Abstract
Localization of mRNAs to postsynaptic sites and their subsequent translation is thought to contribute to synapse-specific plasticity. However, the direct visualization of dendritic RNA transport in living neurons remains a major challenge. Here, we analyze the transport of Alexa-labeled RNAs microinjected into mature hippocampal neurons. We show that microinjected MAP2 and CaMKIIα RNAs form particles that localize into dendrites as their endogenous counterparts. In contrast, nonlocalizing RNAs or truncated CaMKIIα, lacking the dendritic targeting element, remain in the cell body. Furthermore, our microinjection approach allowed us to identify a novel dendritically localized RNA, Septin7. Time-lapse videomicroscopy of neurons injected with CaMKIIα and Septin7 RNAs demonstrates fast directional movement along the dendrites of hippocampal neurons, with similar kinetics to Staufen1 ribonucleoprotein particles (RNPs). Coinjection and simultaneous visualization of two RNAs, as well as double detection of the corresponding endogenous RNAs, reveal that neuronal transcripts are differentially sorted in dendritic RNPs.
Introduction
RNA localization represents a general mechanism to achieve cell polarity in a variety of cells and organisms (St. Johnston, 2005). In the CNS, the localization of mRNAs to synaptic sites and their subsequent local translation is thought to contribute to synapse-specific plasticity (Kiebler and DesGroseillers, 2000; Sutton and Schuman, 2006). Several dendritically localized RNAs have been characterized, including actin-regulated cytoskeletal protein (Arc), the noncoding RNA brain-specific cytoplasmic 1 (BC1), microtubule-associated protein 2 (MAP2), and the α-subunit of calcium/calmodulin-dependent protein kinase II (CaMKIIα), and many more have been reported previously (Bramham and Wells, 2007). However, the underlying mechanism of RNA localization in primary neurons is not well understood (Bramham and Wells, 2007; Martin and Ephrussi, 2009). To study this interesting problem, two principal approaches have been used. First, key RNA-binding proteins (RBPs) [e.g., mammalian Staufen proteins (Köhrmann et al., 1999; Zeitelhofer et al., 2008), zipcode-binding proteins (Fusco et al., 2003), fragile X mental retardation protein (FMRP) (Antar et al., 2004), heteronuclear ribonucleoprotein A2 (hnRNP A2) (Brumwell et al., 2002), and Pur α (Kanai et al., 2004), which have all been implicated in dendritic RNA localization] were tagged with fluorescent protein variants. However, even in the cases when the—direct or indirect—interaction between an RBP and a dendritic transcript has been demonstrated, it is not safe to assume that they travel along the dendrite as a complex, since there is no evidence that their association is either stable or exclusive. Second, several techniques have been used for the visualization of the localized RNA itself. Many groups have routinely assessed RNA localization by in situ hybridization (ISH) in either cultured dissociated neurons or in brain slices. Recently, novel methods have been developed to directly visualize individual dendritic RNAs [e.g., microinjection of fluorescently labeled RNAs in living hippocampal neurons (Shan et al., 2003; Gao et al., 2008) and the MS2 system (Brechbiel and Gavis, 2008; Dictenberg et al., 2008)]. The first two mentioned studies focused on the molecular mechanism of dendritic RNA localization; the latter studied the kinetics of RNA transport in living neurons.
It is currently not known whether different RNAs are targeted to dendrites by independent or by the same trafficking pathway. Carson and colleagues (Gao et al., 2008) showed that Arc, CaMKIIα, and Neurogranin share hnRNP A2 recognition element (A2RE) sequences, assemble in the same particles, and are targeted to neuronal processes by hnRNP A2. However, no other transcripts are shown to share consensus motifs or trans-acting determinants.
In our study, we used a microinjection assay for real-time imaging of RNA in living hippocampal neurons. We show that fluorescently labeled RNAs mimic the localization pattern of the respective endogenous transcripts. Analysis of RNA particle kinetics identifies fast bidirectional motility along the dendrite pointing to motor-dependent active transport along microtubules. Double injection of different transcripts in two colors, as well as double detection of endogenous RNAs, reveal that some RNAs are sorted into distinct ribonucleoprotein particles (RNPs), whereas others might share the same transport pathway. Therefore, our data provide novel insight into dendritic RNA transport in mature neurons.
Materials and Methods
Neuronal cultures and transient transfections
Embryonic day 17 rat hippocampal neurons were cultured and transfected as described previously (Goetze et al., 2004; Zeitelhofer et al., 2007).
Constructs
The following sequences were cloned into pBluescript II KS+ or SK+ (Stratagene) using the indicated restriction sites. The resulting plasmids then served as templates for (1) ISH probes or (2) microinjection as follows: (1) Sept7 [NM_022416, nucleotides 22-533, 512-1040, 1021-1490, and full length (nucleotides 11-1490), NotI/HindIII], MAP2 (NM_013066, nucleotides 2420-3080; U30938, nucleotides 1-1340 and 2532-3738), CaMKIIα [NM_177407, first part of 3′-untranslated region (UTR): nucleotides 1616-3117, NotI/XhoI; second part of 3′-UTR: nucleotides 3156-4756, NotI/XhoI], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (M17701, nucleotides 4-1233, Sac1/XhoI), Histone H3.3 (X73683, 1.1 kb), Tubulin α3 (NM_001040008, nucleotides 420-1069). A control probe, pBluescript, was generated from a ScaI linearized pBluescript KS+ plasmid. (2) Sept7 (NM_022616, nucleotides 11-1490, NotI/HindIII), GAPDH (M17701, nucleotides 4-1233, Sac1/XhoI), MAP2 (U30938, nucleotides 181-3738, EcoRI), Histone H3.3 (X73683, nucleotides 20-1007, NotI/XhoI), CaMKIIα (NM_177407, nucleotides 1556-4799, NotI/BamHI; full-length CaMKIIα 3′-UTR also containing additional 29 nt from the coding region; first part of CaMKIIα 3′-UTR, “1H”: nucleotides 1616-3117 lacking the first 31 nt of the 3′-UTR, NotI/XhoI; second part of CaMKIIα 3′-UTR, “2H”: nucleotides 3156-4756, NotI/XhoI; first part of CaMKIIα 3′-UTR lacking 59 nt of the 3′-UTR, 1644-3117).
Wild-type mouse Stau155 was amplified using PfU Taq polymerase (Promega) with the following primers: 5′-aagctagcatgtataagcccgtggaccc (NheI) and 5′-cgaccggtctcccgcacgctgaaactg (AgeI). The PCR product was digested with the indicated restriction enzymes and then cloned into the pEYFP-N1 expression vector (Clontech). All resulting constructs were sequenced. The shSeptin7 construct has been described previously (Xie et al., 2007).
Microinjection
Templates were linearized by restriction digestion. Subsequently, RNAs were in vitro transcribed (Wilkie and Davis, 2001) in the presence of Alexa 488- or Alexa 546-labeled UTPs (Invitrogen). Unincorporated nucleotides were removed using NucAway Spin Columns (Ambion/Applied Biosystems). The quality of the RNA preparation was assessed by electrophoresis. To increase RNA stability within the neuronal cytoplasm, the microinjected RNA was capped during transcription. To ensure that the somatic restriction of GAPDH and Histone H3.3 (our negative controls for localization) was the result of low localization efficiency and not degradation (because of the lack of potential stability elements), we injected the full-length transcripts.
