Abstract
Activity-dependent changes in synaptic structure and spine morphology are required for learning and memory, and depend on protein translation. We show that the kinase for eukaryotic elongation factor 2 (eEF2K) regulates dendritic spine stability and synaptic structure by modulating activity-dependent dendritic BDNF synthesis. Specifically RNAi knockdown of eEF2K reduces dendritic spine stability and inhibits dendritic BDNF protein expression; whereas overexpression of a constitutively activated eEF2K induces spine maturation and increases expression of dendritic BDNF. Furthermore, BDNF overexpression rescues the spine stability reduced by RNAi knockdown of eEF2K. We also show that synaptic activity-dependent spine maturation and dendritic BDNF protein expression depend on mGluR/EF2K-induced eEF2 phosphorylation. We propose that the eEF2K/eEF2 pathway is a key biochemical sensor that couple neuronal activity to spine plasticity, by controlling the dendritic translation of BDNF.
Introduction
Dendritic spines are essential for nervous system functioning (Bagni and Greenough, 2005; Carlisle and Kennedy, 2005; Hayashi and Majewska, 2005; Tada and Sheng, 2006; Bourne and Harris, 2008). Spine density and morphology are finely tuned during development, and are further modified in response to neuronal activity (Matsuzaki et al., 2004; Nägerl et al., 2004; Zhou et al., 2004; Ethell and Pasquale, 2005; De Roo et al., 2008). It known that protein synthesis is required for LTP, metabotropic glutamate receptor-dependent LTD, and the regulation of homeostatic plasticity (Huber et al., 2000; Kauderer and Kandel, 2000; Schuman et al., 2006). More recently it has been shown that local protein synthesis is necessary for structural changes to occur in dendritic spines (Korte, 2008; Tanaka et al., 2008); such changes may be linked to the formation of long-lasting memories in the hippocampus (Korte, 2008; Tanaka et al., 2008). However the mechanisms whereby neuronal activity is translated into local protein synthesis and long-lasting structural and functional changes in dendritic spines remain largely unknown.
Local protein synthesis might be regulated during nascent peptide elongation by eukaryotic elongation factor 2 (eEF2), which controls the transit of mRNA through the ribosome. eEF2 phosphorylation at Thr56 by the specific eEF2 kinase (eEF2K) usually slows protein translation and elongation (Ryazanov et al., 1988; Ryazanov et al., 1991) but increases the translation of some mRNAs localized in dendrites including αCaMKII and Arc (Marin et al., 1997; Scheetz et al., 2000; Chotiner et al., 2003; Belelovsky et al., 2005; Park et al., 2008).
It has been found that glutamate receptor activation modulates eEF2 phosphorylation, suggesting that the eEF2K-eEF2 axis plays a role in regulating neuronal activity-dependent protein synthesis (Chotiner et al., 2003; Sutton et al., 2007; Park et al., 2008).
We showed previously, using a proteomic approach, that eEF2 phosphorylation is regulated by synaptic activity in hippocampal neurons (Piccoli et al., 2007). In the present paper we show that long-term bicuculline or TTX stimulation (proxies of neuronal activity) modulate eEF2 phosphorylation and stably increase or decrease, respectively, eEF2 phosphorylation. We also show that eEF2 phosphorylation is mediated by mGluR signaling. When we used a siRNA to specifically knockdown eEF2K and abolish eEF2 phosphorylation, the dendritic synthesis of proteins, including brain-derived neurotrophic factor (BDNF), was impaired, and spine morphology was altered. We further show that the dendritic synthesis of BDNF protein is dependent on eEF2 phosphorylation in dendrites: specifically BDNF synthesis is reduced in the absence of eEF2K and increased in neurons overexpressing constitutively active eEF2K.
Together our data indicate that spine plasticity and structure are finely tuned by the eEF2K-eEF2 pathway via the regulation dendritic BNDF expression. Alterations in this pathway are likely to compromise long-lasting spine plasticity and lead to cognitive dysfunction.
Materials and Methods
Hippocampal neuron culture, brain slice culture, and chemicals.
Hippocampal neuron cultures were prepared from 18- to 19-d-old rat embryos from Charles River. High-density (750–1000 cells/mm2) and medium-density (150–200 cells/mm2) neurons were plated and grown as described by (Romorini et al., 2004) using home-made B27. Neurons were plated onto 6-well plastic tissue culture plates (Iwaki, Bibby Sterilin) or 18 mm diameter coverslips, and grown on 12-well plastic tissue culture plates (Iwaki). Cultures were infected with lentivirus-expressing siRNA for eEF2 at 7 d in vitro (DIV) or transfected using calcium phosphate precipitation using the protocol described by Xia et al. (1996). Treatments (comprising bicuculline 40 μm, AP5 100 μm, CNQX 20 μm, MPEP 30 nm, and TTX 2 μm; all from Tocris Bioscience) were started at 16 DIV. Unless otherwise stated, other chemicals were from Applichem GmbH. Neurons were fractionated to obtain the P1 fraction enriched in cell bodies and probably containing large dendritic fragments, and the P2 fraction enriched in presynaptic and postsynaptic structures (Aakalu et al., 2001; An et al., 2008). The enrichment of dendritic components in P2 was assessed by immunoblot.
Hippocampal slices (400 μm) were cut from postnatal 7- to 9-d-old rats, cultured in medium for 8 d (Stoppini et al., 1991) and infected with lentivirus and cultured for a further 15 d before treatments.
Western blotting and antibodies.
Neurons were solubilized in Laemmli buffer and loaded onto 6–12% SDS-PAGE gels. Protein bands were transferred to nitrocellulose membranes (Sigma) at 80 V for 120 min at 4°C. Primary antibodies were applied overnight in blocking buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 0.1% Tween 20, and 3% dried nonfat milk). Secondary antibodies (HRP-conjugated anti-mouse, anti-rabbit or anti-goat) (GE Healthcare) were used at 1:2000 dilution. The signal was detected using an ECL detection system (PerkinElmer Life Sciences) and captured by a Versadoc 1000 digital camera (Bio-Rad). For quantification of immunoblot signals total band intensity was measured with ImageQuant software (Bio-Rad). Proteins were normalized to the actin signal but eEF2 phospho-specific intensity was normalized to the total eEF2 signal in the same lane. Changes in protein levels and eEF2 phosphorylation then were compared with untreated levels and expressed as fold increase. The results are expressed as the mean ± SEM.
The following antibodies and dilutions were used (sources in parentheses): rabbit anti-P-eEF2 1:1000 and rabbit anti-eEF2 1:1000 (gifts from A. C. Nairn, Yale University, New Haven, CT); rabbit anti-CaMKII 1:1000, rabbit anti-eEF2 kinase 1:1000, rabbit anti-ERK 1/2 1:500, rabbit anti P-ERK 1/2 1:500, rabbit anti-S6K 1:500, rabbit anti-4eBP1 1:250 and rabbit anti-GFP 1:500 (Cell Signaling Technology); rabbit anti-BDNF 1:500, rabbit anti-ARC 1:200, anti-NGF 1:1000 (Santa Cruz Biotechnology); rabbit anti-GluR2 1:400 (NeuroMab, UC Davis/NIH NeuroMab Facility); rabbit anti pro-BDNF 1:1000, mouse anti-vimentin 1:1000, mouse anti-β-actin 1:1000, and mouse anti-α-tubulin 1:1000 (Sigma).
Immunocytochemistry.
For immunostaining, neurons were fixed in 4% paraformaldehyde and 4% sucrose at room temperature or in 100% methanol at −20°C. Primary and secondary antibodies were applied in GDB buffer (30 mm phosphate buffer, pH 7.4, containing 0.2% gelatin, 0.5% Triton X-100, and 0.8 m NaCl) for 2 h at room temperature, or overnight at 4°C. The following antibodies and dilutions were used (source in parentheses): rabbit anti-GKAP 1:500, rabbit anti-Shank1 1:200, rabbit anti-PSD-95 1:500 (gifts from E. Kim, KAIST, Daejeon, South Korea); rabbit anti-P-eEF2 1:200 and rabbit anti-eEF2 kinase 1:2000 (gifts from A. C. Nairn, Yale University); rabbit anti-eEF2 kinase 1:2000 (Cell Signaling Technology); rabbit anti-GluR2/3 1:250 (Millipore Bioscience Research Reagents); rabbit anti-BDNF 1:100 (Santa Cruz Biotechnology); mouse anti-synaptophysin 1:1000 (Sigma); FITC-, Cy3- and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch). Texas Red-conjugated phalloidin was purchased from Invitrogen.
Quantification of synaptic proteins.
Neurons were transfected with various constructs on DIV11 using calcium phosphate precipitation, and fixed on DIV18. Labeled transfected or untransfected neurons were chosen randomly for quantification in at least four independent experiments for each condition. Fluorescent images were acquired with a Bio-Rad MRC1024 or Zeiss 510 (donated by the Fondazione Monzino, Milan, Italy) confocal microscope with Nikon 60× or Zeiss 63× objective, respectively, at a resolution of 1024 × 1024 pixels. Each image was a z series projection of ∼7–10 images, each averaged twice and taken at depth intervals of 0.5 μm. The morphometric analysis was done with MetaMorph image analysis software (Universal Imaging). The clusters, localized at the dendrites of transfected neurons, were counted manually using the merge images and divided in two categories, those localized at dendritic protrusions and those not. The percentage of clusters localized at dendritic protrusions relative to total was then calculated (Sala et al., 2001; Romorini et al., 2004). All measurements were expressed means ± SEM.
RNA interference and plasmids.
For plasmid-based RNA inhibition, eEF2K oligonucleotides were annealed and inserted into the HindIII/BglII sites of the pSUPER vector (Oligo Engine30) or the pLVTHM vector for lentivirus production. We used siRNA sequences targeted against rat eEF2K mRNA (GenBank accession number NM_007908): nucleotides 362–380 (siRNA 362), 766–784 (siRNA 766) and 2118–2136 (siRNA 2118) (see supplemental Fig. 2, available at www.jneurosci.org as supplemental material). An eEF2K resistant (eEF2KR) to interference by siRNA 2118 was generated by changing four nucleotides at the siRNA 2118 target site without changing the amino acid sequence of the kinase. The specificity and efficacy of siRNA constructs in interfering with eEF2K expression were first tested against endogenously expressed eEF2K in normal rat kidney [NRK] cells transfected with Lipofectamine reagent (Invitrogen) and starved for 48 h.
