Abstract
Background
Many traits evolve by cis-regulatory modification, by which changes to noncoding sequences affect the binding affinity for available transcription factors and thus modify the expression profile of genes. Multiple examples of cis-regulatory evolution have been described at pattern switch genes responsible for butterfly wing pattern polymorphism, including in the diverse neotropical genus Heliconius, but the identities of the factors that can regulate these switch genes have not been identified.
Results
We investigated the spatial transcriptomic landscape across the wings of three closely related butterfly species, two of which have a convergently evolved co-mimetic pattern and the other having a divergent pattern. We identified candidate factors for regulating the expression of wing patterning genes, including transcription factors with a conserved expression profile in all three species, and others, including both transcription factors and Wnt pathway genes, with markedly different profiles in each of the three species. We verified the conserved expression profile of the transcription factor homothorax by immunofluorescence and showed that its expression profile strongly correlates with that of the selector gene optix in butterflies with the Amazonian forewing pattern element ‘dennis.’
Conclusion
Here we show that, in addition to factors with conserved expression profiles like homothorax, there are also a variety of transcription factors and signaling pathway components that appear to vary in their expression profiles between closely related butterfly species, highlighting the importance of genome-wide regulatory evolution between species.
Electronic supplementary material
The online version of this article (10.1186/s13227-019-0127-4) contains supplementary material, which is available to authorized users.
Keywords: Cis-regulation, Heliconius, Butterfly, Transcription factor, Homothorax, Gene expression, Wnt signaling, Transcriptomics
Background
A major challenge in evolutionary developmental biology is to understand how modifications to gene expression can lead to biological diversity. In particular, variation in cis-regulatory elements has been repeatedly identified as the material source of polymorphism and divergence in physiology, behavior, pigmentation patterns and morphological structures; Gephebase, a database of genotype–phenotype relationships, identifies 323 such examples of cis-regulatory evolution in diverse eukaryote clades [37]. These elements must function by differential binding of regulatory factors, and so to understand the evolution of gene regulatory networks, we must first identify which regulatory factors are present and able to perform this function in a given spatial and temporal context.
The Lepidoptera make up ~ 18% of described animal diversity and have a vast array of wing patterns, both within and between species. Recent advances in genetics, genomics and experimental methods have begun to uncover the underlying genetic and developmental basis of lepidopteran wing pattern variation [22, 33]. One of the most diverse and well-studied groups are the Heliconius butterflies, and it is now understood that much of the variation in wing pattern in this group results from regulatory evolution at just three genes, optix, WntA and cortex [38, 43, 50]. CRISPR/Cas9 mutagenesis has shown that optix and WntA are also involved in the patterning of wings in multiple butterfly lineages [41, 68]. At each of these loci, there are a large diversity of complex regulatory alleles, controlling expression patterns across the wing surface during development [13, 60, 61]. This regulatory diversity that generates the extraordinary variation in wing patterns nonetheless acts against a background of highly conserved regulatory factors that underlie insect wing development. Consistent with this idea, earlier candidate gene studies have shown that many patterning factors previously identified in Drosophila wing development show similar patterns of expression on butterfly wings (Table 1). In some cases, interesting novel expression domains have already been identified in butterfly wings, mainly in association with eyespot elements [9, 44].
Table 1.
A summary of single-gene expression studies performed on developing butterfly wings, indicating whether notable differences in domains of expression have been described in butterflies relative to D. melanogaster
| Gene | Species | Any novel domains? | References |
|---|---|---|---|
| Ultrabithorax | J. coenia | [63, 64] | |
| apterous | J. coenia, B. anynana | Localized reduction in expression associated with ventralization of pattern | [9, 47] |
| engrailed/invected | J. coenia | Eyespot associated | [9] |
| P. rapae, B. anynana, Saturnia pavonia, Antheraea polyphemus | [42] | ||
| scalloped | J. coenia | [9] | |
| wingless | V. cardui, A. vanillae, J. coenia, Battus philenor, B. mori, M. sexta, Z. morio, B. anynana, P. rapae | Expressed in association with basal and marginal pattern elements | [35, 42, 44] |
| cut | V. cardui, A. vanillae, J. coenia, Battus philenor, B. mori, M. sexta, Z. morio | [35] | |
| hedgehog | J. coenia, B. anynana | Eyespot associated | [26] |
| cubitus interruptus | J. coenia, B. anynana | Eyespot associated | [26] |
| patched | J. coenia, B. anynana | Eyespot associated | [26] |
| achaete-scute | J. coenia | [14] | |
| Notch | H. erato, H. melpomene, A. vanillae P. rapae, M. sexta, V. cardui, J. coenia, B. anynana | [49, 51] | |
| Optix | Heliconius spp, J. coenia, V. cardui, | Butterfly specific, in association with specific scale types | [36, 50] |
| WntA | Heliconius spp, Limenitis arthemis, A. vanillae, J. coenia, Danaus plexippus | Butterfly specific, in association with specific pattern elements | [15, 40, 41] |
| doublesex | Papilio, B. anynana | [5, 31] | |
| aristaless1 & aristaless2 | J. coenia, Limenitis arthemis, Spodoptera ornithogalli, Ephestia kuehniella | Wing pattern associated, including Discalis II elements | [39] |
| Distal-less | H. erato, H. melpomene, A. vanillae P. rapae, M. sexta, V. cardui, J. coenia, B. anynana, Saturnia pavonia, Antheraea polyphemus | Dynamic eyespot-associated expression | [6, 9, 51] |
| spalt major | V. cardui, P. rapae, P. oleracea, Colias philodice, C. eurytheme, B. anynana | Eyespot associated | [42, 57, 67] |
| decapentaplegic | J coenia, B. anynana | Parallel to veins, in addition to A-P | [9, 11] |
| Ecdysone receptor | J coenia | Eyespot associated | [28] |
| pSmad | B. anynana, P. rapae | Eyespot associated | [42] |
| Antennapedia | B. anynana, Heteropsis iboina, Pararge aegeria and Melanargia galathea, Lasiommata megera, J. coenia, M. cinxia, Inachis io, Caligo memnon | Eyespot associated | [54] |
| cortex | H. melpomene, H. numata | Butterfly specific, pattern associated | [43] |
