Abstract
Scribble (Scrib) is a key regulator of apicobasal polarity, presynaptic architecture, and short-term synaptic plasticity in Drosophila. In mammals, its homolog Scrib1 has been implicated in cancer, neural tube closure, and planar cell polarity (PCP), but its specific role in the developing and adult nervous system is unclear. Here, we used the circletail mutant, a mouse model for PCP defects, to show that Scrib1 is located in spines where it influences actin cytoskeleton and spine morphing. In the hippocampus of these mutants, we observed an increased synapse pruning associated with an increased number of enlarged spines and postsynaptic density, and a decreased number of perforated synapses. This phenotype was associated with a mislocalization of the signaling pathway downstream of Scrib1, leading to an overall activation of Rac1 and defects in actin dynamic reorganization. Finally, Scrib1-deficient mice exhibit enhanced learning and memory abilities and impaired social behavior, two features relevant to autistic spectrum disorders. Our data identify Scrib1 as a crucial regulator of brain development and spine morphology, and suggest that Scrib1crc/+ mice might be a model for studying synaptic dysfunction and human psychiatric disorders.
Introduction
At excitatory synapses, the postsynaptic compartment often consists of the head of a dendritic spine contacting a presynaptic bouton. A strong relationship exists among the structure, maintenance, and function of these structures. As a result, a broad variety of psychiatric and neurologic disorders are accompanied by patterns of spine disruption, and activity-dependent spine remodeling is believed to underlie learning and memory (Fiala et al., 2002; Kasai et al., 2003; Sala et al., 2008). Consequently, structural and/or functional changes at the level of dendritic spines will be sensed by, and affect, all the local neuronal networks, and ultimately the entire brain.
Synaptic components within spines are highly organized in the postsynaptic density (PSD) by scaffolding proteins. Many of these proteins belong to families that contain PSD-95/Discs-large/ZO-1 (PDZ) domains (Sheng and Hoogenraad, 2007), and have been implicated in trafficking and anchoring of synaptic proteins (Wenthold et al., 2003; Elias and Nicoll, 2007; Lau and Zukin, 2007; Shepherd and Huganir, 2007). Recently, Scribble1 (Scrib1) was shown to interact with several components of PSD via its different interacting domains [leucine-rich repeat (LRR) and PDZ]. Thus, Scrib1 might serve as a scaffold protein for at least two synaptic complexes: the cadherin/catenin complex, and the βPIX/GIT1 complex (Audebert et al., 2004; Navarro et al., 2005; Qin et al., 2005; Sun et al., 2009; Richier et al., 2010). The PDZ domains of Scrib1 bind to βPIX (Audebert et al., 2004), a guanine nucleotide exchange factor (GEF) for Rac and Cdc42 that is tightly associated with p21-activated kinase (PAK) and with GIT1 (Audebert et al., 2004). The Scrib1/βPIX/GIT1 protein complex is implicated in vesicle trafficking in neurons and receptor recycling in thyroid cells (Lahuna et al., 2005), and the Scrib1/βPIX/PAK protein complex is involved in cell migration (Nola et al., 2008). The GIT1/βPIX/Rac/PAK signaling complex may have a role in spine morphogenesis and synapse formation through myosin II regulatory light chain (Zhang et al., 2005) and through a Ca2+ signaling pathway downstream of NMDA receptors (NMDARs) (Saneyoshi et al., 2008).
In this study, we use a spontaneous mutant for Scrib1, the circletail mouse (Scrib1crc) to examine Scrib1 function in hippocampus. We show that Scrib1 is expressed in the soma and dendrites of adult hippocampal pyramidal cells, as well as at the PSD, and that Scrib1 protein levels are reduced by ∼50% in heterozygote mutant mice (Scrib1crc/+). We find a weakened synaptic strength and plasticity in the hippocampus of young Scrib1crc/+ mice, suggesting a delay in synaptic maturation and postsynaptic deficits associated with reduced levels of Scrib1, likely due in part to a disruption of actin cytoskeleton. Analysis of the behavior of these animals shows an increased hippocampal-dependent learning ability in Scrib1crc/+ animals versus wild types (WTs) but an impairment of social interactions. These observations were correlated with altered neuronal morphology in Scrib1crc/+ pyramidal neurons, including an increase in dendritic arborization, a decrease in spine number, and changes in PSD morphology.
Materials and Methods
Circletail animals
The circletail mice were maintained under standard conditions as described previously (Montcouquiol et al., 2003; 2006a). Adult WT and heterozygous (Scrib1crc/+) male mice were housed individually (for behavioral experiments) or collectively in groups of five to eight (for biochemical experiments) in polypropylene cages under controlled conditions (at 23 ± 1°C, with lights on from 8:00 A.M. to 8:00 P.M.). Food and water were available ad libitum. This study was performed in full accordance with the European Communities Council Directives (86/609/EEC) and the French national Committee (87/848) recommendations.
