Abstract
The limited generation of neurons during adulthood is controlled by a balance between quiescence and recruitment of neural stem cells (NSCs). We use here the germinal zone of the zebrafish adult telencephalon to examine how the frequency of NSC divisions is regulated. We show, using several in vivo techniques, that progenitors transit back and forth between the quiescent and dividing state, according to varying levels of Notch activity: Notch induction drives progenitors into quiescence, whereas blocking Notch massively reinitiates NSC division and subsequent commitment toward becoming neurons. Notch activation appears predominantly triggered by newly recruited progenitors onto their neighbors, suggesting an involvement of Notch in a self-limiting mechanism, once neurogenesis is started. These results identify for the first time a lateral inhibition-like mechanism in the context of adult neurogenesis and suggest that the equilibrium between quiescence and neurogenesis in the adult brain is controlled by fluctuations of Notch activity, thereby regulating the amount of adult-born neurons.
Introduction
Adult neural stem cells (NSCs) give rise to newborn neurons in discrete zones of vertebrate brains (Zhao et al., 2008). Although quiescent, i.e., for long periods in the G0 phase of the cell cycle, they can be reactivated in specific circumstances and reenter the G1 phase, allowing neuron generation. The mechanisms regulating the balance between NSC quiescence and recruitment remain incompletely understood.
Several neurogenic zones have been identified in vertebrates in which these mechanisms can be studied. Ependymal cells lining the lateral ventricle in the adult rat brain were proposed to act as NSCs, contributing to the generation of olfactory bulb neurons (Johansson et al., 1999). In the mouse, astrocytes directly underlying this ependymal layer [subependymal zone (SEZ)] have the same function (Doetsch et al., 1999; Mirzadeh et al., 2008). In the adult rodent dentate gyrus, astrocytes with a radial process bordering the subgranular zone (SGZ) serve as the NSCs (Seri et al., 2001, 2004). Finally, throughout the brain of adult zebrafish (Adolf et al., 2006; Chapouton et al., 2006, 2007; Grandel et al., 2006), some radial glial cells lining the ventricular zone (VZ) divide and can generate neurons (Adolf et al., 2006; Pellegrini et al., 2007; März et al., 2010).
Intrinsic regulators, such as cell cycle inhibitors, are involved in maintaining NSCs. Progressive upregulation of p16ink4a during aging lengthens the cell cycle of NSCs (Molofsky et al., 2006), and p21 (Cdkn1a) limits their divisions (Kippin et al., 2005). The polycomb group member Bmi1 favors NSC self-renewal by repressing p16ink4a, p19, and p21 expression (Molofsky et al., 2003; Bruggeman et al., 2005; Fasano et al., 2007). However, the signals instructing NSCs to cycle or remain quiescent are not known. Recently, it was observed that forced activation of Notch signaling blocked the cell cycle reentry of ependymal cells normally observed after injury in the adult mouse telencephalon (Carlén et al., 2009). Notch also limits the regeneration of hair cells in the fish (Ma et al., 2008) and chicken (Daudet et al., 2009). Hence, it appears as an interesting candidate for restricting activation of NSCs in mature tissues.
Notch signaling involves binding of Delta or Jagged ligands, followed by Notch cleavage by the γ-secretase complex to liberate its intracellular domain (NICD). NICD translocates to the nucleus, binds the coactivator MAML (Mastermind), and releases transcriptional repression driven by CBF-1 [RBPJk, Su(H)], inducing the expression of targets such as the Hes/her or Hey genes (Fischer and Gessler, 2007). In the developing neural tube in vertebrates, as well as in the neural epithelium in Drosophila, Notch is used within progenitor pools to drive “lateral inhibition,” enabling ligand-expressing cells to undergo neuronal differentiation, whereas Notch-expressing cells remain in the progenitor mode (Simpson, 1997; Takke et al., 1999). The role of Notch at later stages of development and in the adult is less well understood.
We demonstrate here that the majority of radial glial cells in the zebrafish adult telencephalic VZ are quiescent progenitors, which switch back and forth between quiescence and recruitment after slow oscillating levels of Notch activity. We further identify the dividing progenitors themselves as a Notch signaling source, imposing transitory quiescence to their neighboring glia. We propose that this self-regulatory mechanism controls the number of recruited NSCs in the adult brain.
Materials and Methods
Zebrafish strains and transgenic lines.
Adult fish from 3 to 9 months of age were used. AB/EK crosses were used as wild-type line. The transgenic lines gfap:GFP (Bernardos and Raymond, 2006), uas:nicd and hsp70:gal4 (Scheer et al., 2001), TP1bglob:gfp (Parsons et al., 2009), and deltaA:gfp (R.M. and P.B., unpublished observations) were used. For analysis, fish were anesthetized with 0.4% tricaine, killed on ice, and decapitated. All experiments were performed in accordance with the regulations of the Regierung von Oberbayern on animal welfare.
Heat shocks on adult fish.
Transgenic fish uas:nicd-myc × hsp70:gal4 or wild-type fish were placed in a warm water bath at 38°C for 1, 2, 3, and 4 h, followed by 24 h in normal conditions (28°C), or for 2.5 h, followed by 6 h, 24 h, 48 h, 3 d, 1 week, or 2 weeks at 28°C. As controls, either wild-type fish were heat shocked or transgenic fish were not heat shocked. Because of the mosaic expression of the transgene, the same brains contained NICD–myc-overexpressing and control cells. Myc expression was detectable from 6 h to 3 d after the heat shock (HS), was present only in very few cells after 1 week, and undetectable after 2 weeks.
DAPT treatments.
DAPT (N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester) (Axxora) 10 mm stock solution in DMSO was applied in the swimming water at 28°C at a concentration of 40–50 μm. Control fish were treated with DMSO (1:200). The DAPT solution was exchanged after 18 h. Between the first and the second day of treatment, fish swam for 1–6 h in untreated water.
Electroporation and lipofections.
Zebrafish (3–6 months old) were anesthetized in 0.02% tricaine (3-aminobenzoic acid ethyl ester, Sigma) and immobilized into a piece of foam plastic with a slit, placed into a 6-cm-diameter Petri dish filled with water. Under a stereo microscope, a hole was pierced into the skull right anterior to the epiphysis (supplemental Fig. 3, available at www.jneurosci.org as supplemental material) with a microsurgery tool. An injection capillary was then introduced into the most dorsal part of the diencephalic ventricle (Wullimann et al., 1996), and DNA solution was pressure injected. The colored solution (see below) was visible while propagating into all brain ventricles. The efficiency of transfection was mainly dependent on the quality of the injection, correctly targeting the ventricles. For electroporation, DNA was dissolved in water at 1.5–4 μg/μl, with the same end concentration in control electroporation and coelectroporation of two plasmids. For the lipofection, Lipofectamine 2000 (Invitrogen) was dissolved 1:2 in water or in HBSS. Five microliters of water containing 2–5 μg of DNA (a single plasmid or mix of two plasmids) were mixed to 5 μl of the diluted Lipofectamine. This mix was injected earliest 20 min later. Before loading the capillary, Fast green (Sigma) was added to the injection solution at a final concentration of 0.2 mg/ml. Lipofected fish were released immediately after the injection. For electroporation, the fish hold in the foam plastic were transferred into a box containing 250 ml of water with 0.02% tricaine. Electrodes (Tweezertrodes 520; Harvard Apparatus), connected to an Ovodyne electroporator and current amplifier (TSS20 and EP21; Intracel), were placed close to (2–3 mm) both sides of the head and five pulses (40 V, 50 ms width, 1000 ms space) were given. The following constructs were used: pCS2:egfp; pCS2:myc–dnSu(H) [kindly provided by Chris Kintner, Salk Institute for Biological Studies, La Jolla, CA (Wettstein et al., 1997)]; pCS2:deltastu (Haddon et al., 1998); and pCS2:dn-maml1. The DNA fragment encoding for the highly conserved N-terminal part of zebrafish Maml1 (Nam et al., 2006) was searched in the Ensembl database by blasting the human N-terminal MAML1 (amino acids 13-77) sequence against the zebrafish genome and was found upstream of the annotated maml1 gene (Ensembl database transcript ENSDARG00000076466). The 207 bp fragment was amplified using zebrafish embryonic cDNA and the primers ATGGCGGATTTTGTGGTGCC and CTGCCTGTGTTTGTTGGAGCG. The PCR fragment was subcloned into the pSCA vector (Stratagene cloning kit) and then into the pCS2 vector using the XhoI and Not1 restriction sites.
Bromodeoxyuridine injections.
A fresh solution of 2.5 mg/ml bromodeoxyuridine (BrdU) in saline was injected (5 μl/0.1 g body weight) intraperitoneally into the anesthetized fish, either a single time or repeatedly each day for 5 d for cumulative labelings.
In situ hybridization and immunohistochemistry. Brains were fixed, embedded, and cut (Microm HM 650V) as described previously (Adolf et al., 2006). Primary antibodies directed against the following antigens were incubated 2 h at room temperature: BrdU (Roche), S100β (Dako), green fluorescent protein (GFP) (chicken; Aves Laboratories), proliferating cell nuclear antigen (PCNA) (Santa Cruz Biotechnology), MCM5 (kindly provided by Soojin Ryu), Hu (Invitrogen), polysialic acid-neural cell adhesion molecule (PSA-NCAM) (Millipore Bioscience Research Reagents), brain lipid binding protein (BLBP) (Millipore Bioscience Research Reagents), and Myc 9E10 clone (Sigma). An internal peptide of Ink4b (Ensembl transcript ENSDARP00000054208, CDLRPVDVAQQTGNVDVVELL) was synthesized and coupled to keyhole limpet hemocyanin (KLH) or ovalbumin (Peptide Specialty Laboratories). Rats were immunized with 50 μg of peptide–KLH using CPG 2006 (Tib Molbiol) and incomplete Freunds adjuvant as adjuvant. After a 6 week interval, a final boost without adjuvant was given 3 d before fusion of the rat spleen cells with the murine myeloma cell line P3X63-Ag8.653. Hybridoma supernatants were tested in a differential ELISA with the specific peptide or an irrelevant peptide coupled to ovalbumin. Positive reacting hybridomas (IgG2a) were further analyzed by immunohistochemistry. Secondary antibodies from goat coupled to AlexaFluor dyes (488, 555, or 647; Invitrogen) were incubated 30 min at 1:1000 dilution.
