Abstract
Previous studies have supported the concept that the default network is an intrinsic brain system that participates in internal modes of cognition. Neural activity and connectivity within the default network, which are correlated with cognitive ability even at rest, may be plausible intermediate phenotypes that will enable us to understand the genetic mechanisms of individuals' cognitive function or the risk for genetic brain diseases. Using resting functional magnetic resonance imaging and imaging genetic paradigms, we investigated whether individual default network connectivity was modulated by COMT val158met in 57 healthy young subjects. Compared with COMT heterozygous individuals, homozygous val individuals showed significantly decreased prefrontal-related connectivities, which primarily occurred between prefrontal regions and the posterior cingulate/restrosplenial cortices. Further analyses of the topological characteristics of the default network showed homozygous val individuals had significantly fewer node degrees in the prefrontal regions. This finding may partially elucidate previous reports that the COMT val variant is associated with inefficient prefrontal information processing and poor cognitive performance. Our findings suggest that default network connectivity that involves the prefrontal cortex is modulated by COMT val158met through differential effects on prefrontal dopamine levels.
Introduction
Studies have identified a variety of resting-state networks (RSNs) (Fox and Raichle, 2007). As one of the more interesting RSNs, the brain's default network appears to contribute directly to an internal mentation that is largely detached from the external world, including self-reflective thoughts and judgments, perception of the mental states of others, envisioning the future to conceive alternatives, etc. (Buckner et al., 2008). Additionally, researchers have found that the activity of the default network is damaged in some neuropsychiatric diseases (Broyd et al., 2009). Together, these findings indicate that the intrinsic activity of the default network could play an important role in human cognitive functions.
Neural connections directly reflect the functional activities of the brain. By comparing activation patterns and resting-state brain networks, a recent study suggested a close correspondence of the brain's functional architecture during activation and rest (Smith et al., 2009). Functional connectivity (FC) can be used to explore direct or indirect interactions between brain regions (Friston et al., 1993). The behavioral significance of BOLD FC has been investigated in studies that not only used specific tasks (Hampson et al., 2006; He et al., 2007) but also studied the brain at rest (Seeley et al., 2007; Song et al., 2008). Specifically, some studies have reported that the strength of FC within the default network is correlated with cognitive performance even at rest (Hampson et al., 2006; Sambataro et al., 2008). Therefore, neural activity and connectivity within the default network may be plausible intermediate phenotypes that are helpful in understanding the genetic mechanisms of cognitive function or the risk for genetic brain diseases. Specifically, one recent study (Filippini et al., 2009) reported that distinct patterns of resting brain activity within the default network in young carriers of the APOE-ε4 allele can be detected before any neurophysiological expression of neurodegenerative processes. This suggests that the activity of some brain regions within the default network may be affected by individual genetic variants. So, an interesting question is whether we can identify modulations in the default network connectivity as being caused by specific genetic variants that we studied.
Cathechol-O-methyltransferase (COMT) is a key enzyme which may account for >60% of the dopamine degradation in the prefrontal cortex (PFC) and thus plays a unique role in regulating dopamine levels in the PFC (Karoum et al., 1994). The COMT gene contains a common functional polymorphism in codon 158 (val158met), in which the substitution of methionine (met) for valine (val) can result in three- to four-times-lower thermostability and activity of the COMT enzyme (Männistö and Kaakkola, 1999). Previous studies have demonstrated that the high-activity val allele is associated with “inefficient” prefrontal cortical processing and poorer performance (Egan et al., 2001; Goldberg et al., 2003); however, the specific mechanism remains unclear. In this study, using resting functional magnetic resonance imaging (fMRI) and imaging genetic paradigms, we investigated our hypothesis that COMT val158met impacts the FCs of the prefrontal regions within the default network and thus affects prefrontal function and cognitive performance.
Materials and Methods
Subjects
Fifty-seven healthy, young, right-handed subjects (mean age: 24.7 ± 3.6 years, range: 18.5–33.0; 29 males) were included in this study. All subjects were recruited by advertisement and gave written informed consent. This study was approved by the ethical committee of Xuanwu Hospital of Capital Medical University.
