Abstract
Strains of Monascus filamentous fungal species have been used to produce fermented foods in Asian countries, such as China, Japan, and The Korean Peninsula, for nearly 2,000 years. At present, their fermented products are widely used as food additives and nutraceutical supplements worldwide owing to their production of beneficial secondary metabolites. Heterotrimeric G-protein signaling pathways participate in regulating multiple biological processes in fungi. Previously, we identified three Monascus ruber M7 G-protein α subunits (Mga1–3) and demonstrated that Mga1 can regulate growth, reproduction and some secondary metabolites’ production. Here, we systematically analyzed and compared the roles of mga1–3 by combining single- and double-gene(s) knockouts and their transcriptomic data. First, mga2 and mga3 knock-out mutants and pairwise combinations of mga1–3 deletion strains were generated. Then the changes in growth, development and the main secondary metabolites, Monascus pigments and citrinin, in these mutants were systematically compared with M. ruber M7. Moreover, RNA-Seq analyses of these mutants were performed. All three Gα subunits worked together to regulate biological processes in M. ruber M7, with Mga1 playing a major role, while Mga2 and Mga3 playing supplemental roles. According to the existing literatures which we can find, gene knock-out mutants of the pairwise combination of mga1–3 and their transcriptome analysis are first reported in this study. The current results have clearly demonstrated the functional division of Mga1–3 in M. ruber M7, and could provide a deeper understanding of the effects of different Gα subunits on growth, development and secondary metabolism in other filamentous fungi.
Keywords: Monascus ruber, G-protein α-subunit, development, secondary metabolism, transcriptomic analysis
Introduction
Monascus spp. have been used in food fermentation in Asian countries, such as China, Japan, and The Korean Peninsula, for nearly 2,000 years (Chen et al., 2015; Kim et al., 2016; Rahayu et al., 2017). At present, their fermented products, such as red fermented rice (RFR), also called red yeast rice, Anka, Hongqu, red koji, and red mold rice, are widely used as food coloring, fermentation starters and food supplements worldwide (Chen et al., 2015; Yu et al., 2015; Wei et al., 2017; Derosa et al., 2018; Luo et al., 2018), because Monascus spp. can produce various useful secondary metabolites (SMs), mainly including Monascus pigments (MPs), monacolin K (MK) and γ-amino butyric acid (Feng et al., 2012; Huang et al., 2013; Patakova, 2013; Diana et al., 2014). However, some strains of Monascus spp. may also produce citrinin (CIT), a kind of nephrotoxic mycotoxin (Blanc et al., 1995). Therefore, how to increase the beneficial SMs levels and reduce the CIT level in RFR has become the research hotspot over the last 10 years (Jia et al., 2010; Li et al., 2013; Liu et al., 2014; He and Cox, 2016; Alberti et al., 2017).
The genes involved in SMs biosynthesis in filamentous fungi usually appear as gene clusters (Schwecke et al., 1995; Blin et al., 2015). In the past decade, the gene clusters of MPs, CIT and MK in Monascus spp. have been identified and their biosynthetic pathways have been fully illustrated (Chen et al., 2008; Li et al., 2015; He and Cox, 2016; Liu J. et al., 2016; Chen et al., 2017). The biosynthesis of these SMs cannot only be controlled by the intra-cluster regulating genes but can also be adjusted by the off-cluster global regulating genes, such as LaeA, VeA, and related genes in the G-protein signaling pathway (GPSP) (Fox and Howlett, 2008; Liu Q. et al., 2016; Lin et al., 2018). GPSPs, including the G-protein coupled receptor (GPCR), heterotrimeric G-protein (G-protein) and downstream effectors (Seo and Yu, 2006), play vital roles in growth, differentiation, SMs biosynthesis, pathogenicity and toxicity in filamentous fungi (Yu et al., 2008; Corrochano et al., 2016; Moretti et al., 2017; Liu et al., 2018; van den Hoogen et al., 2018).
Each G-protein generally composes α, β and γ subunits. In most characterized filamentous fungi, the Gα proteins are classified as three groups (Gα1–3) (Li et al., 2007). Although the functions of individual Gα subunits have been well investigated in model and pathogenic fungi (Li et al., 2010; Yang et al., 2012; Hu et al., 2013; Wasil et al., 2013; Garcia-Rico et al., 2017), there is limited research on the interplay among different Gα subunits (Kamerewerd et al., 2008).
In our previous study, the roles of the G protein α subunit gene mga1 (Gα 1 gene) in wild-type M. ruber M7 were analyzed, and mga1 can comprehensively regulate growth, reproduction, and MPs and CIT production (Li et al., 2010). Here, the other two Gα genes, mga2 (Gα 2 gene) and mga3 (Gα 3 gene), and the pairwise combinations of mga1–3 were independently deleted in M. ruber M7. The morphological observations and fermentation experiments of six Gα genes’ mutants, Δmga1, Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3, as well as their RNA-Seq analyses, were conducted to systematically investigate the functions of the Gα subunits in M. ruber M7. And we have found that all three Gα subunits work together to regulate extensive biological processes in M. ruber M7, Mga1 playing a major role and Mga2 and Mga3 as supplementary roles. In detail, during vegetative growth, Mga1 is the essential positive regulator, while Mga2 and Mga3 can enhance the regulatory process when either was double deleted with Mga1. However, a single deletion of Mga2 or Mga3 has little effect. Mga1 contributes the most to the regulation of sexual/asexual reproduction, and the regulation of asexual reproduction may occur prior to the central regulatory pathway. Different Gα subunits can be combined to negatively regulate secondary metabolism. Mga1 and Mga2 can negatively regulate MPs and CIT production individually or jointly, and Mga3 may work in combination with Mga1 to negatively enhance regulation of MPs production. These findings not only illuminate the functions of different Gα subunits in M. ruber M7 but could also provide a deeper understanding of the effects of different Gα subunits on growth, development and secondary metabolism in other filamentous fungi.
Materials and Methods
Strains and Media
Monascus ruber M7 (CCAM 070120, Culture Collection of State Key Laboratory of Agricultural Microbiology, China Center for Type Culture Collection, Wuhan, China) (Chen and Hu, 2005) was used to generate the gene knockout strains Δmga2 and Δmga3. The Δmga1 strain obtained by Li in our laboratory (Li et al., 2010) was used to generate the double-deletion strains Δmga1+2 and Δmga1+3. The Δmga2 strain obtained in this study was used to generate the double-deletion strain Δmga2+3.
Potato dextrose agar (PDA), malt extract agar (MA), czapek yeast extract agar (CYA) and 25% glycerol nitrate agar (G25N) were utilized for phenotypic characterization (He et al., 2013). PDA was used for the analyses of MPs and CIT production. G418 (Sigma-Aldrich, Shanghai, China) or hygromycin (Sigma-Aldrich, Shanghai, China) was added to the medium for transformant selection (Yang et al., 2012).
