Abstract
Background
Coastal marine environments are one of the most productive ecosystems on Earth. However, anthropogenic impacts exert significant pressure on coastal marine biodiversity, contributing to functional shifts in microbial communities and human health risk factors. However, relatively little is known about the impact of eutrophication—human-derived nutrient pollution—on the marine microbial biosphere.
Results
Here, we tested the hypothesis that benthic microbial diversity and function varies along a pollution gradient, with a focus on human pathogens and antibiotic resistance genes. Comprehensive metagenomic analysis including taxonomic investigation, functional detection, and ARG annotation revealed that zinc, lead, total volatile solids, and ammonia nitrogen were correlated with microbial diversity and function. We propose several microbes, including Planctomycetes and sulfate-reducing microbes as candidates to reflect pollution concentration. Annotation of antibiotic resistance genes showed that the highest abundance of efflux pumps was found at the most polluted site, corroborating the relationship between pollution and human health risk factors. This result suggests that sediments at polluted sites harbor microbes with a higher capacity to reduce intracellular levels of antibiotics, heavy metals, or other environmental contaminants.
Conclusions
Our findings suggest a correlation between pollution and the marine sediment microbiome and provide insight into the role of high-turnover microbial communities as well as potential pathogenic organisms as real-time indicators of water quality, with implications for human health and demonstrate the inner functional shifts contributed by the microcommunities.
Electronic supplementary material
The online version of this article (10.1186/s40168-019-0714-6) contains supplementary material, which is available to authorized users.
Keywords: Metagenomics, Pollution concentration, Marine sediments, Antibiotic resistance genes
Background
Over the last two centuries, human activities such as coastal development and nutrient discharge from wastewater have driven major changes in marine biodiversity [1, 2]. Nutrient pollution, or eutrophication, detrimentally affects global marine ecosystems by impacting the diversity and function of a wide range of foundational species such as seagrass, oysters, corals, and other metazoans, and microbes including bacteria and viruses [3–5]. Eutrophication also negatively impacts marine coastal sediments that harbor microbial communities in high abundance and diversity. Not only are marine sediments hot spots for nitrogen and carbon cycling [6], they also serve as a long-term reservoir of terrigenous and aquatic pollutants [7]. Therefore, understanding the mechanisms by which water pollution affects the diversity and function of microbial populations in sediments is paramount.
Until recently, researchers used environmental microbes and their genetic markers as indicators for pollution [8, 9]. These studies began to reveal that microorganisms living inside marine sediments correlated well with pollutants such as trace metals and persistent organic compounds [10] and likely impacted sedimentary niche changes [11]. However, this methodology has not shed light on the overall genetic profiles of the communities.
Advances in high-throughput sequencing have revolutionized the detection of genes in complex environmental communities, offering a more promising avenue for comprehensive genetic profiling [12]. Through shotgun metagenomic and metabarcoding analyses, studies have revealed that sediments in urbanized areas with higher pollution levels may be enriched in human pathogens [13], such as those that cause ciguatera poisoning, avian influenza [14], and gastroenteritis [15]. In addition to changes in taxonomic profiles, changes in gene abundances have also been observed in impacted microbial communities, including genes associated with metabolic processes such as denitrification [16] and genes related to virulence/defense and stress response such as antibiotic resistance genes (ARGs) [17]. Although the aforementioned studies demonstrate that eutrophication alters microbial communities and genetic composition, very few comprehensive studies combining all taxonomic, functional, and resistance profiles have been performed to examine the influence of a well-constrained pollution gradient on the metagenomic profiles of microbial communities and the possible pathogenic risks for both humans and the environment. Moreover, a significant knowledge gap remains in understanding the associated impacts of pollution on influencing microbiome communities.
Furthermore, Hong Kong, which is located within the fastest developing area in southern China (the Greater Bay Area) has a population of approximately 7.5 million, and is situated at the mouth of the Pearl River Delta (PRD) offering one of the greatest opportunities to investigate the influence of pollution on the marine sediment microbiome. The PRD is also regarded as the Pearl River Estuary (PRE), through which the Pearl River enters the South China Sea. During the 1980s to the 1990s, Hong Kong was rapidly developed and subjected to land reclamation and population migration. The pollution footprint increased with a lagging investment in wastewater treatment. Therefore, eutrophication has contributed to losses of foundational species such as hard corals in many areas [18, 19]. Tolo Harbour [20] is a typical area suffering from eutrophication and is an enclosed bay located in northeast Hong Kong. Tolo Harbour was previously coral-dominated but the coral was gradually replaced by other organisms including algae and suspension feeders due to rapid coastal development inshore. However, coral communities still exist in some offshore regions of Tolo Harbour.
Thus, in our study, we examined four field sites in Tolo Harbour that have different degrees of eutrophication, including Centre Island (dead oyster reef), Che Lei Pai (sandy bottom with sparse vegetation), Port Island (50% hard coral cover), and Tung Ping Chau (75% coral cover) (Fig. 1). These sites were selected based on seven water quality gradients (Additional file 1: Table S1) and their coral species richness, which made them ideal regions to study the impact of pollution. By using a shotgun metagenomic approach, we assessed the microbial community composition, the functional characteristics of the microbial community, the prevalence of pathogenic bacteria, and the abundance and dissemination potential of ARGs in marine sediments. Lastly, we evaluated the differences in microbial communities among sampling sites in connection to representative pollution parameters. Our study revealed that the antibiotic resistome composition of marine sediment microbial communities is significantly different depending on the pollution concentration and, moreover, the distribution of microbial communities is highly associated with the pollution parameters.
Fig. 1.