COMT val158met genotyping
We extracted genomic DNA from 250 μl of whole blood using a DNA direct kit (Omega Bio-tek). COMT val158met was genotyped using PCR and restriction digestion techniques as previously described (Qian et al., 2003).
MRI data acquisition and preprocessing
MR imaging was performed using a 3.0 tesla MR scanner (Magnetom Trio, Siemens). Functional images were collected axially using an echo-planar imaging (EPI) sequence sensitive to BOLD contrast. The acquisition parameters were as follows: 32 slices, 2000/30 ms (TR/TE), 3.0/1.0 mm (thickness/gap), 220 × 220 mm (FOV), 64 × 64 (resolution within slice), 90° (flip angle). During the resting-state scanning, the subjects were instructed to keep still with their eyes closed and as motionless as possible and not to think about anything in particular. For each subject, the fMRI scan during the resting state lasted for 9 min and provided 270 volumes.
Several preprocessing steps were used on this fMRI data, including (1) correcting for within-scan acquisition time differences between slices; (2) realigning the volumes to the first volume to correct for interscan movements; (3) spatially normalizing to a standard EPI template and making a resample; (4) spatially smoothing; (5) performing linear regression to remove the influence of head motion, whole-brain signals, and linear trends; and (6) temporal bandpass filtration (0.01–0.08 Hz). We used SPM2 (http://www.fil.ion.ucl.ac.uk/spm/software/spm2/) to carry out preprocessing steps 1–4 and in-house software for steps 5 and 6. Specifically, the parameters obtained during movement correction showed that the maximum displacement in each cardinal direction (x–z) was not >1 mm, and the maximum spin (x–z) was not >1° for each participant.
Multiple region-of-interest-based individual default network functional connectivity graphs
Region-of-interest definition.
In the present study, we used the same regions of interest (ROIs) to define the default network as in previous studies (Fair et al., 2008; Song et al., 2009). The coordinates of these 13 ROIs are shown in Table 1. All ROIs were defined as a spherical region with a radius of 6 mm at the center of the obtained coordinates of a specific ROI. Since the size of each voxel in the present study was 3 × 3 × 3 mm, each ROI was comprised of 33 voxels. (For more detail about the ROI definition, see the supplemental text and supplemental Fig. 1, available at www.jneurosci.org as supplemental material.)
Table 1.
Locations of the ROIs of the default network
| Brain region | MNI coordinates | Talaraich coordinates | BA |
|---|---|---|---|
| aMPFC | −3, 54, 18 | −3, 54, 15 | 10/9 |
| L.Sup.F | −15, 54, 42 | −15, 54, 36 | 9 |
| R.Sup.F | 18, 42, 48 | 18, 42, 42 | 8 |
| vMPFC | −6, 36, −9 | −6, 36, −9 | 32/10/11 |
| L.IT | −60, −9, −24 | −60, −9, −21 | 20/21 |
| R.IT | 57, 0, −27 | 57, 0, −24 | 21/20 |
| L.PHC | −24, −18, −27 | −24, −18, −21 | 35 |
| R.PHC | 27, −18, −24 | 24, −18, −18 | 36/35 |
| PCC | −3, −48, 30 | −3, −45, 30 | 31 |
| Rsp | 9, −54, 12 | 9, −51, 15 | 30 |
| L.LatP | −48, −69, 39 | −48, −66, 39 | 39 |
| R LatP | 48, −66, 36 | 48, −63, 36 | 39 |
| Cereb | −6, −54, −48 | −6, −54, −39 |
All ROIs were defined as a spherical region with a radius of 6 mm at the center of the above seed regions, and then each ROI was comprised of 33 voxels. aMPFC, medial prefrontal cortex (anterior); L.Sup.F, left superior frontal cortex; R.Sup.F, right superior frontal cortex; vMPFC, medial prefrontal cortex (ventral); L.IT, left inferior temporal cortex; R.IT, right inferior temporal cortex; L.PHC, left parahippocampal gyrus; R.PHC, right parahippocampal gyrus; PCC, posterior cingulate cortex; Rsp, retrosplenial; L.LatP, left lateral parietal cortex; R.LatP, right lateral parietal cortex; Cereb, cerebellar tonsils; MNI, Montreal Neurological Institute; BA, Brodmann's area.
Individual functional connectivity graph.