Embryo analysis
Pregnant females were killed by decapitation, and embryos, at stage 18.5 (embryonic day 12.5), were dissected from the uterus in HBSS and washed in PBS. Embryos and brains were photographed using a stereomicroscope and a color camera (Olympus).
cDNA constructs
Scrib1 cDNA samples were generously provided by I. Macara (University of Virginia, Charlottesville, VA) [green fluorescent protein (GFP)-tagged], J. P. Borg (Centre de Recherche en Cancérologie de Marseille, Marseille, France) (HA-tagged), and P. O. Humbert (Peter MacCallum Cancer Centre, East Melbourne, Australia) (CMV-IRES-Scrib1). Stop codons were introduced by direct mutagenesis to generate Scrib1 mutants lacking all the PDZ domains (ScribΔPDZ) or only the last two to generate the crc form of Scrib1 (Scrib1crc). PDZ1-4 were amplified by PCR and subcloned in the pEGFP2 vector from Clontech. The GFP–actin construct was obtained from Clontech, and C. Gauthier-Rouvière (Centre de Recherche en Biochimie Macromoléculaire, Centre National de la Recherche Scientifique Montpellier, France) generously provided the red fluorescent protein (RFP)–actin construct. The SureSilencing short hairpin RNA (shRNA) plasmids were purchase from Tebu-Bio.
In situ hybridization analysis
In situ hybridization was performed using digoxigenin (Dig)-labeled cRNA probes. Transcription was performed from 1 μg of linearized plasmid using Dig-UTP (Roche Diagnostics). Brain sections were postfixed in 4% paraformaldehyde/0.2% glutaraldehyde for 15 min at room temperature (RT), rinsed in PBS, digested in Proteinase K (4 μg/ml in PBS) for 7 min, rinsed, and postfixed in 4% paraformaldehyde/0.2% glutaraldehyde for 15 min at RT. After two washes in PBS, slides were acetylated into freshly prepared 0.1 m triethanolamine/4× SSC, pH 8, for 10 min at RT; 0.25% acetic anhydride acid was added for the last 5 min. Sections were subsequently rinsed twice in PBS, incubated for 2 h at RT in hybridization buffer (50% formamide, 10% dextran sulfate, 20 mm Tris-HCl, 1 mm EDTA, 300 mm NaCl, 1× Denhardt's solution), and hybridized overnight at 60°C with antisense probe in 150 μl of hybridization buffer. The slides were washed in 4× SSC/0.05% Tween20, then in 2× SSC/50% formamide/0.05% Tween20 and placed in 1× SSC/50% formamide/0.05% Tween20 (30 min at 62°C) before being incubated in TBS containing 0.1 m Tris-HCl, pH 9.5, 0.01% Tween20 (TBST) at 30°C (15 min, three times). The slides were saturated with blocking buffer (2% blocking reagent, 20% sheep serum in PBS) and incubated for 5 h at RT with an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Diagnostics) diluted in blocking buffer. After washing in TBST and NTMT (0.1 m NaCl; 0.1 m Tris-HCl, pH 9.5; 0.05 m MgCl2; 0.01% Tween20) three times, the hybridized probes were detected in NTMT solution containing nitro blue tetrazolium and 5-bromo-6-chloro-3-indolyl phosphate (Invitrogen) at RT. The reaction was stopped by rinsing in PBS/1 mm EDTA.
Antibodies
Antibodies used were as follows: anti-Scrib1 antibody (AbMM468) (Montcouquiol et al., 2006b); anti-Scrib1 (C20 and K21), anti-HA (clone Y11) and anti-α-PAK (C19) from Santa Cruz Biotechnology; anti-βPIX (AB3829), anti-synaptophysin (MAB5258), and anti-GAPDH (MAB374) from Millipore; anti-Myc (MMS-150P) and anti-HA (MMS-101P) from Covance; anti-DsRed from Clontech; anti-CaMKII (C265) and anti-Rac1 (R2650) from Sigma-Aldrich; anti-actin (AAN01) from Cytoskeleton; anti-β-catenin (610154), anti-PSD-95 (610495), and anti-N-Cadherin (N-Cad. C70320) from BD Biosciences; anti-GFP (A-11120) from Invitrogen; anti-GIT1 (N39B/8) and anti-Shank (clone N23B/49) from NeuroMab; anti-Erbin, a gift from J. P. Borg (Centre de Recherche en Cancérologie de Marseille, Marseille, France); anti-Lgl1, a gift from V. Vasioukhin (Fred Hutchinson Cancer Research Center, Seattle, WA); and anti-SAP102, a gift from J. Hell (University of California, Davis, Davis, CA). Secondary antibodies used were as follows: anti-rabbit and anti-mouse HRP-conjugated from GE Healthcare; anti-goat HRP-conjugated, anti-mouse or anti-rabbit biotin-conjugated from Vector Laboratories; IRDye (800CW and 680) from LI-COR Biosciences; and Alexa-conjugated antibodies and phalloidin from Invitrogen.
Cell culture, transfection, and immunofluorescence of COS cells
COS-7 cells used for transient transfections were maintained in DMEM, supplemented with 10% (v/v) fetal calf serum (Invitrogen), 2 mm glutamine, and penicillin/streptomycin (50 U/ml), and were kept at 37°C in 5% CO2. Cells were transfected using calcium phosphate and were analyzed 36–48 h after transfection, and immunofluorescence was performed as in Sans et al. (2005).