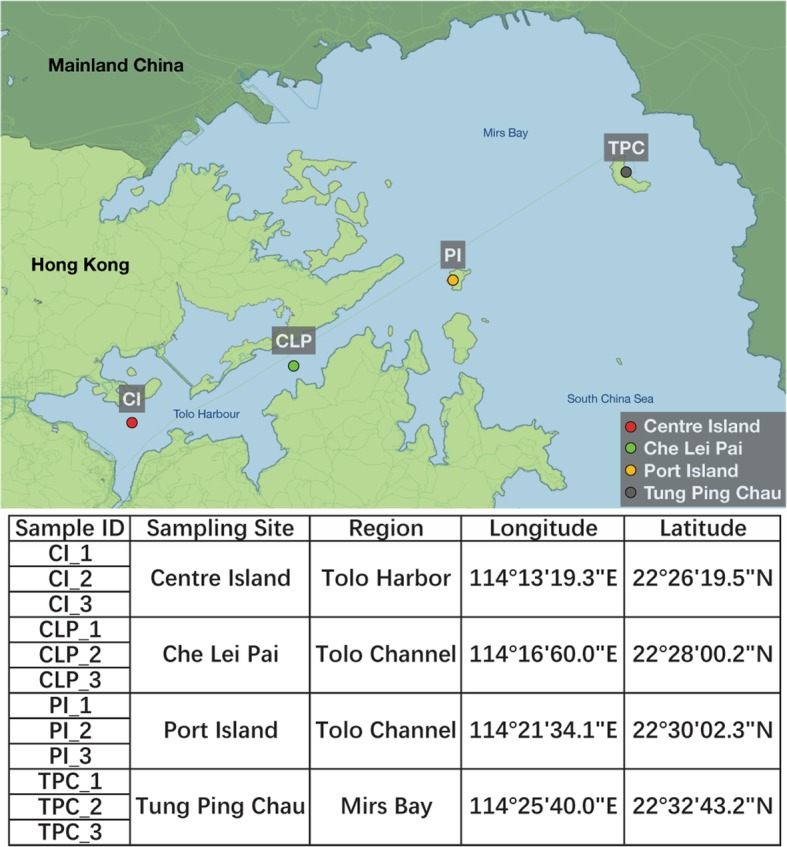
Sampling information. A map of Tolo Channel (including Tolo Harbour) and Mirs Bay shows the four sampling sites marked in circles of different colors: Centre Island, Che Lei Pai, Port Island, and Tung Ping Chau. Samples were collected from the sediments at 6–10-cm depth. The table shows the information of each sample including sampling site, region, longitude, and latitude
Results
Sequencing results
The 12 sediment samples subjected to metagenomic sequencing generated 138.6 Gb of raw data (average of 11.55 Gb per sample). Quality filtering reduced the average raw data to 6.66 Gb of filtered sequencing data per sample. On average, 85.3% of the mapped reads were classified as bacteria per sample, followed by archaea at 12.1% and eukaryotes at 2.6%. The percentage of bacterial reads was significantly higher in samples from Centre Island than those from Tung Ping Chau (P = 0.0172, Student’s t test) whereas the percentage of archaeal and eukaryotic reads were significantly lower (P = 0.0222 and P = 0.0317, respectively). Moreover, the percentage of bacterial reads was also significantly higher in samples from Centre Island compared to those from Port Island (P = 0.0199).
Pollutant concentration profiles
The normalized concentrations of seven geochemical indicators were used to represent environmental pollution levels at each sampling site (Additional file 1: Table S1). Zinc (Zn), lead (Pb), and copper (Cu) are common heavy metal pollutants of marine environments with arsenic regarded as highly toxic. Organic pollution was represented by ammonia nitrogen (NH3-N), chemical oxygen demand (COD) and total volatile solids (TVS). Centre Island, the innermost site within Tolo Harbour, had the highest concentrations of all pollutants with a general trend of decreasing pollutant concentrations with distance from the inner harbor (Fig. 1). Port Island and Tung Ping Chau exhibited similar levels of Zn, Pb, Cu, arsenic, and NH3-N.
Comparative analysis of microbial communities in a pollution gradient
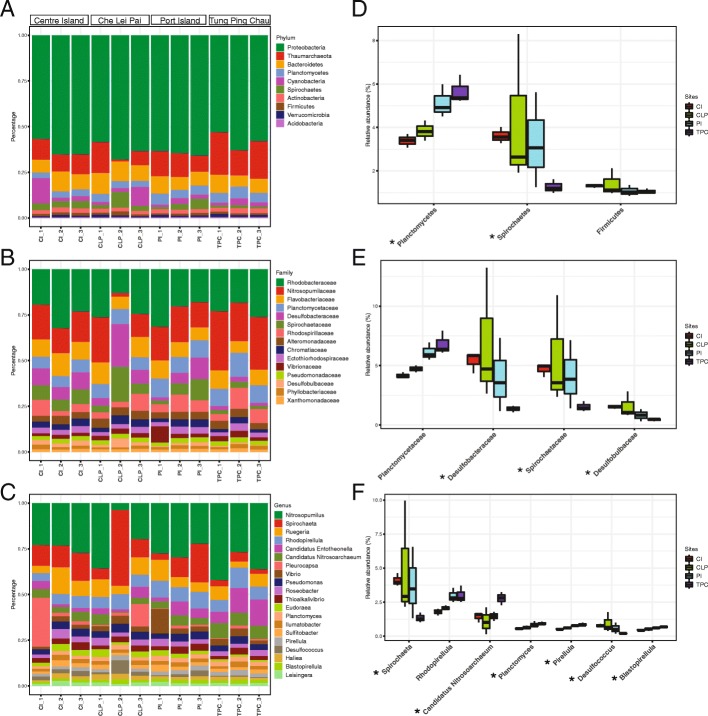
We constructed the total prokaryotic profile by extracting all bacterial and archaeal reads among samples at different taxonomic levels from phylum to genus. The most abundant prokaryotic phyla were Proteobacteria (60.3 ± 4.3%, SD), Thaumarchaeota (12.2 ± 5.7%), and Bacteroidetes (8.46 ± 1.7%) (Fig. 2a). The five most abundant prokaryotic families were Rhodobacteraceae, Nitrosopumilaceae, Flavobacteriaceae, Planctomycetaceae, and Desulfobacteraceae, which comprised on average ~ 42% of the prokaryotic communities (Fig. 2b). At the genus level, Nitrosopumilus (9.1 ± 3.8%) was the most abundant across all communities (Fig. 2c). The comparisons on the relative abundance of prokaryotes among the four sampling sites at the above described taxonomic levels (Additional file 2: Table S2 and Additional file 4: Table S4) revealed that Centre Island and Tung Ping Chau had the most dissimilar microbial communities with the largest number of significantly different prokaryotes (at every taxonomic level). Notably, Planctomycetes was the only phylum showing a significant difference among the four sampling sites (P = 0.030, Kruskal-Wallis test), with an increasing abundance from Centre Island to Tung Ping Chau (Fig. 2d). The abundance of Planctomycetes was significantly different in the comparisons between Centre Island and Tung Ping Chau (P = 0.014, Student’s t test), Centre Island and Port Island (P = 0.043), and Che Lei Pai and Tung Ping Chau (P = 0.020). Further investigation of its major family (Planctomycetaceae) and genus (Planctomyces) revealed the same tendency when comparing Centre Island to Tung Ping Chau (P = 0.022, Kruskal-Wallis test and P = 0.008, Student’s t test, respectively) (Fig. 2e). At the phylum level, we further observed that several human-related prokaryotes had significantly increased abundances in Centre Island compared to Tung Ping Chau, including Spirochaetes, a phylum that includes potential pathogens (P = 0.001, Student’s t test), and Firmicutes, one of the most abundant phyla in the human gut microbiome (P = 0.044) (Fig. 2f).