Deletion of Gα Genes in M. ruber M7
The homologous gene recombination strategy was used to construct the deletion strains (Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3). The hygromycin resistence gene hph was used in the mga2-deletion cassette to construct the Δmga2 strain, while the G418 resistence gene neo was used in the mga3-deletion cassette to construct the Δmga3 strain. The Δmga1 strain had been constructed previously (Li et al., 2010). The Δmga1 strain and another mga2-deletion cassette with the neo gene were used to construct the double-deletion strain Δmga1+2. The Δmga1 strain and the mga3-deletion cassette were used to construct the double-deletion strain Δmga1+3. The Δmga2 strain and the mga3-deletion cassette were used to construct the double-deletion strain Δmga2+3. The gene deletion cassette was constructed by double-joint PCR, as shown in Supplementary Figures S1, S2, using the primers listed in Supplementary Table S1. The construction strategy for the complementary strains is also shown in Supplementary Figure S1. The mutants were generated using an Agrobacterium tumefaciens-mediated transformation method that was previously established in our laboratory (Li et al., 2010). The genotypes of deletion strains were confirmed using PCR amplification and Southern hybridization.
Southern Hybridization
Southern hybridization was performed according to a previously reported method (Liu et al., 2014) using a DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Germany). Fragments of mga2 [open reading frame (ORF), probe 1], hph (selective marker gene, probe 2), mga3 (ORF, probe 3), and neo (selective marker gene, probe 4) were independently amplified to be used as probes. The single-deletion mutants’ DNAs were digested by SacI and XhoI. The double-deletion mutants’ DNAs were digested by KpnI. Primers are listed in Supplementary Table S1.
Phenotypic Analysis
Monascus ruber M7, Δmga1, Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3 strains were cultivated on PDA, CYA, MA and G25N at 28°C to observe their phenotypes. The colony sizes of these strains were measured after cultivated for 12 days, and the cleistothecia or conidia were observed and counted after cultivated for 5 days. Three replicates are for each strain.
Freshly harvested conidiospores (105 conidia mL–1) of M. ruber M7 and Gα-deleted strains were inoculated on PDA medium, covered with cellophane and incubated at 28°C for 11 days. The mycelia and medium were sampled every other day from 3 to 11 days to analyze the intracellular and extracellular MPs and CIT production levels (Li et al., 2014).
Monascus pigments were determined by their UV-Vis spectra (Agilent Cary 60, Australia). CIT was determined by Waters ACQUITY UPLC I-class system (Waters, Milford, MA, United States) with an ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm) according to the established method in our laboratory (Liu J. et al., 2016).
RNA-Seq Analysis
Monascus ruber M7 and Gα mutant strains were independently inoculated on PDA medium, covered with cellophane and incubated at 28°C. Two replicates were conducted for each strain. At 3 and 7 days, mycelia were collected for RNA extraction and then sequenced using the BGIseq-500RS platform (BGI, Wuhan, China1). The expression levels of 11 randomly selected genes in M. ruber M7 and Δmga1+3 were assessed using quantitative real-time PCR (qRT-PCR) to confirm the reliability of the RNA-Seq results.
Monascus ruber M7 genome which contains 8,407 genes, was used as a reference genome (Chen et al., 2015) to calculate the BLAST rate of the genome, and clean data were aligned using Hierarchical Indexing for Spliced Alignment of Transcripts and bowtie2 (Langmead and Salzberg, 2012; Kim et al., 2015). Then, RNA-Seq by Expectation Maximization was used to calculate the expression level of each gene (Li and Dewey, 2011). The genes that possessed an expression differential multiple greater than 1, as well as a Q-value not greater than 0.001 (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003), were selected as differentially expressed genes (DEGs).
Gene ontology (GO)2 and a KEGG pathway3 functional analysis were used to investigate the functions of the DEGs between M. ruber M7 and Gα mutants. Moreover, the DEGs involved in fungal growth, sporulation and secondary metabolism were further analyzed to determine the roles of Gα subunits in development and secondary metabolism of M. ruber M7.
Results
Targeted Deletion of Gα Genes in M. ruber M7
Single-deletion strains Δmga2 with hygromycin resistance and Δmga3 with G418 resistance, as well as double-deletion strains Δmga1+2, Δmga1+3, and Δmga2+3, were obtained. For Δmga2 strain, the PCR analysis confirmed the existence of the hph sequence as well as the absence of the mga2 ORF. Southern hybridization showed a single copy of the hph sequence in the Δmga2 strain. For Δmga3 strain, the PCR analysis confirmed the existence of the neo sequence as well as the absence of the mga3 ORF. Southern hybridization showed a single copy of the neo sequence in the Δmga3 strain. For Δmga1+2 strain, the PCR analysis confirmed the existence of the hph and neo sequence as well as the absence of the mga1 and mga2 ORF. Southern hybridization showed a single copy of the neo sequence in the Δmga1+2 strain. For Δmga1+3 strain, the PCR analysis confirmed the existence of the hph and neo sequence as well as the absence of the mga1 and mga3 ORF. Southern hybridization showed a single copy of the neo sequence in the Δmga1+3 strain. For Δmga2+3 strain, the PCR analysis confirmed the existence of the hph and neo sequence as well as the absence of the mga2 and mga3 ORF. Southern hybridization showed a single copy of the neo sequence in the Δmga2+3 strain. The results of PCR analysis and Southern hybridization were displayed in Supplementary Figures S1, S2. Additionally, the corresponding complementation strains were also obtained. The complementation strains possessed phenotypic characteristics similar to those of M. ruber M7 (Supplementary Figure S3).
