Abstract
Interest in bat-related viruses has increased considerably during the last decade, leading to the discovery of a rising number of new viruses in several bat species. Poxviridae are a large, diverse family of DNA viruses that can infect a wide range of vertebrates and invertebrates. To date, only a few documented detections of poxviruses have been described in bat populations on three different continents (America, Africa, and Australia). These viruses are phylogenetically dissimilar and have diverse clinical impacts on their hosts. Herein, we report the isolation, nearly complete genome sequencing, and annotation of a novel poxvirus detected from an insectivorous bat (Hypsugo savii) in Northern Italy. The virus is tentatively named Hypsugopoxvirus (HYPV) after the bat species from which it was isolated. The nearly complete genome size is 166,600 nt and it encodes 161 genes. Genome analyses suggest that HYPV belongs to the Chordopoxvirinae subfamily, with the highest nucleotide identity (85%) to Eptesipoxvirus (EPTV) detected from a microbat Eptesicus fuscus in WA, USA, in 2011. To date, HYPV represents the first poxvirus detected in bats in Europe; thus, its viral ecology and disease associations should be investigated further.
Keywords: bats, poxvirus, Italy
1. Introduction
Poxviruses are dsDNA viruses with large genomes (130 to 360 kb) that belong to the family Poxviridae. The family is divided into the Entomopoxvirinae and the Chordopoxvirinae subfamilies of viruses, which infect insects and vertebrates, respectively. According to the International Committee on Taxonomy of Viruses (ICTV) 2017 Release [1], 11 genera have been created to classify Chordopoxviruses (Avipoxvirus, Capripoxvirus, Centapoxvirus, Cervidpoxvirus, Crocodylidpoxvirus, Leporipoxvirus, Molluscipoxvirus, Orthopoxvirus, Parapoxvirus, Suipoxvirus, and Yatapoxvirus), but other viruses remain unclassified and new genera are likely to be recognized in the future. Poxviruses show a diverse host range, with some viruses having wide host tropism (e.g., Orthopoxviruses) and thus being consequently associated with greater zoonotic risks [2], and others having strict host specificity.
In recent decades, bats have been increasingly recognized as reservoirs of emerging viral infections, which has important ramifications for animal and public health [3]. However, the majority of bat-borne viruses that can cause severe diseases in humans and other mammals, do not cause apparent clinical signs in bats. Consequently, it has been assumed that bats may have a “special” relationship with viruses based on physiological, ecological, evolutionary, and/or immunological aspects, which allow them to act as special viral reservoirs with exaggerated viral richness [4,5,6,7].
Currently, four poxviruses from the Microchiroptera and Macrochiroptera suborders have been detected in bat populations on three continents (America, Africa, and Australia) [8]. Specifically, Eptesipoxvirus (EPTV) was isolated in North America in 2011 from Eptesicus fuscus [9,10]; Eidolon helvum poxvirus 1 (EHPV1) was detected in West Africa in 2009 from Eidolon helvum [11]; the Pteropox virus (PTPV) was identified in Northwestern Australia in 2015 from Pteropus scapulatus [12]; and a fourth poxvirus was also identified in South Australia from Miniopterus schreibersii bassanii in 2009 [13]. It is remarkable that these viruses are phylogenetically divergent and are associated with variable clinical manifestations.
Virological investigations focused on poxviruses in bat populations may have a positive impact for future ecological studies of bat–pathogen interactions. Moreover, from the perspective of the One Health approach, bats could benefit from these studies, since European bat populations are currently undergoing a global decline that could be linked with so far overlooked viral infections.
In this study, we report the isolation, nearly complete genomic sequencing, and annotation of a novel poxvirus detected from an insectivorous bat (Hypsugo savii) in Northern Italy. The virus was tentatively named Hypsugopoxvirus (HYPV), according to the bat species from which it was isolated. Phylogenetic analyses suggest that HYPV belongs to the Chordopoxvirinae subfamily, revealing the highest similarity (85%) with Eptesipoxvirus (EPTV) detected from the microbat Eptesicus fuscus in WA, USA in 2011, which is associated with bat necrosuppurative osteomyelitis in multiple joints. HYPV is the first poxvirus detected in bats in Europe and its viral ecology and disease associations should be investigated further.
2. Materials and Methods
2.1. Sampling
Dead bats from different species were collected for virological investigations from wild animal rescue/rehabilitation centers in the context of a general surveillance project that has been implemented in Northern Italy since 2009–2010, which focuses on the detection of emerging bat viruses [14,15,16]. The bats were taxonomically identified based on their morphologic characteristics, according to the European bat identification keys [15]. The carcasses were necropsied, and tissue samples were collected for further laboratory exams, particularly for viral detection and isolation.
2.2. Virological Analysis
After necropsy, organ samples (lungs, heart, kidney, brain, and intestines) were mechanically homogenized in minimal essential medium (1 g/10 mL), which contained antibiotics. They were then centrifuged at 3000 g for 15 min. Samples were inoculated in confluent monolayers of VERO and MARC 145 cells (African green monkey), incubated at 37 °C with 5% CO2 and observed daily for seven days to assess their cytopathic effects (CPEs). In the absence of CPEs, the cryolysates were sub-cultured twice onto fresh monolayers. Cell culture supernatants showing CPE were partially purified by ultracentrifugation at 35,000 rpm for 2 h (rotor TST41 Kontron) through a 25% (w/w) sucrose cushion, and the pellet was re-suspended in PBS. This antigen was kept at −70 °C and then submitted for viral identification with the NGS approach and negative-staining electron microscopy (nsEM) by using the Airfuge (Beckman Instruments, Palo Alto, CA, USA) method [17].