Fig. 2.
Comparative analysis of the prokaryotic communities among the sampling sites. a, b, c Relative abundance of the most abundant microbes across sites at the phylum, family, and species levels, respectively. d, e, f Relative abundance of the most abundant microbes which have significantly different abundances among sites. Microbes with asterisk retain strong differentiation after FDR correction (adjust q value < 0.1)
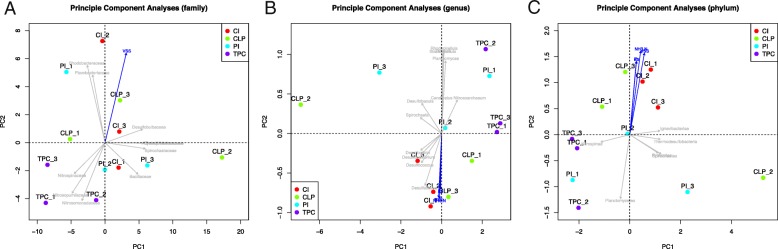
To evaluate the community diversity of each site and the differences in microbial composition among them, we calculated both the alpha and beta diversity using different indices. For the alpha diversity, neither the Shannon nor Simpson diversity indices showed any significant difference among sampling sites (Additional file 7: Figure S2). Notably, the principle component analyses (PCA) based on the most abundant prokaryotes revealed significantly different communities among the four sites at the phylum level (P = 0.048, PERMANOVA). Furthermore, we observed clear separations of the microbial communities between Centre Island and Tung Ping Chau at phylum and genus level (P = 0.047 and P = 0.049, respectively) (Fig. 3). Four pollution parameters (zinc, lead, total volatile solid, and ammonia nitrogen) were significantly correlated with communities at either phylum or genus levels (P < 0.05, Additional file 5: Table S5) (Fig. 3). At the phylum level, the pollution parameters can further serve as potential negative indicators of the distribution of Thaumarchaeota and Planctomycetes, which were significantly enriched in Tung Ping Chau (P = 0.008, Student’s t test). At the genus level, the distribution of Desulfatitalea, Desulfococcus, Desulfobacterium, and Desulfovibrio are positively correlated with the representative parameters to different extents, while four genera of Plactomycetes (Planctomyces, Blastopirellula, Pirellula, and Rhodopirellula) were negatively correlated. At the family level, total volatile solids was the best indicator of the community distribution, and interestingly, was negatively correlated with three nitrifying bacteria (Nitrosomonadaceae, Nitrosopumilaceae, and Nitrospinaceae).
Fig. 3.
Community dissimilarities among sampling sites with pollutant vectors. a, b, c Principal component analyses for the microbes among sites at phylum, family, and species levels, respectively. Vector matrices of the significant pollution parameters are shown by individual black lines with an arrow. The significant microbes are shown in gray
In addition to the abundance of microbes in the sediment, we also investigated the replication activity of different species, which could reflect changes in active communities. Therefore, we selected the 30 most .abundant species among all samples and applied the iRep [21] algorithm for evaluating the replication rate. Among these species, the replication rates of 14 species were estimated based on sufficient genome coverage among all the samples. Ruegeria conchae, which belongs to Rhodobacteraceae, the most abundant family in all samples, had on average the highest replication rate compared to other species across all sites (P < 10−8, Student’s t test) (Additional file 8: Figure S3). By comparing the replication rate among different sampling sites, we observed that Candidatus Nitrosopumilus sp. NF5 had higher replication rates in Port Island and Tung Ping Chau than in Centre Island and Che Lei Pai (P = 0.027). Candidatus Nitrosoarchaeum koreensis also showed a higher replication rate in Tung Ping Chau than in Centre Island (P = 0.031).
Functional shifts contributed by microcommunities
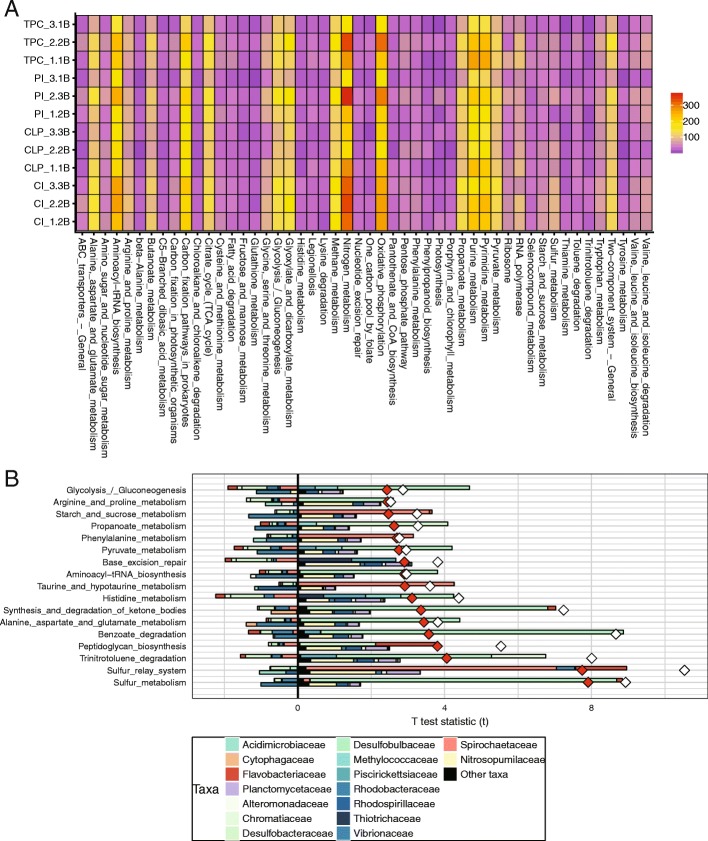
We annotated our metagenomics sequencing data using the KEGG database and constructed the pathway profiles for the microbial communities of each sampling site. “Nitrogen metabolism” was the most abundant functional pathway among all samples (6.1%), followed by “oxidative phosphorylation” (5.2%) and “aminoacyl-tRNA biosynthesis” (4.8%) (Fig. 4a). We implemented FishTaco [22], a novel algorithm for integrating taxonomic and functional comparative analyses to accurately quantify family-level contributions to functional shifts. We performed the analyses by using the microbial communities of Centre Island and Tung Ping Chau, which were the most dissimilar in terms of pollution concentration and were found to have the most diverse prokaryotic composition according to the above analyses.