| armadillo | B. anynana | [11] | |
| BarH1 | Colias | Butterfly specific, pigment associated | [65] |
Many of the genes listed, in addition to having domains of expression that are homologous to those found in D. melanogaster, are also expressed in association with eyespots
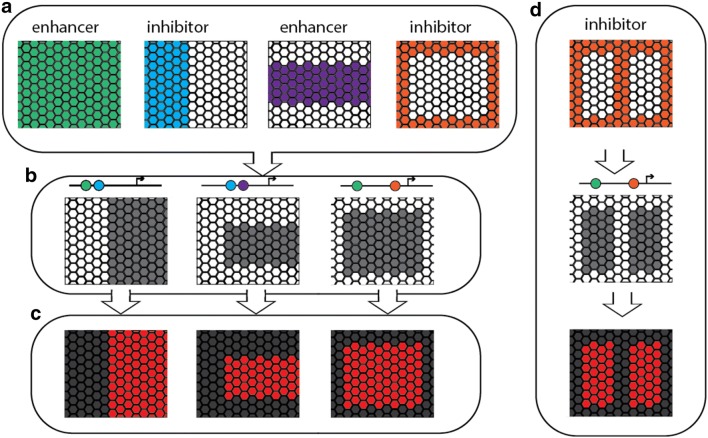
If this is in fact the case, then the pattern variation we observe in Heliconius and other butterflies could be generated by the differential ‘readout’ of a highly conserved set of transcription factors that effectively ‘prepattern’ the wing (Fig. 1a). These conserved expression patterns could then provide input to the regulatory elements of pattern switch genes like optix (Fig. 1b), and in turn, modifications of these elements could lead to production of wing pattern diversity (Fig. 1c), a mechanism that allows for the gain of novel phenotypes with the avoidance of deleterious pleiotropic effects [48]. This hypothesis has previously been examined at the within-species level in Heliconius through transcriptomics [20]. Key examples of this mode of regulatory evolution have been described in evolution of melanic patterns in Drosophila species. For example, the protein Engrailed is a deeply conserved component that specifies the posterior compartment in arthropod segmentation [45, 46]. Some Drosophila species have a melanic spot on the anterior tip of the wing, which is sculpted in part by repression of the yellow gene by en, on the posterior boundary [17]. Here, cis-regulatory evolution at the yellow locus locked onto the conserved spatial information encoded by en. The expression and function of the en gene did not change; rather, a new regulatory connection was established that modified the expression of yellow to generate novel diversity. Likewise, expression of pigmentation genes in the thorax of D. melanogaster is repressed by the expression of the transcription factor stripe, which specifies flight muscle attachment sites. The shape of this thoracic element is thus constrained by factors that specify flight muscle pattern [16]. On the other hand, there are also examples of Drosophila pigmentation evolving by modification to the trans-regulatory landscape, by which conserved components of the regulatory landscape themselves change in expression profile to affect downstream melanin synthesis domains. For example, Dll in D. biarmipes and D. prolongata has gained additional expression profiles to its usual peripheral pattern, in correlation with melanic elements [3]. This mode of trans-regulatory divergence also plays a role in butterfly eyespot evolution (Fig. 1d, [11]). No evidence has been found for trans-regulatory landscape evolution at the intraspecific level, but could occur between more distantly related species for which genetic mapping is not possible.
Fig. 1.
Hypothetical mechanisms of wing pattern development and evolution. In this model, a set of prepatterning factors (a) are expressed early in the developing wing, may pattern the development of the wing and do not vary in their expression profiles in different morphs or species (see Table 1). These factors feed in to the regulation of the wing pattern switch genes and shape their expression profiles accordingly, for example in Heliconius the transcription factor optix (b), which causes scale cells that would otherwise develop to be melanic to express ommochrome pigments (c). It is also possible that changes to the expression of wing pattern switch genes like optix could be caused by changes in expression of prepatterning factors (d)
Here, we begin to test these ideas by using comparative transcriptomic sequencing in two well-described species in the genus Heliconius, as well as an outgroup species Agraulis vanillae. The two Heliconius, H. erato and H. melpomene, are co-mimics that diverged from each other between 10 and 12 mya [30], and recently co-diversified into around 25 different co-occurring wing pattern types. In contrast, A. vanillae, which diverged from Heliconius roughly 25 mya, is largely monomorphic across its extensive geographic distribution. Linkage and association studies of pattern variation in Heliconius and other butterflies have repeatedly identified noncoding regions as primary candidates for the loci of evolution [15, 21, 31, 60, 61]. We hypothesize that these candidate regulatory elements allow for regulatory coupling to upstream patterning transcription factors in the wing. In order to understand the upstream spatial information that provides an input to butterfly wing patterning, we need to understand spatial patterns of gene expression in the developing wing, building on a primarily gene-by-gene, candidate-driven approach, as in many previous studies which have primarily used factors known from Drosophila wing development (Table 1). In particular, these data help us determine which transcription factors show consistent spatial expression profiles in different species and are therefore candidate constituents of a conserved developmental landscape, and which transcription factors show variable patterns and are therefore candidates for the causative regulators of pattern differences. In addition, following on from the discovery that WntA is a key patterning gene in Heliconius and Agraulis [41], we also characterized the expression of Wnt pathway constituents in all three species. Our results highlight both strong conservation and striking flexibility in the gene regulatory landscape in the early wing development in Heliconius.
Results
To gain insights into the regulatory landscape that organizes butterfly wing early patterning, we conducted transcriptomic analysis of 110 samples representing whole larval wings from H. melpomene and H. erato, and pupal wings dissected into 5 sections from H. melpomene, H. erato and A. vanillae. Between 10 and 24 million reads were sequenced per sample, and the average percentage of reads per sample that did not map was 11.8% (Additional file 1: Table S1), compared to a previous RNAseq study in H. melpomene in which more than 50% of reads failed to map [62]. All samples passed quality controls and could be included in the differential expression analyses.
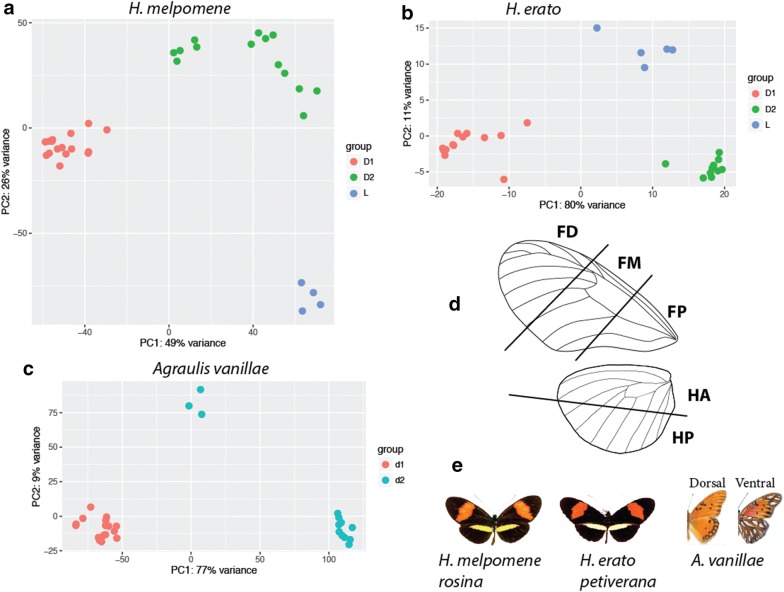
PCA analysis showed clustering of samples within each species by stage (Fig. 2). Sample clustering by wing sector is clear in day 1 samples (Additional file 1: Figures S1 and S2), and in Agraulis and H. melpomene, distinct clusters for forewing and hindwing are also present, indicating that there is sufficient detectible differential expression between wing sections to determine spatial expression differences across the wing. At day 2, clustering by wing sector is not evident, and there is some clustering by individual, indicating that there is less differential expression across the wing at this later time point.
Fig. 2.