2.3. Molecular Analysis
Viral DNA was extracted from 200 μL of positive cell culture supernatants using a BioSprint 96 One-For-All Vet Kit (Qiagen S.p.A., Milan, Italy). Sequencing libraries were made with a Nextera Flex kit (Illumina Inc. San Diego, CA, USA) in accordance with the manufacturer’s instructions. Libraries were sequenced on a MiSeq Instrument (Illumina Inc. San Diego, CA, USA) by using a MiSeq Reagent Kit v2 in a 250 cycle paired-end run. Data were assembled de novo by the CLC Genomic workbench v.11 (Qiagen S.p.A., Milan, Italy).
Genome annotation and analysis was performed with tools from the bioinformatics suite developed at the Viral Bioinformatics Resource Centre [18]. The Genome Annotation Transfer Utility (GATU) [19] uses a reference genome to automatically annotate poxvirus genes with clear orthologs in the reference. Other possible genes were presented to the annotator for further characterization and to make final annotation decisions.
3. Results
3.1. Clinical Case
The case specifically concerned a juvenile Hypsugo savii male that spontaneously died in a wildlife recovery center in Valpredina, Cenate Sopra (BG), Northern Italy after several weeks of hospitalization. The sick bat was originally found alive on July 17, 2017 in Telgate (Bergamo Province, Northern Italy) by a private citizen who brought it to the center. Clinically, the bat had a humerus fracture, sensory depression and a lack of appetite but normal body mass. The death occurred 54 days after admission to the center on September 9, 2017; then, the carcass was sent to the lab for necroscopy and further analyses. Pathological lesions in the internal organs indicative of infectious diseases were not observed, but a soft bone callus due to pathological healing of the humerus fracture associated with osteomalacia and calcium deficiency was detected.
3.2. Virus Isolation and Identification
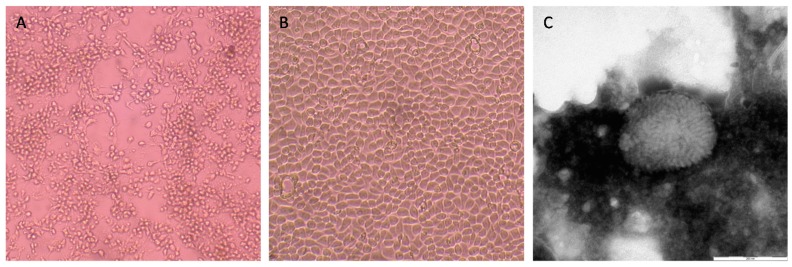
A virus was isolated on MARC 145 cells inoculated with the organ pool composed of the bat’s heart and lungs. The CPE occurred on the third day post-inoculation during the second passage and was characterized by a diffused degeneration of a monolayer with rounded cells floating in the culture medium (Figure 1A,B). The cell culture supernatant showing CPE was submitted to the NGS in order to identify and characterize the unknown isolate. Furthermore, nsEM performed on the purified and concentrated antigen revealed the presence of viral particles that unequivocally morphologically resembled those belonging to the genus Orthopoxvirus (Figure 1C). The virus was tentatively named Hypsugopoxvirus (HYPV), according to the bat species from which it was isolated. Table 1 summarizes the basic information on the HYPV identified in this study in comparison with all known poxviruses detected to date in bats worldwide.
Figure 1.
(A) Cytopathic effects (CPEs) of rounded cells floating in the culture medium of MARC 145 cells infected with the pool of bat organs (heart and lungs) at three days after inoculation (original magnification × 100); (B) mock cells (original magnification ×100); (C) negative-staining electron microscopy showing the presence of a virion morphologically related to the Orthopoxvirus genus from the MARC 145 cell culture.
Table 1.
Basic data on Hypsugopoxvirus (HYPV) in comparison with all known poxviruses detected to date in bats worldwide.
| Poxvirus Strain | Host | Sample Source | Origin | Collection Date | Clinical/Post-Mortem Findings | Laboratory Outcomes | Ref. |
|---|---|---|---|---|---|---|---|
| Hypsugopox virus (HYPV) Id lab: IZSLER 251170-23/2017 |
Hypsugo savii | Pool of viscera (heart and lungs) | Europe (Italy) | 2017 | Humerus fracture and osteomalacia, calcium deficiency | CC, EM, nFGS (166,600 nt), GA (161 genes) | This study |
| Pteropox virus (PTPV) | Pteropus scapulatus | Wing membrane | North Western Australia (Kimberley region) | 2015 | Multiple nodules on the wing membranes | PGS (133,492 nt), GA (143 genes) | [12] |
| Eptesipox virus (EPTV) strain “Washington“ | Eptesicus fuscus | Elbow joint | America (WA, USA) |
2011 | Necro-suppurative osteomyelitis in multiple joints | CC, EM, FGS (176,688 nt), GA (191 genes) | [9,10] |
| Eidolon helvum poxvirus 1 (EHPV1) | Eidolon helvum | Throat swabs | Africa (Ghana) | 2009 | Apparently healthy bats | PGS | [11] |
| NA | Miniopterus schreibersii bassanii | Skin biopsies | South Australia (Naracoorte) | 2009 | Nodular cutaneous lesions | EM | [13] |
NA: not available; CC: cell culture isolation; EM: electron microscopy identification; FGS: full-genome sequence; nFGS: nearly full-genome sequence; PGS: partial genome sequence; GA: genome annotation.
3.3. Genome Characterization
After NGS sequencing, the nearly complete viral genome of a poxvirus was obtained from one contig of 166,600 nucleotides originating from 85,678 reads with an average coverage of 118.53. The nearly full genome sequence of the viral strain was determined and compared with those of other members of the Poxviridae family available on GenBank. For the nearly complete viral genome sequencing, BLAST analysis revealed the highest nucleotide identity (85%) to the Eptesipoxvirus (EPTV) strain “Washington”, a member of the Chordopoxvirinae subfamily identified in microbats in the USA (Table 2). The nearly complete genome sequence for HYPV was submitted to GenBank under accession number MK860688.
Table 2.
Highest nucleotide sequence identities for the nearly complete genome of HYPV.