Mature hippocampal neurons [12–16 d in vitro (DIV)] were microinjected using an AIS2 microinjection system (Cellbiology Trading) assembled on an Axiovert 200M fluorescence microscope (Zeiss). Cells were placed in prewarmed HBSS (37°C) 10 min before microinjection. At the time of mounting and subsequent microinjection, (elevated) room temperature was reached. Microinjection needles with an inner tip diameter between 0.2 and 0.3 μm were used (P-87; Sutter Instruments). After pulling, needles were stored for at least 3 weeks. This way, needles had to be carefully pressure filled, but remained substantially longer unclogged during microinjection. The injection volume was 0.1–0.25 pl; 300 ng/μl or 200 ng/μl of each labeled RNA were used for single or double injections, respectively. Immediately after injection of 10–50 cells per coverslip, successfully injected cells were imaged live or fixed in 4% paraformaldehyde. Only cells that appeared healthy based on their phase contrast (proper shape of the neuron including a halo around the cell body, nonfragmented dendrites without any signs of varicosities, cells that remained attached to the substrate, no granule formation in dendrites) were taken into account.
Probe preparation for ISH
Templates were linearized using the indicated restriction enzymes (see above). Subsequently, probes were generated in the presence of digoxigenin (DIG)-UTP (DIG RNA labeling mix) or fluorescein-UTP (fluorescein RNA labeling mix; both Roche) according to manufacturer's instructions.
Fluorescent ISH with tyramide signal amplification
Single detection was performed as described previously (Vessey et al., 2008). For the simultaneous detection of two transcripts, differentially labeled RNA probes (DIG and fluorescein) are allowed to hybridize together to their targets at 65°C overnight. After extensive washes, the fluorescein-labeled probe is detected first, by an anti-fluorescein antibody coupled to horseradish peroxidase (HRP) (Roche) and Cy3-tyramide [tyramide signal amplification (TSA); PerkinElmer]. To quench peroxidase activity and to allow for an independent second HRP-mediated detection, coverslips are incubated in 3% H2O2 in PBS for 1 h. After extensive washes, the DIG-labeled probe is detected using an anti-DIG antibody coupled to HRP (Roche) followed by TSA using Alexa 488-tyramide (Invitrogen). For detection of Sept7 or MAP2 in two colors, the respective probes were targeting non-overlapping sequences of each transcript. To test specificity, we used DIG- and fluorescein-labeled pBluescript probes (see above). As a negative control for cross-reactivity, we hybridized cells with fluorescein-labeled MAP2 probe but no DIG-labeled probe, and followed the same detection protocol mentioned above.
Imaging
Fluorescent and phase contrast images were acquired using an Axiovert 200M microscope (Zeiss) equipped with 40× Plan-Neofluar [numerical aperture (NA), 1.3] or 63× (NA 1.4) Plan-Apochromat oil-immersion objective (both Zeiss), a Coolsnap HQ CCD camera (Roper Scientific), and MetaMorph 6.3 software (Universal Imaging/Visitron), or with an Axioplan microscope (Zeiss) equipped with an F-view II CCD camera and AnalysisB software (both Olympus). Images were processed with either MetaMorph or AnalysisB and assembled with Adobe Photoshop CS4, version 11.0 (Adobe). Images were not modified other than adjustments of color, magnification, levels, brightness, and contrast.
Data analysis
Single microinjection experiments.
Cells were assigned to the following categories: “cell body only” (RNA signal restricted to dendritic regions less than one cell body diameter from soma), “plus proximal dendrites” (signal up to two cell body diameters from soma), and “plus distal dendrites” (signal extending more than two cell body diameters from soma). At least 30 cells per condition were evaluated by four people in a blind manner. ANOVA analysis (Minitab, version 14; Minitab) was performed on the resulting data set testing (1) whether the rated staining pattern for each injected RNA was independent of the evaluators and (2) whether rating for a specific category depended on the microinjected RNA. The resulting p values were <0.05 in each case indicating that the rated RNA signals were independent of the individual evaluator and depended on the microinjected RNA. (1) Values of p for individual RNAs were as follows: CaMKIIa 3′-UTR (0.010), CaMKIIa 1H 3′-UTR (<0.0005), CaMKIIa 2H 3′-UTR (0.002), CaMKIIa 1H 3′-UTR-60nt (<0.0005), Histone H3.3 (0.006), GAPDH (<0.0005), Septin7 (0.016), MAP2 (0.007); (2) values of p for categories across the data set were as follows: cytoplasmic (<0.001), plus proximal dendrites (0.006), and plus distal dendrites (<0.0005). Alternatively, we measured signal intensities in distal dendrites using MetaMorph 6.3 software (Universal Imaging/Visitron). At least 20 cells per condition were analyzed.
Double microinjection experiments.
Colocalization ratios for each pair of RNAs were determined by dividing the number of particles that contained both RNAs by the total number of particles of the RNA that was the least abundant in dendrites. Error bars represent SEMs of colocalization ratios between individual cells analyzed. The asterisks denote statistical significance as determined using two-tailed Student's t test. Numbers of injected cells were 5, 6, 7, 3, and 2 for Septin7–Septin7, CaMKIIα–Septin7, Septin7–MAP2, CaMKIIα–MAP2, and MAP2–MAP2, respectively. At least 187, 154, 230, 112, and 52 particles of each individual transcript of the respective pairs were analyzed.
Double fluorescence ISH experiments.
Analysis was performed as described above for the double microinjection experiments. Numbers of analyzed cells were 22, 16, 19, 15, and 22 for Sept7–Sept7, CaMKIIα–Sept7, Sept7–MAP2, CaMKIIα–MAP2, and MAP2–MAP2, respectively. At least 181, 211, 290, 304, and 460 particles of each individual transcript of the respective pairs were analyzed. The analysis of colocalization of Sept7 with either CaMKIIα or MAP2, was—in most cases—restricted to proximal dendrites, since in the majority of inspected cells, Sept7 did not localize distally.
Particle tracking.
The average velocities calculated represent absolute values incorporating both anterograde and retrograde particle movement; rarely occurring stationary phases of tracked particles were excluded. Only particles that could be tracked for more than five consecutive frames were analyzed. Maximum velocities are based on the movement of particles between two consecutive frames. Cumulative frequency plots are based on average velocities of the individual particle pools.
Results
To our knowledge, Carson and coworkers (Ainger et al., 1993) were the first to study transport and localization of MBP mRNA in oligodendrocytes in culture. In their seminal study in 1993, they microinjected DIG-labeled mRNA into living oligodendrocytes and analyzed the intracellular distribution of the injected RNA by confocal microscopy. Since then, Wilkie and Davis (2001) significantly improved the method by using Alexa-labeled RNAs to directly visualize RNA transport in Drosophila embryos and provided novel insight into the underlying mechanisms. This approach has then been successfully applied to primary neurons by Smith and colleagues (Shan et al., 2003), who investigated the localization mechanism of chimeric, A2RE-containing RNAs in fixed hippocampal neurons and showed that those reporter RNAs localized in granules in distal dendrites in a microtubule- and hnRNP A2-dependent manner.
