Abstract
Uveal melanoma is a clinically distinct and particularly lethal subtype of melanoma originating from melanocytes in the eye. Here, we performed multi-region DNA sequencing of primary uveal melanomas and their matched metastases from 35 patients. We observed novel driver mutations and established the order in which these and known driver mutations undergo selection. Metastases had genomic alterations distinct from their primary tumors, and metastatic dissemination sometimes occurred early during the development of the primary tumor. Our study offers new insights into the genetics and evolution of this melanoma subtype, providing potential biomarkers for progression and therapy.
Editorial summary:
Multi-region sequencing of 35 primary uveal melanomas and their matched metastases yields new insights into the genetics and evolution of these tumors and provides potential biomarkers for progression and therapy.
Uveal melanomas comprise 5% of melanomas in the US1. They derive from melanocytes in the choroid, iris, or ciliary body of the eye and constitute the most common eye cancer. Ten-year mortality is 50%, markedly higher than most other melanoma subtypes2,3, and there currently are no effective treatment options for patients with metastatic disease4–6. An improved understanding of how uveal melanoma progresses from primary to metastatic disease promises to identify crucial pathogenic steps that could inform the development of new modalities of treatment and prognostication.
Several large-scale sequencing efforts have catalogued the genomic landscape of primary uveal melanoma7–10 and revealed a suite of genetic alterations distinctive from other melanoma subtypes11. Uveal melanomas commonly harbor mutations that activate the G-protein alpha-q signaling cascade, affecting GNAQ12, GNA1113, PLCB414, or CYSLTR215 that, with rare exceptions, occur in a mutually exclusive pattern. They additionally have secondary somatic alterations affecting BAP110,16, SF3B18, or EIF1AX9 that tend to also to occur exclusive of each other. Further, uveal melanomas have recurrent copy number alterations involving gains of chromosomes 6p and 8q as well as losses of chromosomes 1p, 3, 6q, 8p, and 16q. BAP1 constitutes the major tumor suppressor gene on chromosome 3, and MYC is possibly the oncogene on chromosomal arm 8q. It is unclear which genes drive the other copy number changes.
In contrast to primary uveal melanoma, few uveal melanoma metastases have been sequenced to date17- particularly lacking are studies of primary tumors and patient-matched metastases. Uveal melanomas have a propensity to metastasize via hematogenous routes to the liver, a distant site with respect to their origins in the eye. There is a latency between removal of the primary tumor and the emergence of metastases that ranges from months to decades4,18. In order to better understand the evolution of metastatic uveal melanoma, we performed multi-region DNA sequencing of primary uveal melanomas and patient-matched metastases.
Results
We sequenced non-lesional reference tissue, one or more regions of the primary tumor, and corresponding liver metastases from 35 patients with metastatic uveal melanoma (Supplementary Table 1). The majority of tissues were microdissected from archival paraffin blocks. Sequencing libraries from formalin-fixed paraffin-embedded (FFPE) tissues have lower complexity19, and we therefore elected to perform targeted sequencing of a panel of 500 genes commonly involved in cancer (Supplementary Table 2), allowing us to sequence a smaller genomic footprint at substantially higher coverage. On average, each sample was sequenced to 423-fold unique coverage, enabling sensitive mutation detection and helping to overcome the well-documented20 challenge of detecting certain BAP1 mutations.
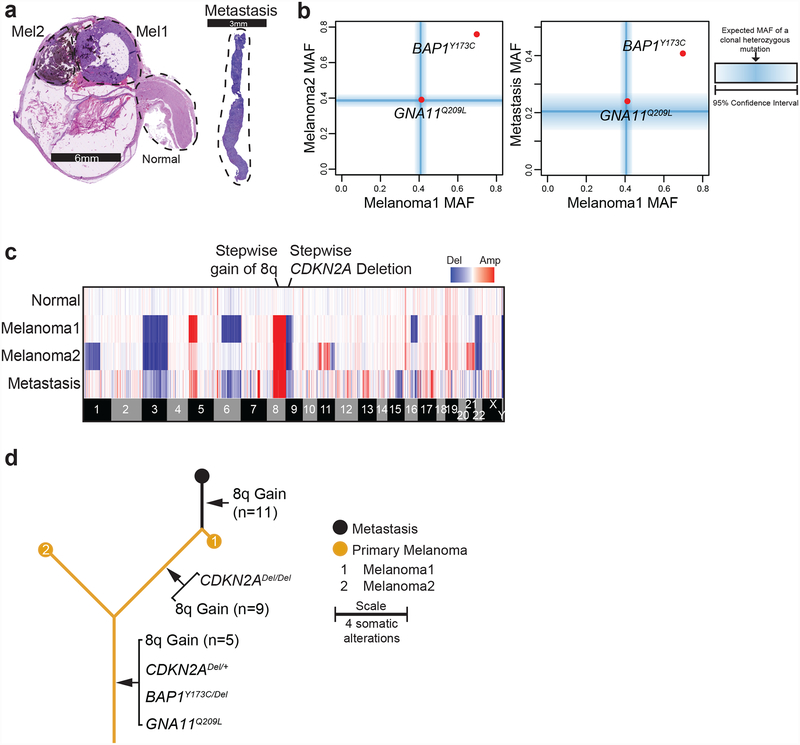
A workflow to delineate the evolution of metastatic uveal melanoma.
Our workflow to deduce the genetic evolution of metastatic uveal melanoma is illustrated by the example case (A11) shown in Figure 1. From this patient, we microdissected two histologically distinct areas of the primary melanoma and part of the optic nerve, which served as a source of non-lesional reference tissue (Fig. 1a). Metastatic tissue was obtained from the patient’s liver core biopsy (Fig. 1a).
Figure 1 |. The patterns of somatic alterations within a patient’s tumor areas provide insights in the sequential order in which they arose (example case A11).
a, H&E sections of the enucleation specimen show morphologically distinct areas. The metastatic tissue stemmed from a liver core biopsy of the patient. Dotted regions were microdissected for genomic analyses. b, Point mutations are stratified by their mutant allele fractions (MAFs) in the two regions of the primary tumor and the metastasis. The blue bars indicate the expected MAFs of fully-clonal, heterozygous mutations after accounting for stromal cell contamination, with shadings indicating confidence intervals where those mutations should reside. Note the presence of a GNA11Q209L mutation that is fully clonal and heterozygous in all samples. By contrast, the BAP1Y173C mutation has an elevated MAF, indicating that it underwent loss-of-heterozygosity. c, Heatmap displaying copy number increases (red) or decreases (blue) for each sample (rows) across the genome with chromosomal boundaries indicated. Additional details related to deletion of CDKN2A and gain of chromosomal arm 8q are shown in Supplementary Figure 1. d, Inferred evolution of case A11 with mutations shared among all samples forming the trunk, and mutations not present in all samples forming the branches. The length of the trunk and branches is scaled by the total number of mutations in the samples.