Phenotypic Characterization of M. ruber M7 and Gα Mutants
Vegetative Growth and Reproduction
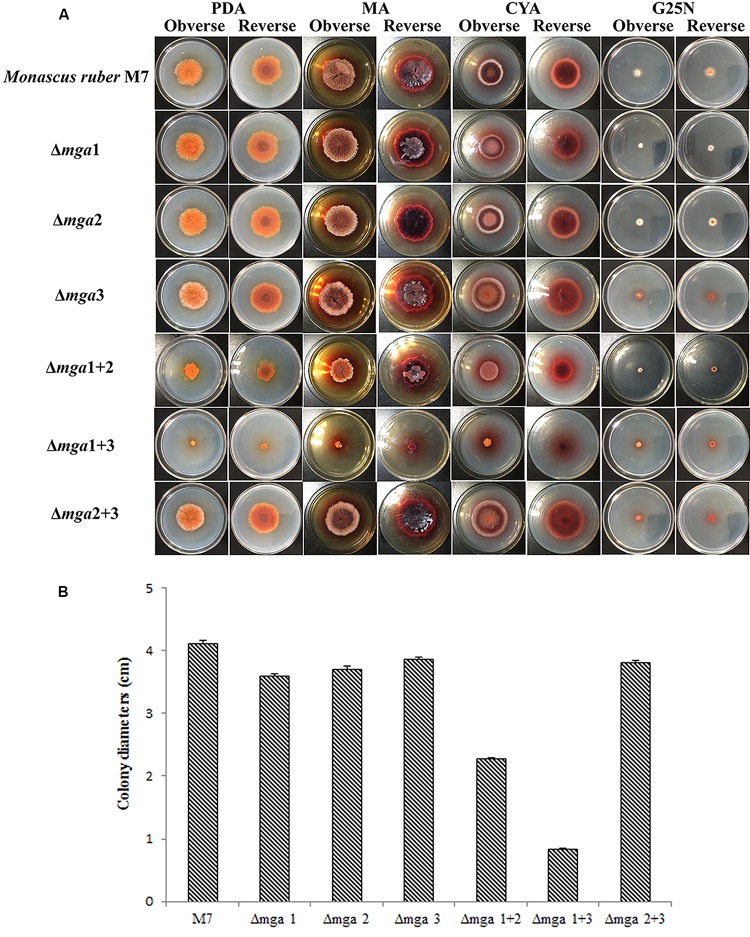
The phenotypes of the six Gα mutants, Δmga1 (prepared by Li et al. (2010)), Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3, were compared with M. ruber M7. As shown in Figure 1, after cultivation on PDA medium for 12 days, the colony sizes of the Δmga1, Δmga2, Δmga3 and Δmga2+3 strains were similar to that of M. ruber M7, while those of Δmga1+2 and Δmga1+3 were about 45% and 80% smaller than M. ruber M7, respectively. We found that when a single Gα gene (mga1, mga2, or mga3) was deleted, the colony sizes did not significantly change. However, when the mga2 or mga3 gene was deleted in the Δmga1 strain, the colony sizes were smaller than other mutants.
FIGURE 1.
Colony morphologies of Gα mutants and Monascus ruber M7. (A) Colony morphologies of M7 and Gα mutants observed on PDA, MA, CYA, and G25N plates and cultured at 28°C for 12 days. (B) Colony sizes of the indicated strains on PDA medium cultured at 28°C for 12 days.
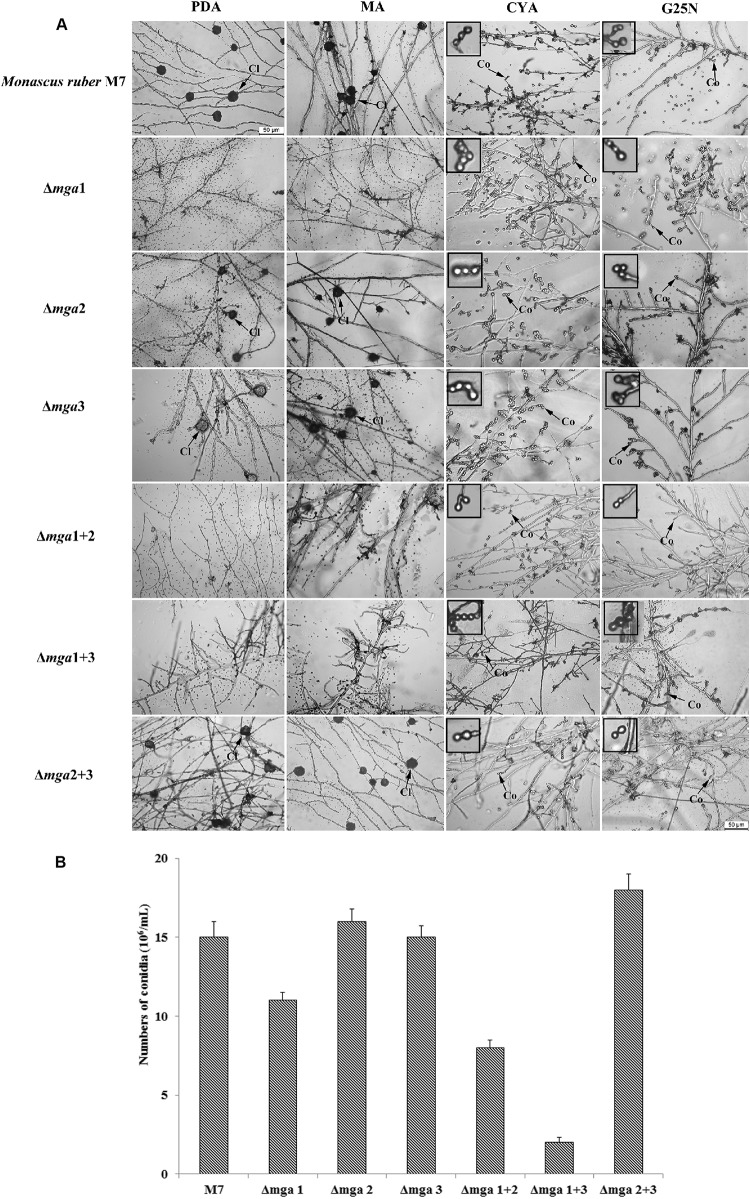
Regarding sexual or asexual reproduction, as shown in Figure 2, cleistothecia were not found in mga1-related mutants (Δmga1, Δmga1+2, and Δmga1+3), and their conidia-forming abilities were also reduced. However, the other mutants (Δmga2, Δmga3, and Δmga2+3) showed no difference in sexual and asexual reproduction compared with M. ruber M7.
FIGURE 2.
Microscopic structures of Gα mutants and Monascus ruber M7. (A) Cleistothecial (Cl) and conidial (Co) morphologies among M7 and Gα mutants were observed on PDA, MA, CYA, and G25N plates cultured at 28°C for 5 days. The enlarged areas are indicated by arrows. Size bar = 50 μm. (B) The numbers of conidia of the indicated strains were measured after growing on PDA medium at 28°C for 5 days.
MPs and CIT Production
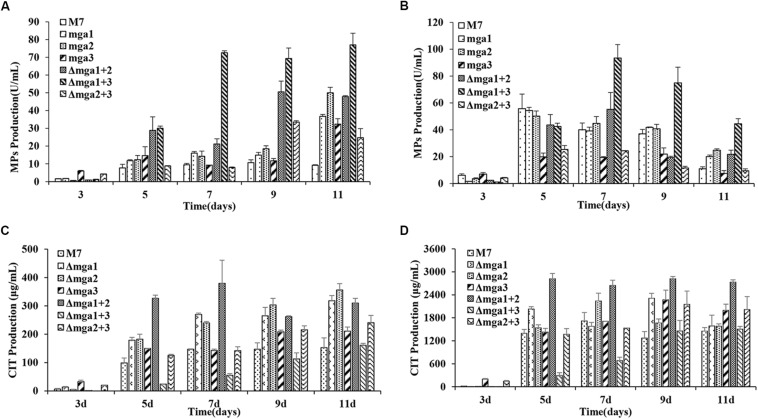
The intracellular and extracellular MPs and CIT production in M. ruber M7 and Gα mutants were analyzed (Figure 3). The intracellular MPs production levels in the Gα mutants, Δmga1, Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3, were 2.1, 2.5, 1.9, 3.9, 6.5, and 2.1 times that of M. ruber M7, respectively. For extracellular MP production, only Δmga1+3 was 1.7 times that of M. ruber M7, while the other five mutants possessed similar yields to M. ruber M7. For CIT, the single-deletion mutants and Δmga1+2 had 1.2- to 2.2-fold increases in CIT production compared with M. ruber M7. Only during the later stage (9–11 d) the CIT production in Δmga2+3 was greater (1.3-fold) than that in M. ruber M7. During the early stage (3–7 days) the CIT production in Δmga1+3 was only 20–40% that of M. ruber M7.