The point (Ser366Ala) mutation to generate an eEF2K (eEF2Kca) with no phosphorylation site was introduced by PCR using the following primers: S366Aup CGCATAAGGACACTCGCTGGCAGCCGGCCC and S366Adown GGGCCGGCTGCCAGCGAGTGTCCTTATGCG.
Measurement of dendritic spine morphology and dendritic protein synthesis.
Neurons were cotransfected with siRNA vector and pEGFP at a ratio of 2:1 (5 μg of total DNA/well) on DIV11, treated with drugs on DIV16, and fixed on DIV18. Labeled transfected neurons were randomly chosen for quantification in at least four independent experiments for each construct. The numbers of neurons used for quantification are shown in the figure legends. Morphometric measurements were made using MetaMorph image analysis software (Universal Imaging). Individual dendrites were selected randomly and their spines were traced manually. The maximum length and head width of each spine were measured and archived automatically. To study dendritic protein synthesis, one of the vectors pcDNA3.1-5′myrdGFP 3′, myrd1GFP-A*B, or myrd1GFP-A, was cotransfected with siRNA and DsRed vectors in the ratio 1:2:1. Terminal dendrites (dendrites without additional branches) and cell bodies were manually traced using MetaMorph software on the DsRed channel. GFP signal intensity (mean pixel intensity) was estimated as the myrdGFP integrated fluorescence intensity divided by the area picked out by the DsReD signal.
Total RNA extraction and real-time quantitative PCR analysis.
Total RNA was isolated from cells using the RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions, treated with DNase I, and then treated with ImProm-II reverse transcriptase (Promega) according to the manufacturer's instructions, to obtain cDNA. Real-time PCR was performed using an iQ5 Real-Time PCR Detection System (Bio-Rad) using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad). Specificities of amplifications were assessed by electrophoretic separation of the products and melting curve analyses. The expression of the genes of interest was quantitated and normalized against β-actin and rpsa gene expression. The primers used for amplification were: forward (F)-BDNF ORF (AGGTGAGAAGAGTGATGACCA), reverse (R)-BDNF ORF (GCTCTCCAGAGTCCCATGG); F-BDNF L (TGGCCTAACAGTGTTTGCAG); R-BDNF L (GGATTTGAGTGTGGTTCTCC); F-bACT (GCCTTCCTTCTTGGGTAATGG), R-bACT (AATGCCTGGGTACATGGTGG); F-RPSA (ACCCAGAGGAGATTGAGAAGG), R-RPSA (TGGGGAACTGCTGAATGGGC).
Statistical analyses.
Data are expressed as means ± SEM. The significance of differences was tested by unpaired Student's t test (two means) or ANOVA with post hoc Tukey test (more than two means). The number of experiments (n) performed and p values are reported in the figure legends. Differences were considered significant for p values <0.05. The SPSS statistical package version 13.0 (SPSS Inc.) was used for the analyses.
Results
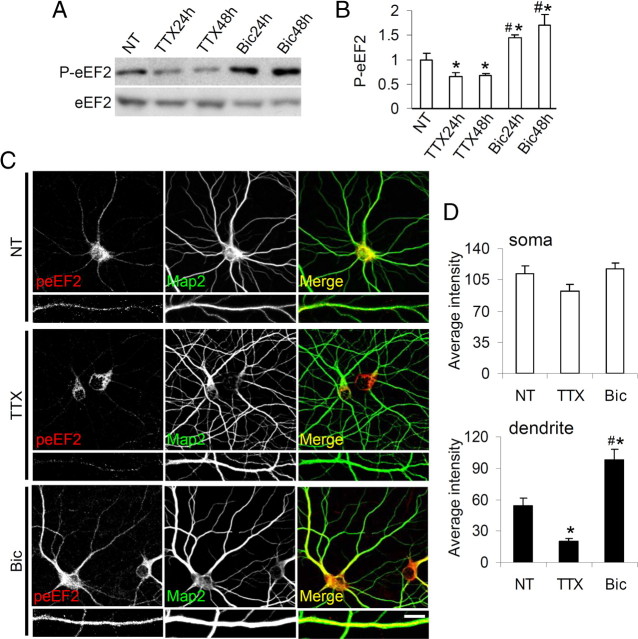
In a previous study (Piccoli et al., 2007) we found that synaptic activity modulated the phosphorylation status of eEF2 in hippocampal neurons. In particular, long-term (24 or 48 h) treatment with TTX (2 μm)—which blocks sodium channels and inhibits propagation of spontaneous action potential (Ehlers, 2003)—reduced eEF2 phosphorylation. By contrast, the GABA receptor antagonist bicuculline (40 μm for 24 or 48 h) increased excitatory synaptic activity and also increased the quantity of phosphorylated eEF2 (P-eEF2) without affecting the total quantity of eEF2 (see Piccoli et al., 2007) (Fig. 1A,B; supplemental Fig. 1, available at www.jneurosci.org as supplemental material). In the present study we found that synaptic activity modulated eEF2 phosphorylation in dendrites but had no significant effect on eEF2 phosphorylation in neuronal soma (Fig. 1C, with quantification in D). In the dendritic sites marked by MAP2 (Fig. 1C), P-eEF2 staining was increased by bicuculline and decreased by TTX (Fig. 1C, with quantification in D).
Figure 1.
Synaptic activity regulates eEF2 phosphorylation in dendrites. A, Western blots of neurons treated with TTX or bicuculline for 24 or 48 h and then solubilized in SDS sample buffer. B, Band intensity of P-eEF2, normalized against total eEF2. The vertical axis shows the mean fold increase over no treatment (NT). Error bars are SEMs; at least four independent experiments were performed per condition. *p < 0.05 versus NT; #p < 0.05 versus TTX samples (ANOVA, Tukey post hoc test). C, Hippocampal neurons untreated (NT), treated with TTX, or bicuculline (Bic) at DIV16, fixed at DIV18, and immunostained to reveal P-eEF2 and MAP2. TTX and Bic treatments modified dendritic but not somatic eEF2 phosphorylation. D, P-eEF2 signals in somatic and dendritic regions expressed as mean pixel intensity (error bars SEM); >15 neurons from three independent experiments were used for condition (soma: NT: 44.8 ± 3.50; TTX: 36.9 ± 3.09; Bic: 47.1 ± 2.53; dendrite: NT: 36.2 ± 4.63; TTX: 13.7 ± 1.83, Bic: 65.2 ± 6.83). *p < 0.01 versus untreated neurons, #p < 0.01 versus TTX-treated neurons (ANOVA, Tukey test). Scale bar, 20 μm for low-magnification images; Scale bar, 10 μm for high-magnification images.
To further probe the role of eEF2 phosphorylation in dendrites and dendritic spines we generated the 362, 766 and 2118 siRNAs which specifically knockdown eEF2K (the eEF2-specific kinase) and reduce eEF2 phosphorylation without affecting a panel of other endogenous proteins, both in NRK cells and in hippocampal neurons (supplemental Fig. 2A–D, available at www.jneurosci.org as supplemental material). We report results on 2118 siRNA (hereafter sieEF2K), but closely similar results were obtained with 362 and 766 (data not shown).
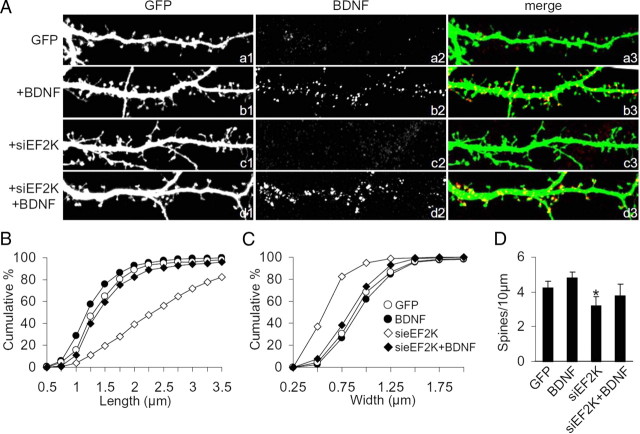
We transfected hippocampal neurons with sieEF2K plus GFP or scrambled siRNA plus GFP at DIV10, and assessed dendrites, dendritic spines and synapses at DIV18. Endogenous eEF2K staining almost completely disappeared from sieEF2K-transfected neurons, but not from those transfected with scrambled siRNA (Fig. 2Aa1–3,Ab1–3). Reduction in eEF2K also resulted in disappearance of mature dendritic spines, which changed into filopodia-like protrusions (Fig. 2Ab1–3) with increased average spine length, and decreased average spine width and density (Fig. 2B–D; supplemental Table 1, available at www.jneurosci.org as supplemental material).
Figure 2.