The primary goal of our study was to investigate the kinetics of dendritically localized RNA in living hippocampal neurons. Previously, this has only been achieved by either focusing on the transport dynamics of key green fluorescent protein (GFP)-labeled RBPs (Köhrmann et al., 1999; Huang et al., 2003; Oleynikov and Singer, 2003; Zeitelhofer et al., 2008) or, alternatively, by labeling an RNA of interest via the MS2 system (Rook et al., 2000; Kanai et al., 2004). To directly visualize the transport of dendritically localized RNAs, we therefore adapted the technique of microinjection of fluorescently labeled RNA into mammalian neurons (Shan et al., 2003) with the following modifications. First, we used 12- to 16-DIV-old mature hippocampal neurons to warrant full and complete dendritic differentiation. Second, neurons were placed in a CO2-independent buffer system, in our case prewarmed HBSS (37°C) 10 min before microinjection, which occurred at elevated room temperature. Third, we generated specifically designed microinjection glass capillaries with a long, tapered shape and small inner tip diameter to minimize denucleation of neurons and immediate cell death. Fourth, we routinely used 300 or 200 ng/μl of each labeled RNA for single and double injections, respectively. Fifth, initial pilot experiments showed it to be essential that microinjected cells have to be very quickly imaged, otherwise most of the transport of the injected RNA has already taken place (data not shown). Therefore, we routinely injected 10–50 cells per coverslip, which took us up to 20 min and started live imaging immediately afterward. Alternatively, cells were immediately fixed.
Microinjected RNAs are specifically distributed into dendrites of living hippocampal neurons
In the first set of experiments, we chose four mRNAs, coding for MAP2, CaMKIIα, Histone H3.3, and GAPDH, respectively, whose localization patterns in neurons are well documented (Kuhl and Skehel, 1998; Mallardo et al., 2003; Poon et al., 2006). These mRNAs were in vitro transcribed in the presence of Alexa-UTP and the resulting purified RNAs microinjected into the soma of mature hippocampal neurons. Since microinjection is an invasive procedure potentially damaging primary neurons and producing experimental artifacts, we only chose neurons for additional analysis that survived microinjection well according to the criteria defined in Materials and Methods. Both MAP2 (Fig. 1A) and CaMKIIα 3′-UTRs (Fig. 2B) yielded consistent particulate, dendritic localization, similar to their endogenous counterparts as shown by ISH (supplemental Fig. 1F,G, available at www.jneurosci.org as supplemental material) and reported previously (Garner et al., 1988; Burgin et al., 1990). To further validate our microinjection assay, we generated two truncated versions representing the first and the second halves of CaMKIIα 3′-UTR. Whereas the first part (CaMKIIα 1H, lacking the first 31 nt of the 3′-UTR) retains full transport capacity comparable with the full-length 3′-UTR (Fig. 1B), the second half of CaMKIIα 3′-UTR (CaMKIIα 2H) is less consistently transported into distal dendrites and was often observed to be restricted in proximal regions (Fig. 1C). This is in agreement with a previous report locating one of the elements necessary for dendritic targeting of the CaMKIIα within the first half of the 3′-UTR (Mori et al., 2000). A truncated transcript lacking the first 59 nt of the 3′-UTR corresponding to nucleotides 1–59 of the Mori element (Mori et al., 2000) did not localize to dendrites on injection (Fig. 1D). Additional dendritic targeting sequences have been identified in other parts of the CaMKIIα 3′-UTR (Blichenberg et al., 2001; Huang et al., 2003). However, it is beyond the scope of our study to address whether all previously mapped—likely redundant—localization signals are necessary or sufficient for the dendritic accumulation of CaMKIIα on injection in the neuronal cytoplasm.
Figure 1.
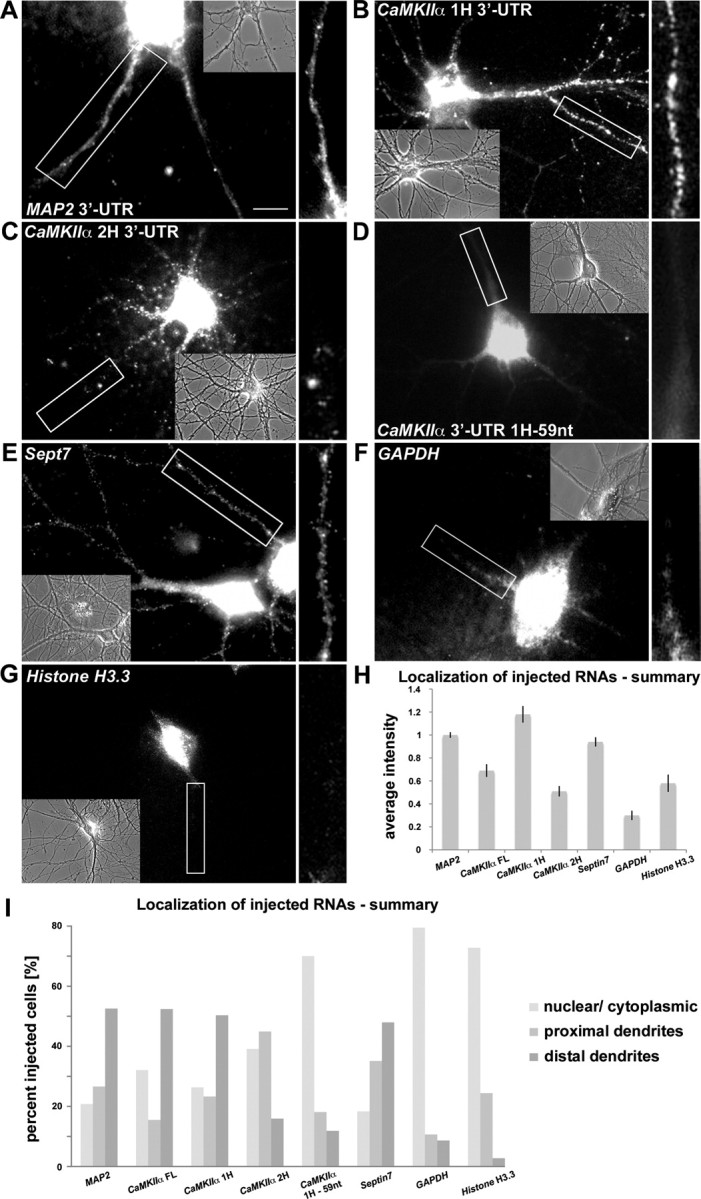
Microinjected RNA forms particles that are sorted to their expected intracellular destination in cultured mature hippocampal neurons. Fifteen DIV neurons were microinjected with the following Alexa-labeled RNAs: MAP2 3′-UTR (A); CaMKIIα 3′-UTR first half (1H) (also lacking the first 31 nt of the 3′-UTR) (B); CaMKIIα 3′-UTR second half (2H) (C); CaMKIIα 3′-UTR first half lacking 59 nt (1H-59 nt) (D); Septin7 (E); GAPDH (F); and Histone H3.3 (G). Particles were imaged 5–10 min after microinjection. Phase contrast images of the respective injected neurons are shown. Insets are magnifications of the indicated dendrites on the right. H, I, Quantification of all microinjection experiments. H, Average intensity of fluorescent signal in distal dendrites. The values are normalized to the signal of microinjected MAP2 RNA. Error bars represent the SEM. I, Evaluation of the localization pattern of microinjected RNAs (>30 cells rated per RNA). Four people evaluated the data independently, in a blind manner. “Distal” reflects fluorescent signals that were found at least two cell body diameters (>25 μm) from the soma. Scale bar, 10 μm.