A conservative approach was taken for genome annotation to avoid over-annotating open reading frames (ORFs) that were unlikely to represent functional genes. ORFs less than 50 codons or overlapping by more than 25% with well-characterized genes were not considered for annotation unless supported by other evidence. A total of 161 genes were annotated for HYPV, showing a percentage value of nt identity with its closest related virus EPTV ranging from 42.5% for the HYPV-2 gene (serpin 2) to 100% for the HYPV-90 gene (VLTF-3) (Table 3).
Table 3.
HYPV genome annotation and nucleotide identities for each gene to the most similar strain Eptesipoxvirus (EPTV). The seven conserved genes used for phylogenetic analysis in previous studies [10,12] are presented in bold.
| Gene Name | Putative Product Identity | Start | Stop | +/− | Size | % Id. to EPTV | Orthologs |
|---|---|---|---|---|---|---|---|
| HYPV-1 | Hypothetical protein | 87 | 557 | − | 471 | 58 | EPTV-001 |
| HYPV-2 | Serpin 2 | 1037 | 1552 | − | 516 | 42.5 | EPTV-002 |
| HYPV-3 | Hypothetical protein | 1581 | 2261 | − | 681 | 82.4 | EPTV-003 |
| HYPV-4 | IL-1 receptor-like protein | 2309 | 3316 | − | 1008 | 65.1 | EPTV-004 |
| HYPV-5 | Hypothetical protein | 3356 | 3835 | − | 480 | 88.8 | EPTV-005 |
| HYPV-6 | Tyrosine protein kinase-like protein | 3872 | 4774 | − | 903 | 91.7 | EPTV-006 |
| HYPV-7 | ER-localized apoptosis regulator | 4842 | 5522 | − | 681 | 63.6 | EPTV-007 |
| HYPV-8 | Hypothetical protein | 7002 | 7481 | − | 480 | 80.5 | EPTV-008 |
| HYPV-9 | Ankyrin repeat-containing protein, host range | 8141 | 9826 | − | 1686 | 63.0 | EPTV-010 |
| HYPV-10 | Monoglyceride lipase | 11,053 | 11,913 | − | 861 | 93.1 | EPTV-014 |
| HYPV-11 | Secreted EGF-like growth factor | 12,340 | 12,588 | − | 249 | 62.4 | EPTV-015 |
| HYPV-12 | Anti-apoptotic factor | 12,594 | 13,100 | − | 507 | 65.7 | EPTV-016 |
| HYPV-13 | dUTPase | 13,144 | 13,569 | − | 426 | 87.2 | EPTV-017 |
| HYPV-14 | IFN-inducible protein | 13,597 | 14,004 | − | 408 | 83.7 | EPTV-018 |
| HYPV-15 | Ribonucleotide reductase small subunit | 14,060 | 15,034 | − | 975 | 93.8 | EPTV-019 |
| HYPV-16 | F5L membrane protein | 15,075 | 16,139 | − | 1065 | 68.4 | EPTV-020 |
| HYPV-17 | Cytoplasmic protein | 16,687 | 16,869 | − | 183 | 71.4 | EPTV-023 |
| HYPV-18 | S–S bond formation pathway protein | 17,361 | 18,008 | − | 648 | 92.6 | EPTV-025 |
| HYPV-19 | Ser|Thr protein kinase | 17,998 | 19,314 | − | 1317 | 94.7 | EPTV-026 |
| HYPV-20 | RhoA signaling inhibitor, virus release protein | 19,334 | 20,626 | − | 1293 | 88.0 | EPTV-07 |
| HYPV-21 | EEV maturation protein | 20,659 | 22,602 | − | 1944 | 89.0 | EPTV-028 |
| HYPV-22 | Palmitylated EEV membrane glycoprotein | 22,640 | 23,755 | − | 1116 | 98.9 | EPTV-029 |
| HYPV-23 | Hypothetical protein | 23,781 | 24,008 | − | 228 | 67.1 | EPTV-031 |
| HYPV-24 | Hypothetical protein | 24,050 | 24,250 | − | 201 | 97.0 | Unique to HYPV |
| HYPV-25 | Hypothetical protein | 24,471 | 24,917 | − | 447 | 92.6 | Unique to HYPV |
| HYPV-26 | Conserved non-functional serine recombinase | 24,992 | 25,654 | − | 663 | 78.8 | EPTV-033 |
| HYPV-27 | DNA-binding phosphoprotein | 25,714 | 26,052 | + | 339 | 86.7 | EPTV-034 |
| HYPV-28 | Poly (A) polymerase catalytic subunit | 26,046 | 27,461 | − | 1416 | 92.6 | EPTV-035 |
| HYPV-29 | IEV morphogenesis | 27,478 | 29,676 | − | 2199 | 93.3 | EPTV-036 |
| HYPV-30 | RNA polymerase subunit | 29,733 | 30,455 | − | 723 | 93.8 | EPTV-038 |
| HYPV-31 | IMV protein, virion morphogenesis | 30,760 | 32,463 | + | 1704 | 95.8 | EPTV-039 |
| HYPV-32 | ER-localized membrane protein, virion core protein | 32,490 | 33,302 | + | 813 | 95.6 | EPTV-040 |
| HYPV-33 | DNA polymerase | 33,299 | 36,319 | − | 3021 | 93.8 | EPTV-041 |
| HYPV-34 | Sulfhydryl oxidase (FAD-linked) | 36,352 | 36,642 | + | 291 | 96.9 | EPTV-042 |
| HYPV-35 | Virion core protein | 36,645 | 37,055 | − | 411 | 87.9 | EPTV-043 |
| HYPV-36 | Virulence, modulates Raf|MEK|ERK pathway | 37,039 | 39,117 | − | 2079 | 91.9 | EPTV-044 |