Hippocampal cultures
Neurons were cultured as described by Sans et al. (2003, 2005) and transfected with the different constructs between 10 and 14 days in vitro (DIV) using the calcium phosphate method. Fifteen micrograms of DNA were mixed with 250 mm CaCl2 and added to the same volume of 2× HBS. The DNA mix was incubated for 20 min and then added to the neurons for 45 min. Neurons were washed with DMEM and cultured in the original medium at 37°C, 5% CO2. After 2–6 d, neurons were fixed with paraformaldehyde, washed, and blocked with NGS before incubating them with primary antibodies at room temperature in 3% normal goat serum (NGS)/PBS, then washed in PBS, and incubated with secondary antibodies in NGS/PBS. All images were acquired on a Leica SP2 laser scanning confocal microscope, and analysis was performed as described by Sans et al. (2005).
Histology and immunohistochemistry
Hematoxylin staining
Brains of embryos were dissected out and postfixed in Carnoy's fixative (Electron Microscopy Sciences) overnight. After inclusion in paraffin, 10 μm coronal sections were prepared using a microtome, deparaffinized, and stained with hematoxylin.
Immunohistochemistry
After terminal anesthesia was induced by brief inhalation of isoflurane (0.05% in air), followed by an intramuscular injection of ketamine (100 mg/kg) and xylazine (10 mg/kg), mice were perfused transcardially with 4% paraformaldehyde (PFA) in 0.1 m PBS, pH 7.4. Brains were fixed and cut into serial sections using a vibratome (40 μm). Sections were processed as described by Driver et al. (2007). Sections were collected in PBS and then incubated in 50 mm ammonium chloride in PBS for 20 min at RT. After extensive washing in PBS, antigen retrieval was performed by incubation of the sections in 10 mm sodium citrate, pH 8.4, for 1 h at 80°C. Sections were once more extensively washed with PBS before being blocked in 0.1% gelatin and 0.1% Triton X-100 in PBS. Sections were incubated with rabbit antibodies to Scrib1 (MM468; 1:100) in the blocking solution at 4°C for 48 h. After rinses, the sections were incubated with biotinylated goat anti-rabbit IgG (heavy and light chain) (1:200, 2 h at RT) (Vector Laboratories) in PBS, followed by ABC Elite Kit (1:200; 2 h at RT) (Vector Laboratories). The peroxidase reaction was revealed by incubating the sections in VIP substrate Kit or DAB substrate (Vector Laboratories). Sections were mounted on glass slides, dehydrated, and coverslipped with VectaMount (Vector Laboratories) before being examined with a standard light microscope. Control experiments, in which the primary antibody was omitted, gave no signal.
For structural analysis of CA1 hippocampal synapses at the electron microscopic level, 10-week-old mice were used for each genotype. Terminal anesthesia was induced by brief inhalation of isoflurane (0.05% in air), followed by an intramuscular injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). Then mice were perfused transcardially with 4% PFA and 2% glutaraldehyde in PBS. Vibratome sections (200 μm thick) were collected in 0.1 m PB buffer, pH 7.2, postfixed in 1% osmium tetroxide, dehydrated in ethanol, infiltrated, and flat embedded in epoxy as previously described (Rubio, 2006). CA1 hippocampi were trimmed and mounted on blocks for further semithin and ultrathin sectioning with a Leica Ultracut. Electron micrographs were analyzed with a TECNAI G2 Spirit Biotwin TEM (FEI) or a Philips CM100 transmission electron microscope (FEI) at 100 kV. The images were captured with an AMT XR40 4 megapixel side-mounted CCD camera at 68,000× magnification. Only identified synapses on dendritic spines of apical dendrites of pyramidal cells were included in the analysis. No tangentially cut or perforated synapses were analyzed. To determine the area of spines, we traced the plasma membrane with ImageJ (http://rsb.info.nih.gov/ij) (Sfakianos et al., 2007). The average thickness of the PSDs was measured as described previously (Rubio, 2006). Briefly, the cytoplasmic outline of a PSD, including the associated dense material, was traced, and this area was then enclosed by tracing the postsynaptic membrane (length of PSD). The area was then divided by the length of the postsynaptic membrane to derive an average thickness for the PSD. The results in WT and Scrib1crc/+ animals are presented as the mean ± SEM. The measurements were all performed by experimenters blind to the genotype.