Fig. 4.
KEGG Orthology annotation and taxonomic drivers of functional shifts between sites. a Heat map of the top 50 abundant KEGG pathway annotations. b Taxon-level shift contribution profiles for significantly different KO pathways between Centre Island (case group) and Tung Ping Chau (control group) based on the top 25 abundant families. Taxonomic contributors to functional pathways differences between Centre Island (case group) and Tung Ping Chau (control group) based on the top 25 abundant families. The white and red diamonds refer to the taxa-based and functional-based shift scores, respectively
After comparing the annotated KEGG pathways between Centre Island and Tung Ping Chau, 17 pathways showed significant enrichment at Centre Island (Additional file 3: Table S3). We further extracted the 25 most abundant families in these two sites that comprised over 72% of all the microbial communities. The Pearson correlation coefficient was calculated based on the pathway profile and the abundant family profile (Additional file 9: Figure S4). The results showed an average Pearson correlation coefficient of 0.91, suggesting a tight correlation between taxon and functional profiles. Desulfobulbaceae was found to be the main driver of the enrichment of the sulfur metabolism and benzoate degradation pathways in Centre Island. Moreover, Spirochaetaceae served as the driver of the sulfur relay system (also known as sulfur transfer system) in Centre Island. In addition, the trinitrotoluene degradation pathway was found to be enriched in Centre Island and was driven by Chromatiaceae and Nitrosopumilaceae. (Fig. 4b). To further confirm our findings, Spearman correlation between the above pathways and families among the four sites were showing overall a relatively good consistency with the Pearson correlation (except the correlation between Spirochaetaceae and sulfur relay system) (Additional file 10: Figure S5).
Putative pathogens and antibiotic resistome
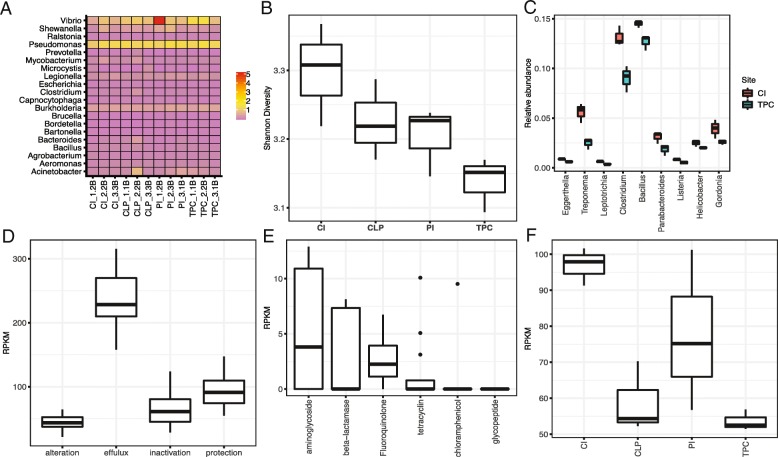
By aligning the taxonomic profiles to a list of potential human pathogens compiled from three different sources [23–25], we discovered opportunistic pathogenic species from 148 genera that comprised 4.92–7.65% of all the microbial communities, among which Vibrio (1.27 ± 1.18%) was the most abundant (Fig. 5a). Typical opportunistic pathogenic species of Vibrio including V. cholera, V. parahaemolyticus, and V. vulnificus were found at all the sampling sites. The potential opportunistic pathogenic species P. aeruginosa was found to have the second highest abundance (1.23 ± 0.10%). We further calculated the alpha diversity of the putative pathogenic communities among sampling sites using both the Shannon and Simpson indices (Fig. 5b). Alpha diversity decreased from Centre Island to Tung Ping Chau significantly, with nine putative pathogenic genera that were significantly more abundant in Centre Island compared to Tung Ping Chau (P < 0.05, Student’s t test) (Fig. 5c). Among them, Bacillus (0.14 ± 0.03%) was the most abundant putative pathogen (P = 0.037). Clostridium, another putative pathogen, was enriched in Centre Island compared to Tung Ping Chau (P = 0.015), mainly due to increased levels of the species C. tetani. In addition, two putative pathogenic species of Treponema (T. denticola and T. pallidum) were found in significantly higher abundances in Centre Island compared to Tung Ping Chau (P = 0.015).
Fig. 5.
Comparisons of abundances of potential pathogens and antibiotic resistant genes (ARGs) among samples. a Heat map based on relative abundances of potential pathogenic genera in different samples. b Comparisons of pathogenic alpha diversity among sampling sites. c Comparisons of significantly different pathogenic genera between Centre Island and Tung Ping Chau. d Total relative abundance of each ARG mechanism among the sampling sites. e Total ARG profile of the samples. f Comparisons of total ARG abundances from each site
Subsequently, we investigated the resistome profiles in the different sampling sites. Among ARG mechanisms, antibiotic efflux pumps (237.24 ± 47.7 RPM—reads per million reads) were the predominant resistance mechanism in the sediment communities, followed by target protection (95.74 ± 29.2 RPM), inactivation (65.29 ± 28.6 RPM) and target alteration (43.52 ± 13.2 RPM) (Fig. 5d). Annotation using ResFams identified 17 ARG families. Among the ARG families, five genes that have activities against specific antibiotics were detected including aminoglycoside, beta-lactamase, fluoroquinolone, tetracycline, and the chloramphenicol resistance gene (Fig. 5e), and there were no significant differences found among the sites. To evaluate the differences in the ARGs among the sediment samples, we compared the total abundances of the ARGs. The results showed that Tung Ping Chau had the lowest abundance of total ARGs, whereas Centre Island had the highest (P = 0.0001, Fig. 5). Further comparisons of the ARG families showed that three efflux pump genes, RND, baeR, and ABC, were significantly enriched in Centre Island compared to the other sites (P = 0.014, 0.047, and 0.033, respectively).
