Abstract
Memory formation is a continuous process composed of multiple phases that can develop independently from each other. These phases depend on signaling pathways initiated after the activation of receptors in different brain regions. The NMDA receptor acts as a sensor of coincident activity between neural inputs, and, as such, its activation during learning is thought to be crucial for various forms of memory. In this study, we inhibited the expression of the NR1 subunit of the NMDA receptor in the honeybee brain using RNA interference. We show that the disruption of the subunit expression in the mushroom body region of the honeybee brain during and shortly after appetitive learning selectively impaired memory. Although the formation of mid-term memory and early long-term memory was impaired, late long-term memory was left intact. This indicates that late long-term memory formation differs in its dependence on NMDA receptor activity from earlier memory phases.
Introduction
Glutamate is the most important excitatory neurotransmitter in mammals (Weinberg, 1999). Its role in the CNS has been mostly studied by focusing on the different types of glutamate receptors. NMDA glutamate receptors (NMDARs) are known to play an important role in memory formation (Kandel, 2001). Their activation depends on the simultaneous release of glutamate and a strong depolarization of the postsynaptic membrane (Mayer et al., 1984). In this manner, NMDARs act as sensors of coincident activity between neural inputs.
In classical appetitive olfactory conditioning in insects, the animal learns to associate an odor, the conditioned stimulus (CS), with sucrose, the unconditioned stimulus (US). It is generally accepted that the neural networks representing the CS in olfactory conditioning are cholinergic (Breer and Sattelle, 1987), whereas octopaminergic neurons mediate the appetitive reinforcer (Hammer and Menzel, 1998; Schwaerzel et al., 2003; Thum et al., 2007). Both networks converge in the antennal lobe (AL) and the mushroom body (MB). The coincident activity between these neural networks plays an important role in memory formation (Hammer and Menzel, 1998).
Interestingly, glutamate immunoreactivity, the localization of glutamate receptors and the recording of inhibitory and excitatory glutamate-induced currents, suggests the existence of a glutamate neurotransmission in the insect CNS (Ultsch et al., 1992; Cully et al., 1996; Barbara et al., 2005; Featherstone et al., 2005; Xia et al., 2005; Zannat et al., 2006; Devaud et al., 2008). In addition, several studies in the honeybee suggest that this neurotransmission is implicated in memory formation, but it is not known how it integrates the neural networks of olfactory coding and learning (Si et al., 2004; Locatelli et al., 2005; Kucharski et al., 2007; El Hassani et al., 2008). Recently, it was shown that NMDARs are implicated in olfactory memory formation in Drosophila (Xia et al., 2005; Wu et al., 2007). However, their function remains to be fully understood.
In this study, we evaluated the role played by NMDARs in olfactory memory formation in the honeybee, Apis mellifera. Multiple conditioning trials induce the formation of an amnesia-resistant mid-term memory (MTM) immediately after learning (Menzel, 1999). In addition, a consolidation process takes place, leading to long-term memory (LTM) (Müller, 2000). LTM can be subdivided into early LTM (eLTM) and late LTM (lLTM); the former occurs 1 and 2 d after conditioning and is dependent on translation of new proteins, whereas the latter occurs after 3 d and is dependent on transcription and translation processes (Wüstenberg et al., 1998; Friedrich et al., 2004).
We inhibited the expression of NMDARs in the MB region of the honeybee brain by inducing RNA interference (RNAi) against the NR1 subunit. We show that the acute disruption of NMDARs during conditioning and shortly after affects the acquisition phase, MTM, and eLTM formation, leaving lLTM intact.
Materials and Methods
Animal treatment and behavioral procedure.
Honeybees (A. mellifera) were caught at the hive entrance, cooled, and restrained in harness (Bitterman et al., 1983). The honeybees were fed to satiation on the first day around 4:00 P.M. with sucrose solution (30% w/v) and left undisturbed until the next day in a dark humid box at 24°C. On the following days, the animals received 6 drops of the sucrose solution around 4:00 P.M. At least 1 h elapsed between any behavioral manipulations and feeding.
On the second day, honeybees were given injections of 260 nl of double-stranded RNA (dsRNA) at a concentration of 50 μm (10 μg/μl) or small-interfering RNA (siRNA) at a concentration of 100 μm (1.5 μg/μl), around 3:00 P.M. In the experiment on the recall of memory, honeybees were given injections on day 4. A hole was pricked in the cornea of the median ocelle with an acupuncture needle to allow the insertion of a fine-glass capillary filled with the dsRNA or siRNA solutions. The solution was injected into the median ocellar tract down to the region of the MBs using a microinjector [Picospritzer II (General Valve) or Pneumatic PicoPump PV820 (WPI)].
Appetitive olfactory conditioning was performed principally in April and September, around noon, 1 or 2 d after the injection or 2 d before the injection. All behavioral procedures were performed blind. The acquisition trial consisted of moving a honeybee from its resting position to the inlet of an exhaust fan. The odor (CS) was delivered after 10 s of accommodation time. Carnation oil from the pharmacy was used as the CS and delivered through a 20 ml syringe that was loaded daily with 4 μl of carnation oil. The odor was presented for 5 s. Three seconds after the odor onset, sucrose (US) was applied by touching the antennae with a sucrose-moistened toothpick, and the honeybees were allowed to lick sucrose for 5 s. Acquisition consisted of three such pairings with an intertrial interval (ITI) of 10 min. A honeybee scored positive when it extended its proboscis between the onset of the CS and the presentation of the US. In the nonassociative procedure, the CS and the US were alternately delivered with a 5 min interval between each stimulation. Animals that did not respond to the US during acquisition or during the final sucrose test (see below), or that responded positively to the first CS presentation, were discarded from the experiment, except for the evaluation of olfactory and gustatory sensitivity in which all honeybees were considered. The recall of memory consisted of the presentation of the CS for 5 s without reinforcement by the US.
In the dsRNA experiment, each honeybee received a single posttraining test, whereas in the siRNA experiment, honeybees received multiple posttraining tests. The sucrose responsiveness of each honeybee was tested directly after the last memory test. Honeybees that did not respond to sucrose stimulation were discarded from the experiment. Statistical analysis was done by performing a χ2 test for each testing trial or a McNemar χ2 test to compare the performances of animals between successive tests (Statistica, 1999 edition; StatSoft).
RNAi.
The sense and antisense RNA were transcribed in vitro from sense and antisense cDNA templates. The cloning of these cDNAs was performed as described previously (Leboulle and Müller, 2004). For the dsNR1 molecule, first-strand cDNA reverse transcribed from brain total RNA was used as the template for the PCR. The amplification of the cDNA template for the transcription of the sense strand was performed with the direct primer 5′-ACACTGACGGTACCGAAGAGGATGC-3′ modified at its 5′ extremity by the addition of a T7 promoter sequence, 5′-TAATACGACTCACTATAGGGCGA-3′, and the reverse primer 5′-AGCAGAACCTTTTACCGTGGCACAT-3′. The amplification of the cDNA template for the transcription of the antisense strand was performed with the reverse primer modified by the addition of a T7 promoter sequence at its 5′ extremity and with the unmodified direct primer. For the dsNEG molecule, the pBlueBac4.5/V5-His/CAT plasmid was used as the template with the direct primer 5′-GTATGGCAATGAAAGACGGTGAGC-3′ and the reverse primer 5′-TGAAAACGGGGGCGAAGAAGT-3′. As for the amplification of the dsNR1 template, modified primers were used to generate sense and antisense strands. The PCR protocol consisted of an initial denaturation step of 3 min at 95°C, 25 cycles of 30 s at 95°C, 1 min at 66°C, 1 min at 72°C, and a final elongation step of 10 min at 72°C.
Several clones were sequenced, and the orientation of the inserts was verified. The selected clones were stored at −80°C.