Pre-embedding immunocytochemistry
After terminal anesthesia was induced by brief inhalation of isoflurane (0.05% in air), followed by an intramuscular injection of ketamine (100 mg/kg) and xylazine (10 mg/kg), mice were perfused with 4% PFA and 0.1% glutaraldehyde in PBS, and vibratome sections (100 μm) were collected in PBS and then incubated in 50 mm ammonium chloride in PBS for 20 min at RT. After extensive washing in PBS, antigen retrieval was performed as above. Sections were once more extensively washed with PBS before being blocked in 0.1% (w/v) gelatin in PBS. Sections were incubated with rabbit antibodies to Scrib1 (MM468, 1:100) in 0.1% (w/v) gelatin, 0.05% Triton X-100 in PBS at 4°C for 48–72 h. Sections were rinsed in PBS, postfixed for 5 min in 4% PFA in PBS, rinsed again, and incubated with biotin-conjugated goat anti-rabbit secondary antibody (1:200) for 12 h in 0.8% bovine serum albumin and 0.2% fish gelatin (Sigma-Aldrich) in PBS at 4°C. The following day, sections were rinsed in PBS, then postfixed in 1% glutaraldehyde in PBS (5 min), and rinsed in PBS followed by ABC Elite Kit (1:200; 2 h at RT) (Vector Laboratories). The peroxidase reaction was revealed by incubating the sections in VIP substrate Kit (Vector Laboratories). After PBS rinses, sections were osmicated, dehydrated, and flat embedded in Durcupan resin (Sigma-Aldrich). Ultrathin sections (70–90 nm) were countercolored with uranyl acetate and lead citrate, and visualized with a Philips CM100 transmission electron microscope (FEI) at 100 kV. The images were captured with an AMT XR40 4 megapixel side mounted CCD camera at a magnification between 7,900 and 92,000×.
Golgi Cox staining and spine density analysis
Animals were perfused transcardially with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 m PBS, pH 7.4. Vibratome sections for Golgi impregnation (70 μm) were treated with 1% osmium tetroxide in PB for 30 min. They were then placed in 3.5% potassium dichromate overnight, followed by 6 h in 2% silver nitrate solution. The sections were finally dehydrated in graded alcohols, infiltrated in epoxy resin, mounted, and coverslipped on glass slides. The Golgi-impregnated pyramidal neurons of the dorsal hippocampus were identified and analyzed. At least 5 neurons in each hemisphere (10–20 neurons per animal) were drawn using camera lucida at a magnification of 100× (DMLS Microscope, Leica). For each neuron, the dendritic tree, including all branches, was reconstructed and the dendritic tracing was quantified using Neurolucida software (MBF). The dendritic surface was quantified by counting the number of branches at each order from the cell body by Sholl analysis and by counting the number of ring intersections using an overlay of concentric rings (10 μm between rings). For quantification of spine density and morphology, 20–100 μm dendritic segments were identified, and protrusions were measured as described previously (WT, n = 526; Scrib1crc/+, n = 514) (Sans et al., 2005).
Biochemistry
Tissue extracts, PSD preparation and immunoblot detection, and glutathione S-transferase (GST) binding assay experiments were performed as described previously (Sans et al., 2000, 2003). For Rac assays, the GST pull-down was performed as previously described by Nola et al. (2008).
The amount of F-actin and G-actin was measured with an actin polymerization assay kit (BK037, Cytoskeleton). To preserve F-actin, experiments were performed at 37°C. Membranes were resuspended in F-actin stabilization buffer with ATP and protease inhibitors, then separated in aliquots and stimulated or not with 20 mm KCl for 60 s, and stopped with incubation on ice for 12 min. Then, F-actin was pelleted by 1 h centrifugation at 100,000 × g and the supernatant (G-actin) separated. The pellet was suspended to the same volume as the supernatant using ice-cold Milli-Q water and 10 μm cytochalasine D (F-depolymerizing solution) for 2 h at 4°C. Actin was quantified by Western blot using an anti-actin antibody (Cytoskeleton). The F-actin/G-actin ratio was determined using the Odyssey infrared imaging system (LI-COR Biosciences).
Whole-cell patch-clamp and extracellular field recordings
Hippocampal slices (300 μm) were prepared from 5-week-old mice after decapitation under isofluorane anesthesia. Slices were perfused into a recording chamber with artificial CSF (aCSF) saturated with 95% O2/5% CO2. The aCSF composition was as follows (in mm): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3 and 12 glucose, pH 7.4 (295–305 mOsm/L). Experiments were performed at room temperature in the presence of 1.3 mm Mg2+, 2.5 mm Ca2+, and 50 μm picrotoxin, and a cut between CA3 and CA1 was made to avoid epileptiform activity. Schaffer collaterals were electrically stimulated at 0.05 Hz with a concentric electrode placed in the stratum radiatum. For input–output, paired-pulse (50, 100, and 200 ms intervals), and long-term potentiation (LTP) experiments, evoked field EPSPs (fEPSPs) were recorded using field electrodes filled with aCSF and placed in the stratum radiatum of CA1 region. Patch-clamp recordings were also obtained from CA1 pyramidal cells, with patch electrodes (2–4 MΩ) filled with cesium methane–sulfonate solution containing the following (in mm): 150 Cs+ methane-sulfonate, 1.3 MgCl2, 1 EGTA, 10 HEPES, 0.1 CaCl2 (295–300 mOsm/L), pH ∼7.2. The LTP protocol consisted of a 100 Hz, 1 s train of stimuli repeated three times at 20 s interval, and the averages of the last 10 min (30–40 min) were compared between Scrib1crc/+ and WT mice. Cells were voltage clamped at +40 mV for AMPA/NMDA ratio experiments or at −70 mV for miniature EPSC recordings. TTX (1 μm), D-AP5 (50 μm), or NBQX (10 μm) were used to block respectively voltage-gated sodium channels, NMDA receptors, and AMPA receptors. All drugs were purchased from Tocris Bioscience. The data were recorded with a Multiclamp 700A amplifier (Molecular Devices), and data were collected and analyzed using pClamp9 software (Molecular Devices). Data, reported as the mean ± SEM, were compared using the Student's t test. Significance was assessed at p < 0.05. Experiments and analysis were performed without knowledge of mice genotype.
