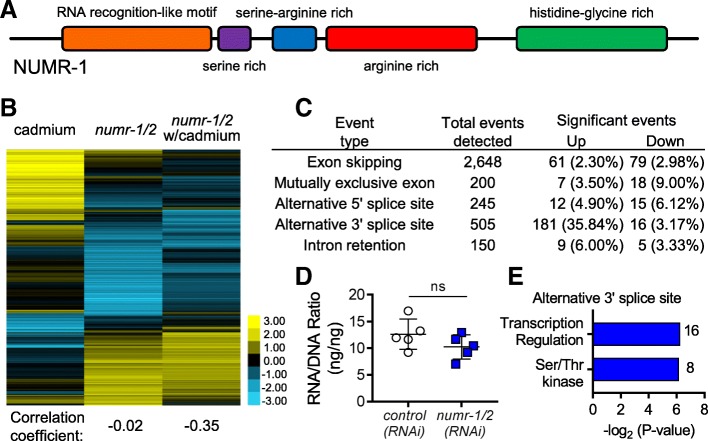
Protein BLAST searches fail to identify NUMR-1 homologs outside of nematodes, which may be caused by high sequence divergence and stretches of low complexity. However, while inspecting primary sequence and secondary structure predictions, we identified motifs in NUMR-1 (Fig. 1a, Additional file 1: Figure S1) that resemble those found in serine-/arginine-rich (SR) proteins, a family of eukaryotic proteins with well-established roles in multiple steps of RNA metabolism [25]. Jpred4 secondary structure modeling [22] predicts a sequence of beta sheets and alpha helices at the N-terminus of NUMR-1 the same as RNA-recognition motifs (RRM) found in canonical SR proteins (Fig. 1a, Additional file 1: Figure S1). NUMR-1 also has serine- and arginine-rich regions that are common in SR proteins, and a C-terminus tail rich in histidine and glycine (Fig. 1a, Additional file 1: Figure S1). These features suggest that NUMR-1 might function in RNA metabolism.
Fig. 1.
The numr-1 gene encodes an SR-like protein that influences RNA splicing. a NUMR-1 resembles an SR-like protein with an N-terminal RNA recognition-like motif, serine- and arginine-rich regions, and a C-terminal histidine-glycine-rich region [22–24]. b Clustered heat map of log2 gene expression changes caused by 300 μM cadmium relative to no cadmium, numr-1/2(RNAi) relative to control(RNAi) on agar without cadmium, or numr-1/2(RNAi) relative to control(RNAi) on agar with 300 μM cadmium. Correlation coefficients relative to the first column of data are shown below. A list by gene is provided in Additional file 2: Table S1. c Summary of alternative splicing events caused by numr-1/2(RNAi) relative to control(RNAi). A list by gene is provided in Additional file 4: Table S3. Percentage of events that were significantly changed are provided in parentheses. d RNA to DNA ratios of worms fed with control(RNAi) or numr-1/2(RNAi). e Enriched categories within genes alternatively spliced by numr-1/2(RNAi)
NUMR-1 influences RNA splicing
NUMR-1 was previously shown to reside in nuclei and promote longevity and survival of cadmium in adult worms, but nothing is known about its biochemical function [18]. The numr-1 gene is immediately adjacent to a duplicate named numr-2 that is controlled by the same promoter [18]. A deletion of both copies does not exist, but RNAi will target both because they are 100% identical at the nucleotide level. To explore the function of numr-1/2 in RNA metabolism, we performed whole-genome RNA sequencing in worms fed numr-1/2 dsRNA and compared it with worms fed control dsRNA with and without 12-h exposure to 300 μM cadmium.
A heat map of genes differentially expressed by cadmium or numr-1/2(RNAi) is shown in Fig. 1b with correlation coefficients relative to cadmium listed at the bottom. Cadmium changed the expression of 999 genes (771 up and 228 down) (Fig. 1b; Additional file 2: Table S1). As expected, genes at least fourfold upregulated by cadmium are enriched for functions in stress response, including drug metabolism, heat shock proteins, and c-type lectins [19] (Additional file 3: Table S2). RNAi of numr-1/2(RNAi) changed the expression of 1387 genes on control agar (518 up and 869 down) and 980 on agar with cadmium (538 up and 442 down) (Fig. 1b; Additional file 2: Table S1). Genes upregulated at least fourfold by numr-1/2(RNAi) with and without cadmium are only enriched for cuticle collagen, and there is no enrichment for genes downregulated by numr-1/2(RNAi) (Additional file 3: Table S2). Expression changes caused by numr-1/2(RNAi) were not correlated with expression changes caused by cadmium and only weakly negatively correlated on agar with cadmium (Fig. 1b). These results suggest that NUMR-1 does not function to regulate the expression of any specific type of genes associated with environmental stress responses.
More interestingly, numr-1/2(RNAi) led to 404 statistically significant changes to alternative splicing events (Fig. 1c, Additional file 4: Table S3). We did not detect a statistically significant decrease in total RNA to DNA ratios (Fig. 1d); with our sample size and standard deviations, we had a power of 0.8 to detect a 36% change. Of the five different types of alternate splicing events, significant changes in 3′ splice sites were the most numerous at 197 total and were the most enriched when normalized by the total number of events detected at 39%. Using DAVID analysis, we found that genes with significant changes in 3′ splice sites caused by numr-1/2(RNAi) were enriched for functions in transcription regulation and serine/threonine kinase (Fig. 1e). Examples of read maps for alternate 3′ splice site events are shown in Additional file 5: Figure S2.
NUMR-1 promotes larval development in cadmium
Alternative mRNA splicing is essential for cell differentiation and tissue development [3]. To investigate the role of numr-1 in development, we tested whether it influences larval growth of C. elegans in cadmium. Cadmium arrests larval development and decreases body size, and these effects were exacerbated by numr-1/2 RNAi (Additional file 6: Figure S3). RNAi of numr-1/2 did not affect body size under basal conditions indicating that the effects were specific to cadmium (Additional file 6: Figure S3). Therefore, NUMR-1 promotes larval development and growth in cadmium in addition to its previously reported role in promoting adult survival of cadmium and longevity [18].
Silencing of spliceosome and integrator genes induces numr-1
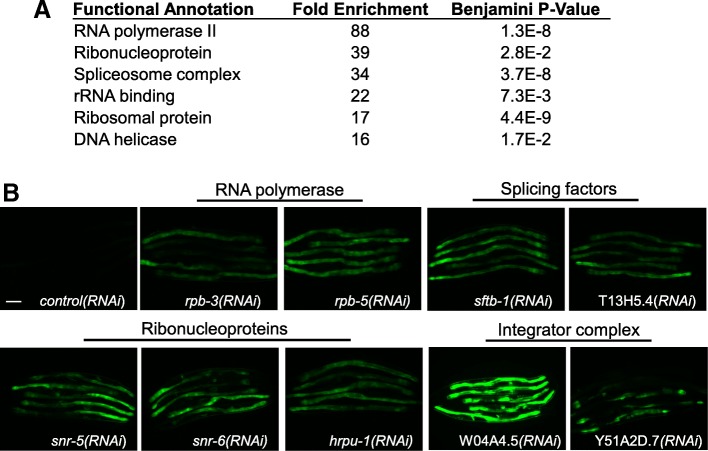
Figure 1, Additional file 1: Figure S1, Additional file 5: Figure S2, and Additional file 6: Figure S3 provide evidence that NUMR-1 functions to influence RNA splicing and promotes tolerance of cadmium. To our knowledge, a stress-responsive gene that functions in RNA metabolism and splicing has not been described in metazoans [26, 27]. To gain insights into regulation of numr-1, we generated an integrated GFP reporter driven by the numr-1 promoter (numr-1p::GFP). Fluorescence of numr-1p::GFP is increased by cadmium similar to endogenous numr-1 as measured by qPCR (Additional file 7: Figure S4). We performed a genome-wide RNAi screen to identify genes that are required to maintain low basal numr-1 expression and identified 170 dsRNA clones that activated numr-1p::GFP reproducibly (Additional file 8: Table S4). Using DAVID functional annotation analysis, we found a striking enrichment for RNA metabolic processes (Fig. 2a) [28]. Examples include RNA polymerase subunits, ribonucleoproteins, splicing factors, and subunits of the integrator complex (Fig. 2b).