Principal component analyses of RNA samples for each species clustered by stage, with the exception of three samples of Agraulis vanillae form the day 2 stage, which formed a separate cluster. d The dissection scheme used for tissue collection: FP proximal forewing, FM medial forewing, FD distal forewing, HA anterior hindwing, HP posterior hindwing. e depicts the three butterfly species used: H. melpomene rosina, H. erato demophoon (race formerly designated petiverana) and Agraulis vanillae
Differential expression analysis revealed 209 differentially expressed genes between H. melpomene larval forewings and hindwings, versus 77 in H. erato. In total, 28 of these genes are differentially expressed in both species (Additional file 1: Table S2). This includes the transcription factor Ubx, the notch pathway repressor and microtubule binding protein pigs, and 9 genes with no homology to known transcripts. At day 1, we identified 2848 genes differentially expressed between the five wing sectors in H. melpomene, 1713 in H. erato and 1780 in A. vanillae; 617 transcripts were differentially expressed in all three species at day 1 (Additional file 1: Table S3). At day 2, 319 transcripts were differentially expressed in H. melpomene, 2663 in H. erato and 167 in A. vanillae, with no genes differentially expressed in all three species, and 30 genes differentially expressed in a pair of species (Additional file 1: Table S4), including the pigmentation genes Ddc and tan.
Transcription factors
In order to investigate the nature of the regulatory landscape of the developing wing, we focused on the expression of the 237 identified transcription factor orthology groups, determined by reciprocal best BLAST hits between the three species and homology to known transcription factors from other insect genomes. No expression was detected in any species for 37 of these genes; of the 200 TFs that were expressed, 6 were differentially expressed in all three species, 16 were differentially expressed in 2 species, and an additional 28 were differentially expressed in one species (Fig. 3). This confirms the presence of TFs that are expressed in a patterned way across the proximal–distal axis of the wing. Minimal differential expression of transcription factors was detected between the anterior and posterior sections of hindwings.
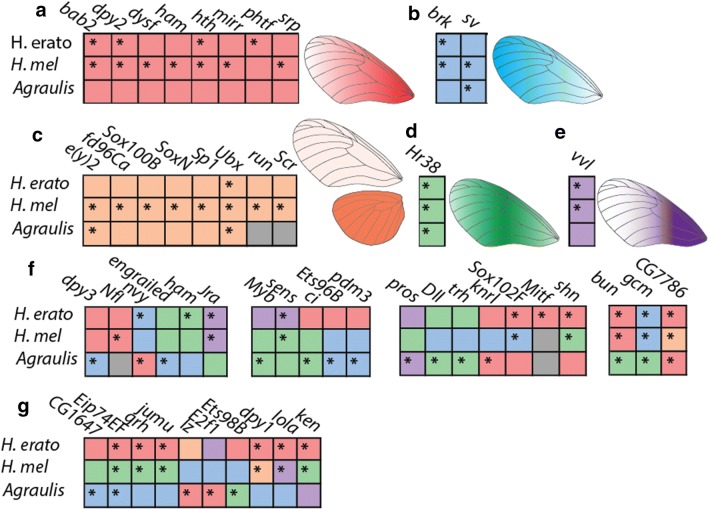
Fig. 3.
Differential expression of transcription factors in day 1 pupae. Transcription factors are color-coded for their pattern of differential expression—a, in red, indicates factors that are highly expressed in the proximal forewing and expressed in a falling gradient in the medial and distal forewing; b, in blue, indicates factors that are highly expressed in the distal forewing; c, in orange, indicates factors that are highly expressed in the hindwing relative to the forewing; d, in green, indicates factors that are highly expressed in the medial forewing; and e, in purple, indicates factors that are highly expressed in the proximal forewing but low in the rest of the forewing. Gray indicates no detectible expression. f lists factors with expression patterns shared between two species, and g lists factors with different expression patterns in all three species. Asterisks indicate genes which are significantly differentially expressed; all depicted genes are differentially expressed in at least one species
To examine the relationships between spatial domains of expression between the species, expression profiles were clustered into 5 classes by similarity. A total of 20 TFs shared the same expression profile in all three species, and an additional 21 shared the same expression profile in two species, with an additional 10 factors showing a different expression profile in all three species. Of the 37 factors which were differentially expressed in either H. melpomene or H. erato, 26 (70%) were expressed in the same pattern, whereas of the 27 factors differentially expressed in Agraulis, only 13 TFs (48%) were differentially expressed in the same pattern between Agraulis and one species of Heliconius, indicating that while some factors have conserved ancestral expression profiles, others vary in their expression along the proximal–distal axis.
A number of TFs have expression profiles that match known profiles either from immunohistochemistry of butterfly wings or by analogy with gene expression in Drosophila wings (Table 1). This includes Ultrabithorax (Ubx), expressed only in the hindwing; homothorax (hth), expressed only in the proximal forewing and anterior hindwing; distal-less (dll), expressed in an increasing gradient from proximal to distal; and mirror (mirr), expressed in the proximal forewing and anterior hindwing (Fig. 3c, f, Table 1). The recapitulation of these expression profiles, along with that of cubitus interruptus and invected in hindwings, confirms the presence of conserved expression of genes involved with wing pattern specification between insects and also serves as validation that our experimental design can detect differential expression of transcription factors, which are typically expressed at relatively low levels (Additional file 1: Figure S4).
Multiple additional factors with conserved expression profiles along the proximal–distal axis were identified, including brinker, a negative regulator of Dpp signaling [8]; bric a brac 2 (bab2), part of a proximal–distal gene regulatory module linked to abdominal pigmentation pattern in Drosophila [52]; ventral veins lacking (vvl), linked to vein development in Drosophila, here highly expressed in the proximal forewing, and previously suggested as a candidate wing pattern regulator gene in H. erato [12, 60]; Hr38, a hormone receptor upregulated in the medial forewing; and shaven (sv), related to the development of sensory structures and upregulated in the distal forewing [25]. These factors serve as additional candidate prepattern regulators in heliconiine wings.
Several transcription factors are differentially expressed in all three species, but with different profiles. bunched (bun) (Dpp pathway, [58]), glial cells missing (gcm) (related to neurogenesis, [23]) and jun-related antigen (a transcription factor in the JNK pathway, [29]) are expressed in the same profiles in Heliconius but in a different profile in Agraulis, whereas the functionally undescribed TF CG7786 is expressed in a similar profile in H. erato and Agraulis but differently in H. melpomene, and the Wnt pathway component factors sens and Sox102F are differentially expressed in different patterns in H. erato and H. melpomene. In all three species, Ecdysone-induced protein 74EF Eip74EF is differentially expressed in a different pattern [59]. The divergent expression of these factors in this set of species may indicate a role downstream of the prepatterning factors, for example as targets of Wnt signaling or in scale cell differentiation.