| HYPV-37 | Nonessential glutaredoxin | 39,173 | 39,487 | − | 315 | 91.3 | EPTV-045 |
| HYPV-38 | DNA-binding core protein | 39,613 | 40,545 | − | 933 | 90.3 | EPTV-046 |
| HYPV-39 | IMV membrane protein | 40,546 | 40,767 | − | 222 | 83.6 | EPTV-047 |
| HYPV-40 | ssDNA-binding phosphoprotein | 40,768 | 41,577 | − | 810 | 87.5 | EPTV-048 |
| HYPV-41 | Ribonucleotide reductase large subunit | 41,640 | 43,925 | − | 2286 | 95.2 | EPTV-049 |
| HYPV-42 | IMV protein (VP13) | 43,966 | 44,202 | − | 237 | 88.5 | EPTV-050 |
| HYPV-43 | Telomere-binding protein | 44,220 | 45,371 | − | 1152 | 90.9 | EPTV-051 |
| HYPV-44 | Viral core cysteine proteinase | 45,364 | 46,650 | − | 1287 | 94.6 | EPTV-052 |
| HYPV-45 | RNA-helicase, DExH-NPH-II | 46,656 | 48,686 | + | 2031 | 94.3 | EPTV-053 |
| HYPV-46 | Insulin metalloproteinase-like protein | 48,678 | 50,465 | − | 1788 | 92.3 | EPTV-054 |
| HYPV-47 | Entry|fusion complex component | 50,462 | 50,794 | − | 333 | 97.3 | EPTV-055 |
| HYPV-48 | Late transcription elongation factor (VLTF) | 50,788 | 51,456 | + | 669 | 90.5 | EPTV-056 |
| HYPV-49 | Thioredoxin-like protein | 51,423 | 51,800 | − | 378 | 89.6 | EPTV-057 |
| HYPV-50 | FEN1-like nuclease | 51,803 | 53,140 | + | 1338 | 87.0 | EPTV-058 |
| HYPV-51 | RNA polymerase subunit | 53,142 | 53,333 | + | 192 | 96.8 | EPTV-059 |
| HYPV-52 | NLPc|P60 superfamily protein | 53,337 | 53,870 | + | 534 | 87.7 | EPTV-060 |
| HYPV-53 | Virion structural phosphoprotein, early morphogenesis | 53,836 | 54,933 | − | 1098 | 91.3 | EPTV-061 |
| HYPV-54 | Late transcription factor | 54,962 | 55,744 | + | 783 | 98.5 | EPTV-062 |
| HYPV-55 | Myristylated entry|fusion protein | 55,760 | 56,782 | + | 1023 | 93.8 | EPTV-063 |
| HYPV-56 | Myristylated IMV envelope protein | 56,783 | 57,532 | + | 750 | 96.4 | EPTV-064 |
| HYPV-57 | Crescent membrane|immature virion protein | 57,558 | 57,833 | + | 276 | 84.6 | EPTV-065 |
| HYPV-58 | Internal virion protein | 57,825 | 58,790 | − | 966 | 92.1 | EPTV-066 |
| HYPV-59 | DNA-binding virion protein | 58,815 | 59,573 | + | 759 | 98.4 | EPTV-067 |
| HYPV-60 | IMV protein, entry|fusion | 59,588 | 59,992 | + | 405 | 94.0 | EPTV-068 |
| HYPV-61 | IMV membrane protein, virion morphogenesis | 59,934 | 60,380 | + | 447 | 95.9 | EPTV-069 |
| HYPV-62 | Thymidine kinase | 60,402 | 60,932 | + | 531 | 93.8 | EPTV-070 |
| HYPV-63 | Type I IFN inhibitor | 61,026 | 61,625 | + | 600 | 73.6 | EPTV-071 |
| HYPV-64 | Poly (A) polymerase small subunit | 61,692 | 62,693 | + | 1002 | 94.3 | EPTV-072 |
| HYPV-65 | RNA polymerase subunit (RPO22) | 62,608 | 63,165 | + | 558 | 96.8 | EPTV-073 |
| HYPV-66 | IMV membrane protein, entry|fusion | 63,170 | 63,580 | − | 411 | 94.1 | EPTV-074 |
| HYPV-67 | RNA polymerase subunit (RPO147) | 63,688 | 67,545 | + | 3858 | 98.5 | EPTV-075 |
| HYPV-68 | Tyr|Ser kinase, virus assembly, IFN-gamma inhibitor | 67,542 | 68,060 | − | 519 | 97.7 | EPTV-076 |
| HYPV-69 | Entry|fusion IMV protein | 68,074 | 68,646 | + | 573 | 98.9 | EPTV-077 |
| HYPV-70 | IMV heparin-binding surface protein | 68,654 | 69,667 | − | 1014 | 90.6 | EPTV-078 |
| HYPV-71 | RNA polymerase-associated protein (RAP94) | 69,671 | 72,058 | − | 2388 | 97.5 | EPTV-079 |
| HYPV-72 | Late transcription factor | 72,228 | 72,872 | + | 645 | 71.9 | EPTV-080 |
| HYPV-73 | DNA topoisomerase type I | 72,894 | 73,829 | + | 936 | 93.9 | EPTV-081 |
| HYPV-74 | Crescent membrane|immature virion protein | 73,868 | 74,314 | + | 447 | 88.6 | EPTV-082 |
| HYPV-75 | mRNA capping enzyme large subunit | 74,355 | 76,889 | + | 2535 | 95.7 | EPTV-083 |
| HYPV-76 | Virion core protein | 76,851 | 77,288 | − | 438 | 89.8 | EPTV-084 |
| HYPV-77 | Virion core protein | 77,287 | 78,030 | + | 744 | 83.0 | EPTV-085 |
| HYPV-78 | Uracil DNA glycosylase, DNA pol processivity factor | 78,027 | 78,683 | + | 657 | 96.8 | EPTV-086 |
| HYPV-79 | NTPase, DNA primase | 78,717 | 81,080 | + | 2364 | 96.6 | EPTV-087 |