Discussion
Microbial communities inhabiting marine sediments are extremely diverse due to the complex physicochemical gradients therein [26]. Community dynamics are affected by human activities, particularly those related to water chemistry and quality [13]. Hong Kong has a diverse distribution of highly urbanized and relatively undisturbed coastal environments [27] and provides a perfect opportunity to assess the relationship between microbial communities and different levels of perturbation. Earlier studies have investigated the community of bacteria and ARGs in marine sediments of Hong Kong using both clone libraries and metagenomics [7, 28, 29]. However, they were unable to identify clear patterns in either bacterial communities or ARGs. To fill this gap, we constructed a comprehensive pipeline for the proof-of-concept analysis of the microbiome communities, their functional diversity, and the resistome as affected by human activities and pollution levels. Throughout our analysis, we provided evidence of the impact of different pollution levels on the diversity of microbial communities and their functional-associated genes as well as the risks of increased levels of ARGs and pathogens.
Regarding the prokaryotic profile, we observed an enrichment of a number of phyla tightly associated with the human gut and terrestrial biomes in the most polluted areas. These phyla include Spirochaetes, a potential pathogenic phylum causing a wide range of diseases including leptospirosis, Lyme disease, and Alzheimer’s disease [30–32], and Firmicutes, which make up the largest portion of the human gut microbiome [33]. While the majority of sewage generated from the local population is treated and then redirected for discharge elsewhere (https://www.epd.gov.hk/epd/wqo_review/en/watQua.htm), the presence of Firmicutes may indicate that inputs remain and Planctomycetes, a widely distributed phylum found in a variety of marine environments [34, 35], exhibits a dose-response sensitivity effect to heavy metals [36] while generally resistant to high concentrations of inorganic nitrogen compounds. In our study, Planctomycetes was the fourth most abundant phylum among the samples, and its abundance was significantly decreased at Centre Island compared to Tung Ping Chau. Combined with the pollution data from the sampling sites, our study suggests that Planctomycetes may also serve as an important potential water quality indicator.
By evaluating the microbial diversity at the community level, we observed clear differentiation among the sampling sites—especially between Centre Island and Tung Ping Chau—and coherence with the distribution of representative pollutants. Interestingly, at the genus level, the four sulfate-reducing microbes (SRM) Desulfatitalea, Desulfococcus, Desulfobacterium, and Desulfovibrio were found to be positively correlated with pollution levels, suggesting that eutrophication increases the abundance of SRM. Previous studies in marine sediments of Hong Kong had also reported the relationship between SRM and pollution levels. For instance, pollutants may influence the relative abundance of SRMs to total microbes under a unimodal paradigm while low concentrations of pollutants will decrease relative abundance of SRMs in total microbes [7]. Moreover, with the clone library methods, unique SRM members related to the polluted harbor environment and estimated SRM richness correlated with several environment factors [29].
In our study, we also implemented a novel algorithm, based on sequencing coverage, which was used to calculate an index of replication (iRep) for estimating changes in the active microbial communities [21]. Ruegeria conchae had the highest replication rate among all sites, which is consistent with previous studies [37, 38]. These studies have shown that Ruegeria is present in a wide range of marine habitats, from coastal regions to deep-sea sediments, and constitute up to 25% of the total bacterial community. Interestingly, the active community analysis revealed additional changes in bacterial spp. replication rates along the water quality gradient. Two species of Thaumarchaeota—Candidatus Nitrosopumilus sp. NF5 and Candidatus Nitrosoarchaeum koreensis—which are responsible for the major ammonia-oxidizing process [39, 40] in sediment, replicated faster in sites with high water quality (i.e., low ammonia; Additional file 1: Table S1). The concordance of replication rate with relative abundance not only confirms the adaptation of Thaumarchaeota to low-ammonia conditions but also reveals the mechanism by which it can maintain a high abundance.
When investigating pathogenicity among all sites, we observed that Vibrio, including its three opportunistic pathogenic species, V. cholera, V. parahaemolyticus, and V. vulnificus, were the most abundant. These disease-causing strains of Vibrio are associated with infectious diarrhea and are commonly carried by marine animals including crustaceans [41]. Pseudomonas aeruginosa, the second most abundant potential pathogenic species in our analysis, is a multidrug resistant pathogen and causes a wide variety of serious infectious diseases including meningitis, pneumonia, and septicemia [42–44]. The alpha diversity calculated from only putative pathogenic microbes decreased significantly from Centre Island to Tung Ping Chau, indicating that pollution inputs influence the local potential pathogen composition. Furthermore, nine opportunistic pathogenic genera were enriched at the most polluted site. Considering that coastal waters are the most impacted by human activities including recreation (e.g., swimming) and food supply (e.g., fishing and aquaculture), surface or water column sampling to assess public safety may overlook potential pathogen reservoirs in the sediment.
Based on the evidence above, we observed a connection between the extent of pollution and the abundance and diversity of pathogenic organisms in marine sediments. Pinpointing the drivers of these patterns is complex as feedback loops between both biotic and abiotic process function across multiple spatial and temporal scales. For example, healthy and biodiverse marine ecosystems buffer against the proliferation of bacterial pathogens [5, 45] but foundational species can be sensitive to pollutants. The persistence of diverse coral assemblages at Tung Ping Chau may contribute to increased pathogen resistance while the eutrophication of Tolo Harbour and disappearance of coral communities [27] may have reduced this important ecosystem service. More direct links between human activity and pollution stress can be inferred from our antibiotic resistance analysis which shows an increase in the dominant resistance mechanisms and abundance of ARGs. It is well known that efflux pumps are the predominant resistance mechanism in sediments [17], and our results further provide the direct evidence that an increase in efflux pump-related genes is correlated to an increase in pollution [46]. These data suggest the increased capability of microbes to reduce intracellular concentrations of antibiotics, heavy metals, or other toxins/environmental stresses [17] in polluted sediments. In other words, the increasing efflux pump mechanisms in the microbes would greatly contribute to antibiotic resistance and further presented a growing threat to antibiotic therapy and a major challenge for antibiotic development.