Fig. 2.
Silencing of genes functioning in RNA metabolism activates numr-1. a DAVID enrichment analysis of dsRNA clones that increased numr-1p::GFP fluorescence consistently. Complete list of RNAi hits shown in Additional file 8: Table S4. b Representative fluorescence micrographs of worms fed dsRNA targeting RNA metabolism genes that strongly activated the numr-1p::GFP; five worms are shown in each image; scale bar is 100 μm
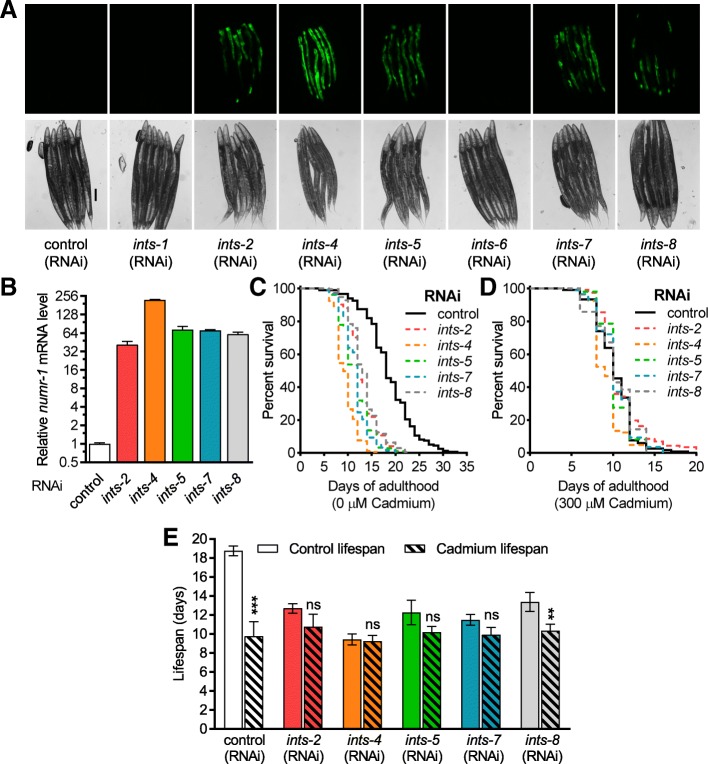
W04A4.5 is the dsRNA clone that caused the greatest increase in numr-1p::GFP fluorescence (Fig. 2b), and this gene encodes a homolog of human integrator complex subunit-4 (ints-4). Integrator mediates 3′ end processing of snRNA molecules that function in the spliceosome. The snRNA molecules are transcribed by RNA polymerase II but are not polyadenylated after transcription; instead, snRNA molecules are post-transcriptionally processed and cleaved at the 3′ end by the integrator [5, 29, 30]. Using BLAST, we found C. elegans homologs for 10/14 human integrator subunits (Additional file 9: Table S5), which match those recently reported by others [7]. C. elegans ints genes have been named based on their human homologs (Additional file 9: Table S5). RNAi of five out of seven of these integrator homologs available in our library increased numr-1p::GFP fluorescence (Fig. 3a), and these results were confirmed for endogenous numr-1 with qPCR (Fig. 3b). Therefore, in the absence of exogenous stress, numr-1 is induced by disruption of well-characterized RNA metabolism genes and the integrator complex.
Fig. 3.
RNAi of integrator complex subunits induces numr-1 and reduces survival of cadmium and longevity. a Representative fluorescence and DIC micrographs of worms fed dsRNA for C. elegans integrator complex subunits. Six worms are shown in each image, scale bar is 100 μm. N2 worms fed with control, ints-2, ints-4, ints-5, ints-7, or ints-8 dsRNA were assessed for b fold changes in numr-1 mRNA levels assessed by qPCR, c lifespan, and d survival of 300 μM cadmium from day 1 of adulthood. N = 4 replicates of 200–300 worms in b and two to three independent trials for (c, d). Statistics for individual trials in c and d are listed in Additional file 10: Table S6. In b, all integrator RNAi significantly increased numr-1 mRNA with P < 0.001 compared to control dsRNA as determined by one-way ANOVA with Tukey post hoc tests. In c, all integrator RNAi significantly decreased the longevity of worms compared to control dsRNA as determined by the Log-rank test. e Median lifespans of individual trials from data in (c, d) and Additional file 10: Table S6. **P < 0.01 and ***P < 0.001 compared to corresponding control (0 μM cadmium) lifespan as determined by two-way ANOVA with Bonferroni post hoc tests
Disruption of integrator complex decreases lifespan
Loss of integrator homologs was previously reported to have detrimental effects on embryonic and larval development [31]. To further investigate the physiological functions of the integrator complex, we used RNAi to knockdown subunits identified from the RNAi screen to evaluate their effects on C. elegans lifespan and cadmium resistance. On control plates without cadmium, RNAi of each of the five integrator subunits identified in our screen reduced the lifespan of N2 worms by 5 to 8 days (Fig. 3c–e, Additional file 10: Table S6). Alternatively, RNAi of four out of five of these integrator subunits had no effect on survival in the presence of 300 μM cadmium (Fig. 3d, e, Additional file 10: Table S6). This lack of an additive effect raises the possibility that integrator complex RNAi and cadmium reduce lifespan by similar mechanisms.
Cadmium alters 3′ processing of snRNAs
Data in Figs. 2 and 3 establish disruption of RNA metabolism and integrator as strong endogenous stimuli for numr-1. We next tested if disruption of RNA metabolism could be a signal for inducing numr-1 when integrator subunits are silenced. Loss of integrator function in human cell lines, mice, and Drosophila causes the failure of transcription termination of snRNA and aberrant 3′ polyadenylation [29, 30, 32, 33]. A recent study also reported the same effect when C. elegans integrator homologs are disrupted [7]. To verify altered snRNA processing in C. elegans under our conditions, we silenced the five integrator complex genes that influence numr-1 expression (Fig. 3) and measured snRNAs. To differentiate between total and polyadenylated (misprocessed) snRNAs, we synthesized two sets of cDNA from each RNA sample: one using random primers and one using oligo-dT primers [29]. Of the five C. elegans snRNA transcripts (U1, U2, U4, U5, and U6), polyadenylated U2 and U4 were increased the most by integrator homolog RNAi (Fig. 4a); there was little effect on the other snRNAs. These results confirm that 3′ processing of some snRNA by integrator is conserved in C. elegans.
Fig. 4.