Behavioral testing
Behavioral experiments were conducted with WT and Scribcrc/+ male littermate mice of 10–11 weeks of age at the start of behavioral tests. All animals had been weighed twice a week from 6 to 11 weeks of age. All behavioral experiments were performed during the light phase (between 9:00 A.M. and 7.00 P.M.) of a 12 h light/dark cycle, under conditions of dim light and low noise. To look for behavioral abnormalities, mice were tested in activity cages (to measure locomotor activity), an open field (to measure exploratory activity and anxiety-like behavior), the Morris water maze (to measure spatial learning and memory performances), and the sociability test (to measure social interaction) (Crawley, 2008).
Locomotor activity
Locomotor activity in response to novelty and daily rhythm of activity were assessed in photocell-based activity chambers under light/dark environmental conditions, using individual Plexiglas chambers (18.2 × 12 × 22 cm) equipped with infrared sensors, allowing measurement of ambulatory locomotor activity. Chambers were interconnected and linked to a computer using an electronic interface (Imetronic). Activity data were collected for each mouse over a 180 min time course for habituation to the apparatus (response to novelty in 5 min bouts) followed by 72 h of recording to measure circadian activities (in 1 h bouts).
Test of sociability
Mice were tested in a transparent plastic cage divided into three chambers (each 20 × 40 × 22 cm) as previously described (Nadler et al., 2004; Jamain et al., 2008). The partitions have openings that allow the animal to move freely from one chamber to another.
Habituation.
During the habituation period, mice were placed in the middle chamber and were allowed to explore the different chambers for 10 min.
Sociability test.
After the habituation, mice were placed in their home cage. A mesh box containing an unknown stranger male mouse was placed in one of the side of chambers (stranger). A similar empty mesh box was placed in the other side chamber (novel object). The tested mice were replaced in the middle chamber and permitted more time to explore the three chambers for 10 min. During this period, the time spent and the number of entries into each chamber, as well as the time spent sniffing in contact with the stranger and the empty box, were measured. At the end of each session, chambers were cleaned with paper containing 70% ethanol.
Morris water maze
Testing took place in a circular pool (diameter, 150 cm) filled with water (19−20°C) rendered opaque by a nontoxic white cosmetic adjuvant. Mice were trained to swim to a submerged platform (14 cm diameter, 1.5 cm below the water surface) using spatial cues located in the room (posters on the walls) to escape from the water. Data were collected using a video camera fixed to the ceiling of the room and connected to a computerized tracking system (Viewpoint) located in an adjacent room. To measure performance in hidden trials, the escape latency, distance to the target, and swim speed were analyzed as outcome measures for each session. One day before training, the animals were habituated to swimming for 60 s in the pool without a platform.
Acquisition training.
Mice were trained for three daily trials with a cutoff of 60 s intertrial intervals. If mice found the platform, they were allowed to remain on it for 15 s; mice that failed to find the platform within 60 s were led by the experimenter to the platform, where they were allowed to stay for 15 s. Mice were released from different starting points at each trial, and different sequences of starting points were used day to day.
Probe testing.
The probe trials were designed to examine the extent of spatial discrimination learning (spatial bias) at two different time points during the acquisition platform training (day 10 and day 28). To do this, the platform was removed from the pool 24 h after the last hidden platform training trial. The number of crossings of the platform location, and the time that mice spent swimming in the target quadrant (where the platform was located during hidden platform training) were measured over a 60 s trial.
Statistical analysis
For the quantitative analysis of PSD, statistical analyses were performed using Sigmaplot11 (Systat Software) for Windows with Mann–Whitney rank sum test followed by Student's t test or Kolmogorov–Smirnov test to assess statistical significance between groups. All calculations for behavioral study were performed with Statistica 5.0 for Windows. Behavioral data were analyzed by ANOVA. Repeated-measures ANOVA was used to analyze locomotor activity and water maze learning data. All data are presented as mean ± SEM; p < 0.05 was considered statistically significant.