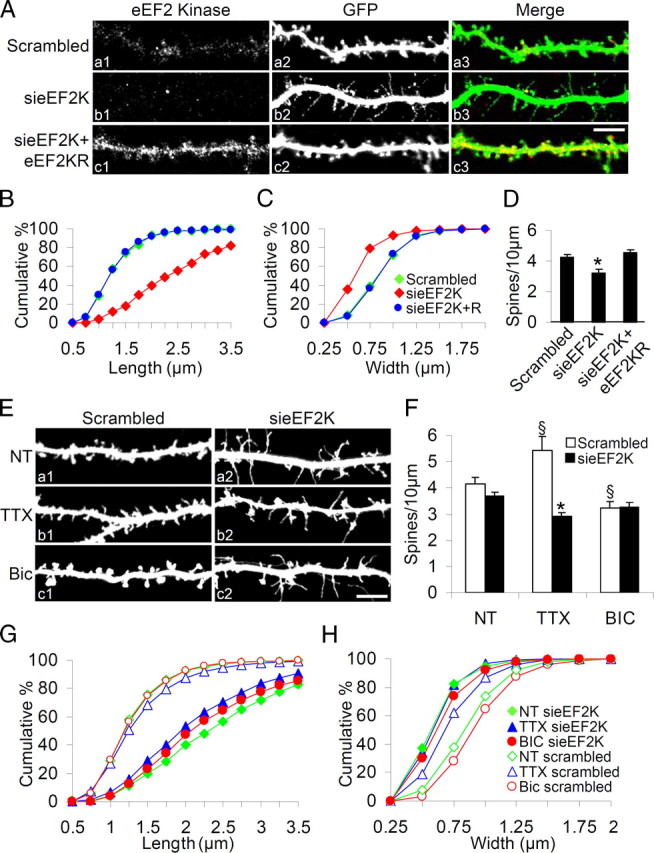
eEF2 phosphorylation by eEF2K plays a key role in remodeling spine morphology. A, Hippocampal neurons were transfected with scrambled siRNA, sieEF2K or sieEF2K plus eEF2K insensitive to siRNA (eEF2KR) together with GFP at DIV10 and fixed at DIV18. Inhibition of eEF2K expression caused disappearance of mature dendritic spines and production of protrusions resembling filopodia; whereas coexpression of eEF2KR restored eEF2K staining and also spine morphology. B, C, Cumulative frequency plots showing distribution of spine length and width in neurons transfected with the constructs indicated. D, Quantification of spine number (errors bars SEM) in neurons transfected with the constructs indicated. Over 14 transfected neurons from four independent experiments (corresponding to >7000 μm in dendrite length) were examined for each condition. *p < 0.05 versus scrambled and sieEF2K+eEF2KR transfected neurons. E, Hippocampal neurons were transfected with sieEF2K or scrambled siRNA together with GFP on DIV10, left untreated (NT) or treated with TTX or bicuculline on DIV16, and fixed on DIV18. Knockdown of eEF2K disrupted spine structure despite continuing discharges. F, Quantification of spine number (error bars SEM) in neurons transfected with indicated constructs and treated with indicated drugs. *p < 0.05 versus scramble-transfected neurons same treatment; §p < 0.05 versus untreated neurons same transfection (ANOVA, Tukey test). G, H, Cumulative frequency plots of distribution of spine length and width in neurons transfected with the constructs indicated. Over 14 transfected neurons from four independent experiments were measured for each condition. Scale bar, 10 μm.
The morphological effect was specific to dendritic spines: eEF2K knockdown did not change the mean number of branches or branch length in the dendritic arbor (numerical data in supplemental Table 2, available at www.jneurosci.org as supplemental material). Furthermore, the effect of sieEF2K on spine morphology was rescued by cotransfection with a vector encoding eEF2KR—an eEF2K containing four silent mutations that abolish interference by siRNA (Figs. 2Ac1–3,B–D; supplemental Fig. 2E, available at www.jneurosci.org as supplemental material, shows eEF2K levels in cells transfected and not with eEF2KR).
Absence of organized spine structure in sieEF2K-transfected neurons also markedly disturbed the expression and localization of a panel of presynaptic and postsynaptic markers: GluR2/3 AMPA subunit, PSD-95, GKAP, Shank, F-actin and Synaptophysin. Figure 3A shows that scramble-transfected neurons had synaptic proteins highly localized at dendritic spines, whereas eEF2K silencing massively depleted protein clusters present at the degenerate dendritic protrusions (Fig. 3A,B) without affecting total expression (supplemental Fig. 2B,D, available at www.jneurosci.org as supplemental material).
Figure 3.
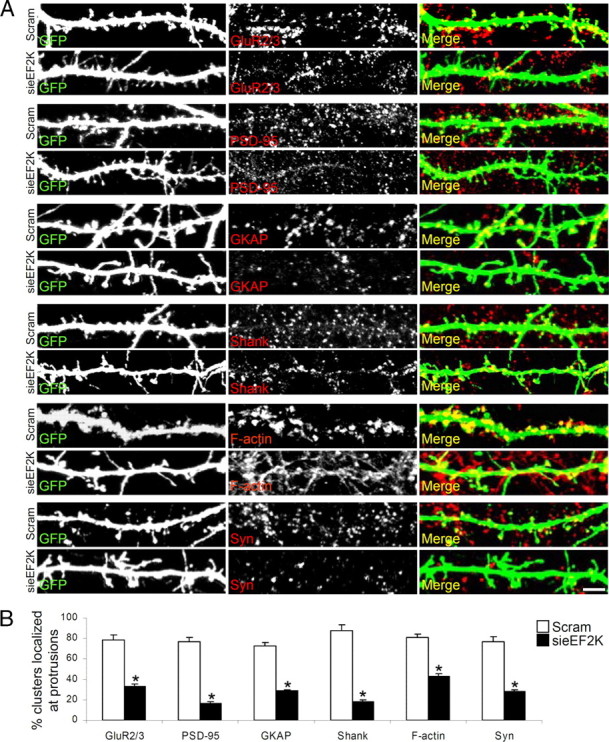
eEF2 phosphorylation by eEF2K regulates synapse morphology. A, Hippocampal neurons were transfected with scrambled siRNA or sieEF2K together with GFP on DIV10, and fixed on DIV18. The distribution of various presynaptic and postsynaptic markers (GluR2/3, PSD-95, GKAP, Shank, F-actin and synaptophysin) at the dendritic sites, were analyzed after staining with specific antibodies. B, Quantification of stained protein clusters localizing at dendritic protrusions in scrambled and sieEF2K neurons (mean ± SEM), from at least 10 neurons from three independent experiments per condition for each staining. *p < 0.05 versus scrambled transfected neurons (Student's t test). Scale bar, 10 μm.
The next step was to determine whether disturbed eEF2 signaling interferes with the activity-dependent modification of dendritic spine morphology previously demonstrated (Piccoli et al., 2007). We transfected hippocampal neurons with sieEF2K or scrambled siRNA and then treated them with bicuculline or TTX. Forty-eight hours of stimulation with bicuculline decreased the number of spines and increased their width, whereas 48 h activity depression by TTX increased the number and length of spines so they became filopodia-like (Fig. 2E–H; supplemental Table 1, available at www.jneurosci.org as supplemental material) (see also Piccoli et al., 2007). Thus the morphological changes due to chronic activity or inactivity were prevented by sieEF2K transfection: inhibition of eEF2 phosphorylation induced mature dendritic spines to change into filopodia-like protrusions (Fig. 2E–H; supplemental Table 1, available at www.jneurosci.org as supplemental material) while average spine length increased, and average width decreased, regardless of the pharmacological treatments (Fig. 2F–H; supplemental Table 1, available at www.jneurosci.org as supplemental material). These data suggest that eEF2K, and therefore eEF2 phosphorylation, is required for activity-dependent regulation of dendritic spines morphology.
Synaptic activity regulates dendritic spine maturation by regulating eEF2 phosphorylation
To further probe the role of eEF2K in shaping spines, neurons were transfected with eEF2K or empty vector (and GFP), treated with TTX or bicuculline on DIV16, and fixed on DIV18 (Fig. 4A). Overexpression of eEF2K (Fig. 4A, TTX panel) did not change spine length, width or density in neurons chronically depressed by TTX (Fig. 4E–G, columns TTX and TTX+eEF2K; supplemental Table 1, available at www.jneurosci.org as supplemental material) but, in bicuculline-stimulated neurons (Fig. 4A, Bic panel), eEF2K overexpression accentuated changes in spine morphology further increasing width and reducing density (Fig. 4E–G, columns Bic and Bic+eEF2K; supplemental Table 1, available at www.jneurosci.org as supplemental material).
Figure 4.
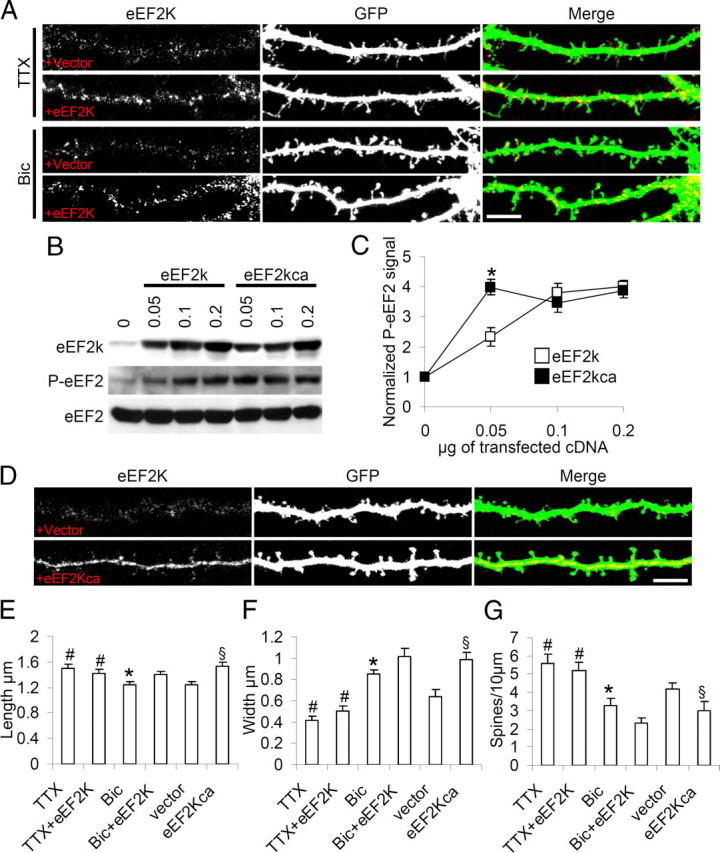
Activity regulates eEF2K-dependent spine morphogenesis. A, Neurons were transfected or not with eEF2K together with GFP on DIV10, treated with TTX or bicuculline on DIV16, fixed on DIV18 and immunostained to reveal eEF2K. Overexpression of eEF2K (lower panels in both treatments) modified spine morphology only in bicuculline-treated neurons. B, COS-7 cells were transfected or not with the indicated amounts of eEF2K or eEF2KSer366Ala (eEF2Kca), and solubilized in Laemmli buffer after 48 h. Levels of eEF2K, P-eEF2 and total eEF2 were analyzed by Western blot. C, Mean (error bars SEM) P-eEF2 levels (normalized against total eEF2 signal) in cells transfected with the indicated amounts of each construct (data collected from four independent experiments). *p < 0.05 (Student's t test). D, Neurons were transfected with an empty vector or eEF2Kca together with GFP on DIV10, fixed on DIV18 and immunostained to reveal eEF2K. eEF2Kca overexpression changed spine morphology even in the absence of stimulation. E–G, Mean length, width and number of dendritic spines (error bars SEM) in neurons transfected with indicated constructs at reported treatments. Over 10 neurons from three independent experiments were measured at each condition; *p < 0.05 versus Bic+eEF2K; #p < 0.05 versus Bic; §p < 0.05 versus vector transfected neurons (ANOVA, Tukey test). Scale bar, 10 μm.