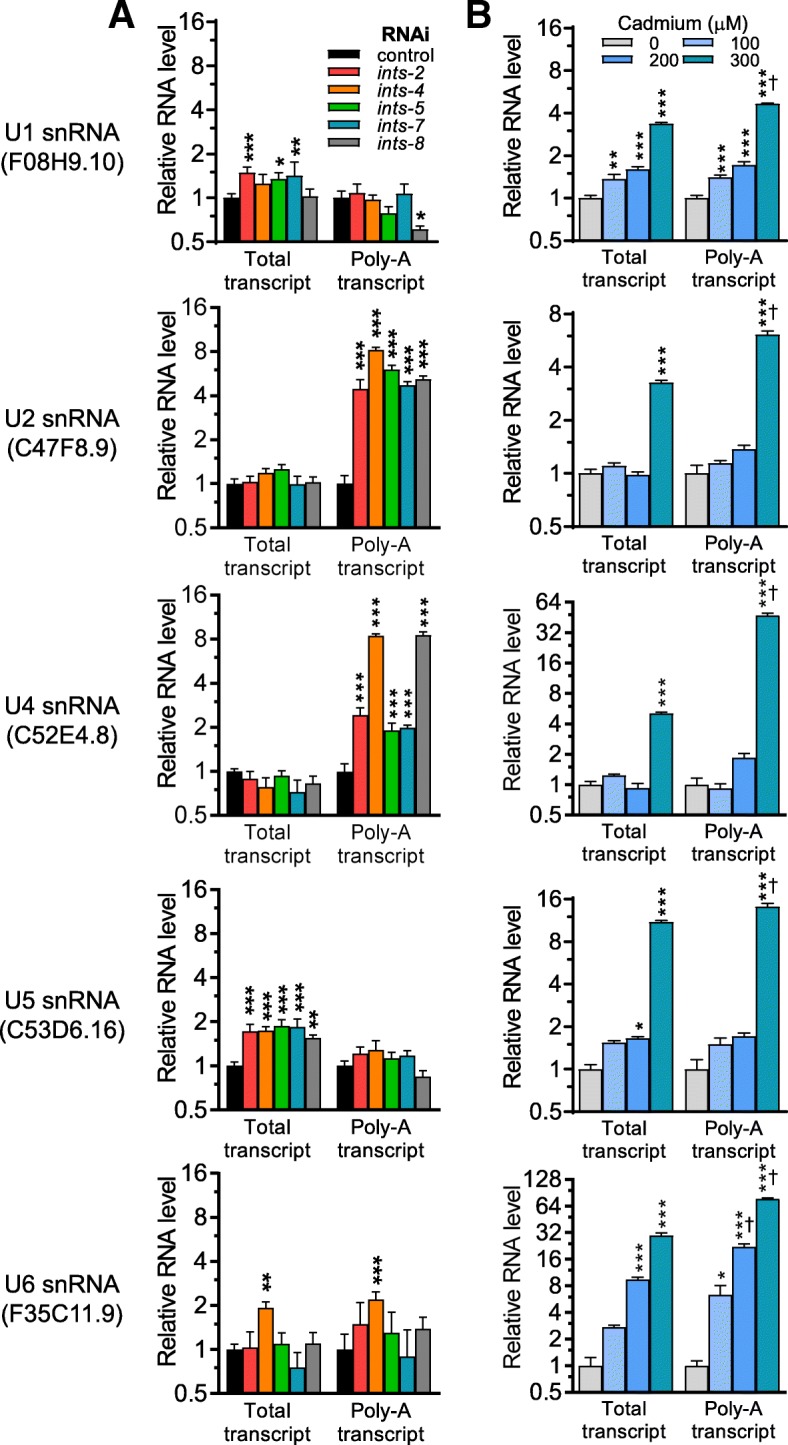
Cadmium disrupts snRNA 3′ processing. Fold change in total or polyadenylated (Poly-A) U1 snRNA (F08H9.10), U2 snRNA (C47F8.9), U4 snRNA (C52E4.8), U5 snRNA (C53D6.16), and U6 snRNA (F35C11.9) in N2 worms measured with qPCR. Worms were fed control, ints-2, ints-4, ints-5, ints-7, or ints-8 dsRNA clones in a or exposed to 0, 100, 200, and 300 μM cadmium in b. Random or oligo-dT primers were used to synthesize total or polyadenylated cDNA from each total RNA sample. N = 4 replicates of 200–300 worms. a *P < 0.05, **P < 0.01, or ***P < 0.001 compared to control dsRNA. b *P < 0.05, **P < 0.01, or ***P < 0.001 compared to 0 μM cadmium and †P < 0.001 compared to the total transcript levels of the same cadmium concentration as determined by two-way ANOVA with Bonferroni post hoc tests
We next tested if cadmium also disrupts snRNA processing. Exposure of worms to 300 μM cadmium for 24 h increased all total and polyadenylated snRNAs that we measured, but with a consistently larger effect on the latter (Fig. 4b). Importantly, this exposure to cadmium is not acutely lethal as worms survive for multiple days (Fig. 3d). These results indicate that cadmium and integrator RNAi both disrupt 3′ processing of snRNAs.
Disruption of integrator complex and cadmium alter RNA splicing
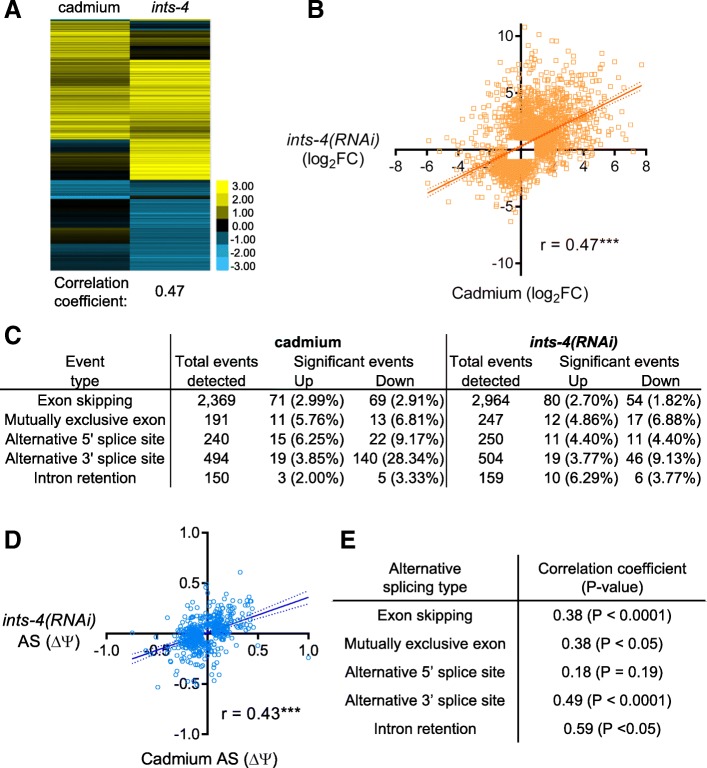
After snRNA transcripts are processed by the integrator complex, they interact with small nuclear ribonucleoproteins (snRNP) to form essential components of the spliceosome [1, 5]. Human cells with mutations in integrator complex subunits have genome-wide splicing defects consistent with malfunctioning snRNAs [34]. Given that cadmium and integrator disruption both alter 3′ processing of snRNAs (Fig. 4), we next tested for global changes in RNA processing using RNAseq. We used RNAi of ints-4 because it caused the greatest induction of numr-1/2 and misprocessing of snRNAs (Figs. 3 and 4); subunit-4 is also the most critical component of the Drosophila and human integrator complexes [35, 36]. RNAi of ints-4 changed the expression of 1978 genes (1161 up and 817 down) (Additional file 2: Table S1). A clustered heat map of expression changes demonstrates striking similarity in the changes caused by 300 μM cadmium and ints-4(RNAi) (Fig. 5a; correlation coefficient is 0.47), particularly for genes upregulated. The 662 genes that were upregulated at least fourfold by ints-4(RNAi) are enriched for membrane proteins and heat shock proteins (Additional file 3: Table S2), the latter consistent with activation of protein homeostasis responses. There is no correlation between the gene expression changes caused by ints-4(RNAi) and numr-1/2(RNAi) (Additional file 11: Figure S5A-B).
Fig. 5.