After extracting the 13 ROIs for each subject, we computed the functional connectivity between each pair of the 13 ROIs. The functional connectivity was generated by averaging the BOLD time series separately in the two regions and then computing the Pearson's correlation coefficient between the two averaged time series. The resulting correlation was then transformed to approximate a Gaussian distribution using Fisher's r-to-z transformation. Thus, for each subject, we obtained a 13 × 13 matrix, with each element representing the strength of the functional connectivity between the corresponding two brain regions within the default network.
In the present study, using a one-sample t test, we found that all the functional connectivities between any pair of brain regions within the default network were significantly greater than 0 at a group level [p < 0.05, false discovery rate (FDR) corrected]. In addition, although we found that some of the individual functional connectivities in some subjects were negative, the proportion of the negative functional connectivities was <5% of all the functional connectivities from the entire group. To adopt the node degree so that we could investigate the total connections of each ROI within the default network, we set the negative functional connectivity to 0. This allowed us to use an undirected weighted graph to model the default network. That is, each node of the graph was used to denote a brain region within the default network, and the weight of the edge between any two nodes represented the z-valued strength of the functional connectivity between the two corresponding brain regions. Thus, we constructed a complete undirected weighted graph to model the topology of the default network for each subject.
The degree si of a node i is the number of edges linking to the node, and is defined as follows:
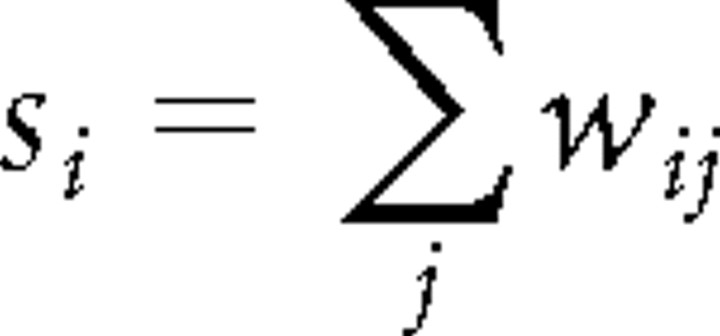 |
where wij denotes the weighted edge that connects node i and node j, that is, in the present study, the z-valued strength of the functional connectivity between brain region i and brain region j. The degree si can be used to qualify the extent to which a given node is central in the graph. From the node degree, we can define hub nodes, which are nodes with high degrees in a graph.
Voxel-based individual default network functional connectivity graphs
To validate our findings, we separately extracted some brain areas as seed regions and then performed voxel-based whole-brain functional connectivity analyses. These regions included the medial PFC (ventral) (vMPFC), the posterior cingulate cortex (PCC), the striatum, and the primary visual area (PVA). The vMPFC and the PCC, two important nodes within the default network, were selected to validate our ROI-based results. We also selected the striatum and the PVA, two regions that are outside the default network, to determine whether or not the FCs from these areas are affected by the COMT polymorphism. The functional connectivity analysis was performed separately for each of the four seed regions. For more details, see the supplemental text, available at www.jneurosci.org as supplemental material.
Statistical analyses
A Pearson χ2 test was used to test for differences in gender and education level and a two-sample t test was used to test for differences in age between the two genotype groups. We used a two-sample t test to examine differences in the strength of connectivity and node degree between the two genotype groups. We used a permutation-based correction for multiple comparisons using Ptest software by 10,000 permutations (Westfall and Young, 1993; Belmonte and Yurgelun-Todd, 2001; Conneely and Boehnke, 2007; Camargo et al., 2008; Sowell et al., 2008). For each seed region in whole-brain analyses, we performed a two-sample t test to examine the significance of the differences in the voxel-based default network between val-homozygous and heterozygous individuals.
Results
We categorized 57 young healthy subjects according to their COMT genotypes, into 26 heterozygous (met/val) and 31 val-homozygous (val/val) individuals. The two groups showed no significant differences in age, gender, and education level (Table 2).
Table 2.