For in situ hybridization, gelatin albumin-embedded brains were cross sectioned at 80 μm before hybridization, as described previously (Topp et al., 2008). For fluorescent in situ hybridizations, whole brains were hybridized with a digoxigenin (DIG)-labeled probe, washed, subsequently embedded in agarose and sectioned, as described previously (Adolf et al., 2006), incubated with anti-DIG–HRP Fab fragments (Roche), and processed with a tyramide–cyanine 3 TSA kit (PerkinElmer Life and Analytical Sciences). The following antisense digoxigenin-labeled probes were used: notch1a, deltaA, deltaB, deltaD, her4, and ascl1a. The maml1 probe (Ensembl database transcript ENSDART00000108868) was amplified by PCR from zebrafish embryonic cDNA using the primers CTGTACCCTACGAGGACATCCAGG and GGAGGGTAGGAGCCTGAATCCT and cloned into the Stratagene pSCA vector. notch 1b, notch2, notch 3, delta like4, jagged1a, and jagged1b were kindly provided by the Julian Lewis Laboratory.
Image acquisition, cell countings, and statistics.
Immunostainings were documented with a confocal microscope (Carl Zeiss LSM510 meta) using the 20× and 40× water-immersion and 63× oil-immersion objectives. Live images of the TP1bglob:gfp fish were taken with a Leica stereomicroscope (MZ1600). Cells were counted manually using the LSM Image Examiner software, separately in each channel (488, 543, and 633 nm) on single optical sections (1 μm thickness), to assess the colocalization of two or three markers. Several vibratome sections per brain (three to six) were photographed as stacks of 10–25 optical sections. Single optical sections are shown in the figures, if not differently stated. The proportions of distinct cell types were calculated in several brains, and averages ± SD or SEM are given as a result. Methods for grouped data (repeated measurements data) were applied in the statistical analysis, taking into account data collected from several sections of several brains. Group comparisons and ANOVAs were calculated by fitting appropriate linear mixed-effects models with brain as the grouping variable. For paired comparisons, the grouping variable was section nested within brain. The R and Excel software were used [R Development Core Team (2008), http://www.R-project.org, Jose Pinheiro, Douglas Bates, Saikat DebRoy, Deepayan Sarkar, and the R Core team; nlme: Linear and Nonlinear Mixed Effects Models, R package version 3.1–89].
Results
The ventricular zone of the adult telencephalon contains three cell states
With the aim of identifying quiescent progenitors within the neurogenic domain of the adult zebrafish telencephalon, we examined the cellular composition of the ventricular zone using radial glia markers such as the gfap:GFP transgenic line (Bernardos and Raymond, 2006; Bernardos et al., 2007) and S100β, proliferation markers such as PCNA and MCM5, expressed in late G1 phase to G2 but not in G0 (Gonzalez et al., 2005), and markers of neurogenesis such as the neuroblast specific epitope PSA-NCAM or the proneural gene ascl1a (Fig. 1). We found three different cell types, which together accounted for virtually all ventricular zone cells: (1) nondividing radial glial cells, expressing gfap:GFP, S100β, and BLBP (BLBP not shown) (März et al., 2010) but not PCNA (state I cells) (Fig. 1a–d, green arrows); (2) dividing radial glial cells that expressed gfap:GFP and S100β together with PCNA (state II cells) (Fig. 1a–d, yellow arrows); and (3) cells that expressed PCNA in the absence of radial glia markers (Fig. 1a–d, red arrows) but with PSA-NCAM and ascl1a (state III cells) (Fig. 1e–g, red arrows in g), which most likely correspond to committed progenitors (dividing neuroblasts) on the way to becoming neurons and also possibly oligodendrocytes.
Figure 1.
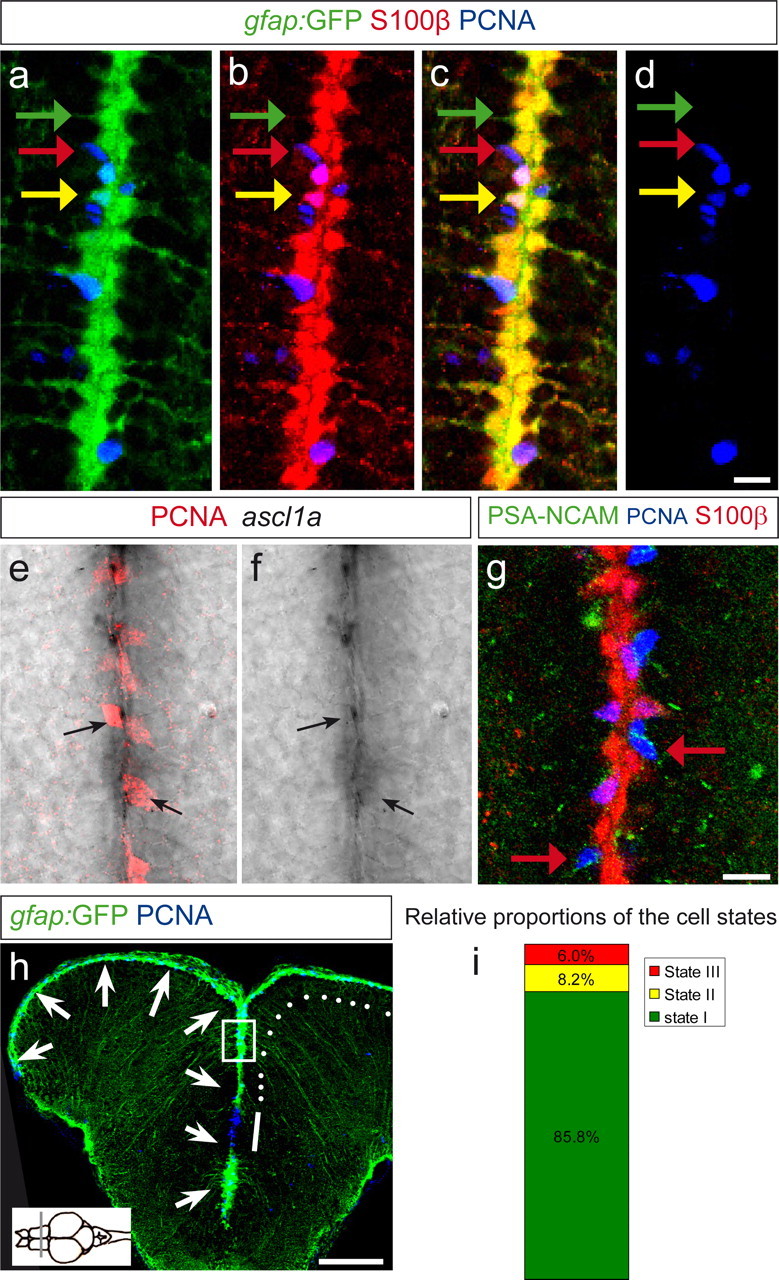
The telencephalon neurogenic zone contains three cell states. Telencephalic cross sections at the anteroposterior level indicated in the inset on the bottom left in h. a–d, Single optical section in a gfap:GFP transgenic brain section, immunostained for GFP (green), S100β (red), and PCNA (blue), depicting a close-up view on the ventricular surface at the midline between the two brain hemispheres. gfap:GFP and S100β are coexpressed in all radial glial cells (yellow in c). Three cell states are visible: the majority of radial glial cells are noncycling (PCNA-negative; green arrow, state I cells). Some radial glial cells are PCNA positive (yellow arrow, state II cells). Some cells express only PCNA and no radial glial marker (red arrow, state III cells). e–g, State III cells express markers of neuroblasts. e, f, Expression of ascl1a revealed by in situ hybridization (black signal), revealing colocalization with PCNA (red). g, PSA-NCAM antibody staining (green) labels many state III cells, as shown by red arrows, revealing their identity of migrating neuroblasts. h, Overview of a gfap:GFP telencephalic section in a confocal stack projection, stained for GFP (green) and PCNA (blue). The arrows depict the telencephalic ventricle, located at the surface and at the midline. The boxed area depicts the region shown in a–g. The white line depicts the pallial–subpallial border region, in which mostly state III cells are located. The white dots depict the pallial ventricular region in which states I, II, and III are intermingled. This latter region is quantified in i and examined further in the next experiments. i, Relative proportions of state I, II, and III cells, counted in 2845 VZ cells of four brains. Scale bars: (in d) a–d, 10 μm; (in g) e–g, 10 μm; h, 100 μm.
The relative distributions of these cell types varied along the dorsoventral axis of the ventricular zone, in agreement with previous studies reporting the spatial organization of ventricular proliferation domains (Adolf et al., 2006). At the pallial–subpallial border, mostly state III cells were present and clustered in a stripe of active proliferation extending toward the olfactory bulb (Fig. 1h, white bar). More dorsally, in the pallium (Fig. 1h, white dots), the three cell states were intermingled in a scattered manner, with a majority of state I cells. We further examined this pallial domain and quantified the proportions of cells in states I, II, and III from four brains. State I cells represented 86 ± 5%, state II 8 ± 5%, and state III 6 ± 2% of the total ventricular zone cells (Fig. 1i). Most radial glial cells of the adult zebrafish telencephalon, including state I cells, express the progenitor marker Nestin (Lam et al., 2009), but the exact status of nondividing glial cells has not been defined. In particular, it remains unknown whether this population has irreversibly exited the cell cycle or is in the quiescent, G0 phase and thereby could include potential NSCs of the adult zebrafish brain. To address this issue, we tested whether these cells could be brought back into the cell cycle during stimulation.