FIGURE 3.
MPs and CIT levels in Gα mutants and Monascus ruber M7. (A) The intracellular MP production of Gα mutants and M7. (B) The extracellular MP production levels of Gα mutants and M7. (C) The intracellular CIT production levels of Gα mutants and M7. (D) The extracellular CIT production levels of Gα mutants and M7. The error bars indicate the standard deviations of three independent cultures. Significantly different at P < 0.01.
DEG Analysis, Annotation and Functional Classification
The RNAs of M. ruber M7 and six deletion mutants (Δmga1, Δmga2, Δmga3, Δmga1+2, Δmga1+3, and Δmga2+3) were independently extracted for a further transcriptomic analysis. The obtained clean sequence reads of the 14 samples were validated by qRT-PCR. In total, 11 genes in the M. ruber M7 genome were randomly selected for relative gene expression comparisons between M7 and Δmga 1+3 strain, the selected genes are listed in Supplementary Table S2. As shown in Supplementary Figure S4, the relative expression levels of these 11 random genes had the same trends as in the RNA-Seq, which indicated that the transcriptome sequencing was reliable.
DEGs Analyses and Transcriptome Classification
The genes that possessed an expression differential multiple greater than 1, as well as a Q-value not greater than 0.001, were selected as DEGs. Compared with M. ruber M7, different mutants at different time points have diverse trends in their numbers of DEGs. There were greater numbers of DEGs in double-deletion strains than in single-deletion strains. In particular, in the Δmga1+3 strain, 1,858 and 2,000 genes showed down-regulated expression levels at 3rd day and 7th day, respectively, which were much greater numbers than those in the corresponding single-deletion strains Δmga1 and Δmga3. This may explain the distinctive phenotype of the Δmga1+3 strain (Figure 2).
A GO enrichment analysis of DEGs was performed. GO has three ontologies: molecular biological function, cellular component and biological process. For each ontology, the functional enrichment was determined. Compared with M. ruber M7, metabolic process possessed the most DEGs in all the mutants at both 3 and 7 days, and most genes in this GO ontology were down-regulated, such as in cellular process, cell part and catalytic activity. Among the KEGG pathways, the metabolic pathway possessed the most DEGs in all the mutants at both 3rd and 7th day, and most genes in this KEGG pathway were down-regulated, including those involved in meiosis–yeast, SMs biosynthesis and carbon metabolism.
Gα Genes Positively Regulate Vegetative Growth
The DEGs of carbon and nitrogen source metabolism are listed in Supplementary Table S3. The regulation of carbon source metabolism mostly focuses on the tricarboxylic acid cycle (TCA cycle), meanwhile many major facilitator superfamily (MFS) transporters were down-regulated. RNA-Seq results revealed that the absence of Gα subunits generally depressed the TCA cycle, especially reducing the biosynthesis of citric acid and succinyl CoA. The absence of both Mga1 and Mga3 regulated most genes in the TCA cycle. Data on the DEGs related to the TCA cycle are presented in Supplementary Figure S5. On the basis of the GO and KEGG analyses, we also analyzed the influence of different Gα genes on nitrogen metabolism. The expression levels of genes related to nitrogen metabolism mostly decreased in the mutants, with Δmga1+2 and Δmga1+3 possessing the greatest numbers of DEGs related to nitrogen metabolism. This indicates that all the Gα genes positively regulated nitrogen metabolism. Data on DEGs related to nitrogen source metabolism are presented in Supplementary Figure S6. The decreased expression of vegetative growth-related genes corresponded to the repressed colony sizes of the Δmga1+2 and Δmga1+3 strains (Figure 1).
Gα Genes Play Different Roles in Sexual and Asexual Reproduction
In filamentous fungi, the central regulatory pathway of conidiospore formation generally consists of abaA, brlA and wetA genes (Yu, 2006). The most reported sexual reproduction- related genes are mating type (MAT)-related genes (Varga et al., 2014). In addition, the velvet family genes are related to sexual/asexual reproduction (Yu et al., 2008; Liu Q. et al., 2016). The expression changes in all these genes as determined by the DEGs analysis are listed in Supplementary Figure S7.
Compared with M. ruber M7, the expression level of the cleistothecia-related gene MAT1-2 was decreased only in mga1-deleted strains (Δmga1, Δmga1+2, and Δmga1+3), while their expression levels increased in Δmga3 at 3 days. They were not changed in the Δmga2 and Δmga2+3 strains. This explained why cleistothecia were not found in Δmga1, Δmga1+2, and Δmga1+3 strains (Figure 2), and it suggested that Mga1 positively regulates sexual reproduction while Mga2 and Mga3 have slight effect in sexual reproduction. However, most genes involved in conidial production, including the conidiospore formation genes brlA and wetA, had increased expression levels in the Gα mutants, except in Δmga1+3. Only the expression levels of velvet regulators were decreased in almost all the mutants. This is different from the phenotypic analysis (Figure 2) that the conidia-forming ability was reduced in mga1-related mutants (Δmga1, Δmga1+2, and Δmga1+3) but not in the other mutants.
Gα Genes Negatively Regulate MPs and CIT Biosynthesis
RNA-Seq results revealed that the expression levels of MPs biosynthetic genes (Chen et al., 2017), except MpigL, were increased in all the mutants at 7th day. However, these genes in Δmga1+2 and Δmga1+3 were up-regulated at 3rd and 7th day. In addition, in the mga1-deleted strains (Δmga1, Δmga1+2, and Δmga1+3) more genes were up-regulated than those in the other mutants. The DEGs involved in MPs biosynthesis are shown in Supplementary Figure S8. This result matches the increased MPs yields in Gα mutants (Figure 3) and indicates that Gα negatively regulates MPs production by regulating the MPs biosynthetic gene cluster.
According to the RNA-Seq results, genes in the CIT gene cluster (He and Cox, 2016) showed different trends on different days. Gα subunits mainly regulated the expression of the CIT gene cluster at 3rd day. Most genes in the CIT biosynthetic gene cluster were up-regulated in the single-deletion mutants and Δmga1+2, and CIT production also increased in these mutants (Figure 3). In Δmga1+3, although the pksCT gene was up-regulated, most other genes (MRR1–4 and MRR7–8) in the cluster were down-regulated, and the early stage (3–7 days) CIT production in the Δmga1+3 strain was lower than that in M. ruber M7. In Δmga2+3, only pksCT and MRL2 were up-regulated, and only in the later stage (9–11 days) the CIT production was greater than that in M. ruber M7 (Figure 3). Data on DEGs involved in CIT biosynthesis are provided in Supplementary Figure S9. This result indicates that Gα genes (mainly mga1 and mga2) negatively influenced CIT production by regulating the CIT biosynthetic gene cluster.