Subject demographics for the two genotype groups
| Met/Val | Val/Val | Difference (P value) | |
|---|---|---|---|
| n | 26 | 31 | |
| Gender (male:female ratio) | 13:13 | 16:15 | 0.903 |
| Age (mean ± SD, years) | 25.2 ± 3.3 | 24.2 ± 3.8 | 0.329 |
| Age range (years) | 18.5–33.3 | 19.5–31.5 | |
| Education (1:2:3 ratio) | 9:4:13 | 8:13:10 | 0.091 |
| Handedness (% right) | 100% | 100% |
Education level was determined on a discrete scale with three levels: low = 1, middle = 2, high = 3.
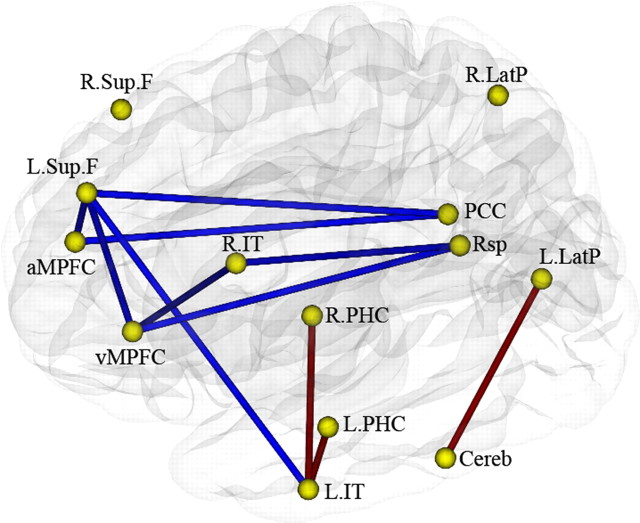
Using the multiple ROI-based connectivity analysis, we obtained an undirected weighted network with 13 nodes and 78 edges that globally described the default network connectivity pattern for each subject. As can be seen in the mean connectivity maps of the two genotype groups, we found much less connectivity between multiple regions in val homozygotes than in COMT heterozygotes (supplemental text and supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Then we examined the group differences in the default network connectivity for the two different COMT genotypes. Compared with the heterozygous individuals, the val homozygotes had significantly decreased connectivities. Two of the decreases were within the prefrontal lobules [L.Sup.F–aMPFC, L.Sup.F–vMPFC]; five of the other six decreases were between the prefrontal lobules and other regions [L.Sup.F–PCC, aMPFC–PCC, vMPFC–Rsp, vMPFC–R.IT, L.Sup.F–L.IT], and only one decreased connectivity was unrelated to the PFC (R.IT–Rsp) (Fig. 1; supplemental Table 1, available at www.jneurosci.org as supplemental material). Specifically, the decreased connectivities were primarily concentrated in the connectivities between the prefrontal regions and the PCC/Rsp. Three increased connectivities were also found between the L.IT and the L.PHC between the L.IT and the R.PHC, and between the L.LatP and the Cereb.
Figure 1.
Significantly different functional connectivity in the default network in COMT Val-homozygous and heterozygous groups. The blue line indicates decreased and the red line indicates increased connectivity in Val-homozygous individuals compared with the heterozygous group (p < 0.05, permutation-based multiple-comparison correction).
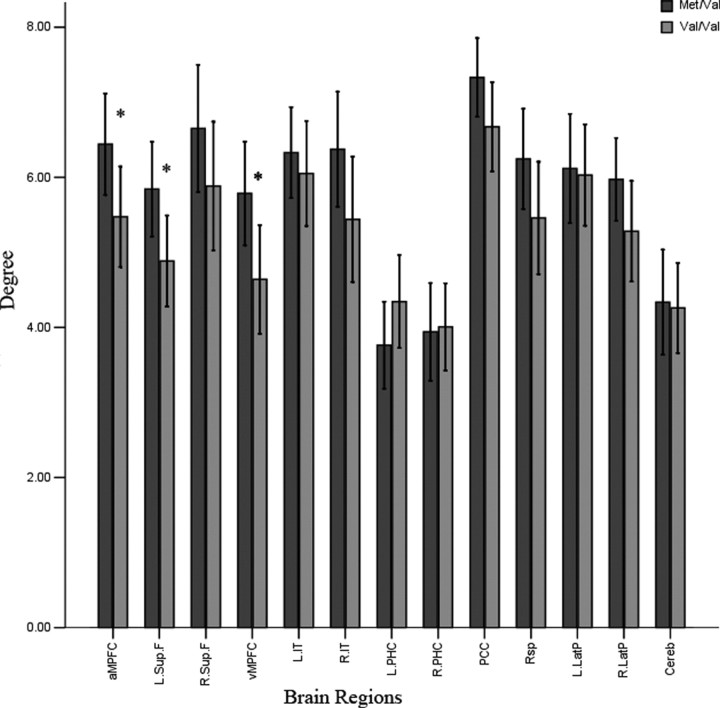
For each individual's connectivity graph of the default network, we also computed the degree of each brain region. Then we tested the differences in the connectivity degree of each region between individuals who were COMT heterozygous and those who were val homozygous (Fig. 2). Those who were val homozygous showed significantly lower degrees of connectivity than did those who were COMT heterozygous in the vMPFC, aMPFC, and L.Sup.F (p < 0.05, permutation-based multiple-comparison correction). Thus we demonstrated lower degrees of connectivity in three of the four prefrontal regions in the default network. We did not find any significantly increased node degree in any of the 13 ROIs.
