This case-control study examines neuronal-enriched extracellular vesicles as predictors of Alzheimer disease.
Key Points
Question
Can blood extracellular vesicle biomarkers diagnose Alzheimer disease at the preclinical and clinical stages?
Findings
In a large case-control study examining 887 longitudinal samples from 128 Baltimore Longitudinal Study of Aging participants (split into training and test sets), combining extracellular vesicle biomarkers predicted Alzheimer disease with high discrimination accuracy and specificity about 4 years before symptom onset; individual biomarkers were associated with cognitive performance. Biomarkers were further validated in a case-control cohort from Johns Hopkins.
Meaning
Further development of extracellular vesicle biomarkers may establish them as a blood test for Alzheimer disease.
Abstract
Importance
Blood biomarkers able to diagnose Alzheimer disease (AD) at the preclinical stage would enable trial enrollment when the disease is potentially reversible. Plasma neuronal-enriched extracellular vesicles (nEVs) of patients with AD were reported to exhibit elevated levels of phosphorylated (p) tau, Aβ42, and phosphorylated insulin receptor substrate 1 (IRS-1).
Objective
To validate nEV biomarkers as AD predictors.
Design, Setting, Participants
This case-control study included longitudinal plasma samples from cognitively normal participants in the Baltimore Longitudinal Study of Aging (BLSA) cohort who developed AD up to January 2015 and age- and sex-matched controls who remained cognitively normal over a similar length of follow-up. Repeated samples were blindly analyzed over 1 year from participants with clinical AD and controls from the Johns Hopkins Alzheimer Disease Research Center (JHADRC). Data were collected from September 2016 to January 2018. Analyses were conducted in March 2019.
Main Outcomes and Measures
Neuronal-enriched extracellular vesicles were immunoprecipitated; tau, Aβ42, and IRS-1 biomarkers were quantified by immunoassays; and nEV concentration and diameter were determined by nanoparticle tracking analysis. Levels and longitudinal trajectories of nEV biomarkers between participants with future AD and control participants were compared.
Results
Overall, 887 longitudinal plasma samples from 128 BLSA participants who eventually developed AD and 222 age and sex-matched controls who remained cognitively normal were analyzed. Participants were followed up (from earliest sample to AD symptom onset) for a mean (SD) of 3.5 (2.31) years (range, 0-9.73 years). Overall, 161 participants were included in the training set, and 80 were in the test set. Participants in the BLSA cohort with future AD (mean [SD] age, 79.09 [7.02] years; 68 women [53.13%]) had longitudinally higher p-tau181, p-tau231, pSer312-IRS-1, pY-IRS-1, and nEV diameter than controls (mean [SD] age, 76.2 [7.36] years; 110 women [50.45%]) but had similar Aβ42, total tau, TSG101, and nEV concentration. In the training BLSA set, a model combining preclinical longitudinal data achieved 89.6% area under curve (AUC), 81.8% sensitivity, and 85.8% specificity for predicting AD. The model was validated in the test BLSA set (80% AUC, 55.6% sensitivity, 88.7% specificity). Preclinical levels of nEV biomarkers were associated with cognitive performance. In addition, 128 repeated samples over 1 year from 64 JHADRC participants with clinical AD and controls were analyzed. In the JHADRC cohort (35 participants with AD: mean [SD] age, 74.03 [8.73] years; 18 women [51.43%] and 29 controls: mean [SD] age, 72.14 [7.86] years; 23 women [79.31%]), nEV biomarkers achieved discrimination with 98.9% AUC, 100% sensitivity, and 94.7% specificity in the training set and 76.7% AUC, 91.7% sensitivity, and 60% specificity in the test set.
Conclusions and Relevance
We validated nEV biomarker candidates and further demonstrated that their preclinical longitudinal trajectories can predict AD diagnosis. These findings motivate further development of nEV biomarkers toward a clinical blood test for AD.
Introduction
Alzheimer disease (AD) is a neurodegenerative disease characterized by a long preclinical phase with evolving and progressively irreversible pathology. Therefore, biomarkers are essential for identifying patients early in the course of the disease, when disease-modifying interventions may have the greatest chance of success. Existing AD biomarkers provide limited sensitivity and specificity for clinical and preclinical diagnosis and have been adopted in research but not in clinical practice.1,2,3,4 Among the best performing modalities, positron emission tomography biomarkers are expensive and involve radiation, whereas cerebrospinal fluid (CSF) biomarkers are invasive. For biomarkers to become part of clinical practice, low cost, wide availability, and noninvasiveness are required—criteria that can only be satisfied by blood biomarkers.5 It is widely accepted that the development of reliable predictive blood biomarkers is a crucial step in the pursuit of disease-modifying interventions for AD and their widespread implementation.6
A limitation of biomarkers measured in the soluble phase of blood is their tenuous link to brain pathology because they are often produced by multiple tissues and their brain-derived fraction has to cross multiple barriers before reaching the circulation. Our team has developed a new approach to biomarker discovery in AD that addresses this limitation by harvesting extracellular vesicles (EVs) enriched for neuronal origin from blood. Given their origin, neuronal-enriched EVs (nEVs) can be used to interrogate intraneuronal pathogenic processes previously inaccessible in vivo, akin to a liquid biopsy.7
Extracellular vesicles are membranous particles shed by all cells and found in all biofluids; they include exosomes (30 nm to 150 nm) originating from endosomes/multivesicular bodies and microvesicles (150 nm to 1000 nm) produced through budding of the plasma membrane. Extracellular vesicles have been implicated in the pathogenesis of neurodegenerative diseases and have gained interest as biomarkers.8 Circulating EVs manifest a constitutive protein signature9 and cargoes reflecting physiological and pathological states. Some cargo proteins are shared by all EVs,10 other proteins specify a distinct cellular origin, and a few proteins appear or change in amount in pathogenic processes, so that they may be considered disease biomarkers.7
In previous case-control studies,11,12,13,14 we and others implemented a methodology for enriching plasma EVs for neuronal origin to identify protein biomarkers for AD. The first such study focused on pathogenic proteins by measuring total tau, phosphorylated (p)-tau181, pSer396, and β-amyloid 42 (Aβ42), and showed higher levels in participants with AD compared with control participants (except for total tau), achieving high classification accuracy; moreover, in a few cases with available samples, levels were already abnormal in preclinical AD.14 Subsequently, other groups showed similar elevations in progression from mild cognitive impairment to dementia15 and in Down syndrome16 and low levels with normal aging.17 Given the implication of insulin resistance in AD pathogenesis,18 a second nEV study investigated insulin signaling effectors showing phosphorylation changes in insulin receptor substrate 1 (IRS-1) in AD.12 The classification accuracy achieved through p-tau, Aβ42, and p-IRS-112,14 was not exceeded by other markers13,19; therefore, this set was selected for validation.
