Abstract
Biological lipids are a structurally diverse and historically vexing group of hydrophobic metabolites. Here, we review recent advances in chemical imaging techniques that reveal changes in lipid biosynthesis, metabolism, dynamics, and interactions. We highlight tools for tagging many lipid classes via metabolic incorporation of bioorthogonally functionalized precursors, detectable via click chemistry, and photocaged, photoswitchable, and photocrosslinkable variants of different lipids. Certain lipid probes can supplant traditional protein-based markers of organelle membranes in super-resolution microscopy, and emerging vibrational imaging methods, such as stimulated Raman spectroscopy (SRS), enable simultaneous imaging of more than a dozen different types of target molecule, including lipids. Collectively, these chemical imaging techniques will illuminate, in living color, previously hidden aspects of lipid biology.
Lipids: The ‘Other’ Biological Molecules
Lipids (see Glossary) are a large and diverse class of metabolites with myriad roles within biological systems. These hydrophobic molecules serve as structural components of cellular membranes, energy stores, and potent signaling agents that affect physiological processes from the subcellular to the organismal level. Due to the broad impact of lipid signaling, many diseases are associated with disruptions in lipid metabolism and function, including several cancers, neurological disorders, autoimmune diseases, and pathogenic infections [1,2]. Despite these important roles in physiology and pathology, lipids have perennially been regarded as among the most challenging of biological molecules to study. Their hydrophobicity complicates in vitro biochemistry, and all too often, redundant biosynthetic pathways confound their study by reverse genetics.
Historically, major advances in imaging methods have fueled waves of biological discovery in lipid and membrane biology. The first wave primarily involved the use of electron microscopy for revealing the morphology of membranes and membrane-bound compartments within cells [3]. The advent of genetically encoded fluorophores ushered in an era where almost any protein target could be visualized, making molecular information accessible by optical microscopy within live cells [4]. However, because individual lipids are not directly encoded in the genome, their visualization using fluorescent protein-based probes is less straightforward and subject to caveats and complications in data interpretation (Box 1). Given the need to visualize the localization of lipid biosynthesis, metabolism, and trafficking, as well as the effects of lipids on signaling pathways, chemists have taken up the challenge to develop a diverse collection of tools and probes for these purposes. In this review, we discuss recent exciting advances that use chemistry to illuminate lipids and membranes throughout eukaryotes.
Box 1. Lipid-Binding Domains as Biosensors: A Cautionary Tale.
Phosphatidylinostitol-4-phosphate (PI4P) is one of seven phosphoinositide lipids that decorate the cytosolic leaflet of organelle membranes. The prevailing understanding of which membranes contain PI4P has shifted over the decades based on advances in tool development. PI4P was first isolated and characterized from brain white matter during the 1940–1960s [72,73]. Given that myelin within white matter mainly comprises plasma membranes, it became widely accepted that PI4P was located, along with its downstream metabolite, PI(4,5)P2, in the plasma membrane. When the first PI4P-recognizing protein domains were characterized during the late 1990s, they were surprisingly found within pleckstrin homology (PH) domains of the Golgi-resident proteins OSBP and FAPP1. These PH domains, which localized to the Golgi apparatus dependent on PI4P, became widely used as PI4P biosensors and contributed to a general consensus that this organelle contained a major pool of PI4P. However, additional PI4P probes were developed, such as the PH domain of yeast Osh2, which demonstrated strong plasma membrane localization in mammalian cells.
Why did these different PI4P biosensors localize to different organelle membranes? In addition to recognizing PI4P, these PH domains are coincidence detectors, meaning that they bind to other factors, such as the GTPase Arf1 for OSBP and FAPP1 and plasma membrane-localized PI(4,5)P2 for Osh2. This property biased their localizations. Today, unbiased PI4P biosensors have been developed (P4M and P4C domains from the Legionella proteins SidM [74] and SidC [75]) that detect both Golgi and plasma membrane PI4P pools simultaneously and reveal an additional pool of PI4P at endosomes, finally accounting for the localizations of all PI 4-kinase isoforms.
A final word of caution: even with these far more specific probes, it is not possible to know with complete certainty that other minor PI4P pools are not being visualized or if the relative affinity of the probes is equivalent for PI4P in each compartment.
Lipid-Binding Protein Domains Are a Double-Edged Sword
The most widely used approach for visualizing lipids within cells is genetically encoded, fluorescently tagged lipid-binding proteins. These probes, often referred to as biosensors, are based on small peptide fragments or well-folded protein domains tethered to a fluorescent protein to enable visualization by fluorescence microscopy [5]. Lipid-binding probes are introduced to cells by transient transfection. Critically, these biosensors do not image the lipid molecules directly; instead, what is being visualized is the protein biosensor itself. The advantages of this approach are obvious. They capitalize upon naturally occurring lipid-binding domains, obviating the need to evolve a specific lipid binder in vitro. In addition, they can be easily delivered to a variety of cell types by transfection or viral transduction and come in many colors to enable multiplexing.
However, this indirect approach to detecting lipids does have drawbacks. First, some lipids do not have a known corresponding binding domain with suitable affinity or specificity. In particular, abundant lipids, such as phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol, and some low-abundance signaling lipids, lack binding probes and have proven to be elusive targets for antibody labeling [6,7]. Second, these biosensors typically reveal only qualitative information about relative amounts of the lipid in target membranes, and extracting quantitative information is challenging. For example, one can deplete a lipid or stimulate its synthesis and watch the localization of the binding probe shift, but it is difficult to measure exact concentrations of target lipids or even quantify percent changes in lipid concentrations, because the binding of such probes to their targets typically does not scale linearly with concentration.
Third, these probes report on total populations of a given lipid and do not enable any measure of flux through lipid biosynthetic pathways. Fourth, as part of their function, biosensors necessarily bind to and mask the lipid, preventing any native interactions [5]. While this property can be useful because it functions like a ‘lipid knockdown’, it is perturbative. Fifth, many of the biosensors that were initially characterized to bind to a single target in fact exhibit affinity for multiple species, which can complicate analysis (Box 1).
Despite these limitations, fluorescently tagged lipid binding probes have major roles in lipid and membrane biology. However, their drawbacks highlight the need for complementary tools, creating an opportunity for chemical innovation to enable the study of this unique class of biomolecules.