The plasmidic DNA of the selected clones was digested with EcoRI restriction enzyme and reamplified to generate the template for the in vitro transcription. The sense and antisense strands were transcribed separately with the T7 Ribomax Express large-scale production system (Promega) by following the manufacturer's instructions and resuspended in 1× PBS (137 mm NaCl, 2.7 mm KCl, 10.1 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4). The corresponding sense and antisense RNAs were mixed in equal amounts, incubated 15 min at 68°C, cooled to 37°C, and incubated 45 min at this temperature for the hybridization of the dsRNAs. The dsNR1 dsRNA is 290 bp long and is specific for the region located between the positions 1907 and 2196 of the AmNR1.1 mRNA (GenBank accession number AM040211). The dsNEG dsRNA is 294 bp long and is specific for the region located between the positions 411 and 704 of the pBlueBac4.5/V5-His/CAT plasmid.
The siRNAs were designed by Qiagen and resuspended in siRNA suspension buffer as mentioned by the manufacturer (Qiagen). siNR1 [sense, r(GGCGUGAACUGUUAACAAA)dTdT; antisense, r(UUUGUUAACAGUUCACGCC)dCdT] is specific for the region located between the positions 10 and 30 of AmNR1.1. siNEG [sense, r(GGCAGUUUCUACACAUAUA)dTdT; antisense, r(UAUAUGUGUAGAAACUGCC)dGdG] is specific for the region of the CAT insert of the pBlueBac4.5/V5-His/CAT located between the positions 529 and 549.
Western blot analysis.
Honeybees were anesthetized on ice and decapitated. Head capsules were fixed on melted wax, and brains were dissected. The optic lobes of the animals were removed, and the central part of the brain was cut between the ALs and the α-lobes to separate the MB and the AL regions. In some preparations, the samples consisted of the central part of the brain (MB and AL regions) excluding the optic lobes. The samples were homogenized in 1× PBS, 2 mm EDTA, and 2 mm EGTA (PBS-EE) with a Teflon–glass homogenizer. Homogenates were sonicated for 10 min and centrifuged at 4°C at 20,000 × g for 10 min. The supernatants were discarded, and the pellets were resuspended at a concentration of 1 brain/20 μl and 1 MB or 1 AL region/10 μl in PBS-EE. Sample buffer (0.225 m Tris, pH 6.8, 50% glycerol, 5% SDS, 0.05% bromophenol blue, 0.25 m dithiothreitol) was added, and the samples were sonicated for 10 min and stored at −80°C until analysis. The rat brain extract was a gift from Dr. F. Locatelli (University of Buenos Aires, Buenos Aires, Argentina). To examine the expression levels of the NR1 subunit and of α-tubulin, the protein samples were subjected to SDS-PAGE and transferred onto nitrocellulose or polyvinylidene difluoride membranes. The membranes were blocked in 1× PBS, 0.1% Tween 20, and 5% nonfat milk powder (blocking solution) for 1 h at room temperature (RT) and cut at the level of the ∼79 kDa band of the molecular weight marker. The membranes were probed with the polyclonal antibody NR1-pan (1:2000; Millipore and Biomol), the AmNR1 serum (1:1000), or the anti-α-tubulin antibody (1:10,000; DM1A; Merck) dissolved in the blocking solution, overnight at 4°C. The AmNR1 serum was obtained from rabbits immunized with the peptide N-term CASNAPEGLLGLKLINAENET C-term, at Inbiolabs. In the peptide competition assay, the antigenic peptide N-term CASNAPEGLLGLKLINAENET C-term was incubated at a 150 nm concentration with the antibody solution (10 times molar excess of peptide) before the incubation on the membrane. Note that NR1-pan could only be used at the beginning of the experiments, because its production was stopped during the course of this study. The membranes were washed with 1× PBS and 0.1% Tween 20 (PBS-T) four times for 10 min at RT and incubated with the secondary antibodies directed against mouse or rabbit IgG coupled to horseradish peroxidase (1:10,000; Sigma) dissolved in the blocking solution for 1 h at RT. The membranes were washed with PBS-T four times for 10 min at RT and developed by enhanced chemiluminescence (ECL) detection (100 mm Tris, pH 8.6, 625 μm Luminol, 15 μm p-Coumaric acid, 0.0175% H2O2), and the signals were acquired with a LAS1000 camera and the software Image Reader LAS1000 2.60 (Fujifilm), or they were developed by the ECL system (PerkinElmer), and the signals were acquired on Kodak X-Omat AR films (VWR). For the quantification of the NR1 levels, in each experiment, 15 honeybees were given injections of dsNR1 or siNR1, and 15 honeybees were given injections of dsNEG or siNEG. Depending on the conditions tested, these experiments were repeated between 5 and 11 times. The AL or MB regions of these injected groups were pooled and repeatedly analyzed. Eighteen microliters of these probes were loaded in each lane that constituted one measurement. Repeated measurements of several experiments were used to calculate the mean values. The quantification of the NR1 subunit levels was done by relating the intensities of the NR1 subunit signals to the α-tubulin signals by using the program NIH ImageJ version 1.36b or MultiGauge version 3.0 (Fujifilm). For each Western blot analysis, the relative NR1 levels were normalized to the highest NR1/α-tubulin ratio for each considered brain region (AL or MB regions). The normality of the data for each tested condition was tested with the Shapiro-Wilk's W test implemented in Statistica (1999 edition). In every case, the W value was >0.05, indicating normality. The difference between dsNR1- or siNR1-injected and dsNEG- or siNEG-injected honeybees was tested for significance with the Student's t test for independent samples implemented in Statistica (1999 edition).
Results
Characterization of the NR1 subunit of the NMDARs by Western blot
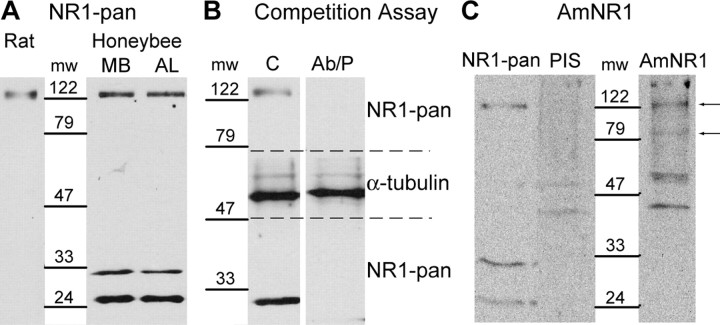
In a previous study, the expression sites of the NR1 subunit of NMDARs were localized in the brain of the honeybee with two different antibodies (Zannat et al., 2006). One of them, NR1-pan, specific for the 259–278 portion of the rat NR1 subunit, was used to detect the NR1 subunit in a rat brain extract and in the MB and AL regions of the honeybee brain by Western blot (Fig. 1A). The MB region dissected in our experiments comprised the MBs, the central body, and the dorsal part of the protocerebral lobe, and the AL region consisted of the ALs, the suboesophageal ganglion, and the ventral part of the protocerebral lobe. In all extracts, a protein migrating at an apparent molecular weight of ∼122 kDa was detected by the antibody. In addition, two additional proteins migrating to an apparent molecular weight of ∼33 and ∼24 kDa were detected in the AL and MB regions of the honeybee brain. The specificity of the interaction of NR1-pan with the NR1 subunit of the honeybee brain was challenged in a peptide competition assay (Fig. 1B). In this assay, the antibody was incubated with a peptide before the detection of the NR1 subunit. The peptide is specific for the portion of the honeybee subunit that should be recognized by NR1-pan. When incubated with a 10 times molar excess of peptide, NR1-pan did not detect the ∼122 kDa protein in the honeybee brain extract. To verify that the inhibition of the detection was not induced by an unspecific inhibitory factor contained in the antigenic peptide solution, the same assay was repeated with an antibody specific for α-tubulin, a housekeeping gene. This antibody detected a protein migrating at an apparent molecular weight of ∼50 kDa. The preincubation of the antibody with the peptide did not impair the detection of α-tubulin (Fig. 1B).