Notch receptors and a Notch activity reporter are expressed along the ventricular zone in the nondividing radial glia population
We searched for signaling mechanisms active in state I cells and which could control their cycling status. Notch receptors and ligands were expressed in the adult neurogenic domains. Specifically, notch1a, notch1b, notch2, and notch3 as well as maml1, encoding the Notch coactivator Maml1 (Mastermind), were expressed along the telencephalic ventricular zone, within the first cell rows containing the three cell states described above (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). notch3 was the most strongly expressed notch gene in the VZ (Fig. 2a) and displays an expression domain complementary to the cell cycle markers PCNA and MCM5, excluding MCM5-expressing cells (Fig. 2b,c) [both PCNA and MCM5 are expressed in nearly the same set of cells, with a slightly higher number of MCM5-expressing cells because of its earlier onset of expression in late G1 phase (Gonzalez et al., 2005)] (data not shown).
Figure 2.
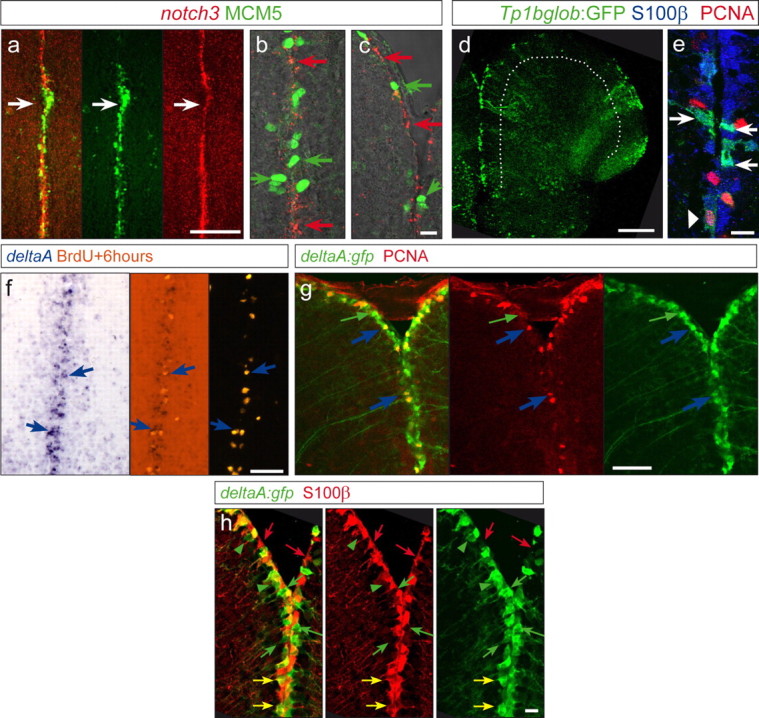
Expression of notch3 and the Notch reporter transgenic line highlight state I cells, whereas proliferating progenitors express deltaA. a–c, notch3a in situ hybridization (red signal) and MCM5 (green, proliferation marker) in the midline of the telencephalon showing a complementary expression: regions with dense expression of MCM5 express lower levels of notch3 (white arrow in a), and high magnifications (b, c) reveal a complementary expression at the single-cell level. d, Cross sections, as a projection of a confocal stack, of a TP1bglob:gfp transgenic telencephalon, highlighting activation of RBPJ targets/canonical Notch signaling (green) along the VZ (the VZ is underlined by the white dotted line). e, GFP-positive cells colocalize with S100β (blue). Only a few (6%) weak GFP-positive cells (arrowhead) colocalize with S100β/PCNA double-positive cells (PCNA in red). f, g, Dividing cells marked by a BrdU pulse 6 h before analysis (orange, f) or by PCNA (red, g) express the Notch-ligand DeltaA (blue arrows), as visible by in situ hybridization (f, blue signal) and in the deltaA:gfp transgenic line (g, green; 95% of the PCNA-positive cells are GFP positive). The green arrow in g depicts also deltaA:GFP cells that are PCNA negative (64% of the GFP-positive cells are PCNA negative). h, Expression of deltaA:GFP (green) compared with S100β-positive radial glia (red). Green arrows point to single GFP-positive cells, and red arrows point to cells expressing S100β only. The majority of labeled cells are double-positive cells and appear yellow in the first panel (yellow arrows, 75% of deltaA:GFP cells are S100β positive). Scale bars: a, d, 100 μm; c, e, h, 10 μm; f, g, 50 μm.
To determine specifically which ventricular cell state displays active Notch signaling, we analyzed a transgenic line expressing GFP under the control of concatamerized Su(H)/RBPJk binding sites (TP1bglob:gfp transgenic line) (Fig. 2d,e). All GFP-positive cells coexpressed S100β along the adult telencephalic ventricle, indicating that they are radial glia (Fig. 2e), in keeping with the expression of notch genes. GFP from this line was however expressed in only 16.5% of the S100β-positive cells, likely because of the site of transgene integration, in addition to possible restrictions of Notch activity to a subset of notch-expressing cells. This line nevertheless permitted analyzing the cycling activity of GFP-positive cells in relation to the intensity of GFP expression. We found that most GFP-positive cells were PCNA negative, whereas the small proportion (6% of GFP;PCNA double-labeled cells) was characterized by either weak levels of GFP or weak levels of PCNA (Fig. 2e, arrowhead pointing to a PCNA-positive cell that is weakly GFP positive), suggesting that coexpression of these two markers is not a stable state. The ligand-encoding genes deltaA, deltaB, deltaD, delta-like4, jagged1a, jagged1b, and jagged2 (data not shown) were also expressed in the telencephalic VZ (supplemental Fig. 1e–j, available at www.jneurosci.org as supplemental material), and we analyzed more closely which type of ventricular zone cell expressed Notch ligands. In contrast to notch3, deltaA could be found in dividing cells, as visible by in situ hybridization (Fig. 2f) and in a deltaA:gfp transgenic line (Fig. 2g,h): specifically, the deltaA:GFP-positive population distributed in 64% of PCNA-negative cells (state I cells) and 36% of PCNA-positive cells. Virtually all (95%) dividing cells at the VZ were expressing deltaA:GFP. Together, we found a distinct expression pattern of Notch receptors and DeltaA, depending on the dividing status of the VZ cells: Notch is predominantly active in nondividing cells, whereas almost all dividing cells express DeltaA.
State I cells are progenitors maintained in a quiescent state by Notch signaling
The expression patterns observed above indicate a possible role of Notch signaling in the quiescence of state I cells. To test this hypothesis, we first blocked Notch activity pharmacologically in vivo by applying to the swimming water the small-molecule DAPT, an inhibitor of the γ-secretase complex preventing the generation of NICD, hence Notch signaling (Geling et al., 2002). To verify that this treatment lead to a successful block of Notch signaling, we analyzed the live expression of GFP in the TP1bglob:gfp Notch reporter line before and after treatment, using the olfactory epithelium, a structure visible in the live fish under simple stereomicroscopical observation. GFP was expressed in scattered cells and showed a rapid decrease of expression already after 30 h of treatment, indicating that the transgene was successfully silenced by DAPT treatment (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). We then analyzed the expression of PCNA in the telencephalon after DAPT treatment. Remarkably, blocking Notch elicited a massive increase in the number of PCNA-positive cells (Fig. 3a,b), representing a relative change from 14.2 to 57.5% within the ventricular population (see percentages in Fig. 3e, states II + III), a fourfold increase.
Figure 3.
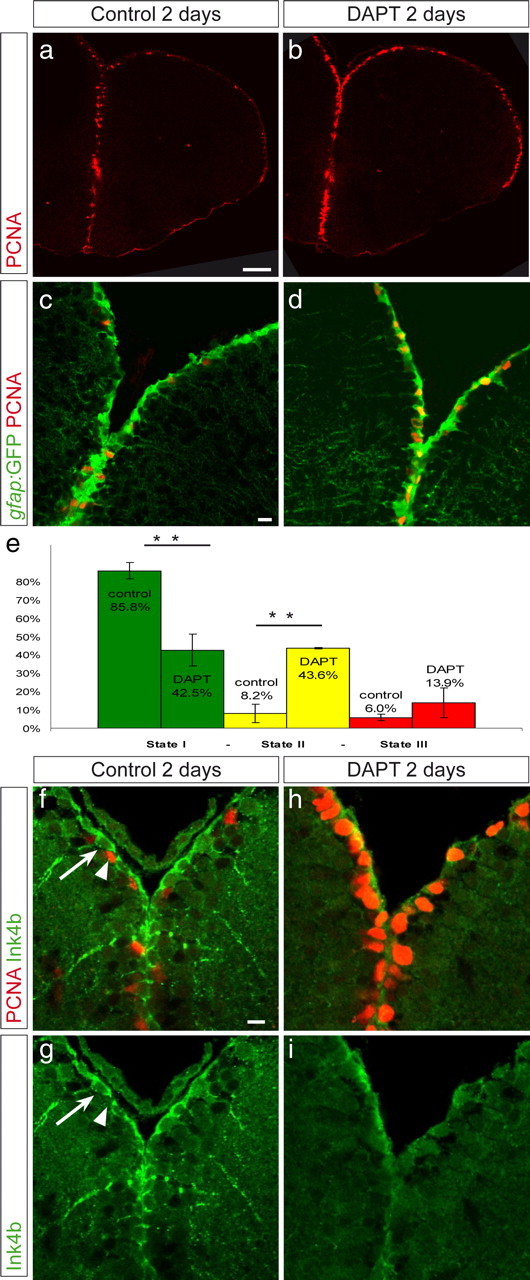
Inhibition of Notch activity triggers a state I to II switch. Vehicle-treated fish (a, c, f, g) and fish treated with 40 μm DAPT (b, d, h, i) were compared. a, b, Overview on a telencephalic cross section, as a single confocal plane, of a gfap:GFP fish stained for PCNA (red), revealing the increased density of PCNA-positive cells along the ventricular zone in DAPT-treated fish. c, d, Dorsal telencephalic ventricular zone of gfap:GFP, control (c), or DAPT-treated (d) fish, revealing an increase in state II cells (PCNA, red; GFP, green) within the GFAP-positive cell population. e, The relative proportions of states I, II, and III cells were calculated in n = 4 control brains (2845 cells) and n = 3 DAPT-treated brains (3199 cells). The difference between control and DAPT is highly significant for state I and state II cells (t test, **p < 0.001). The density of GFAP-positive cells per ventricular surface was unchanged. Error bars show the SEM (n = 4 and 3). f–i, DAPT-treated (h, i) and vehicle-treated (f, g) brains were immunostained for Ink4b (green). Ink4b is found in noncycling cells (arrow) but not in PCNA-positive cells. Ink4b expression disappeared during DAPT treatment. Scale bars: a, 50 μm; c, f, 10 μm.