Results
Scrib1 is strongly expressed in the brain and is localized in dendritic spines
We first examined Scrib1 mRNA expression in the brain. Parasagittal sections of 10-week-old mouse brains revealed high levels of Scrib1 mRNA in several areas, including the olfactory bulb, cerebral cortex, thalamus, amygdala, mesencephalon, pons, and cerebellum, consistent with the information available in the Allen Mouse Brain Atlas (http://mouse.brain-map.org). The hippocampus showed very high expression of Scrib1 mRNA, with transcript detected in CA1–CA3 pyramidal neurons, dentate gyrus granule cells, and interneurons (supplemental Fig. S1A–C, available at www.jneurosci.org as supplemental material). To analyze in detail the distribution of the Scrib1 protein in hippocampus, we used a previously characterized Scrib1 antibody (Montcouquiol et al., 2006a). Scrib1 protein was expressed in both septal and temporal hippocampus, suggesting a uniform expression of the protein along the longitudinal axis of the hippocampus (data not shown). Scrib1 labeling was present in pyramidal neurons and dentate gyrus granule cells, and in scattered interneurons (Fig. 1A,D). The protein was also expressed in neocortical pyramidal cells and interneurons. Scrib1 did not appear significantly expressed in oligodendrocytes, as we observed no staining in the white matter region, but was present in astrocytes (both in vivo and in vitro, data not shown). Finally, using pre-embedding immunocytochemistry at the electron microscopy (EM) level, we determined that Scrib1 was present within the soma and neurites, associated with membrane-bound compartments and other organelles in the cytoplasm of CA1 pyramidal neurons (Fig. 1E–G). Scrib1 staining was also observed in 58.8 ± 1.9% of spine heads, often associated with membrane and closely associated to the PSD (Fig. 1E–G). Some presynaptic staining was also observed (39.2 ± 2.9% of boutons). After fractionation of brain homogenate, Scrib1 was found in all fractions including membrane fractions (P2 (crude synaptosome), P3 (microsomes), LP1 (synaptosomal membrane), and PSD fraction (supplemental Fig. S2, available at www.jneurosci.org as supplemental material; see Fig. 8B). The strong presence of Scrib1 in dendrites and spines suggests that it could play a role at the postsynaptic level and regulate the development, maintenance, and function of these structures.
Figure 1.
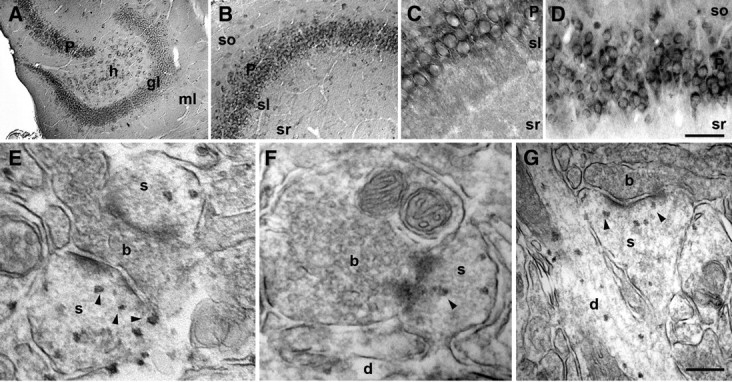
Scrib1 localization in brain. A–D, Horizontal sections showing the distribution of Scrib1 protein in hippocampus. A, Expression of Scrib1 in the dentate gyrus. Signal was present within granule cells (gl) as well as putative interneurons in the hilus (h) and molecular layer (ml). B, Scrib1 labeling in the CA3-CA1 region (left to right, respectively), within pyramidal cells and putative interneurons, scattered in the various strata (sl, stratum lucidum; so, stratum oriens). C, D, High-magnification images of CA3 and CA1 neurons expressing Scrib1 in strata pyramidale (P) in the hippocampus, respectively. Scale bar (in D): A, 165 μm; B, 120 μm; C, 50 μm; D, 45 μm. E–G, Pre-embedding immunoperoxidase EM labeling of Scrib1 in the hippocampus CA1 stratum radiatum (sr). Examples of axonal boutons (b) making asymmetric synapses onto spines (s) labeled for Scrib1. Note the presence of labeling apposed to the PSD in dendritic spines (arrowheads), and within dendrites (d). Scale bar (in G): E, 140 nm; F, 235 nm; G, 260 nm.
Figure 8.
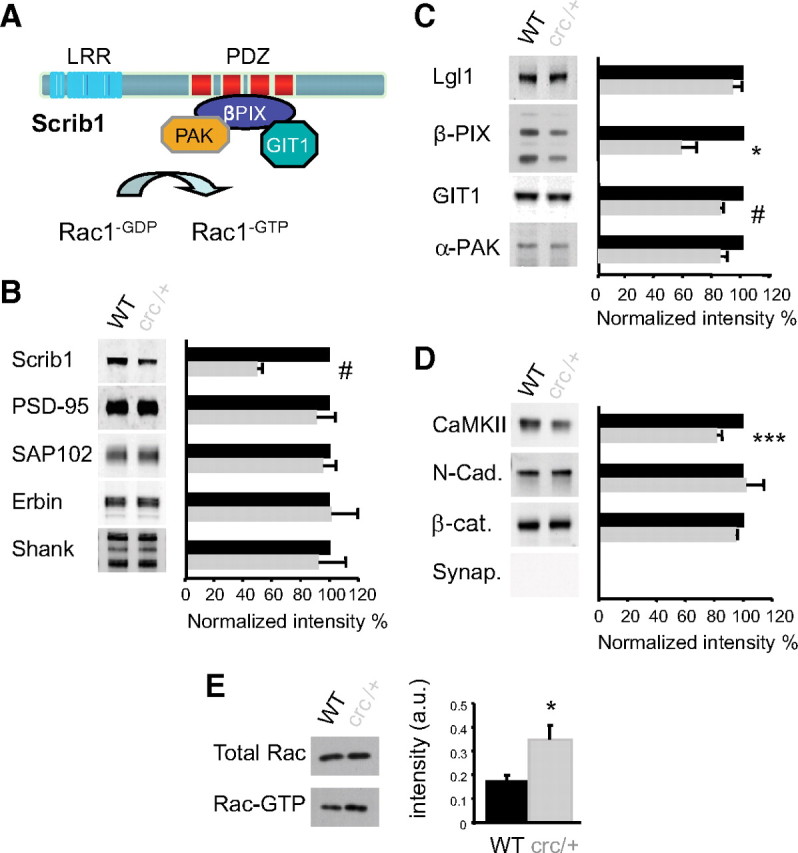