Figure 1.
Detection of the NR1 subunit by Western blot. A, Detection of the NR1 subunit in the MB and the AL regions of the honeybee brain and in rat brain with NR1-pan. B, The specificity of NR1-pan was evaluated in a peptide competition assay, on a honeybee brain extract. The membrane was cut between ∼79 and ∼47 kDa (dashed lines). In the control experiment, the membranes were incubated with NR1-pan or α-tubulin antibodies (C). In the peptide competition assay, the NR1-pan and α-tubulin antibodies were preincubated with the antigenic peptide (Ab/P) before the incubation on the membranes. C, Detection of the NR1 subunit in the honeybee brain with AmNR1. The NR1 subunit was detected with NR1-pan, with the AmNR1 preimmune serum (PIS), and with the AmNR1 serum (AmNR1). Arrows indicate proteins detected with AmNR1 that were not detected with PIS. mw, Molecular weight in kilodaltons.
A second polyclonal antibody, AmNR1, was raised against an antigenic peptide specific for the honeybee subunit. This peptide has the sequence of the honeybee epitope homologous to the one used to generate NR1-pan. A search in the honeybee genome showed that this polypeptide was only found in the NR1 subunit (http://www.ncbi.nlm.nih.gov/genome/guide/bee/). The preimmune serum detected a double-band pattern around ∼47 kDa and a protein that migrated at an approximate molecular weight of 200 kDa (Fig. 1C). These proteins were also detected with the AmNR1 serum. The AmNR1 serum detected additional proteins that migrated to an apparent molecular weight of ∼122 and ∼79 kDa. The proteins detected at ∼122 kDa with NR1-pan and AmNR1 have exactly the same apparent molecular weights. The proteins detected at ∼33 and ∼24 kDa with NR1-pan were not detected with AmNR1.
It is most probable that the ∼122 kDa protein corresponds to the honeybee NR1 subunit. For this reason, only this protein was considered in the quantitative analysis. The ∼24, ∼33, and ∼79 kDa proteins were detected only with one of the two antibodies, therefore they are probably the result of unspecific interactions.
Induction of RNAi against the NR1 subunit
The NR1 subunit is common to all NMDARs in mammals. Electrophysiological analysis of NMDARs showed that the NR1 subunit is necessary for the formation of functional receptors both in mammals and in Drosophila (Monyer et al., 1992; Xia et al., 2005). For this reason, we decided to target the NR1 subunit with RNAi to downregulate the expression of functional NMDARs. The molecules were injected through the median ocellar tract, a method that does not interfere with olfactory conditioning and memory formation (Fiala et al., 1999). The RNAi technique was already successfully used to inhibit the expression of an octopamine receptor subunit in the honeybee brain (Farooqui et al., 2003).
RNAi was first induced by dsNR1, a 290-bp-long dsRNA specific for the AmNR1.1 mRNA. In nonmammal species, dsRNA molecules do not activate defense mechanisms, characterized among others by a general inhibition of protein translation (Holen and Mobbs, 2004). dsRNAs interact with the Dicer enzymatic complex, which reduces them to siRNAs that are 21 bp long. Then, siRNAs are incorporated into an enzymatic complex to form the RNA-induced silencing complex that induces the degradation of mRNA species matching the sequence of the siRNA (Hannon, 2002). The comparison of dsNR1 against the honeybee genome showed that fully identical stretches of sequences of dsNR1 longer than 19 bp are only found in the nmdar1 gene. Hence, it can be assumed that the siRNAs generated from dsNR1 are only specific for the AmNR1 mRNA.
To avoid unspecific effects caused by the injection of dsRNAs in the brain, a dsRNA specific for the chloramphenicol acetyl transferase gene (dsNEG), which is expressed only in prokaryotes, was used as the negative control. The comparison of the dsNEG sequence to the honeybee genome allowed us to exclude the possibility that siRNA specific for any honeybee gene could be generated from this dsRNA.
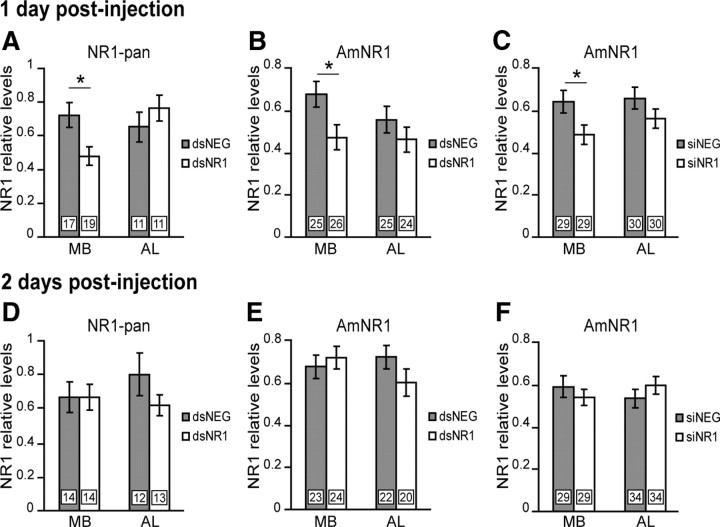
The RNAi effect on NR1 subunit levels was evaluated in the MB and AL regions 1 and 2 d after the injection of dsRNA, by using the NR1-pan and AmNR1 antibodies in independent experiments. One day after the injection, a significant reduction of 33.6% was found in the MB region as quantified with NR1-pan (Student's t test, 2.64; df = 34; p = 0.01) and of 30.2% with AmNR1 (Student's t test, 2.41; df = 49; p = 0.02) (Fig. 2A,B). In contrast, in the AL region, the NR1 subunit levels were not significantly different between dsNR1- and dsNEG-injected animals (NR1-pan: Student's t test, −0.97; df = 20; p = 0.34; AmNR1: Student's t test, 1.09; df = 47; p = 0.28) (Fig. 2A,B). Two days after the injection, the NR1 subunit levels were unchanged between dsNEG- and dsNR1-injected animals, both in the MB region (NR1-pan: Student's t test, −0.02; df = 26; p = 0.98; AmNR1: Student's t test, −0.55; df = 45; p = 0.58) and in the AL region (NR1-pan: Student's t test, 1.33; df = 23; p = 0.20; AmNR1: Student's t test, 1.43; df = 40; p = 0.16), although a small nonsignificant reduction of 22.6% with NR1-pan and 16.7% with AmNR1 was observed in the AL region of dsNR1-injected groups (Fig. 2D,E).
Figure 2.
Evaluation of the RNAi effect. Quantification of relative NR1 subunit levels in the MB and AL regions 1 or 2 d after injection of dsRNA (gray bars, dsNEG; white bars, dsNR1) or siRNA (gray bars, siNEG; white bars, siNR1) is shown. The expression levels were evaluated by quantitative Western blot, using NR1-pan (A, D) or AmNR1 (B, C, E, F). Each column represents the mean ± SEM of n measurements as indicated by the numbers on the bars. Asterisks indicate a significant reduction (*p < 0.05, Student's t test).