| HYPV-80 | Early transcription factor small subunit (VETF-s) | 81,077 | 82,984 | + | 1908 | 99.1 | EPTV-088 |
| HYPV-81 | RNA polymerase subunit | 83,017 | 83,532 | + | 516 | 90.1 | EPTV-089 |
| HYPV-82 | Carbonic anhydrase, GAG-binding MV membrane protein | 83,464 | 84,339 | − | 876 | 82.5 | EPTV-090 |
| HYPV-83 | mRNA decapping enzyme | 84,397 | 85,068 | + | 672 | 85.7 | EPTV-091 |
| HYPV-84 | mRNA decapping enzyme | 85,043 | 85,822 | + | 780 | 92.0 | EPTV-092 |
| HYPV-85 | ATPase, NPH1 | 85,796 | 87,703 | − | 1908 | 98.1 | EPTV-093 |
| HYPV-86 | mRNA capping enzyme small subunit | 87,746 | 88,609 | − | 864 | 96.5 | EPTV-094 |
| HYPV-87 | Trimeric virion coat protein | 88,643 | 90,295 | − | 1653 | 94.6 | EPTV-095 |
| HYPV-88 | Late transcription factor (VLTF-2) | 90,321 | 90,776 | − | 456 | 93.4 | EPTV-096 |
| HYPV-89 | Late transcription factor (VLTF-3) | 90,805 | 91,479 | − | 675 | 100.0 | EPTV-097 |
| HYPV-90 | S-S bond formation pathway protein | 91,476 | 91,706 | − | 231 | 93.4 | EPTV-098 |
| HYPV-91 | P4b precursor | 91,726 | 93,726 | − | 2001 | 93.3 | EPTV-099 |
| HYPV-92 | RNA polymerase subunit (RPO19) | 94,462 | 94,992 | + | 531 | 87.5 | EPTV-101 |
| HYPV-93 | Virion morphogenesis core protein | 94,989 | 96,107 | − | 1119 | 94.1 | EPTV-102 |
| HYPV-94 | Early transcription factor large subunit (VETF-L) | 96,131 | 98,275 | − | 2145 | 97.8 | EPTV-103 |
| HYPV-95 | Intermediate transcription factor (VITF-3s) | 98,338 | 99,213 | + | 876 | 94.2 | EPTV-104 |
| HYPV-96 | IMV membrane protein, early morphogenesis | 99,223 | 99,459 | − | 237 | 92.5 | EPTV-105 |
| HYPV-97 | P4a precursor | 99,460 | 102,192 | − | 2733 | 90.5 | EPTV-106 |
| HYPV-98 | Viral membrane formation | 102,207 | 103,142 | + | 936 | 96.1 | EPTV-107 |
| HYPV-99 | Virion core and cleavage processing protein | 103,139 | 103,705 | − | 567 | 76.7 | EPTV-108 |
| HYPV-100 | IMV membrane protein, virion maturation | 103,799 | 104,002 | − | 204 | 71.6 | EPTV-109 |
| HYPV-101 | IMV membrane protein, essential | 104,067 | 104,348 | − | 282 | 96.8 | EPTV-110 |
| HYPV-102 | IMV membrane protein, non-essential | 104,365 | 104,526 | − | 162 | 98.1 | EPTV-111 |
| HYPV-103 | Core protein | 104,516 | 104,809 | − | 294 | 95.9 | EPTV-112 |
| HYPV-104 | Myristylated protein, essential for entry | 104,793 | 105,935 | − | 1143 | 91.8 | EPTV-113 |
| HYPV-105 | IMV membrane protein | 105,936 | 106,526 | − | 591 | 96.9 | EPTV-114 |
| HYPV-106 | DNA helicase, transcript release factor | 106,541 | 107,995 | + | 1455 | 90.1 | EPTV-115 |
| HYPV-107 | Zn finger-like protein, late morphogenesis | 107,967 | 108,188 | − | 222 | 93.2 | EPTV-116 |
| HYPV-108 | IMV membrane protein, entry|fusion | 108,189 | 108,533 | − | 345 | 95.6 | EPTV-117 |
| HYPV-109 | DNA polymerase processivity factor | 108,532 | 109,809 | + | 1278 | 89.4 | EPTV-118 |
| HYPV-110 | Holliday junction resolvase | 109,793 | 110,338 | + | 546 | 91.3 | EPTV-119 |
| HYPV-111 | Intermediate transcription factor (VITF-3L) | 110,335 | 111,495 | + | 1161 | 91.5 | EPTV-120 |
| HYPV-112 | RNA polymerase subunit (RPO132) | 111,492 | 115,010 | + | 3519 | 97.7 | EPTV-121 |
| HYPV-113 | A-type inclusion protein | 114,996 | 117,869 | − | 2874 | 77.4 | EPTV-122 |
| HYPV-114 | P4c precursor | 117,926 | 119,800 | − | 1875 | 82.1 | EPTV-123 |
| HYPV-115 | IMV membrane protein, fusion | 119,856 | 120,206 | − | 351 | 86.2 | EPTV-124 |
| HYPV-116 | IMV membrane protein, entry | 120,207 | 120,623 | − | 417 | 94.2 | EPTV-125 |
| HYPV-117 | RNA polymerase subunit (RPO35) | 120,637 | 121,539 | − | 903 | 94.7 | EPTV-126 |
| HYPV-118 | IMV protein | 121,523 | 121,750 | − | 228 | 92.0 | EPTV-127 |
| HYPV-119 | Hypothetical protein | 121,953 | 122,435 | + | 483 | 80.7 | EPTV-128 |
| HYPV-120 | ATPase|DNA packaging protein | 122,465 | 123,235 | − | 771 | 95.3 | EPTV-129 |
| HYPV-121 | C-type lectin-like EEV membrane phosphoglycoprotein | 123,371 | 123,922 | + | 552 | 77.2 | EPTV-130 |
| HYPV-122 | C-type lectin like IEV|EEV membrane glycoprotein | 123,970 | 124,470 | + | 501 | 91.0 | EPTV-131 |