Conclusion and Discussion
G-protein signaling pathways play important roles in fungal reproduction and SMs production, and the functions of different Gα subunits (Gα1–3) have been analyzed in some fungi using single gene modification (Xu et al., 2015; Yoda et al., 2015; Zhang et al., 2016). The positively regulatory function of the Gα1 subunit on colony growth and asexual reproduction, which is conserved and extensive in most reported fungi, has been extensively researched (Li et al., 2010; Yang et al., 2012; Hu et al., 2013; Wasil et al., 2013; Garcia-Rico et al., 2017). However, until now, there has been no literature regarding double deletions combined with RNA-Seq of Gα subunit genes. In the current study, single- and double-gene(s) deletion mutants of the three Gα subunits were first systematically analyzed to determine the effects of different Gα subunits on M. ruber M7 according to the phenotypic characteristics combined with RNA-Seq analyses. The results show that all three Gα subunits (Mga1-3) in M. ruber M7 work together to regulate biological processes. Briefly, Mga1 comprehensively regulates the growth, development and secondary metabolism, while Mga2 and Mga3 act as supplementary regulators on growth and secondary metabolism. These findings not only illuminate the functions of different Gα subunits in M. ruber M7, but also provide a deeper understanding of the functional connections among different Gα subunits that involve regulating growth, development and secondary metabolism in other filamentous fungi.
Different Gα subunits (Gα1–3) regulate different biological processes in fungi (Li et al., 2010; Yang et al., 2012; Hu et al., 2013; Wasil et al., 2013; Garcia-Rico et al., 2017). For vegetative growth, Gα1 positively regulate the related processes in fungi such as Penicillium camembertii and Fusarium oxysporum (Guo et al., 2016b; Garcia-Rico et al., 2017), and Gα2 has no significant influence on fungal vegetative growth in Valsa mali and F. oxysporum (Guo et al., 2016a; Song et al., 2017), while Gα3 possesses different regulatory functions in different fungi. For example, PGA3 (Gα3) in P. camembertii and Gvm3 (Gα3) in V. mali positively regulate vegetative growth (Hu et al., 2013; Song et al., 2017), while FGA3 (Gα3) in F. oxysporum has no influence on vegetative growth (Guo et al., 2016b). In the current study, we find that Mga1(Gα1) has slightly effects on the vegetative growth of M. ruber M7, while Mga2 and Mga3 have no significant effects, which is similar to the results in P. camembertii and F. oxysporum (Guo et al., 2016b; Garcia-Rico et al., 2017), and the colony sizes of Δmga1+2 and Δmga1+3 are much smaller than those of M. ruber M7 and Δmga1, which suggests that the Mga2 and Mga3 subunits enhance this regulatory process when either is deleted along with Mga1. In addition, a group of MFS transporters involved in carbon source metabolism are more down-regulated in the Δmga1+2 and Δmga1+3 strains than those in M. ruber M7 according to RNA-Seq analyses (Supplementary Table S3), which implies that the transportation of carbon sources may be essential for Monascus growth and that Gα subunits may directly regulate MFS transporters to affect Monascus vegetative growth. Thus, further investigations of these transporters could contribute to determining the key elements involved in Monascus and other fungi vegetative growth.
Asexual reproduction, in many filamentous fungi, is mainly positively regulated by the sporogenesis central regulatory genes, including abaA, brlA and wetA (de Vries et al., 2017; Wu et al., 2018). However, in this study, the increased expression levels of brlA and wetA (no abaA in Monascus genome) in mga1-related mutants (Δmga1, Δmga1+2, and Δmga1+3) do not enhance conidial reproduction. This implies that a new asexual reproduction-related regulatory pathway might exist in M. ruber M7. Further studies on reproduction related regulatory pathways which we are doing, might find a new asexual reproduction regulatory pathway in Monascus spp.
The Gα regulation of SMs biosynthesis has been verified by single gene deletions, indicating that the negative regulation of Gα1 is conserved in most fungi (Yu et al., 2008; Guo et al., 2016a), and Gα2’s regulatory roles are diverse. For example, Gvm2 (Gα2) in V. mali negatively regulates SMs biosynthesis (Song et al., 2017), while GanA (Gα2) in Aspergillus nidulans has no influence on SMs biosynthesis (Yu, 2006). Additionally, Gα3 has no significant influence on SMs biosynthesis (Chang et al., 2004; Guo et al., 2016b). In our study, the single gene deletions have revealed that Mga1 (Gα1) and Mga2 (Gα2) can negatively regulate MPs and CIT production and that Mga3 (Gα3) has no significant effect. These results are similar to those of studies in V. mali and F. oxysporum (Guo et al., 2016b; Song et al., 2017). Moreover, double-gene deletions of Gα1–3 subunits can jointly regulate SMs. For instance, Mga2 and Mga3 combined with Mga1 can negatively regulate MPs production, since according to the phenotypic and transcriptomics analyses, Δmga1+2 and Δmga1+3 strains have much greater MPs yields (Figure 3) as well as greater numbers of up-regulated MPs biosynthesis-related DEGs compared with the other mutants (Supplementary Figure S8).
The RNA-Seq results (Supplementary Table S4) show that, besides MPs and CIT polyketide synthase (PKS) genes, many other PKS and non-ribosomal peptide synthetase genes are also regulated by Gα subunits. This is especially true of the Δmga1+3 strain in which nearly all the PKS and non-ribosomal peptide synthetase genes are differentially expressed. The analyses of related SMs in Δmga1+3 may help to improve our understanding of Monascus SMs.
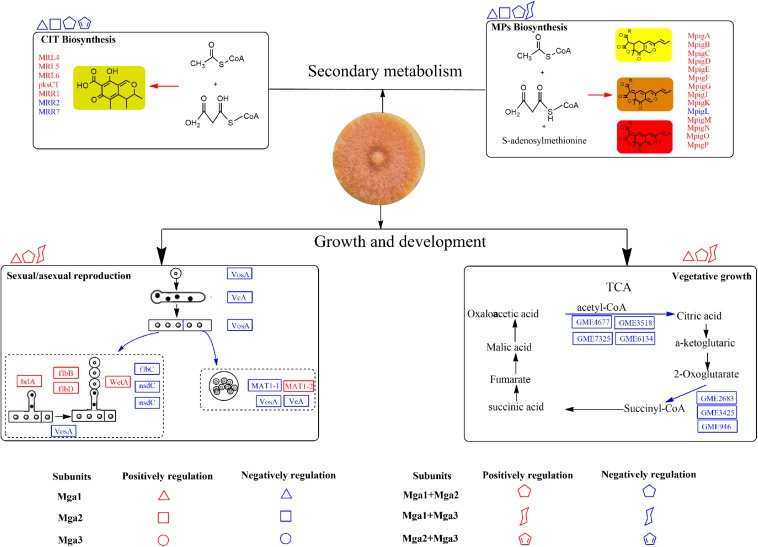
Based on the above findings, a Gα regulatory system in M. ruber M7 is proposed in Figure 4. First, vegetative growth is mainly positively regulated by Mga1, and Mga2 and Mga3 can improve this regulatory process when either is deleted along with Mga1. All the Gα subunits positively regulate carbon and nitrogen metabolism (Supplementary Table S3) to affect vegetative growth. Second, Mga1 contributes the most to the regulation of sexual/asexual reproduction compared with Mga2 and Mga3, and the regulation of asexual reproduction may occur prior to the central regulatory pathway. The regulation of sexual reproduction is reflected in the regulation of MAT1-2 gene, which is down-regulated in mga1-deleted strains (Supplementary Figure S7). Third, Gα subunits in M. ruber M7 negatively regulate the SMs. In detail, Mga1 and Mga2 can negatively regulate MPs and CIT production individually or jointly, while Mga3 may combine with Mga1 to only negatively regulate MPs yields.