Metabolic Labeling to Produce Lipid Reporters
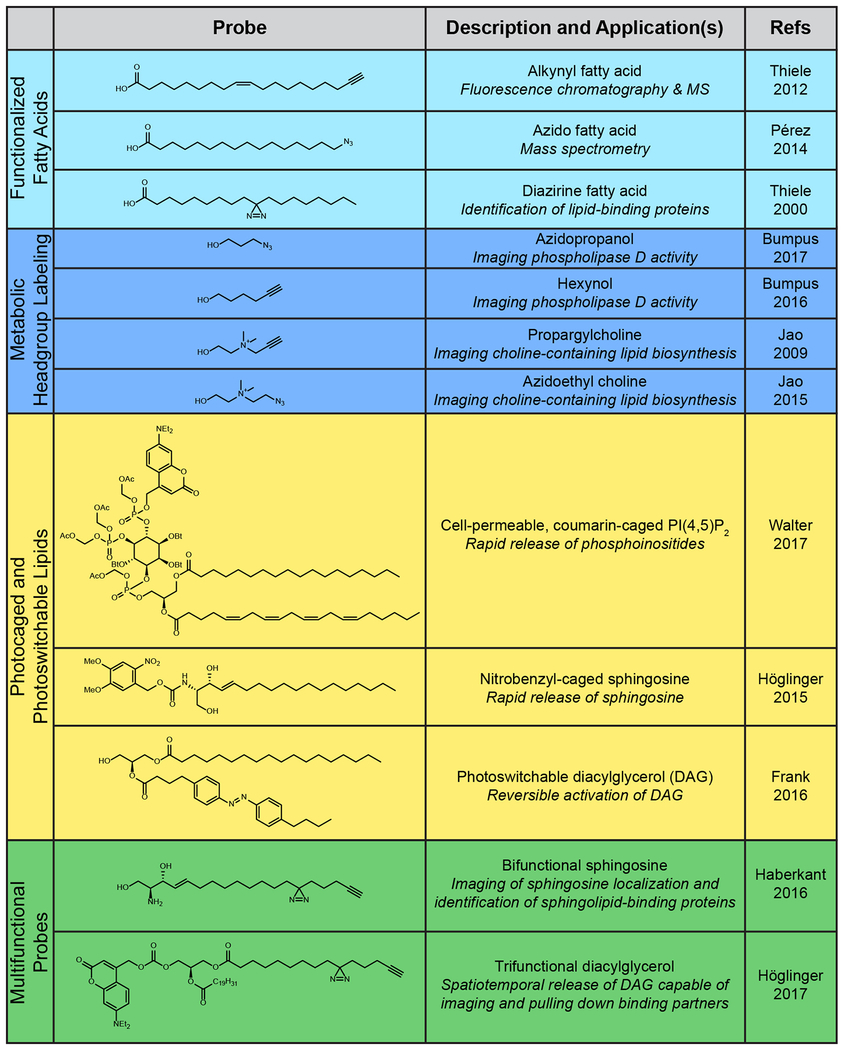
One of the most promising avenues for overcoming the inherent challenges of visualizing lipids, and metabolites in general, within their cellular environment is to feed cells a chemically functionalized and ultimately detectable biosynthetic precursor. This small-molecule reporter is taken up by the native biosynthetic machinery, allowing the metabolite of interest to be fluorescently tagged using a selective bioorthogonal, or click chemistry, reaction. This approach takes its cues from classic radiolabeling studies using 3H- and 14C-labeled biosynthetic precursors that were used to map biosynthetic pathways, but eliminates the hazards of using radioactive isotopes and further enables visualization by fluorescence microscopy directly within cells. For tagging lipids, the functional handle can be incorporated either into the hydrophobic tail or the hydrophilic head group of the lipid (Table 1).
Table 1.
Chemical Probes for Studying Diverse Cellular Lipid Processes
|
Labeling Lipid Tails: Don’t Let the Tail Wag the Dog
A defining characteristic of lipids is their fatty acyl tails. The terminus of the acyl chain provides a synthetically convenient position for attachment of bioorthogonal handles, such as alkynes [8] or azides [9] (Table 1). However, all two-tailed lipids, such as phospholipids and sphingolipids, use fatty acids as metabolic precursors; thus, when cells are labeled with alkynyl or azido fatty acids, the bioorthogonal group is incorporated into a variety of lipid classes. Thus, this approach is useful for tagging large swaths of the lipidome, but other approaches are necessary to gain specificity for individual classes of lipids (vide infra). To further complicate matters, these fatty acid probes are also incorporated into proteins co-translationally and post-translationally. In fact, the study of protein lipidation with these probes has proven successful and is expertly described and reviewed elsewhere [10–14]. Thus, while fatty acids can be used effectively to label the whole lipidome for ex vivo analysis, including chromatography [8] and mass spectrometry [8,15], in a radioisotope-free manner, they have limited utility for experiments that necessitate selectivity for a class of lipids or even for lipids over other classes of biomolecule without the ability to physically separate undesired, off-target signals.
To overcome the issue of general metabolic precursors, such as fatty acids, being incorporated into undesired pathways, researchers have also prepared bioorthogonally functionalized analogs of several lipid species, including sterols [16,17] and sphingolipids [18,19] (Table 1). Given that these lipids are bound for a narrower realm of metabolic fates, they are more useful for revealing precise lipid localizations within cells. However, bolus dosing of cells with these probes introduces a degree of uncertainty in the localization of the signal in imaging experiments, because these probes are not biosynthesized and trafficked identically to their unlabeled counterparts.
Nonetheless, bioorthogonal fatty acids are versatile probes. By clicking on an affinity tag, such as biotin, instead of a fluorophore, the fatty acid can be enriched instead of visualized. To best take advantage of this property, an additional chemical modification is required to irreversibly crosslink the lipid to protein-binding partners to preserve transient interactions. Whereas it has long been appreciated that much of the proteome acts at or within membranes, it has been difficult to capture these interactions due to detergents and other lipid-solubilizing additives commonly used in protein purification and analysis. This challenge motivated the development of bifunctional fatty acids containing both an alkyne, which allows for sample enrichment or visualization, and a photoactivatable diazirine group that covalently crosslinks to adjacent proteins and, thus, preserves the lipid–protein interaction even through detergent-based enrichment [20]. Similar to the monofunctionalized lipids described earlier, selectivity for lipid classes can be achieved by replacing the fatty acid chain with a more advanced biosynthetic intermediate. In particular, a bifunctional sphingosine containing an alkyne and a diazirine was developed for probing sphingolipid–protein interactions [15] (Table 1 and Figure 1).
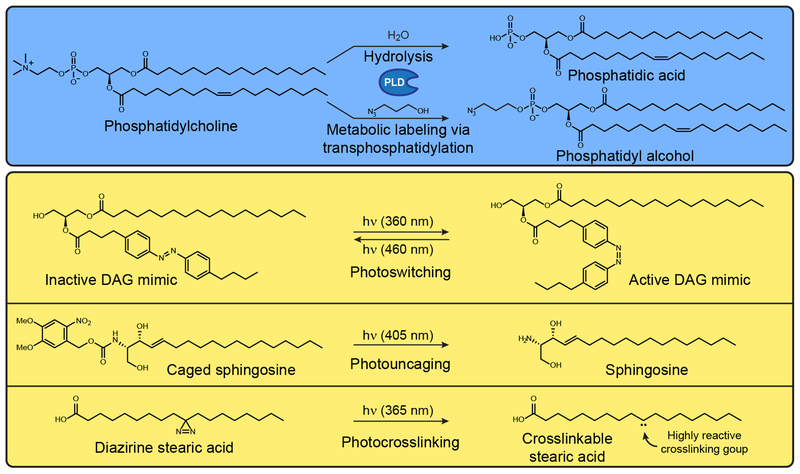
Figure 1. Biochemical and Photochemical Reactions Used in Conjunction with Lipid Probes.