| HYPV-123 | MHC class II antigen presentation inhibitor | 124,509 | 125,033 | + | 525 | 81.0 | EPTV-132 |
| HYPV-124 | Concanavalin-like precursor | 125,073 | 125,918 | + | 846 | 78.0 | EPTV-133 |
| HYPV-125 | EEV glycoprotein | 125,953 | 126,681 | + | 729 | 68.7 | EPTV-134 |
| HYPV-126 | Hypothetical protein | 126,724 | 127,554 | + | 831 | 79.4 | EPTV-135 |
| HYPV-127 | Hypothetical protein | 127,578 | 127,817 | + | 240 | 68.2 | EPTV-136 |
| HYPV-128 | Truncated CD47-like protein, integral membrane protein | 127,814 | 128,404 | − | 591 | 80.3 | EPTV-137 |
| HYPV-129 | Myristylated protein | 128,422 | 128,829 | + | 408 | 73.3 | EPTV-138 |
| HYPV-130 | Hypothetical protein | 128,826 | 129,587 | + | 762 | 78.3 | EPTV-139 |
| HYPV-131 | Chemokine binding protein | 129,575 | 130,438 | − | 864 | 69.0 | EPTV-140 |
| HYPV-132 | Profilin-like protein, ATI-localized | 130,558 | 130,959 | + | 402 | 98.5 | EPTV-141 |
| HYPV-133 | Hypothetical protein | 130,956 | 131,339 | − | 384 | 76.9 | EPTV-142 |
| HYPV-134 | 3 beta-hydroxysteroid dehydrogenase|delta 5->4 isomerase | 131,948 | 133,015 | + | 1068 | 84.2 | EPTV-144 |
| HYPV-135 | Thymidylate kinase | 133,646 | 134,233 | + | 588 | 85.2 | EPTV-147 |
| HYPV-136 | DNA ligase-like protein | 134,265 | 135,944 | + | 1680 | 87.2 | EPTV-148 |
| HYPV-137 | A52R-like family protein | 137,441 | 138,046 | + | 606 | 77.9 | EPTV-150 |
| HYPV-138 | Hypothetical protein | 138,641 | 139,717 | + | 1077 | 65.2 | EPTV-151 |
| HYPV-139 | Toll|IL-1 receptor-like protein, IL-1, NFkB signaling inhibitor | 139,781 | 140,428 | + | 648 | 89.8 | EPTV-153 |
| HYPV-140 | Hypothetical protein | 140,538 | 140,954 | − | 417 | 70.5 | EPTV-154 |
| HYPV-141 | BTB kelch-domain protein | 141,056 | 142,690 | + | 1635 | 76.8 | EPTV-155 |
| HYPV-142 | Hemagglutinin | 142,737 | 143,477 | + | 741 | 70.2 | EPTV-156 |
| HYPV-143 | Ser|Thr protein kinase | 143,532 | 144,467 | + | 936 | 91.0 | EPTV-157 |
| HYPV-144 | IL-1 receptor antagonist | 144,505 | 145,470 | + | 966 | 71.5 | EPTV-158 |
| HYPV-145 | RING finger protein, host range | 145,503 | 146,378 | + | 876 | 68.4 | EPTV-159 |
| HYPV-146 | Partial schlafen-like protein | 146,424 | 147,011 | + | 588 | 81.1 | EPTV-160 |
| HYPV-147 | EEV type-1 membrane glycoprotein | 147,108 | 147,815 | + | 708 | 75.3 | EPTV-161 |
| HYPV-148 | Anti-apoptotic Bcl-2-like protein | 147,851 | 148,294 | + | 444 | 73.5 | EPTV-162 |
| HYPV-149 | Serpin 1 | 149,551 | 150,555 | + | 1005 | 76.9 | EPTV-164 |
| HYPV-150 | Hypothetical protein | 150,662 | 151,132 | + | 471 | 69.4 | EPTV-165 |
| HYPV-151 | Tyrosine protein kinase-like protein | 151,173 | 152,036 | + | 864 | 77.7 | EPTV-166 |
| HYPV-152 | IL-1 beta-receptor | 152,065 | 153,093 | + | 1029 | 71.9 | EPTV-167 |
| HYPV-153 | Ankyrin repeat protein | 153,125 | 155,104 | + | 1980 | 68.2 | EPTV-168 |
| HYPV-154 | Ankyrin repeat protein | 157,276 | 157,827 | + | 552 | 60.6 | EPTV-169 |
| HYPV-155 | Alpha-amanitin target protein | 158,306 | 159,013 | + | 708 | 82.3 | EPTV-170 |
| HYPV-156 | NFkB inhibitor | 159,072 | 159,752 | + | 681 | 81.1 | EPTV-171 |
| HYPV-157 | Endothelin precursor | 159,796 | 160,020 | + | 225 | 64.9 | EPTV-172 |
| HYPV-158 | NFkB inhibitor | 160,059 | 160,712 | + | 654 | 65.0 | EPTV-173 |
| HYPV-159 | Secreted complement binding protein C3b|C4b | 160,747 | 161,529 | + | 783 | 74.6 | EPTV-174 |
| HYPV-160 | IL-18 binding protein | 162,096 | 162,521 | + | 426 | 83.2 | EPTV-175 |
| HYPV-161 | Ankyrin repeat protein | 164,706 | 166,571 | + | 1866 | 60.3 | EPTV-179 |
+/−: direction of open reading frame (ORF) (“+” (5′–3′) or “−” (3′–5′)).
When the seven conserved genes—RPO147, RAP94, mRNA capping enzyme large subunit, P4a precursor, RPO132, VETF-L, and DNA primase—were considered individually, the value of nt similarity with EPTV ranged from 90.5% to 98.5%. The above conserved genes that have been used for phylogenetic analysis in previous studies [10,12] are presented in bold in Table 3.