FIGURE 4.
The possible gene regulatory network of Gα subunits in Monascus. The proteins and arrows marked in red indicate that they are up-regulated, the proteins and arrows marked in blue indicate that they are down-regulated.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
FC managed the project. ML, JL, YS, and YF conducted the transformants construction, secondary metabolites analysis, and transcriptome results analysis in this work. LL constructed the Δmga1 strain. ML conducted the phenotypic characterization, and interpreted the analysis results and wrote the manuscript. J-HY and YF contributed to the revision of the manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the Major Program of the National Natural Science Foundation of China (Nos. 31730068 and 31330059 to FC), the National Key Research and Development Program of China (No. 2018YFD0400404 to FC), the Program of Chengdu Science and Technology Bureau (2016-XT00-00033-GX to YF), and the Young Scientist Program of the National Natural Science Foundation of China (No. 31701583 to JL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01555/full#supplementary-material
Deletion strategy and confirmation of Δmga2 and Δmga3 mutants. (A) The strategy to construct the Δmga2 strain and mga2 complementary strain (Rmga2). (B) The strategy to construct the Δmga3 strain and mga3 complementary strain (Rmga3). (C) PCR identification of Δmga2. PCR products of M7, Δmga2, and Rmga2 with different primers; M, Trans 2K plus II marker. (D) PCR identification of Δmga3. PCR products of M7, Δmga3, and Rmga3 with different primers; M, Trans 2K plus II marker. (E) Southern hybridization analysis. SacI and XhoI are used in double-digesting genomic DNA, M: λDNA/HindIII marker. Probe 1, mga2 ORF; Probe 2, hph gene; Probe 3, mga3 ORF; Probe 4, neo gene.
Deletion strategy and confirmation of double-deletion strains. (A) The strategy to construct the Δmga1+2 strain. (B) The strategy to construct the Δmga1+3 and Δmga2+3 strains. (C) PCR identification of double-deletion strains; M, Trans 2K plus II marker. (D) Southern hybridization analysis. KpnI is used in digesting genomic DNA; M, λDNA/HindIII marker; Probe 1, mga2 ORF; Probe 3, mga3 ORF; Probe 4, neo gene.
Colony morphologies and microscopic structures of Rmga2 (Δmga2::mga2) and Rmga3 (Δmga3::mga3) strains. (A) Colony morphologies of M7, Δmga2 strain and Rmga2 mutants observed on PDA, MA, CYA, and G25N plates and cultured at 28°C for 12 days. (B) Cl and Co morphologies among M7, Δmga2 strain and Rmga2 mutants were observed on PDA, MA, CYA, and G25N plates cultured at 28°C for 5 days. The enlarged areas are indicated by arrows. Size bar = 100 μm. (C) Colony morphologies of M7, Δmga3 strain and Rmga3 mutants observed on PDA, MA, CYA, and G25N plates and cultured at 28°C for 12 days. (D) Cl and Co morphologies among M7, Δmga3 strain and Rmga3 mutants were observed on PDA, MA, CYA, and G25N plates cultured at 28°C for 5 days. The enlarged areas are indicated by arrows. Size bar = 50 μm.
Gene expression levels analyzed by RNA-Seq and qRT-PCR. The x-axis represents the selected 11 genes; the y-axis on the left side represents the gene expression levels as assessed by RNA-Seq; the y-axis on the right side represents the relative gene expression level as assessed by qRT-PCR.
The influence of Gα mutants on the TCA cycle. Blue indicates decreased enzyme expression; red indicates increased enzyme expression.
The DEGs involved in the nitrogen metabolism of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the sporulation of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the MP biosynthesis of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the CIT biosynthesis of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates regulated in the Gα mutants.
References
- Alberti F., Foster G. D., Bailey A. M. (2017). Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Bio. 101 493–500. 10.1007/s00253-016-8034-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Blanc P. J., Laussac J. P., Le Bars J., Le Bars P., Loret M. O., Pareilleux A., et al. (1995). Characterization of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 27 201–213. 10.1016/0168-1605(94)00167-5 [DOI] [PubMed] [Google Scholar]
- Blin K., Weber T., Kim H. U., Lee S. Y., Medema M. H., Duddela S., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43 W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. H., Chae K. S., Han D. M., Jahng K. Y. (2004). The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 167:1305. 10.1534/genetics.103.025379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Hu X. (2005). Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int. J. Food Microbiol. 103 331–337. 10.1016/j.ijfoodmicro.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Chen W., Chen R., Liu Q., He Y., He K., Ding X., et al. (2017). Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 8 4917–4925. 10.1039/c7sc00475c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., He Y., Zhou Y., Shao Y., Feng Y., Li M., et al. (2015). Edible filamentous fungi from the speciesmonascus: rarly traditional fermentations, modern molecular biology, and future genomics. Comp. Rev. Food Sci. F. 14 555–567. 10.1111/1541-4337.12145 [DOI] [Google Scholar]
- Chen Y. P., Tseng C. P., Liaw L. L., Wang C. L., Chen I. C., Wu W. J., et al. (2008). Cloning and characterization of monacolin K biosynthetic gene cluster from Monascus pilosus. Food Chem. 56 5639–5646. 10.1021/jf800595k [DOI] [PubMed] [Google Scholar]
- Corrochano L. M., Kuo A., Marcet-Houben M., Polaino S., Salamov A., Villalobos-Escobedo J. M., et al. (2016). Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 26 1577–1584. 10.1016/j.cub.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P., Riley R., Wiebenga A., Aguilar-Osorio G., Amillis S., Uchima C. A., et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18:28. 10.1186/s13059-017-1151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa G., Catena G., Raddino R., Gaudio G., Maggi A., D’Angelo A., et al. (2018). Effects on oral fat load of a nutraceutical combination of fermented red rice, sterol esters and stanols, curcumin, and olive polyphenols: a randomized, placebo controlled trial. Phytomedicine 42 75–82. 10.1016/j.phymed.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Diana M., Quílez J., Rafecas M. (2014). Gamma-aminobutyric acid as a bioactive compound in foods: a review. J. Funct. Foods 10 407–420. 10.1016/j.jff.2014.07.004 [DOI] [Google Scholar]