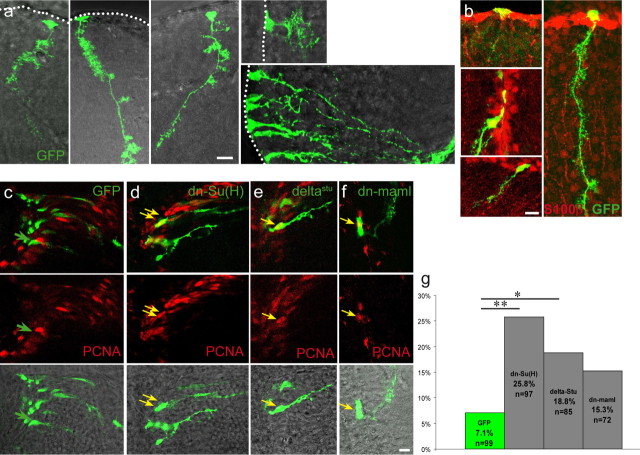
To add support to this result, we also aimed to block the Notch signaling pathway using genetic approaches. We developed live electroporation and lipofection techniques (supplemental Fig. 3, available at www.jneurosci.org as supplemental material), allowing the transfection of VZ cells with DNA constructs (Fig. 4a,b). Using two reporter genes, we first verified that these were efficiently cotransfected in the same cells (supplemental Fig. 3c, available at www.jneurosci.org as supplemental material), permitting to use a GFP construct as tracer of transfected cells in subsequent experiments. Next, three dominant-negative constructs impairing the Notch pathway were electroporated or lipofected into the VZ, and the cycling activity of transfected cells was analyzed 24–48 h later (Fig. 4c–f). Specifically, we chose dominant-negative constructs of the ligand Delta (deltastu), of the transcription factor Suppressor of Hairless [dn-Su(H)], and of the coactivator Mastermind (dn-maml1), which all block Notch signaling in a cell-autonomous manner. deltastu encodes the extracellular domain of Delta and renders its expressing cells insensitive to Notch (Haddon et al., 1998), whereas both dn-Su(H) and dn-Maml act in a complex with NICD and block its transcriptional activity (Wettstein et al., 1997; Weng et al., 2003). We observed a significant increase in the proportion of PCNA-positive cells (3.6-fold and 2.6-fold, respectively) among GFP-positive cells when dn-Su(H) or deltastu were transfected, as well as a smaller increase in PCNA-positive cells transfected with dn-maml1 compared with cells transfected with GFP alone (Fig. 4g). Thus, genetic interference with the Notch pathway using dominant-negative constructs led to increased entry of VZ cells into the cell cycle, much like in response to the DAPT treatment.
Figure 4.
In vivo transfection via lipofection and electroporation of dominant-negative constructs of the Notch pathway induces an increase of PCNA-positive cells. The ventricular zone of the telencephalon in adult fish was lipofected or electroporated with constructs in the pCS2 vector expressing eGFP (a–c) or was coelectroporated with pCS2–eGFP together with pCS2–dn-Su(H) (d), pCS2–deltastu (e), or pCS2–dn-maml1 (f). a and b depict the morphology and S100β expression (b) of transfected radial glia, which can be evaluated as individual cells. Transfected cells in c–f were assessed for their expression of PCNA (red) 2 d after electroporation; green arrows point to single GFP-positive cells, and yellow arrows point to GFP and PCNA double-positive cells. The percentage of GFP-positive cells also expressing PCNA is plotted in g for the different constructs. Note that all three dominant-negative proteins with Notch blockade activity increase cell cycle entry. In the case of dn-Su(H) and DeltaStu, this increase reaches significance (p = **0.0004 and *0.02, respectively, Fisher's exact test). n is the total number of cells counted of 5–10 brains per construct. Scale bars, 10 μm.
This increase in the proportion of cycling cells after Notch signaling inhibition might be attributable to an acceleration of the cycle speed of previously PCNA-positive cells or to the activation of quiescent cells. To distinguish between these possibilities, we calculated the percentage of nondividing radial glial cells in control and DAPT-treated animals (Fig. 3c–e). During DAPT treatment, the proportion of nondividing radial glial cells was significantly reduced, decreasing from 85.8 to 42.5% (Fig. 3e, state I cells). This change primarily occurred within the radial glia population because the proportion of dividing glial cells increased in the same relative amount (Fig. 3e, state II cells), whereas the total amount of radial glial cells remained unchanged (data not shown). Hence, blocking Notch activity induces a switch from state I to state II. We conclude that a large proportion of state I cells are maintained in quiescence by Notch signaling. To add support to these results, we analyzed the expression of the cell cycle inhibitor Ink4b (Cdkn2b, p15ink), a close homolog of p16ink, which inhibit CDK4 and CDK6 (Cánepa et al., 2007). We found that it was expressed in noncycling radial glia, with a cytoplasmic localization, as observed in other systems (Reynisdóttir and Massagué, 1997) (Fig. 3f,g, arrow). Ink4a protein was almost undetectable after DAPT treatment (Fig. 3h,i), in agreement with the entry of the radial glial cells into cell cycle.
Notch activity regulates quiescence next to dividing progenitors
Although a significant proportion of the state I cells were converted to state II cells during DAPT treatment, this switch was most noticeable in regions already containing cycling cells (Fig. 5a–e). This finding led us to speculate that Notch might maintain quiescence (state I) via contact with dividing and/or differentiating progenitors (state II and III cells). Three sets of data support this hypothesis. First, as described above, almost all (95%) PCNA-positive cells express the Notch ligand Delta (Fig. 2f,g). Second, PCNA-positive cells were repeatedly found adjacent to GFP-positive cells in the Notch transgenic reporter TP1bglob:gfp (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Finally, we reasoned that, if PCNA-positive cells keep neighboring cells quiescent by activating Notch signaling, then DAPT-reactivated cells should be found more often adjacent to recently labeled BrdU-positive cells. To address this, we analyzed PCNA expression in cells neighboring BrdU-labeled cells in both control and DAPT-treated fish (Fig. 5f–l). In vehicle-treated fish 3 d after BrdU injection (Fig. 5f–h), dividing cells are spaced from each other by several cell diameters, and most newly dividing cells (PCNA-positive, BrdU-negative) were not in contact with BrdU-labeled cells. In contrast, when DAPT was administered (Fig. 5i–k), many newly dividing cells (white arrows) were located close to BrdU-labeled cells (green arrows). To quantify these observations, we categorized BrdU-labeled cells into three groups according to their newly dividing (PCNA-positive, BrdU-negative) neighbors: no dividing neighbor, one dividing neighbor, and two and more dividing neighbors (Fig. 5l). We found that DAPT treatment induced a massive drop in the proportion of BrdU-labeled cells without dividing neighbor (70 to 19%; n = 2 brains for DAPT, 2 brains for vehicle) and a correspondingly massive increase in the proportion of BrdU-labeled cells with two and more dividing neighbors (1.3 to 40.2%). The overall increase in the number of PCNA-positive cells after DAPT treatment, however, makes it more likely for BrdU-positive cells to have dividing cells as neighbors. To exclude this as the sole explanation of our results, we analyzed the neighbors of ventricular zone cell having incorporated BrdU 3 weeks earlier. Although the proportion of such BrdU-labeled cells without PCNA-positive neighbor was also decreased after a DAPT treatment (68 vs 45%), and the proportion of those with two or more neighbors was increased (13 vs 8%), both changes were not significant (Fig. 5m). These results demonstrate that one mechanism by which progenitors are maintained quiescent in the adult brain involves Notch signaling activated by dividing neighbors. Additional, yet unknown, signaling mechanisms may be responsible for maintaining the remaining noncycling radial glia located farther away (see Discussion).
Figure 5.