In the present study, we sought to validate nEV biomarker candidates for AD prediction by analyzing longitudinal samples from Baltimore Longitudinal Study of Aging (BLSA) participants. The BLSA is an ideal cohort for this pursuit because it enrolls cognitively normal participants at different ages, performs longitudinal cognitive assessments, and uses consistent criteria for AD diagnosis via consensus research diagnostic conferences. Moreover, the BLSA implements uniform procedures for blood sample collection and storage spanning decades. The BLSA case-control sample assembled for this study is much larger than any previously analyzed, to our knowledge, and offers the opportunity for validation of nEV biomarkers for preclinical AD. Moreover, to concurrently validate nEV biomarkers for clinical AD, we analyzed repeated samples from Johns Hopkins Alzheimer Disease Research Center (JHADRC) participants with clinical AD and controls.
Methods
Participants and Study Design
Reporting of participants, methods, and results follows the reporting standards for the reporting of diagnostic accuracy studies–dementia (the STARDdem Initiative).20We blindly analyzed plasma or serum samples collected from BLSA participants and from JHADRC participants. The BLSA and JHADRC protocols were approved by the National Institute of Environmental Health Sciences and the Johns Hopkins University Institutional Review Boards, respectively; all participants gave written informed consent. All investigators involved in EV isolation and biomarker quantification were blinded until all measurements were made and the data set was locked for analysis. Unblinded investigators were responsible for identifying BLSA and JHADRC participants and visits but were not involved in any experimental procedures.
In the BLSA cohort, the Blessed Information Memory Concentration (BIMC) Test and the Clinical Dementia Rating (CDR) are used to screen participants for cognitive impairment; all participants receive the BIMC and a subset receives the CDR at every visit, including all with a BIMC score greater than 3. If participants score more than 3 on the BIMC or 0.5 or more on the CDR, they undergo more extensive neuropsychological testing and results are evaluated at consensus conference. During these consensus conferences, AD diagnosis is determined based on the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) criteria, using all available clinical and neuropsychological data.21 For the purpose of this study, unblinded investigators identified BLSA participants diagnosed as having AD and pulled samples from 1 to 8 visits prior to AD symptom onset. For instance, if a given participant was diagnosed as having amnestic mild cognitive impairment on visit 9 and probable AD on visit 10, symptom onset was considered on visit 9 and samples were drawn from visit 8 and earlier; if another participant was cognitively normal on visit 9 and was diagnosed as having probable AD on visit 10, symptom onset was on visit 10. All participants who developed AD over the course of the BLSA until January 2015 and had available samples were included. In the JHADRC cohort, cognitively normal controls were participants with CDR score of 0, Mini-Mental State Examination score more than 28, and no reported memory impairments by history. Diagnosis of probable AD was based on NINCDS-ADRDA criteria. Annual cognitive diagnoses for JHADRC participants were derived from review of history, examination, and neuropsychological testing at consensus conferences.
nEV Isolation
In the BLSA group, blood draws were conducted between 7 am to 10 am after a 12-hour fast. Blood samples were collected from JHADRC participants during their annual visit. All blood draws and processing followed established BLSA and JHADRC protocols using standard venipuncture procedures. For both cohorts, blood was collected in EDTA polypropylene tubes, centrifuged at 3000 rpm for 15 minutes at 4°C; supernatant plasma was divided into 0.5-mL aliquots and stored at −80°C until analysis. Hemolysis was ruled out using spectrophotometry (data not shown). Preanalytical factors for blood collection and storage comply with guidelines for EV biomarkers.22,23 For nEV isolation we used the protocol detailed by Mustapic et al7 with modifications (eMethods in the Supplement). Briefly, we defibrinated plasma samples using thromboplastin-D, precipitated total EVs using particle precipitation by Exoquick and immunoprecipitated nEVs expressing L1 cell adhesion molecule (L1CAM). Neuronal-enriched EVs (L1CAM positive) were lysed and stored at −80°C until prior to assays.
nEV Characterization
To calculate nEV concentration and average diameter, we used nanoparticle tracking analysis (NanoSight NS500) (eFigure 1 in the Supplement). To further characterize nEVs we performed transmission electron microscopy with negative staining revealing a population of round, predominantly smaller (<100 nm) particles, consistent with exosomes and fewer microvesicles (eFigure 2 in the Supplement). eFigure 3 in the Supplement demonstrates the presence of typical EV markers CD81, CD9, and Alix (positive controls) and the absence of GM130 (negative control). eFigures 4 and 5 in the Supplement provide evidence for neuronal origin enrichment in terms of levels of neurofilament light, synaptophysin, L1CAM, and neural cell adhesion molecule.
nEV Protein Quantification
To assure data quality, we implemented rigorous quality control assessments that are presented in eFigures 6, 7, and 8 and eResults in the Supplement. All assays were conducted in duplicates. All samples from repeated visits of a given participant were included on the same plate to minimize within-participant variability. We used Mesoscale Discovery electrochemiluminescence assays to quantify total-tau, p-tau181, p-tau231, pSer312-IRS-1, pY-IRS-1, and TSG101 as well as SIMOA assay for Aβ42 (eTable 2 and eResults in the Supplement).
Statistical Analyses
Data were not normally distributed. Therefore, analyses were performed on log-transformed values and results (ie, means over time) were back-transformed (exponentiated). Wilcoxon rank sum tests were used to compare individual EV biomarkers between future AD and control participants. For each biomarker, we compared values from the last visit prior to symptom onset, within-individual means, and slopes over time. In addition, we performed the same comparisons after adjustment for nEV concentration and average diameter, as a means for normalization for differential EV yield.24,25 (Our approach to normalization is discussed in the eDiscussion in the Supplement.) To visualize age-specific retrospective longitudinal biomarker changes for future participants with AD and control participants, we performed locally weighted scatterplot smoothing regression and plotted locally weighted sum of squares smoothing splines.