Conclusions
Our investigation of the microbiome composition and functionality in marine sediments revealed that microbe communities are likely influenced by human activity and pollution discharge. Through our analysis, we also identified several microbes within the Planctomycetes phylum that may serve as useful bio-indicators of water quality. The relationships among sediment microbial communities and water quality presented here set the stage for a valuable reference database for environmental assessments and public policy. Therefore, we strive for the continuous sampling of these as well as additional sites in Hong Kong to provide new insight into the interactions between humans and coastal marine ecosystems.
Materials and methods
Sampling sites and sample collection
Sampling was conducted in August 2016 from four field sites (Centre Island (CI), Che Lei Pai (CLP), Port Island (PI), and Tung Ping Chau (TPC)) in Tolo Harbour. At each sampling site, 50 g of sediment from the top 10 cm were collected via scuba using sterile 15-mL falcon tubes and were further divided into three aliquots as replicates (Fig. 1). Within minutes after sampling, sediment samples were flash frozen in liquid nitrogen and stored at − 80 °C for DNA extraction and metagenomic shotgun sequencing.
Pollution data collection
Seven geochemical indicators (zinc (Zn), lead (Pb), copper (Cu), chemical oxygen demand (COD), arsenic (As), total volatile solid (TVS), and ammonia nitrogen (NH3-N)) were obtained from the Hong Kong Environmental Protection Department’s extensive periodic water and sediment monitoring database [47] (Additional file 1: Table S1) representing the environmental pollution in the sediment including heavy metals and organic matter. These data were obtained from the sediment samples collected in August 2016 at the benthic sediment sampling locations. Pollution data were normalized using the minimum-maximum normalization procedure for further analyses (Additional file 6: Figure S1).
DNA extraction and metagenomic sequencing
DNA from the sediment samples was extracted using the MoBio PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA) following the manufacturer’s instructions. DNA concentrations (2.41 ± 1.25 μg) were verified using the Qubit Fluorometer and DNA quality was checked via agarose gel electrophoresis (using Takara λ-Hind III digest and the Tiangen D2000 marker) prior to library construction. DNA libraries were constructed using the Nextera XT kit. Metagenomic shotgun sequencing was then performed on an Illumina HiSeq 1500 (101 bp PE) platform. The raw sequence data were uploaded into the Sequence Read Archive of NCBI (accession numbers SRR8361706-17).
Taxonomic profiling
The raw sequencing paired-end reads passed standard quality control after the adapter regions and low-quality reads were removed, as per Li et al. [48]. The filtered reads were mapped to the nonredundant database using DIAMOND [49] and the default settings. The aligned reads were filtered using an e-value < 1e−10 and a 95% cutoff. The lowest common ancestor (LCA) algorithm was implemented with the LCA mapper from mtools of MEGAN5 [50] for the taxonomic assignment of each aligned read. The relative abundances of each taxa were further distilled from the LCA results for each taxonomic level. Prokaryotic community profiles were constructed at phylum, family, and genus levels for further statistical analysis. Additionally, potential pathogenic species and genera were taxonomically identified using three publicly available lists of putative pathogens [23–25] and further summarized to genus level for statistical comparisons. The organisms appearing on the list were regarded as opportunistic pathogens and the species containing at least one opportunistic pathogenic strain were marked as putative pathogenic species.
Microbial community composition
Alpha diversity indices detailing microbial community composition within each sample was calculated using vegan [51] in R. Both the Shannon and Simpson indices were used for alpha diversity evaluation based on the relative abundance of each taxonomic level. For estimating community dissimilarities, both Bray-Curtis and Euclidean distances were calculated by phyloseq [52] and vegan [51] based on the relative abundance of each taxon at different levels.
Estimation of species cellular replication rate
To estimate the replication rate of the 30 most abundant species, an in-house genome database was constructed manually by obtaining the draft or complete genomes of the target species (either contig or scaffold) from the NCBI Genome database. The high-quality metagenomic reads were mapped to the in-house genome database by Bowtie2 [53]. The mapped results of each target species were further manipulated with SAMtools [54] and were used for measuring replication rates with iRep [21].
Functional annotation
From each sample’s filtered sequencing data, one million reads were subsampled randomly and were aligned to the KEGG Orthology (KO) database [55] built by KOBAS 2.0 [56] using DIAMOND BLASTX (−e 1e−10, best hits reserved). The identified KO genes were further annotated into different pathways based on predefined collections in the KEGG database and were quantified by read counts. The prokaryotic taxonomic profiles of the 25 most abundant families among samples and the pathway profile were integrated to quantify taxa-specific contributions to functional shifts using FishTaco [22].
Antibiotic resistance genes (ARGs)
Subsampled reads were mapped against an in-house ARG database based on CARD [57], ARDB [58], and ResFams [59] using BLASTX. Aligned reads were filtered (best hit from BLASTX, identity > 70% and coverage > 70%), classified into different ARG mechanisms, and further annotated into ARG families using ResFams. The abundance of ARG genes was calculated as RPKM (reads per million mapped reads).
Data analysis
Three replicate samples were analyzed. Statistical comparisons of prokaryotic taxonomic profiles, potential pathogens, and ARGs (including clinically important ARGs) were performed by using Kruskal-Wallis tests between the four sampling sites and Student’s t tests between each pairwise site in R. The Benjamini-Hochberg FDR correction [60] was applied to adjust the P value for multiple t test comparisons in R (Additional file 4). We performed principal component analysis (PCA) on the relative abundance of taxonomic profiles to evaluate the community dissimilarities using the vegan and ape [61] packages in R (Additional file 11). The pollution parameters were served as fitted vectors in the above multidimensional scaling for testing the correlation between community distributions and pollution levels. Adonis from vegan package in R was used as PERMANOVA test to evaluate the significance of a variable in determining variation of distances (the number of permutations were set as 999). The homogeneity of dispersions test was calculated using PERMDISP from vegan package in R. Pearson’s correlation coefficients were calculated between the taxa-based and KO function-based profiles using FishTaco. Spearman correlations were further performed between taxonomic abundance and KO pathways among the four sampling sites based on the FishTaco results using the correlation function in R.
Data visualization
Packages including ggplot2 and gplots in R and matplotlib in Python were used for visualization purposes.