Only two somatic point mutations were identified, reflecting the low mutation burden that is typical of this melanoma subtype. Both mutations, a heterozygous, gain-of-function GNA11 variant and a hemizygous, loss-of-function BAP1 variant, were shared between all three melanoma samples (Fig. 1b). To determine the allelic status of mutations, we bioinformatically estimated the fraction of neoplastic cells in each melanoma region. The blue lines in Figure 1b denote the expected mutant allele fractions where a heterozygous mutation would be expected to reside with fading zones that indicate the 95% confidence intervals. The loss of heterozygosity of the BAP1 mutation is evident by its mutant allele fraction falling outside those zones in all three samples.
We next inferred copy number alterations using the software package CNVkit21 (Fig. 1c). In the example case, some copy number alterations were shared between all samples, including a deletion of chromosome 3 that encompassed the BAP1 gene and thus explains how the BAP1 mutation became hemizygous. A loss of chromosomal arm 9p and a gain of chromosomal arm 8q were also present in each melanoma region (Fig. 1c).
By contrast, some copy number alterations were exclusive to certain tissues. For instance, there was a superimposed homozygous deletion encompassing the CDKN2A gene that was only present in the unpigmented area of the primary melanoma (Mel1) and the metastasis (Supplementary Fig. 1a). Also, the amplitudes of 8q gain differed among the melanoma tissues (Supplementary Fig. 1b) - the pigmented area of the primary (Mel2) had 5 absolute copies, while the unpigmented area (Mel1) had 9 absolute copies, and the metastasis had 11 absolute copies. These observations indicate stepwise deletion of CDKN2A paralleled by stepwise amplification of 8q during the evolution of this case.
The distribution of somatic mutations and copy number changes among the three tumor areas made it possible to delineate the sequential order in which they arose during tumor evolution. All three tumor samples shared the GNA11Q209L mutation, bi-allelic BAP1 alterations, mono-allelic deletion of CDKN2A, and gain of at least three copies of chromosomal arm 8q – this indicates that these alterations arose comparatively early and thus reside on the trunk of the evolutionary tree (Fig. 1d). The Mel1 area of the primary tumor and the metastasis shared a homozygous deletion of CDKN2A and additional levels of 8q gain, placing these somatic alterations on a shared branch of the evolutionary tree (Fig. 1d). Finally, the metastasis harbored the highest amplitude of 8q gain, placing these extra levels of 8q gain on the terminal branch of the evolutionary tree (Fig. 1d).
In aggregate, activation of GNA11, bi-allelic loss of BAP1, low-level gain of 8q, and mono-allelic loss of CDKN2A occurred in the most recent common ancestral clone. A subclone with higher levels of 8q gain and bi-allelic loss of CDKN2A emerged in the primary (Mel1), and this clone gave rise to a liver metastasis with even higher levels of 8q gain (Fig. 1d). The equivalent detailed evolution of the remaining 34 cases is available in the supplementary dataset on FigShare (https://doi.org/10.6084/m9.figshare.6845675.v1).
The genomic landscapes of primary and metastatic uveal melanoma.
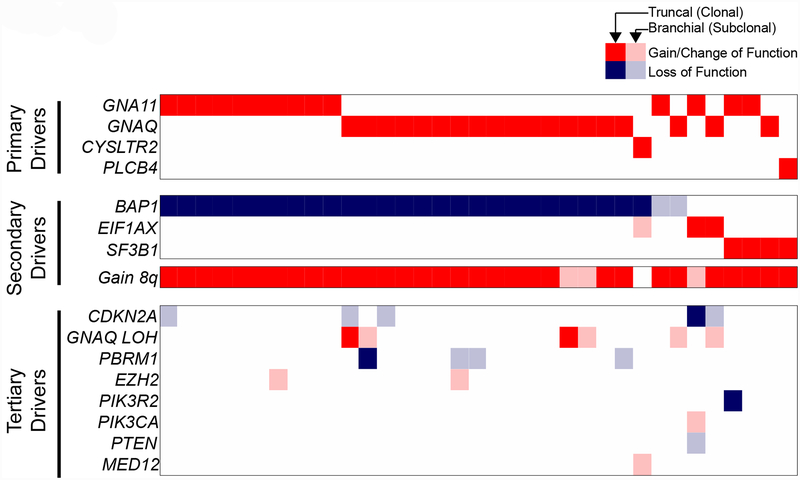
We next annotated the driver mutations in each sequenced region from all 35 patients’ tumors. As expected, based on prior studies, we observed mutually exclusive mutations involving genes in the Gαq signaling cascade as well as alterations affecting BAP1, SF3B1, and EIF1AX that tended to not overlap. However, in contrast to prior studies, we found a long tail of additional mutations (Fig. 2) that included loss-of-function mutations affecting CDKN2A, PBRM1, PIK3R2, and PTEN, loss-of-heterozygosity over the GNAQ locus, and gain-of-function mutations affecting EZH2, PIK3CA, and MED12. The majority of these alterations were in just one region of the primary tumor or private to their metastases (designated in Figure 2 by the faded tiles), indicating that they arose later during progression, after mutational activation of Gαq and mutations of BAP1, SF3B1, and EIF1AX. These mutations are thus considered tertiary driver mutations, likely explaining why they were not apparent in prior bulk sequencing of primary uveal melanomas.
Figure 2 |. The spectrum of driver mutations in uveal melanoma.
Multi-region sequencing of primary and matched metastases was performed on tumors from 35 patients. The spectrum of driver mutations (rows) in all patients (columns) is shown. Gain/change-of-function and loss-of-function mutations are respectively indicated as red or blue tiles. Mutations present in every sequenced region from a given patient are portrayed by solid tiles, and mutations present in only a subset of tissues from a given patient are portrayed by faded tiles. Mutations are grouped into three categories based on their deduced order of emergence during progression: primary drivers (shared and fully clonal), secondary drivers (shared and usually clonal), and tertiary drivers (private to later stages and typically subclonal).
Early mutational activation of the Gαq signaling pathway is pathognomonic of uveal melanoma.