Using data from 887 samples from 350 BLSA participants, we used mixed-models to compute person-specific slopes and means for each nEV biomarker. Participants with complete information for all predictors (complete demographic data and mean, slope, and last visit data for all nEV biomarkers) contributed to model building. We assessed the influence of missing data by comparing AD status and demographic characteristics between participants who were included vs excluded from prediction model building (eTable 8 in the Supplement). We performed a random split of the data to create a training set (two-thirds of the BLSA cohort) and a test set (one-third of the BLSA cohort). In the training set, we performed stepwise logistic regression with internal cross-validation and receiver operating characteristic (ROC) analysis to identify a model discriminating participants with future AD from controls; model fit was assessed in the test set. To assess nEV biomarkers performance in AD prediction individually and collectively, we built 10 models to predict AD as functions of the following predictors: model 1: age, sex, and sample type (to account for the fact that 83 samples were serum rather than plasma); model 2: model 1 predictors plus measures of EV concentration and mean diameter (to assess whether nanoparticle tracking analysis parameters inform AD classification in their own right and as a normalizers for subsequent models); models 3 to 9: model 2 predictors plus measures of individual nEV biomarkers; and model 10: model 2 predictors plus the most predictive measures of multiple EV biomarkers. We considered 12 measures of each biomarker as candidate predictors: last preclinical visit measurement, within-individual mean; within-individual slope; interactions of last visit, mean, and slope with sex; interactions with age; and interactions with age and sex. Models were fit using logistic regression to appropriately handle the case-control design.
We computed area under the ROC curve and performed internal leave-10%-out cross-validation to compute a cross-validated area under the curve (AUC). Model 10 was built by identifying the set of single-EV biomarker model measures that maximized cross-validation AUC, then recursively adding proteins until cross-validation AUC no longer increased, followed by a reduction step that determined whether a submodel produced higher cross-validation AUC. We chose to optimize cross-validation AUC to avoid overfitting and enhance validity. Model 10 included measures of age, sex, sample type, nEV concentration, nEV mean diameter, TSG101, total tau, p-tau181, p-tau231, pY-IRS-1, pSer312-IRS-1, and Aβ42. To compare the models’ performance in discrimination, we plotted ROC curves and included side-by-side boxplots of participants’ risk scores. Nonparameteric tests compared AUC and cross-validation AUC between models.26Model 10 performance was further evaluated by selecting the threshold risk score that minimizes the distance from the ROC curve from the top left corner and computing sensitivity (proportion above the threshold risk score among participants with future AD), specificity (proportion below the threshold risk score among controls), and odds ratio to address the case-control design. Performance statistics (ROC, sensitivity, specificity, odds ratio) were calculated separately for the BLSA training set, the BLSA test set, and the JHADRC set.
Results
We blindly analyzed plasma or serum samples collected over 887 person-visits from 350 BLSA participants (participants with future AD: mean [SD] age, 79.09 [7.02] years; 68 women [53.13%]; controls: mean [SD] age, 76.2 [7.36] years; 110 women [50.45%]) comprising a case-control sample selected from the BLSA. We also blindly analyzed plasma samples collected over 128 person-visits from 64 participants from the JHADRC, which included 29 participants with normal cognition (mean [SD] age, 72.14 [7.86] years; 23 women [79.31%]) and 35 with AD (mean [SD] age, 74.03 [8.73] years; 18 women [51.43%]) (eTable 1 in the Supplement). The median (interquartile range) time interval between AD symptom onset and the earliest preclinical sample was 4.07 (3.06-5.37) years (range, 0.03-9.94 years). Unblinded investigators identified 222 age- and sex- matched controls with similar number of visits over the same time interval, 128 of whom remained cognitively normal for the course of their participation, whereas 94 were actively enrolled and remained cognitively normal to date. Overall, 241 BLSA participants contributed to model building. The training set included 161 BLSA participants, and the test set included 80 BLSA participants. Participants were followed up (from earliest sample to AD symptom onset) for a mean (SD) time of 3.5 (2.31) years (range, 0-9.73 years).
Longitudinal Trajectories for nEV Biomarkers in Future AD and Control BLSA Participants
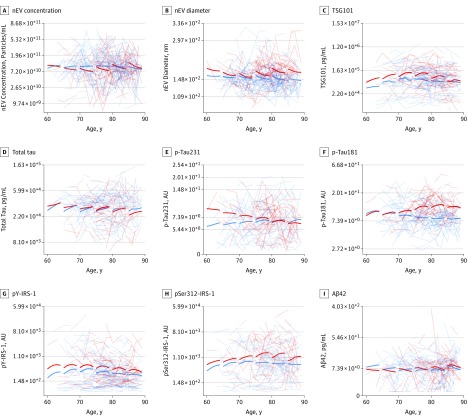
Figure 1 shows age-specific retrospective longitudinal biomarker changes for participants with future AD and control participants from locally weighted scatterplot smoothing regression. These trajectories reveal that preclinical longitudinal changes for nEV average diameter, p-tau181, pSer312-IRS-1, and perhaps pY-IRS-1 tend to diverge between participants with future AD and control participants in older age groups; longitudinal changes for p-tau231 tend to diverge in younger age groups; whereas longitudinal changes for nEV concentration, TSG101, total tau, and Aβ42 show overlap in all age groups. eTable 3 in the Supplement shows results on the stability of L1CAM+ EV biomarkers over time; eTable 4 in the Supplement shows intercorrelations between means of EV biomarkers across all visits; eTable 5 in the Supplement shows intercorrelations between EV biomarkers at the last preclinical visit; eTable 6 in the Supplement shows intercorrelations between the slopes of EV biomarkers; and eTable 8 in the Supplement shows missing values assessment.
Figure 1. Age-Specific Retrospective Longitudinal Neuronal-Enriched Extracellular Vesicle (nEV) Biomarker Changes in Participants With Future Alzheimer Disease (AD) and Control Participants .