Additional files
Table S1. Representative water quality data from the Hong Kong Environmental Protection Department. (CSV 264 bytes)
Table S2. Significant statistical comparison result on the top 50 abundant prokaryotes at phylum/family/genus levels. (XLSX 46 kb)
Table S3. Significantly enriched KEGG pathways in Centre Island comparing to Tung Ping Chau. (XLSX 9 kb)
Table S4. Taxonomic significant statistical comparison result between Centre Island and Tung Ping Chau with FDR correction. (XLSX 51 kb)
Table S5. PERMANOVA and PERMDISP results on microbial communities among the four sampling sites. (XLSX 35 kb)
Figure S1. Representative water quality data among the four sampling sites. (PDF 4 kb)
Figure S2. The comparison of alpha diversity in the Shannon index among sampling sites. (PDF 4 kb)
Figure S3. Comparisons of bacterial replication rate among the four sampling sites. (PDF 8 kb)
Figure S4. Pearson’s correlation coefficients between the taxa-based and KO function-based profiles for the significantly different pathways. (PDF 5 kb)
Figure S5. Spearman’s correlation results between the correlated pathways and families among the four sampling sites. (PDF 31 kb)
File S1. Statistical analysis scripts performed in R. (PDF 72 kb)
Acknowledgements
The authors would like to thank Dr. Jun Li (City University of Hong Kong, China), Dr. Till Röthig (The University of Hong Kong, China), Dr. Yueqiong Ni, and Dr. Kang Kang (Leibniz Institute for Natural Product Research and Infection Biology, Hans Knoll Institute, Jena, Germany) for valuable discussions. GP would like to thank Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) CRC/Transregio 124 “Pathogenic fungi and their human host: Networks of interaction,” subproject B5 and INF and DFG under Germany’s Excellence Strategy—EXC 2051—Project-ID 390713860 for intellectual input. Funding was provided from the School of Biological Sciences of the University of Hong Kong and an Environment and Conservation Fund award (#67-2016) to GP and DB. This paper is contribution 43 to the Smithsonian’s MarineGEO-TMON program.
Abbreviations
- ARGs
Antibiotic resistance genes
- COD
Chemical oxygen demand
- iRep
Index of replication
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCA
Lowest common ancestor
- PCA
Principle component analyses
- PRD
Pearl river delta
- RPKM
Reads per million mapped reads
- RPM
Reads per million mapped reads
- SRM
Sulfur-reducing microbe
- TVS
Total volatile solid
Authors’ contributions
DMB and GP designed this study. AA and SEM collected the sediment samples from sampling locations in Hong Kong. JC analyzed the data. JC and GP wrote the manuscript and AA, SEM, and DB revised it. All authors read and approved the final manuscript.
Funding
This work was supported by Environment and Conservation Fund (ECF 2016-67).
Availability of data and materials
The raw Illumina sequence data of metagenomic data have been deposited in the sequence read archive (SRA accession: SRR8361706-17) at NCBI under Bioproject accession #PRJNA511137. The scripts for all statistical analysis are available as Additional file 11.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiarui Chen and Shelby E. McIlroy contributed equally to this work.
Contributor Information
David M. Baker, Email: dmbaker@hku.hk
Gianni Panagiotou, Phone: 49-3641532-1000, Email: Gianni.Panagiotou@hki-jena.de.
References
- 1.McKee JK, Sciulli PW, Fooce CD, Waite TA. Forecasting global biodiversity threats associated with human population growth. Biol Conserv. 2004;115:161–164. doi: 10.1016/S0006-3207(03)00099-5. [DOI] [Google Scholar]
- 2.Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS, et al. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314:787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- 3.Phillips DJ. The use of biological indicator organisms to monitor trace metal pollution in marine and estuarine environments—a review. Environ Pollution (1970) 1977;13:281–317. doi: 10.1016/0013-9327(77)90047-7. [DOI] [Google Scholar]
- 4.Knowlton N, Jackson JB. Shifting baselines, local impacts, and global change on coral reefs. PLoS Biol. 2008;6:e54. doi: 10.1371/journal.pbio.0060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb JB, van de Water JA, Bourne DG, Altier C, Hein MY, Fiorenza EA, et al. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science. 2017;355:731–733. doi: 10.1126/science.aal1956. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman JA, Hewson I. Chapter 25 – Viruses, bacteria, and the microbial loop. In: Capone DG, Bronk DA, Mulholland MR, Carpenter EJ, editors. Nitrogen in the marine environment (2nd edition). Amsterdam: Elsevier; 2008. p. 1097–134.
- 7.Guo F, Li B, Yang Y, Deng Y, Qiu JW, Li X, et al. Impacts of human activities on distribution of sulfate-reducing prokaryotes and antibiotic resistance genes in marine coastal sediments of Hong Kong. FEMS Microbiol Ecol. 2016;92:9. doi: 10.1093/femsec/fiw128. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen LD, Sørensen SJ. The effect of longterm exposure to mercury on the bacterial community in marine sediment. Curr Microbiol. 1998;36:291–297. doi: 10.1007/s002849900312. [DOI] [PubMed] [Google Scholar]
- 9.Gillings MR, Gaze WH, Pruden A, et al. Using the class 1 integronintegrase gene as a proxy for anthropogenic pollution. ISMEJ. 2015;9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Zhang Q, Xia C, et al. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISMEJ. 2014;8:1932–1944. doi: 10.1038/ismej.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyzer G, Stams AJ. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 12.Li LG, Yin X, Zhang T. Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome. 2018;6:93. doi: 10.1186/s40168-018-0480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medeiros JD, Cantão ME, Cesar DE, Nicolás MF, Diniz CG, Silva VL, et al. Comparative metagenome of a stream impacted by the urbanization phenomenon. Braz J Microbiol. 2016;47:835–845. doi: 10.1016/j.bjm.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis TM, Barry Bousfield R, Bissett LA, Dyrting KC, Luk GS, Tsim ST, et al. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 2004;33:492–505. doi: 10.1080/03079450400003601. [DOI] [PubMed] [Google Scholar]
- 15.Lau SK, Woo PC, Fan RY, Lee RC, Teng JL, Yuen KY. Seasonal and tissue distribution of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis, in retail freshwater fish in Hong Kong. Int J Food Microbiol. 2017;113:62–66. doi: 10.1016/j.ijfoodmicro.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Scott NM, Hess M, Bouskill NJ, Mason OU, Jansson JK, Gilbert JA. The microbial nitrogen cycling potential is impacted by polyaromatic hydrocarbon pollution of marine sediments. Front Microbiol. 2014;5:108. doi: 10.3389/fmicb.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Yang Y, Liang X, Yu K, Zhang T, Li X. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ Sci Technol. 2013;47:12753–12760. doi: 10.1021/es403818e. [DOI] [PubMed] [Google Scholar]