Since multiple signaling pathways control eEF2K activity (Browne et al., 2004; Browne and Proud, 2004; Lenz and Avruch, 2005) and since phosphorylation of Ser366 by p70S6K or p90RSK inhibits eEF2K activity (Wang et al., 2001), we introduced the point mutation Ser366Ala in eEF2K (producing eEF2Kca) that abolishes the phosphorylation site. eEF2Kca had greater baseline activity than wild- type eEF2K: in a COS cell line transfected with increasing amounts of wild-type eEF2K or eEF2Kca plasmid, eEF2Kca saturated P-eEF2 levels even with the smallest amounts of DNA (Fig. 4B,C, which shows P-eEF2 levels normalized against total eEF2 levels).
Neurons were transfected with eEF2Kca or an empty vector plus GFP on DIV10 and fixed on DIV18 (Fig. 4D). Overexpression of eEF2Kca resulted in a significant increase in spine width and reduction spine density in the absence of chronic stimulation (Fig. 4E–G, columns vector and eEF2Kca; supplemental Table 1, available at www.jneurosci.org as supplemental material).
Together, these findings indicate that the synaptic activity controlling dendritic spine morphology involves positive modulation of eEF2K activity.
eEF2K regulates dendritic protein synthesis
Recent studies have shown that neuronal activity can modulate the dendritic synthesis of Arc/Arg3.1 and CaMKII—key proteins involved in synaptic plasticity—by regulating eEF2K activity and eEF2 phosphorylation (Scheetz et al., 2000; Park et al., 2008). We therefore investigated whether the perturbation of spine morphology induced by eEF2K knockdown, had a negative effect on local protein synthesis. We used the protein produced by a GFP reporter construct and monitored its synthesis in dendrites (Aakalu et al., 2001; Meyer-Luehmann et al., 2009). The construct expresses a myristoylated (and destabilized form) of the GFP protein, and contains the 3′UTR sequence of CaMKIIα which target the mRNA to dendrites (Mayford et al., 1996; Mori et al., 2000).
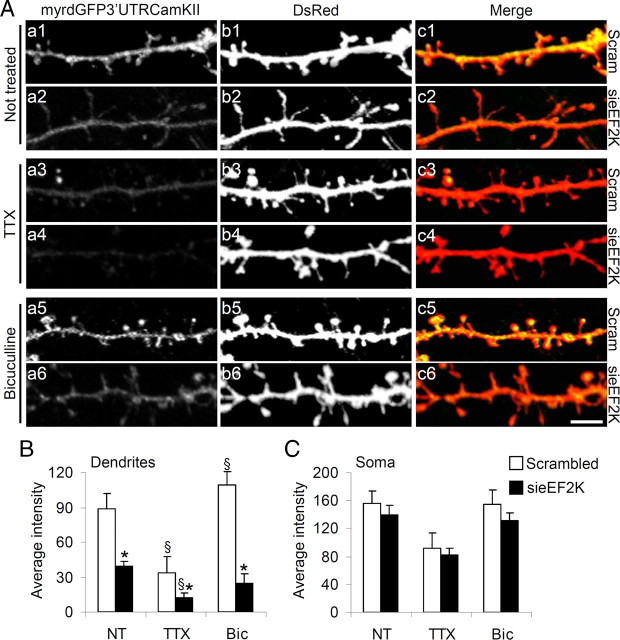
Neurons were transfected with sieEF2K or scrambled siRNA together with DsRed (to label transfected neurons) and the myrdGFP3′UTRCaMKIIα (myrdGFP) vector on DIV10, treated or not with synaptic activity modifying drugs on DIV16, and fixed on DIV18. The myrdGFP signal, but not the DsRed signal, in terminal dendrites was regulated by synaptic activity in scramble-transfected neurons (Fig. 5Aa1,Aa3,Aa5). Quantification of the myrdGFP signal (as GFP intensity divided by the dendritic area under consideration as picked out by the DsRed signal) showed that TTX treatment reduced, while bicuculline increased, local synthesis (Fig. 5B). When, however, eEF2K was knocked-down (Fig. 5Aa2,Aa4,Aa6), dendritic synthesis of the reporter was impaired both in nontreated and pharmacologically treated (including Bic-treated) neurons (Fig. 5B). eEF2K knockdown had no significant effect on myrdGFP expression in the soma (Fig. 5C; supplemental Fig. 3, available at www.jneurosci.org as supplemental material). These findings suggest that synaptic activity regulates dendritic protein synthesis. Together the results reported so far indicate that synaptic activity regulates dendritic protein synthesis as well as dendritic spine morphology and maturation, by modulating the phosphorylation and hence activity of eEF2.
Figure 5.
eEF2 phosphorylation by eEF2K regulates local protein synthesis. A, Neurons were transfected with sieEF2K or scrambled (Scram) siRNA on DIV10 in both cases together with DsRed and also the pcDNA3.1–5′myrdGFP 3′ vector, and treated with TTX, bicuculline or untreated on DIV16, and fixed at DIV18. The intensity of the myrdGFP signal (indicating extent of local myrdGFP protein synthesis) in terminal dendrites was proportional to, and regulated by, synaptic activity in scrambled (Scram, labeling on right of panels) transfected neurons (a1, a3, a5). When eEF2K was knocked down, local myrdGFP synthesis was impaired despite pharmacological treatment (a2, a4, a6). Dendritic process areas (b1–b6) were used to normalize myrdGFP intensity. The merged channels are shown in c1–c6. B, C, The signal intensity of myrdGFP was calculated as the ratio between localized GFP intensity and dendritic (B) or soma (C) areas, and expressed as mean pixel intensity (error bars SEM). At least 14 neurons per condition from four independent experiments were analyzed [mean ± SEM control neurons: 59.1 ± 9.0 (untreated), 22.2 ± 9.6 (TTX), and 72.8 ± 7.8 (bicuculline); sieEF2K: 26.4 ± 2.5 (untreated), 8.2 ± 2.9 (TTX), and 16.3 ± 5.4 (bicuculline)]. *p < 0.05 versus scramble-transfected neurons for each treatment; §p < 0.05 versus untreated neurons transfected with the same siRNA (ANOVA, Tukey test). Scale bar, 5 μm.
Dendritic synthesis of BDNF is controlled by the eEF2K-eEF2 pathway
Neuronal activity has been shown to enhance the local synthesis and secretion of BDNF, which in turn plays a crucial role in synaptic transmission and plasticity. (An et al., 2008; Tanaka et al., 2008) We investigated whether synaptic activity modulates BDNF expression in our model. Neurons were treated for 48 h with TTX or bicuculline and tested for BDNF expression by Western blot (supplemental Fig. 4A, available at www.jneurosci.org as supplemental material). We found that BDNF expression was significantly reduced by TTX treatment and strongly upregulated by bicuculline (supplemental Fig. 4E, available at www.jneurosci.org as supplemental material), whereas expression of the postsynaptic proteins CaMKII, Arc and the GluR2 subunit was less strongly affected by these treatments (supplemental Fig. 4D,E, available at www.jneurosci.org as supplemental material). As shown in supplemental Figure 4A (available at www.jneurosci.org as supplemental material) we used antibodies that mainly label BDNF protein (14 kDa), less pro-BDNF (29 kDa). This antibody recognized several other bands of higher molecular weight not modified by TTX and bicuculline, we therefore monitored the 14 kDa band only (supplemental Fig. 4A, available at www.jneurosci.org as supplemental material). As expected, bicuculline treatment also increased the expression of pro-BDNF (supplemental Fig. 4B, available at www.jneurosci.org as supplemental material) but not NGF (supplemental Fig. 4C, available at www.jneurosci.org as supplemental material).
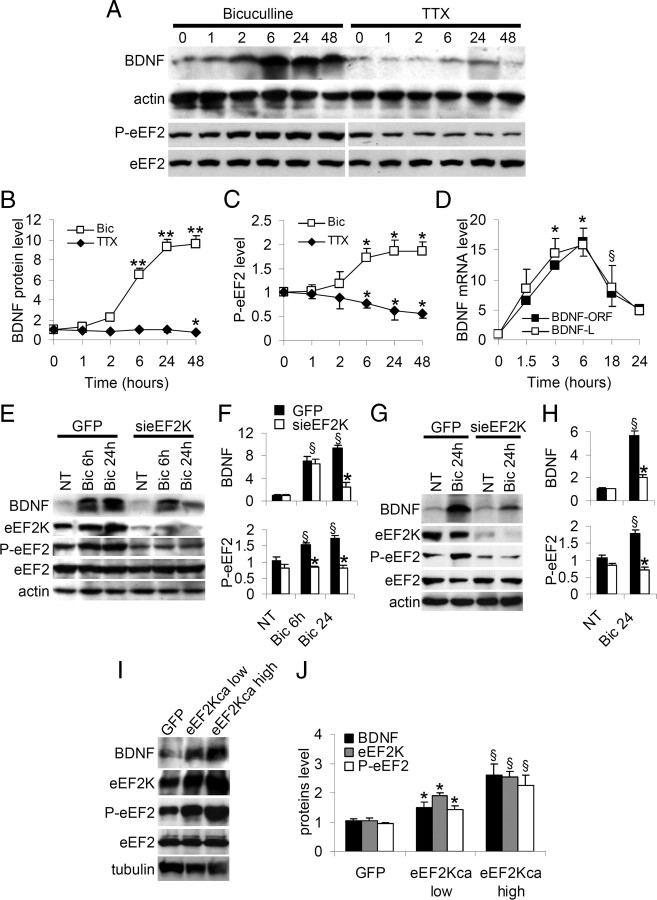
We next monitored the induction of BDNF protein expression over time. DIV16 neurons were treated with TTX or bicuculline for 1, 2, 6, 24 and 48 h, and BDNF and actin levels were determined by Western blot at each time point (Fig. 6A). BDNF levels were increased by bicuculline and reduced by TTX (Fig. 6B shows BDNF levels normalized against no treatment and actin expression). In particular, BDNF expression was substantially increased by bicuculline stimulated activity by 6 h and reached a plateau at 24 h (Fig. 6A,B). At the same time points we measured levels of phosphorylated eEF2, finding that bicuculline increased eEF2 and TTX reduced eEF2 phosphorylation with changes over time similar to BDNF protein expression changes (Fig. 6A,C; in Fig. 6C P-eEF2 levels are normalized against no treatment and total eEF2).