It is known that RNAi can induce the degradation of nontargeted mRNA, a phenomenon reported as off-targets effects (Ma et al., 2006). Therefore, the NR1 subunit was also inhibited with siRNAs. The molecules were designed by using an siRNA resource for the selection of functional molecules. The siNR1 molecule was specific for another region of the AmNR1.1 mRNA, and the negative control (siNEG) was again specific for the CAT gene. The specificity of siNR1 and siNEG was assessed by comparison to the honeybee genome resource. The RNAi effect was evaluated in the MB and AL regions, 1 and 2 d after the injection of siRNA, by using the AmNR1 antibody. One day after the injection, a significant reduction of 24.5% was found in the MB region (Student's t test, 2.24; df = 56; p = 0.03) (Fig. 2C), but not in the AL region (Student's t test, 1.40; df = 58; p = 0.166) (Fig. 2C). Two days after the injection, the NR1 subunit levels were unchanged in the MB region (Student's t test, 0.76; df = 56; p = 0.448) and in the AL region (Student's t test, −1.05; df = 66; p = 0.296) (Fig. 2F).
The inhibition of the NR1 subunit by the injection of dsNR1 or siNR1 is not detrimental for the honeybee (Table 1). The mortality rate in the dsRNA- and siRNA-injected animals was not different between test and control animals (dsRNA: dsNEG, 37%; dsNR1, 35%; χ2 = 1.18, df = 1, p = 0.278; siRNA: siNEG, 56%; siNR1, 56%; χ2 = 0.01, df = 1, p = 0.924). The higher mortality rate in the siRNA-injected animals is probably attributable to the fact that all animals had to survive until day 6 or 7 after the experiment had began. The observed mortality rate is not unusual for proboscis extension response (PER) conditioning. However, to exclude that the injection in the median ocellar tract induced a higher mortality, we compared the mortality rate between honeybees injected with dsRNA and noninjected honeybees 6 d after the beginning of the experiment and found no differences (χ2 = 0.00; df = 1; p = 1).
Table 1.
Mortality rates of honeybees after an injection of dsRNA or siRNA or no injection
| Injection | Dead bees | Total number | Mortality rate | χ2 (df = 1), p value |
|---|---|---|---|---|
| dsNEG | 441 | 1185 | 37% | 1.18, p = 0.2783 |
| dsNR1 | 416 | 1186 | 35% | |
| siNEG | 157 | 280 | 56% | 0.01, p = 0.9235 |
| siNR1 | 157 | 278 | 56% | |
| dsNEG | 23 | 57 | 40% | 0.00, p = 1 |
| No injection | 23 | 57 | 40% |
Attenuation of the RNAi effect after classical conditioning
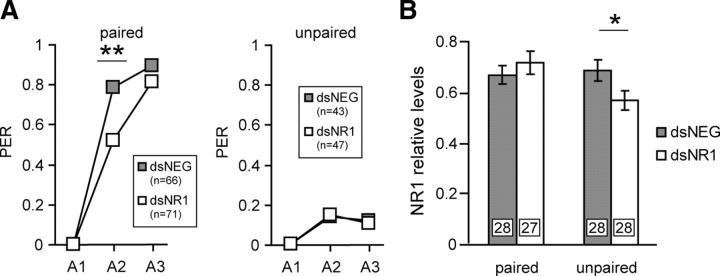
The persistence of the inhibitory effect was evaluated when combined with the behavioral procedure (Fig. 3A). One day after the injection of dsRNA, one group was trained in the PER paradigm, consisting of three CS–US pairings with an ITI of 10 min, a protocol known to induce the formation of LTM (Stollhoff et al., 2005). Another group was stimulated with a nonassociative procedure in which each animal had the same sensory experience as the conditioned group, except that the CS and the US were unpaired. During acquisition, the learning scores of the paired group increased rapidly as the reaction of the unpaired group to the CS was not different across trials (dsNR1, McNemar: χ2A2-A3 = 0, p = 1; dsNEG, McNemar: χ2A2-A3 = 0, p = 1). In the paired group, the dsNR1-injected animals had a significantly reduced response to the CS on the second acquisition trial (χ2 = 10.61; df = 1; p = 0.001), and there was no difference between the dsNEG and dsNR1 groups on the third acquisition trial (χ2 = 1.63; df = 1; p = 0.202). This behavioral effect attests that RNAi was effectively induced in the paired group. In the unpaired group, there was no difference between dsNEG- and dsNR1-injected animals (A2: χ2 = 0.03, df = 1, p = 0.868; A3: χ2 = 0.02, df = 1, p = 0.882). The NR1 subunit levels were analyzed 2 h after conditioning in the MB region (Fig. 3B). Interestingly, there was no differences between the dsNEG and dsNR1 groups of the paired group (Student's t test, −0.83; df = 53; p = 0.408) as the NR1 levels were significantly reduced in the dsNR1 honeybees of the unpaired group (Student's t test, 2.10; df = 54; p = 0.039).
Figure 3.
Evaluation of the RNAi effect in conditioned honeybees. A, One day after the injection of dsRNA (gray squares, dsNEG; white squares, dsNR1), animals were subjected to three CS–US pairings (paired, A1–A3) or to three unpaired CS–US presentations (unpaired, A1–A3). n represents the number of animals that participated in the experiment. Brains were dissected 2 h later. PER, Percentage of animals that showed a PER during the CS presentation. B, The relative NR1 subunit levels were quantified in the MB region after injection of dsRNA (gray bars, dsNEG; white bars, dsNR1). The expression levels were evaluated by quantitative Western blot, using AmNR1. Each column represents the mean ± SEM of n measurements as indicated by the numbers on the bars. Asterisks indicate significant differences between groups (**p < 0.01, χ2 test; *p < 0.05, Student's t test).
In this manner, it can be concluded that the conditioning procedure induced a process that overwhelmed the RNAi effect within a time window of maximum 2 h after the conditioning.
The inhibition of the NR1 subunit in the MB region disturbs specific phases of memory formation
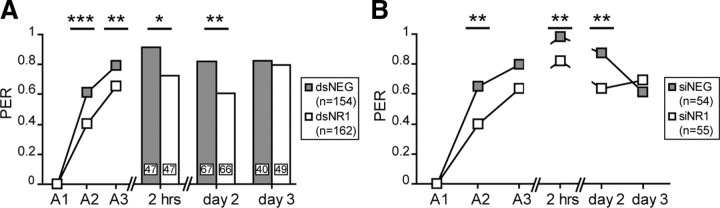
Honeybees were trained in the PER paradigm 1 d after the injection of dsRNA or siRNA. At this time, the NR1 subunit levels of dsNR1- and siNR1-injected animals are reduced in the MB region compared with dsNEG- and siNEG-injected animals (Fig. 2A–C). Behavioral differences were observed during the acquisition phase (Fig. 4). On the second acquisition trial, the conditioned responses of dsNR1- and siNR1-injected animals were significantly reduced compared with dsNEG- and siNEG-injected animals (dsRNA: χ2 = 13.82, df = 1, p = 0.0002; siRNA: χ2 = 6.73, df = 1, p = 0.0095). On the third acquisition trial, the dsNR1- and siNR1-injected animals also showed a reduced performance compared with the negative controls; however, the difference was significant only for dsRNA-injected animals (dsRNA: χ2 = 7.47, df = 1, p = 0.0063; siRNA: χ2 = 3.42, df = 1, p = 0.064), probably because of the higher number of animals that participated in this experiment.
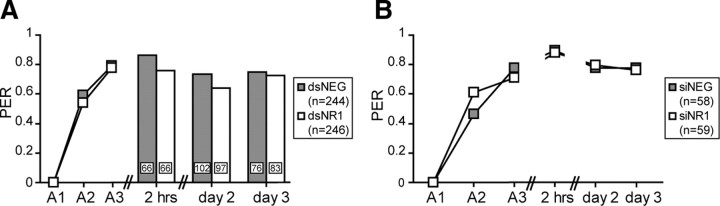
Figure 4.