- Feng Y., Shao Y., Chen F. (2012). Monascus pigments. Appl. Microbiol. Biot. 96 1421–1440. 10.1007/s00253-012-4504-3 [DOI] [PubMed] [Google Scholar]
- Fox E. M., Howlett B. J. (2008). Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11 481–487. 10.1016/j.mib.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Garcia-Rico R. O., Gil-Duran C., Rojas-Aedo J. F., Vaca I., Figueroa L., Levican G., et al. (2017). Heterotrimeric G protein alpha subunit controls growth, stress response, extracellular protease activity, and cyclopiazonic acid production in Penicillium camemberti. Fungal Biol. 121 754–762. 10.1016/j.funbio.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Guo L., Yang L., Liang C., Wang J., Liu L., Huang J. (2016a). The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 93 29–38. 10.1016/j.pmpp.2015.12.003 [DOI] [Google Scholar]
- Guo L., Yang Y., Yang L., Wang F., Wang G., Huang J. (2016b). Functional analysis of the G-protein α subunits FGA1 and FGA3 in the banana pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 94 75–82. 10.1016/j.pmpp.2016.04.003 [DOI] [Google Scholar]
- He Y., Cox R. J. (2016). The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 7 2119–2127. 10.1039/c5sc04027b [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Liu Q., Shao Y., Chen F. (2013). Ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M 7. Appl. Microbiol. Biotechnol. 97 4965–4976. 10.1007/s00253-013-4851-8 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu G., Li Z., Qin Y., Qu Y., Song X. (2013). G protein-cAMP signaling pathway mediated by PGA3 plays different roles in regulating the expressions of amylases and cellulases in Penicillium decumbens. Fungal Genet. Biol. 5 62–70. 10.1016/j.fgb.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Huang C. H., Shiu S. M., Wu M. T., Chen W. L., Wang S. G., Lee H. M. (2013). Monacolin K affects lipid metabolism through SIRT1/AMPK pathway in HepG2 cells. Arch. Pharm.Res. 36 1541–1551. 10.1007/s12272-013-0150-2 [DOI] [PubMed] [Google Scholar]
- Jia X. Q., Xu Z. N., Zhou L. P., Sung C. K. (2010). Elimination of the mycotoxin citrinin production in the industrial important strain Monascus purpureus SM001. Metab. Eng. 12 1–7. 10.1016/j.ymben.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Kamerewerd J., Jansson M., Nowrousian M., Poggeler S., Kuck U. (2008). Three alpha-subunits of heterotrimeric G proteins and an adenylyl cyclase have distinct roles in fruiting body development in the homothallic fungus Sordaria macrospora. Genetics 180 191–206. 10.1534/genetics.108.091603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12:357. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C., Lee G. D., Choi I. (2016). Breast meat quality of broilers fed fermented red ginseng marc powder mixed with red-koji during storage. Emir. J. Food Agric. 28 283–287. [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., He L., Lai Y., Shao Y., Chen F. (2014). Cloning and functional analysis of the Gbeta gene Mgb1 and the Ggamma gene Mgg1 in Monascus ruber. J. Microbiol. 52 35–43. 10.1007/s12275-014-3072-x [DOI] [PubMed] [Google Scholar]
- Li L., Shao Y., Li Q., Yang S., Chen F. (2010). Identification of Mga1, a G-protein alpha-subunit gene involved in regulating citrinin and pigment production in Monascus ruber M 7. FEMS Microbiol. Lett. 308 108–114. 10.1111/j.1574-6968.2010.01992.x [DOI] [PubMed] [Google Scholar]
- Li L., Wright S. J., Krystofova S., Park G., Borkovich K. A. (2007). Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61 423–452. 10.1146/annurev.micro.61.080706.093432 [DOI] [PubMed] [Google Scholar]
- Li Y. P., Pan Y. F., Zou L. H., Xu Y., Huang Z. B., He Q. H. (2013). Lower citrinin production by gene disruption of ctnB involved in citrinin biosynthesis in Monascus aurantiacus Li AS3.4384. J. Agric. Food Chem. 61 7397–7402. 10.1021/jf400879s [DOI] [PubMed] [Google Scholar]
- Li Y. P., Tang X., Wu W., Xu Y., Huang Z. B., He Q. H. (2015). The ctnG gene encodes carbonic anhydrase involved in mycotoxin citrinin biosynthesis from Monascus aurantiacus. Food Add. Contam. Part A 32 577–583. 10.1080/19440049.2014.990993 [DOI] [PubMed] [Google Scholar]
- Lin H., Lyu H., Zhou S., Yu J., Keller N. P., Chen L., et al. (2018). Deletion of a global regulator LaeB leads to the discovery of novel polyketides in Aspergillus nidulans. Org. Biomol. Chem. 16 4973–4976. 10.1039/C8OB01326H [DOI] [PubMed] [Google Scholar]
- Liu Q., Xie N., He Y., Wang L., Shao Y., Zhao H., et al. (2014). MpigE, a gene involved in pigment biosynthesis in Monascus ruber M 7. Appl. Microbiol. Biotechnol. 98 285–296. 10.1007/s00253-013-5289-8 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhou Y., Yi T., Zhao M., Xie N., Lei M., et al. (2016). Identification and role analysis of an intermediate produced by a polygenic mutant of Monascus pigments cluster in Monascus ruber M 7. Appl. Microbiol. Biotechnol. 100 7037–7049. 10.1007/s00253-016-7397-8 [DOI] [PubMed] [Google Scholar]
- Liu Q., Cai L., Shao Y., Zhou Y., Li M., Wang X., et al. (2016). Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 120 297–305. 10.1016/j.funbio.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Liu Y., Yang K., Qin Q., Lin G., Hu T., Xu Z., et al. (2018). G protein α subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 10:117. 10.3390/toxins10030117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Li L., Wu Z., Yang J., Yi X., Zhang W. (2018). Development of new red mold rice and determination of their properties. LWT 87 259–265. 10.1016/j.lwt.2017.08.088 [DOI] [Google Scholar]
- Moretti M., Wang L., Grognet P., Lanver D., Link H., Kahmann R. (2017). Three regulators of G protein signaling differentially affect mating, morphology and virulence in the smut fungus Ustilago maydis. Mol. Microbiol. 105 901–921. 10.1111/mmi.13745 [DOI] [PubMed] [Google Scholar]
- Patakova P. (2013). Monascus secondary metabolites: production and biological activity. J. Ind. Microbiol. Biotechnol. 40 169–181. 10.1007/s10295-012-1216-8 [DOI] [PubMed] [Google Scholar]