(A) Metabolic labeling of phospholipase D (PLD) activity. PLD functions naturally as a hydrolase that produces phosphatidic acid from phosphatidylcholine, but the addition of low concentrations of exogenous primary alcohols, such as 3-azido-1-propanol, causes PLDs to catalyze a transphosphatidylation reaction to form a functionalized phosphatidyl alcohol. (B) Photochemical reactions that can be used to turn on inactive precursors, including those that can activate photoswitchable (i), photocaged (ii), and photocrosslinkable (iii) lipids. Abbreviation: DAG, diaglycerol.
Labeling Lipid Head Groups: Head of the Class
Whereas lipids in general are defined by their long fatty acyl tails, individual classes of lipid are often categorized by the identity of their head group, which imparts unique biophysical and biochemical properties to the lipid molecule. Importantly, head groups are chemically diverse and installed through dedicated biosynthetic pathways, making them attractive targets for bioorthogonal metabolic labeling (Table 1 and Figure 1).
This approach was first used to target the de novo synthesis of lipids containing the head group choline [21]. Here, an alkyne-containing choline analog called propargylcholine was used to incorporate a bioorthogonal handle into choline-containing lipids, including phosphatidylcholine and sphingomyelin, enabling their visualization following click chemistry tagging by fluorescence and electron microscopy [21,22]. This approach can monitor bulk lipid synthesis and visualize many intracellular membrane compartments, and it functions well even beyond mammalian cells, labeling choline lipids in yeast [22] and plants [23]. Later, this strategy was updated to include an azido choline analog that enables tagging within live cells using a nontoxic, copper-free click chemistry tagging reaction [24]. The abundance of choline-containing lipids within the cell also made azidoethylcholine an attractive probe of choice for visualizing metabolically labeled nonprotein biomolecules via electron microscopy[25]. Propargylcholine labeling can also be combined with functionalized fatty acid labeling, such as diazirine fatty acids, to allow cellular metabolism to produce multifunctional lipid probes [26].
An important and complex class of lipids are those containing glycan head groups. Collectively, glycolipids, many of which reside at the cell surface, have critical roles in signaling and cell–cell communication. Metabolic oligosaccharide engineering with bioorthogonal probes was first introduced in 1997 to target cell-surface, sialic acid-containing glycans with ketone groups, which can be conjugated with a variety of reagents [27]. Since this landmark study, many bioorthogonal monosaccharide reporters have been developed to target different glycan classes, reviewed elsewhere [28].
While the bulk of the analytical efforts have focused on the use of these probes for studying protein glycosylation, some of the same monosaccharide probes are appreciated to also target lipid glycosylation [29–32]. Within intact cells, it can be challenging to obtain selectivity for lipid glycosylation over protein glycosylation, because glycosyltransferase specificity is often determined by the identity of the acceptor sugar at the nonreducing end of the glycan rather than the protein or lipid scaffold. However, these probes tag the totality of a particular type of glycosylation, and the tagged lipids can then be separated from glycoproteins for in vitro analysis. Glycolipids can also be synthesized ex vivo with bioorthogonal functionality, applied to cells and then be tagged for fluorescence imaging [33]. This approach is less perturbative than one involving direct attachment of the fluorophore during the initial synthesis, affording a better chance to observe proper lipid localization relative to previous methods while ensuring adequate specificity for glycolipids.
Phosphatidic acid (PA) is both a central intermediate in phospholipid biosynthesis and a potent lipid second messenger [34]. Given that it is generated by multiple enzymatic activities for these various purposes (acylation of glycerol-3-phosphate, phosphorylation of diacylglycerol, and hydrolysis of phosphatidylcholine), it is a classic case where even an ideal lipid-binding probe would not enable one to distinguish signaling from biosynthetic pools of the lipid. The challenge to label pools of PA used for acute signaling functions motivated us to develop a metabolic reporter-based strategy for its visualization.
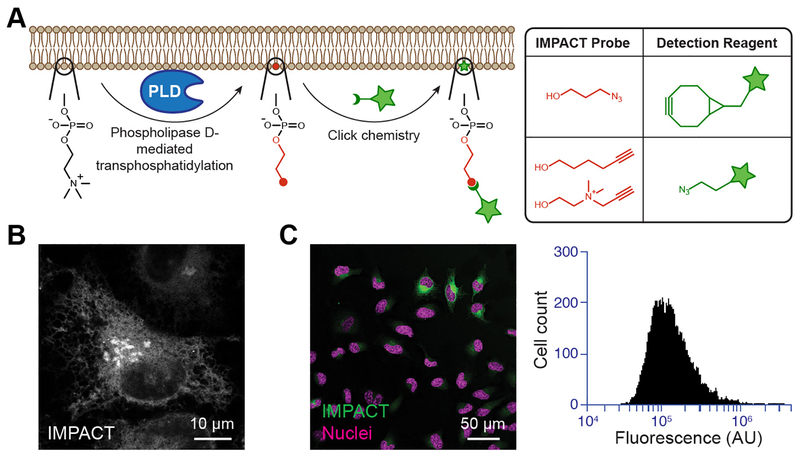
We focused on PA produced for signaling by hydrolysis of phosphatidylcholine by phospholipase D (PLD) enzymes (Figure 1). Our method takes advantage of a second but related enzymatic activity of PLDs, namely transphosphatidylation with primary alcohols, such as ethanol or butanol, to generate phosphatidyl alcohols, which are traditionally detected in bulk biochemical measurements, such as chromatography or mass spectrometry, and do not enable visualization by microscopy. To address this need, we developed a method termed Imaging phospholipase D activity with clickable alcohols via transphosphatidylation (IMPACT). In IMPACT, instead of unfunctionalized primary alcohols, alkynyl or azido alcohols are used as PLD substrates to generate corresponding alkynyl [35] or azido [36] phosphatidyl alcohols, which are lipid reporters of PLD activity and mark membranes where PLD enzymes are activated to produce PA (Table 1 and Figures 1 and 2). These clickable lipids are then tagged with fluorophores using a click chemistry reaction, and the resultant fluorescent lipids enable visualization of cellular membranes bearing active PLD enzymes (Figure 2). IMPACT labeling revealed a heterogeneity in PLD activities across cell populations and can be used in conjunction with fluorescence-activated cell sorting to stratify such subpopulations according to their PLD activity for downstream biochemical analysis (Figure 2). It was previously not possible to achieve this kind of cellular and subcellular resolution of PLD activity, and we envision many applications of IMPACT to study PLD signaling in complex physiological and pathological settings.