Scrib1 mutation causes alteration of the PSD composition and activation of Rac1 in Scrib1crc/+ mice. A, Schematic representation of the domain structure of Scrib1 and of the identified binding partners of Scrib1. B, Immunoblot analysis of PSD fractions from WT and Scrib1crc/+ mice for PDZ proteins are indicated on the left, and quantitation of the various proteins normalized to WT on the right (100% = WT black histograms). Histograms show mean ± SEM (n = 14 for each genotype). Levels were measured using the Odyssey infrared imaging system. There is a significant difference in total Scrib1 (reduction of 50.17 ± 3.60% in the Scrib1crc/+ PSD compared with the WT). C, Immunoblot analysis of PSD fractions from WT and Scrib1crc/+ mice for direct known interactors of Scrib1 are indicated on the left, and quantitation of the various proteins normalized to WT on the right (100%; black histograms). Histograms show mean ± SEM (n = 14 for each genotype). There is a significant difference in βPIX and GIT1 (reduction of 42.85 ± 10.76% and 15.60 ± 2.1%, respectively, in the Scrib1crc/+ compared with the WT). D, Immunoblot analysis of PSD fractions from WT and Scrib1crc/+ mice for other PSD proteins are indicated on the left, and quantitation of the various proteins normalized to WT on the right (100%; black histograms). Histograms show mean ± SEM (n = 14 for each genotype). There is a significant difference in CaMKII (reduction of 18.33 ± 3.61%, in the Scrib1crc/+ PSD compared with the WT). Synaptophysin was used as a negative control for our PSD preps. *p < 0.05, ***p < 0.001, #p < 0.0001. E, Immunoblots showing the amount of total and activated Rac1 (Rac-GTP) in WT and Scrib1crc/+ mouse hippocampus. The level of Rac1-GTP in the Scrib1crc/+ hippocampus was approximately twofold greater than in WT [WT, 0.17 ± 0.04 arbitrary units (a.u.); Scrib1crc/+, 0.33 ± 0.07 a.u.; p < 0.05, n = 6 for each genotype].
The Scrib1crc/crc mutation leads to severe embryonic defects but Scrib1crc/+ mutant mice develop normally
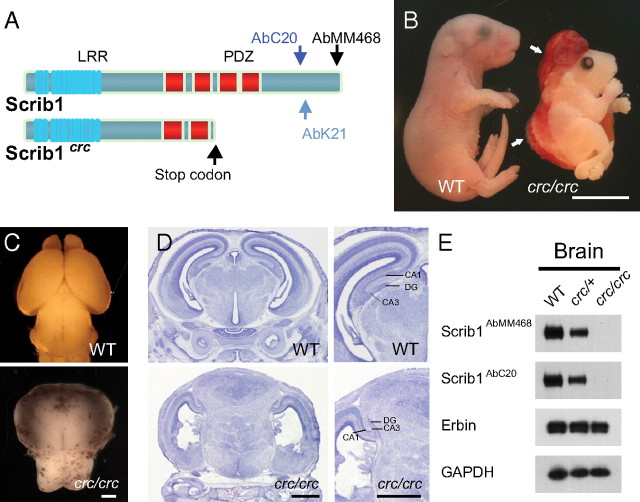
To understand the role of Scrib1 in neurons, we used the crc mutant mouse. In 2003, Murdoch et al. demonstrated that the Scrib1 gene was mutated in these animals, and that an insertion of a base within the gene introduced a stop codon in the reading frame. The putative Scrib1 protein resulting from this mutation was truncated and lacked the last two of its four PDZ domains and the C-terminal part (Fig. 2A). As a result, homozygous Scrib1crc/crc embryos suffered from the most severe form of neural tube defect, craniorachischisis (Fig. 2B), and died shortly before or at birth (Rachel et al., 2000; Murdoch et al., 2003). The severity of this phenotype was variable, but the structures of the brain were always severely damaged. Typically, a large mass of forebrain tissue was present at the anterior surface of the brain (Fig. 2C,D). The morphology of the hippocampus was also affected, and displayed misfolding of the overall structure and delayed regional development (Fig. 2D). Using two different antibodies targeting the C terminus of Scrib1, we showed in WT forebrain extracts, the presence of a major band of ∼180 kDa corresponding to the full-length protein. The intensity of this band decreased by 50% in the Scrib1crc/+ brain and was absent in the Scrib1crc/crc brain from E18.5 animals (Fig. 2E). By comparison, analyzing the pattern of expression of other members of the LAP family of proteins in brains revealed that Erbin (Fig. 2E) and Lano (data not shown) levels were unchanged in both homozygote Scrib1crc/crc and heterozygote Scrib1crc/+ E18.5 brain samples. These data indicate that only half the amount of the full-length Scrib1 protein is produced or maintained in Scrib1crc/+ compared with WT, and that no compensatory increase of other members of the LAP protein family occurred.