Figure 2.
Degrees of connectivity of 13 brain regions in the two different COMT genotype groups. *Significant differences in the two groups (p < 0.05, permutation-based multiple-comparison correction).
The whole-brain analyses obtained by taking vMPFC as the seed region revealed that val homozygotes had significantly lower connectivity than COMT heterozygotes in the aMPFC, PCC/Rsp, and R.IT (supplemental text, available at www.jneurosci.org as supplemental material; Fig. 3). In addition, we found no significantly increased connectivities. Similar analyses in which we took the PCC as a seed region also found decreased connectivity in an area located in the aMPFC region (supplemental text and supplemental Fig. 3, available at www.jneurosci.org as supplemental material). We also tested the differences in the resting-state functional connectivity of the striatum and the PVA, and did not find any significantly different connectivity between the COMT heterozygotes and the val homozygotes.
Figure 3.
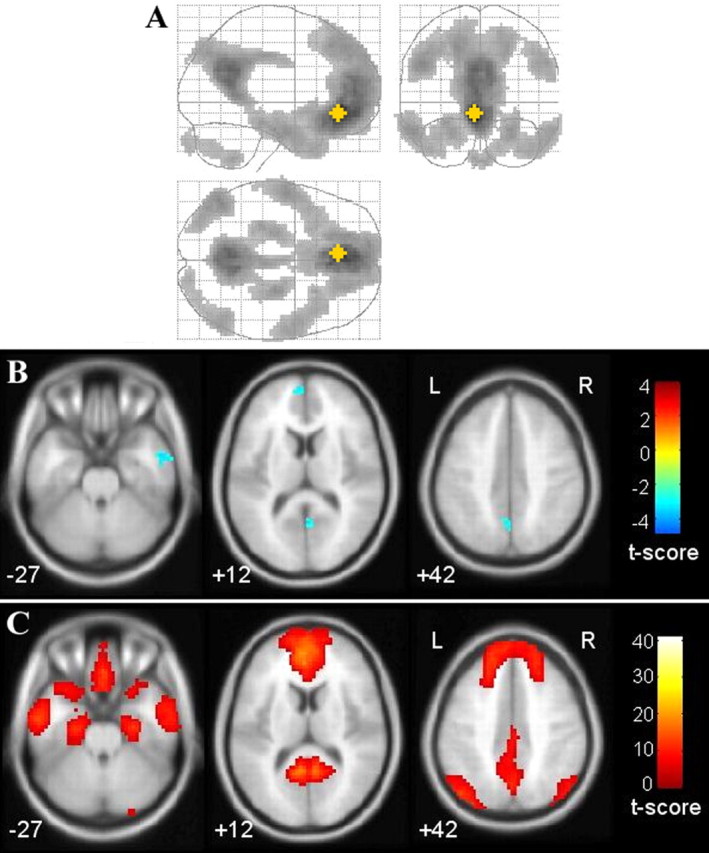
Significantly different connectivities in two genotype groups resulting from a voxel-based functional connectivity analysis, taking the vMPFC as the seed region. A, Overall default network illustration, in which the yellow star represents the position of the vMPFC seed region (1-sample t test, df = 56; p = 0.01, FDR corrected; cluster size = 20 voxels). B, Significantly decreased connectivity in the COMT val homozygotes compared with the heterozygotes (2-sample t test; p = 0.005; cluster size = 20 voxels). C, The corresponding default network in the same layer with B.