Figure 6.
Neuronal activity modulates BDNF protein and transcripts in a biphasic manner, through the eEF2-eEF2K pathway. A, Neurons were treated with TTX or bicuculline for up to 48 h, solubilized in SDS sample buffer, and levels of BDNF, P-eEF2 and total eEF2 analyzed by Western blot. B, Mean BDNF levels (error bars SEM) normalized against actin are shown as fold increases relative to time 0; *p < 0.05 versus untreated neurons; **p < 0.01 versus untreated neurons (ANOVA, Tukey test, n = 6). C, Mean P-eEF2 levels (error bars SEM) normalized against actin and shown as fold increase relative to time 0; *p < 0.05 versus untreated neurons (ANOVA, Tukey test, n = 6). D, Whole-cell levels of BDNF-ORF and BDNF-L mRNA were measured by RT-quantitative PCR after bicuculline treatment. Transcripts increased during the first 6 h of bicuculline stimulation but decreased subsequently (fold expression relative to untreated neurons normalized against actin mRNA: 1.5 h = 7.5 ± 0.60, 3 h = 12.5 ± 0.32, 6 h = 16.3 ± 2.32 for BDNF-ORF; 1.5 h = 8.6 ± 3.20, 3 h = 14.4 ± 2.57, 6 h = 15.9 ± 2.76 for BDNF-L, 18 h = 7.8 ± 2.09; 24 h = 5.2 ± 0.71 for BDNF-ORF; 18 h = 8.6 ± 3.70; 24 h = 4.8 ± 0.35 for BDNF-L), *p < 0.01 versus time 0, §p < 0.05 versus time 6 h. (ANOVA, Tukey test, n = 7). E, Neurons were infected with sieEF2K lentivirus or control virus on DIV8, treated as shown, and BDNF, eEF2K, P-eEF2 and eEF2 levels analyzed by Western blot. F, BDNF and P-eEF2 levels (error bars SEM) after treatments in neurons infected with GFP virus or sieEF2K virus (n = 7) (P-eFF2 levels normalized against total eEF2: NT+GFP 1.0 ± 0.09; Bic 6 h+GFP 1.5 ± 0.07; Bic 24 h+GFP 1.7 ± 0.09; NT+sieEF2K 0.8 ± 0.09; Bic 6 h+ sieEF2K 0.8 ± 0.07; Bic 24 h+ sieEF2K 0.8 ± 0.10; BDNF levels normalized against actin: NT+GFP 1.0 ± 0.10; Bic 6 h+GFP 7.1 ± 0.76; Bic 24 h+GFP 9.4 ± 0.57; NT+sieEF2K 1.0 ± 0.09; Bic 6 h+ sieEF2K 6.5 ± 0.90; Bic 24 h+sieEF2K 2.5 ± 0.77). *p < 0.05 versus GFP-infected neurons, §p < 0.01 versus untreated neurons. G, Hippocampal slices were infected with sieEF2K lentivirus or control virus on DIV8, treated as shown, and BDNF, eEF2K, P-eEF2 and eEF2 levels determined by Western blot. H, BDNF and P-eEF2 levels (error bars SEM) after the treatment of hippocampal slices infected with GFP virus or sieEF2K virus (n = 7) (BDNF, NT+GFP 1.0 ± 0.12, Bic 24 h+GFP 5.6 ± 0.41, NT+sieEF2K 1.0 ± 0.02, Bic 24 h+sieEF2K 2.1 ± 0.19; P-eEF2, NT+GFP 1.0 ± 0.09; Bic 24 h+GFP 1.8 ± 0.10; NT+sieEF2K 0.8 ± 0.09; Bic 24 h+ sieEF2K 0.7 ± 0.07). *p < 0.01 versus GFP-infected neurons, same treatment; §p < 0.05 versus untreated neurons same transfection (ANOVA, Tukey test). I, Neurons were infected with low and high titer lentivirus preparations expressing eEF2Kca and BDNF; eEF2K, P-eEF2 and eEF2 levels were determined by Western blot. J, BDNF, eEF2K and P-eEF2 levels (error bars SEM) after infection (n = 5) (BDNF: GFP = 1.0 ± 0.80, eEF2Kca low = 1.5 ± 0.20, eEF2Kca high = 2.6 ± 0.36; eEF2K: GFP = 1.0 ± 0.12, eEF2Kca low = 1.9 ± 0.09, eEF2Kca high = 2.5 ± 0.20; P-eEF2: GFP = 0.9 ± 0.05, eEF2Kca low = 1.4 ± 0.11, eEF2Kca high = 2.2 ± 0.35). *p < 0.05 versus GFP-infected neurons, §p < 0.01 versus GFP-infected.
The transcription of BDNF mRNA is regulated in a time-dependent manner by neuronal activity (Zafra et al., 1991; Tao et al., 1998; Lauterborn et al., 2000). BDNF mRNAs are transcribed from multiple promoters regulated by different signaling pathways (Tabuchi et al., 2000; Sakata et al., 2009) and are polyadenylated at two alternative sites, leading to mRNA populations with either short or long 3′ untranslated regions (3′UTRs) (Ghosh et al., 1994). Short 3′ UTR mRNAs are restricted to soma, whereas long 3′ UTR mRNAs also localize in dendrites (An et al., 2008). Using primers that specifically amplify either the long UTR of BDNF, or the open reading frame (BDNF-ORF), we determined BDNF mRNA levels (by quantitative RT-PCR) in neurons chronically stimulated with bicuculline and assess whether the increase in BDNF protein was due to increased transcription. Neurons were bicuculline-treated for 1.5, 3, 6, 18 and 24 h and the BDNF mRNA quantified. BDNF mRNA peaked at 6 h (Fig. 6D) but gradually decreased with longer bicuculline treatment, thus the kinetics of expression of BDNF mRNA differed from that of the protein (Fig. 6B). The kinetics of BDNF-L and BDNF-ORF mRNA expression were closely similar (Fig. 6D). These findings are compatible with BDNF protein expression at 24 h and beyond being sustained by translational control of BDNF mRNA. We therefore hypothesized that eEF2 phosphorylation was involved in controlling BDNF translation following transient induction of BDNF mRNA transcription by neuron activity. To investigate this we knocked down eEF2K in hippocampal neurons and hippocampal slice cultures, using lentivirus encoding sieEF2K or GFP as control. In eEF2K-knocked down cells, BNDF protein expression was increased after 6 h of bicuculline stimulation but had decreased almost to control levels after 24 h (Fig. 6E,F). In GFP-infected neurons, BDNF protein expression after 6 and 24 h did not differ from that in uninfected neurons (Fig. 6E,F). eEF2K knockdown in brain slices also significantly inhibited eEF2 phosphorylation and the BDNF production induced by 24 h of bicuculline treatment (Fig. 6G,H). These findings indicate that eEF2 phosphorylation is required for the sustained activity-dependent expression of BDNF protein.
To provide further evidence of eEF2K-eEF2 pathway involvement in regulating BDNF translation, we overexpressed eEF2Kca in hippocampal neurons. As shown in Figure 6, I and J, BDNF and P-eEF2 protein levels were higher in neurons infected with lentivirus expressing eEF2Kca than in neurons infected with GFP. Furthermore the BDNF and P-eEF2 increases obtained were related to the titer (low versus high) of the lentivirus preparation, and the level of eEF2Kca expression (Fig. 6J).
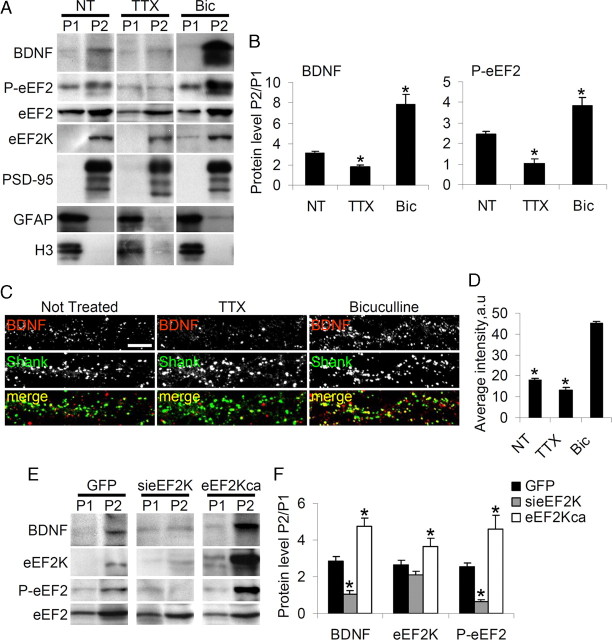
To demonstrate that neuronal activity modulates BDNF protein expression specifically in dendrites, treated and untreated neurons were fractionated into P1 enriched in cell bodies, and P2 enriched in synaptosomes and dendrites (An et al., 2008). We found that in untreated neurons BDNF protein was mainly localized in P2 in which the synaptic marker PSD-95 was also enriched (Fig. 7A,B). Interestingly BDNF protein was reduced in the P2 fraction in TTX-treated neurons, and increased in the P2 fraction in bicuculline-treated neurons (Fig. 7A,B). Similarly, both P-eEF2 (reduced by TTX, induced by bicuculline) and eEF2K were enriched in the P2 fraction (Fig. 7A,B; B shows the P2/P1 ratios of P-eEF2 and BDNF in untreated, TTX- and bicuculline-treated neurons). BDNF immunostaining also indicated that neuronal activity modulates BDNF expression in dendrites (Fig. 7C,D): activity stimulation with bicuculline significantly increased BDNF staining in dendrites (picked out by Shank), while activity depression with TTX reduced BDNF staining relative to untreated (Fig. 7D).