Activation of Gαq signaling was ubiquitous with mutations affecting GNA11, GNAQ, CYSTLR2, or PLCB4 in a mutually exclusive pattern in all 35 cases in our cohort, consistent with prior findings. The mutations activating the Gαq signaling pathway were always shared among all samples from a given patient, indicating that they undergo selection early and are required for tumor formation (Supplementary Fig. 2a). This is also consistent with the recent finding that these mutations are common in choroidal nevi, which is likely a precursor for a subset of uveal melanomas22. Gαq-signaling-pathway mutations were heterozygous in the majority of cases. However, in six cases, GNAQ mutations were hemizygous or homozygous (Supplementary Fig. 2b) as a consequence of additional genetic alterations that eliminated the wild-type allele. Loss-of-heterozygosity of mutant GNAQ tended to take place later during progression (Supplementary Fig. 2b). Preferential expression of mutant over wild-type GNAQ has been observed previously23, and our data offer a mechanism as to how this can occur.
Uveal melanomas with SF3B1 or EIF1AX mutations acquire additional oncogenic mutations during metastatic progression.
We next sought to investigate the relative timing of BAP1, SF3B1, and EIF1AX mutations. Patients whose uveal melanomas have SF3B1 or EIF1AX mutations tend to have a better prognosis than those with BAP1 mutations7. Likely because our cohort consisted entirely of metastatic uveal melanomas, SF3B1- and EIF1AX- mutant cases were underrepresented (n = 7). In general, the BAP1, SF3B1, and EIF1AX mutations did not co-occur. However, one EIF1AX-mutant tumor also had bi-allelic BAP1 mutations (Supplementary Fig. 3a). This case was further unusual because the EIF1AX mutation resided at codon 70, a site that is recurrently mutated but less frequently affected than the N-terminal region of EIF1AX. Finally, the Gq-pathway mutation in this atypical case involved CYSTLR2, which is rare compared to GNAQ and GNA11 mutations.
The cases with SF3B1 or EIF1AX mutations tended to have additional oncogenic alterations. Two had homozygous CDKN2A deletions, three had PI3-kinase pathway mutations (PTENC218*, PIK3CAH1047R, and PIK3R2N485S), and one had a MED12 hotspot mutation (Supplementary Fig. 3). In one EIF1AX-mutant case, we sequenced two areas of primary tumor, and the more histologically advanced area acquired a deletion of chromosome 3 (Supplementary Fig. 3b) – this was notable because it has been proposed that primary tumors without monosomy 3 may progress towards a more malignant stage by losing chromosome 324.
The SF3B1 and EIF1AX mutations were predominately truncal, whereas the additional oncogenic mutations were more often branchial. This pattern suggests that SF3B1 and EIF1AX mutations undergo selection early but may require additional oncogenic alterations to complement their likely limited ability to drive disease progression and metastatic dissemination.
Selection to disrupt chromatin remodeling factors begins early and continues throughout the evolution of metastatic uveal melanoma.
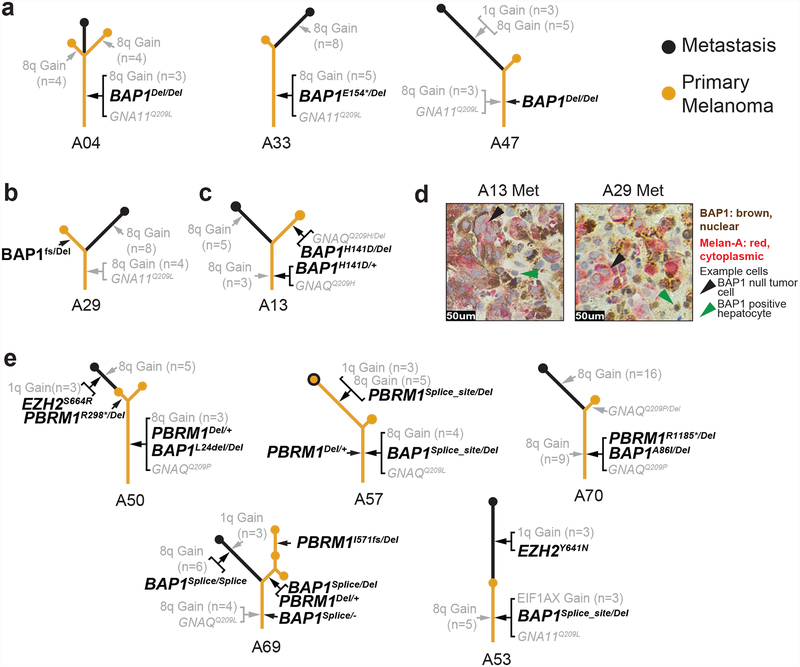
BAP1 mutations were expectedly common in our cohort of uveal melanomas that became metastatic and were usually shared among all samples from a given patient, placing them on the trunks of evolutionary trees (Fig. 3a). These observations suggest that bi-allelic BAP1 mutations tend to precede metastatic dissemination.
Figure 3 |. Mutations disrupting chromatin remodeling factors emerge throughout the progression to metastatic disease.
a–c, Exemplary phylogenetic trees in which BAP1 loss emerged early (a) and late (b,c). In cases with early loss, bi-allelic loss-of-function mutations reside on the trunks of the trees, whereas in cases where it arose later, at least one hit is situated on a branch. d, Dual immunostaining for BAP1 (brown) and melan-A (red) demonstrates that A13 and A29 metastases are null or BAP1 protein. e, Cases in which mutations in other chromatin remodeling factors were detected. These alterations tended to arise later, positioning them on the branches.
There were, however, two cases in which BAP1 alterations had a different distribution between primary tumors and their metastases (Fig. 3b,c). In case A29, the primary tumor had a frameshift mutation and a deletion affecting BAP1, but its metastasis had only very few reads of the frameshift mutation and did not have a deletion on chromosome 3 (Fig. 3b). In case A13 (Fig. 3c), the primary tumor and the metastasis shared a point mutation in BAP1, but only the primary had eliminated the second allele of BAP1, whereas the mutation in the metastasis remained heterozygous. The metastases from cases A29 and A13 were both negative for BAP1 protein by immunohistochemistry (Fig. 3d), suggesting that either BAP1 was silenced epigenetically, or the mutation escaped detection by our methods. Overall, these two cases raise the possibility that bi-allelic loss of BAP1 is not absolutely required for metastatic dissemination. This would be consistent with anecdotal reports of monosomy 3 occurring later in the evolution of uveal melanoma25,26; however, further studies will be necessary to confirm whether this is true.