Figure 2. Imaging Phospholipase D (PLD) Activity with Clickable Alcohols via Transphosphatidylation (IMPACT).
(A) (i) Schematic of IMPACT labeling. Functionalized alcohols are installed into reporter lipids by PLD transphosphatidylation. These unnatural reporter lipids can then be visualized with fluorescent click reagents. (ii) Examples of fluorescent click reagents. (B) IMPACT can be used to visualize intracellular organelle membranes bearing PLD activity. (C) IMPACT can also be used to visualize PLD activity at the single-cell level within a population by fluorescence microscopy (i) or flow cytometry (ii).
During the development of IMPACT, we noted that the choline mimics (e.g., propargylcholine) used to label de novo phosphatidylcholine biosynthesis were also clickable primary alcohols that could in principle be PLD transphosphatidylation substrates. Thus, we asked whether a single bioorthogonal probe, propargylcholine, could be used to study two distinct pathways, namely de novo phospholipid synthesis via the Kennedy Pathway and PLD signaling. Propargylcholine is indeed a substrate for transphosphatidylation by PLD enzymes, and, by controlling the cellular activation state and the length of time that the cells are incubated with propargylcholine, this chemical probe could be used to report on two different biosynthetic pathways by generation of the same clickable lipid reporter [37]. This approach is an example of probe economy, in which the probe exhibits a one-to-many relationship with downstream biological applications.
Unleashing Native Lipids with Light
Whereas dosing cells with metabolic precursors to the lipid of interest is an elegant method to label lipids as a function of their natural biosynthetic pathways, because it helps to ensure proper localization and trafficking of the lipid, it is not without its drawbacks. The temporal resolution provided by the natural synthetic pathway may be low, with diffusion and trafficking of the labeled lipid as competing and complicating factors. In addition, the lipid of interest may lack any unique or chemically modifiable functionality that can serve as the basis for developing a suitable metabolic reporter probe. To overcome these limitations, substantial effort has also been expended to develop lipid-based probes that are synthesized chemically in an inactive form, administered to cells, and then activated on demand to remove the inactivating group.
The most widely used strategy for unleashing an active lipid on demand in cells is illumination with UV or visible light, because it allows for temporal as well as spatial control of activation within cells. A variety of photocaged lipids have been synthesized and applied (Table 1 and Figure 1). Key recent examples include caged diacylglycerols (DAG) for uncovering the relationship between DAG acyl chain length and signaling patterns [38], caged sphingosine for studying the roles of sphingosine in regulating calcium release [39], caged phosphoinositides for studying their myriad functions in cell signaling [40,41], and caged lyso-phosphatidic acid for studying effects on chemotaxis [42]. Adding a further layer of control, synthetic incorporation of an additional organelle-targeting group can enable the study of organellespecific metabolism [43,44].
Beyond caging the headgroup of the lipid to render it inactive, photoresponsive groups (i.e., those that change their structure in response to light, such as azobenzene) can be placed within the hydrophobic lipid tail region. Reversible, light-induced isomerization of azobenzenes dramatically changes the 3D shape of this hydrophobic group, mimicking different conformations of saturated, monounsaturated, and polyunsaturated lipid fatty acyl tails. This strategy was successfully demonstrated by using synthetic, photoswitchable variants of fatty acids[45] and the lipid-derived metabolite capsazepine [46] (an analog of the natural product capsaicin, which occurs in hot peppers) to control the activity of TRPV1 channels (Table 1 and Figure 1). Azobenzene groups can also be incorporated into an acyl tail of DAG, creating so-called ‘PhoDAG’ probes. These tools have enabled the reversible activation of protein kinase C, for which DAG serves as an agonist, by illuminating cells with UV light [47]. Building on these results, second-generation PhoDAG probes were constructed by replacing both acyl chains with the photoswitchable azobenzene, which in one of its two isomeric forms mimics the polyunsaturated fatty acid arachidonic acid. These new probes were used to control the activity and uncover new aspects of the regulatory mechanism of TRPC3, a cation channel that is also regulated by DAG binding [48].
A photoswitchable ceramide probe exhibited isomer-dependent differences in biophysical properties in artificial membranes, suggesting that photoswitchable lipids would be useful for modeling and understanding membrane fluidity [49]. Photoswitchable lipids have the added advantage that their activation is reversible, because the azobenzene will return to the lower energy trans isomer slowly in the absence of 365-nm illumination or can be rapidly reconverted from the cis isomer by illumination with lower energy 470-nm light. Thus, the levels of these potent signaling agents can be rapidly tuned in response to biocompatible intensities of light, giving researchers an unprecedented level of spatiotemporal control. In addition to ‘monofunctional’ photocaged lipids, photoresponsive groups have been combined with other strategies to create bifunctional and even trifunctional fatty acids, phospholipids, and sphingolipids [50].
Using Lipid Probes to Image Organelle Membranes
Thus far, we have focused on tools for studying individual lipid species and classes within cells, which are used to study their biological functions. As an ensemble, lipids comprise membrane bilayers and act to define the boundaries and barriers of cells and intracellular organelle compartments. Often, it is desirable to visualize not a particular lipid species but instead an individual organelle, which can be accomplished by expressing fland act to tagged membrane-resident or membrane-associated proteins. The small size of some organelle compartments and/or proximity to other organelles can necessitate advanced super-resolution microscopy techniques to distinguish the desired structures (e.g., Golgi stacks or membrane contact sites). However, super-resolution microscopy techniques, which give subdiffraction resolution, place high demands on fluorophores. The labels must withstand powerful irradiation, which typically leads to photobleaching of sparsely labeled fluorescent protein targets. Lipids have a higher molar density within membranes compared with proteins, leading a team from Yale to develop a lipid-labeling strategy rather than a genetically encoded protein-labeling method to enable extended timeframe super-resolution imaging of membrane compartment dynamics within live cells (Figure 3).
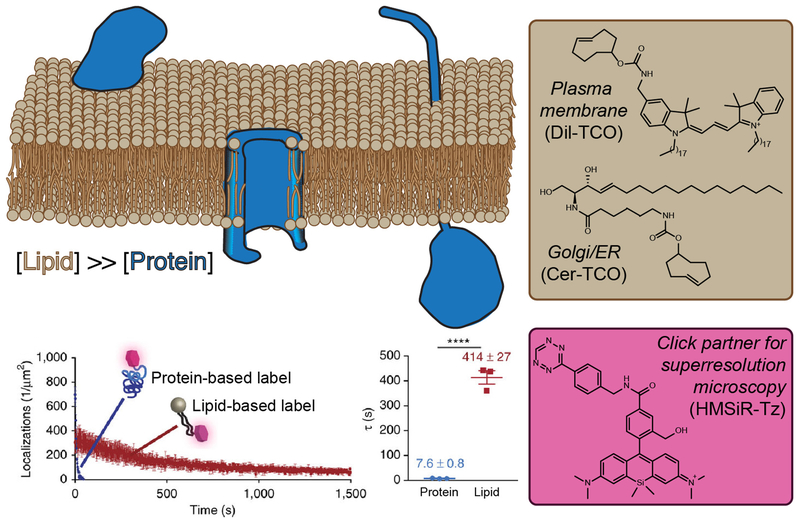
Figure 3. Targeting Lipids Enables Time-Lapse Super-Resolution Microscopy Visualization of Organelles.