- Rahayu Y. Y. S., Yoshizaki Y., Yamaguchi K., Okutsu K., Futagami T., Tamaki H., et al. (2017). Key volatile compounds in red koji-shochu, a Monascus-fermented product, and their formation steps during fermentation. Food Chem. 224 398–406. 10.1016/j.foodchem.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Schwecke T., Aparicio J. F., Molnár I., König A., Khaw L. E., Haydock S. F., et al. (1995). The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci U.S.A. 92:7839. 10.1073/pnas.92.17.7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. A., Yu J. H. (2006). The phosducin-like protein PhnA is required for Gβγ-mediated signaling for vegetative growth, developmental control, and toxin biosynthesis in Aspergillus nidulans. Eukaryot. Cell 5 400–410. 10.1128/EC.5.2.400-410.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N., Dai Q., Zhu B., Wu Y., Xu M., Voegele R. T., et al. (2017). Gα proteins Gvm2 and Gvm3 regulate vegetative growth, asexual development, and pathogenicityon apple in Valsa mali. PLoS One 12:e0173141. 10.1371/journal.pone.0173141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100 9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen J., Verbeek-de Kruif N., Govers F. (2018). The G-protein γ subunit of Phytophthora infestans is involved in sporangial development. Fungal Genet. Biol. 116 73–82. 10.1016/j.fgb.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Varga J., Szigeti G., Baranyi N., Kocsubé S., O’Gorman C. M., Dyer P. S. (2014). Aspergillus: sex and recombination. Mycopathologia 178 349–362. 10.1007/s11046-014-9795-8 [DOI] [PubMed] [Google Scholar]
- Wasil Z., Pahirulzaman K. A. K., Butts C., Simpson T. J., Lazarus C. M., Cox R. J. (2013). One pathway, many compounds: heterologous expression of a fungal biosynthetic pathway reveals its intrinsic potential for diversity. Chem. Sci. 4:3845 10.1039/c3sc51785c [DOI] [Google Scholar]
- Wei W., Lin S., Chen M., Liu T., Wang A., Li J., et al. (2017). Monascustin, an unusual γ-lactam from red yeast rice. J. Nat. Prod. 80 201–204. 10.1021/acs.jnatprod.6b00493 [DOI] [PubMed] [Google Scholar]
- Wu M.-Y., Mead M. E., Lee M.-K., Ostrem Loss E. M., Kim S.-C., Rokas A., et al. (2018). Systematic dissection of the evolutionarily conserved wetA developmental regulator across a genus of filamentous fungi. mBio 9:e1130-18. 10.1128/mBio.01130-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D. B., Chen M., Ma Y. N., Xu Z. S., Li L. C., Chen Y. F., et al. (2015). A G-protein beta subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS One 10:e0116385. 10.1371/journal.pone.0116385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li L., Li X., Shao Y., Chen F. (2012). mrflbA, encoding a putative FlbA, is involved in aerial hyphal development and secondary metabolite production in Monascus ruber M-7. Fungal Biol. 116 225–233. 10.1016/j.funbio.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Yoda A., Adelmant G., Tamburini J., Chapuy B., Shindoh N., Yoda Y., et al. (2015). Mutations in G protein beta subunits promote transformation and kinase inhibitor resistance. Nat. Med. 21 71–75. 10.1038/nm.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Y., Seo J. A., Kim J. E., Han K. H., Shim W. B., Yun S. H., et al. (2008). Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae. Microbiology 154 392–401. 10.1099/mic.0.2007/012260-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-H. (2006). Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44 145–154. 10.1021/la803176d [DOI] [PubMed] [Google Scholar]
- Yu X., Wu H., Zhang J. (2015). Effect of Monascus as a nitrite substitute on color, lipid oxidation, and proteolysis of fermented meat mince. Food Sci. Biotechnol. 24 575–581. 10.1007/s10068-015-0075-2 [DOI] [Google Scholar]
- Zhang X., Zhai C., Hua C., Qiu M., Hao Y., Nie P., et al. (2016). PsHint1, associated with the G-protein alpha subunit PsGPA1, is required for the chemotaxis and pathogenicity of Phytophthora sojae. Mol. Plant Pathol. 17 272–285. 10.1111/mpp.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deletion strategy and confirmation of Δmga2 and Δmga3 mutants. (A) The strategy to construct the Δmga2 strain and mga2 complementary strain (Rmga2). (B) The strategy to construct the Δmga3 strain and mga3 complementary strain (Rmga3). (C) PCR identification of Δmga2. PCR products of M7, Δmga2, and Rmga2 with different primers; M, Trans 2K plus II marker. (D) PCR identification of Δmga3. PCR products of M7, Δmga3, and Rmga3 with different primers; M, Trans 2K plus II marker. (E) Southern hybridization analysis. SacI and XhoI are used in double-digesting genomic DNA, M: λDNA/HindIII marker. Probe 1, mga2 ORF; Probe 2, hph gene; Probe 3, mga3 ORF; Probe 4, neo gene.
Deletion strategy and confirmation of double-deletion strains. (A) The strategy to construct the Δmga1+2 strain. (B) The strategy to construct the Δmga1+3 and Δmga2+3 strains. (C) PCR identification of double-deletion strains; M, Trans 2K plus II marker. (D) Southern hybridization analysis. KpnI is used in digesting genomic DNA; M, λDNA/HindIII marker; Probe 1, mga2 ORF; Probe 3, mga3 ORF; Probe 4, neo gene.
Colony morphologies and microscopic structures of Rmga2 (Δmga2::mga2) and Rmga3 (Δmga3::mga3) strains. (A) Colony morphologies of M7, Δmga2 strain and Rmga2 mutants observed on PDA, MA, CYA, and G25N plates and cultured at 28°C for 12 days. (B) Cl and Co morphologies among M7, Δmga2 strain and Rmga2 mutants were observed on PDA, MA, CYA, and G25N plates cultured at 28°C for 5 days. The enlarged areas are indicated by arrows. Size bar = 100 μm. (C) Colony morphologies of M7, Δmga3 strain and Rmga3 mutants observed on PDA, MA, CYA, and G25N plates and cultured at 28°C for 12 days. (D) Cl and Co morphologies among M7, Δmga3 strain and Rmga3 mutants were observed on PDA, MA, CYA, and G25N plates cultured at 28°C for 5 days. The enlarged areas are indicated by arrows. Size bar = 50 μm.
Gene expression levels analyzed by RNA-Seq and qRT-PCR. The x-axis represents the selected 11 genes; the y-axis on the left side represents the gene expression levels as assessed by RNA-Seq; the y-axis on the right side represents the relative gene expression level as assessed by qRT-PCR.
The influence of Gα mutants on the TCA cycle. Blue indicates decreased enzyme expression; red indicates increased enzyme expression.
The DEGs involved in the nitrogen metabolism of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the sporulation of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the MP biosynthesis of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates not regulated in the Gα mutants.
The DEGs involved in the CIT biosynthesis of Gα mutants. Blue indicates down-regulation in the Gα mutants; red indicates up-regulation in the Gα mutants; gray indicates regulated in the Gα mutants.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.