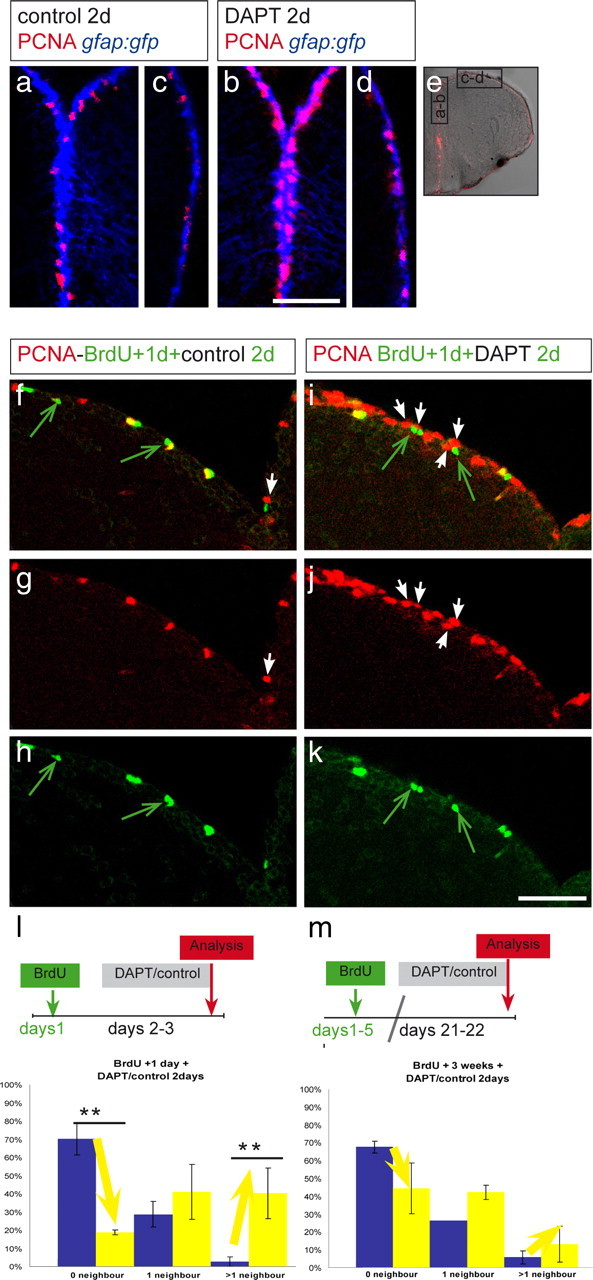
Notch maintains quiescence in cells neighboring active progenitors. DAPT-treated (b, d, i–k) or vehicle-treated (a, c, f–h) fish were compared. e, Overview of a telencephalic cross section. Black rectangles depict regions enlarged in a–d. a, b, Medial ventricular area, containing dividing progenitors (PCNA, red) under normal conditions (a) and a higher density of PCNA-positive cells after DAPT (b). c, d, Dorsolateral area of the telencephalon (as boxed in e; medial is to the top and lateral to the bottom) containing few PCNA-positive cells under normal conditions (c) and a slight increase of PCNA-positive cells after DAPT (d). The proportion of PCNA-positive cells reaches a higher level in medial regions under Notch-blocking conditions. f–k, Dorsomedial region harboring a density of PCNA-positive cells comparable with the region in a. BrdU was administered twice within 3 h, and 1 d later, the fish were treated with vehicle (f–h) or DAPT water (i–k) for 2 d. Cells neighboring BrdU-positive cells were analyzed for PCNA expression. Few BrdU-labeled cells in control brains are in contact with newly dividing, PCNA-positive cells (white arrow, f, g), and many BrdU-positive cells (some of them are still PCNA positive; yellow) have no PCNA-positive (red) neighbor (green arrows, f, h). Most BrdU-labeled cells after DAPT are surrounded by PCNA-positive cells (arrows in i–k), suggesting that Notch prevents neighbors of an already dividing cell to enter cell cycle. The amount of BrdU-positive cells is unchanged, suggesting that the cell cycle speed has not been changed by the DAPT treatment. l, m, BrdU-labeled cells, counted in two brains for each condition, were categorized in three groups according to their neighbors: no dividing neighbor, one dividing neighbor, and more than one dividing neighbor. The proportion of cells belonging to each category is represented, and the error bars represent the SEM, in which n is number of brains. The blue bar of each category represents vehicle-treated and the yellow bar DAPT-treated animals. l, In DAPT-treated animals, the proportion of BrdU-positive, 3-d-labeled cells without dividing neighbor is decreased, whereas the proportion of BrdU-labeled cells with more than one neighbor is increased (one-way ANOVA for repeated measurements; 6 sections from 2 control brains and 4 sections from 2 DAPT brains; **p < 0.01 for 0 neighbors, **p = 0.02 for 2 or more neighbors). m, Fish were injected for 5 d with BrdU and treated 3 weeks after the last injection with DAPT or with DMSO. The neighbors from these BrdU-positive, 3-week-labeled cells did not reveal a significant difference after DAPT treatment compared with control treatment. The nonsignificant changes observed are probably attributable to the general increase of dividing cells along the VZ. Scale bars: in a for a–d, in f for f–k, 100 μm.
Notch activity is sufficient to reinstall quiescence within the radial glia population
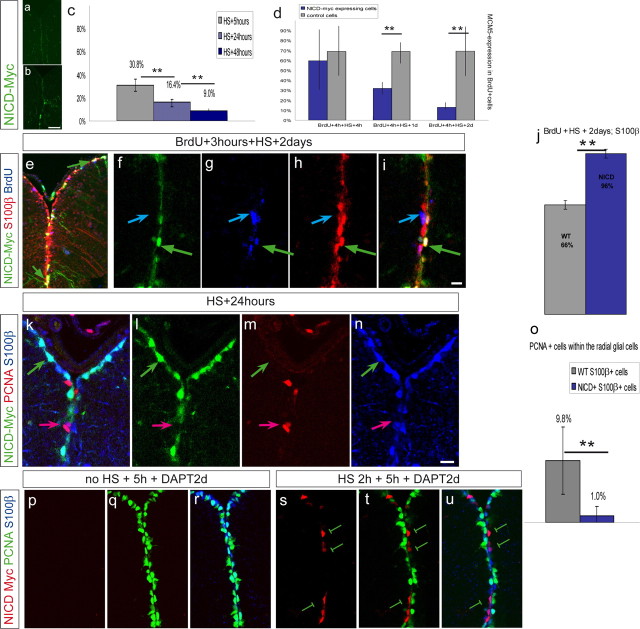
To substantiate the conclusion that endogenous Notch activity is necessary for the maintenance of quiescence within radial glial cells, we assessed the effect of constitutive Notch activation in the adult ventricular zone. Specifically, we asked whether ectopic Notch activation in state II cells would be sufficient to drive these cells toward state I. Hence, we overexpressed the intracellular domain of Notch in the ventricular zone using the double-transgenic line hsp:gal4 [expression of galactosidase-4 (Gal4) driven by the heat shock promoter) × uas:nicd–myc (expression of Notch intracellular domain–Myc driven by the binding of Gal4 to the upstream activating sequence (UAS) promoter] (Scheer et al., 2001). A HS of the fish at 38°C promoted a mosaic expression of NICD–Myc in the adult brain (Fig. 6b,f,l,s). Expression was confined to the area of interest, along ventricular areas of the brain. Fish without HS revealed no or very weak Myc staining (Fig. 6a,p), indicating that the transgene expression was not leaky.
Figure 6.
NICD expression induces a switch from state II to state I. Double-transgenic fish carrying hsp70:gal4 and uas:nicd–myc were submitted to HS for 2.5 h at 38°C. Expression of NICD–Myc fusion protein is revealed by Myc antibody staining on telencephalic cross sections, as depicted in green (a–n) or red (p–u). a, b, Anti-Myc staining in a transgenic fish without HS (a) and with HS (b), indicating that the transgene is not or is only weakly expressed before HS. c, Fish were killed 5, 24, and 48 h after the end of the HS. The percentage of dividing cells (expressing MCM5) was calculated, in three independent experiments. The proportion of dividing cells significantly decreases with increasing time of NICD expression (n = 3 brains each with a total of 444, 1884, and 1540 counted cells for the respective time points; **p = 0.012, one-way ANOVA). d, In two independent experiments, BrdU was administered 3 h before HS to trace NICD–Myc-expressing (blue bars) and neighboring wild-type (gray bars) cells that had just divided. The proportion of such cells that remained positive for MCM5 was calculated. Control BrdU-labeled cells essentially remained MCM5-positive over 2 d, but NICD-expressing BrdU-labeled cells rapidly exited the cell cycle (5–11 sections per brain from 2 brains for each time point; significant decrease for the second and third time point, paired t test for repeated measurements, **p < 0.01). The three time points are significantly different from each other (interaction term in repeated measurement two-way ANOVA, p < 0.01). e–j, BrdU was administered 3 h before HS. At 2 d after HS, the proportion of BrdU-positive cells (blue) expressing S100β (red) within the NICD–Myc-positive population (green), quantified in i, is higher than within the neighboring control population. The green arrow in f–i depicts a Myc+ BrdU+ cell that is S100β positive, while the Myc-negative BrdU+ cell depicted by the blue arrow is S100β positive. j, Quantification (n = 4 sections, 178 Myc+S100β+ cells, 407 Myc(−)S100β+ cells, **p < 0.01, paired t test). k–o, One day after HS, the proportion of S100β+ (red), PCNA+ (blue) cells (state II cells) within the total S100β+ population was significantly decreased in the NICD-Myc+ (green) compared to the NICD-Myc-negative population (n = 2 brains, total of 572 Myc+ and 1701 Myc-negative cells counted in 11 sections, **p < 0.01, paired t test for repeated measurements). Scale bars: b, 50 μm; i, m, 10 μm. The error bars represent the SEM, with n = number of brains (c, d, o) or number of sections (j). p–u, NICD expression can rescue the effect of DAPT. Transgenic animals were subject to a 2 h HS (s–u) or no HS (p–r) followed by a 5 h pause and a 2 d DAPT treatment. The division status (PCNA expression, green) of Myc-negative radial glia (expressing S100β, blue) or Myc-positive radial glia (anti-Myc immunocytochemistry, red) was then assessed. Note that all Myc-positive glia are negative for PCNA (n = 113 Myc-positive cells counted out of 8 sections from three brains; some examples are indicated by the green bars). Hence, they fail to induce cell cycle upon DAPT treatment, in contrast to Myc-negative glia in non-heat-shocked (p–r) or heat-shocked (s–u) animals.
We analyzed the proportion of dividing NICD–Myc-positive cells, using the markers PCNA or MCM5, as a function of time after HS or duration of HS. We consistently observed a decrease in the proportion of cells maintained in cycle. Using MCM5, this proportion gradually and significantly declined from 30 to 9% (Fig. 6c). To assess whether actively cycling cells could enter quiescence during NICD expression, we counted the cells that had incorporated BrdU 3 h before the HS and that remained MCM5 positive at several time points after the HS. As an internal control, these values were compared at each time point with the proportion of MCM5-positive, BrdU-positive cells in the neighboring, NICD–Myc-negative population. Although a high proportion of wild-type cells remained in cycle 8 h, 1 d, and 2 d after BrdU incorporation, NICD-expressing cells exited the cell cycle much quicker (Fig. 6d). Thus, expressing NICD was sufficient to induce a noncycling state.
Next, to assess the fate of cells exiting the cell cycle during Notch activation, we counted how many of the NICD–BrdU double-positive cells expressed S100β 2 d after HS (Fig. 6e–i). This proportion was much higher (97%) than for neighboring wild-type cells (65%) (Fig. 6i), which acquire after 2–3 d an early neuronal identity [∼50% of Hu expression (Adolf et al., 2006)]. In agreement with this conclusion, only 1% of NICD–Myc-positive, S100β-positive cells expressed PCNA 1 d after HS compared with 10% of the control cells (Fig. 6j–n). Thus, overexpressing Notch induced proliferating progenitors to exit the cell cycle and acquire an S100β-positive, state I identity. The specificity of these results was further supported by our observation that NICD expression was sufficient to restore radial glia quiescence in the presence of DAPT, as expected from a Notch component acting downstream of DAPT action. To address this point, HS-induced NICD expression was followed by a 2 d DAPT treatment, and the division status of NICD–Myc-positive radial glia was assessed. Although the vast majority of Myc-negative, S100β-positive cells expressed PCNA in both non-heat-shocked or heat-shocked fish (Fig. 6p–u), none of the Myc-positive cells were PCNA positive in heat-shocked fish (Fig. 6s–u).