Locally weighted sum of squares smoothing splines were computed by locally weighted scatterplot smoothing regression. Transparent lines are individual trajectories. Aβ42 indicates β-amyloid 42; AU, arbitrary units; IRS-1, insulin receptor substrate 1; p-tau; phosphorylated tau.
nEV Biomarker Comparisons Between Future AD and Control BLSA Participants and Associations With Cognition
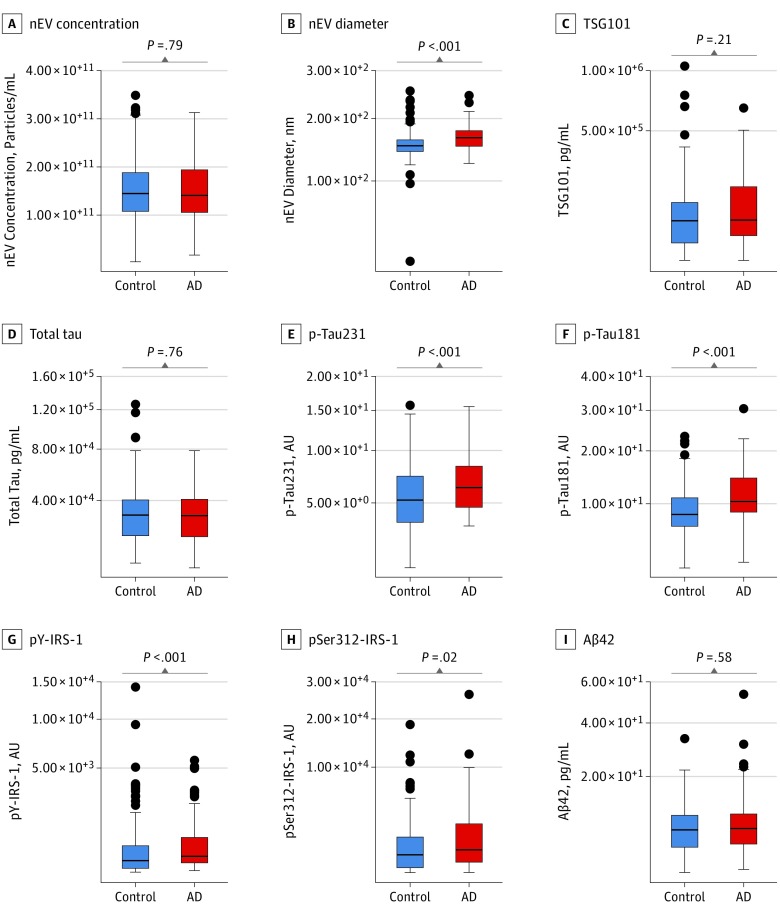
Participants with future AD compared with control participants had on average (across all preclinical visits) higher nEV average diameter (166 nm vs 152 nm; difference = 14 nm; 95% CI, 7-17; P < .001); higher p-tau231 (6.3 AU vs 5.2 AU; difference = 1.1 AU; 95% CI, 0.3-1.5; P = .004); higher p-tau181 (10.4 AU vs 8.4 AU; difference = 2 AU; 95% CI, 1.5-3.3; P < .001); higher pY-IRS-1 (3.2 AU vs 2.3 AU; difference = 0.9 AU; 95% CI, 0.3-1.5; P = .003); and higher pSer312-IRS-1 (8.0 AU vs 6.1 AU; difference = 1.9 AU; 95% CI, 0.2-4; P = .02) (Figure 2). Similar differences were obtained for the last preclinical visit and slopes (eFigure 9, eFigure 10, and eResults in the Supplement). Neuronal-enriched EV concentration, TSG101, total tau, and Aβ42 showed no statistically significant group differences for the last preclinical visit on average or for slopes. Higher p-tau181 was associated with worse verbal memory, attention, executive function, and visuospatial function cross-sectionally. Higher pSer312-IRS-1 was associated with worse verbal memory and executive function cross-sectionally. Higher Aβ42 at the first preclinical visit was associated with better verbal memory and language longitudinally. Cross-sectional and longitudinal associations of nEV biomarkers and composite cognitive scores are shown in eTable 7 in the Supplement.
Figure 2. Within-Person Mean Neuronal-Enriched Extracellular Vesicle (nEV) Biomarker Levels for Participants With Future Alzheimer Disease (AD) and Control Participants.
Within-person means were computed on the log-transformed scale and then exponentiated. Boxes depict the median and upper and lower quartiles; error bars, 1.5 × interquartile range; and single dots, outliers. P values from Wilcoxon rank sum tests comparing participants with AD and control participants are shown separately in each panel. Aβ42 indicates β-amyloid 42; AU, arbitrary units; IRS-1, insulin receptor substrate 1; p-tau; phosphorylated tau.
Performance in AD Preclinical Prediction (BLSA)
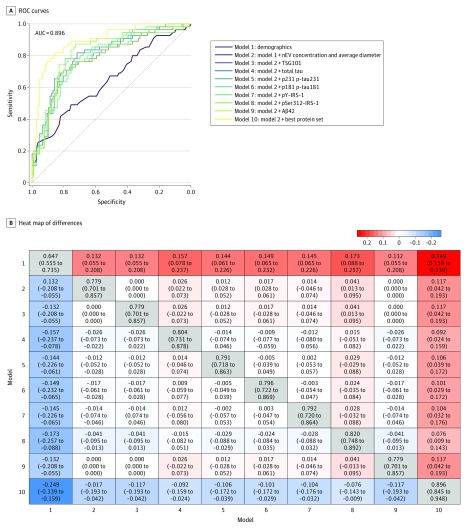
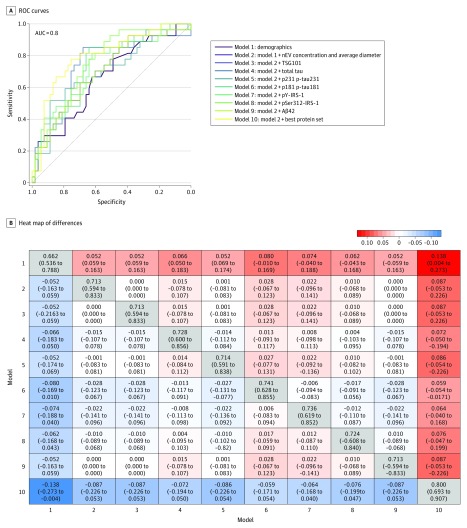
In the training set (161 BLSA participants [67%]), the best model for AD prediction was model 10, which included nEV concentration and mean diameter and all measured proteins, except for TSG101 and Aβ42 (ie, total tau, p-tau181, p-tau231, pY-IRS-1, and pSer312-IRS-1), achieving 89.6% AUC (95% CI, 84.5%-94.8%) for predicting AD (Figure 3). eFigure 11 in the Supplement presents model performance internally cross-validated by the leave-10%-out method in the training set. In the test set (80 BLSA participants [33%]), model 10 was again the best model achieving 80% AUC (95% CI, 63.9%-90.7%) for predicting AD (Figure 3). Moreover, model 10 outperformed all individual biomarker models in both sets (Figure 3 and Figure 4 depicts heat maps of AUC differences between all models for the training and test sets, respectively).