The inhibition of the NR1 subunit selectively affects memory formation. One day after the injection of dsRNA (A; gray, dsNEG; white, dsNR1) or siRNA (B; gray, siNEG; white, siNR1), animals were subjected to three CS–US pairings (A1–A3). Memory was retrieved 2 h, 2 and 3 d after conditioning. In the dsRNA experiment, the animals were tested only once. Data from the acquisition phase were pooled for all subgroups (inset, n). The numbers on the bars represent the number of animals tested for each time point. In the siRNA experiment, n animals received multiple posttraining tests. Asterisks indicate significant differences between groups (*p < 0.05; **p < 0.01; ***p < 0.001; χ2 test). PER, Percentage of animals that showed a PER during the CS presentation.
The memory was evaluated 2 h, 2 d, and 3 d after conditioning by determining the retention scores. Honeybees given injections of dsRNA received only a single posttraining test at 2 h, 2 d, or 3 d after training, to exclude the possibility that extinction or reconsolidation processes might have influenced the behavioral effect (Stollhoff et al., 2005, 2008). Although the inhibition of the NR1 subunit did not induce an increased mortality (Table 1), RNAi is probably not induced with the same efficiency in each animal, and we cannot exclude that the majority of animals that survived until the memory test on day 3 were not efficiently treated. For this reason, animals received multiple posttraining tests in the siRNA experiment (Fig. 4). Retention scores evaluated 2 h after training were increased compared with the last acquisition trial for each experimental group. However, dsNR1- and siNR1-injected animals showed significantly reduced performances compared with their respective negative controls (dsRNA: χ2 = 5.82, df = 1, p = 0.016; siRNA: χ2 = 8.01, df = 1, p = 0.005). Two days after training, dsNR1- and siNR1-injected animals showed significantly reduced performances compared with their respective negative controls (dsRNA: χ2 = 7.52, df = 1, p = 0.0061; siRNA: χ2 = 8.01, df = 1, p = 0.005). On day 3, all animals performed equally well both in the dsRNA and siRNA groups (dsRNA: χ2 = 0.12, df = 1, p = 0.7285; siRNA: χ2 = 0.76, df = 1, p = 0.382). In both experiments, retention scores of the test groups increased on day 3 compared with the recall on day 2; the increase was significant in the dsNR1 group (χ2 = 4.71; df = 1; p = 0.03) but not in the siNR1 group (McNemar: χ2day 2-day 3 = 0.24, df = 1, p = 0.628).
The inhibition of the NR1 subunit affects specifically MTM
To exclude the possibility that the conditioned responses measured 2 h after conditioning are influenced by the defect observed during the acquisition, the animals that did not show a conditioned PER after presentation of the CS in the third acquisition trial, in the experiment presented in Figure 4, were removed from the analysis (Fig. 5). In this manner, the 2 h retention scores for animals that showed a conditioned response at the end of the training were still significantly reduced in the dsNR1 and siNR1 groups (dsRNA: χ2 = 4.67, df = 1, p = 0.03; siRNA: χ2 = 3.57, df = 1, p = 0.023).
Figure 5.
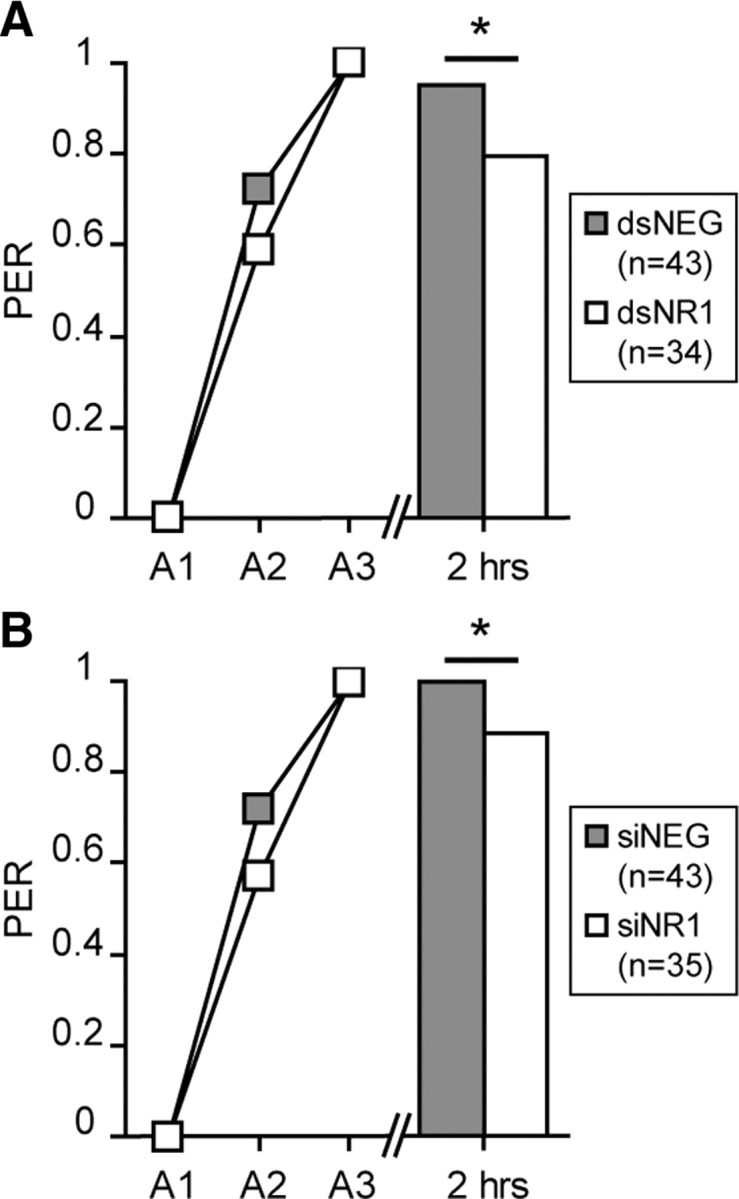
Evaluation of the behavioral effect by considering only learner honeybees. The behavioral data of honeybees conditioned 1 d after the injection of dsRNA (A; gray, dsNEG; white, dsNR1) or siRNA (B; gray, siNEG; white, siNR1) were analyzed by considering only animals that showed a PER during the CS presentation of the third acquisition trial (A3). Their memory performances were evaluated 2 h after conditioning. The number of considered animals is indicated in the inset. Asterisks indicate significant differences between groups (*p < 0.05, χ2 test). PER, Percentage of animals that showed a PER during the CS presentation.
The inhibition of the NR1 subunit does not affect olfactory and gustatory sensitivity
To control that the behavioral effect does not result from differences in gustatory sensitivity, we evaluated the response of the animals to sucrose during the acquisition phase and at the sucrose responsiveness test performed at the end of each behavioral protocol. No differences between the different groups were found (Table 2). Therefore, it can be concluded that the downregulation of the NR1 subunit does not alter gustatory sensitivity at the sucrose concentration used for the US.
Table 2.