Figure 7.
Activity modulates BDNF synthesis in dendrites. A, Neurons were untreated, TTX-treated, or Bic-treated for 48 h, and fractionated into cell body-rich and dendrite-rich fractions. By Western blots, the dendrite-rich (P2) fraction appeared free of nuclear protein H3 and glial marker GFAP, but was enriched in the postsynaptic marker PSD-95. B, BDNF and P-eEF2 were also enriched in P2 as shown by the ratios (P2/P1) of band intensities (means with SEM error bars, n = 6) for BDNF and P-eEF2 in untreated (NT), TTX and bicuculline-treated (Bic) neurons (BDNF: NT = 3.1 ± 0.22, TTX = 1.8 ± 0.19, Bic = 7.9 ± 0.91; P-eEF2: NT = 2.4 ± 0.16, TTX = 1.0 ± 0.21, Bic = 3.8 ± 0.43). *p < 0.05 versus untreated neurons (Student's t test). C, Neurons were untreated, TTX-treated, or Bic-treated for 48 h, then immunoprocessed to reveal BDNF and Shank. The panels show dendrites at high magnification. D, Quantification (mean pixel intensity with SEM error bars) of BDNF signal in dendritic fraction. Over 20 neurons from four independent experiments were assessed for each condition (NT = 17.8 ± 2.50, TTX = 13.3 ± 1.95, Bic = 45.0 ± 3.28). *p < 0.01 versus bicuculline-treated neurons (ANOVA, Tukey test). Scale bar, 10 μm. E, Neurons were infected with lentivirus expressing either GFP, sieEF2K or eEF2Kca, and the P1 and P2 fractions obtained: BDNF, eEF2K and P-eEF2 are enriched in the dendrite-rich P2 fraction. F, The histograms show by the ratios (P2/P1) of band intensities (means with SEM error bars, n = 6) for BDNF, eEF2K and P-eEF2 in GFP, sieEF2K and eEF2Kca-infected neurons (BDNF, GFP 2.8 ± 0.23, sieEF2K 1.1 ± 0.17, eEF2Kca 4.7 ± 0.46; eEF2K, GFP 2.7 ± 0.26, sieEF2K 2.1 ± 0.19, eEF2Kca 3.6 ± 0.48; P-eEF2, GFP 2.5 ± 0.24, sieEF2K 0.7 ± 0.10 eEF2Kca 4.7 ± 0.46). *p < 0.05 versus GFP-infected neurons (ANOVA, Tukey test).
To provide further evidence that eEF2K activity regulates BDNF expression in dendrites, neuronal cell cultures were infected with either GFP, sieEF2K or eEF2Kca and the P1 and P2 fractions produced. As shown in Figure 7, E and F, in the absence of sieEF2K, levels of both BDNF and P-eEF2 were preferentially reduced in the P2 fraction, while overexpression of eEF2Kca increased the level of P-eEF2 and BDNF in the P2 fraction (Fig. 7E,F, ratio P2/P1 ± SEM). These findings strongly support the hypothesis that eEF2 phosphorylation is required for the activity-dependent expression of BDNF protein in the dendritic compartment.
BDNF overexpression can rescue the effect of sieEF2K on spine morphology
We finally tested whether the effect of eEF2K knock-down on spine morphology can be rescued by means of postsynaptic BNDF overexpression. Neurons were transfected with GFP alone or GFP plus sieEF2K or sieEF2K and the BDNF-expressing plasmid (pcDNA3mycBDNF, containing a CMV promoter and an SV40 3′ UTR) on DIV10, and fixed on DIV18 (Fig. 8A). Morphological analysis showed that, although the overexpression of BDNF (confirmed by antibody staining) induces moderate modifications in the dendritic protrusions, the coexpression of BDNF and sieEF2K completely reverted the spine phenotype associated with eEF2K knock-down, as demonstrated by the measurements of spine length and width (Fig. 8B–D; Table 1).
Figure 8.
BDFN overexpression rescues spine phenotype due to eEF2K knock-down. A, Neurons were transfected with GFP, BDNF, sieEF2K, or sieEF2K+BDNF vectors on DIV10, fixed on DIV18, and stained with BDNF antibody. BDNF coexpression increased the BDNF signal along the dendrites and positively affected spine number and morphology in comparison with sieEF2K alone. B–D, Mean length, width and number of dendritic spines (±SEM) in neurons transfected with the indicated constructs; >10 transfected neurons (corresponding to >5000 μm in dendrite length), were measured for every condition; *p < 0.05 versus GFP, BDNF and sieEF2K + BDNF, Student t test. Scale bar, 10 μm.
mGluR receptor activity regulates long-lasting eEF2 phosphorylation
Glutamate receptor activation modulates eEF2 phosphorylation (Marin et al., 1997; Scheetz et al., 2000; Cossenza et al., 2006; Kanhema et al., 2006; Sutton et al., 2007), and physical and functional interactions between eEF2K and mGluR1 have been documented (Park et al., 2008). We therefore investigated the glutamate receptor subtypes involved in the eEF2K activation (that sustains eEF2 phosphorylation) caused by long-term stimulation with bicuculline. On DIV16, neurons were treated for 24 h with bicuculline with or without one or more of the following: AP5 100 μm (A), CNQX 40 μm (C) and MPEP 30 nm (M)—antagonists of NMDA, AMPA and metabotropic glutamate receptors respectively—and analyzed by Western blot (Fig. 9A). Blockade of metabotropic glutamate receptors almost completely abolished the eEF2 phosphorylation induced by bicuculline (Fig. 9B, which shows P-eEF2 induction normalized against total eEF2) whereas antagonizing NMDAR or AMPAR did not significantly reduce eEF2 phosphorylation (Fig. 9B). Blockade of all three GluRs reduced bicuculline-induced eEF2 phosphorylation only slightly more than the mGluR antagonist alone (Fig. 9B).
Figure 9.
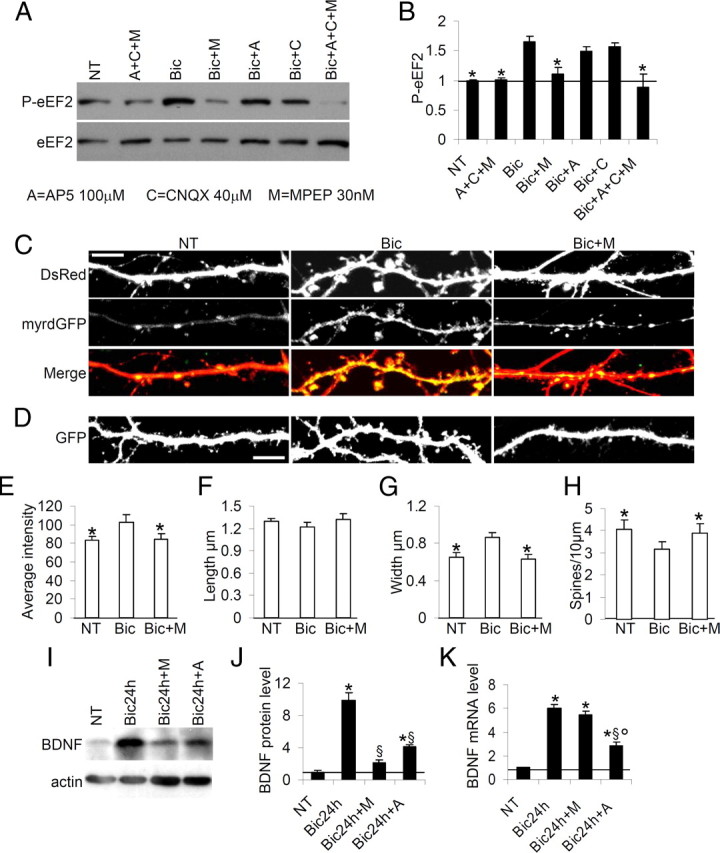
Activity-dependent phosphorylation of eEF2, local protein synthesis and dendritic spine maturation is regulated by mGluR signaling. A, Neurons were treated or not with the indicated drugs for 48 h, then solubilized in SDS sample buffer. Blockade of metabotropic glutamate receptors with MPEP markedly impairs activity-driven eEF2 phosphorylation. Blockade of all three glutamate receptors appears to deplete P-eEF2. B, Mean P-eEF2 levels (error bars SEM), normalized against total eEF2 and shown as fold increase relative to untreated (NT) condition; n = 7 (Bic = 1.7 ± 0.08, Bic+M = 1.1 ± 0.10, Bic+A = 1.5 ± 0.07, Bic+C = 1.6 ± 0.07 Bic+A+C+M = 0.9 ± 0.22). *p < 0.05 versus bicuculline-treated neurons. C, Neurons were transfected with DsRed and pcDNA3.1–5′myrdGFP 3′ vector on DIV10, untreated, treated with bicuculline or treated with bicuculline plus MPEP (Bic+M) on DIV16, and fixed on DIV18. MPEP coadministration antagonized bicuculline-induced local protein synthesis as shown by impaired myrGFP expression in dendrites. D, In neurons transfected with GFP on DIV10 and treated as above, MPEP appeared to reduce spine modifications induced by bicuculline. E, Mean myrdGFP signal intensity (error bars SEM), expressed as mean pixel intensity divided by dendritic area considered, is reduced by MPEP compared with Bic alone: NT = 55.3 ± 2.98; Bic = 69.7 ± 5.48; Bic+M = 56.3 ± 4.22, average intensity. *p < 0.05 versus bicuculline-treated neurons. At least 10 neurons analyzed per condition, from three independent experiments. F–H, Mean length, width and number of dendritic spines (error bars SEM) in neurons untreated or treated with bicuculline or bicuculline plus MPEP; at least 8 neurons were analyzed per condition, from three independent experiments; *p < 0.05 versus bicuculline-treated neurons (ANOVA, Tukey test). Scale bar, 10 μm. I, Neurons were treated or not with the indicated drugs for 48 h, and then solubilized in SDS sample buffer. J, Mean BDNF protein levels (error bars SEM) normalized against actin and shown as fold increase relative to untreated conditions after indicated stimulations, n = 8 (Bic = 9.9 ± 1.24-fold relative to untreated neurons, Bic+M = 2.1 ± 0.51 Bic+A = 4.2 ± 0.19). *p < 0.01 versus untreated neurons; §p < 0.01 versus Bic 24 h neurons. K, BDNF mRNA, normalized against β-actin were measured by RT-quantitative PCR after the indicated treatments, n = 7 (Bic = 6.0 ± 0.30-fold relative to untreated neurons, Bic+M = 5.5 ± 0.37, Bic+A = 2.9 ± 0.27 for BDNF mRNA). *p < 0.01 versus untreated; §p < 0.01 versus Bic24 h neurons; °p < 0.01 versus Bic24 h+M-treated neurons (ANOVA, Tukey test).