We also observed mutations in other chromatin remodeling factors, including hemizygous, loss-of-function mutations in PBRM1 and gain-of-function EZH2 mutations (Fig. 3e). PBRM1 encodes a critical subunit of the SWI/SNF chromatin remodeling complex, and EZH2 encodes the enzymatic subunit of Polycomb repressive complex 2 (PRC2)27. BAP1 is a negative regulator of PRC1 which activates PRC2, so BAP1 loss is thought to promote PRC2 activation. The SWI/SNF chromatin remodeling complex also opposes PRC2 activity in maintaining cell-state appropriate chromatin modifications28–30; therefore, the alterations in PBRM1 and EZH2 likely shift the balance of chromatin remodeling activity further towards PRC2 dominance. In contrast to BAP1 mutations, which occurred earlier in most cases, mutations affecting PBRM1 or EZH2 occurred later, as they were generally not shared among all tumor areas of a given patient.
Copy number gains of chromosome 8q arise in the primary tumor and ramp up during metastatic progression.
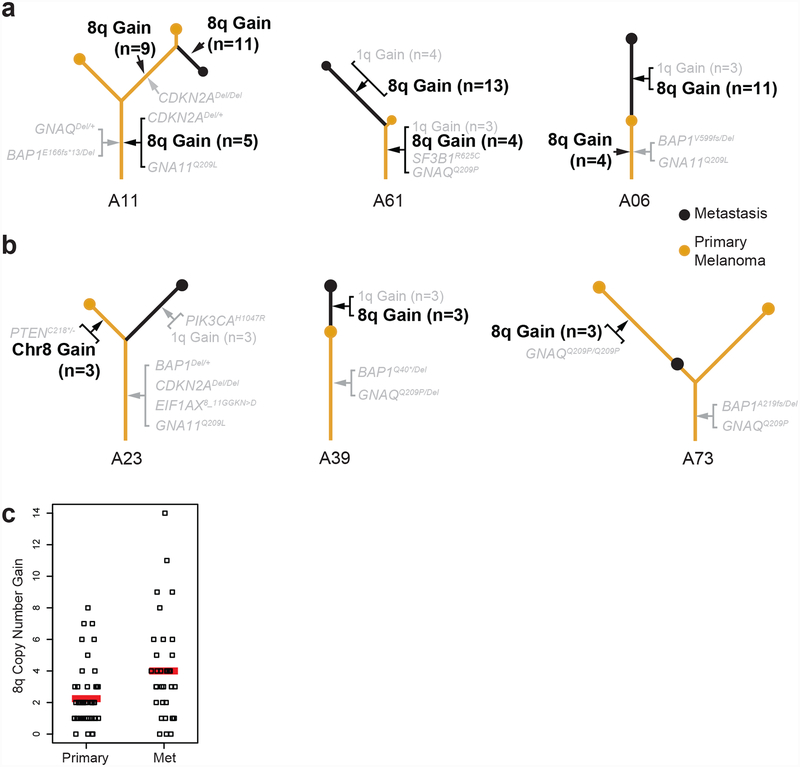
Copy number gains of chromosome 8q were present in nearly every case and were typically shared among the different tumor areas of a given patient (Fig. 4a), suggesting that they occur early during evolution of the primary tumor, prior to metastatic dissemination, as previous data have suggested31. There were three exceptions to this pattern in which copy number gains were not shared in every tumor area (Fig. 4b), indicating that it is at least possible for metastatic dissemination to precede 8q gain.
Figure 4 |. Gene dosage of chromosome 8q increases during progression.
a, Examples of phylogenetic trees in which gains of 8q occur early and subsequently increase (see Supplementary Figs. 1b and 4 for more details on these copy number alterations). b, Examples of phylogenetic trees of tumors in which metastatic dissemination preceded gain of 8q. The detailed evolution of each case is available in the supplementary dataset (see Methods). c, Copy number increase of 8q in metastases and primary tumors is shown on the y-axis, with 0 reflecting the normal, diploid state. Red bars denote averages, indicating that in our cohort the level of 8q copy number increase doubled from primaries to metastases (P = 0.002, two-tailed t-test).
We delved deeper into the amplitudes of 8q gain and observed that the copy number of 8q tended to increase from primaries to metastases. For instance, as described in case A11, one region of the primary tumor had 5 copies of 8q, while another region of the primary tumor had 9 copies, and the metastasis had 11 copies (Fig. 4a and Supplementary Fig. 1b). In other examples, the primary tumor for case A61 had 4 copies of 8q, and the metastasis had 11 copies (Fig. 4a and Supplementary Fig. 4a), and in case A06, the primary tumor had 4 copies of chromosomal arm 8q, while the metastasis had 9 copies (Fig. 4a and Supplementary Fig. 4b). Overall, metastases had more copies of 8q than their corresponding primary tumors in most patients, with an average of 6 versus 4 copies (P = 0.002 two-tailed t-test) (Fig. 4c). We validated our sequencing-based copy number estimates by measuring allelic imbalance and assessing copy number with independent technologies, including multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH) (Supplementary Fig. 4c–e). The findings of these methods were concurrent and indicate progressive increase of 8q copy number during progression to metastatic disease.
The sequential order of genetic alterations during metastatic progression.