To test whether NICD-induced cell cycle exit is indeed reversible (quiescent state) or whether NICD-expressing cells have acquired a postmitotic S100β-positive state, we analyzed a series of brains at increasing time points after HS, up to 2 weeks (supplemental Fig. 5, available at www.jneurosci.org as supplemental material). Quantification of state II cells reveals a significant increase from 3 d to 1 week and from 1 week to 2 weeks after HS, coinciding with the steady decrease of the number of Myc–NICD-positive cells (from 24 to 10 to 0% of the total ventricular zone cells; data not shown). Thus, after the peak of exogenous Notch signaling induced by HS, S100β-positive cells are able to reenter the cell cycle, indicating that they did not acquire a postmitotic fate. Together, these results demonstrate that dividing radial glial cells reversibly enter quiescence when Notch is activated.
Endogenous Notch activity controls the equilibrium between state I and II
Experimental manipulations of Notch activity indicate that states I and II are plastic and interrelated. However, do cells switch back and forth between these states endogenously, and, if so, does this involve Notch? To address this question, we analyzed the fates of cells that had been in cycle several weeks earlier. Accordingly, BrdU was injected repetitively for 5 d in the gfap:GFP transgenic background, followed by a chase period of several weeks. We expect BrdU-labeled cells remaining at the VZ to have originated from cycling state II cells, because the descendants of state III cells will most probably have differentiated and entered the parenchyma. We then analyzed label-retaining BrdU-positive cells that had remained in the germinal area. The gfap:GFP transgenic line revealed ventricular BrdU-positive cells that were GFP positive but PCNA negative (Fig. 7a–e), indicative of a state II to state I transition. Likewise, in the TP1bglob:gfp Notch reporter line, after 60 d, some (8.5%) of the BrdU-positive PCNA-negative cells at the ventricle were GFP positive (Fig. 7f–j). Together, these results demonstrate that state II cells can endogenously return to the state I and that at least a subpopulation of these cells displays a Notch signaling response.
Figure 7.
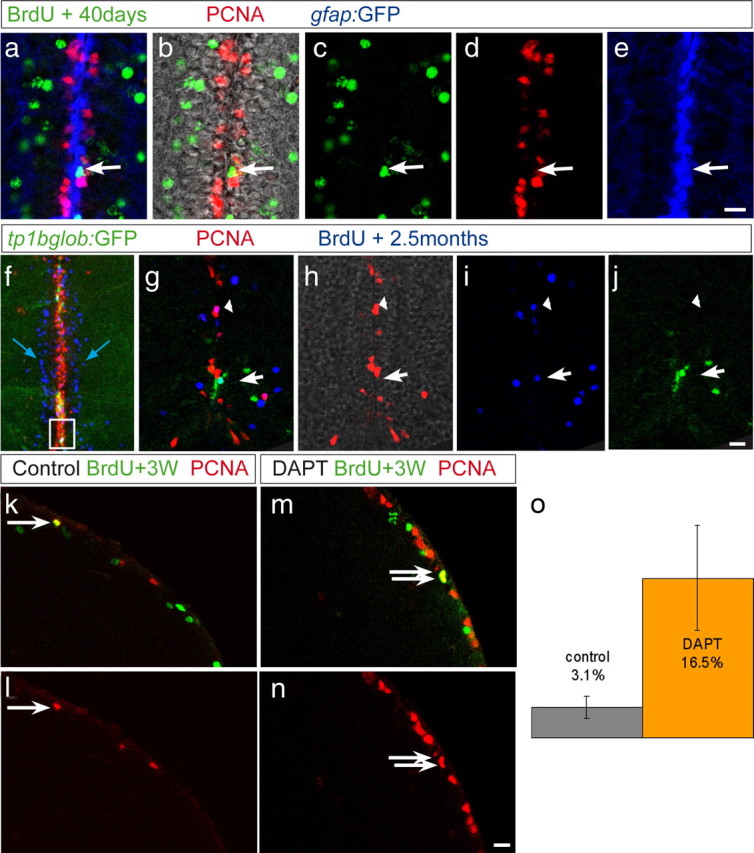
State II cells endogenously transit to state I and back to state II, according to Notch activity. a–e, Cross section of a gfap:GFP transgenic fish (GFP in blue) treated with BrdU for 5 d and killed 40 d after the last injection. Some BrdU-labeled cells (green) remain in the ventricular area but are PCNA (red) negative, as pointed to by the arrow, indicating that they have entered state I after having divided. f–j, TP1bglob:gfp transgenic fish treated with BrdU for 5 d and killed 2.5 months later. f, Projection of several confocal planes, overview of the telencephalic midline: Many BrdU+ cells (blue) have exited from the ventricular zone containing PCNA+ cells (red) and have entered the parenchyma (blue arrow). g–j, Single optical section showing the area boxed in f. Some BrdU+, PCNA-negative cells remaining in the ventricular area are GFP+ (white arrow), indicating that they have entered state I and are under high Notch signaling. k–o, WT fish were injected with BrdU for 5 d and treated 3 weeks later for 2 d with DAPT or control water. Brains were stained for BrdU (green) and PCNA (red). o, The proportion of BrdU+, PCNA+ cells within the ventricular BrdU+ population was calculated in two control and two DAPT-treated brains. After DAPT treatment, this proportion is increased by 5 times compared to the control, indicating that Notch signaling had kept label-retaining cells quiescent. Error bars represent SEM, n = 2 with a total number of 217 control and 235 DAPT-treated BrdU+ cells, p < 0.01, Rao-Scott test (modified χ2 test for clustered data). Scale bars (e, j, n), 10 μm.
Next, we determined whether state I cells naturally derived from state II were quiescent (rather than differentiated) and if they were kept so by Notch activity. DAPT was applied 3 weeks after the end of a repetitive BrdU labeling, and the percentage of PCNA/BrdU double-positive cells within the BrdU population was counted (Fig. 7c–g). During DAPT treatment, the double-labeled population was indeed increased compared with control (16 ± 5.4% in DAPT-treated compared with 3.1 ± 1.2% in control animals, i.e., a 5.3-fold increase), in proportions identical to those observed on the whole radial glia population (Fig. 3e, yellow bars). Thus, at least some state I cells that have reverted from state II require endogenous Notch activity to remain quiescent. We note that not all ventricular BrdU-positive cells were PCNA positive after DAPT treatment, suggesting again that some additional mechanisms account for their quiescent state. Nonetheless, our results demonstrate that progenitor cells oscillate between division and quiescence as a function of endogenous Notch activity.
Blocking Notch in the adult brain induces an increased production of newborn neurons
The transition of state I progenitors to cycling state II progenitors induced by decreased Notch activity prompted us to ask whether these state II progenitors were able to proceed through the cell cycle and then exit it toward a neuroblast fate. Hence, we analyzed the expression of the proneural gene ascl1a (Fig. 8a,b,f,g) and the neuroblast marker PSA-NCAM (Fig. 8c–j). Cells expressing these markers were increased after DAPT treatment compared with control brains. Unexpectedly, after DAPT treatment, PSA-NCAM expression was detected in many PCNA-positive cells that also expressed S100β, suggesting an accelerated transition from state II to state III cells (Fig. 8e,j). Thus, inhibiting Notch signaling leads to an increase in the number of differentiating neuroblasts.
Figure 8.
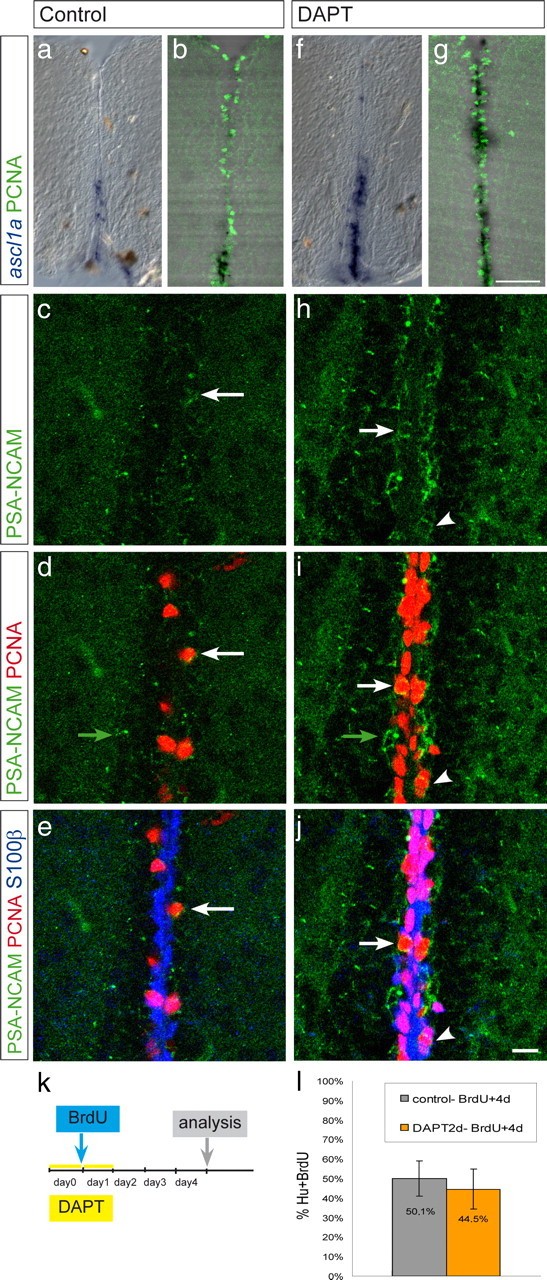
Notch blocking induces an increased number of neuroblasts. Cross sections through the telencephalon of wild-type animals, treated with vehicle only (a–e) or with DAPT (f–j). a, b, f, g, In situ hybridization with the ascl1a antisense probe (blue in a and f and black in b and g). DAPT treatment induces an increased number of dividing cells undergoing neurogenesis. c–e, h–j, PSA-NCAM (green), PCNA (red), and S100β (blue), merged in e and j. DAPT treatment induces an increased number of PCNA+, PSA-NCAM+ cells (state III cells, white arrow) and of cells only positive for PSA-NCAM (likely differentiating neurons, green arrow). S100β, PCNA, PSA-NCAM triple-labeled cells are also increased under DAPT conditions (arrowhead), suggesting an accelerated switch from state II to state III. k, Dividing cells in 2 control- and 2 DAPT-treated fish were traced for 4 d after BrdU injection. The proportion of BrdU+, Hu+ cells is similar in control and treated brains, indicating that the reactivation of state I cells by DAPT is followed by normal neuronal maturation, and that consequently the net number of generated neurons is increased. Scale bars: g, 50 μm; j, 10 μm.