Figure 2.
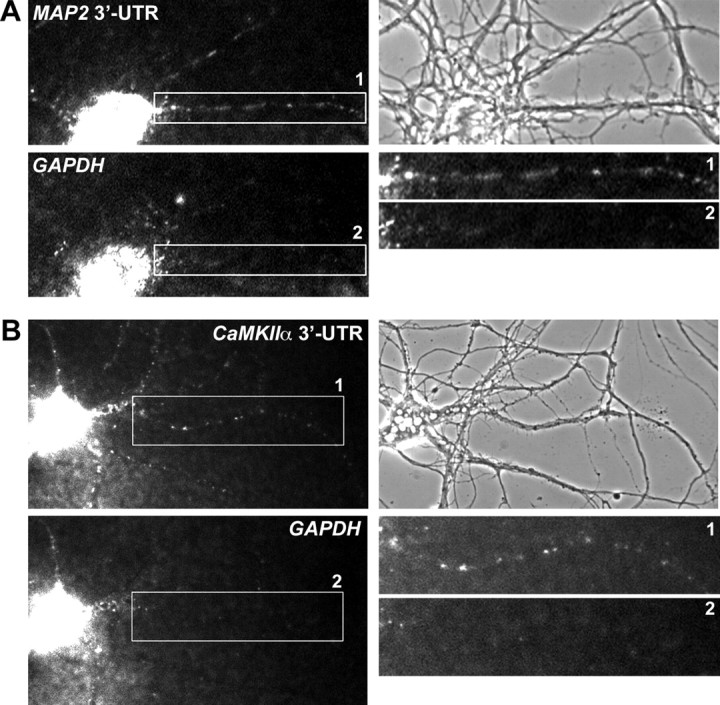
Coinjected GAPDH RNA and MAP2 or CaMKIIα RNA are sorted to their expected destinations in cultured hippocampal neurons. Coinjection of Alexa 488-labeled MAP2 3′-UTR RNA and Alexa 546-labeled GAPDH RNA (A) or Alexa 488-labeled CaMKIIα 3′-UTR and Alexa 546-labeled GAPDH RNA (B), respectively, results in dendritic localization of either MAP2 RNA or CaMKIIα 3′-UTR, whereas GAPDH RNA remains in the cell soma. Magnifications of the boxed dendritic regions and phase contrast images of the respective injected neurons are shown on the right. The size of the cell body of a typical pyramidal neuron is 10–15 μm.
To exclude the possibility that the observed dendritic localization is an artifact resulting from the excess of RNA introduced into the cells, we injected GAPDH and Histone H3.3, which are found to be low abundant in dendrites or restricted to the cell body, respectively (supplemental Fig. 1E,C, available at www.jneurosci.org as supplemental material). GAPDH and Histone H3.3 do not localize into distal dendrites on injection (Fig. 1F,G), further confirming the specificity of the assay. A summary of the distribution of all injected transcripts is shown in Figure 1, H and I, as determined by the average signal intensity in dendrites or by visual inspection of the localization pattern.
Since the RNA was microinjected into the cytoplasm, our data suggest that dendritic targeting of mRNAs does not require nuclear events, as reported previously for oskar mRNA (Hachet and Ephrussi, 2004), and is in agreement with data on the localization of other Drosophila transcripts (Wilkie and Davis, 2001; Bullock et al., 2003; MacDougall et al., 2003) and reporter RNAs in hippocampal neurons (Shan et al., 2003).
We next investigated whether a microinjected neuron would maintain the capacity to sort two RNAs with different subcellular localization patterns to their expected destination. When MAP2 3′-UTR/GAPDH and CaMKIIα 3′-UTR/GAPDH pairs (Fig. 2A,B) were coinjected in mature neurons, MAP2 and CaMKIIα 3′-UTRs were transported as particles into dendrites, whereas GAPDH was retained in the soma. This indicates that GAPDH does not hitchhike on dendritic RNAs when they are simultaneously injected in the neuronal cytoplasm.
Microinjected Septin7 RNA localizes into dendrites of mature neurons
In an attempt to identify RNAs interacting with the double-stranded RBP Staufen1 (Stau1) (Kiebler et al., 1999), we previously isolated Stau1-containing RNPs from rat brain (Mallardo et al., 2003). In addition to the two known dendritically localized RNAs, CaMKIIα and BC1, 15 RNAs were found to be present in a Stau1-containing particle fraction (Mallardo et al., 2003) (M. Mallardo and M. A. Kiebler, unpublished data). One of these, Septin7 (Sept7) (also called CDC10), was of particular interest, since it codes for a component of the postsynaptic density of mammalian synapses (Walikonis et al., 2000). We recently showed that Sept7 exists in a trimeric complex consisting of Sept5/7/11, localizes to the base of dendritic spines, and is critical for dendrite branching and dendritic spine morphology (Xie et al., 2007).
Based on the synaptic localization of Sept7 protein, we investigated whether the corresponding Sept7 mRNA is dendritically localized. When Sept7 RNA was microinjected into mature neurons, distinct RNA particles were detected in the cell body as well as in proximal and distal dendrites (Fig. 1E). Detailed analysis of the localization pattern of Sept7 and its comparison with that of the RNAs presented in Figure 1 showed a sorting behavior comparable with that of MAP2 and CaMKIIα 3′-UTRs and significantly different than cell body-restricted RNAs (Fig. 1). We then investigated whether endogenous Sept7 mRNA localizes to distal dendrites of hippocampal neurons by ISH. In some neurons—in consistence with the localization of Sept7 on microinjection—the distribution pattern of the endogenous transcript (supplemental Fig. 1H,I, available at www.jneurosci.org as supplemental material) was similar to that of MAP2 and CaMKIIα mRNAs (supplemental Fig. 1F,G, available at www.jneurosci.org as supplemental material). In the majority of neurons, however, the distribution of Sept7 (supplemental Fig. 1D, available at www.jneurosci.org as supplemental material) resembled more the pattern of GAPDH (supplemental Fig. 1E, available at www.jneurosci.org as supplemental material), whose abundance in distal dendrites is low, or the cell body-restricted markers Tubulin α3 and Histone H3.3 (supplemental Fig. 1B,C, available at www.jneurosci.org as supplemental material). Summarizing, although we were able to detect Sept7 RNA particles in dendrites, most cells contained very little Sept7 RNA at distal dendritic sites. To confirm the specificity of the signal, we transfected primary hippocampal neurons with a pSuperior plasmid expressing a short hairpin RNA that targets Sept7 (Xie et al., 2007). The ISH signal in transfected neurons was significantly reduced compared with nontransfected, wild-type control neurons (supplemental Fig. 1I,I′, available at www.jneurosci.org as supplemental material).