These findings therefore suggested that mGluR plays a major role in regulating activity-driven P-eEF2, suggesting further investigation as to whether mGluR is also involved in modulating the local protein synthesis and dendritic spine maturation arising from bicuculline stimulation. Neurons were transfected with DsRed and myrdGFP vector on DIV10, treated with drugs on DIV16, and fixed on DIV18 (Fig. 9C). Quantification of myrdGFP signal intensity showed that, in the presence of MPEP (Bic+M), bicuculline stimulation failed to increase myrdGFP synthesis (Fig. 9E). Similarly, bicuculline did not induce spine alterations in the presence of MPEP as shown by spine length, width and density, in fact spine morphology with Bic+M was similar to in untreated neurons (NT) (Fig. 9F–H; supplemental Table 1, available at www.jneurosci.org as supplemental material).
To ascertain whether mGluR stimulation is also required for bicuculline-induced BDNF expression, we treated neurons for 24 h with bicuculline in the presence and absence of MPEP (30 nm) and AP5 (100 μm), and then analyzed BDNF protein and mRNA expression (Fig. 9I–K). MPEP (M, mGluR antagonist) significantly reduced Bic-induced BDNF protein (Fig. 9I,J), but not BDNF mRNA expression (Fig. 9K), while antagonizing NMDAR with AP5 (A) significantly reduced both BDNF protein and transcript (Fig. 9I–K).
Together these results suggest that during bicuculline treatment, mGluR activation regulates local protein translation, BDNF protein translation and dendritic spine morphology, and does so by promoting eEF2 phosphorylation.
Discussion
The present paper has delineated a causal link between synaptic activity and dendritic spine morphology in rat hippocampal neurons. We have shown that synaptic activity (as modulated pharmacologically by long-term treatment with bicuculline or TTX) regulates eEF2 phosphorylation and hence the activity of eEF2. We provide evidence that synaptic activity influences eEF2 phosphorylation via the activation of mGluR receptors. We have further shown that activity-dependent eEF2 phosphorylation occurs in the dendrites and correlates with protein translation in the dendrites. In particular we showed that eEF2 phosphorylation is required for dendritic BDNF synthesis, with indications that control is exerted at the level of BDNF mRNA translation. In turn BDNF—known to be involved in LTP, synaptic remodeling, homeostatic plasticity and neuronal survival—directly regulates dendritic spine morphology.
eEF2 phosphorylation regulates spine morphology via an effect on protein synthesis
We found that reducing eEF2 phosphorylation—by eEF2K knockdown or by action potential silencing with TTX—abolished spine maturation, shifting morphology to immature filopodia-like protrusions. By contrast, increasing eEF2 phosphorylation—by overexpression of constitutively active eEF2K, or by synaptic stimulation with bicuculline—induced dendritic spine maturation.
We were able to correlate these effects with the regulation of dendritic, but not somatic, protein synthesis, using a previously characterized reporters of dendritic protein synthesis (Aakalu et al., 2001; Meyer-Luehmann et al., 2009) (Fig. 5; supplemental Fig. 3, available at www.jneurosci.org as supplemental material). It is noteworthy that eEF2 kinase was enriched in the dendritic compared with the somatic compartment, so phosphorylated eEF2 would control protein translation mainly in dendrites.
Dendritic protein synthesis can be regulated at the translation level by activation of the translation initiation complex (eIF4F) (Banko et al., 2004; Richter and Sonenberg, 2005; Banko et al., 2006) or during the elongation of nascent peptides by eEF2 activity (Scheetz et al., 1997; Sutton et al., 2007; Park et al., 2008). In fact it was initially shown (Ryazanov et al., 1988) that phosphorylation eEF2 by eEF2K inhibited protein synthesis. However, phosphorylated eEF2 also reduces translation errors and promotes the translation of specific mRNAs (Walden and Thach, 1986; Scheetz et al., 2000) necessary for sustaining LTP (Chotiner et al., 2003), for mGluR-induced LTD (Park et al., 2008) and for memory formation (Belelovsky et al., 2005). So far eEF2 phosphorylation has also been directly correlated with the dendritic upregulation of Arc/Arg3.1 (Park et al., 2008) and CaMKIIα (Scheetz et al., 2000).
Recent data indicate that dendritic protein synthesis could be regulated by mTOR activity (Jaworski et al., 2005; Kumar et al., 2005). Interestingly, inhibition of mTOR by rapamycin mainly altered dendrite morphogenesis, while in our study eEF2K knock down did not alter dendrite morphology (supplemental Table 2, available at www.jneurosci.org as supplemental material) but dendritic spine morphology. These considerations suggest that the two kinases may control the translation of different subsets of mRNA. However the two pathways could also be connected because mTOR is able to phosphorylate and activate p70S6 kinase which in turn phosphorylates and inactivates eEF2K (Wang et al., 2001).
Involvement of mGluR activation
How does synaptic activity regulate eEF2K activity? Subsequent experiments we performed suggested that mGluR activation was required for bicuculline to induce greater eEF2 phosphorylation at steady state. Other findings add support to this idea, thus mGluR is known to directly modulate two of the pathways regulating eEF2K activity: ERK (which activates p70 S6KA and p90rsk by phosphorylation) and local Ca2+ levels (Wang et al., 2001; Lenz and Avruch, 2005). There is also a close association between eEF2K activity and mGluR-mediated endocytosis of AMPAR, as a result of the regulation of dendritic synthesis of MAP1B (Davidkova and Carroll, 2007). It is noteworthy that a physical interaction between eEF2K and type I mGluR has been demonstrated: DHPG stimulation induces eEF2K disassociation from mGluR and eEF2K activation (Park et al., 2008). It is plausible that one of these mechanisms is involved in mGluR-dependent eEF2K activation.
eEF2 phosphorylation and BDNF protein expression in dendrites
We next found that BDNF protein expression in dendrites and eEF2 phosphorylation status were related. In particular we showed that BDNF protein in the P2 dendrite-rich fraction was reduced in the absence of eEF2K and in TTX-treated neurons (characterized by low P-eEF2 levels) and increased in neurons overexpressing constitutively active eEF2K, as well as in bicuculline-treated neurons (high levels of P-eEF2). We also showed that mGluR activation is required for the synthesis of BDNF protein that occurs after the transient increase in BDNF mRNA transcription mediated by NMDA receptor activation (Jia et al., 2008). In particular we found that AP5 blocked BDNF mRNA and protein synthesis but not eEF2 phosphorylation induced by bicuculline (Fig. 9A,B,J–L) and MPEP blocked bicuculline-induced eEF2 phosphorylation and BDNF protein synthesis, but not BDNF mRNA transcription (Figs. 9A,B,J–L).
It is known that BDNF is involved in synaptic plasticity (Levine et al., 1995; Thoenen, 1995; Korte et al., 1996). In particular, BDNF scales up synaptic transmission and is involved in hippocampal LTP (Korte et al., 1996; Patterson et al., 1996; Ying et al., 2002); while neuronal activity increases the distal localization of BDNF mRNA in dendrites (Tongiorgi et al., 1997) where it is required for pruning and enlarging dendritic spines (Shimada et al., 1998; An et al., 2008; Tanaka et al., 2008).
It has been shown that in neurons exogenous BDNF decreases the eEF2 phosphorylation and enhances global protein synthesis (Takei et al., 2009), while BDNF-induced LTP in live rats increases eEF2 phosphorylation (Kanhema et al., 2006) suggesting that BDNF signaling in dendrites might contribute to eEF2 phosphorylation. However the major difference between our studies and those of Kanhema et al. and Takei et al., is that while they looked at the effect of exogenous BDNF on neurons, we induced the expression of endogenous BDNF by increasing synaptic activity with bicuculline and the locally produced BDNF may have a different effect on eEF2 phosphorylation than added BDNF. For example mGluR activation was required for synaptic activity-induced eEF2 phosphorylation in our model. However it is possible that endogenous BDNF may also counteract mGluR-induced eEF2 phosphorylation at some point. Takei et al. also showed that eEF2 phosphorylation is reduced in mice overexpressing BDNF and increased in BDNF knock-out mice. Thus in their model BDNF levels were inversely proportional to P-eEF2 levels in vivo. Another a possible explanation for this difference is that Takei et al. were looking at eEF2 phosphorylation in basal conditions and not after synaptic stimulation.
Our data suggest that the lack of BDNF protein in dendrites induced by sieEF2K correlates with reduced spines maturation and number, a hypothesis that is strongly supported by the finding that overexpressing BDNF restored spine morphology after it had been altered by eEF2K knockdown. We used a cDNA coding for BDNF mRNA that contains a CMV promoter and an SV40 30 UTR. This mRNA should remains localized in the cell body (An et al., 2008) and that it might be translated efficiently even in absence of eEF2K and hence rescue the effect of eEF2K knockdown on dendritic spines. However it has recently been shown that in transfected neurons the coding sequence for BDNF contains a dendritic targeting signal (Chiaruttini et al., 2009). Thus it is possible that the mRNA for BDNF constructs we used in this study may have localized to the dendrites. If this is the case, then to account for our finding—the rescue effect of eEF2K knockdown on dendritic spines—we suggest that the translation of this mRNA is not controlled by the eEF2K-eEF2 axis because is missing regulatory elements.