Sucrose sensitivity of honeybees given injections of dsRNA or siRNA
| % Responding bees (number of bees) |
||||
|---|---|---|---|---|
| A1 | A2 | A3 | Sucrose test | |
| siNR1 | 98.5 (65) | 98.5 (65) | 98.5 (65) | 90.8 (65) |
| siNEG | 100 (61) | 96.7 (61) | 98.3 (61) | 90.2 (61) |
| χ2 (df = 1), p value | 0.95, 0.3308 | 0.41, 0.522 | 0, 0.9639 | 0.01, 0.9079 |
| dsNR1 | 96.6 (175) | 95.4 (175) | 97.1 (175) | 96.6 (175) |
| dsNEG | 95.9 (171) | 94.7 (171) | 95.3 (171) | 94.1 (171) |
| χ2 (df = 1), p value | 0.11, 0.745 | 0.09, 0.766 | 0.79, 0.3731 | 1.15, 0.2840 |
To control that there was no difference in olfactory sensitivity in treated animals, we reevaluated the results presented in Figures 3 and 4. In olfactory conditioning, a small proportion of honeybees react spontaneously to the presentation of the odor by showing a PER. A difference in spontaneous response to the presentation of the odor indicates a difference in odor sensitivity. For this reason, we analyzed the data by including animals that reacted to the odor at the first acquisition trial. In Figure 3, one group of honeybees was stimulated with a nonassociative protocol, and the reaction of the dsNEG and dsNR1 groups to the CS was not different across trials (Table 3). However, in this experiment, the animals were sensitized with the US, and this might have influenced the olfactory sensitivity of the honeybees. For this reason, we reevaluated the behavioral experiments presented in Figure 4. There was no difference between dsRNA and siRNA injected in the first acquisition trial (Table 3). For this reason, it can be considered that at the odor concentration used, the olfactory sensitivity of the honeybees was not altered.
Table 3.
Spontaneous PER to the odor of honeybees given injections of dsRNA or siRNA
| % Responding bees (number of bees) |
|||
|---|---|---|---|
| A1 | A2 | A3 | |
| Experiment 1a | |||
| dsNR1 | 7.8 (51) | 19.6 (51) | 13.7 (51) |
| dsNEG | 12.2 (49) | 20.4 (49) | 14.3 (49) |
| χ2 (df = 1), p value | 0.11, 0.745 | 0.01, 0.932 | 0.03, 0.854 |
| Experiment 2b | |||
| dsNR1 | 15.2 (191) | ||
| dsNEG | 13.5 (178) | ||
| χ2 (df = 1), p value | 0.46, 0.498 | ||
| Experiment 3c | |||
| siNR1 | 3.5 (57) | ||
| siNEG | 0 (54) | ||
| χ2 (df = 1), p value | 1.93, 0.165 | ||
The RNAi effect is reversible
We took advantage of the fact that the inhibitory effect is reversible to correlate it with the behavioral effect. Honeybees were trained 2 d after the injection of dsRNA or siRNA. At this time point, no inhibition of the NR1 subunit expression was detected in brains injected with dsRNA and siRNA (Fig. 2D–F). Honeybees given injections of dsRNA received a single posttraining test at 2 h, 2 d, or 3 d after training, whereas the honeybees given injections of siRNA received multiple posttraining tests. There were no significant differences in the acquisition phases between the dsNEG and dsNR1 groups (A2: χ2 = 1.43, df = 1, p = 0.231; A3: χ2 = 0.15, df = 1, p = 0.695) and between the siNEG and siNR1 groups (A2: χ2 = 2.46, df = 1, p = 0.117; A3: χ2 = 0.63, df = 1, p = 0.428) (Fig. 6). Conditioned responses at any retention test were not significantly different between the dsNEG and dsNR1 groups (2 h: χ2 = 2.42, df = 1, p = 0.12; day 2: χ2 = 2.14, df = 1, p = 0.143; day 3: χ2 = 0.15, df = 1, p = 0.698) and between the siNEG and siNR1 groups (2 h: χ2 = 0.07, df = 1, p = 0.794; day 2: χ2 = 0.07, df = 1, p = 0.784; day 3: χ2 = 0.03, df = 1, p = 0.866) (Fig. 6). The dsNR1 group had lower retention scores than the dsNEG group for the 2 h and the day 2 memory tests, but these differences were not significant (Fig. 6A).
Figure 6.
The RNAi effect is reversible. Two days after the injection of dsRNA (A; gray, dsNEG; white, dsNR1) or siRNA (B; gray, siNEG; white, siNR1), animals were subjected to three CS–US pairings (A1–A3). Memory was retrieved 2 h, 2 and 3 d after conditioning. In the dsRNA experiment, the animals were tested only once. Data from the acquisition phase were pooled for all subgroups (inset, n). The numbers on the bars represent the number of animals tested for each time point. In the siRNA experiment, n animals received multiple posttraining tests. PER, Percentage of animals that showed a PER during the CS presentation.
NMDARs are not required for the recall of memory
To further characterize the behavioral effect, we examined whether the inhibition of the NR1 subunit impaired memory recall. First, we tested whether RNAi against the NR1 subunit can be induced in honeybees that were conditioned before the injection of dsRNA. Hence, honeybees were given injections 2 d after training with either dsNR1 or dsNEG. One day later, on day 3 after conditioning, NR1 subunit levels were significantly reduced in the MB region (Student's t test, 2.47; df = 44; p = 0.017) and unchanged in the AL region (Student's t test, 1.36; df = 49; p = 0.180) (Fig. 7A). In a separate experiment, the same procedure was repeated. Untreated animals received three training trials. As expected, there was no difference in the acquisition phase between the dsNEG and the dsNR1 groups (Fig. 7B). Two days after conditioning, the honeybees were given injections. On day 3, 1 d after the injection, retention scores were not different between dsNEG- and dsNR1-injected animals (Fig. 7B).
Figure 7.
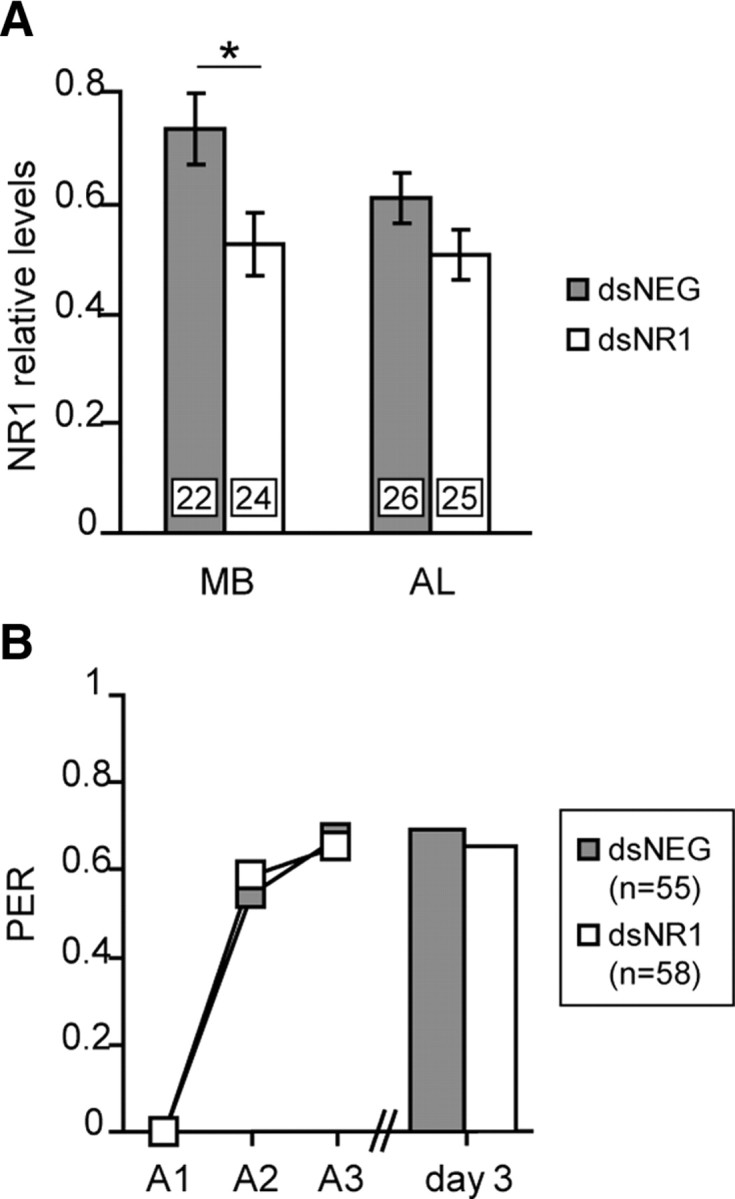
The inhibition of the NR1 subunit does not affect the recall of memory. A, Two days after conditioning, animals were given injections of dsRNA (gray, dsNEG; white, dsNR1). One day later, the NR1 expression levels were evaluated in the MB and AL regions. The NR1 levels were evaluated by quantitative Western blot, using AmNR1. Each column represents the mean ± SEM of n measurements as indicated by the numbers on the bars. The asterisk indicates a significant reduction (*p < 0.05, Student's t test). B, Animals were subjected to three CS–US pairings (A1–A3). Two days after conditioning, they were given injections of dsRNA, and memory was retrieved on day 3. The number of animals is indicated in the inset. PER, Percentage of animals that showed a PER during the CS presentation.