(A) Lipids (tan) have a higher molar density than proteins (blue) within a given surface area of a membrane bilayer and, therefore, intracellular membranes can be densely tagged with lipid probes bearing reactive handles capable of rapid ligation with a flA) Lipids bearing the opposing click partner. (B) Clickable lipid-based probes for targeting specific organelles. (C) An appropriately functionalized click partner bearing a hydroxymethylene silicon rhodamine dye. This probe switches between nonfluorescent and fluorescent forms within a hydrophobic environment, enabling the extended imaging times relative to identically labeled proteins. Graphs show photobleaching time course (D) and fluorescence half-life (E) of lipid and protein probes targeted to the same organelle and bearing the same silicon rhodamine dye.
Their investigation took advantage of a bioorthogonally functionalized ceramide probe bearing a trans-cyclooctene that localized to the Golgi apparatus [51,52]. The trans-cyclooctene is then rapidly ligated with a tetrazine-containing silicon-rhodamine dye via inverse electron-demand Diels–Alder click chemistry and visualized by stimulated emission-depletion (STED) microscopy, a super-resolution technique that enables an increase in resolution verses standard confocal methodology. Importantly, the high laser intensities required for STED imaging did not result in rapid photobleaching for the lipid label compared with protein markers tagged with the identical fluorophore, demonstrating the virtue of choosing lipids as the labeling target.
This technique was further refined and generalized. Regarding fluorophore development, a second silicon-rhodamine dye containing an environment-sensitive, internal quenching mechanism [53] was used for single-molecule switching (SMS) nanoscopy, an alternate set of techniques to STED that afford a further increase in spatial and temporal resolution. The technique was also expanded to other organelle locations by switching the Golgi-resident ceramide-localizing tag for a dialkylindocarbocyanine, DiI, to target the plasma membrane marker [54,55] and removing the temperature block from the ceramide to label the endoplasmic reticulum [54], enabling super-resolution visualization of membrane dynamics across several cellular compartments in live cells and without the need for transient transfection, membrane permeabilization, or oxygen scavenging (Figure 3). Collectively, these super-resolution microscopy-capable, lipid-based organelle vital stains will allow us to better study the dynamics of membrane-bound compartments [56].
Seeing Lipids beyond ROYGBIV
Up to this point, most of the work discussed here has taken advantage of small-molecule fluorophore tags to visualize lipids using fluorescence microscopy. A limitation of this general approach is the broad excitation and emission bands of common fltags to vi, which limits the number of labels that can be simultaneously installed and detected without the signals bleeding into one another. While specialized microscopy setups can enable upwards of six channels to be visualized independently in real time [57], a typical microscope setup will enable visualization of only three or four channels at once. A separate limitation is that the large size of the fluorophore tag relative to the lipid target may perturb localization and trafficking patterns.
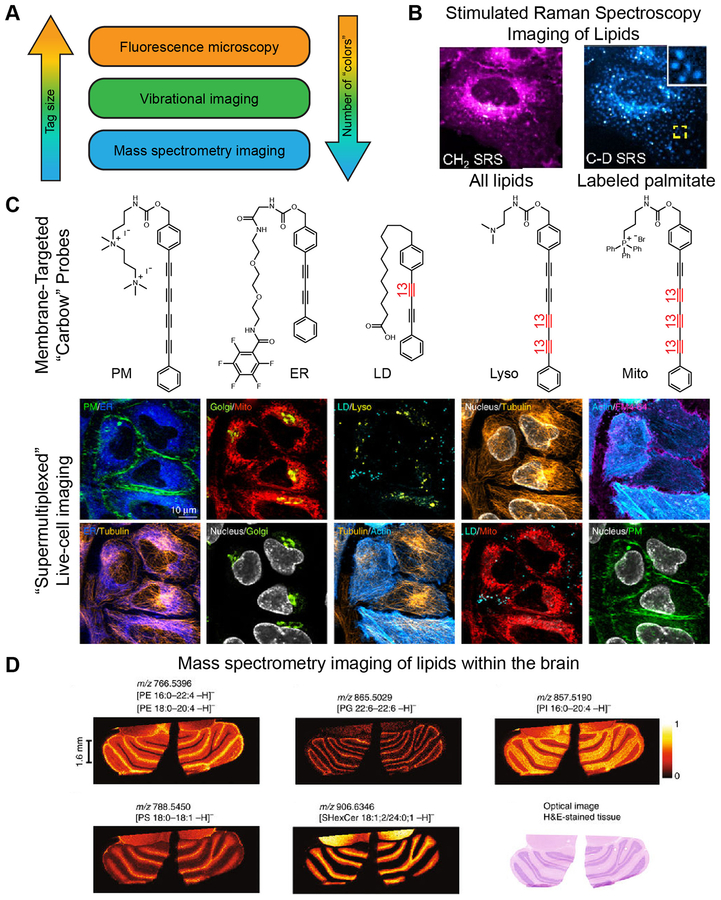
Stimulated Raman scattering (SRS) imaging represents a solution to these problems. Using vibrational rather than electronic excitation, vibrational imaging techniques, such as SRS imaging, can use contrast agents as small as single carbon–deuterium or carbon–carbon bonds. This technique can be carried out in live cells [58] and tissues [59] across a variety of cellular targets and lipid-targeting modalities (Figure 4). One of the first targets described was the terminal alkyne. Although alkynes have commonly been included in probes due to their ability to participate in a copper-catalyzed click chemistry reaction with azides, their carbon–carbon triple bonds also provide a unique spectroscopic handle for Raman scattering and allow for selective detection without the need for a click reaction or cell fixation [60]. This technique can also be adapted for multi-’color’ imaging by isotopically labeling one or both carbons in the triple bond, because this modification shifts the vibrational frequency of the bond sufficiently to allow for separate detection (i.e., 12C–12C, 12C–13C, and 13C–13C triple bonds are all spectroscopically distinct) [61]. Further ‘supermultiplexing’, or the ability to detect even larger numbers of distinct labels, can be achieved by introducing other functional groups, such as nitriles, which contain a carbon–nitrogen triple bond [62], or polyynes, which contain stretches of conjugated carbon–carbon triple bonds [63] (Figure 4). SRS imaging has been used to image choline-containing lipids using propargylcholine [21,60] without the need for cell fixation and click labeling. Replacing carbon–hydrogen bonds with carbon–deuterium bonds within sterols [64] or fatty acids [65] enables visualization of biological molecules with truly minimal structural perturbations. Fatty acid probes containing differing degrees of saturation have been used to interrogate phase separation within the endoplasmic reticulum [65]. Excitingly, SRS imaging is also compatible with standard fluorescence microscopy in the same cell, meaning that one can take full advantage of different regions of the electromagnetic spectrum, combined with different labeling strategies, for multiplexed imaging of lipids, organelle membranes, and protein targets [63] (Figure 4).