We speculate that Sept7 RNA has low affinity for the dendritic targeting machinery and therefore localizes with low efficiency to the dendrites of some neurons, mainly at proximal sites. The injection of Sept7 RNA at high concentrations might further facilitate the recruitment of transport determinants and its subsequent dendritic localization. We have indeed identified a motif within the first 40 nt of the 3′-UTR of Sept7 RNA (Y. Xie, P. Macchi, and M. A. Kiebler, unpublished data) that is similar in sequence and secondary structure (as predicted by Mfold; http://mfold.burnet.edu.au) with the A2RE element (Munro et al., 1999). However, we have no evidence whether it may be recognized by the hnRNP A2 transport machinery. Alternatively, Sept7 RNA might hitchhike on localizing RNPs—on injection—and travel from the cell body to neuronal dendrites.
It is worth mentioning that we were unable to detect Sept7 mRNA in the molecular layer of the hippocampus (X. Mo, M. A. Kiebler, and D. Kuhl, unpublished data). We believe that Sept7 escaped detection because of its low expression level (significantly lower than that of CaMKIIα mRNA) (Allan brain atlas; http://mouse.brain-map.org/welcome.do%3bjsessionid=3EA00E3FB39E35598C97A5B00F32274B) in combination with its restriction to proximal dendritic sites in the majority of neurons.
Real-time RNA imaging in mature neurons reveals fast bidirectional motility of CaMKIIα RNA particles along the dendrite
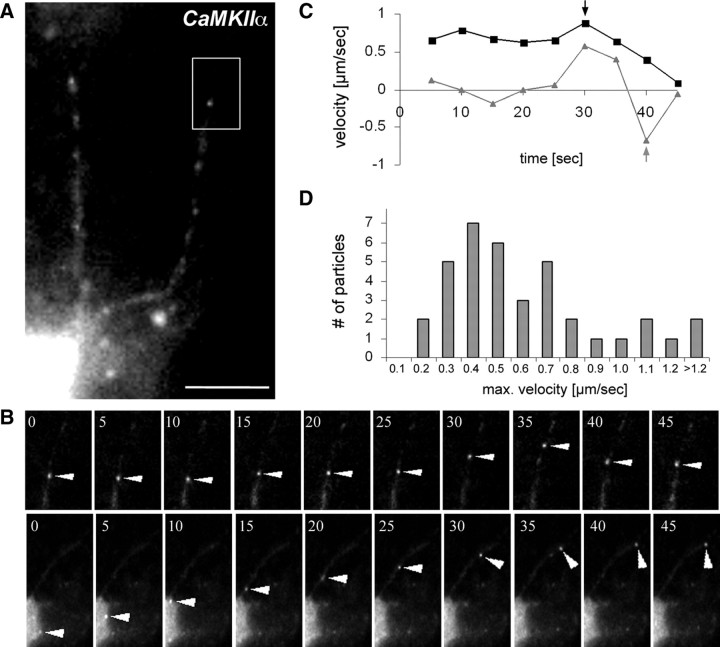
To study the kinetics of CaMKIIα RNA, fluorescently labeled, full-length CaMKIIα 3′-UTR was microinjected into mature neurons and RNA transport was monitored by time-lapse videomicroscopy (Fig. 3). Motile particles generally displayed unidirectional or bidirectional movements, occasionally interrupted by pauses (Fig. 3). As in previous studies, however, the majority of RNA particles remained stationary in the observed time periods (Köhrmann et al., 1999; Rook et al., 2000). The motile particles travel with an average velocity of 0.39 ± 0.24 μm/s (Fig. 3D; supplemental Movie SM1, available at www.jneurosci.org as supplemental material). Transport velocities >1.2 μm/s were observed. The observed transport rates are 5- to 10-fold higher than the previously reported rates of Stau1 motility along dendrites (Köhrmann et al., 1999). We speculate that this is most likely attributable to the higher frame rate that has been applied in the present study, which enabled us to monitor faster particles that previously escaped detection.
Figure 3.
CaMKIIα RNA particles move bidirectionally into dendrites of hippocampal neurons. A, A cultured neuron microinjected with Alexa 488-labeled CaMKIIα 3′-UTR. B, Time series of individual particles. The top panel shows bidirectional transport of a CaMKIIα RNA-containing particle (arrowheads) indicated in the boxed region of A. The bottom panel shows unidirectional (anterograde) transport of a CaMKIIα particle starting in the cell body (arrowheads). Images were taken every 5 s (supplemental Movie SM1, available at www.jneurosci.org as supplemental material). C, Kinetics of the two presented CaMKIIα particles. Negative velocities correspond to retrograde movement, and positive velocities correspond to anterograde transport. The arrows indicate the respective maximum velocities. D, Histogram of the maximum velocities of a total of 41 CaMKIIα particles. Scale bar, 10 μm.
To test this hypothesis, we remeasured transport of Stau1-EYFP in mature hippocampal neurons with a faster frame rate than in our own preceding study (Köhrmann et al., 1999) (supplemental Movie SM2, available at www.jneurosci.org as supplemental material). We now observed a significantly faster average transport velocity of 0.40 ± 0.24 μm/s. This is in good agreement with our data on RNA transport velocities. Furthermore, our measurements correlate with maximal velocities that were recently reported in the following experimental systems: an average velocity of 2.1 μm/s with maximum rates of 4.5 μm/s of GFP-tagged ZBP1 in hippocampal neurons (Tiruchinapalli et al., 2003) and velocities of up to 1.29 μm/s for FMRP-GFP in 10 DIV hippocampal neurons (Dictenberg et al., 2008). In conclusion, the independent visualization of RNA and protein transport in the same experimental system resulted in very similar transport velocities, providing reciprocal confirmation for the individual measurements. Our data support that CaMKIIα exhibits fast directional motility. Although the distribution of microinjected CaMKIIα has been studied previously (Gao et al., 2008), its kinetics has not been reported. The motion of MS2-GFP-tagged CaMKIIα 3′-UTR in hippocampal neurons has also been analyzed recently (Dictenberg et al., 2008), but only upon neuronal stimulation shown to trigger its accumulation in dendrites or with a slow acquisition rate (Rook et al., 2000) that has only revealed much slower motion than our study. Our data would argue for a motor-dependent process as previously suggested (Kanai et al., 2004). However, additional work, which is beyond the scope of this study, is required to dissect the molecular machinery that mediates the transport of CaMKIIα to dendrites.
Transport dynamics of injected Sept7 RNA in living neurons
To investigate the dynamics of Sept7 mRNA transport, fluorescently labeled Sept7 RNA was microinjected into mature neurons and RNA transport was monitored by time-lapse videomicroscopy. Although the majority of Sept7 RNA—like CaMKIIα—is stationary or displays corralled motion (Fig. 3B), directed movements were also identified. Figure 4A shows a time series of an anterogradely moving Sept7 particle (supplemental Movie SM3, available at www.jneurosci.org as supplemental material) (see also Rook et al., 2000). We analyzed transport velocities for 21 individual RNA particles independently of their direction. Sept7, similarly to CaMKIIα RNA and Stau1, traveled with an average transport velocity of 0.43 ± 0.23 μm/s.
Figure 4.