To trace the later fate of these cells, we administered a BrdU pulse after the first day of 2 d DAPT treatment. The treatment was followed by 3 d of chase (Fig. 8k). The same proportion of BrdU-labeled cells expressed Hu (an early neuronal postmitotic marker) after 4 d in DAPT-treated and control animals (Fig. 8l), leading to a net increase of neurons newly generated and indicating that reactivated cells matured to neurons following the same schedule as normal progenitors. Thus, reactivating quiescent glia by inhibiting Notch is sufficient to trigger the entire neurogenic cascade. Together, our data reveal a key role of endogenous Notch activity in controlling the frequency of radial glia divisions and thereby neuroblast generation and neuron production in the adult brain. Thus, Notch appears to be an important gatekeeper of the transition between neural stem cell maintenance and their recruitment toward neurogenesis.
Discussion
In this study, we have uncovered a mechanism controlling the frequency of adult neural stem cells recruitment, using the zebrafish telencephalon as a model system. We have described three states of progenitors: quiescent radial glial cells (state I), dividing radial glial cells (state II), and cycling neuroblasts committed toward differentiation (state III). We have shown that the Notch pathway is endogenously active in state I progenitors and that activation of the pathway results in a transition from state II to state I, whereas blocking endogenous Notch activity induces a transition from state I to state II. Furthermore, we have demonstrated that the Notch pathway is activated in the immediate vicinity of cycling cells, suggesting that one mechanism driving quiescence is a feedback control arising from active progenitors. Finally, radial glia reactivated by blocking the Notch pathway proceed toward the neuroblast and postmitotic neuronal states, suggesting that, through maintaining the equilibrium between quiescent and proliferating progenitors, Notch activity regulates neuronal production during adulthood.
Uncovering quiescent progenitors and their Notch-controlled recruitment
The presence of PCNA-positive radial glial cells in the adult ventricular zone of the fish brain has been reported in several studies (Adolf et al., 2006; Pellegrini et al., 2007; Lam et al., 2009; März et al., 2010). However, the implication of their nondividing neighbors in the neurogenesis process has not been examined so far. The experiments presented here strongly suggest that radial glia in the adult germinal area of the telencephalon switch back and forth between a dividing (PCNA-or MCM5-positive, state II) and a quiescent (PCNA-and MCM5-negative, state I) state and are thus lineage related. For their expression of glial markers and their cycling status, type I and II cells are reminiscent of the quiescent and activated astrocytes recently described and generated in sequence from the subependymal zone of the adult mouse telencephalon (Pastrana et al., 2009). Our results further demonstrate the existence of oscillations taking place between these states. Indeed, we show that state II cells, labeled at one time point with BrdU, can be found in the VZ in state I (GFAP-positive, PCNA-negative) several weeks after division (Fig. 7a–e). This entry into quiescence can also be triggered by expression of NICD (Fig. 6c–o). In turn, state I cells, including label-retaining cells, can return into cycle when Notch activity is downregulated (Figs. 6p–u, 7j–o). We therefore propose that, along the adult VZ, the radial glia population is composed of equivalent slow dividing progenitors, in equilibrium between the G0 phase (state I, most of the time) and the cycling phase (state II, more rarely). Our observations also strongly suggest that cycling neuroblasts (state III) are generated from state II progenitors. Indeed, the presence of very few cells triple-labeled gfap:GFP, PCNA, and PSA-NCAM positive suggests a direct lineage between dividing radial glia and neuroblasts. This clear cellular organization of the adult germinal area allowed us to further address how quiescence and cell cycle entry are coordinated between neighboring NSCs, an issue that had not been considered previously.
Balance mechanism between recruitment and quiescence
The sum of our results not only suggests a switch back and forth between state I and state II, implying varying levels of Notch activity, but also a reciprocal control of these states between neighboring NSCs. First, state II cells are scattered in a regular manner throughout the VZ, with state I cells in between (Fig. 1), suggesting a coordinated activity of the progenitors. Our results further indicate that state I cells neighboring dividing cells are particularly sensitive to Notch inhibition, because they are reactivated with higher frequency than randomly picked state I cells (Fig. 5l,m). We also observed that almost all PCNA-positive cells express DeltaA (Fig. 2); thus, they are able to trigger Notch signaling into their neighbors and, accordingly, GFP in the Notch reporter line often highlights state I quiescent progenitors in the direct vicinity of PCNA-positive cells (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). We therefore propose that one mechanism of Notch activation is generated within the VZ through a lateral inhibition-like process originating from state II and/or state III cells, triggering Notch into their neighbors and rendering them refractory to cell cycle entry. The division and eventual migration of the neuroblasts, which eventually loose Delta expression (Delta is only expressed in the first two layers of cells lining the VZ) should in turn lower Notch signaling in the VZ, permitting a reentry into the cell cycle. Overall, this mechanism would result in self-sustained oscillations of Notch activity within the radial glia population, accounting for the entry into cycle of each cell once in a while. Lateral inhibition mechanisms by Notch signaling have been described in many different contexts and are found for example in the embryonic mouse forebrain, in which Notch signaling generated by newborn neurons instructs remaining radial glial progenitors to either remain progenitor (Yoon et al., 2008) or to acquire an astrocytic fate (Namihira et al., 2009). Our present work demonstrates for the first time the relevance of such “lateral inhibition” models to restrain neurogenesis in adult germinal zones. Another mechanism, however, likely accounts for the reactivation by Notch blockade of state I cells that are not located in the immediate vicinity of dividing progenitors. Because we also found a high proportion of cells expressing DeltaA:GFP (Fig. 2h) or other Notch ligands (see jagged1b, supplemental Fig. 2j, available at www.jneurosci.org as supplemental material) within the state I population, Notch stimulation among quiescent cells is a possibility. The mechanism of such interactions remains to be defined.
Maintaining quiescence in large areas
The fact that Notch blockade does not reactivate the entire radial glia population (Figs. 3e, 4c–g, 5a–e) suggests the existence of additional signaling pathways maintaining quiescence at the VZ. In the zebrafish lateral line, similarly, Notch activity limits the proliferation of supporting cells during regeneration but has no effect in a steady-state situation, indicating that an additional mechanism maintains quiescence (Ma et al., 2008). Likewise, the ependymal layer in the adult mouse seems to be subject to high Notch signaling as soon as it has been injured, and it appears that only a minority of ependymal cells are brought back into cycle by Notch blocking without injury (Carlén et al., 2009). One important player in quiescence is the family of Hes or their her orthologs in zebrafish genes (Curry et al., 2006; Sang et al., 2008). Members of the her family are indeed expressed in the entire nondividing radial glia population of the zebrafish telencephalon, as well as in other quiescent populations of the adult brain (supplemental Fig. 6, available at www.jneurosci.org as supplemental material) (P. Chapouton, K. J. Webb, C. Stigloher, and L. Bally-Cuif, unpublished observations) (Chapouton et al., 2006), and several signaling pathways other than Notch have been shown to drive the expression of Hes genes (Curry et al., 2006; Nakayama et al., 2008; Wall et al., 2009). In the adult zebrafish, the phospho-extracellular signal-regulated kinase, a downstream target of FGF, is also expressed in all noncycling radial glial cells (Topp et al., 2008). Hence, expression of Hes or her genes independently of Notch activity could be involved in maintaining a widespread quiescence in large groups of cells.
Distinct outcomes of Notch activity
Our results demonstrate that Notch activity along the adult telencephalic VZ controls the choice between cellular states I and II, with consequences for cell fate. Notch function is implicated in binary fate choice in many developing systems. In the CNS alone, this is the case for selecting between spinal cord interneuron subtypes (Peng et al., 2007), interneurons/motoneurons (Shin et al., 2007), motoneurons/oligodendrocytes (Park and Appel, 2003), habenular subnuclei (Aizawa et al., 2007), epiphysis photoreceptors/projection neurons (Cau et al., 2008), pituitary secretory cell types (Zhu et al., 2006), or between retinal neuronal subtypes and Mueller glia (Scheer et al., 2001; Bernardos et al., 2005; Jadhav et al., 2006). These fate choices often implicate the maintenance by Notch of a progenitor state as opposed to differentiation, and such a mechanism fits with our results. In the embryonic CNS, however, the progenitor state is generally proliferating. Loss of Notch function or abrogation of its effectors Hes1, Hes3, and Hes5 at early developmental stages promotes depletion of the cycling progenitor pools and premature neuronal differentiation (Hatakeyama et al., 2004; Park et al., 2005; Yeo and Chitnis, 2007). In contrast, Notch has been observed to have the opposite effect, promoting quiescence, in other systems. It is the case, for example, in the regenerating lateral line in zebrafish (Ma et al., 2008), the postnatal ependymal and subependymal zone in the mouse (Chambers et al., 2001; Carlén et al., 2009), or in developing blood vessels (Leslie et al., 2007; Siekmann and Lawson, 2007). The different outcomes of Notch signaling, promoting either a cycling state or a quiescent state, are surprising but might be explained by distinct modes of Notch activity in distinct progenitors, as suggested in different studies. A characteristic of Notch activity in the embryonic CNS is rapid oscillations (Cisneros et al., 2008; Shimojo et al., 2008). It is possible that, by preventing the accumulation of commitment factors, rapidly fluctuating Notch activity levels might maintain cells in a labile state, prone to division. In contrast, sustained Hes1 expression at boundary regions between brain compartments correlates with low proliferation rates, and forced Hes1 expression in embryonic neural progenitors in culture reduces proliferation (Baek et al., 2006). We observed high expression of her genes in all state I cells (supplemental Fig. 6, available at www.jneurosci.org as supplemental material) and propose that Notch activity might oscillate at a low frequency within the zebrafish adult VZ, supporting a quiescent state in part by permitting high levels of its target proteins and/or quiescence factors to accumulate. One possible target identified here is the cell cycle inhibitor Ink4b (Cdkn2b) that contains Suppressor of Hairless binding sites on its enhancer region (data not shown), although we do not know whether its regulation by Notch is direct. It is also possible that Notch target genes differ, at least in part, in the embryonic and adult contexts, and identifying these targets and their function on cell division and/or quiescence remains an important issue.