Figure 4. Less Is More: Non-Fluorescence Methods for ‘Supermultiplexed’ Imaging with Smaller Tags.
(A) Comparison of fluorescence imaging with vibrational imaging and mass spectrometry imaging in terms of tag size and number of species that can be detected simultaneously (‘colors’). (B) Stimulated Raman spectroscopy (SRS) imaging can be used to visualize lipids by exciting the vibrational modes of the CH2 groups within lipid tails (left) without any exogenous labels. Deuterated palmitate allows for pulse-chase imaging of the subset of lipids containing this particular lipid tail (right). (C) SRS imaging can be combined with fluorescence microscopy to enable ten-color imaging in live cells. Multiple vibrational ‘colors’ are created by extending the length of the dyes in polyyne chain and by introducing 13C–12C pairs into the chain, indicated in red. Such probes are dubbed ‘carbow’ (for carbon rainbow) and can target different organelles, including the plasma membrane (PM), endoplasmic reticulum (ER), lipid droplets (LD), lysosomes (lyso), and mitochondria (mito). (D) Mass spectrometry enables label-free imaging of lipids within tissues. Shown are images of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), and sulfatide (SHexCer) within a brain section taken from a mouse.
Other promising modern, nonfluorescent imaging techniques include mid-infrared optoacoustic microscopy (MiROM), which is a bond vibrational imaging modality similar to SRS that allows for higher sensitivity and deeper tissue penetration by limiting the effect of the ubiquitous water found in biological samples. MiROM was recently used to monitor the dynamics of lipids and proteins in live, label-free mode in adipocytes [66].
Finally, mass spectrometry (MS) is a nonspectroscopic method that is widely used to identify and profile cellular lipid populations, or lipidomes [67,68]. The combination of traditional MS-based lipidomics with subcellular fractionation can reveal information about lipid localization with organelle-level resolution. To achieve greater spatial resolution and visualize individual molecules within intact cells and tissues, various forms of mass spectrometry imaging have emerged [69] (Figure 4). These techniques enable incredibly sensitive detection of molecules in intact, fixed cells and tissues with nearly unlimited ‘multiplexing’, because detection occurs at the molecule of interest as opposed to detecting an imaging contrast agent as a proxy for the molecule of interest, as is typically done with flxed cells and chromophores. These emerging techniques have been applied to image cellular lipids [70,71], speaking to their promise for visualizing these important molecular components of the cell.
Concluding Remarks
Lipids and cellular membranes are a diverse and dynamic group of molecules and structures that have been historically challenging to study. However, we believe that the future is bright for those interested in lipids and their roles in biology. New tools abound to study ever-increasing sectors of the lipidome with greater precision, and these tools will help enable researchers to address major outstanding questions in the regulation and dynamics of lipid metabolism and signaling (see Outstanding Questions).
Outstanding Questions.
What is the balance between vesicle-dependent and vesicle-independent transport of lipids in between different organelles? How is such a balance regulated? What chemical tools can allow dynamic monitoring and quantification of these processes?
How is lipid bilayer asymmetry maintained in cellular membranes? Can chemical tools help to uncover these regulatory mechanisms?
What roles can chemical probes have in helping to understand biophysical properties of cellular membranes, such as curvature, potential, rigidity?
How does the introduction of bioorthogonal handles into lipids affect the localization of the resultant unnatural lipid reporters?
How long does it take for the turnover of lipid probes within cells? What, if any, are the effects of these degradation products?
Can lipid-based chemical probes be designed to label every organelle compartment in the cell to enable long-term visualization at super-resolution, with minimal photobleaching?
What are the most efficient and effective methods to use multiply labeled (i.e., supermultiplexed) samples to study the dynamics of lipid metabolism and lipid signaling?
Lipid-binding probes, where suitable, can be used to understand the subcellular localization of a lipid of interest. These biosensors are particularly useful when it is desirable to observe depletion of a lipid or rapid changes in the localization of a large pool of the target lipid, either by migration, trafficking, or stimulated biosynthesis at a distant location.
Metabolic reporter probes enable visualization of active pools of biosynthetic enzymes, provided that the temporal resolution of the labeling is high enough to compete with the diffusion and trafficking of the functionalized lipids. This approach also allows for populations to be stratified by the level of flux through a specific lipid biosynthetic pathway, because signal strength directly correlates to reporter lipid levels and not to external factors, such as transfection level.
Multifunctional, including photocaged, variants of natural lipids can be used to directly observe the responses arising from rapid production of potent signaling lipids. These techniques can then be combined with the increasing array of detection modalities to enable an amazing array of experimental conditions depending on the availability of techniques, from standard epifluorescence and confocal microscopes to the increasingly widely available super-resolution fluorescence microscopes and the more rarified setups for SRS, depending on the number of types of molecules that must be tracked and analyzed simultaneously.
As new probes and more systems for multiplexing data collection continue to emerge, we envision that the portion of biochemical space accessible to live-cell imaging will continue to increase. Researchers will be able to gather information throughout the cell simultaneously, both as single time-point snapshots and tracking of whole-cell dynamics in real time. Thus, we believe that these new chemical methodologies will ‘grease the wheels of discovery’ in lipid biology for years to come.
Highlights.
New chemical tools are enabling a clearer vision of the many roles of lipids in biology.
Metabolic probes act as substrate mimics that hijack endogenous bio-synthetic enzymes to produce bioorthogonally functionalized reporter lipids, enabling visualization of flux through lipid biosynthetic pathways following tagging via a bioorthogonal click chemistry reaction.
Photoactivatable and photoswitchable variants of lipids enable the rapid turning on and turning off of lipid signaling molecules in live cells.
Specialized tagged lipids, the concentrations of which within membranes dwarf those of membrane protein tags, enable long-term, continuous imaging of dynamic processes at super-resolution in live cells with little photo-bleaching.
New vibrational imaging tools for ‘supermultiplexed’ tagging enable the simultaneous visualization of lipids and many other tags.
Acknowledgments
Work in the Baskin laboratory is supported by the NIH (R00GM110121 and R01GM131101, J.M.B.), the Arnold and Mabel Beckman Foundation (Beckman Young Investigator, J.M.B.), and the NSF (CAREER, CHE-1749919, J.M.B.; GRFP, DGE-165-0441, T.W.B.).