Figure 3. Alzheimer Disease Risk Prediction Models for Training Data.
Receiver operating characteristic (ROC) curves for 10 models based on training data (A). In model 1, demographics include age, sex, and plasma/serum sample type. In model 10, best protein set includes measures of TSG101, total tau, pY-IRS-1, pSer312-IRS-1, p-tau181, and β-amyloid 42 (Aβ42). Heat map of differences in area under the curve (AUC) between models with 95% CIs based on training data (B). Off diagonal elements show column model minus row model, and diagonal elements show model AUC with 95% CIs. IRS-1 indicates insulin receptor substrate 1; nEV, neuronal-enriched extracellular vesicle; p-tau; phosphorylated tau.
Figure 4. Comparison of Alzheimer Disease Risk Prediction Models for Test Data.
Receiver operating characteristic (ROC) curves for 10 models based on test data (A). In model 1, demographics include age, sex, and plasma/serum sample type. In model 10, best protein set includes measures of TSG101, total tau, pY-IRS-1, pSer312-IRS-1, p-tau181, and β-amyloid 42 (Aβ42). Heat map of differences in area under the curve (AUC) between models with 95% CIs based on test data (B). Off diagonal elements show column model minus row model, and diagonal elements show model AUC with 95% CIs. IRS-1 indicates insulin receptor substrate 1; nEV, neuronal-enriched extracellular vesicle; p-tau; phosphorylated tau.
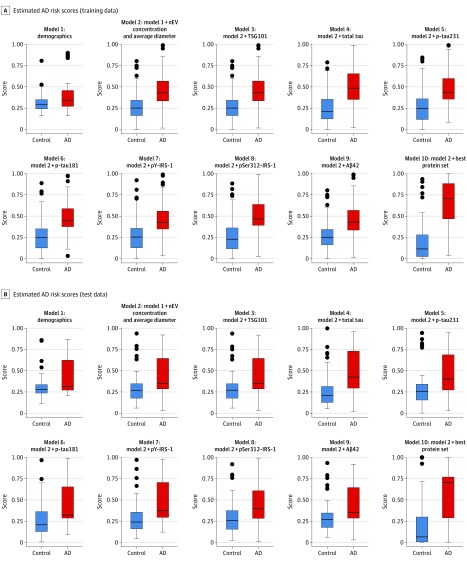
To further visualize and compare the models’ ability to discriminate between participants with future AD and control participants, we provide side-by-side boxplots of participants’ risk scores for the training and test sets (Figure 5). eFigure 12 in the Supplement depicts internally cross-validated risk scores.
Figure 5. Risk Scores from the Prediction Models.
Boxplots of risk scores from the prediction models for the training data (A) and test data (B). The y-axis depicts estimated risk score estimates using all preclinical data for participants with future AD and control participants based on each model (range, 0-1). Risk scores were computed by converting logistic regression linear predictors to a scale of 0 to 1 via the exit transformation: expit(x) = exp(x)/[1 + exp(x)]. Boxes depict the median and upper and lower quartiles; error bars, 1.5 × interquartile range; and single dots, outliers. Aβ42 indicates β-amyloid 42; AD, Alzheimer disease; IRS-1, insulin receptor substrate 1; nEV, neuronal-enriched extracellular vesicle; p-tau; phosphorylated tau.
Model 10 performance in the training data was evaluated by selecting the threshold estimated risk score (>0.407) and computing sensitivity, specificity, and odds ratio. The odds of over-threshold risk score were 27.3 (95% CI, 11.8-68.9) times higher for participants with future AD than control participants; sensitivity was 81.8% (95% CI, 68.6%-90.5%), and specificity was 85.8% (95% CI, 77.4%-91.6%) (eTable 9 in the Supplement). The threshold risk score estimated from the training data (>0.407) achieved even higher specificity in the test data (88.7%), at the expense of lower sensitivity (55.6%). In the test data, the odds of over-threshold risk score were 9.8 (95% CI, 3.3-32.8) times higher for participants with future AD than control participants (eTable 9 in the Supplement).
Performance of nEV Biomarkers in Clinical AD Discrimination (JHADRC)
We concurrently validated the measures selected in the BLSA training using the JHADRC data. Owing to the different study design between BLSA and JHADRC, we split the JHADRC group into training (43 participants [67%]) and test sets (21 participants [33%]), fit a model in the training set, and assessed its performance in the test set. In the training set, the model achieved 98.9% AUC (95% CI, 96.4%-100%) for discriminating participants with AD from controls (eFigure 13 in the Supplement). The threshold risk score estimated in the training set (>0.412) achieved 100% sensitivity (95% CI, 82.2%-100%) and 94.7% specificity (95% CI, 71.9%-99.7%). In the test set, the same model achieved 76.7% AUC (95% CI, 54%-99.4%), with 91.7% sensitivity (95% CI, 59.8%-99.6%) and 60% specificity (95% CI, 27.4%-86.3%) (eFigure 13 in the Supplement). The odds of over-threshold risk score were more than 999 times higher for participants with AD than control participants in the training set and 16.5 times higher in the test set.