Similarly An et al. (2008) have found that in vivo maturation and pruning of the spines is impaired in neurons lacking the long 3′UTR and dendritic BDNF synthesis. Thus our study and that by An et al. (2008) suggest that the level of BDNF mRNA translation in dendrites (controlled by eEF2 phosphorylation) correlates with the maturation of dendritic spines. However the two studies show a difference in dendritic spine number, which was reduced in our study, and increased in the study by An et al. (2008). The discrepancy might be due to the difference between in vitro versus in vivo models or by the fact that eEF2 phosphorylation regulates the synthesis of other proteins controlling dendritic spine number. We used embryonic neurons in culture, and cannot exclude that the control mechanisms elucidated occur mainly during development, and eEF2K-eEF2 regulation and function may differ in adults (Panja et al., 2009).
Recently it has been shown that eEF2 undergoes phosphorylation during LTP in the dentate gyrus, but such phosphorylation is not required for LTP consolidation or for enhanced Arc synthesis (Panja et al., 2009). At present it is unclear whether the pathways we have elucidated are concerned with LTP consolidation or whether they are mainly involved in the regulation of spine morphology.
To conclude we have delineated a causal chain between linking synaptic activity to spine morphology, which represents an important advance in our understanding of the molecular mechanisms of synaptic plasticity in general and dendritic spine morphology in particular. It seems likely that eEF2 is a key biochemical sensor controlling spine plasticity and hence memory and learning.
Footnotes
This work was supported by grants from Telethon, Italy (Grant No. GGP06208), Fondazione Cariplo (Project number 2006-0779), Compagnia di San Paolo (Project number 2005.1964), the European Community [LSHM-CT-2004-511995, SYNSCAFF (Synaptic Scaffolding Proteins Orchestrating Cortical Synapse Organisation during Development)], PRIN (Progetto di Ricerca di Interesse Nazionale) 2007, CNR RTSL (Consiglio Nazionale delle Ricerche Ricerca Scientifica a Tema Libero), and the Giovanni Armenise-Harvard Foundation Career Development Program. We thank Rossana Pisani and Elisa Zianni for technical assistance and comments on the manuscript. We thank A. Nairn, Yale University, for providing the P-eEF2 antibody and Erin M. Schuman, California Institute of Technology, for the pcDNA3.1-5′myrdGFP 3′ vector. Donald Ward helped with the English.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Banko JL, Hou L, Klann E. NMDA receptor activation results in PKA- and ERK-dependent Mnk1 activation and increased eIF4E phosphorylation in hippocampal area CA1. J Neurochem. 2004;91:462–470. doi: 10.1111/j.1471-4159.2004.02734.x. [DOI] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belelovsky K, Elkobi A, Kaphzan H, Nairn AC, Rosenblum K. A molecular switch for translational control in taste memory consolidation. Eur J Neurosci. 2005;22:2560–2568. doi: 10.1111/j.1460-9568.2005.04428.x. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci U S A. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiner JK, Khorasani H, Nairn AC, O'Dell TJ, Watson JB. Adenylyl cyclase-dependent form of chemical long-term potentiation triggers translational regulation at the elongation step. Neuroscience. 2003;116:743–752. doi: 10.1016/s0306-4522(02)00797-2. [DOI] [PubMed] [Google Scholar]
- Cossenza M, Cadilhe DV, Coutinho RN, Paes-de-Carvalho R. Inhibition of protein synthesis by activation of NMDA receptors in cultured retinal cells: a new mechanism for the regulation of nitric oxide production. J Neurochem. 2006;97:1481–1493. doi: 10.1111/j.1471-4159.2006.03843.x. [DOI] [PubMed] [Google Scholar]
- Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27:13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Garcia PM, Poglia L, Muller D. Spine dynamics and synapse remodeling during LTP and memory processes. Prog Brain Res. 2008;169:199–207. doi: 10.1016/S0079-6123(07)00011-8. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Majewska AK. Dendritic spine geometry: functional implication and regulation. Neuron. 2005;46:529–532. doi: 10.1016/j.neuron.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JM, Chen Q, Zhou Y, Miao S, Zheng J, Zhang C, Xiong ZQ. Brain-derived neurotrophic factor-tropomyosin-related kinase B signaling contributes to activity-dependent changes in synaptic proteins. J Biol Chem. 2008;283:21242–21250. doi: 10.1074/jbc.M800282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- Kauderer BS, Kandel ER. Capture of a protein synthesis-dependent component of long-term depression. Proc Natl Acad Sci U S A. 2000;97:13342–13347. doi: 10.1073/pnas.97.24.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M. Neuroscience. A protoplasmic kiss to remember. Science. 2008;319:1627–1628. doi: 10.1126/science.1155748. [DOI] [PubMed] [Google Scholar]
- Korte M, Staiger V, Griesbeck O, Thoenen H, Bonhoeffer T. The involvement of brain-derived neurotrophic factor in hippocampal long-term potentiation revealed by gene targeting experiments. J Physiol Paris. 1996;90:157–164. doi: 10.1016/s0928-4257(97)81415-5. [DOI] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JC, Lynch G, Vanderklish P, Arai A, Gall CM. Positive modulation of AMPA receptors increases neurotrophin expression by hippocampal and cortical neurons. J Neurosci. 2000;20:8–21. doi: 10.1523/JNEUROSCI.20-01-00008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Avruch J. Glutamatergic regulation of the p70S6 kinase in primary mouse neurons. J Biol Chem. 2005;280:38121–38124. doi: 10.1074/jbc.C500363200. [DOI] [PubMed] [Google Scholar]
- Levine ES, Dreyfus CF, Black IB, Plummer MR. Brain-derived neurotrophic factor rapidly enhances synaptic transmission in hippocampal neurons via postsynaptic tyrosine kinase receptors. Proc Natl Acad Sci U S A. 1995;92:8074–8077. doi: 10.1073/pnas.92.17.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin P, Nastiuk KL, Daniel N, Girault JA, Czernik AJ, Glowinski J, Nairn AC, Prémont J. Glutamate-dependent phosphorylation of elongation factor-2 and inhibition of protein synthesis in neurons. J Neurosci. 1997;17:3445–3454. doi: 10.1523/JNEUROSCI.17-10-03445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci U S A. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Mielke M, Spires-Jones TL, Stoothoff W, Jones P, Bacskai BJ, Hyman BT. A reporter of local dendritic translocation shows plaque-related loss of neural system function in APP-transgenic mice. J Neurosci. 2009;29:12636–12640. doi: 10.1523/JNEUROSCI.1948-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Nägerl UV, Eberhorn N, Cambridge SB, Bonhoeffer T. Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron. 2004;44:759–767. doi: 10.1016/j.neuron.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem. 2009;284:31498–31511. doi: 10.1074/jbc.M109.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Piccoli G, Verpelli C, Tonna N, Romorini S, Alessio M, Nairn AC, Bachi A, Sala C. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J Proteome Res. 2007;6:3203–3215. doi: 10.1021/pr0701308. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Romorini S, Piccoli G, Jiang M, Grossano P, Tonna N, Passafaro M, Zhang M, Sala C. A functional role of postsynaptic density-95-guanylate kinase-associated protein complex in regulating Shank assembly and stability to synapses. J Neurosci. 2004;24:9391–9404. doi: 10.1523/JNEUROSCI.3314-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Rudkin BB, Spirin AS. Regulation of protein synthesis at the elongation stage. New insights into the control of gene expression in eukaryotes. FEBS Lett. 1991;285:170–175. doi: 10.1016/0014-5793(91)80798-8. [DOI] [PubMed] [Google Scholar]
- Sakata K, Woo NH, Martinowich K, Greene JS, Schloesser RJ, Shen L, Lu B. Critical role of promoter IV-driven BDNF transcription in GABAergic transmission and synaptic plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2009;106:5942–5947. doi: 10.1073/pnas.0811431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piëch V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. N-methyl-d-aspartate receptor activation and visual activity induce elongation factor-2 phosphorylation in amphibian tecta: a role for N-methyl-d-aspartate receptors in controlling protein synthesis. Proc Natl Acad Sci U S A. 1997;94:14770–14775. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Mason CA, Morrison ME. TrkB signaling modulates spine density and morphology independent of dendrite structure in cultured neonatal Purkinje cells. J Neurosci. 1998;18:8559–8570. doi: 10.1523/JNEUROSCI.18-21-08559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Nakaoka R, Amano K, Yukimine M, Andoh T, Kuraishi Y, Tsuda M. Differential activation of brain-derived neurotrophic factor gene promoters I and III by Ca2+ signals evoked via L-type voltage-dependent and N-methyl-d-aspartate receptor Ca2+ channels. J Biol Chem. 2000;275:17269–17275. doi: 10.1074/jbc.M909538199. [DOI] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Ishizuka Y, Kakiya N, Inamura N, Namba H, Nawa H. Brain-derived neurotrophic factor enhances the basal rate of protein synthesis by increasing active eukaryotic elongation factor 2 levels and promoting translation elongation in cortical neurons. J Biol Chem. 2009;284:26340–26348. doi: 10.1074/jbc.M109.023010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Thach RE. Translational control of gene expression in a normal fibroblast. Characterization of a subclass of mRNAs with unusual kinetic properties. Biochemistry. 1986;25:2033–2041. doi: 10.1021/bi00356a030. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra F, Castrén E, Thoenen H, Lindholm D. Interplay between glutamate and gamma-aminobutyric acid transmitter systems in the physiological regulation of brain-derived neurotrophic factor and nerve growth factor synthesis in hippocampal neurons. Proc Natl Acad Sci U S A. 1991;88:10037–10041. doi: 10.1073/pnas.88.22.10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]