Several differentially expressed TFs are associated with the development of imaginal disks generally, or are specifically associated with other imaginal disks, in particular related to the eye and the genitals suggesting the possible evolution of novel functions in the Lepidoptera, for example bunched (bun, eye development), lozenge (lz, compound eye development and genital morphogenesis), ken and barbie (ken, genital morphogenesis). Many differentially expressed TFs have known roles in neurogenesis and the nervous system, including senseless (sens), gilal cells missing (gcm), nervy (nvy, axon guidance and chetae morphogenesis). Other factors have specific associations with cuticle or bristle development such as nvy, grainy head (grh), as well as multiple copies of dumpy (dpy). The factor pdm3 is upregulated in Agraulis distal hindwing and is a hotspot for abdominal pigmentation evolution in Drosophila [66]. This may implicate a novel or undescribed role for these factors in the pupal development of insect wings, or specifically in the wings of butterflies.
homothorax
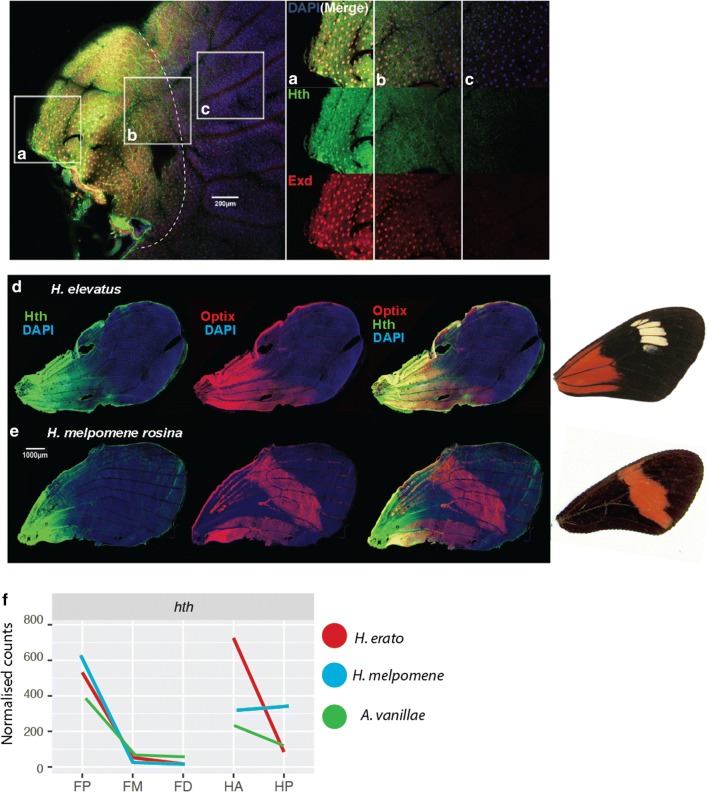
The gene homothorax (hth) was significantly upregulated in the proximal forewing of H. erato and H. melpomene and found to be expressed in the same pattern in Agraulis. This replicates its previously detected expression in early pupal wings of H. erato [20]. To confirm patterning of the hth protein, pupal butterfly wings were fluorescently stained against an anti-hth antibody raised against Drosophila hth. The DNA binding homeodomain of Heliconius, Danaus (Monarch butterfly), Tribolium and Drosophila hth is highly conserved (Additional file 1: Figure S5). Staining with anti-Hth highlighted a gradient of hth from the basal to the medial region of the wing in H. melpomene. Expression was most strongly detected in presumptive scale cell nuclei (Fig. 4).
Fig. 4.
Immunohistochemistry shows pattern of Hth expression in the butterfly wing is replicated by RNAseq analysis. Immunohistochemistry confirmed the expression of Homothorax in a proximal–distal gradient across the basal third of the Heliconius wing, in larvae (a–c) and pupae (d, e) of Heliconius butterflies. a–c Highlight three regions along the proximal–distal axis of the larval wing, showing coincident expression of Homothorax and its cofactor Extradenticle. d Homothorax expression in a region coincident with the expression of Optix in a dennis-ray butterfly, Heliconius elevatus. The same expression pattern of Hth is conserved in a red-banded butterfly (e), but is not associated with Optix expression. All Heliconius show Optix expression in the overlapping fore and hindwing region, associated with wing coupling scales as documented previously [36]. The expression profile observed in pupal wings here recapitulates the levels of hth transcript observed in the RNAseq analysis of all three species examined here (f)
Some Heliconius wing patterns show a red patch in the proximal forewing that correlates with this hth expression domain, known as the dennis patch. This patch of red scales is known to be specified by the optix gene, and we therefore hypothesized that the evolution of this patch might have arisen through a novel regulatory link between hth and optix. To explore this possibility, we co-stained wings for both hth and optix from taxa both with and without the dennis patch. In H. elevatus which has the dennis phenotype, hth expression was strongly coincident with Optix. In contrast, in H. melpomene rosina which has a medial red forewing band, there was no coincident expression of hth with Optix, except in the region of forewing and hindwing overlap where Optix expression is known to be widely conserved among butterflies. Importantly, hth expression was detected from the start of pupation up to and during optix expression at 12–60 h post-pupation. Together, the coincident timing, position and nuclear staining suggest that hth is a potential interacting factor of the dennis enhancer in Heliconius butterflies. hth was not coincident with the dennis bar in the hindwing, however, suggesting that other factors are also involved (Additional file 1: Figure S6).
Wnt pathway
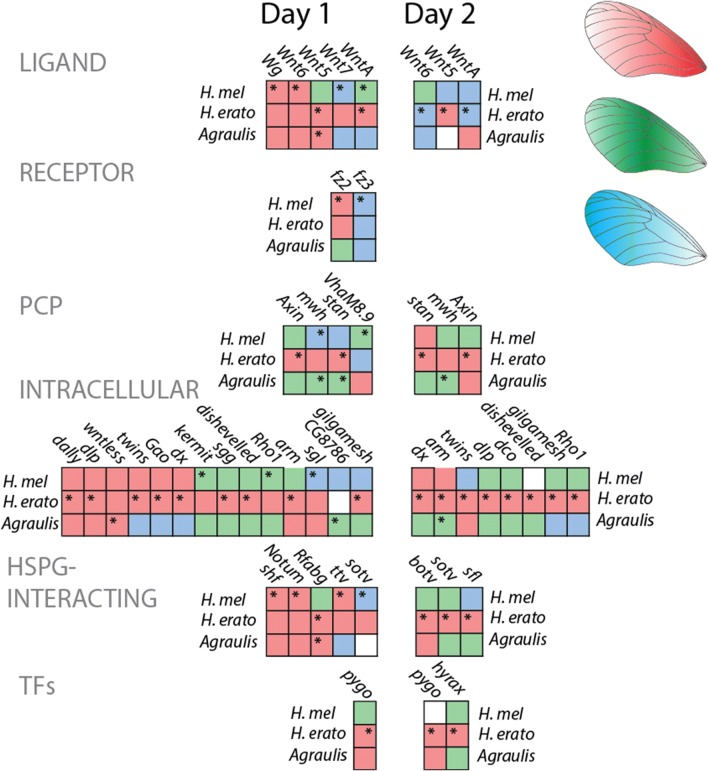
Recent studies have highlighted the importance of the ligand WntA in butterfly wing patterning, so we next focused on the expression domains of other Wnt pathway constituents. We identified 52 Wnt pathway constituents in the genomes of our three species (Additional file 1: Table S5). Expression profiles were split into three groups based on similarity. In H. erato pupal wings, most Wnt pathway genes showed a very similar pattern of the highest expression in the proximal forewing and lower expression in the medial and distal forewing, whereas in H. melpomene and A. vanillae, a variety of different expression profiles for these genes were observed (Fig. 5). The variance in Wnt pathway gene expression is reflected in the prototypical Wnt target transcription factor senseless (Fig. 3). In contrast to the transcription factors, of the 32 genes differentially expressed at day 1, only 8 (25%) had shared expression profiles in all three species, with an additional 4 (12.5%) sharing a profile between the Heliconius species. At day 2, further divergence in expression profiles between species occurred, with none of 19 differentially expressed genes sharing an expression profile between all three species.