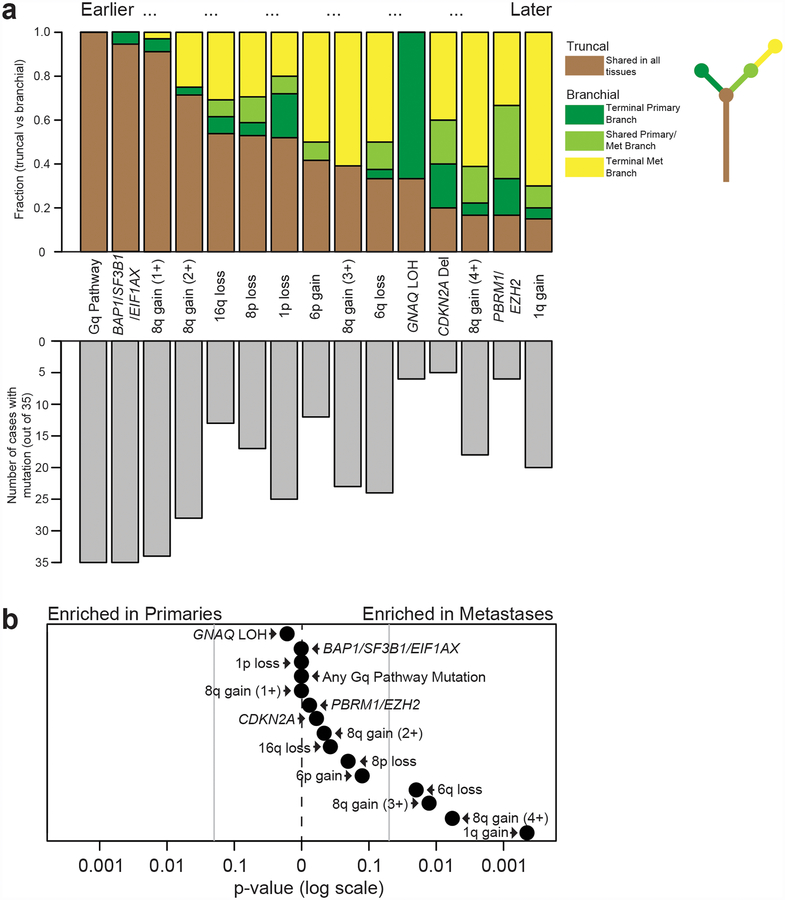
We deduced the sequential order in which somatic alterations undergo selection during the evolution of metastatic uveal melanoma by calculating how often a given alteration resided on the trunk versus the branch of each evolutionary tree (Fig. 5a). This analysis confirmed that Gαq-pathway mutations are the earliest mutations to undergo selection, followed by gain of chromosomal arm 8q as well as BAP1, SF3B1, or EIF1AX mutations. Selective pressures continue to operate on these pathways at comparatively later points in the progression cascade by way of GNAQ loss-of-heterozygosity, additional chromatin remodeling mutations, and further ramp-up of 8q copy number (Fig. 5a). Copy number alterations affecting chromosomal arms 16q, 8p, 1p, 6p, and 6q underwent selection at intermediate points in the evolutionary cascade (Fig. 5a).
Figure 5 |. The sequential order of genetic alterations during metastatic progression.
a, Top panel: the y-axis indicates the relative fraction of patients in which the indicated somatic alterations occupied the trunk or branches of their respective phylogenetic trees. Somatic alterations that were more common in the trunks of phylogenetic trees were assumed to undergo selection earlier than those situated on their branches. Bottom panel: the number of patients with each of the indicated somatic alterations. b, Enrichment scores (P-values) were calculated using a two-tailed binomial test (see Methods for details) for the 15 genetic alterations highlighted in a to determine whether they were more common in primary tumors (n = 53) or in metastases (n = 36). Vertical gray lines indicate P-values of 0.05. Scores were calculated under the assumption (null hypothesis) that somatic alterations are equally distributed between primaries and metastases. Most somatic alterations were enriched in metastases, though only 6q loss, gain of at least 3 copies of 8q, and gain of 1q reached statistical significance.
Finally, we compared the frequency of somatic alterations between primaries and metastases. Most somatic alterations were somewhat enriched in metastases, but only copy number changes of 6q, 1q, and high-level gains of 8q (copy number increase by at least 3 copies) reached statistical significance (Fig. 5b). In particular, gains of 1q were frequent in our cohort compared to prior studies on primary uveal melanomas, consistent with their emergence later during progression (Fig. 5 and Supplementary Fig. 5). Further studies will be needed to elucidate the gene(s) driving the selection of these aberrations.
Correlations with clinical outcomes.
Genomic studies of primary uveal melanomas have revealed several mutations with prognostic value8–10. Our cohort was underpowered to discover new biomarkers of this type, but capitalizing on a unique aspect of our study, we instead investigated whether specific evolutionary routes of progression correlated with clinical outcomes.
As all of our patients ultimately developed metastatic disease, we assessed correlations between clinical outcomes (disease-free survival, survival after metastasis, and overall survival) and features of their genetic evolution (i.e. metrics of the phylogenetic trees, such as individual and combined lengths of trunk and branches as well as branch separation; see Methods) (Supplementary Fig. 6a,b). The overarching goal was to investigate whether certain evolutionary trajectories were associated with clinical outcomes, as was recently shown to be true in renal cell carcinoma32.
Patients whose trees had longer metastasis branches and shorter primary branches tended to have better disease-free survival (Supplementary Fig. 6c) – this comparison was statistically significant by itself (R = 0.34, P = 0.046), though it did not remain significant after correcting for multiple-hypothesis testing. If this finding is validated in larger cohorts, it likely reflects the fact that some uveal melanomas require more genetic alterations to develop clinically detectable metastases, and these uveal melanomas have better survival metrics reflecting the time required to incrementally acquire the necessary set of alterations.
Discussion
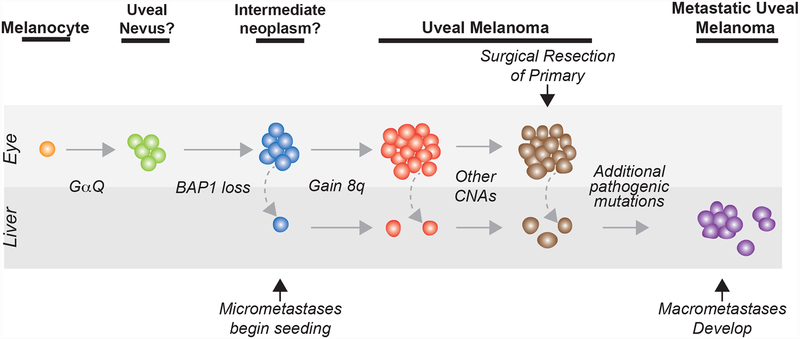
Our study shows that uveal melanomas continue to genetically evolve as they progress from primary to metastatic disease, as indicated by the fact that metastases tend to have more oncogenic mutations than primary tumors. This contrasts with most other tumor types for which metastases are genetically similar to primary tumors33,34. One peculiar aspect of uveal melanoma is that in many cases metastases develop after a long period of latency after successful eradication of the primary tumor – this latency period may partially reflect the additional time needed to acquire the necessary mutations for growth at the distant site (Fig. 6). In addition to having more oncogenic alterations, metastases also acquire distinct oncogenic alterations, including additional copies of chromosomes 1q and 8q. This may reflect different selective pressures operating in the liver, the major organ in which metastases develop in uveal melanoma.
Figure 6 |. The evolution of metastatic uveal melanoma.