RNA splicing is disrupted by cadmium or RNAi of integrator complex. a Clustered heat map of log2 gene expression changes caused by 300 μM cadmium relative to no cadmium and ints-4(RNAi) relative to control(RNAi) on agar without cadmium. Correlation coefficient is shown below. A list by gene is provided in Additional file 2: Table S1. b Linear regression analysis of fold changes in panel a; ***P < 0.001 by linear regression F test. c Number of genes alternatively spliced within each category by cadmium or ints-4(RNAi). Lists by gene are provided in Additional file 12: Table S7 and Additional file 13: Table S8. Percentage of events that were significantly changed are provided in parentheses. d Linear regression analysis of ΔΨ values for all significantly altered splicing events caused by ints-4(RNAi) or cadmium, ***P < 0.001 by linear regression F test. e Correlation coefficients and linear regression P values for each type of splicing event change caused by cadmium and ints-4(RNAi)
Cadmium and ints-4(RNAi) caused 368 and 266 statistically significant alternative splicing events, respectively (Fig. 5c, Additional file 12: Table S7, Additional file 13: Table S8). To test if a correlation exists for the extent of alternative splicing, we performed linear regression analysis on relative percentage spliced in index scores (PSI, ΔΨ), which are a measure of the degree of alternative splicing. As shown in Fig. 5d, ΔΨ values for cadmium and ints-4(RNAi) alternate splicing events are highly correlated with a correlation coefficient of 0.43 (P < 0.001). We also analyzed the five different alternative splicing types separately and found significant ΔΨ correlation for four out of five splicing categories (Fig. 5e). In contrast, no correlation of ΔΨ values was found between spliced event changes caused by numr-1/2(RNAi) and ints-4(RNAi), and ΔΨ values for numr-1/2(RNAi) and cadmium were only weakly negatively correlated (r = − 0.09; P = 0.02) (Additional file 11: Figure S5C-D). Taken together, results in Fig. 5 demonstrate that cadmium and integrator knockdown lead to overlapping changes in gene expression and mRNA splicing.
HSF-1 regulates numr-1
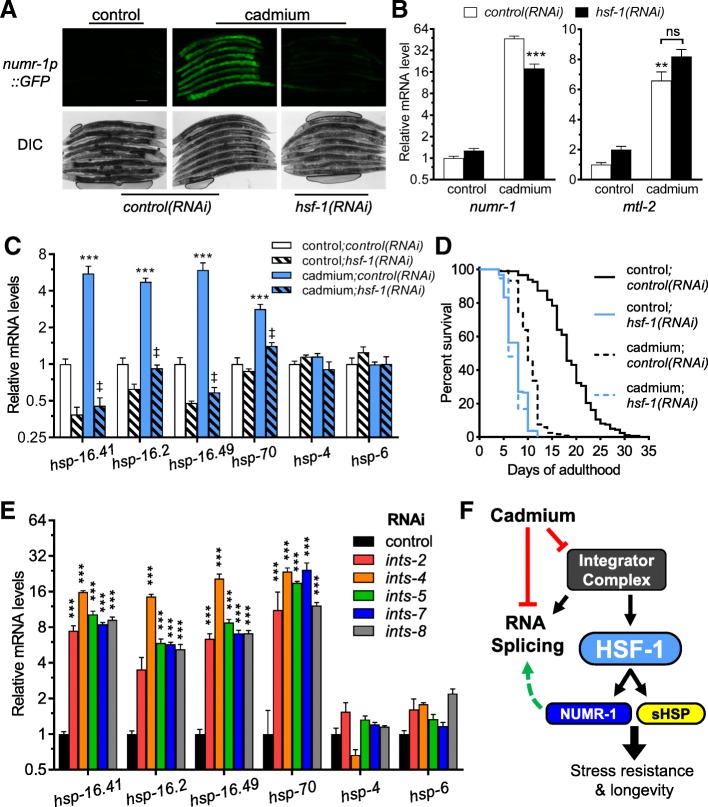
To gain insights into the signaling mechanism that activates numr-1, we performed an RNAi screen to identify transcription factors required for numr-1p::GFP fluorescence. RNAi of three C. elegans transcription factors, ceh-24 (C. elegans homeobox-24), fkh-6 (forkhead transcription factor family-6), and hsf-1 (heat shock factor-1), led to a significant reduction of numr-1 mRNA (Additional file 14: Figure S6) with hsf-1 the strongest and most consistent (Fig. 6a, b). Using qPCR, we next found that four cytosolic heat shock protein genes regulated by HSF-1 were induced by 100 μM cadmium indicating that the canonical heat shock response is activated in addition to numr-1 (Fig. 6c); heat shock protein mRNA was also increased by 300 μM cadmium in RNAseq experiments (Additional file 2: Table S1, Additional file 3: Table S2). As others have reported, loss of hsf-1 strongly decreases lifespan (Fig. 6d) [37]. Similar to ints subunit RNAi (Fig. 3), we found that 300 μM cadmium had no additive effect with hsf-1(RNAi) consistent with cadmium causing a type of cellular damage that overlaps with loss of HSF-1.
Fig. 6.
HSF-1 is required for full induction of numr-1. a Representative fluorescence and DIC micrographs of worms expressing numr-1p::GFP fed control or hsf-1 dsRNA and treated with 0 (control) or 100 μM cadmium for 24 h. Eight worms are shown in each image; scale bar is 100 μm. Fold changes in numr-1 (b), mtl-2 (b), or heat shock protein gene (c) mRNA levels in N2 worms fed control or hsf-1 dsRNA and treated with 0 (control) or 100 μM cadmium for 24 h as measured by qPCR. For b and c, N = 4 replicates of 200–300 worms. **P< 0.01 or ***P < 0.001 compared to control;control(RNAi) and ‡P < 0.001 compared to cadmium;control(RNAi) as determined by two-way ANOVA with Bonferroni post hoc tests. d Lifespan of N2 worms fed control or hsf-1 dsRNA grown in the absence (control) or presence of 300 μM cadmium starting on day 1 of adulthood. Combined results are shown and statistics for individual trials are listed in Additional file 15: Table S9. The control data are the same as in Fig. 3c, d and listed in Additional file 4: Table S3. e Fold change in heat shock protein gene mRNAs in N2 worms fed with control, ints-2, ints-4, ints-5, ints-7, or ints-8 dsRNA. ***P < 0.001 compared to control dsRNA as determined by two-way ANOVA with Bonferroni post hoc tests, N = 4 replicates with each replicate containing 200–300 worms. f Proposed model. Cadmium disrupts RNA splicing in part via RNA processing by the integrator complex as evident by a change in snRNA. Following integrator complex disruption, an hsf-1-dependent transcriptional response is activated to induce the expression of small heat shock protein genes and an RRM-like domain containing gene named numr-1 that promotes RNA splicing, longevity, and stress resistance
Heat shock protein genes of the endoplasmic reticulum (hsp-4) and mitochondrial (hsp-6) unfolded protein responses were not induced by 100 μM cadmium (Fig. 6c), suggesting specificity to the cytosolic response. Furthermore, RNAi of hsf-1 did not reduce activation of the canonical heavy metal response gene mtl-2 (metallothionein-2) (Fig. 6b), which was previously shown to be regulated by transcription factor DAF-16/FOXO [38, 39]. Therefore, during cadmium exposure, HSF-1 is required for full activation of numr-1 and the cytosolic heat shock response without affecting metal chelator or broader organelle-specific protein damage responses.
Disruption of integrator complex induces the heat shock response
A previous study observed an increase in hsp70p::GFP fluorescence with RNAi of integrator complex subunit 4 (W04A4.5) [40] suggesting that, like cadmium, integrator disruption activates the heat shock response. As shown in Fig. 6e, RNAi of integrator subunits induces the same four cytosolic heat shock protein genes that were induced by cadmium (Fig. 6c). Taken together, these results are consistent with HSF-1 functioning to activate numr-1 and cytosolic heat shock proteins when RNA metabolism is disrupted by cadmium or integrator silencing.