Discussion
In this large validation study primarily based on a case-control sample from a well-characterized longitudinal cohort, we demonstrated the reproducibility of the methodology for isolating nEVs, blindly replicated findings for many lead candidate nEV biomarkers for AD, and uncovered longitudinal trajectories of nEV biomarkers, which differed between participants with future AD and controls and depended on age and sex. We further demonstrated that, by leveraging repeated measures as long-term averages and rates of changes and optimally combining them in a prediction model, nEV biomarkers predict AD with high specificity. The discriminant ability of the final model outperformed both classic and recently proposed blood biomarkers6,27,28,29 (with the possible exception of combinations of Aβ and amyloid precursor protein measured through immunoprecipitation/mass spectrometry30) rivaling CSF biomarkers.6,31,32 Importantly, for predicting AD preclinically, nEV biomarkers outperformed other blood33 and CSF biomarkers34 and even amyloid imaging.34,35,36 (For context, refer to Lewczuk et al6 for an extensive review.) Moreover, the same set of nEV biomarkers was accurate in classifying participants with clinical AD and controls in an external cohort from JHADRC (albeit with higher sensitivity than specificity). For a field plagued by the failure to replicate results of index studies,37 these results reaffirm the validity of EV biomarkers and open the road toward their further development as a clinical blood test for AD. (Additional discussion regarding individual nEV biomarkers is available in the eDiscussion in the Supplement.)
Our findings on tau biomarkers confirmed the pattern seen in the first study using nEVs14: participants with future AD had higher p-tau231 and p-tau181, but not total tau, than controls. Functions of repeated measures of all 3 tau biomarkers offered strong prediction of AD (models 4, 5, and 6; Figure 4) and contributed to the final model.
An unexpected result of this study was the absence of significant differences in Aβ42 levels between participants with future AD and control participants, even though we quantified it using a more sensitive immunoassay27 than in our previous study.14 Moreover, Aβ42 did not contribute to the final model. The ability of plasma Aβ to assist in diagnostic classification has remained questionable over the years,27,38 although this may change with recent innovations.30 Future biomarker studies should measure both EV-bound and -unbound Aβ42 fractions (as further suggested by a 2019 study using a novel EV analytic platform39) to disentangle their differential contributions in AD prediction.
Brain insulin resistance is important for AD pathogenesis, potentially linking amyloid and tau pathologies.18 Tau hyperphosphorylation induces brain insulin resistance,40 and this may be reflected in the strong associations between p-tau231 and p-tau181 with pSer312-IRS-1 and pY-IRS-1. The present study reaffirmed the importance of IRS-1 phosphotypes as predictive biomarkers for AD: pSer312-IRS-1 and pY-IRS-1 were among the strongest individual predictors and contributed to the final model. Moreover, the negative associations of pSer312-IRS-1 and pY-IRS-1 with cognition represent an impressive in vivo replication of the reported associations between these markers in autopsied brains of participants with AD and ante-mortem cognition.41
Strengths and Limitations
The strengths of this study include its large sample size, which far exceeds any previous nEV biomarker studies to our knowledge, and the availability of repeated preclinical samples, which allowed us to uncover previously unobserved age- and sex-related longitudinal trajectories of EV biomarkers, determine their performance in predicting AD and distinguish between cross-sectional and longitudinal associations with cognition. Another strength is that all experimental procedures were performed blindly.
Limitations include cohort representativeness, reduced sensitivity of the threshold estimated risk score at 55.6% in the BLSA test data compared with 81.8% in the training data (with comparable high specificities), which suggests that the optimal threshold may vary between cohorts, and the fact that predictive performance of EV biomarkers was gauged in comparison with future clinical AD diagnosis by NINCDS-ADRDA criteria, which allows for a degree of diagnostic misclassification.42 In contrast, earlier case-control studies of nEV biomarkers12,14 were based on cases with high probability AD based on CSF biomarkers (low Aβ42, high tau, and p-tau181).1,2,4 The higher specificity over sensitivity in the BLSA preclinical cohort suggests that the clinical context of use for presently examined nEV biomarkers may be in ruling out preclinical AD (ie, accurately identifying participants without preclinical AD), rather than in screening for it. Biomarkers with high specificity could help reassure older adults concerned about their risk for AD and improve misclassification accuracy in secondary prevention clinical trials by minimizing enrollment of participants without preclinical AD pathology; biomarkers with high sensitivity could be used as a screening test and could improve misclassification in clinical trials by enriching for the presence of AD pathology. The ever-expanding list of candidate nEV (and recently astrocytic EV43,44) biomarkers for AD includes markers of synaptic pathology,11,45 complement,43 and RNA species.46 Future research should identify sets of EV markers with high sensitivity in par with high specificity.
Conclusions
In summary, among BLSA participants, nEV biomarker data from up to 9 years prior to symptom onset were used to build a model that was able to predict future AD diagnosis with high level of accuracy. The same set of nEV biomarkers were effective in correctly classifying participants with clinical AD in a separate cohort of JHADRC participants. By splitting the BLSA data into training and test sets we assessed the predictive validity of the model, whereas by analyzing JHADRC data, we assessed the concurrent validity of the model. These findings reaffirm the validity of nEV biomarkers and motivate their further development toward a clinical blood test for AD. Additional validation studies are required to definitively define the most effective set of nEV biomarkers for prognosis and diagnosis and specify cutoff values for individual biomarkers and model-derived risk score. The definitive assessment of the utility of nEV biomarkers in AD prediction and diagnosis should be based on large prospective longitudinal studies in diverse cohorts that leverage nEV biomarker histories. The ultimate motivation for nEV biomarker studies is the hope that they may enable researchers to identify older individuals at the preclinical stage of AD and select them for clinical trials, augmenting our ability to test hypotheses and spearheading therapeutic discovery for the disease.
eMethods.
eTable 1. BLSA and JH ADRC Study sample characteristics
eTable 2. Electrochemiluminescence units for p231-tau and p181- tau and corresponding concentration of phosphorylated full length 441 tau standard used for calibration
eTable 3. Stability of L1CAM+ EV biomarkers over time
eTable 4. Inter-correlations between means of EV biomarkers across all visits
eTable 5. Inter-correlations between EV biomarkers at the last preclinical visit
eTable 6. Inter-correlations between the slopes of EV biomarkers
eTable 7. Cross-sectional and longitudinal associations of nEV biomarkers and composite cognitive scores across all BLSA participants
eTable 8. Missing values assessment
eTable 9. Performance statistics of Final Model 10 for Alzheimer Disease Prediction
eFigure 1. Nanoparticle tracking analysis (NTA) of total EVs isolated by Exoquick® alone and neuronal-enriched EVs isolated through Exoquick® followed by immunoprecipitation with antibodies against L1CAM (L1CAM+ EVs) from a typical plasma sample.