Discussion
In a previous study, we identified the honeybee NR1 subunit. Sequence analysis showed that it shares the common features of NR1 subunits from other species and it is encoded by a single gene (Zannat et al., 2006). The NR1 subunit is necessary for the formation of functional receptors both in vertebrates and in Drosophila (Monyer et al., 1992; Xia et al., 2005). For this reason, we decided to target the NR1 subunit to impair the formation of functional NMDARs.
The honeybee NR1 subunit was specifically detected by Western blot by using two antibodies at a molecular weight of ∼122 kDa in the honeybee brain (Fig. 1). RNAi was induced against the NR1 subunit with dsRNA and siRNA specific for different portions of the AmNR1 mRNA. We used appropriate negative controls to exclude unspecific effects. The separate analysis of brain regions allowed us to discriminate between different neuropiles. One day after the induction of RNAi, NR1 levels were reduced in the MB region but not in the AL region (Fig. 2). The conditioning procedure induced a process overwhelming the RNAi effect such that no inhibition was observed 2 h after conditioning (Fig. 3). Therefore, the time window of the inhibition was precisely defined. In addition, the treatment was not detrimental for the animals (Table 1) and did not alter olfactory or gustatory perceptions (Tables 2, 3).
Our results provide strong evidence that the inhibition of the NR1 subunit effectively reduced the formation of functional receptors and that the effect on learning and memory formation is highly specific. Indeed, we showed that the inhibition of the NR1 subunit 1 d after the injection is associated with behavioral effects (Figs. 2, 4). Two days after the injection, the downregulation of the NR1 subunit by RNAi was no longer detected, and neither acquisition nor retention was affected in animals trained at this moment (Figs. 2, 6).
The inhibition of functional NMDARs in the MB region during and shortly after learning impaired several phases of memory formation (Figs. 4, 5). During acquisition, a significant reduction in learned performance is already observed during the second acquisition trial. On the third acquisition trial, the conditioned responses of NR1-RNAi-treated honeybees are still reduced compared with negative controls, but the difference was significant only in the dsRNA experiment. The retention scores measured 2 h after conditioning were also reduced in NR1-RNAi-treated animals (Fig. 4), at which time MTM is formed (Menzel, 1999). MTM is probably directly affected because the effect was still present in the group of animals that showed a conditioned PER on the third acquisition trial (Fig. 5). NR1 levels were reduced during the acquisition and MTM test, possibly indicating that NMDARs are important for memory retrieval. However, we demonstrated that memory retrieval was not affected when RNAi was induced after conditioning before the retention test (Fig. 7). Hence, we can conclude that NMDARs of the MB region are required for memory formation but not for memory retrieval.
We demonstrated that the inhibition of NMDARs in the MB region affected eLTM but not lLTM (Fig. 4). In addition, the retention scores increased from day 2 to day 3 in dsNR1-treated animals. This indicates that lLTM is intact and suggests that it consolidates independently from earlier memory phases. Our results are supported by those of Locatelli et al. (2005), who showed that the photorelease of glutamate on the honeybee MB just after, but not before, training improved eLTM. Unfortunately, the effect on lLTM was not tested in this work. LTM formation depends on a prolonged activation of protein kinase A (PKA) in the range of minutes after conditioning (Müller, 2000). Friedrich et al. (2004) showed that feeding honeybees 4 h before conditioning affected the training-induced PKA activation and all memory phases. Interestingly, the rescue of basal PKA activity just after the training of fed honeybees lead to a specific rescue of lLTM but had no effect on eLTM, suggesting that eLTM and lLTM develop from molecular cascades acting in parallel in memory formation. Our results also suggest that eLTM and lLTM develop independently from each other, and we propose that in our experimental conditions, NMDARs are not involved in the PKA-dependent process leading to the formation of the transcription-dependent lLTM. It was already shown in the honeybee and in other species that memory phases can develop in parallel (Squire, 1987; Grünbaum and Muller, 1998; Sutton et al., 2001; Wu et al., 2007).
It is surprising that in our study lLTM was not affected by the treatment. Indeed, studies in other model systems claim that the receptor is required during the acquisition of the association and subsequently for the formation of all memory phases (Martin et al., 2000; Roberts and Glanzman, 2003; Wu et al., 2007).
On one hand, it might be that NMDARs are coincidence detectors, but that the MB is not important for lLTM formation. In the honeybee as well as in Drosophila, both the AL and the MB contribute to the formation of the memory trace in appetitive learning (Menzel and Muller, 1996; Thum et al., 2007). In addition, it was recently shown in Drosophila that NMDARs of the MB are important for MTM and those of the ellipsoid body are important for LTM (Wu et al., 2007). Hence, the activation of NMDARs during appetitive conditioning in the MB region might be important for the CS–US association leading to MTM and eLTM. Then, the effect observed during conditioning would reflect a specific impairment of this association. The activation of the receptor in other neuropiles, probably in the AL region, could be important for CS–US association underlying lLTM.
On the other hand, it can be hypothesized that NMDARs are not important for the acquisition of the association but that they, rather, play a role in memory formation. In this case, the effect observed during the acquisition reflects an impairment of short-term memory (STM), and we see three possibilities to explain that lLTM was not affected. (1) lLTM is independent of NMDARs. (2) It might be that a moderate reduction of NMDARs, like the one achieved in this work, is sufficient to impair STM, MTM, and eLTM and that a stronger inhibition is required to inhibit lLTM. In the honeybee, eLTM depends on translation, and lLTM depends on translation and transcription (Friedrich et al., 2004). In Aplysia, local translation taking place in synaptic processes mediates intermediate-term facilitation at that synapse. Transcription and translation in the soma induce long-term facilitation (Martin et al., 1997; Casadio et al., 1999; Liu et al., 2003). Thus, it might be that NMDAR-dependent local translation mediates eLTM formation in the honeybee. If this holds true, the moderate reduction of NMDAR in our experiment would have directly affected synaptic events underlying early memories (STM, MTM, and eLTM). In contrast, lLTM formation could depend on several signaling cascades converging to the nucleus to induce translation and transcription. It is conceivable that the NMDAR signaling cascade is not the predominant one and that only a massive inhibition of NMDAR activation would impair lLTM formation. (3) It is conceivable that the activation of the receptor during the 2 h after conditioning is sufficient for the formation of STM, MTM, and eLTM but that a longer activation or a reactivation of the receptor is required for the consolidation of lLTM. There is evidence that memory consolidation depends on the reactivation of NMDARs after conditioning, probably during sleep (Shimizu et al., 2000; Aton et al., 2009). In addition, Wu et al. (2007) showed that the inhibition of NMDARs during conditioning and 1 d after impaired 4 d memory in Drosophila.
In conclusion, our study showed that the acute inhibition of NMDARs in the MB region of the honeybee brain during conditioning and shortly after impaired selectively the acquisition phase, MTM, and eLTM formation, leaving lLTM intact. Thus, our results suggest that lLTM consolidates independently from earlier memory phases.