‘CNA’, copy number alteration. ‘Other CNAs’ include combinations of 16q loss, 8p loss, 6q loss, or 6p gain, among others. ‘Additional pathogenic mutations’ include various combinations of 8q amplification, 1q gain, CDKN2A loss, SWI/SNF mutations, and EZH2 mutations, among others. The precursors to primary uveal melanoma are not well understood, as reflected here by the questions marks adjacent to those clinical stages in our model.
Prior sequencing studies of uveal melanoma mostly analyzed a single tissue sample from the primary tumor. Our multi-region sequencing of primaries and metastases revealed multiple novel mutations, expanding the catalog of driver mutations for this aggressive melanoma subtype. These novel mutations were more common in uveal melanomas with SF3B1 or EIF1AX mutations, and they possibly complement the otherwise limited ability of SF3B1 and EIF1AX mutations to drive metastatic disease. The earliest driver mutations in primary uveal melanoma are not easily druggable, but there is hope that some of the newly recognized mutations found in some metastases can be targeted therapeutically.
We also observed loss-of-heterozygosity of mutant GNAQ, but not GNA11, in multiple samples, indicating hidden complexity in oncogenic signaling through mutant Gαq subunits. The Q209 alterations in GNAQ and GNA11 abrogate their GTPase activity, similar to mutant RAS. RAS mutations also undergo loss-of-heterozygosity by a variety of mechanisms that shift the allelic balance towards the mutant allele35. Earlier studies using Sanger sequencing have reported increased frequency of GNA11 mutations in metastatic uveal melanomas, whereas GNAQ mutations are more common in benign lesions, such as blue nevi13. This led to the proposition that GNA11 mutations are more potent. The finding of recurrent loss-of-heterozygosity of GNAQ mutations later in progression may indicate that GNAQ, but not GNA11, requires a second hit to fully activate the pathway, which could explain the association with GNA11 mutations with more aggressive disease.
Uveal melanomas have been proposed to arise after an early, punctuated burst of mutations followed by a period of neutral evolution20. In our cohort, we found three cases in which metastatic dissemination preceded 8q gain and potentially two cases in which metastatic dissemination preceded bi-allelic loss of BAP1. These cases suggest that it is at least possible for metastatic dissemination to occur before the neoplasm has developed the full complement of oncogenic mutations (Fig. 6), though additional studies, focused on the earlier stages of neoplasia, will be needed to fully resolve the incipient events involved in the evolution of uveal melanoma. We also found multiple added driver mutations in the branches of phylogenetic trees, arguing against neutral evolution in the latter phases of evolution.
In summary, our study reveals the selective pressures operating on primary tumors and metastases during the evolution of uveal melanoma and offers candidate biomarkers for staging and prognosis.
Online Methods
Patient samples.
Primary and patient-matched metastatic uveal melanoma tissues were from the Ocular Tumor Division in the Copenhagen University Hospital. Most primary uveal melanomas were from archival (formalin-fixed paraffin-embedded) blocks of enucleated eyes, though some were derived from fresh biopsies, as indicated (Supplementary Table 1). All metastases were liver metastases. Among the metastatic tissues, most were derived from archival blocks of liver core biopsies with a subset derived from archival blocks of liver resections, as indicated (Supplementary Table 1). All metastases were treatment naïve. Most reference (normal) tissues were microdissected from non-lesional areas of the primary tumor blocks with some originating from patient-matched blood, as indicated (Supplementary Table 1).
This retrospective study was approved by the regional ethical committee in Copenhagen, Denmark (H-2-2013-064) and the Danish Data Protection agency (RH-2015–167). All tissues were collected in accordance with the institutional review board with regard to informed consent.
The overarching design of this study is also described in the Life Sciences Reporting Summary.
Microdissection and DNA extraction.
All microdissections were performed with a scalpel under a dissection scope and guided by a pathologist with the intent to maximize tumor cell content. 10-μm unstained sections (between 4–10 levels) were used. Genomic DNA was isolated with Qiagen DNA FFPE Tissue Kit (p/n 56404). When possible, we microdissected multiple regions of the primary tumor. In total, we collected and sequenced 53 pieces of primary uveal melanoma, 36 pieces of uveal melanoma metastasis, and 35 normal (i.e. non-lesional) tissues.
DNA sequencing and analysis.
25–250 ng of genomic DNA was sheared (250 bp target fragment size) and prepared for Illumina sequencing as previously described36,37. As expected, the smaller FFPE-derived neoplasms had lower library complexities. We therefore elected to perform targeted sequencing of a panel of 538 genes commonly involved in cancer (see Supplementary Table 2 for baits), favoring more samples and more coverage over a larger sequencing footprint. This decision did not hinder the overarching goal of the study, which was to delineate the order in which known driver mutations arise during the evolution of metastatic uveal melanoma. On average, each sample was sequenced to 423-fold de-duplicated coverage (see Supplementary Table 1 for more details). Baits were developed with the customized Nimblegen SeqCap EZdeveloper platform. Sequencing was performed on an Illumina HiSeq 2500 instrument.
Alignment, grooming, and mutation calling were performed with the following software packages: Burrows-Wheeler Aligner (BWA), Genome Analysis Tool-Kit (GATK), Picard, and MuTect. Sequencing metrics for each sample are available in Supplementary Table 1.
CNVkit was used to infer copy number information. This software can be run in reference or reference-free mode. We elected to run CNVkit in reference mode, as this consistently produces higher quality copy number calls when references (i.e. normal tissues) are available. Two separate reference pools were generated for this study – non-neoplastic eye tissue and non-neoplastic liver tissue. The eye reference was used to infer copy number information from primary tumors whereas liver reference tissue was used to infer copy number information from metastases.
Driver gene calling.
The driver genes in each case are summarized in Figure 2. We include any mutation affecting GNAQ, GNA11, PLCB4, CYSTLR2, BAP1, EIF1AX, or SF3B1 – this is based on these genes’ previously described roles in driving uveal melanoma. We also include alterations affecting CDKN2A, PBRM1, and EZH2 due to their recurrence in our study and known roles as cancer genes in other tumor types. Moreover, we include hotspot mutations affecting PIK3CA and MED12, which were observed once in our study but are common in pan-tumor analyses. Finally, we include inactivating mutations affecting PTEN and PIK3R2, which are prominent tumor suppressors in other cancers and are lost in a bi-allelic fashion in our study.
Copy number estimation.