eFigure 2. Electron Microscopy of neuronal-enriched plasma EVs
eFigure 3. Western Blot characterization of neuronal-enriched plasma EVs
eFigure 4. Neuronal enrichment of L1CAM+ EVs
eFigure 5. Degree of Neuronal enrichment of L1CAM+ EVs compared to Total plasma EVs
eFigure 6. Representative standard curves and determinations of the Lowest Limit of Quantification (LLoQ) for the A) p181-tau and B) TSG101 electrochemiluminescence assays
eFigure 7. Within-Operator variability for assays and the entire methodology
eFigure 8. Between-Operator variability for the entire methodology
eFigure 9. EV biomarker levels at the last visit before AD onset for AD and Control BLSA participants
eFigure 10. EV biomarker slopes (percent change in levels of log-transformed biomarker per year) for future AD and Control BLSA participants
eFigure 11. Comparison of Alzheimer’s Disease risk prediction models using internal leave-10%-out cross-validation
eFigure 12. Boxplots of risk scores from the internally leave-10%-out cross-validated prediction models (BLSA training set)
eFigure 13. Receiver operating characteristic analysis for classification of JH ADRC participants into AD cases or Controls
eResults.
eDiscussion.
eReferences.
References
- 1.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263-269. doi: 10.1016/j.jalz.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubois B, Feldman HH, Jacova C, et al. Advancing research diagnostic criteria for Alzheimer’s disease: the IWG-2 criteria. Lancet Neurol. 2014;13(6):614-629. doi: 10.1016/S1474-4422(14)70090-0 [DOI] [PubMed] [Google Scholar]
- 3.Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280-292. doi: 10.1016/j.jalz.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270-279. doi: 10.1016/j.jalz.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiman EM, Brinton RD, Katz R, et al. Considerations in the design of clinical trials for cognitive aging. J Gerontol A Biol Sci Med Sci. 2012;67(7):766-772. doi: 10.1093/gerona/gls124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewczuk P, Riederer P, O’Bryant SE, et al. ; Members of the WFSBP Task Force Working on this Topic: Peter Riederer, Carla Gallo, Dimitrios Kapogiannis, Andrea Lopez Mato, Florence Thibaut . Cerebrospinal fluid and blood biomarkers for neurodegenerative dementias: an update of the Consensus of the Task Force on Biological Markers in Psychiatry of the World Federation of Societies of Biological Psychiatry. World J Biol Psychiatry. 2018;19(4):244-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mustapic M, Eitan E, Werner JK Jr, et al. Plasma extracellular vesicles enriched for neuronal origin: a potential window into brain pathologic processes. Front Neurosci. 2017;11:278. doi: 10.3389/fnins.2017.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol. 2012;3:124. doi: 10.3389/fphys.2012.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eitan E, Green J, Bodogai M, et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci Rep. 2017;7(1):1342. doi: 10.1038/s41598-017-01386-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968-E977. doi: 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goetzl EJ, Kapogiannis D, Schwartz JB, et al. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016;30(12):4141-4148. doi: 10.1096/fj.201600816R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapogiannis D, Boxer A, Schwartz JB, et al. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer’s disease. FASEB J. 2015;29(2):589-596. doi: 10.1096/fj.14-262048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goetzl EJ, Boxer A, Schwartz JB, et al. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology. 2015;85(1):40-47. doi: 10.1212/WNL.0000000000001702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiandaca MS, Kapogiannis D, Mapstone M, et al. Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement. 2015;11(6):600-607. doi: 10.1016/j.jalz.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winston CN, Goetzl EJ, Akers JC, et al. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement (Amst). 2016;3:63-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamlett ED, Goetzl EJ, Ledreux A, et al. Neuronal exosomes reveal Alzheimer’s disease biomarkers in Down syndrome. Alzheimers Dement. 2017;13(5):541-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abner EL, Jicha GA, Shaw LM, Trojanowski JQ, Goetzl EJ. Plasma neuronal exosomal levels of Alzheimer’s disease biomarkers in normal aging. Ann Clin Transl Neurol. 2016;3(5):399-403. doi: 10.1002/acn3.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins RJ, Diehl TC, Chia CW, Kapogiannis D. Insulin resistance as a link between amyloid-beta and tau pathologies in Alzheimer’s disease. Front Aging Neurosci. 2017;9:118. doi: 10.3389/fnagi.2017.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goetzl EJ, Boxer A, Schwartz JB, et al. Low neural exosomal levels of cellular survival factors in Alzheimer’s disease. Ann Clin Transl Neurol. 2015;2(7):769-773. doi: 10.1002/acn3.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noel-Storr AH, McCleery JM, Richard E, et al. Reporting standards for studies of diagnostic test accuracy in dementia: the STARDdem Initiative. Neurology. 2014;83(4):364-373. doi: 10.1212/WNL.0000000000000621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawas C, Gray S, Brookmeyer R, Fozard J, Zonderman A. Age-specific incidence rates of Alzheimer’s disease: the Baltimore Longitudinal Study of Aging. Neurology. 2000;54(11):2072-2077. doi: 10.1212/WNL.54.11.2072 [DOI] [PubMed] [Google Scholar]
- 22.Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:2. doi: 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coumans FAW, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120(10):1632-1648. doi: 10.1161/CIRCRESAHA.117.309417 [DOI] [PubMed] [Google Scholar]
- 24.Eitan E, Tosti V, Suire CN, et al. In a randomized trial in prostate cancer patients, dietary protein restriction modifies markers of leptin and insulin signaling in plasma extracellular vesicles. Aging Cell. 2017;16(6):1430-1433. doi: 10.1111/acel.12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Althauda D, Gulyani S, Karnati HK, et al. Utility of neuronal-derived exosomes to examine molecular mechanisms that affect motor function in patients with Parkinson Disease: a secondary analysis of the Exenatide-PD Trial [published online January 14, 2019]. JAMA Neurol. doi: 10.1001/jamaneurol.2018.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837-845. doi: 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 27.Janelidze S, Stomrud E, Palmqvist S, et al. Plasma β-amyloid in Alzheimer’s disease and vascular disease. Sci Rep. 2016;6:26801. doi: 10.1038/srep26801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nabers A, Perna L, Lange J, et al. Amyloid blood biomarker detects Alzheimer’s disease. EMBO Mol Med. 2018;10(5):e8763. doi: 10.15252/emmm.201708763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varma VR, Oommen AM, Varma S, et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: a targeted metabolomics study. PLoS Med. 2018;15(1):e1002482. doi: 10.1371/journal.pmed.1002482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554(7691):249-254. doi: 10.1038/nature25456 [DOI] [PubMed] [Google Scholar]