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft Grants LE1809/1-1 and LE1809/1-2. We thank Olga Ganeshina for supporting the project, Lena Sünke Mortensen for technical assistance, Julien Colomb for critical comments about this manuscript, and Lise Bruyneel for intensive support.
The authors declare no competing financial interests.
References
- Aton SJ, Seibt J, Dumoulin M, Jha SK, Steinmetz N, Coleman T, Naidoo N, Frank MG. Mechanisms of sleep-dependent consolidation of cortical plasticity. Neuron. 2009;61:454–466. doi: 10.1016/j.neuron.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara GS, Zube C, Rybak J, Gauthier M, Grunewald B. Acetylcholine, GABA and glutamate induce ionic currents in cultured antennal lobe neurons of the honeybee, Apis mellifera. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191:823–836. doi: 10.1007/s00359-005-0007-3. [DOI] [PubMed] [Google Scholar]
- Bitterman ME, Menzel R, Fietz A, Schafer S. Classical conditioning of proboscis extension in honeybees (Apis mellifera) J Comp Psychol. 1983;97:107–119. [PubMed] [Google Scholar]
- Breer H, Sattelle DB. Molecular properties and functions of insect acetylcholine receptors. J Insect Physiol. 1987;33:771–790. [Google Scholar]
- Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey CH, Kandel ER. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–237. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- Cully DF, Paress PS, Liu KK, Schaeffer JM, Arena JP. Identification of a Drosophila melanogaster glutamate-gated chloride channel sensitive to the antiparasitic agent avermectin. J Biol Chem. 1996;271:20187–20191. doi: 10.1074/jbc.271.33.20187. [DOI] [PubMed] [Google Scholar]
- Devaud JM, Clouet-Redt C, Bockaert J, Grau Y, Parmentier ML. Widespread brain distribution of the Drosophila metabotropic glutamate receptor. Neuroreport. 2008;19:367–371. doi: 10.1097/WNR.0b013e3282f524c7. [DOI] [PubMed] [Google Scholar]
- El Hassani AK, Giurfa M, Gauthier M, Armengaud C. Inhibitory neurotransmission and olfactory memory in honeybees. Neurobiol Learn Mem. 2008;90:589–595. doi: 10.1016/j.nlm.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Farooqui T, Robinson K, Vaessin H, Smith BH. Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee. J Neurosci. 2003;23:5370–5380. doi: 10.1523/JNEUROSCI.23-12-05370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Rushton E, Rohrbough J, Liebl F, Karr J, Sheng Q, Rodesch CK, Broadie K. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J Neurosci. 2005;25:3199–3208. doi: 10.1523/JNEUROSCI.4201-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A, Muller U, Menzel R. Reversible downregulation of protein kinase A during olfactory learning using antisense technique impairs long-term memory formation in the honeybee, Apis mellifera. J Neurosci. 1999;19:10125–10134. doi: 10.1523/JNEUROSCI.19-22-10125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A, Thomas U, Muller U. Learning at different satiation levels reveals parallel functions for the cAMP-protein kinase A cascade in formation of long-term memory. J Neurosci. 2004;24:4460–4468. doi: 10.1523/JNEUROSCI.0669-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünbaum L, Muller U. Induction of a specific olfactory memory leads to a long-lasting activation of protein kinase C in the antennal lobe of the honeybee. J Neurosci. 1998;18:4384–4392. doi: 10.1523/JNEUROSCI.18-11-04384.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Menzel R. Multiple sites of associative odor learning as revealed by local brain microinjections of octopamine in honeybees. Learn Mem. 1998;5:146–156. [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Holen T, Mobbs CV. Lobotomy of genes: use of RNA interference in neuroscience. Neuroscience. 2004;126:1–7. doi: 10.1016/j.neuroscience.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Mitri C, Grau Y, Maleszka R. Characterization of a metabotropic glutamate receptor in the honeybee (Apis mellifera): implications for memory formation. Invert Neurosci. 2007;7:99–108. doi: 10.1007/s10158-007-0045-3. [DOI] [PubMed] [Google Scholar]
- Leboulle G, Müller U. Synergistic activation of insect cAMP-dependent protein kinase A (type II) by cyclicAMP and cyclicGMP. FEBS Lett. 2004;576:216–220. doi: 10.1016/j.febslet.2004.08.079. [DOI] [PubMed] [Google Scholar]
- Liu K, Hu JY, Wang D, Schacher S. Protein synthesis at synapse versus cell body: enhanced but transient expression of long-term facilitation at isolated synapses. J Neurobiol. 2003;56:275–286. doi: 10.1002/neu.10242. [DOI] [PubMed] [Google Scholar]
- Locatelli F, Bundrock G, Muller U. Focal and temporal release of glutamate in the mushroom bodies improves olfactory memory in Apis mellifera. J Neurosci. 2005;25:11614–11618. doi: 10.1523/JNEUROSCI.3180-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- Menzel R. Memory dynamics in the honeybee. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1999;185:323–340. [Google Scholar]
- Menzel R, Muller U. Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Müller U. Prolonged activation of cAMP-dependent protein kinase during conditioning induces long-term memory in honeybees. Neuron. 2000;27:159–168. doi: 10.1016/s0896-6273(00)00017-9. [DOI] [PubMed] [Google Scholar]
- Roberts AC, Glanzman DL. Learning in Aplysia: looking at synaptic plasticity from both sides. Trends Neurosci. 2003;26:662–670. doi: 10.1016/j.tins.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290:1170–1174. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- Si A, Helliwell P, Maleszka R. Effects of NMDA receptor antagonists on olfactory learning and memory in the honeybee (Apis mellifera) Pharmacol Biochem Behav. 2004;77:191–197. doi: 10.1016/j.pbb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Squire LR. Memory and brain. New York: Oxford UP; 1987. [Google Scholar]
- Stollhoff N, Menzel R, Eisenhardt D. Spontaneous recovery from extinction depends on the reconsolidation of the acquisition memory in an appetitive learning paradigm in the honeybee (Apis mellifera) J Neurosci. 2005;25:4485–4492. doi: 10.1523/JNEUROSCI.0117-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollhoff N, Menzel R, Eisenhardt D. One retrieval trial induces reconsolidation in an appetitive learning paradigm in honeybees (Apis mellifera) Neurobiol Learn Mem. 2008;89:419–425. doi: 10.1016/j.nlm.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Masters SE, Bagnall MW, Carew TJ. Molecular mechanisms underlying a unique intermediate phase of memory in Aplysia. Neuron. 2001;31:143–154. doi: 10.1016/s0896-6273(01)00342-7. [DOI] [PubMed] [Google Scholar]
- Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. J Neurosci. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster: cloning of a kainate-selective subunit expressed in the central nervous system. Proc Natl Acad Sci U S A. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RJ. Glutamate: an excitatory neurotransmitter in the mammalian CNS. Brain Res Bull. 1999;50:353–354. doi: 10.1016/s0361-9230(99)00102-1. [DOI] [PubMed] [Google Scholar]
- Wu CL, Xia S, Fu TF, Wang H, Chen YH, Leong D, Chiang AS, Tully T. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10:1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüstenberg D, Gerber B, Menzel R. Short communication: long- but not medium-term retention of olfactory memories in honeybees is impaired by actinomycin D and anisomycin. Eur J Neurosci. 1998;10:2742–2745. doi: 10.1046/j.1460-9568.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannat T, Locatelli F, Rybak J, Menzel R, Leboulle G. Identification and localisation of the NR1 sub-unit homologue of the NMDA glutamate receptor in the honeybee brain. Neurosci Lett. 2006;398:274–279. doi: 10.1016/j.neulet.2006.01.007. [DOI] [PubMed] [Google Scholar]