HYPV showed nucleotide divergence from its closest relative, EPTV. The smaller genome size with 166,600 nt encoding 161 genes for HYPV in comparison to 176,688 nt and 191 genes for EPTV, is likely due to the omission of the ITRs from the analysis and therefore, is not possible to establish the exact length of its the viral genome. Two ORFs (HYPV-24 and HYPV-25, Table 3), whose function is still unknown, appear to be unique to HYPV.
4. Discussion
The potential zoonotic risks associated with bats and their fascinating and special relationship with viruses have attracted the attention of many researchers worldwide. Consequently, general and target surveillance on bat populations has increased in the last decade with the purpose of clarifying the genetic diversity of bat-associated viruses as well as acquiring comprehensive information on bat–pathogen interactions. In fact, viral disease prevention and biological conservation issues could both benefit from such research.
Virological surveillance of bat populations in Italy is a relative novelty and has only recently been extensively applied, but almost immediately, a great heterogeneity of virus identifications has been observed. Viruses belonging to several viral families, such as Reoviridae [14], Coronaviridae [15,20,21,22,23,24], Paramyxoviridae [24], Rhabdoviridae [16,25], and Astroviridae [26], have been detected, allowing the identification of some novel/previously unknown viral agents. The results of the general surveillance of bats, which have been randomly applied so far as pilot virus discovery studies, may drive future activity to more specific longitudinal and target studies aimed at understanding the epidemiology of potential new pathogens.
In this study, a novel poxvirus, HYPV, was detected from the microbat Hypsugo savii in Italy. This likely represents the first poxvirus detection in bats in Europe. In fact, only four poxviruses have been documented to date in bat populations worldwide, and these and these have diverse and somehow incomplete descriptions, with just some common aspects. Firstly, EHPV1 was detected in 2009 with a high-prevalence in throat swabs from apparently healthy African megabats (Eidolon helvum), and metagenomic analysis identified poxvirus sequences that were most closely related with Molluscum contagiosum (MOCV), a human-only pathogen [11]. In the same year of 2009, another bat poxvirus was incidentally detected in South Australia during the investigation of an outbreak of parasitic skin disease in a population of the microbat species, Miniopterus schreibersii bassanii. In one of the twenty-one bats examined, an independent (non-nematode-associated) lesion containing intracytoplasmic inclusion bodies indicative of poxvirus infection was observed, and this was confirmed with electron microscopy [13]. Between 2009 and 2011, EPTV was detected in adult big brown bats (Eptesicus fuscus) with severe joint disease (tenosynovitis and osteoarthritis) at a wildlife center in Northwestern United States. Phylogenetic analysis revealed that Eptesipoxvirus is most closely related to the Cotia virus, a virus detected in sentinel suckling mice in Sao Paulo, Brazil in 1961 [27,28]. PTPV was detected from an Australian little red flying fox (Pteropus scapulatus) that died following entrapment on a fence. Post-mortem examination revealed multiple nodules on the wing membranes. Phylogenetic analysis indicated that PTPV is not closely related to any other poxvirus isolated from bats or other species, and that it likely should be placed in a new genus [12].
It is noteworthy that PTPV and EHPV were isolated from megabat hosts (Pteropus scapulatus and Eidolon helvum, respectively), whereas EPTV and HYPV were isolated from microbats (Eptesicus fuscus and Hypsugo savii, respectively). While EHPV was detected in apparently healthy bats, the other viruses were identified in sick bats and their association with the pathological condition was assumed. Specifically, clinical symptoms of EPTV in Eptesicus fuscus manifested in the form of joint swelling and increased lethargy [10]. PTPV-infected Pteropus scapulatus presented vesicular to nodular skin lesions on the wing membranes that are typical of poxvirus infections [13]. HYPV was detected in a bat showing pathological healing of the humerus fracture associated with osteomalacia and calcium deficiency. Neither symptom was directly linked to fatality and thus the capability of these viruses still needs to be ascertained, including the role of HYPV in causing deadly disease in bats.
The results of our study indicate that HYPV presents the typical morphology of the Orthopoxvirus genus and that it could be isolated in cell culture. Indeed, its final identification was obtained by genomic characterization. The nearly complete genomic sequencing clearly demonstrated that HYPV is a new virus that is distantly related to its closest known relative EPTV (WA, USA, 2011) with a nucleotide identity of 85% (almost whole genome). Indeed, the percentage value of the nt identity of HYPV with EPTV ranged from 42.5% for the HYPV-2 gene (serpin 2) to 100% for the HYPV-90 gene (VLTF-3). Regarding ORFs annotation the HYPV was shown to be defective in particular in the ITR genes i.e., 12 out of 13 described in EPTV, but this should be not a real structural defect but more likely due to the omission of the ITRs from the analysis. On the contrary, two ORFs, whose function is still unknown, appear to be unique to HYPV.
To conclude, a new poxvirus, HYPV, was detected in bats in Europe and its viral ecology and disease associations should be investigated further.
Acknowledgments
Special thanks to Anna Tirelli, Loredana Zingarello, Giovanni Bozzoni and all technicians in the IZSLER virology section for their valuable technical work and support in virological analysis.
Author Contributions
D.L. designed the study and wrote the manuscript; A.L. performed electron microscopy, participated in study coordination, and helped to draft the manuscript; C.C. and L.B. performed the next-generation sequencing and data analysis; A.M.G. and M.M. performed the sampling and data collection; G.L.C. performed the clinical investigations; A.P. and F.F. performed the necropsies and molecular tests; E.S. and T.T. were involved in the virological analysis and interpretation of the results; A.M. performed the molecular genetic studies and helped to draft the manuscript. All of the authors have read and approved the final manuscript.