Fig. 5.
Differential expression of Wnt pathway components in pupal development. Wnt pathway components are color-coded for their pattern of expression. Red indicates transcripts that were highly expressed in the proximal forewing, green indicates transcripts that were highly expressed in the medial forewing, and blue indicates transcripts that were highly expressed in the distal forewing. Asterisks indicate transcripts that were identified as significantly differentially expressed. Note the low discordance of expression profile between species, in contrast to the transcription factors indicated in Fig. 3. HSPG heparin sulfate proteoglycan
The ligand WntA was differentially expressed in the Heliconius species in profiles that correlate with that observed in the Panamanian pattern forms in Martin et al. [38], and multiple other Wnt ligands were differentially expressed in the three species, including Wnt2 and Wnt6 in Heliconius, which in other Nymphalid species have redundant expression profiles and have been correlated with the discalis pattern elements, and which was also previously reported in H. erato [20, 36]. Two Wnt receptors, fz2 and fz3, are expressed in opposition to each other; fz2 is the primary Wg receptor in Drosophila, while fz3 plays an inhibitory role [4, 55]. Wnt pathway components involved with planar cell polarity including multiple wing hairs (mwh), starry night (stan) and Axin were differentially expressed at both stages, along with other intracellular components of the Wnt pathway including armadillo/β-catenin and wntless, a transmembrane factor required for Wnt ligand secretion. In H. erato, two TFS downstream of Wnt signaling were differentially expressed: pygo and hyrax.
Discussion
We have explored the patterns of gene expression both through development, across evolutionary divergence and across the developing wing. Our results paint a vivid picture of how wing patterns develop and evolve across the three butterfly species. Broadly, H. melpomene, H. erato and A. vanillae share a common spatial transcriptomic landscape in the developing wing with other Lepidoptera and with Drosophila, implying the existence of a shared insect wing gene regulatory network (cf. Table 1). The results for H. erato here also broadly replicate those from a previous transcriptomic analysis there [20]. However, our data also highlight considerable flexibility in the transcriptional landscape. More than half of the transcription factors that are differentially expressed across the developing wing surface have different expression profiles within the heliconiines. This flexibility is most evident within the Wnt signaling pathway constituents, where H. erato has a derived pattern of strongly correlated Wnt pathway gene expression that is not seen in the other two species, despite strong convergence in pattern between the two Heliconius species studied. Recent work has shown genome-wide selection on regulatory elements at the between-population level in Heliconius [32], and it is likely that in the ancestral lineages of each species, many functional changes could be accrued that would lead to many differences in patterns of gene expression in the wing.
Transcription factors expressed in butterfly wings
Transcription factors provide the physical interactions that lead to differential regulation in gene regulatory networks. Several transcription factors that are known to be involved in development of wings in Drosophila and Junonia were identified in this experiment in their expected expression profiles, including Ubx and hth. Several other factors were expressed in similar patterns in all three species in this experiment; these additional factors could delineate the developmental morphospace along the forewing proximal–distal axis and hindwing anteroposterior axis in pupal wings. We identified an additional cohort of transcription factors with non-conserved expression profiles between the three studied species; many more of these factors had shared expression profiles between H. melpomene and erato than between Agraulis and Heliconius. The two Heliconius species are more closely related to one another, but are also convergent in their wing patterns, so we cannot currently disentangle whether the share expression patterns are due to common ancestry or are convergent due to shared selection pressures. Others were different in all three species, implying developmental drift, or a lack of constraint, on the regulation of these factors. Such factors have the potential to act as the substrate for functional diversification.
hth implicated in mimetic pattern evolution
One of the most strongly and consistently differentiated transcription factors was hth, and we therefore followed up on the expression patterns using immunohistochemistry. This confirmed that the hth protein shows a conserved pattern of expression restricted to the proximal wing region in all species examined. Furthermore, co-staining with anti-optix demonstrated a strong correlation of hth with the expression of optix protein in butterflies with the red proximal dennis patch. Expression of both proteins was localized to scale cell nuclei, and spatial patterns were tightly correlated between the two factors. In contrast, butterflies lacking the dennis patch showed a conserved expression of hth, but no correlated expression of optix, indicating that hth is a candidate regulator of optix in dennis + butterflies. A possible mechanism for the evolution of the dennis pattern is therefore that an optix regulatory region gained transcription factor binding sites for hth, allowing the development of a novel pattern without the requirement for changes to the expression or function of hth itself, similar to the roles of en and sr in the development of melanic patterns in Drosophila [16, 17]. Alternatively, hth might regulate optix through intermediate factors, and a number of other candidates upregulated in the proximal region are evident from our results. Future analyses of the dennis regulatory element will be required to determine the precise mechanisms of interaction with upstream regulators.
Wnt pathway variance implies different functions
Variance in WntA expression in correlation with wing pattern has previously been shown in many butterfly clades, including between races and species of Heliconius, and in Agraulis [38, 40]. We found that other Wnt pathway constituents also vary in their expression domains between species.
Surprisingly, expression patterns of Wnt pathway constituents were completely different between the two Heliconius co-mimics. In particular, the Wnt pathway constituents in H. erato were mainly expressed in a correlated pattern—high in the proximal forewing and low everywhere else, whereas the same factors were expressed in a variety of patterns in Agraulis and H. melpomene.
Notably, the correlated pattern in H. erato closely mirrors the expression profile of WntA in larval wing disks of H. erato demophoon from Panama, the pattern form used in this study [38], while the WntA expression profile for Panamanian H. melpomene rosina is notably different from its co-mimic. Here, expression is present in the distal forewing as well as the proximal forewing, and the boundary of proximal WntA expression does not correlate well with the proximal boundary of the red pattern element in the adult wing.
Together, these differences in both WntA function and Wnt pathway component expression could suggest that regulatory diversification of the Wnt pathway has occurred between Heliconius species, implying that this aspect of the wing gene regulatory network has diverged in the lineages leading to these mimetic forms, requiring the utilization of different functional mechanisms for building an identical wing pattern. This suggests extensive genome-wide regulatory divergence at the between-species level between Heliconius butterflies. Alternatively, the differences in expression of Wnt pathway components could entirely be a consequence of the different expression profiles of WntA between the two species. Wnt ligands in Drosophila effect their own autoregulation through modulation of receptor, ligand and transcription factor expression levels [7, 56]. If WntA is capable of directly regulating the expression of Wnt pathway components, this could explain all the differences we observed here while requiring between-species regulatory divergence at just one genomic locus. Separating these two models will require the functional exploration of the effects of WntA signaling.
Conclusion
Our understanding of the regulatory evolution of wing pattern in butterflies is dependent on a clear picture of the expression of developmental factors around the time of wing pattern specification. This study has provided a picture of gene expression along one axis of developing wings in a manner unbiased by our understanding of wing development in non-lepidopteran systems. At the within-species level, we can broadly rule out the hypothesis that trans-regulatory factors change their expression profiles in different pattern forms (Fig. 1d) based on genetic mapping, but we are not able to rule out this phenomenon at the between-species level—it is likely that both processes play a role, either through selection or drift. Our deeper understanding of factors that are expressed in the wing in correlation with pattern elements will permit us to decode the regulatory linkages that lead to the differential expression of pattern switch genes like optix, WntA and cortex, and it is clear that we should look to both conserved and diverging regulatory factors as the causative agents of cis-regulatory evolution.