Discussion
This study investigated the potential role of COMT val158met in modulating default network connectivity in healthy young subjects. Using the multiple ROI-based resting functional connectivity approach, we found that, compared with heterozygous individuals, COMT val homozygotes showed significantly decreased prefrontal-related connectivities, which primarily occurred between prefrontal regions and the posterior cingulate/restrosplenial cortices (Fig. 1). Further analyses of the topological characteristics of the default network showed homozygous val individuals had significantly fewer node degrees in three (vMPFC, aMPFC, and L.Sup.F) of the four prefrontal regions (Fig. 2).
In this study, our major finding is that the default network connectivities, especially the connectivities within the prefrontal lobes and the ones from the prefrontal lobes to the posterior regions, were modulated by the COMT functional genetic variation. Evidence from anatomical and functional studies of the default network has demonstrated that the brain's default network can be distinguished from other brain systems by the fact that it is an intrinsic brain system that participates in internal modes of cognition (Buckner et al., 2008). Moreover, disrupted default network activity or connectivity has been extensively reported in various brain diseases, including Alzheimer's disease, schizophrenia, depression, ADHD, and so forth. (Broyd et al., 2009). However, to our knowledge only one study (Filippini et al., 2009) investigated the genetic basis for the neural functions of the default network. Additionally, evidence suggests that prefrontal dopamine plays a critical role in the normal cognitive process, especially in working memory, planning and attention (Seamans and Yang, 2004). Some studies have found that dysfunctions in prefrontal DA may underlie neuropsychiatric pathologies, such as the cognitive deficits associated with schizophrenia and bipolar disorder (Egan et al., 2001; Seamans and Yang, 2004; Tunbridge et al., 2006; Goghari and Sponheim, 2008). Moreover, one recent study suggests a reciprocal interplay between working memory capacity and cortical dopamine neurotransmission (McNab et al., 2009). Additionally, the COMT enzyme encoded by the COMT gene influences dopamine degradation by inactivating released dopamine (Männistö and Kaakkola, 1999). COMT val158met, a common genetic polymorphism in humans, is associated with a three- to fourfold variation in COMT enzyme activity and is also associated with individual variation in COMT thermal instability (Lachman et al., 1996). The substitution of met for val results in a more sluggish COMT enzyme (Lotta et al., 1995; Lachman et al., 1996). Thus, the val variant of the COMT gene corresponds to lower dopamine levels in the prefrontal cortex. Previous studies indicated that the high-activity COMT val variant is associated with worse performance on working memory (Egan et al., 2001; Goldberg et al., 2003) and attention tasks (Blasi et al., 2005). In this study, we found decreased prefrontal-related connectivities within the default network in young COMT val-homozygous individuals using fMRI. We speculate that the val variant is associated with lower dopamine levels in the PFC and thus with decreased prefrontal-related FCs within the default network, thus causing poor cognitive performance (Barnett et al., 2008). In fact, several fMRI studies have reported that the strength of certain specific connectivities within the default network, in particular the FCs between the PCC and the MPFC, are positively correlated with working memory performance, not only during the task state, but also at rest (Hampson et al., 2006; Sambataro et al., 2008). Therefore, we suggest that the prefrontal dopamine availability modulated by the COMT genotype affects the prefrontally mediated cognitive function in healthy individuals, presumably by impacting prefrontal-related functional integration within the default network.
The COMT gene is expressed in many regions of the human CNS (Hong et al., 1998; Matsumoto et al., 2003). However, COMT plays a unique role only in regulating the dopamine system of the prefrontal cortex. This is apparently because the prefrontal cortex contains less dopamine transporter protein than other brain regions and is thus more dependent on the COMT enzyme for terminating the action of released dopamine (Männistö and Kaakkola, 1999; Seamans and Yang, 2004). An earlier study showed that the COMT enzyme accounted for >60% of the dopamine degradation in the rat prefrontal cortex, but for <15% of the dopamine degradation in the striatum (Karoum et al., 1994). In fact, our study found that the functional genetic variation in the COMT gene disproportionately affects connectivities that involve the prefrontal cortex within the default network, leaving other brain regions, such as the striatum and the PVA, relatively unaffected.