Discussion
RNA metabolism is an extremely complex process with broad relevance to fundamental eukaryotic cell functions, pathogenesis, and aging. Relative to protein homeostasis, we know very little about how animal cells sense and respond to the disruption of RNA metabolism by environmental stress [10, 17, 26]. The intron-less gene numr-1 was previously shown to encode a small nuclear protein that promotes longevity and is induced strongly by cadmium [18]. We find that NUMR-1 contains motifs similar to splicing factors, affects splicing in vivo, and promotes larval development in cadmium (Fig. 1, Additional file 1: Figure S1, Additional file 5: Figure S2, Additional file 6: Figure S3). Although stress-inducible splicing factors have not been previously identified in animals, a stress-inducible splicing factor named SR34b in the plant model Arabidopsis is also induced by cadmium, and SR34b mutation sensitizes plants to cadmium [41]. The presence of stress-inducible SR-like proteins in two different kingdoms is consistent with their fundamental importance to eukaryotic cell function during conditions that disrupt RNA metabolism.
Our genome-wide RNAi screen revealed that numr-1 is strongly and specifically induced by disruption of endogenous RNA metabolic processes including the metazoan-specific integrator complex, which processes snRNA 3′ ends for functioning in the spliceosome [5, 6]. RNAi of integrator in C. elegans alters 3′ end processing of snRNA and RNA splicing in a manner similar to cadmium (Figs. 4 and 5). Because numr-1 and the cytosolic heat shock response are adaptive responses to cadmium that are also activated by RNAi of integrator (Figs. 3 and 6), we propose that integrator processing of snRNAs may act as a surveillance mechanism for cadmium-induced molecular damage (Fig. 6f). Cadmium has broad effects in cells that are often attributed to oxidative stress [42]; direct effects on dsDNA structure have also been reported [43]. It remains to be determined how cadmium alters integrator processing of snRNAs, but it is conceivable that one or more of the protein-protein or protein-RNA interactions within integrator are altered directly by cadmium or redox changes, or indirectly by a signaling mechanism [6]. Interestingly, numr-1 expression is far more sensitive to cadmium than other heavy metals suggesting specificity in the mechanism [18]. Given that integrator function is highly conserved [44], it could also be a potential target of cadmium in mammalian cells.
Other core cell processes are also used to detect stress. For example, disruption of protein translation is a signal for induction of osmotic response genes, protein miss-folding is a signal for the heat shock response, and loss of ribosome, mitochondria, and vesicle transport is a signal for detoxification responses [45–49]. This general strategy also extends to the tissue level as we recently identified a specific type of extracellular matrix disruption that induces three distinct stress responses in C. elegans [50].
Interestingly, RNAi of integrator subunit 4 had the largest effects on RNA splicing, numr-1 expression, hsp expression, stress resistance, and longevity(Figs. 2, 3, and 6). Although some of these differences could be caused by variation in RNAi sensitivity, disruption of subunit 4 also has the largest effects on RNA processing and development in Drosophila and subunit 4 was recently shown to function as a scaffold for the catalytic core of human integrator complex [35, 36].
The full composition of integrator, interactions between subunits and accessory proteins, and regulation of integrator are active areas of research with relevance to transcription, cell cycle control, and a growing list of pathologies [6, 7]. Integrator mutation or silencing disrupts multiple steps of development and cell differentiation in C. elegans, Drosophila, zebrafish, and mouse cells [33, 51–54]. The human INTS6 (originally named deleted in cancer 1, DICE1) was first identified as a tumor suppressor in lung and squamous cell carcinomas and was recently shown to also act as a tumor suppressor in hepatocellular carcinomas [55–57]. INTS2 and INTS8 are also miss-expressed or mutated in many cancers [6]. Our study establishes cadmium regulation of numr-1 in C. elegans as a paradigm for understanding interactions between integrator and the environment that is amenable to genetic analysis in a whole-animal context.
HSF-1 and the cadmium stress response
Heat shock factors are highly conserved master regulators of heat shock protein chaperone genes under many conditions that perturb protein homeostasis. Cadmium was previously shown to activate heat shock factor in cultured mammalian cells [58–60]. As mentioned above, heat shock factors were also previously shown to respond to RNA splicing disruption by promoting expression of intronless hsp genes [15]. Our results demonstrate that the C. elegans cytosolic heat shock response is also activated by cadmium and is required for full induction of numr-1 (Fig. 6 and Additional file 14: Figure S6). Unlike our results during exposure to cadmium (Fig. 6), numr-1 is not induced by heat shock and is not dependent on HSF-1 under control or heat shock conditions in C. elegans [61]. The upstream regulatory region of numr-1 does not contain a heat shock binding element [62]. Therefore, regulation of numr-1 by HSF-1 is likely to be indirect and specific to conditions that target RNA metabolism.
HSF-1 has recently been shown to co-localize with pre-mRNA splicing factors in the nucleus of stressed cells within loci termed nuclear stress bodies (nSBs). These nSBs are thought to be sites of transcription and splicing of stress-induced transcripts [63, 64]. Interestingly, NUMR-1::GFP fusion proteins were previously shown to localize to nuclear loci with HSF-1::mCherry in C. elegans in response to cadmium [18]. It remains to be determined whether NUMR-1 functions to promote processing of transcripts at these loci.
Materials and methods
C. elegans strains
All C. elegans strains were cultured at 20 °C using standard methods [65]. The following strains were used in this study: N2 Bristol and QV151 qvIs4[numr-1p::GFP].
Analysis of NUMR-1 sequence
NUMR-1 secondary structure was predicted with JPred4 [22]. Integrator subunit BLAST searches were conducted within WormBase [31].
RNAi experiments and genome-wide RNAi screen
A genome-wide RNAi screen was performed by feeding worms with Escherichia coli [HT115(DE3)] engineered to transcribe double-stranded RNA (dsRNA) homologous to C. elegans target genes using methods described previously [66, 67]. The complete ORFeome RNAi feeding library (Open Biosystems, Huntsville, AL) was screened in addition to clones supplemented from the original MRC genomic RNAi feeding library (Geneservice, Cambridge, UK). Briefly, synchronized L1 larvae of QV151 C. elegans were grown in liquid medium supplemented with dsRNA-producing bacteria for 3 days and screened manually for numr-1p::GFP fluorescence with a Zeiss Stemi SV12 microscope. Positive dsRNA clones were rescreened for three additional trials. dsRNA clones with positive results from all three trials are listed in Additional file 8: Table S4. To identify transcription factors that regulated numr-1p::GFP, C. elegans were grown in the same manner as described above using a transcription factor specific RNAi sub-library created from the ORFeome and MRC RNAi collections. After 3 days of dsRNA feeding, C. elegans were incubated with 100 μM of cadmium for 24 h to induce numr-1p::GFP, followed by manual fluorescent screening to identify transcription factor knockdowns that blocked cadmium-induced numr-1p::GFP activation. All subsequent RNAi experiments were performed with nematode growth medium (NGM) agar plates in the presence of 50 μg mL−1 carbenicillin and 0.2% β-lactose. RNAi feeding experiments on agar plates were all initiated at the synchronized L1 larval stage for 48–72 h before worms were used for subsequent experiments. The pPD129.36 (LH4440) clone was used as a control RNAi; it expresses a 202-bp dsRNA that is not homologous to any C. elegans genes.
Fluorescent reporter analysis and PCR
To visualize the C. elegans fluorescent reporters under cadmium stress, worms were treated with either 100 μM of cadmium diluted in NGM buffer or NGM buffer alone as a control for 24 h at the L4/young adult (YA) stage. For RNAi experiments, worms were fed with appropriate dsRNA from L1 larvae to YA, or to the indicated worm age, before being imaged. Worms were mounted on 2% agarose pads with 5 mM levamisole and imaged using an Olympus BX60 microscope with a Zeiss AxioCam MRm camera fitted with either a GFP or a RFP filter (Center Valley, PA). To image whole worms that exceed the viewing plane under × 10 magnifications, multiple images were taken and merged into a single composite (Additional file 7: Figure S4).