Methods
Tissue sampling and dissection
Heliconius melpomene rosina and Heliconius erato demophoon were collected from stocks maintained at the Smithsonian Tropical Research Institute in Gamboa, Panama, in February and July 2014. Adults were provided with an artificial diet of pollen/glucose solution supplemented with flowers of Psiguria, Lantana and/or Psychotria alata according to availability. Females were provided with Passiflora plants for egg laying (P. menispermifolia for H. melpomene, P. biflora for H. erato). Eggs were collected daily, and caterpillars reared on fresh shoots of P. williamsi (melpomene) or P. biflora (erato) until late 5th (final) instar, when they were separated into individual pots in a temperature-monitored room, and closely observed for the purpose of accurate developmental staging. Agraulis vanillae larvae were collected from P. edulis located near the insectary in March 2014.
Pre-pupation larvae were identified for dissection. Late 5th instar larvae of Heliconius undergo color changes from white to purple on the last larval day, followed by an additional change to pink–orange in the hours before pupation. Additionally, several behavioral changes accompany the pre-pupation period; the larvae stop eating and clear their digestive tract and then undertake a period of rapid locomotion and wandering until they find an appropriate perch for pupation—preferably the underside of a leaf or a sturdy twig—at which point they settle in place and produce a strong silk attachment. Gradually, over a period of 30–120 min, they suspend themselves form their perch in a J shape and then pupate. Larvae that were post-locomotion but pre-J-shape were dissected in cold PBS and the wing disks removed.
Pupae were allowed to develop until 36 h (± 1.5 h), or to 60 h (± 1.5 h). These time points are referred to as day 1 and day 2 throughout. In the hours immediately post- pupation (day 0), the pupal carapace is soft and the membranous structures of the pupa are thin, weak, transparent and sticky; hence, the effective dissection of unfixed, intact pupal structures is very challenging at the earliest pupal time points. The time of pupal development is approximately equal in the three species.
Pupae were dissected in cold PBS. Wings were removed from the pupa and cleared of peripodial membrane. The wings were then cut with microdissection scissors into 5 sections: forewing proximal, medial and distal, and hindwing anterior and posterior (Fig. 2). The developing veins were used as landmarks for dissection.
Whole larval wing disks and pupal wing sections were immediately placed into RNAlater (Thermo Fisher, Waltham, MA) and subsequently frozen and stored in liquid nitrogen in Gamboa, Panama, and subsequently transported to the UK on dry ice. Samples were then stored at − 80 °C on arrival in Cambridge until RNA extraction.
RNA extraction and sequencing
RNA extraction was carried out using a standard hybrid protocol. Briefly, wing tissue sections were transferred into Trizol Reagent (Invitrogen, Carlsbad, CA) and disassociated using stainless steel beads in a tissue lyser. Chloroform phase extraction was performed, followed by purification with the RNeasy kit (Qiagen, Valencia, CA). RNA was eluted into distilled water and treated with DNAseI (Ambion, Naugatuck, CT), then quantified and stored at − 80 °C. Left and right wings and wing sections were pooled.
cDNA synthesis, library preparation and sequencing were carried out by Beijing Genomics Institute (Beijing, China). Samples were sequenced at either 75 PE on Illumina HiSeq 3000 or at 150 PE on Illumina HiSeq 4000.
Transcriptome assembly—Agraulis
All paired end sequence data for Agraulis were assembled with the transcriptome assembler Trinity [19]. This generated 87,214 contigs. Next the Trinity output was passed through TransDecoder (Haas & Papanicolaou, in prep), which annotates the transcript contigs based on the likelihood that they contain reading frames and also based on similarity by BLAST of transcripts to reference assemblies, in this case H. erato and H. melpomene. This annotation (a GFF3 annotation of the Trinity contigs) contained 24,984 genes, which compares to 20,102 annotated genes in H. melpomene v2.1 and 13,676 in H. erato v1.
Mapping and quantification
Reads were aligned with Hisat2 aligner to the genome of the respective species [27, 53]. The highest percentage of unique mappings was achieved using default parameters. Alignments were then quantified using GFF annotations of each genome with HTSeqCount, union mode [2]. Genomes and annotations are publicly available at www.lepbase.org [10].
Data analysis
Statistical analysis of counts was carried out using the R package DESeq 2 [34] using the following generalized linear model (GLM):
where ‘compartment’ == wing sector. In larvae, the wing sectors were forewing (FW) and hindwing (HW). In pupae, the wing sectors were as follows: proximal Forewing (FP), medial forewing (FM), distal forewing (FD), anterior hindwing (HA), posterior hindwing (HPo) (Fig. 2). Genes with an adjusted P value of > 0.05 and a Log2FoldChange of at least 1 were considered as significantly differentially expressed. Three biological replicates were generated.
Determining homology
Homology between differentially expressed genes in the three species was determined in two ways. First, a small percentage of genes have been assigned homologs by comparison with other lepidopteran genomes on LepBase using InterProScan [24]. This allowed recovery of one-to-one homologs and gene families with distinct insect lineages. However, in a number of cases where similar copies of a gene are present, for example the Wnt ligands, some genes were assigned to the incorrect orthogroup and were manually curated. For the rest of the genes, as well as all genes in Agraulis, amino acid sequences were reciprocally searched with BLASTp, and the top hit was taken as the homolog [1]. Genes with no assigned orthogroup were compared by pBLAST against the polypeptide library of D. melanogaster genes retrieved from FlyBase, associating them with a FBpp number and gene code based on homology with Drosophila genes [18].
Immunohistochemistry
Pupae of H. m rosina and H. elevatus were dissected 60–80 h after pupation in chilled PBS, or at an estimated 12 h before pupation for final instar larvae. H. elevatus was not used in RNAseq analysis, but also has a similar developmental time to the other species used here. Wings were fixed in 4% MeOH-free formaldehyde (Pierce) in PBSTw [PBS plus 0.01% Tween-20 (Sigma)] on ice for 40 min. They were then washed and permeabilized 6 × 5 min in PBSTx [PBS plus 0.5% Triton-X (Sigma)] and blocked for 2 h room temperature in PBSTx plus 5% goat serum (Sigma). Rabbit anti-Homothorax antibody (gift from Prof. Adi Salzberg, Technion-Israel Institute of Technology) or Rat anti-Optix (gift from Prof. Robert Reed, Cornell University) antibody was diluted to 1/1000 in PBSTx plus 0.5% goat serum and applied to wings overnight at 4 °C. Wings were washed 6 x 5 mins in PBSTx and incubated with 1/1000 goat anti-rabbit alexa-488 conjugated antibody (Abcam) in PBSTx for 3 h at room temperature. Wings were washed 4 x 5 mins with PBSTx and incubated 10 min in 1 µg/ml DAPI (Thermo Scientific) PBSTx. Wings were mounted in Fluoromount-G (SouthernBiotech) and imaged using a Leica DM6000B SP5 confocal microscope. An average of 60–80 images were taken to cover the entire wing, each a stack of 40 2 µm thick slices and composed of three channels: 408 nm, 488 nm and 568 nm. Each stack was converted to single images of maximum intensity using FIJI ImageJ 1.47 m and then split by channel. Images were then compiled into a single image and formatted using Adobe Photoshop CS5.1.