From an imaging genetics perspective, neuronal connectivity may be regarded as a plausible neural intermediate phenotype and is attracting more and more attention in recent imaging genetic studies (Esslinger et al., 2009; Meyer-Lindenberg, 2009). These studies investigated the modulation by specific genetic variants of the activity and functional coupling of certain brain regions under specific cognitive tasks. In this study, we investigated resting-state functional connectivity within the default network and reported that some resting-state connectivities are affected by individual genetic variants.
In addition to the significant decreased functional connectivities, three increased connectivities (L.IT–L.PHC, L.IT–R.PHC, L.LatP–Cereb) were also found when we compared the val-homozygous with the heterozygous individuals. These increased connectivities were not located within the prefrontal cortex. We speculate that the increased connectivities in the val-homozygous group could possibly reflect a compensatory effect for the decreased prefrontal related connectivities or the recruitment of cognitive resources to maintain normal cognitive function. However, the mechanism of the compensatory reallocation remains unclear and deserves further study.
Our findings demonstrated that prefrontal-related connectivities within the default network were modulated by a COMT functional genetic variation, presumably because this polymorphism plays a unique role in regulating prefrontal dopamine levels (Egan et al., 2001). However, COMT val158met is not the only functional variation. Previous studies have reported that the COMT gene may contain at least three functional polymorphisms (rs737865, rs4680, rs165599) which impact its biological activity (Mukherjee et al., 2009). Other studies have shown that certain commonly found haplotypes may regulate the gene even more strongly than does val158met (Bray et al., 2003; Nackley et al., 2006). Thus, the effects of other several single nucleotide polymorphisms and their common haplotypes need further study. Moreover, since we speculate that dopamine levels influence default network connectivity, in future studies we also need to investigate whether and how other dopaminergic system related genes, such as the dopamine transporter gene (DAT) and dopamine receptor genes, modulate default network connectivity. In this way, we can more systematically investigate how the dopaminergic system affects the connectivity pattern of the default network. In addition, investigating the genetic control of neural connectivity within other brain systems or within the whole brain network should be an important research direction in the near future.
Footnotes
This work received support from the National Key Basic Research and Development Program (973), Grant 2007CB512305, the Natural Science Foundation of China (NSFC), Grant 30730035, the Excellent State Key Laboratory Project of NSFC, Grant 60723005, and the Program for New Century Excellent Talents in University, Grant NCET-07-0568. We thank the subjects involved in this study and their families. We also thank Drs. Edmund F. and Rhoda E. Perozzi for editing assistance.
References
- Barnett JH, Scoriels L, Munafò MR. Meta-analysis of the cognitive effects of the catechol-O-methyltransferase gene Val158/108Met polymorphism. Biol Psychiatry. 2008;64:137–144. doi: 10.1016/j.biopsych.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Belmonte M, Yurgelun-Todd D. Permutation testing made practical for functional magnetic resonance image analysis. IEEE Trans Med Imaging. 2001;20:243–248. doi: 10.1109/42.918475. [DOI] [PubMed] [Google Scholar]
- Blasi G, Mattay VS, Bertolino A, Elvevåg B, Callicott JH, Das S, Kolachana BS, Egan MF, Goldberg TE, Weinberger DR. Effect of catechol-O-methyltransferase val158met genotype on attentional control. J Neurosci. 2005;25:5038–5045. doi: 10.1523/JNEUROSCI.0476-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NJ, Buckland PR, Williams NM, Williams HJ, Norton N, Owen MJ, O'Donovan MC. A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am J Hum Genet. 2003;73:152–161. doi: 10.1086/376578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJ. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Camargo A, Azuaje F, Wang H, Zheng H. Permutation-based statistical tests for multiple hypotheses. Source Code Biol Med. 2008;3:15. doi: 10.1186/1751-0473-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, Haddad L, Mier D, Opitz von Boberfeld C, Raab K, Witt SH, Rietschel M, Cichon S, Meyer-Lindenberg A. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A. 2008;105:4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-e4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Frith CD, Liddle PF, Frackowiak RS. Functional connectivity: the principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993;13:5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- Goghari VM, Sponheim SR. Differential association of the COMT Val158Met polymorphism with clinical phenotypes in schizophrenia and bipolar disorder. Schizophr Res. 2008;103:186–191. doi: 10.1016/j.schres.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TE, Egan MF, Gscheidle T, Coppola R, Weickert T, Kolachana BS, Goldman D, Weinberger DR. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889–896. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- Hampson M, Driesen NR, Skudlarski P, Gore JC, Constable RT. Brain connectivity related to working memory performance. J Neurosci. 2006;26:13338–13343. doi: 10.1523/JNEUROSCI.3408-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BJ, Snyder AZ, Vincent JL, Epstein A, Shulman GL, Corbetta M. Breakdown of functional connectivity in frontoparietal networks underlies behavioral deficits in spatial neglect. Neuron. 2007;53:905–918. doi: 10.1016/j.neuron.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Hong J, Shu-Leong H, Tao X, Lap-Ping Y. Distribution of catechol-O-methyltransferase expression in human central nervous system. Neuroreport. 1998;9:2861–2864. doi: 10.1097/00001756-199808240-00033. [DOI] [PubMed] [Google Scholar]
- Karoum F, Chrapusta SJ, Egan MF. 3-Methoxytyramine is the major metabolite of released dopamine in the rat frontal cortex: reassessment of the effects of antipsychotics on the dynamics of dopamine release and metabolism in the frontal cortex, nucleus accumbens, and striatum by a simple two pool model. J Neurochem. 1994;63:972–979. doi: 10.1046/j.1471-4159.1994.63030972.x. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melén K, Julkunen I, Taskinen J. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- Männistö PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51:593–628. [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Akil M, Lipska BK, Hyde TM, Herman MM, Kleinman JE, Weinberger DR. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience. 2003;116:127–137. doi: 10.1016/s0306-4522(02)00556-0. [DOI] [PubMed] [Google Scholar]
- McNab F, Varrone A, Farde L, Jucaite A, Bystritsky P, Forssberg H, Klingberg T. Changes in cortical dopamine D1 receptor binding associated with cognitive training. Science. 2009;323:800–802. doi: 10.1126/science.1166102. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–1946. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Kidd KK, Pakstis AJ, Speed WC, Li H, Tarnok Z, Barta C, Kajuna SL, Kidd JR. The complex global pattern of genetic variation and linkage disequilibrium at catechol-O-methyltransferase. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.64. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Qian Q, Wang Y, Zhou R, Li J, Wang B, Glatt S, Faraone SV. Family-based and case-control association studies of catechol-O-methyltransferase in attention deficit hyperactivity disorder suggest genetic sexual dimorphism. Am J Med Genet B Neuropsychiatr Genet. 2003;118B:103–109. doi: 10.1002/ajmg.b.10064. [DOI] [PubMed] [Google Scholar]
- Sambataro F, Murty VP, Callicott JH, Tan HY, Das S, Weinberger DR, Mattay VS. Age-related alterations in default mode network: impact on working memory performance. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.05.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamans JK, Yang CR. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol. 2004;74:1–58. doi: 10.1016/j.pneurobio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Zhou Y, Li J, Liu Y, Tian L, Yu C, Jiang T. Brain spontaneous functional connectivity and intelligence. Neuroimage. 2008;41:1168–1176. doi: 10.1016/j.neuroimage.2008.02.036. [DOI] [PubMed] [Google Scholar]
- Song M, Liu Y, Zhou Y, Wang K, Yu CS, Jiang TZ. Default network and intelligence difference. IEEE Trans Auton Ment Dev. 2009;1:101–109. doi: 10.1109/IEMBS.2009.5334874. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Kan E, Yoshii J, Thompson PM, Bansal R, Xu D, Toga AW, Peterson BS. Thinning of sensorimotor cortices in children with Tourette syndrome. Nat Neurosci. 2008;11:637–639. doi: 10.1038/nn.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-o-methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol Psychiatry. 2006;60:141–151. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing. New York: Wiley; 1993. [Google Scholar]