Quantitative PCR (qPCR) assays were carried out as described previously using the delta-delta Ct method [68]. For all PCR assays, four replicate RNA samples were extracted per condition with each sample containing 100–300 synchronized worms. For samples from RNAi experiments, RNA was extracted from worms fed with the corresponding dsRNA from L1 larvae to young adult stage. For samples from cadmium exposure experiments, RNA was extracted from worms after exposure to cadmium for 24 h at the young adult stage. For analysis of snRNA transcripts, either random primers or oligo-dT primers were used to synthesize cDNA. All qPCR reactions were performed in 10 μL reaction volumes with a realplex2 (Eppendorf AG, Hamburg, Germany) with all data analyzed by normalization to the rpl-2 gene as an internal reference. Primers used for qPCR are available on request.
RNA/DNA assay
Worms were grown and treated the same as for whole-transcriptome RNA sequencing. For each biological replicate, five worms were picked to 10 μl of lysis buffer (5 mM Tris pH 8.0, 0.5% Triton X-100, 0.5% Tween 20, 0.25 mM EDTA, and 1 mg/ml proteinase K) and frozen at − 80 °C. Tubes containing frozen worms were placed immediately into a thermocycler and incubated 10 min at 65 °C and 2 min at 85 °C. Concentrations of RNA and DNA were determined with Promega Quantifluor dsDNA System (E2670) and Quantifluor RNA System (E3310). N = 5 biological replicates were used.
Whole-transcriptome RNA sequencing
N2 worms were synchronized at the L1 larval stage and grown on RNAi agar plates seeded with either control, numr-1/2, or ints-4 dsRNA-expressing bacteria until the late L3 stage. Worms were then collected in liquid NGM buffer and transferred to new RNAi agar plates containing either 0 or 300 μM cadmium for 12 h. For each of the six treatment groups, total RNA was extracted from three biological replicates of late L4 to young adult worms using an RNAqueous-Micro Total RNA isolation kit (ThermoFisher Scientific, AM1931) and sent to Novogene (Sacramento, CA) for oligo(dT) library construction, sequencing, mapping, and statistical analysis. HISAT2 was used to map sequences to the C. elegans reference genome [69]. Between 75 and 87 million clean reads and 11.4 and 13.1 Gbp were obtained from each sample; raw sequence data is available at Gene Expression Omnibus (GSE129970). Alternative splicing analysis was performed with rMATS using the total number of junction counts and reads on targets to quantify alternative splicing events [70, 71]. Gene Ontology enrichment was identified with DAVID functional analysis using high stringency, and we report Benjamini adjusted P values [28].
Differential expression analysis was carried out using DEGSeq, and we only considered transcripts detected in all treatment groups with changes of at least twofold. Because we are focused on environmental response and wanted to avoid differential gene expression changes caused indirectly by development, we also filtered out genes previously identified from ModEncode data to be enriched in one of the developmental stages potentially present in our samples (L4 larval, young adult, early embryo stages) [72]. To generate heat maps, differentially expressed genes were clustered with Gene Cluster 3.0 using correlation (uncentered) average linkage and mapped with Java Treeview 1.1.6r4.
Lifespan, stress resistance, and developmental assays
For lifespan assays, synchronized L1 larvae were grown on NGM agar plates seeded dsRNA and scored for survival every 1 to 2 days starting at the third day of adulthood. For cadmium resistance assays, synchronized worms were grown under standard conditions from L1 to YA and then transferred onto appropriately seeded dsRNA NGM agar plates containing 300 μM cadmium. For both lifespan and stress resistance assays, adult worms were picked manually while gravid to avoid offspring. Worms with protruding gonad or intestine were censored, and worms were counted dead if they did not respond to gentle prodding with pick.
For developmental assays, synchronized L1 worms were fed dsRNA on NGM agar plates containing 0 or 250 μM of cadmium for 70 h before being imaged and quantified for body length using ImageJ. F1 offspring obtained from P0 worms fed with numr-1/2 dsRNA were used for this assay to ensure silencing in early larvae. At least three independent trials were performed for all assays (with the exception of ints-5 cadmium survival where only two trials were recorded), with the number of animals used in each experiment described in the corresponding supplementary table.
Statistical analyses
With the exception of expression heat maps, data were analyzed and graphed using the Prism software 5.04 (La Jolla, CA) with Student’s t test performed when two means were compared, one-way ANOVA with Tukey’s post hoc test when multiple comparisons were performed with one factor, and two-way ANOVA with Bonferroni post hoc test in comparison over two factors. Longevity and survival assays were analyzed by the Log-rank test using the OASIS online statistic tool [73]. Linear regressions were analyzed by the F test. Statistical significance is indicated in each figure legend with *P < 0.05, **P < 0.01, and ‡/***P < 0.001. All P values for RNAseq analyses were corrected for FDR.
Additional files
Figure S1. NUMR-1 secondary protein structure prediction. (A) Jpred 4 analysis of NUMR-1 protein sequence reveals an N-terminal RRM-like domain from amino acids 23-87 defined by four beta strands (green) and two alpha helices (red) arranged in the order of β-α-β-β-α-β [22]. Similar structural arrangement of the RRM motif is found in a canonical SR protein and splicing factor, human serine- and arginine-rich splicing factor 2 (SRSF2) protein. (B) Complete amino acid sequence of the NUMR-1 protein highlighted with motifs identified in Fig. 1. (PDF 72 kb)
Table S1. Differential gene expression. (XLSX 3482 kb)
Table S2. DAVID gene enrichments. (XLSX 10 kb)
Table S3. numr-1 RNAi alternative splicing. (XLSX 796 kb)
Figure S2. RNAi of numr-1/2 affects alternative 3′ splice site selection. Representative genome coverage tracks for alternative 3′ splice acceptor sites in cdka-1, M4.1, and R02F2.1 for control and numr-1/2(RNAi) samples. (PDF 186 kb)
Figure S3. numr-1 influences larval growth in cadmium. Effects of numr-1/2(RNAi) on body length of C. elegans grown with 0 or 250 μM cadmium assessed 70 h after hatching from L1 larvae. L1 numr-1/2(RNAi) worms were the F1 population obtained from P0 that were fed with numr-1/2 RNAi. ***P < 0.001 compared to control(RNAi) as determined by Student’s t test, with 72–75 animals measured per condition from 3 trials. Scale bar is 100 μm. (PDF 79 kb)
Figure S4. The numr-1 GFP reporter and endogenous mRNA are activated by cadmium. (A) numr-1p::GFP representative fluorescence and DIC micrographs of worms exposure to control NGM buffer or NGM buffer with 100 μM of cadmium for 24 h at the L4/YA stage. Six worms are shown in each image, scale bar is 100 μm. (B) Relative mRNA levels of the numr-1 gene in control and 100-μM-cadmium-treated N2 worms for 24 h as assessed by qPCR. This data is replotted from Fig. 6b. N = 4 replicates of 200–300 worms. ***P < 0.001 compared to control as determined by Student’s t test. (PDF 101 kb)
Table S4. Gene dsRNAs that induce numr-1p::GFP. (XLSX 20 kb)
Table S5. Integrator complex subunit homology. (XLSX 9 kb)
Table S6. Integrator longevity and cadmium survival. (XLSX 13 kb)
Figure S5. ints-4 and numr-1 gene expression and alternative splicing effects are not correlated. (A) Clustered heat map of log2 gene expression changes caused by numr-1/2(RNAi) and ints-4(RNAi) relative to control(RNAi) without cadmium. Correlation coefficient is shown below. A list by gene is provided in Additional file 2: Table S1. (B) Linear regression analysis of fold changes shown in panel A. Linear regression analysis of ΔΨ values for all significantly altered splicing events caused by numr-1/2 (RNAi) or ints-4(RNAi) in (C) and by numr-1/2(RNAi) and cadmium in (D). (PDF 109 kb)
Table S7. Cadmium exposure alternative splicing. (XLSX 757 kb)
Table S8. ints-4 RNAi alternative splicing. (XLSX 893 kb)
Figure S6. Transcription factors required for numr-1 activation. (A) Workflow of the transcription factor RNAi screen. (B) Representative fluorescence micrographs of worms fed dsRNA that inhibited cadmium-induced numr-1p::GFP activation, 8 worms are shown in each image, and scale bar is 100 μm. Images shown for control and hsf-1(RNAi) are the same as Fig. 6a. (C) Gene names and descriptions of transcription factors required for full numr-1 induction. (D) qPCR analysis of numr-1 mRNA levels fed with control, ceh-24, fkh-6, and hsf-1 dsRNA under control and cadmium exposed conditions. ***P < 0.001 as determined by two-way ANOVA with Bonferroni post hoc tests, N = 4 replicates with each replicate containing 200–300 worms. (PDF 423 kb)
Table S9. hsf-1 longevity and cadmium survival. (XLSX 8 kb)
Acknowledgements
Some of the C. elegans strains were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) which is supported by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank Danilo Polanco for assistance with preliminary analysis of numr-1/2 function.