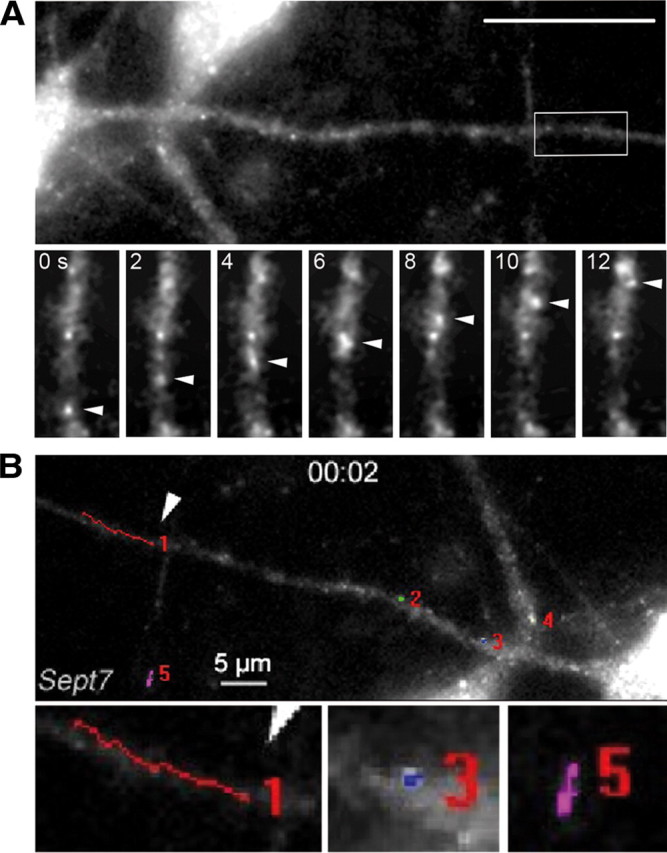
Analysis of Sept7 RNA kinetics. A, Sept7 RNA particles can exhibit fast directed movement into dendrites of hippocampal neurons. Dendrite of a cultured hippocampal neuron after microinjection with Alexa-labeled Sept7 RNA. Below are images from a time-lapse video showing the movement of a Sept7 RNA particle (arrowheads) in the distal dendrite (boxed). The time span between panels corresponds to 2 s (supplemental Movie SM3, available at www.jneurosci.org as supplemental material). B, Individual Sept7 particles were tracked manually. We detected directed movement along dendrites (see particle 1). The majority of dendritic Sept7 particles either appear stationary (see particles 2, 3, 4) or seem to display diffusion (see particle 5). Scale bar, 10 μm.
Differential sorting of dendritically localized RNAs into distinct particles
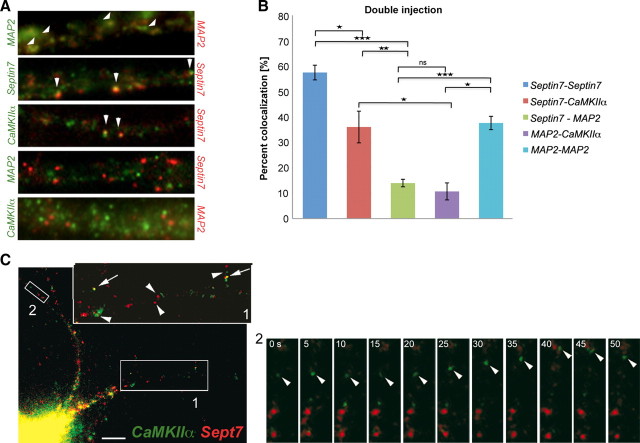
A major unresolved question is whether different localized RNAs are sorted into the same or into distinct RNPs. We focused on the following transcripts: MAP2, CaMKIIα, and Sept7. As a first control, we coinjected Sept7 RNA labeled by either Alexa 488 or Alexa 546. In that case, 36.5% of observed particles contained both Alexa 488- and Alexa 546-labeled Sept7 RNA, or 57.5% of Alexa 488-labeled particles also contained Alexa 546-labeled RNA (Fig. 5A,B). Subsequently, we coinjected Sept7 and CaMKIIα RNAs into mature neurons. We used the first part of the CaMKIIα 3′-UTR that exhibits a comparable localization pattern to full-length CaMKIIα 3′-UTR (CaMKIIα 1H) (Fig. 1B). As expected, both RNAs were transported into the dendritic compartment (Fig. 5A,C, and boxed insets). In the time series [supplemental Movie SM4 (available at www.jneurosci.org as supplemental material) from Fig. 5C, inset 2], a CaMKIIα 1H RNA particle is transported anterogradely in a distal dendrite, whereas several surrounding Sept7 RNA particles remain stationary. This result originally indicated that these two RNAs can be transported independently from each other into dendrites. We then quantified the colocalization of dendritic particles in six coinjected neurons and found that 21.0% of dendritic particles contained both CaMKIIα and Sept7 RNAs, or 36.0% of Sept7 particles also contained CaMKIIα RNA (Fig. 5A,B). In contrast, only 6.3% of the particles were positive for both MAP2 3′-UTR and Sept7 in seven coinjected neurons, or 13.4% of Sept7 particles contained MAP2 3′-UTR (Fig. 5A,B). In conclusion, our experiments lead us to the hypothesis that some localized RNAs that possibly share common sorting signals (e.g., CaMKIIα and Sept7 RNA) can be sorted into the same particles to dendrites, whereas other RNAs that are localized via a different pathway (e.g., MAP2 RNA) are transported in distinct particles to dendrites. It has been shown before that CaMKIIα RNA can reach more distal dendritic sites in the hippocampus than MAP2 (Paradies and Steward, 1997), suggesting that the two transcripts are targeted to dendrites by different localization signals. In consistence to this, we observe low colocalization rates between coinjected CaMKIIα 1H and MAP2 3′-UTRs, with only 10.7% of CaMKIIα particles containing MAP2 RNA (Fig. 5A,B). To exclude that the differential sorting we observe is an artifact introduced by the injection of a high excess of RNA, we confirmed the findings of the double injection experiments with double ISH against the endogenous transcripts (Fig. 6). The observed colocalization ratios for all three pairs of endogenous RNAs (CaMKIIα–Sept7, Sept7–MAP2, CaMKIIα– MAP2) were lower than the ones for the coinjected RNAs, as expected and reported previously for other transcripts (Gao et al., 2008). It is clear, however, that endogenous Sept7 preferentially colocalizes with CaMKIIα (in 16 cells analyzed, 11.8% of Sept7 particles contain CaMKIIα) than with MAP2 RNA (6.4% of Sept7 particles from 19 analyzed cells contain MAP2 RNA). Consistently, we observe that only a small number of MAP2 particles contain CaMKIIα RNA (6.7% of particles from 15 cells), suggesting that the two transcripts are transported to dendrites by independent pathways.
Figure 5.