- 31.Mufson EJ, Ikonomovic MD, Counts SE, et al. Molecular and cellular pathophysiology of preclinical Alzheimer’s disease. Behav Brain Res. 2016;311:54-69. doi: 10.1016/j.bbr.2016.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansson O, Seibyl J, Stomrud E, et al. ; Swedish BioFINDER study group; Alzheimer’s Disease Neuroimaging Initiative . CSF biomarkers of Alzheimer’s disease concord with amyloid-β PET and predict clinical progression: a study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimers Dement. 2018;14(11):1470-1481. doi: 10.1016/j.jalz.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnham SC, Rowe CC, Baker D, et al. Predicting Alzheimer disease from a blood-based biomarker profile: a 54-month follow-up. Neurology. 2016;87(11):1093-1101. doi: 10.1212/WNL.0000000000003094 [DOI] [PubMed] [Google Scholar]
- 34.Dubois B, Epelbaum S, Nyasse F, et al. ; INSIGHT-preAD study group . Cognitive and neuroimaging features and brain β-amyloidosis in individuals at risk of Alzheimer’s disease (INSIGHT-preAD): a longitudinal observational study. Lancet Neurol. 2018;17(4):335-346. doi: 10.1016/S1474-4422(18)30029-2 [DOI] [PubMed] [Google Scholar]
- 35.Rowe CC, Bourgeat P, Ellis KA, et al. Predicting Alzheimer disease with β-amyloid imaging: results from the Australian imaging, biomarkers, and lifestyle study of ageing. Ann Neurol. 2013;74(6):905-913. doi: 10.1002/ana.24040 [DOI] [PubMed] [Google Scholar]
- 36.Villemagne VL, Pike KE, Chételat G, et al. Longitudinal assessment of Aβ and cognition in aging and Alzheimer disease. Ann Neurol. 2011;69(1):181-192. doi: 10.1002/ana.22248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casanova R, Varma S, Simpson B, et al. Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement. 2016;12(7):815-822. doi: 10.1016/j.jalz.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsson B, Lautner R, Andreasson U, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol. 2016;15(7):673-684. doi: 10.1016/S1474-4422(16)00070-3 [DOI] [PubMed] [Google Scholar]
- 39.Lim CZJ, Zhang Y, Chen Y, et al. Subtyping of circulating exosome-bound amyloid β reflects brain plaque deposition. Nat Commun. 2019;10(1):1144. doi: 10.1038/s41467-019-09030-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Rodriguez P, Sandebring-Matton A, Merino-Serrais P, et al. Tau hyperphosphorylation induces oligomeric insulin accumulation and insulin resistance in neurons. Brain. 2017;140(12):3269-3285. doi: 10.1093/brain/awx256 [DOI] [PubMed] [Google Scholar]
- 41.Talbot K, Wang HY, Kazi H, et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J Clin Invest. 2012;122(4):1316-1338. doi: 10.1172/JCI59903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6(8):734-746. doi: 10.1016/S1474-4422(07)70178-3 [DOI] [PubMed] [Google Scholar]
- 43.Goetzl EJ, Schwartz JB, Abner EL, Jicha GA, Kapogiannis D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann Neurol. 2018;83(3):544-552. doi: 10.1002/ana.25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goetzl EJ, Mustapic M, Kapogiannis D, et al. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer’s disease. FASEB J. 2016;30(11):3853-3859. doi: 10.1096/fj.201600756R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetzl EJ, Abner EL, Jicha GA, Kapogiannis D, Schwartz JB. Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of Alzheimer’s disease. FASEB J. 2018;32(2):888-893. doi: 10.1096/fj.201700731R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng L, Doecke JD, Sharples RA, et al. ; Australian Imaging, Biomarkers and Lifestyle (AIBL) Research Group . Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2015;20(10):1188-1196. doi: 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eTable 1. BLSA and JH ADRC Study sample characteristics
eTable 2. Electrochemiluminescence units for p231-tau and p181- tau and corresponding concentration of phosphorylated full length 441 tau standard used for calibration
eTable 3. Stability of L1CAM+ EV biomarkers over time
eTable 4. Inter-correlations between means of EV biomarkers across all visits
eTable 5. Inter-correlations between EV biomarkers at the last preclinical visit
eTable 6. Inter-correlations between the slopes of EV biomarkers
eTable 7. Cross-sectional and longitudinal associations of nEV biomarkers and composite cognitive scores across all BLSA participants
eTable 8. Missing values assessment
eTable 9. Performance statistics of Final Model 10 for Alzheimer Disease Prediction
eFigure 1. Nanoparticle tracking analysis (NTA) of total EVs isolated by Exoquick® alone and neuronal-enriched EVs isolated through Exoquick® followed by immunoprecipitation with antibodies against L1CAM (L1CAM+ EVs) from a typical plasma sample.
eFigure 2. Electron Microscopy of neuronal-enriched plasma EVs
eFigure 3. Western Blot characterization of neuronal-enriched plasma EVs
eFigure 4. Neuronal enrichment of L1CAM+ EVs
eFigure 5. Degree of Neuronal enrichment of L1CAM+ EVs compared to Total plasma EVs
eFigure 6. Representative standard curves and determinations of the Lowest Limit of Quantification (LLoQ) for the A) p181-tau and B) TSG101 electrochemiluminescence assays
eFigure 7. Within-Operator variability for assays and the entire methodology
eFigure 8. Between-Operator variability for the entire methodology
eFigure 9. EV biomarker levels at the last visit before AD onset for AD and Control BLSA participants
eFigure 10. EV biomarker slopes (percent change in levels of log-transformed biomarker per year) for future AD and Control BLSA participants
eFigure 11. Comparison of Alzheimer’s Disease risk prediction models using internal leave-10%-out cross-validation
eFigure 12. Boxplots of risk scores from the internally leave-10%-out cross-validated prediction models (BLSA training set)
eFigure 13. Receiver operating characteristic analysis for classification of JH ADRC participants into AD cases or Controls
eResults.
eDiscussion.
eReferences.