To deduce the discrete levels for each copy number alteration, we first calculated the expected log ratios for each level of gain or loss. For example, a mono-allelic deletion should have a log2(tumor/reference) segmentation value of −1 (because log2(1/2) = −1); however, in practice, this is never observed because each tumor has some level of stromal cell contamination, and stromal cells do not harbor copy number alterations. We took into account stromal cell contamination for each sample by proportionally weighting the expected segment value for a given copy number alteration with the expected log2(stroma/reference) from stromal cells (assumed to be 0) in that sample.
Supplementary dataset covering the evolution of all 35 cases.
In Figure 1, we present the detailed evolution of an example case, including the distribution of point mutations, copy number alterations, and allelic imbalances over each tumor region from a single patient. We analyzed all 35 cases in this cohort to the same level of detail, and we include the detailed evolution of the other 34 cases in the following supplementary dataset38 hosted at Figshare (https://doi.org/10.6084/m9.figshare.6845675.v1).
Phylogenetic tree construction.
Point mutation calls are available in Supplementary Table 3, and copy number calls are available in Supplementary Table 4. Phylogenetic trees were constructed as described for Figure 1 from the shared and private somatic alterations. All phylogenetic trees are rooted to the germline state. The trunk and branch lengths for each phylogenetic tree are provided in Supplementary Table 1. When multiple mutations occurred on the same segment of the phylogenetic tree, they are bracketed, serving as an indication that it is not possible to deduce the relative order in which they occurred.
Tumor cellularity inference.
Most tumors are infiltrated with stromal cells, resulting in a mixture of sequencing reads derived from stromal and tumor cells. It is important to accurately measure tumor cell content in order to ensure that sequencing depth is sufficiently powered to detect mutations and also to measure the zygosity of somatic mutations. We employed a series of bioinformatic approaches to estimate tumor cell content36,39. Specifically, copy number alterations induce allelic imbalances over germline heterozygous SNPs, and we used the extent of allelic imbalance to measure tumor cellularity. We also calculated tumor cell content based on somatic mutation allele fractions after taking into account the copy number status of the locus. When possible, tumor cell content was calculated from both germline and somatic variant allele fractions to produce a consensus estimate (see Supplementary Table 1).
8q copy number inference.
In Supplementary Figure 4a,b, we inferred discrete levels of copy number gain over chromosomal arm 8q as described in the “Copy number estimation” section above. We validated these estimates several ways.
In Supplementary Figure 4c, we calculated the ratio of reads mapping to the major versus minor allele over heterozygous SNPs situated on chromosomal arm 8q. Here, the term “major allele” refers to the more abundant, or amplified, allele in the tumor. To calculate this ratio, we first subtracted out the reads emanating from stromal cells – these reads would have allelic ratios of 1:1, thereby diluting the overall ratio. The specific formula to determine the ratio of reads in the tumors cells that we used was as follows:
For higher level gains, there are several potential allelic combinations (see Supplementary Fig. 4c for a schematic); we therefore show the expected relationship between absolute copy number and major to minor allele ratios after considering all of these possibilities (Supplementary Fig. 4c, left panel). We next plotted the observed relationship between absolute copy number and major to minor allele ratios from our data (Supplementary Fig. 4c, right panel). Note that the observed relationship closely mirrors the expected relationship. In particular, tumors with high levels of 8q gain were more likely to have greater ratios of major to minor allelic reads. In aggregate, the allelic ratios over chromosomal arm 8q support our absolute copy number inferences.
In Supplementary Figure 4d, we used multiplex ligation-dependent probe amplification (MLPA)40 to measure 8q levels in 8 primary tumors predicted to harbor between 2–6 copies of chromosomal arm 8q. DNA was extracted using the QIAamp DNA Micro Kit (Qiagen), followed by standard sodium acetate/ethanol precipitation for optimal purity. Probes were ligated to specific DNA sequences on chromosomes 1, 3, 6 and 8 using the SALSA MLPA KIT P027-B1 Uveal Melanoma (MRC-Holland). In addition, 12 reference probes were ligated to chromosomes 2, 4, 5, 10, 11, 12 13, 14, 15 and 18. The signal intensities of the various probes were measured using capillary electrophoresis on a Genetic Analyzer 3130xl (Applied Biosystems). The signal intensity of each probe was normalized to the signal intensity of each reference probe and the relative amount of each target sequence was calculated using the Coffalyzer. Net data analysis software from MRC Holland. The y-axis of Supplementary Figure 4d depicts the ratio of 8q signal to other parts of the genome without copy number alterations. It is important to note that MLPA signals are subject to saturation at higher level gains, providing a relative (rather than an absolute) estimate of copy number. We observed a near perfect correlation (R = 0.97) between MLPA ratios and absolute copy number estimates from sequencing data, supporting our absolute copy number inferences.
In Supplementary Figure 4e, we performed fluorescence in situ hybridization (FISH) to validate copy number of chromosomal arm 8q. In the right panel, FISH was performed using probes targeting c-MYC (Vysis LSI MYC spectrum orange probe) and CEP12 (Vysis CEP12 spectrum green probe) (both probes from Abbott Molecular Inc). Slides were scanned with a fluorescence microscope (Carl Zeiss AxioImager.Z2) and analyzed using the Metafer4 v 3.8.12 software (Metasystems) per manufacturer’s instructions. Tumor-bearing areas were identified by a pathologist using the DAPI filter at low magnification (10× lens) with the aid of the H&E. All areas involving tumor cells were scanned and within this area cells, which did not overlap and had bright signals for the probes were used for enumeration. Nuclei that had complete absence of signals for the control probe (CEP12) were excluded. At least 30 cells were analyzed per case. In the left panel, the tumor specimens were placed in a culture medium immediately after surgery and analyzed. The following fluorescent probes were hybridized to the tissues: Telomeric probe for chromosome 1p (Vysis TelVysion 1p) and centromeric probes for chromosome 3 (CEP3 D3Z1), 6 (CEP6 D6Z1), and 8 (CEP8) (all probes from Abbott Molecular, Inc.; www.abbottmolecular.com). The probes were visualized and counted using a fluorescence microscope. At least 100 cells were evaluated by two technicians and copy number variations in more than 10% of the cells were used as a cut off. The six tumors analyzed with a centromeric 8 probe were chosen because they did not have copy number transitions across the centromere of chromosome 8. The copy number estimates from our sequencing data typically aligned with the mode counts from the FISH data (Supplementary Fig. 4e).
Statistics.