Authors’ contributions
AP identified numr-1 as a stress-inducible gene with uncharacterized regulation and generated the numr-1p::GFP transgene. WD integrated the numr-1p::GFP transgene and first identified the interactions with hsf-1. KW prepared the RNA for RNA sequencing and helped analyze the RNAseq data. MBA conducted the RNA/DNA ratio measurements. KPC conducted the longevity and cadmium survival assays. CWW conducted and designed the remaining experiments; KPC and CWW analyzed and interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by NSF grants IOS-1120130 and IOS-1452948 to KPC, a University of Florida Research Opportunity Seed Fund to KPC and CWW, and a Natural Sciences and Engineering Research Council (NSERC) of Canada postdoctoral fellowship to CWW.
Availability of data and materials
All datasets supporting the conclusions of the manuscript are included within the article and its additional files. Raw data used to generate individual graphs in this manuscript are available at https://figshare.com/s/9cae6f74962d85ceab6a [74]. The accession number for the RNA-sequencing data presented in this paper is GEO: GSE129970 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129970 [75].
Ethics approval and consent to participate
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cheng-Wei Wu, Email: michael.wu@usask.ca.
Keith P. Choe, Email: kchoe@ufl.edu
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3. [DOI] [PMC free article] [PubMed]
- 2.Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsen TW. The spliceosome: The most complex macromolecular machine in the cell? BioEssays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 5.Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Rienzo M, Casamassimi A. Integrator complex and transcription regulation: recent findings and pathophysiology. Biochim Biophys Acta Gene Regul Mech. 2016;1859:1296–1280. doi: 10.1016/j.bbagrm.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Gómez-Orte E, Sáenz-Narciso B, Zheleva A, Ezcurra B, de Toro M, López R, et al. Disruption of the Caenorhabditis elegans Integrator complex triggers a non-conventional transcriptional mechanism beyond snRNA genes. PLoS Genet. 2019;15:e1007981. doi: 10.1371/journal.pgen.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 9.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Heintz C, Doktor TK, Lanjuin A, Escoubas CC, Zhang Y, Weir HJ, et al. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature. 2017;541:102–106. doi: 10.1038/nature20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschênes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017;16:918–933. doi: 10.1111/acel.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Jolly C, Vourc’h C, Robert-Nicoud M, Morimoto RI. Intron-independent association of splicing factors with active genes. J Cell Biol. 1999;145:1133–1143. doi: 10.1083/jcb.145.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- 17.Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-X. [DOI] [PubMed] [Google Scholar]
- 18.Tvermoes BE, Boyd WA, Freedman JH. Molecular characterization of numr-1 and numr-2: genes that increase both resistance to metal-induced stress and lifespan in Caenorhabditis elegans. J Cell Sci. 2010;123:2124–2134. doi: 10.1242/jcs.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Y, McBride SJ, Boyd WA, Alper S, Freedman JH. Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 2007;8:R122. doi: 10.1186/gb-2007-8-6-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metal toxicity and the environment. EXS. 2012;101:88–98. doi: 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamer D. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- 22.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott MS, Troshin PV, Barton GJ. NoD: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics. 2011;12:317. doi: 10.1186/1471-2105-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigrist CJA, De Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 2018;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutertre M, Sanchez G, Barbier J, Corcos L, Auboeuf D. The emerging role of pre-messenger RNA splicing in stress responses: sending alternative messages and silent messengers. RNA Biol. 2011;8:740–747. doi: 10.4161/rna.8.5.16016. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, et al. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015;25:288–305. doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Ezzeddine N, Waltenspiel B, Albrecht TR, Warren WD, Marzluff WF, et al. An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3′-end formation. RNA. 2012;18:2148–2156. doi: 10.1261/rna.035725.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wormbase. www.wormbase.org. Accessed 28 June 2019.
- 32.Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, et al. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- 33.Hata T, Nakayama M. Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim Biophys Acta Mol Cell Res. 2007;1773:1039–1051. doi: 10.1016/j.bbamcr.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Oegema R, Baillat D, Schot R, van Unen LM, Brooks A, Kia SK, et al. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13:e1006809. doi: 10.1371/journal.pgen.1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, et al. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3’-end formation. Mol Cell Biol. 2010;31:328–341. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrecht TR, Shevtsov SP, Wu Y, Mascibroda LG, Peart NJ, K-LL H, et al. Integrator subunit 4 is a ‘Symplekin-like’ scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Res. 2018;46:4241–4255. doi: 10.1093/nar/gky100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.e03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 39.Barsyte D, Lovejoy DA, Lithgow GJ. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 2002;15:627–634. doi: 10.1096/fj.99-0966com. [DOI] [PubMed] [Google Scholar]
- 40.Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Du B, Liu D, Qi X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem Biophys Res Commun. 2014;455:312–317. doi: 10.1016/j.bbrc.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 42.Rani A, Kumar A, Lal A, Pant M. Cellular mechanisms of cadmium-induced toxicity: a review. Int J Environ Health Res. 2014;24:378–399. doi: 10.1080/09603123.2013.835032. [DOI] [PubMed] [Google Scholar]
- 43.Gao H, Zhu Z, Jiang X, Guo J, Song B, Zhang F. Denaturation of dsDNA induced by specific major groove binding of cadmium ion to thymine. ACS Omega. 2017;2:8490–8494. doi: 10.1021/acsomega.7b01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Wagner EJ. snRNA 3′ end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–1087. doi: 10.1042/BST0381082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 46.Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee EC-H, Strange K. GCN-2 dependent inhibition of protein synthesis activates osmosensitive gene transcription via WNK and Ste20 kinase signaling. Am J Physiol Physiol. 2012;303:C1269–C1277. doi: 10.1152/ajpcell.00294.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11:375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, et al. A damage sensor associated with the cuticle coordinates three core environmental stress responses in Caenorhabditis elegans. Genetics. 2018;208:1467–1482. doi: 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao S, Cai Y, Sampath K. The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development. 2009;136:2757–2765. doi: 10.1242/dev.034959. [DOI] [PubMed] [Google Scholar]
- 52.Rutkowski RJ, Warren WD. Phenotypic analysis of deflated/Ints7 function in Drosophila development. Dev Dyn. 2009;238:1131–1139. doi: 10.1002/dvdy.21922. [DOI] [PubMed] [Google Scholar]