Does the mechanism described here apply for mammalian brains?
The two adult mouse adult neurogenic domains, SEZ and SGZ, like the zebrafish adult telencephalic VZ, contain slow-cycling progenitors and undergo neurogenesis under normal conditions. Our results demonstrate that manipulating Notch can dramatically impact on progenitor proliferation and neurogenesis in a comparable region, the zebrafish telencephalon. As Notch receptors, ligands and target transcripts are also expressed in the neurogenic areas of the postnatal mouse brain (Stump et al., 2002; Givogri et al., 2006; Ohtsuka et al., 2006), Notch signaling might play a similar role in these neurogenic domains. Thus, variations in Notch signaling might enhance the generation of adult-born neurons, either in neurogenic domains or in injured regions normally devoid of neurogenic activity, as suggested by studies in the adult rat spinal cord (Yamamoto et al., 2001), or in the mouse ependymal layer of the lateral ventricle (Carlén et al., 2009). In one study, apparently contradictory with these findings, overexpression of NICD using Cre-mediated recombination in GFAP-positive astrocytes of the mouse postnatal dentate gyrus did not induce quiescence but rather pushed progenitors toward the transient amplifying state, whereas γ-secretase inhibition of Notch or genetic invalidation of Notch1 induced cell cycle exit (Breunig et al., 2007). How to reconcile these observations is unclear at present, but manipulations of Notch signaling in the mouse adult DG also go along the lines of our results (O. Ehm and C. Lie, personal communication). An important observation of our work is that quiescent progenitors reactivated by blocking Notch signaling further commit toward becoming neurons (Fig. 8). Hence, the active maintenance of quiescence by Notch activity is a limiting step toward the formation of newly born neurons. Taken as a whole, it is therefore interesting to consider the study results of ours and others in a clinical perspective, because the administration of Notch-blocking drugs in adult brains might provide an approach to enhance the recruitment and neurogenic fate of dormant NSCs in repair mechanisms. In the future, it will be important to assess the late fate of the newborn neurons generated from the ependymal zone during Notch blockade after stroke (Carlén et al., 2009) and in the zebrafish model.
Footnotes
Work in the Zebrafish Neurogenetics Department at the Helmholtz Zentrum München was funded by the Volkswagen Association, European Union Sixth Framework Integrated Project ZF-Models Contract LSHC-CT-2003-503466, Life Science Association Grant GSF 2005/01, and the Center for Protein Science–Munich. For work at the Centre National de la Recherche Scientifique (CNRS), funds from the European Union Seventh Framework Integrated Project NeuroXsys, the Agence Nationale de la Recherche, the Ecole des Neurosciences de Paris, the PIME program, and the Schlumberger Association are greatly acknowledged. Financial support to R.M. was from the French Ministère de la Recherche and to P.B. from Université de Toulouse, Université Paul Sabatier, CNRS, Inserm, Fondation pour la Recherche Médicale, and Fédération pour la Recherche sur le Cerveau. We thank Magdalena Götz, Leanne Godinho, and William Norton for their critical reading of this manuscript. notch2, notch3, jagged1a, jagged1b, and jagged2 in situ probes were kindly provided by the Julian Lewis laboratory. We are also grateful to Andrea Wizenmann for providing initial insights into the electroporation technique.
References
- Adolf B, Chapouton P, Lam CS, Topp S, Tannhäuser B, Strähle U, Götz M, Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol. 2006;295:278–293. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Goto M, Sato T, Okamoto H. Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev Cell. 2007;12:87–98. doi: 10.1016/j.devcel.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59:419–426. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- Carlén M, Meletis K, Göritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabé-Heider F, Yeung MS, Naldini L, Honjo T, Kokaia Z, Shupliakov O, Cassidy RM, Lindvall O, Frisén J. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- Cau E, Quillien A, Blader P. Notch resolves mixed neural identities in the zebrafish epiphysis. Development. 2008;135:2391–2401. doi: 10.1242/dev.013482. [DOI] [PubMed] [Google Scholar]
- Chambers CB, Peng Y, Nguyen H, Gaiano N, Fishell G, Nye JS. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development. 2001;128:689–702. doi: 10.1242/dev.128.5.689. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Adolf B, Leucht C, Tannhäuser B, Ryu S, Driever W, Bally-Cuif L. her5 expression reveals a pool of neural stem cells in the adult zebrafish midbrain. Development. 2006;133:4293–4303. doi: 10.1242/dev.02573. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Jagasia R, Bally-Cuif L. Adult neurogenesis in non-mammalian vertebrates. Bioessays. 2007;29:745–757. doi: 10.1002/bies.20615. [DOI] [PubMed] [Google Scholar]
- Cisneros E, Latasa MJ, García-Flores M, Frade JM. Instability of Notch1 and Delta1 mRNAs and reduced Notch activity in vertebrate neuroepithelial cells undergoing S-phase. Mol Cell Neurosci. 2008;37:820–831. doi: 10.1016/j.mcn.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- Daudet N, Gibson R, Shang J, Bernard A, Lewis J, Stone J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev Biol. 2009;326:86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givogri MI, de Planell M, Galbiati F, Superchi D, Gritti A, Vescovi A, de Vellis J, Bongarzone ER. Notch signaling in astrocytes and neuroblasts of the adult subventricular zone in health and after cortical injury. Dev Neurosci. 2006;28:81–91. doi: 10.1159/000090755. [DOI] [PubMed] [Google Scholar]
- Gonzalez MA, Tachibana KE, Laskey RA, Coleman N. Control of DNA replication and its potential clinical exploitation. Nat Rev Cancer. 2005;5:135–141. doi: 10.1038/nrc1548. [DOI] [PubMed] [Google Scholar]
- Grandel H, Kaslin J, Ganz J, Wenzel I, Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295:263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Haddon C, Smithers L, Schneider-Maunoury S, Coche T, Henrique D, Lewis J. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998;125:359–370. doi: 10.1242/dev.125.3.359. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, März M, Strähle U. gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn. 2009;238:475–486. doi: 10.1002/dvdy.21853. [DOI] [PubMed] [Google Scholar]
- Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28:2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- März M, Chapouton P, Diotel N, Vaillant C, Hesl B, Takamiya M, Lam CS, Kah O, Bally-Cuif L, Strähle U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia. 2010;58:870–888. doi: 10.1002/glia.20971. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Satoh T, Igari A, Kageyama R, Nishida E. FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr Biol. 2008;18:R332–R334. doi: 10.1016/j.cub.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol Cell Neurosci. 2006;31:109–122. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Park HC, Appel B. Delta-Notch signaling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini E, Mouriec K, Anglade I, Menuet A, Le Page Y, Gueguen MM, Marmignon MH, Brion F, Pakdel F, Kah O. Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J Comp Neurol. 2007;501:150–167. doi: 10.1002/cne.21222. [DOI] [PubMed] [Google Scholar]
- Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53:813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisdóttir I, Massagué J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Seri B, García-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seri B, García-Verdugo JM, Collado-Morente L, McEwen BS, Alvarez-Buylla A. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J Comp Neurol. 2004;478:359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shin J, Poling J, Park HC, Appel B. Notch signaling regulates neural precursor allocation and binary neuronal fate decisions in zebrafish. Development. 2007;134:1911–1920. doi: 10.1242/dev.001602. [DOI] [PubMed] [Google Scholar]
- Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- Simpson P. Notch signalling in development: on equivalence groups and asymmetric developmental potential. Curr Opin Genet Dev. 1997;7:537–542. doi: 10.1016/s0959-437x(97)80083-4. [DOI] [PubMed] [Google Scholar]
- Stump G, Durrer A, Klein AL, Lütolf S, Suter U, Taylor V. Notch1 and its ligands Delta-like and Jagged are expressed and active in distinct cell populations in the postnatal mouse brain. Mech Dev. 2002;114:153–159. doi: 10.1016/s0925-4773(02)00043-6. [DOI] [PubMed] [Google Scholar]
- Takke C, Dornseifer P, v Weizsäcker E, Campos-Ortega JA. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Topp S, Stigloher C, Komisarczuk AZ, Adolf B, Becker TS, Bally-Cuif L. Fgf signaling in the zebrafish adult brain: association of Fgf activity with ventricular zones but not cell proliferation. J Comp Neurol. 2008;510:422–439. doi: 10.1002/cne.21802. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H. Neuroanatomy of the zebrafish brain. Basel: Birkhäuser Verlag; 1996. [Google Scholar]
- Yamamoto N, Takebayashi H, Nabeshima Y, Kitamura T, Weinmaster G, Nakamura K, Nakafuku M. Transcription factor expression and Notch-dependent regulation of neural progenitors in the adult rat spinal cord. J Neurosci. 2001;21:9814–9823. doi: 10.1523/JNEUROSCI.21-24-09814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SY, Chitnis AB. Jagged-mediated Notch signaling maintains proliferating neural progenitors and regulates cell diversity in the ventral spinal cord. Proc Natl Acad Sci U S A. 2007;104:5913–5918. doi: 10.1073/pnas.0607062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]