In Figure 1b, we calculated the fraction of tumor cells in each microdissected tissue (described above) – the blue lines are anchored at half of that value (i.e. the expected mutant allele fraction of a fully clonal, heterozygous somatic mutation). The fading zones represent the 95% confident intervals. This was calculated under the assumption that mutant reads are sampled randomly from the total pool of mutant alleles in our DNA input, and therefore the final mutant allele fraction should fall within a binomial distribution around the expected mutant allele fraction of a fully clonal, heterozygous somatic mutation. Under this assumption the standard deviation of this distribution is dependent upon the sequencing depth (SD) in that sample and tumor cellularity (TC) as follows: ((0.5*TC) – (0.5*TC)2) × SD)0.5. The 95% confidence interval was calculated by multiplying the standard deviation by 1.96. Finally, these confidence intervals were converted to fractions by dividing them by the total sequencing depth in that sample.
In Figure 4c, we performed a two-tailed t-test, assuming equal variance, to compare the level of copy number gain of chromosomal arm 8q in primary tumors (n = 53) versus metastases (n = 36) (degrees of freedom = 87).
In Figure 5b, for the genomic alterations shown in Figure 5, we tabulated the number of times each occurred in metastases and primaries. We tested the probability that genomic alterations were enriched in metastases or primaries against the null hypothesis that the alterations should be equally distributed in each compartment. The P-values shown in Figure 5 reflect two-tailed probabilities under the assumption that our sampling of genomic alterations fits a binomial distribution of enrichments dictated by sampling size. Adjustments for multiple-hypothesis testing were not performed.
In Supplementary Figure 6, we sought to identify significant associations between clinical outcomes and phylogenetic-tree features. We took several measurements of phylogenetic trees from each case (see Supplementary Fig. 6b), including tree height, trunk length, branch lengths, and branch separation. Next, we investigated whether any of these measurements correlated with disease-free survival, survival after metastasis, or overall survival. In total, we made 18 pairwise comparisons between the 6 phylogenetic-tree measurements and the 3 clinical outcome datatypes. Each comparison comprised 35 data points, corresponding to each patient in the study. The correlations are shown as R-values (Pearson correlation coefficients) in Supplementary Figure 6b. We also performed a linear regression analysis to produce P-values for each correlation against the null hypothesis that the correlation coefficient (R-value) should be 0. Lastly, we report adjusted P-values, which were corrected for multiple-hypothesis testing using the Bonferroni correction.
Data availability.
The raw sequencing data are available at the European Genome Phenome Archive under accession EGAD00001004453.
Supplementary Material
Acknowldegements
We wish to acknowledge Ben Vainer, who passed away during the course of the study, and Heidi Ugleholdt, for the collection and preparation of tissue samples used in this study. We thank Morten Tolstrup Andersen and Mette Klarskov Andersen for genetic assistance. We also thank Sonia Mirza for help with fluorescence in situ hybridization. We also acknowledge funding support from the following: Melanoma Research Foundation (A.H.S.), National Cancer Institute K22 (A.H.S.), Melanoma Research Alliance (A.H.S.), Dermatology Foundation (A.H.S.), Fight for Sight Denmark (M.M.B.), the Danish Eye Research Foundation (M.M.B.), the Research Fund of Rigshospitalet (M.M.B.), the Danish Medical Research Grant/Højmosegård grant (M.M.B.), Research to Prevent Blindness (B.C.B.), National Cancer Institute 1R35CA220481 (B.C.B.), and the Terri Patters Foundation (B.C.B.).
Footnotes
Competing Interests
B.C.B. is a consultant for Lilly Inc.
References
- 1.Bastian BC The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu. Rev. Pathol 9, 239–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh AD & Topham A Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology 110, 962–965 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Singh AD, Turell ME & Topham AK Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology 118, 1881–1885 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Kujala E, Mäkitie T & Kivelä T Very long-term prognosis of patients with malignant uveal melanoma. Invest. Ophthalmol. Vis. Sci 44, 4651–4659 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Dogrusöz M et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Invest. Ophthalmol. Vis. Sci 58, 833–842 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Augsburger JJ, Corrêa ZM & Shaikh AH Effectiveness of treatments for metastatic uveal melanoma. Am. J. Ophthalmol 148, 119–127 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Robertson AG et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbour JW et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet 45, 133–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet 45, 933–936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtin JA et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med 353, 2135–2147 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Raamsdonk CDV et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457, 599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Raamsdonk CD et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med 363, 2191–2199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson P et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 7, 4624–4631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore AR et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet 48, 675–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesner T et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet 43, 1018–1021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Royer-Bertrand B et al. Comprehensive genetic landscape of uveal melanoma by whole-genome sequencing. Am. J. Hum. Genet 99, 1190–1198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagger M et al. Long-term metastatic risk after biopsy of posterior uveal melanoma. Ophthalmology 125, 1969–1976 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Kerick M et al. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med. Genomics 4, 68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field MG et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun 9, 116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talevich E, Shain AH, Botton T & Bastian BC CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol 12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vader MJC et al. GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. Br. J. Cancer 117, 884–887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang JL-Y, Urtatiz O & Van Raamsdonk CD Oncogenic G protein GNAQ induces uveal melanoma and intravasation in mice. Cancer Res 75, 3384–3397 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Callejo SA, Dopierala J, Coupland SE & Damato B Sudden growth of a choroidal melanoma and multiplex ligation-dependent probe amplification findings suggesting late transformation to monosomy 3 type. Arch. Ophthalmol. Chic. Ill 1960 129, 958–960 (2011). [DOI] [PubMed] [Google Scholar]
- 25.de Lange MJ et al. Heterogeneity revealed by integrated genomic analysis uncovers a molecular switch in malignant uveal melanoma. Oncotarget 6, 37824–37835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh N, Singh AD & Hide W Inferring an evolutionary tree of uveal melanoma from genomic copy number aberrations. Invest. Ophthalmol. Vis. Sci 56, 6801–6809 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Shain AH & Pollack JR The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PloS One 8, e55119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zingg D et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun 6, 6051 (2015). [DOI] [PubMed] [Google Scholar]
- 29.LaFave LM et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med 21, 1344–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson BG et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassoux N et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br. J. Ophthalmol 98, 769–774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turajlic S et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter JG et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bremner R & Balmain A Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 61, 407–417 (1990). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 36.Shain AH et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med 373, 1926–1936 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Shain AH et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet 47, 1194–1199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shain H Compiled genetic summary of the evolution of 35 metastatic uveal melanomas (2019). doi: 10.6084/m9.figshare.6845675.v1 [DOI]
- 39.Shain AH et al. Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell 34, 45–55.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schouten JP et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30, e57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available at the European Genome Phenome Archive under accession EGAD00001004453.