Differential sorting of microinjected dendritic transcripts in neuronal RNPs. A, Coinjection of Alexa 546- and Alexa 488-labeled MAP2 3′-UTR–MAP2 3′-UTR, Sept7–Sept7, Sept7–CaMKIIα 1H 3′-UTR, Sept7–MAP2 3′-UTR, and MAP2 3′-UTR–CaMKIIα 1H 3′-UTR pairs. B, Quantification of the double-injection results. Colocalization ratios of Sept7–Sept7, Sept7–CaMKIIα 1H 3′-UTR, Sept7–MAP2 3′-UTR, MAP2 3′-UTR–CaMKIIα 1H 3′-UTR, and MAP2 3′-UTR–MAP2 3′-UTR were 57.48 ± 2.89, 36.04 ± 6.25, 13.98 ± 1.49, 10.69 ± 3.35, and 37.63 ± 2.63%, respectively. Error bars represent the SEM of colocalization ratios in individual cells. The asterisks denote statistical significance as determined using the two-tailed Student's t test for the complete data set. *p < 0.05; **p < 0.01; ***p < 0.001. C, Coinjection of Sept7 RNA (Alexa 546) and CaMKIIα RNA (Alexa 488; the first part of the 3′-UTR, 1H) (Fig. 1) into a representative hippocampal neuron. The arrowheads indicate particles in the enlargement (top right) of box 1 that contain either Sept7 or CaMKIIα RNA; the arrows denote colocalization of Sept7 and CaMKIIα RNA in a single particle. Movement of an individual particle found in box 2 is shown in supplemental Movie SM4 (available at www.jneurosci.org as supplemental material). The time span between individual frames is 5 s.
Figure 6.
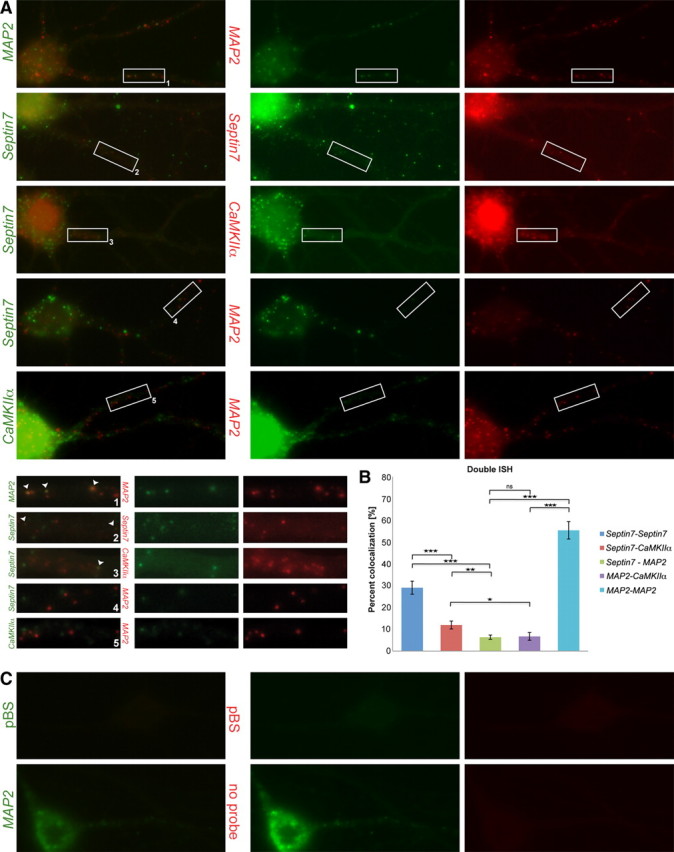
Differential sorting of endogenous dendritic transcripts in neuronal RNPs. A, Double detection with Alexa 488 and Cy3 tyramide of endogenous MAP2–MAP2, Sept7–Sept7, Sept7–CaMKIIα, Sept7–MAP2, and CaMKIIα–MAP2 pairs by fluorescence ISH. B, Quantification of the double ISH results. Colocalization ratios of Sept7–Sept7, Sept7–CaMKIIα, Sept7–MAP2, MAP2–CaMKIIα, and MAP2–MAP2 were 29.09 ± 3.00, 11.96 ± 1.82, 6.37 ± 0.98, 6.74 ± 1.77, and 55.75 ± 4.02%, respectively. Error bars represent the SEM of colocalization ratios in individual cells. The asterisks denote statistical significance as determined using two-tailed Student's t test for the complete data set. C, Double ISH specificity control with pBluescript sequences used labeled with DIG and fluorescein probes (top panel). Negative control for cross-reactivity. Cell hybridized with a fluorescein probe against MAP2 RNA shows signal only using anti-fluorescein-HRP tyramide detection and not with the subsequent anti-DIG-HRP tyramide detection (bottom panel).
Discussion
In this study, we demonstrate that in vitro-transcribed, microinjected fluorescently labeled RNAs form transport-competent particles that become sorted into dendrites, in a manner analogous to the corresponding endogenous RNAs. Furthermore, despite the excess of injected transcripts over the endogenous counterparts and the cytoplasmic injections bypassing nuclear processes (Ainger et al., 1993; Muslimov et al., 1997) (for review, see Wharton, 2009), the neurons retain their capacity to distinguish between dendritic and somatic transcripts, restricting the latter to the cell body compartment. In addition, our microinjection approach allowed us to identify a novel dendritically localized RNA, Sept7, and characterize its transport characteristics in detail in hippocampal neurons.
Microinjection of RNA into different cell types (e.g., Drosophila oocytes and embryos, oligodendrocytes, and primary neurons) has yielded important new insights (Ainger et al., 1993; Wilkie and Davis, 2001). In their landmark paper in 1993, Carson and coworkers (Ainger et al., 1993) were the first to observe the intracellular movement of MBP mRNA in oligodendrocytes. Interestingly, they described several distinct modes of granule movement in oligodendrocytes: (1) granules in the processes undergoing sustained directional movement with a velocity of ∼0.2 μm/s, (2) granules at branch points showing oscillatory motion with a mean displacement of 0.1 μm/s, and (3) granules in the periphery of the cell circulating randomly with a mean displacement of ∼1 μm/s. Based on their data, they proposed a multistep model for transport and localization [see also the accompanying News & Views by Wilhelm and Vale (1993)]. The first ones who successfully applied microinjection of 35S-labeled RNAs to primary rat sympathetic neurons were Muslimov and Tiedge in 1997 (Muslimov et al., 1997). They generated chimeric RNAs containing fragments of a dendritically localized, noncoding RNA, BC1, and revealed that a 5′-BC1 segment of 62 nt was delivered to distal dendrites with an average dendritic delivery rate of 242 ± 25 μm/h (0.07 μm/s), as shown by a series of sympathetic neurons that were fixed at different time points after microinjection and subjected to autoradiography.
How do these observed transport rates compare with our reported results? As mentioned above, we detected motile CaMKIIα particles with an average velocity of 0.39 ± 0.24 μm/s (Fig. 3D; supplemental Movie SM1, available at www.jneurosci.org as supplemental material); the fastest transport velocities were >1.2 μm/s. Whereas our data concur within the measured velocities of Carson and coworkers, the corresponding values in rat sympathetic neurons were approximately sixfold slower than ours, since Muslimov and Tiedge were assessing the overall distribution of RNA in neurons and not that of individual moving granules.
Furthermore, the kinetics of our CaMKIIα granules is in the same range as those recently reported that identified bidirectional movement of 0.13 μm/s of the MS2-GFP-tagged CaMKIIα 3′-UTR in hippocampal neurons (Rook et al., 2000), of 0.12 μm/s of SYTO14-labeled RNA granules in hippocampal neurons (Knowles and Kosik, 1997), of 0.08 μm/s of endogenous Arc RNA in the dentate gyrus determined by ISH (Wallace et al., 1998), of up to 1.1 μm/s of MS2-GFP-tagged Arc RNA in cortical neurons (Dynes and Steward, 2007), of 1.25 μm/s maximum velocities of injected, Alexa-labeled pair rule RNAs in Drosophila embryos (Wilkie and Davis, 2001), and up to 3 μm/s in fibroblasts (GFP-MS2) (Fusco et al., 2003).