Glossary
- Bioorthogonal
the chemical property of being stable, or inert, in biological fluids but prone to specific reactivity with appropriately functionalized exogenous chemical reagents to enable subsequent tagging
- Click chemistry
a group of bond-forming chemical reactions wherein two reactive partners react selectively with one another, even in complex milieus containing competing functionality, to rapidly form a stable, covalent product in high yield with few side-products at ambient temperatures
- Hydrophobic
the property of repelling water; similar to lipophilic, which means fat-loving, or having a tendency to dissolve or associate with greasy substances
- Imaging phospholipase D activity with clickable alcohols via transphosphatidylation (IMPACT)
a chemical method for visualizing cellular membranes bearing active phospholipase D enzymes, which physiologically generate the lipid second messenger phosphatidic acid
- Lipid
a member of the large group of hydrophobic compounds that occur in biological systems and are typically soluble in organic solvents but only sparingly soluble in water. In addition to their mostly hydrophobic structures, many biological lipids also have a water-loving, or hydrophilic, head group
- Membrane bilayer
a cellular structure comprising two layers of lipids with a hydrophobic core and hydrophilic surfaces with embedded integral membrane proteins. These structures serve as barriers to protect cells and organelles from their surroundings, regulating the influx and efflux of nutrients and other substances
- Metabolic labeling
a strategy wherein cells are fed radiolabeled or bioorthogonally functionalized substrates for biosynthetic pathways to detect flux through such pathways
- Photocage
a group that is used to mask a bioactive molecule, making it biologically inert, that can be removed by illumination with visible or UV light
- Photoswitchable
the ability to be interconverted between two different 3D structures (isomers) by the application of visible or ultraviolet light
- Super-resolution microscopy/nanoscopy
a collection of fluorescence microscopy techniques enabling images to be acquired with a resolution greater than the diffraction limit, which in practice is 250–350 nm, representing approximately a half of the wavelength of the emitted fluorescent light
- Vibrational imaging
an imaging modality wherein specific chemical bonds (i.e., carbon–deuterium single bond, carbon–carbon triple bond, etc.) serve as the contrast agent, and contrast is generated via transitions to vibrationally rather than electronically excited states, as occurs in traditional fluorescence imaging. An example of vibrational imaging is SRS imaging
References
- 1.Wymann MP and Schneiter R (2008) Lipid signalling in disease. Nat. Rev. Mol. Cell Biol 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 2.Brown HA et al. (2017) Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat. Rev. Drug Discov 16, 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afzelius BA and Maunsbach AB (2004) Biological ultrastructure research; the first 50 years. Tissue Cell 36, 83–94 [DOI] [PubMed] [Google Scholar]
- 4.Cubitt AB et al. (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci 20, 448–455 [DOI] [PubMed] [Google Scholar]
- 5.Platre MP and Jaillais Y (2016) Guidelines for the use of protein domains in acidic phospholipid imaging. Methods Mol. Biol 1376, 175–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doshi R (2013) Uncharted territory: the specific detection and quantification of lipids. J. Anal. Bioanal. Tech 4, 10–11 [Google Scholar]
- 7.Maekawa M and Fairn GD (2014) Molecular probes to visualize the location, organization and dynamics of lipids. J. Cell Sci 127, 4801–4812 [DOI] [PubMed] [Google Scholar]
- 8.Thiele C et al. (2012) Tracing fatty acid metabolism by click chemistry. ACS Chem. Biol 7, 2004–2011 [DOI] [PubMed] [Google Scholar]
- 9.Pérez AJ and Bode HB (2014) ω-Azido fatty acids as probes to detect fatty acid biosynthesis, degradation, and modification.J. Lipid Res 55, 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng T and Hang HC (2015) Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein-protein interactions in mammalian cells. J. Am. Chem. Soc 137, 556–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Won SJ and Martin BR (2018) Temporal profiling establishes a dynamic S-palmitoylation cycle. ACS Chem. Biol 13, 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thinon E and Hang HC (2015) Chemical reporters for exploring protein acylation. Biochem. Soc. Trans 43, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davda D et al. (2013) Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol 8, 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X and Hannoush RN (2018) A decade of click chemistry in protein palmitoylation: impact on discovery and new biology. Cell Chem. Biol 25, 236–246 [DOI] [PubMed] [Google Scholar]
- 15.Haberkant P et al. (2016) Bifunctional sphingosine for cell-based analysis of protein-sphingolipid interactions. ACS Chem. Biol 11, 222–230 [DOI] [PubMed] [Google Scholar]
- 16.Jao CY et al. (2015) Bioorthogonal probes for imaging sterols in cells. Chembiochem 16, 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakers L et al. (2018) Addressable cholesterol analogs for live imaging of cellular membranes. Cell Chem. Biol 25, 1–10 [DOI] [PubMed] [Google Scholar]
- 18.Collenburg L et al. (2016) A functionalized sphingolipid analogue for studying redistribution during activation in living T cells. J. Immunol 196, 3951–3962 [DOI] [PubMed] [Google Scholar]
- 19.Garrido M et al. (2015) Azide-tagged sphingolipids: new tools for metabolic flux analysis. Chembiochem 16, 641–650 [DOI] [PubMed] [Google Scholar]
- 20.Haberkant P et al. (2013) In vivo profiling and visualization of cellular protein-lipid interactions using bifunctional fatty acids. Angew. Chem. Int. Ed 52, 4033–4038 [DOI] [PubMed] [Google Scholar]
- 21.Jao CY et al. (2009) Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. U. S. A 106, 15332–15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyoshi S et al. (2014) Asymmetrical distribution of choline phospholipids revealed by click chemistry and freeze-fracture electron microscopy. ACS Chem. Biol 9, 2217–2222 [DOI] [PubMed] [Google Scholar]
- 23.Paper JM et al. (2018) Bioorthogonal click chemistry for fluorescence imaging of choline phospholipids in plants. Plant Methods 14, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jao CY et al. (2015) Biosynthetic labeling and two-color imaging of phospholipids in cells. Chembiochem 16, 472–476 [DOI] [PubMed] [Google Scholar]
- 25.Ngo JT et al. (2016) Click-EM for imaging metabolically tagged nonprotein biomolecules. Nat. Chem. Biol 12, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D et al. (2017) Global mapping of protein – lipid interactions by using modified choline-containing phospholipids metabolically synthesized in live cells. Angew. Chem. Int. Ed. Engl 129, 5923–5927 [DOI] [PubMed] [Google Scholar]
- 27.Mahal LK et al. (1997) Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis engineering chemical reactivity on cell surfaces through oligosaccharide bio-synthesis. Science 276, 1125–1128 [DOI] [PubMed] [Google Scholar]
- 28.Sminia TJ et al. (2016) Getting a grip on glycans: a current overview of the metabolic oligosaccharide engineering toolbox. Carbohydr. Res 435, 121–141 [DOI] [PubMed] [Google Scholar]