- 53.Otani Y, Nakatsu Y, Sakoda H, Fukushima T, Fujishiro M, Kushiyama A, et al. Integrator complex plays an essential role in adipose differentiation. Biochem Biophys Res Commun. 2013;434:197–202. doi: 10.1016/j.bbrc.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 54.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 55.Wieland I, Röpke A, Stumm M, Sell C, Weidle UH, Wieacker PF. Molecular characterization of the DICE1 (DDX26) tumor suppressor gene in lung carcinoma cells. Oncol Res. 2001;12:491–500. doi: 10.3727/096504001108747503. [DOI] [PubMed] [Google Scholar]
- 56.Wieland I, Arden KC, Michels D, Klein-Hitpass L, Böhm M, Viars CS, et al. Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene. 1999;18:4530–4537. doi: 10.1038/sj.onc.1202806. [DOI] [PubMed] [Google Scholar]
- 57.Li WJ, Hu N, Su H, Wang C, Goldstein AM, Wang Y, et al. Allelic loss on chromosome 13q14 and mutation in deleted in cancer 1 gene in esophageal squamous cell carcinoma. Oncogene. 2003;22:314–318. doi: 10.1038/sj.onc.1206098. [DOI] [PubMed] [Google Scholar]
- 58.Wirth D, Christians E, Li X, Benjamin IJ, Gustin P. Use of Hsf1-/-mice reveals an essential role for HSF1 to protect lung against cadmium-induced injury. Toxicol Appl Pharmacol. 2003;192:12–20. doi: 10.1016/S0041-008X(03)00256-4. [DOI] [PubMed] [Google Scholar]
- 59.Shinkai Y, Masuda A, Akiyama M, Xian M, Kumagai Y. Cadmium-mediated activation of the HSP90/HSF1 pathway regulated by reactive persulfides/polysulfides. Toxicol Sci. 2017;156:412–421. doi: 10.1093/toxsci/kfw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/MCB.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guhathakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, et al. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–712. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahl P, Lutz Y, Puvion E, Fuchs JP. Rapid effect of heat shock on two heterogeneous nuclear ribonucleoprotein-associated antigens in HeLa cells. J Cell Biol. 1989;109:1921–1935. doi: 10.1083/jcb.109.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metz A, Soret J, Vourc’h C, Tazi J, Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci. 2004;117:4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- 65.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu CW, Deonarine A, Przybysz A, Strange K, Choe KP. The Skp1 homologs SKR-1/2 are required for the Caenorhabditis elegans SKN-1 antioxidant/detoxification response independently of p38 MAPK. PLoS Genet. 2016;12:e1006361. doi: 10.1371/journal.pgen.1006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu CW, Wang Y, Choe KP. F-box protein XREP-4 is a new regulator of the oxidative stress response in Caenorhabditis elegans. Genetics. 2017;206:859–871. doi: 10.1534/genetics.117.200592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 71.Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li JJ, Huang H, Bickel PJ, Brenner SE. Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data. Genome Res. 2014. [DOI] [PMC free article] [PubMed]
- 73.Kim S, Lee D, Lee SJ V., Kim D, Son HG, Lee H, et al. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016;7:56147–56152. [DOI] [PMC free article] [PubMed]
- 74.Wu CW, Wimberly K, Pietras A, Atlas MB, Choe KP. Raw data for: RNA processing errors triggered by cadmium and integrator complex disruption are signals for environmental stress. Figshare. https://figshare.com/articles/_/7221326. Accessed 28 June 2019. [DOI] [PMC free article] [PubMed]
- 75.Wu CW, Wimberly K, Pietras A, Atlas MB, Choe KP. RNA-sequencing data deposit for: RNA processing errors triggered by cadmium and integrator complex disruption are signals for environmental stress. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129970. Accessed 28 June 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NUMR-1 secondary protein structure prediction. (A) Jpred 4 analysis of NUMR-1 protein sequence reveals an N-terminal RRM-like domain from amino acids 23-87 defined by four beta strands (green) and two alpha helices (red) arranged in the order of β-α-β-β-α-β [22]. Similar structural arrangement of the RRM motif is found in a canonical SR protein and splicing factor, human serine- and arginine-rich splicing factor 2 (SRSF2) protein. (B) Complete amino acid sequence of the NUMR-1 protein highlighted with motifs identified in Fig. 1. (PDF 72 kb)
Table S1. Differential gene expression. (XLSX 3482 kb)
Table S2. DAVID gene enrichments. (XLSX 10 kb)
Table S3. numr-1 RNAi alternative splicing. (XLSX 796 kb)
Figure S2. RNAi of numr-1/2 affects alternative 3′ splice site selection. Representative genome coverage tracks for alternative 3′ splice acceptor sites in cdka-1, M4.1, and R02F2.1 for control and numr-1/2(RNAi) samples. (PDF 186 kb)
Figure S3. numr-1 influences larval growth in cadmium. Effects of numr-1/2(RNAi) on body length of C. elegans grown with 0 or 250 μM cadmium assessed 70 h after hatching from L1 larvae. L1 numr-1/2(RNAi) worms were the F1 population obtained from P0 that were fed with numr-1/2 RNAi. ***P < 0.001 compared to control(RNAi) as determined by Student’s t test, with 72–75 animals measured per condition from 3 trials. Scale bar is 100 μm. (PDF 79 kb)
Figure S4. The numr-1 GFP reporter and endogenous mRNA are activated by cadmium. (A) numr-1p::GFP representative fluorescence and DIC micrographs of worms exposure to control NGM buffer or NGM buffer with 100 μM of cadmium for 24 h at the L4/YA stage. Six worms are shown in each image, scale bar is 100 μm. (B) Relative mRNA levels of the numr-1 gene in control and 100-μM-cadmium-treated N2 worms for 24 h as assessed by qPCR. This data is replotted from Fig. 6b. N = 4 replicates of 200–300 worms. ***P < 0.001 compared to control as determined by Student’s t test. (PDF 101 kb)
Table S4. Gene dsRNAs that induce numr-1p::GFP. (XLSX 20 kb)
Table S5. Integrator complex subunit homology. (XLSX 9 kb)
Table S6. Integrator longevity and cadmium survival. (XLSX 13 kb)
Figure S5. ints-4 and numr-1 gene expression and alternative splicing effects are not correlated. (A) Clustered heat map of log2 gene expression changes caused by numr-1/2(RNAi) and ints-4(RNAi) relative to control(RNAi) without cadmium. Correlation coefficient is shown below. A list by gene is provided in Additional file 2: Table S1. (B) Linear regression analysis of fold changes shown in panel A. Linear regression analysis of ΔΨ values for all significantly altered splicing events caused by numr-1/2 (RNAi) or ints-4(RNAi) in (C) and by numr-1/2(RNAi) and cadmium in (D). (PDF 109 kb)
Table S7. Cadmium exposure alternative splicing. (XLSX 757 kb)
Table S8. ints-4 RNAi alternative splicing. (XLSX 893 kb)
Figure S6. Transcription factors required for numr-1 activation. (A) Workflow of the transcription factor RNAi screen. (B) Representative fluorescence micrographs of worms fed dsRNA that inhibited cadmium-induced numr-1p::GFP activation, 8 worms are shown in each image, and scale bar is 100 μm. Images shown for control and hsf-1(RNAi) are the same as Fig. 6a. (C) Gene names and descriptions of transcription factors required for full numr-1 induction. (D) qPCR analysis of numr-1 mRNA levels fed with control, ceh-24, fkh-6, and hsf-1 dsRNA under control and cadmium exposed conditions. ***P < 0.001 as determined by two-way ANOVA with Bonferroni post hoc tests, N = 4 replicates with each replicate containing 200–300 worms. (PDF 423 kb)
Table S9. hsf-1 longevity and cadmium survival. (XLSX 8 kb)
Data Availability Statement
All datasets supporting the conclusions of the manuscript are included within the article and its additional files. Raw data used to generate individual graphs in this manuscript are available at https://figshare.com/s/9cae6f74962d85ceab6a [74]. The accession number for the RNA-sequencing data presented in this paper is GEO: GSE129970 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129970 [75].