In a number of previous studies, maximum velocities of 0.1–0.3 μm/s have been measured for GFP fusions of RBPs in hippocampal neurons (Köhrmann et al., 1999; Rook et al., 2000; Kanai et al., 2004). Our 5- to 10-fold faster transport rates compared with our own previous data (Köhrmann et al., 1999) as well as of recent data by Bassell and coworkers (Dictenberg et al., 2008) are most likely attributable to the higher frame rate that has been applied. We speculate that this enabled us to monitor faster particles that previously escaped detection, thereby yielding significantly faster transport rates.
What are the main advantages of this technique over other methods such as ISH or transfection of exogenous reporters? Microinjection of RNA in neurons is by no means advantageous over ISH in investigating the localization patterns and the steady-state levels of RNA in dendrites, as long as endogenous RNA levels are high enough to obtain specific ISH signals. It is also certainly more laborious and therefore probably not suitable for large-scale screens for the identification of novel dendritically localized RNAs. Transfection of truncated reporters followed by ISH has been successfully used to identify dendritic targeting elements in RNAs (e.g., CaMKIIα or MAP2 RNA in primary neurons) (Blichenberg et al., 1999, 2001). However, it does not allow the analysis of particle dynamics. The injection assay, in contrast, allows us to visualize the assembly of individual RNA particles in real time and to assess the kinetics of RNA transport in living neurons. Since the average and maximum velocities of transport determined in our experiments were higher than previously reported for MS2-GFP CaMKIIα (see above), this points toward motor-dependent transport and therefore nicely complements a recent study on the role of kinesin in neurons (Kanai et al., 2004). It was beyond the scope of this study to investigate this dependence in more detail. We feel that microinjection of fluorescently labeled RNA will be instrumental in the future in further dissecting the molecular machinery underlying dendritic targeting of RNAs, as it can reveal subtle effects of regulatory factors that ISH might fail to detect.
The MS2 system has the obvious advantage of tagging endogenous—yet overexpressed—RNA, and in this respect it is preferable over microinjection. It cannot be easily used, however, for dual-color imaging of RNA and proteins in neurons, as it would either require simultaneous transfection of single neurons with at least three plasmids (a feature that we found to be very difficult to achieve) or with a multipromoter plasmid. Another limitation of the MS2 system is that it cannot be applied for the simultaneous detection of two transcripts. The λN-GFP system, recently established in NRK cells (Daigle and Ellenberg, 2007), might overcome those limitations when successfully applied to primary neurons. Improvement of the sensitivity of the assay will also be necessary, as it does currently not allow the visualization of RNA particles, but rather yields diffuse signal in the cytoplasm.
Our direct labeling approach of RNAs enabled us to perform dual-color imaging of RNAs in primary neurons. Coinjection of two differently labeled transcripts in neurons, as well as double detection of the respective endogenous transcripts, allowed us to covisualize two RNAs and address whether different mRNAs are transported in the same or distinct RNPs to dendrites. We provide quantitative evidence that the dendritically localized CaMKIIα and MAP2 RNAs rarely colocalize in the same dendritic RNA granules. This result confirms data by Paradies and Steward (1997), who observed a different localization pattern of these RNAs in the hippocampus. We observe a low degree of colocalization between Sept7 and CaMKIIα RNA. It appears, however, that their association is rather transient and/or restricted at proximal sites, since we fail to detect the two transcripts being cotransported along dendrites.
Our work, together with a recent study (Gao et al., 2008), demonstrating the coassembly of CaMKIIα, Neurogranin, and Arc RNAs in hnRNP A2-containing RNPs, covisualize different dendritic transcripts at the level of individual particles and investigate their degree of association. Our findings suggest differential sorting of neuronal RNAs, arguing for independent dendritic trafficking pathways. Our approach will also have applications in dual imaging of micro-RNAs with their target mRNAs, as we initially attempted (Schratt et al., 2006). Future dual-color imaging of injected RNA and P-body or transport RNP protein markers will give insight into the interplay between translational regulation/RNA storage or degradation and RNA transport, under conditions of synaptic activity, translational silencing, or stress (Kiebler and Bassell, 2006).
Footnotes
This work was supported by a predoctoral fellowship from the Boehringer Ingelheim Fonds (F.T.), two Austrian Science Funds (P.M., M.A.K.), and grants from SFB446 (University of Tübingen, Tübingen, Germany), Hertie-Stiftung, Schram-Stiftung im Stifterverband der Deutschen Wissenschaft, the European Science Foundation Program RNAQuality, two Human Frontier Science Program networks, and the Max Planck Institute for Developmental Biology (all to M.A.K.). We thank Barbara Grunewald, and Drs. John Carson, Ralf Dahm, Jim Deshler, Bernhard Goetze, Rudolf Kern, Dietmar Kuhl, Massimo Mallardo, Verena Meyer, Xiasong Mo, Oswald Steward, John Vessey, Yunli Xie, and Manuel Zeitelhofer for advice or assistance in experiments; Drs. Claudia Bagni, Jim Deshler, and Oswald Steward for plasmids; Jacki Heraud, Ilham Muslimov, and Alexandre Raposo for critically reading this manuscript; and Sebastian Butter for assistance in preparing the figures. We are very thankful to Dr. Friedrich Bonhoeffer for gracious support and advice.
References
- Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci. 1999;19:8818–8829. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Müller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol. 2008;18:745–750. doi: 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumwell C, Antolik C, Carson JH, Barbarese E. Intracellular trafficking of hnRNP A2 in oligodendrocytes. Exp Cell Res. 2002;279:310–320. doi: 10.1006/excr.2002.5604. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods. 2007;4:633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- Goetze B, Grunewald B, Baldassa S, Kiebler M. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: application for transfection of mature hippocampal neurons. J Neurobiol. 2004;60:517–525. doi: 10.1002/neu.20073. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Kosik KS. Neurotrophin-3 signals redistribute RNA in neurons. Proc Natl Acad Sci U S A. 1997;94:14804–14808. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl D, Skehel P. Dendritic localization of mRNAs. Curr Opin Neurobiol. 1998;8:600–606. doi: 10.1016/s0959-4388(98)80087-1. [DOI] [PubMed] [Google Scholar]
- MacDougall N, Clark A, MacDougall E, Davis I. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell. 2003;4:307–319. doi: 10.1016/s1534-5807(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Müller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci U S A. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies MA, Steward O. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J Neurobiol. 1997;33:473–493. doi: 10.1002/(sici)1097-4695(199710)33:4<473::aid-neu10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIα 3′-untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, Aparicio SA, Abel T, Kiebler MA. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci U S A. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace CS, Lyford GL, Worley PF, Steward O. Differential intracellular sorting of immediate early gene mRNAs depends on signals in the mRNA sequence. J Neurosci. 1998;18:26–35. doi: 10.1523/JNEUROSCI.18-01-00026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP. A splicer that represses (translation) Genes Dev. 2009;23:133–137. doi: 10.1101/gad.1768509. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, Kiebler MA. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M, Vessey JP, Xie Y, Tübing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protoc. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]