- 29.Lu L et al. (2015) Labeling cell surface GPIs and GPI-anchored proteins through metabolic engineering with artificial inositol derivatives. Angew. Chem. Int. Ed 54, 9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dehnert KW et al. (2012) Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem 13, 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X et al. (2016) Exploring the binding proteins of glycolipids with bifunctional chemical probes. Angew. Chem. Int. Ed 55, 14330–14334 [DOI] [PubMed] [Google Scholar]
- 32.Xie R et al. (2016) In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl. Acad. Sci 113, 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dauner M et al. (2016) Synthetic glycosphingolipids for live-cell labeling. Bioconjug. Chem 27, 1624–1637 [DOI] [PubMed] [Google Scholar]
- 34.Selvy PE et al. (2011) Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev 111, 6064–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bumpus TW and Baskin JM (2016) A chemoenzymatic strategy for imaging cellular phosphatidic acid synthesis. Angew. Chem. Int. Ed. Engl 55, 13155–13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bumpus TW and Baskin JM (2017) Clickable substrate mimics enable imaging of phospholipase D activity. ACS Cent. Sci 3, 1070–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bumpus TW et al. (2018) Ex uno plura: differential labeling of phospholipid biosynthetic pathways with a single bioorthogonal alcohol. Biochemistry 57, 226–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadler A et al. (2013) The fatty acid composition of diacylglycerols determines local signaling patterns. Angew. Chem. Int. Ed 52, 6330–6334 [DOI] [PubMed] [Google Scholar]
- 39.Höglinger D et al. (2015) Intracellular sphingosine releases calcium from lysosomes. eLife 4, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter AM et al. (2017) Phosphatidylinositol 4,5-bisphosphate optical uncaging potentiates exocytosis. eLife 6, 1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mentel M et al. (2011) Photoactivatable and cell-membrane-permeable phosphatidylinositol 3,4,5-trisphosphate. Angew. Chem. Int. Ed 50, 3811–3814 [DOI] [PubMed] [Google Scholar]
- 42.Hövelmann F et al. (2016) Optotaxis: caged lysophosphatidic acid enables optical control of a chemotactic gradient. Cell Chem. Biol 23, 629–634 [DOI] [PubMed] [Google Scholar]
- 43.Feng S et al. (2018) Mitochondria-specific photoactivation to monitor local sphingosine metabolism and function. eLife 7, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadler A et al. (2015) Exclusive photorelease of signalling lipids at the plasma membrane. Nat. Commun 6, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank JA et al. (2015) Photoswitchable fatty acids enable optical control of TRPV1. Nat. Commun 6, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein M et al. (2013) Optical control of TRPV1 channels. Angew. Chem. Int. Ed 52, 9845–9848 [DOI] [PubMed] [Google Scholar]
- 47.Frank JA et al. (2016) Photoswitchable diacylglycerols enable optical control of protein kinase C. Nat. Chem. Biol 12, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lichtenegger M et al. (2018) An optically controlled probe identifies lipid-gating fenestrations within the TRPC3 channel article. Nat. Chem. Biol 14, 396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank JA et al. (2016) Optical control of lipid rafts with photo-switchable ceramides. J. Am. Chem. Soc 138, 12981–12986 [DOI] [PubMed] [Google Scholar]
- 50.Höglinger D et al. (2017) Trifunctional lipid probes for comprehensive studies of single lipid species in living cells. Proc. Natl. Acad. Sci 114, 1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erdmann RS et al. (2014) Super-resolution imaging of the Golgi in live cells with a bioorthogonal ceramide probe. Angew. Chem. Int. Ed 53, 10242–10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erdmann RS et al. (2017) STED Imaging of golgi dynamics with Cer-SiR: a two-component, photostable, high-density lipid probe for live cells. In Methods in Molecular Biology (1663), pp. 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uno SN et al. (2014) A spontaneously blinking fluorophore based on intramolecular spirocyclization for live-cell super-resolution imaging. Nat. Chem 6, 681–689 [DOI] [PubMed] [Google Scholar]
- 54.Takakura H et al. (2017) Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat. Biotechnol 35, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson AD et al. (2017) Long-term live-cell STED nanoscopy of primary and cultured cells with the plasma membrane HIDE probe DiI-SiR. Angew. Chem. Int. Ed 56, 10408–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu AP et al. (2017) The big and intricate dreams of little organelles: Embracing complexity in the study of membrane traffic. Traffic 18, 567–579 [DOI] [PubMed] [Google Scholar]
- 57.Valm AM et al. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong S et al. (2014) Live-cell stimulated Raman scattering imaging of alkyne-tagged biomolecules. Angew. Chem. Int. Ed 53, 5827–5831 [DOI] [PubMed] [Google Scholar]
- 59.Hu F et al. (2016) Bioorthogonal chemical imaging of metabolic activities in live mammalian hippocampal tissues with stimulated Raman scattering. Sci. Rep 6, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei L et al. (2014) Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 11, 410–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z et al. (2014) Multicolor live-cell chemical imaging by isotopically edited alkyne vibrational palette. J. Am. Chem. Soc 136, 8027–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei L et al. (2017) Super-multiplex vibrational imaging. Nature 544, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu F et al. (2018) Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat. Methods 15, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villareal VA et al. (2016) Hepatitis C virus selectively alters the intracellular localization of desmosterol. ACS Chem. Biol 11, 1827–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y et al. (2017) Metabolic activity induces membrane phase separation in endoplasmic reticulum. Proc. Natl. Acad. Sci 114, 13394–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pleitez MA et al. (2018) Mid-infrared optoacoustic microscopy with label-free chemical contrast in living cells and tissues. BioRxiv Published online February 22, 2018. 10.1101/270082 [DOI] [Google Scholar]
- 67.Topp S and Gallivan JP (2010) Exploring disease through metabolomics. ACS Chem. Biol 5, 91–103 [DOI] [PubMed] [Google Scholar]
- 68.Lizardo DY et al. (2018) Noncanonical roles of lipids in different cellular fates. Biochemistry 57, 22–29 [DOI] [PubMed] [Google Scholar]
- 69.Buchberger AR et al. (2018) Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem 90, 240–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellis SR et al. (2018) Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat. Methods 15, 515–518 [DOI] [PubMed] [Google Scholar]
- 71.Nielsen MMB et al. (2016) Mass spectrometry imaging of biomarker lipids for phagocytosis and signalling during focal cerebral ischaemia. Sci. Rep 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Várnai P and Balla T (2006) Live cell imaging of phosphoinosi-tide dynamics with fluorescent protein domains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1761, 957–967 [DOI] [PubMed] [Google Scholar]
- 74.Hammond GRV et al. (2014) A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol 205, 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo X et al. (2015) Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog 11, 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]