Funding
This research was funded by the Italian Ministry of Health (WFR GR-2011-023505919).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016) Arch. Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shchelkunov S.N. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog. 2013;9:e1003756. doi: 10.1371/journal.ppat.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: Important reservoir hosts of emerging virus. Clin. Microbiol. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L.F., Walker P.J., Poon L.L. Mass extinctions, biodiversity and mitochondrial function: Are bats “special” as reservoirs for emerging viruses? Curr. Opin. Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: A review. Zoonoses Public Health. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luis A.D., Hayman D.T., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A., et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker K.S., Murcia P.R. Poxviruses in bats … so what? Viruses. 2014;6:1564–1577. doi: 10.3390/v6041564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson G.L., Nordhausen R., Garner M.M., Huckabee J.R., Johnson S., Wohrle R.D., Davidson W.B., Wilkins K., Li Y., Doty J.B., et al. Novel poxvirus in big brown bats, northwestern United States. Emerg. Infect. Dis. 2013;19:1002–1004. doi: 10.3201/eid1906.121713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu S.L., Nakazawa Y., Gao J., Wilkins K., Gallardo-Romero N., Li Y., Emerson G.L., Carroll D.S., Upton C. Characterization of Eptesipoxvirus, a novel poxvirus from a microchiropteran bat. Virus Genes. 2017;53:856–867. doi: 10.1007/s11262-017-1485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker K.S., Leggett R.M., Bexfield N.H., Alston M., Daly G., Todd S., Tachedjian M., Holmes C.E., Crameri S., Wang L.F., et al. Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Dea M.A., Tu S.L., Pang S., de Ridder T., Jackson B., Upton C. Genomic characterization of a novel poxvirus from a flying fox: Evidence for a new genus? J. Gen. Virol. 2016;97:2363–2375. doi: 10.1099/jgv.0.000538. [DOI] [PubMed] [Google Scholar]
- 13.McLelland D.J., Reardon T., Bourne S., Dickason C., Kessell A., Boardman W. Outbreak of Skin Nodules associated with Riouxgolvania beveridgei (Nematoda: Muspiceida) in the Southern Bentwing Bat (Miniopterus schreibersii bassanii), South Australia. J. Wildl. Dis. 2013;49:1009–1013. doi: 10.7589/2012-11-288. [DOI] [PubMed] [Google Scholar]
- 14.Lelli D., Moreno A., Lavazza A., Bresaola M., Canelli E., Boniotti M.B., Cordioli P. Identification of Mammalian Orthoreovirus type 3 in Italian bats. Zoonosis Public Health. 2013;60:84–92. doi: 10.1111/zph.12001. [DOI] [PubMed] [Google Scholar]
- 15.Lelli D., Papetti A., Sabelli C., Rosti E., Moreno A., Boniotti M.B. Detection of coronaviruses in bats of various species in Italy. Viruses. 2013;5:2679–2689. doi: 10.3390/v5112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lelli D., Prosperi A., Moreno A., Chiapponi C., Gibellini A.M., De Benedictis P., Leopardi S., Sozzi E., Lavazza A. Isolation of a novel Rhabdovirus from an insectivorous bat (Pipistrellus kuhlii) in Italy. Virol. J. 2018;15:37. doi: 10.1186/s12985-018-0949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavazza A., Pascucci S., Gelmetti D. Rod-shaped virus-like particles in intestinal contents of three avian species. Vet. Rec. 1990;126:581. [PubMed] [Google Scholar]
- 18.Da Silva M., Upton C. Bioinformatics for analysis of poxvirus genomes. Methods Mol. Biol. 2012;890:233–258. doi: 10.1007/978-1-61779-876-4_14. [DOI] [PubMed] [Google Scholar]
- 19.Tcherepanov V., Ehlers A., Upton C. Genome Annotation Transfer Utility (GATU): Rapid annotation of viral genomes using a closely related reference genome. BMC Genom. 2006;7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balboni A., Gallina L., Palladini A., Prosperi S., Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci. World J. 2012;2012:989514. doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Benedictis P., Marciano S., Scaravelli D., Priori P., Zecchin B., Capua I., Monne I., Cattoli G. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes. 2014;48:366–371. doi: 10.1007/s11262-013-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno A., Lelli D., de Sabato L., Zaccaria G., Boni A., Sozzi E., Prosperi A., Lavazza A., Cella E., Castrucci M.R., et al. Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy. Virol. J. 2017;14:239. doi: 10.1186/s12985-017-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Sabato L., Lelli D., Faccin F., Canziani S., Di Bartolo I., Vaccari G., Moreno A. Full genome characterization of two novel Alpha-coronavirus species from Italian bats. Virus Res. 2018;260:60–66. doi: 10.1016/j.virusres.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo F., Edenborough K.M., Toffoli R., Culasso P., Zoppi S., Dondo A., Robetto S., Rosati S., Lander A., Kurth A., et al. Coronavirus and paramyxovirus in bats from Northwest Italy. BMC Vet. Res. 2017;13:396. doi: 10.1186/s12917-017-1307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopardi S., Priori P., Zecchin B., Poglayen G., Trevisiol K., Lelli D., Zoppi S., Scicluna M.T., D’Avino N., Schiavon E., et al. Active and passive surveillance for bat lyssaviruses in Italy revealed serological evidence for their circulation in three bat species. Epidemiol. Infect. 2018;4:1–6. doi: 10.1017/S0950268818003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amoroso M.G., Russo D., Lanave G., Cistrone L., Pratelli A., Martella V., Galiero G., Decaro N., Fusco G. Detection and phylogenetic characterization of astroviruses in insectivorous bats from Central-Southern Italy. Zoonoses Public Health. 2018;65:702–710. doi: 10.1111/zph.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopesode S., Lacerda J.P., Fonseca I.E., Castro D.P., Forattini O.P., Rabello E.X. Cotia virus: A new agent isolated from sentinel mice in Sao Paulo, Brazil. Am. J. Trop. Med. Hyg. 1965;14:156–157. doi: 10.4269/ajtmh.1965.14.156. [DOI] [PubMed] [Google Scholar]
- 28.Afonso P.P., Silva P.M., Schnellrath L.C., Jesus D.M., Hu J., Yang Y., Renne R., Attias M., Condit R.C., Moussatche N., et al. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae) J. Virol. 2012;86:5039–5054. doi: 10.1128/JVI.07162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]