- 18.Morton B. Hong Kong’s coral communities: status, threats and management plans. Mar Pollut Bull. 1994;29:74–83. doi: 10.1016/0025-326X(94)90429-4. [DOI] [Google Scholar]
- 19.Scott PJB. Chronic pollution recorded in coral skeletons in Hong Kong. J Exp Mar Biol Ecol. 1990;139:51–64. doi: 10.1016/0022-0981(90)90038-E. [DOI] [Google Scholar]
- 20.Xu F, Lam K, Zhao Z, Zhan W, Chen Y, Tao S. Marine coastal ecosystem health assessment: a case study of the Tolo Harbour, Hong Kong, China. Ecol Model. 2004;173:355–370. doi: 10.1016/j.ecolmodel.2003.07.010. [DOI] [Google Scholar]
- 21.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016;34:1256. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manor O, Borenstein E. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe. 2017;21:254–267. doi: 10.1016/j.chom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2013;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duprey NN, Yasuhara M, Baker DM. Reefs of tomorrow: eutrophication reduces coral biodiversity in an urbanized seascape. Glob Chang Biol. 2016;22:3550–3565. doi: 10.1111/gcb.13432. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Ki JS, Qian PY. Microbial diversity in polluted harbor sediments I: bacterial community assessment based on four clone libraries of 16S rDNA. Estuar Coast Shelf Sci. 2008;76:668–681. doi: 10.1016/j.ecss.2007.07.040. [DOI] [Google Scholar]
- 29.Zhang W, Song LS, Ki JS, Lau CK, Li XD, Qian PY. Microbial diversity in polluted harbor sediments II: sulfate-reducing bacterial community assessment using terminal restriction fragment length polymorphism and clone library of dsrAB gene. Estuar Coast Shelf Sci. 2008;76:682–691. doi: 10.1016/j.ecss.2007.07.039. [DOI] [Google Scholar]
- 30.Palaniappan RU, Ramanujam S, Chang YF. Leptospirosis: pathogenesis, immunity, and diagnosis. Curr Opin Infect Dis. 2007;20:284–292. doi: 10.1097/QCO.0b013e32814a5729. [DOI] [PubMed] [Google Scholar]
- 31.Schwan TG. Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect Agents Dis. 1996;5:167–181. [PubMed] [Google Scholar]
- 32.Miklossy J. Alzheimer’s disease-a neurospirochetosis. Analysis of the evidence following Koch’s and Hill’s criteria. J Neuroinflammation. 2011;8:90. doi: 10.1186/1742-2094-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Fuerst JA. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141:1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- 35.Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol. 2011;9:403. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 36.Flores C, Catita JA, Lage OM. Assessment of planctomycetes cell viability after pollutants exposure. Antonie Van Leeuwenhoek. 2014;106:399–411. doi: 10.1007/s10482-014-0206-4. [DOI] [PubMed] [Google Scholar]
- 37.Brinkhoff T, Giebel HA, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenschein EC, Nielsen KF, D'Alvise P, Porsby CH, Melchiorsen J, Heilmann J, et al. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 2017;11(2):569. doi: 10.1038/ismej.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa T, Stahl DA. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol. 2013;79:6911–6916. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim BK, Jung MY, Yu DS, Park SJ, Oh TK, Rhee SK, et al. Genome sequence of an ammonia-oxidizing soil archaeon,“Candidatus Nitrosoarchaeum koreensis” MY1. J Bacteriol. 2011;193:5539–5540. doi: 10.1128/JB.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan CM. Bacteria. Encyclopedia of Earth. Washington DC: National Council for Science and the Environment; 2010. [Google Scholar]
- 42.Diekema DJ, Pfaller MA, Jones RN, Doern GV, Winokur PL, Gales AC, et al. Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis. 1999;29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 43.Prithiviraj B, Bais HP, Weir T, Suresh B, Najarro EH, Dayakar BV, et al. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect Immun. 2005;73:5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirienko NV, Ausubel FM, Ruvkun G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc Natl Acad Sci. 2015;112:1821–1826. doi: 10.1073/pnas.1424954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raymundo LJ, Halford AR, Maypa AP, Kerr AM. Functionally diverse reed-fish communities ameliorate coral disease. Proc Natl Acad Sci U S A. 2009;106:17067–17070. doi: 10.1073/pnas.0900365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu YG, Zhao Y, Li B, Huang CL, Zhang SY, Yu S, et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat Microbiol. 2017;2(4):16270. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- 47.Hong Kong Environmental Protection Department (EPD). Hong Kong. http://www.epd.gov.hk/epd/english/resources_pub/publications/pub_reports_ap.html. Accessed 24 Oct 2016.
- 48.Li J, Sung CYJ, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci. 2016;113:E1306–E1315. doi: 10.1073/pnas.1518189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 50.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2013;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 52.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Suppl 1):316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2008;37(Suppl1):443–447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9:207. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Yoav, Hochberg Yosef. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 61.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Representative water quality data from the Hong Kong Environmental Protection Department. (CSV 264 bytes)
Table S2. Significant statistical comparison result on the top 50 abundant prokaryotes at phylum/family/genus levels. (XLSX 46 kb)
Table S3. Significantly enriched KEGG pathways in Centre Island comparing to Tung Ping Chau. (XLSX 9 kb)
Table S4. Taxonomic significant statistical comparison result between Centre Island and Tung Ping Chau with FDR correction. (XLSX 51 kb)
Table S5. PERMANOVA and PERMDISP results on microbial communities among the four sampling sites. (XLSX 35 kb)
Figure S1. Representative water quality data among the four sampling sites. (PDF 4 kb)
Figure S2. The comparison of alpha diversity in the Shannon index among sampling sites. (PDF 4 kb)
Figure S3. Comparisons of bacterial replication rate among the four sampling sites. (PDF 8 kb)
Figure S4. Pearson’s correlation coefficients between the taxa-based and KO function-based profiles for the significantly different pathways. (PDF 5 kb)
Figure S5. Spearman’s correlation results between the correlated pathways and families among the four sampling sites. (PDF 31 kb)
File S1. Statistical analysis scripts performed in R. (PDF 72 kb)
Data Availability Statement
The raw Illumina sequence data of metagenomic data have been deposited in the sequence read archive (SRA accession: SRR8361706-17) at NCBI under Bioproject accession #PRJNA511137. The scripts for all statistical analysis are available as Additional file 11.