Additional file
Additional file 1. Supplementary Tables and Figures.
Acknowledgements
We thank Lucas Brenes, Henry Arenas-Castro, Oscar Paneso and Elizabeth Evans for assistance with insect rearing and tissue dissection, and the Beijing Genomics Institute for library preparation and sequencing. We thank Arnaud Martin for comments on an earlier draft of this text. We thank Adi Salzberg for use of the Hth antibody. We thank the Smithsonian Tropical Research Institute for support for tissue collection and work with butterflies in Panama, and ANAM in Panama for permission to collect butterflies.
Authors’ contributions
JJH, WOM and CDJ planned the research and wrote the paper, JJH performed experiments and analysis, RWRW performed immunofluorescence and imaging. All authors read and approved the final manuscript.
Funding
This research was funded by a Ph.D. Studentship from the Wellcome Trust to JJH, European Research Council Grant No. 339873 and Leverhulme trust Grant No RPG-2014-167 to RWRW.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the SRA repository [PRJNA552081].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnoult L, Su KF, Manoel D, Minervino C, Magrina J, Gompel N, Prud’homme B. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science. 2013;339:1423–1426. doi: 10.1126/science.1233749. [DOI] [PubMed] [Google Scholar]
- 4.Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 5.Bhardwaj S, Prudic KL, Bear A, Dasgupta M, Wasik BR, Tong X, Cheong WF, Wenk MR, Monteiro A. Sex differences in 20-hydroxyecdysone hormone levels control sexual dimorphism in Bicyclus anynana wing patterns. Mol Biol Evol. 2018;35:465–472. doi: 10.1093/molbev/msx301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- 7.Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/S0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- 8.Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/S0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- 9.Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GE, Selegue JE, Williams JA. Pattern formation and eyespot determination in butterfly wings. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- 10.Challis RJ, Kumar S, Dasmahapatra KKK, Jiggins CD, Blaxter M. Lepbase: the Lepidopteran genome database. BioRxiv. 2016:056994. https://www.biorxiv.org/content/10.1101/056994v1.abstract.
- 11.Connahs H, Tlili S, van Creij J, Loo TYJ, Banerjee TD, Saunders TE, Monteiro A. Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process. Development. 2019;146(9):dev169367. doi: 10.1242/dev.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Celis JF, Llimargas M, Casanova J. Ventral veinless, the gene encoding the Cf1a transcription factor, links positional information and cell differentiation during embryonic and imaginal development in Drosophila melanogaster. Development. 1995;121:3405–3416. doi: 10.1242/dev.121.10.3405. [DOI] [PubMed] [Google Scholar]
- 13.Enciso-Romero J, Pardo-Diaz C, Martin SH, Arias CF, Linares M, Mcmillan WO, Jiggins CD, Salazar C. Evolution of novel mimicry rings facilitated by adaptive introgression in tropical butterflies. Mol Ecol. 2017;26:5160–5172. doi: 10.1111/mec.14277. [DOI] [PubMed] [Google Scholar]
- 14.Galant R, Skeath JB, Paddock S, Lewis DL, Carroll SB. Expression pattern of a butterfly achaete-scute homolog reveals the homology of butterfly wing scales and insect sensory bristles. Curr Biol. 1998;8:807–813. doi: 10.1016/S0960-9822(98)70322-7. [DOI] [PubMed] [Google Scholar]
- 15.Gallant JR, Imhoff VE, Martin A, Savage WK, Chamberlain NL, Pote BL, Peterson C, Smith GE, Evans B, Reed RD, Kronforst MR, Mullen SP. Ancient homology underlies adaptive mimetic diversity across butterflies. Nat Commun. 2014;5:4817. doi: 10.1038/ncomms5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibert JM, Mouchel-Vielh E, Peronnet F. Pigmentation pattern and developmental constraints: flight muscle attachment sites delimit the thoracic trident of Drosophila melanogaster. Sci Rep. 2018;8:5328. doi: 10.1038/s41598-018-23741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 18.Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, The Flybase C. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45:D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hines HM, Papa R, Ruiz M, Papanicolaou A, Wang C, Nijhout HF, Mcmillan WO, Reed RD. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genom. 2012;13:288. doi: 10.1186/1471-2164-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iijima T, Kajitani R, Komata S, Lin CP, Sota T, Itoh T, Fujiwara H. Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon. Sci Adv. 2018;4:eaao5416. doi: 10.1126/sciadv.aao5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiggins CD, Wallbank RW, Hanly JJ. Waiting in the wings: what can we learn about gene co-option from the diversification of butterfly wing patterns? Philos Trans R Soc Lond B Biol Sci. 2017;372(1713):20150485. doi: 10.1098/rstb.2015.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones BW, Fetter RD, Tear G, Goodman CS. Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 24.Jones P, Binns D, Chang HY, Fraser M, Li W, Mcanulla C, Mcwilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavaler J, Fu W, Duan H, Noll M, Posakony JW. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development. 1999;126:2261–2272. doi: 10.1242/dev.126.10.2261. [DOI] [PubMed] [Google Scholar]
- 26.Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RL, Gates J, Scott MP, Carroll SB. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–534. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch PB, Merk R, Reinhardt R, Weber P. Localization of ecdysone receptor protein during colour pattern formation in wings of the butterfly Precis coenia (Lepidoptera: Nymphalidae) and co-expression with Distal-less protein. Dev Genes Evol. 2003;212:571–584. doi: 10.1007/s00427-002-0277-5. [DOI] [PubMed] [Google Scholar]
- 29.Kockel L, Zeitlinger J, Staszewski LM, Mlodzik M, Bohmann D. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. [DOI] [PubMed] [Google Scholar]
- 30.Kozak KM, Wahlberg N, Neild AF, Dasmahapatra KK, Mallet J, Jiggins CD. Multilocus species trees show the recent adaptive radiation of the mimetic heliconius butterflies. Syst Biol. 2015;64:505–524. doi: 10.1093/sysbio/syv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, Mullen SP, Kronforst MR. Doublesex is a mimicry supergene. Nature. 2014;507:229–232. doi: 10.1038/nature13112. [DOI] [PubMed] [Google Scholar]
- 32.Lewis JJ, Reed RD. Genome-wide regulatory adaptation shapes population-level genomic landscapes in Heliconius. Mol Biol Evol. 2018;36:159–173. doi: 10.1093/molbev/msy209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livraghi L, Martin A, Gibbs M, Braak N, Arif S. Breuker CJ. CRISPR/Cas9 as the key to unlocking the secrets of butterfly wing pattern development and its evolution. In: Advances in insect physiology, vol. 54. Academic Press; 2017. p. 85–115.
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macdonald WP, Martin A, Reed RD. Butterfly wings shaped by a molecular cookie cutter: evolutionary radiation of lepidopteran wing shapes associated with a derived Cut/wingless wing margin boundary system. Evol Dev. 2010;12:296–304. doi: 10.1111/j.1525-142X.2010.00415.x. [DOI] [PubMed] [Google Scholar]
- 36.Martin A, Mcculloch KJ, Patel NH, Briscoe AD, Gilbert LE, Reed RD. Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. Evodevo. 2014;5:7. doi: 10.1186/2041-9139-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 38.Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, Mcmillan WO, Reed RD. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA. 2012;109:12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin A, Reed RD. Wingless and aristaless2 define a developmental ground plan for moth and butterfly wing pattern evolution. Mol Biol Evol. 2010;27:2864–2878. doi: 10.1093/molbev/msq173. [DOI] [PubMed] [Google Scholar]
- 40.Martin A, Reed RD. Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev Biol. 2014;395:367–378. doi: 10.1016/j.ydbio.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 41.Mazo-vargas A, Concha C, Livraghi L, Massardo D, Wallbank RWR, Zhang L, Papador JD, Martinez-Najera D, Jiggins CD, Kronforst MR, Breuker CJ, Reed RD, Patel NH, Mcmillan WO, Martin A. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci USA. 2017;114:10701–10706. doi: 10.1073/pnas.1708149114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro A, Glaser G, Stockslager S, Glansdorp N, Ramos D. Comparative insights into questions of lepidopteran wing pattern homology. BMC Dev Biol. 2006;6:52. doi: 10.1186/1471-213X-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadeau NJ, Pardo-Diaz C, Whibley A, Supple MA, Saenko SV, Wallbank RW, Wu GC, Maroja L, Ferguson L, Hanly JJ, Hines H, Salazar C, Merrill RM, Dowling AJ, Ffrench-Constant RH, Llaurens V, Joron M, Mcmillan WO, Jiggins CD. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature. 2016;534:106–110. doi: 10.1038/nature17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozsu N, Chan QY, Chen B, Gupta MD, Monteiro A. Wingless is a positive regulator of eyespot color patterns in Bicyclus anynana butterflies. Dev Biol. 2017;429:177–185. doi: 10.1016/j.ydbio.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 45.Patel NH, Kornberg TB, Goodman CS. Expression of engrailed during segmentation in grasshopper and crayfish. Development. 1989;107:201–212. doi: 10.1242/dev.107.2.201. [DOI] [PubMed] [Google Scholar]
- 46.Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 47.Prakash A, Monteiro A. apterous A specifies dorsal wing patterns and sexual traits in butterflies. BioRxiv. 2018:131011. https://royalsocietypublishing.org/doi/full/10.1098/rspb.2017.2685. [DOI] [PMC free article] [PubMed]
- 48.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed RD. Evidence for Notch-mediated lateral inhibition in organizing butterfly wing scales. Dev Genes Evol. 2004;214:43–46. doi: 10.1007/s00427-003-0366-0. [DOI] [PubMed] [Google Scholar]
- 50.Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, Halder G, Nijhout HF, Mcmillan WO. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 51.Reed RD, Serfas MS. Butterfly wing pattern evolution is associated with changes in a Notch/Distal-less temporal pattern formation process. Curr Biol. 2004;14:1159–1166. doi: 10.1016/j.cub.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 52.Rogers WA, Salomone JR, Tacy DJ, Camino EM, Davis KA, Rebeiz M, Williams TM. Recurrent modification of a conserved cis-regulatory element underlies fruit fly pigmentation diversity. PLoS Genet. 2013;9:e1003740. doi: 10.1371/journal.pgen.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rutledge W, Gibson R, Siegel E, Duke K, Jones R, Rucinski D, Nunn G, Torrence WA, Lewellen-Williams C, Stewart C, Blann K, Belleton L, Fincher L, Klimberg VS, Greene P, Thomas B, Erwin D, Henry-TillmaN R. Arkansas special populations access network perception versus reality—cancer screening in primary care clinics. Cancer. 2006;107:2052–2060. doi: 10.1002/cncr.22147. [DOI] [PubMed] [Google Scholar]
- 54.Saenko SV, Marialva MS, Beldade P. Involvement of the conserved Hox gene Antennapedia in the development and evolution of a novel trait. Evodevo. 2011;2:9. doi: 10.1186/2041-9139-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato A, Kojima T, Ui-Tei K, Miyata Y, Saigo K. Dfrizzled-3, a new Drosophila Wnt receptor, acting as an attenuator of Wingless signaling in wingless hypomorphic mutants. Development. 1999;126:4421–4430. doi: 10.1242/dev.126.20.4421. [DOI] [PubMed] [Google Scholar]
- 56.Schilling S, Steiner S, Zimmerli D, Basler K. A regulatory receptor network directs the range and output of the Wingless signal. Development. 2014;141:2483–2493. doi: 10.1242/dev.108662. [DOI] [PubMed] [Google Scholar]
- 57.Stoehr AM, Walker JF, Monteiro A. Spalt expression and the development of melanic color patterns in pierid butterflies. Evodevo. 2013;4:6. doi: 10.1186/2041-9139-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treisman JE, Lai ZC, Rubin GM. Shortsighted acts in the decapentaplegic pathway in Drosophila eye development and has homology to a mouse TGF-beta-responsive gene. Development. 1995;121:2835–2845. doi: 10.1242/dev.121.9.2835. [DOI] [PubMed] [Google Scholar]
- 59.Urness LD, Thummel CS. Molecular analysis of a steroid-induced regulatory hierarchy: the Drosophila E74A protein directly regulates L71-6 transcription. EMBO J. 1995;14:6239–6246. doi: 10.1002/j.1460-2075.1995.tb00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Belleghem SM, Rastas P, Papanicolaou A, Martin SH, Arias CF, Supple MA, Hanly JJ, Mallet J, Lewis JJ, Hines HM, Ruiz M. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 2017;1(3):0052. doi: 10.1038/s41559-016-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallbank RW, Baxter SW, Pardo-Diaz C, Hanly JJ, Martin SH, Mallet J, Dasmahapatra KK, Salazar C, Joron M, Nadeau N, Mcmillan WO, Jiggins CD. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 2016;14:e1002353. doi: 10.1371/journal.pbio.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walters JR, Hardcastle TJ, Jiggins CD. Sex chromosome dosage compensation in heliconius butterflies: global yet still incomplete? Genome Biol Evol. 2015;7:2545–2559. doi: 10.1093/gbe/evv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren RW, Nagy L, Selegue J, Gates J, Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- 64.Weatherbee SD, Nijhout HF, Grunert LW, Halder G, Galant R, Selegue J, Carroll S. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 1999;9:109–115. doi: 10.1016/S0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 65.Woronik A, Tunström K, Perry MW, Neethiraj R, Stefanescu C, de la Paz Celorio-Mancera M, Brattström O, Hill J, Lehmann P, Käkelä R, Wheat CW (2018) A homeobox gene, BarH-1, underlies a female alternative life-history strategy. BioRxiv. 424879 https://www.biorxiv.org/content/10.1101/424879v3.full.
- 66.Yassin A, Delaney EK, Reddiex AJ, Seher TD, Bastide H, Appleton NC, Lack JB, David JR, Chenoweth SF, Pool JE, Kopp A. The pdm3 locus is a hotspot for recurrent evolution of female-limited color dimorphism in Drosophila. Curr Biol. 2016;26:2412–2422. doi: 10.1016/j.cub.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Reed RD. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 2016;7:11769. doi: 10.1038/ncomms11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang LL, Mazo-Vargas A, Reed RD. Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc Natl Acad Sci USA. 2017;114:10707–10712. doi: 10.1073/pnas.1709058114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Tables and Figures.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the SRA repository [PRJNA552081].