Figure 2.
The brain of the homozygote Scrib1crc/crc is dramatically affected. A, Schematic representation of the domain structure of normal and mutated Scrib1 protein. The epitopes of the different Scrib1 antibodies are indicated. B, Gross anatomic anomalies in Scrib1crc/crc mutant mice at E18.5. Scrib1crc/crc mice have an open cephalic neural tube from the midbrain throughout the spine (arrow). Scale bar, 4 mm. C, D, Brain morphology in WT and Scrib1crc/crc littermates at E18.5 and coronal sections stained with hematoxylin. Scale bars, 100 μm. E, Total forebrain extracts of WT, heterozygote (Scrib1crc/+), and homozygote Scrib1 (Scrib1crc/crc) mutant mice (10 μg per lane) were analyzed by SDS-PAGE and immunoblotted with the antibody indicated. Samples analyzed with the different antibodies were obtained from the same preparation of brain tissues.
Since the Scrib1crc/crc animals did not survive beyond birth, we analyzed heterozygote Scrib1crc/+ mice, which appeared to develop normally, with similar weights when compared with WT adults and no gross abnormalities in the size or structure of the adult brain (data not shown). However, we did observe that ∼70% of Scrib1crc/+ mice exhibited enlarged lateral ventricles when compared with WT littermates (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material). This dilatation of lateral ventricles seemed more pronounced in 5-week-old animals. At 10 weeks, hematoxylin-stained brain sections from heterozygote Scrib1crc/+ mice revealed no gross abnormalities, and showed an overall normal cytoarchitecture and cell layering in hippocampus or cortex when compared with WT (supplemental Fig. 3B, available at www.jneurosci.org as supplemental material). We also used SCIP, KA1, and GAD65 as neuronal subpopulation markers for the CA1 and CA3 pyramidal cells and for inhibitory neurons, and, again, did not detect major differences between Scrib1crc/+ and WT adult tissues (data not shown). Finally, to search for possible alterations in the organization of the neuronal network of Scrib1crc/+ mice brain, we performed an immunohistochemical screening using the neuronal markers TuJ1, MAP2, and NeuroH, or the presynaptic markers Bassoon and synaptophysin. We observed the same pattern of staining in the hippocampus of Scrib1crc/+ mice and WT (data not shown). Consistent with our immunoblot results, Scrib1 staining decreased in the Scrib1crc/+ mice (supplemental Fig. 4, available at www.jneurosci.org as supplemental material).
Synaptic transmission and plasticity are reduced in Scrib1crc/+ mice
We used Scrib1crc/+ mice to address the functional consequences of the 50% decrease in Scrib1 levels in the CA1 region of the hippocampus and investigated, first, synaptic transmission. Field responses were recorded in the stratum radiatum of the CA1 region, in response to increasing intensities of stimulation. This input–output relationship revealed a reduced synaptic strength in Scrib1crc/+ animals (n = 16/5 mice) compared with WT animals (n = 15/4 mice), thereby revealing an overall impairment of glutamatergic transmission in Scrib1crc/+ animals (Fig. 3A). Measurement of paired-pulse ratio at different intervals of stimulation (50, 100, and 200 ms) revealed no significant difference between Scrib1crc/+ and WT mice (n = 19/5 WT, n = 26/8 Scrib1crc/+; p > 0.05 for each interval), suggesting that the probability of glutamate release was similar in both groups of animals (Fig. 3B). Analysis of miniature EPSCs (mEPSCs) indicated a mild but significant decrease of the average amplitude of monoquantal events in Scrib1crc/+ animals compared with WT animals (Scrib1crc/+, −4.8 ± 0.3 pA, n = 22/5; WT, −6.5 ± 0.4 pA, n = 16/5; p = 0.0012) (Fig. 3C,D). More strikingly, mEPSC frequency was also significantly reduced from 1.7 ± 0.1 Hz in WT mice (n = 16/5) to 1.2 ± 0.1 Hz in Scrib1crc/+ mice (n = 22/5, p = 0.002). Finally, we examined the ability of Scrib1crc/+ mice to display hippocampal long-term synaptic plasticity using a classic tetanic LTP-inducing protocol. In WT animals, such a protocol produced a long-lasting strengthening of synaptic transmission (165.4 ± 7.4% of baseline, n = 16/4) (Fig. 3E) that was significantly reduced in the Scrib1crc/+ mutants (140.5 ± 3.6%, n = 24/7 mice; p < 0.001). Because LTP in the CA1 region depends on NMDARs (Malenka and Bear, 2004), we next investigated whether the activity of these receptors was altered in Scrib1crc/+ mice. Measurement of AMPA/NMDA ratios, however, revealed no significant difference between Scrib1crc/+ (0.62 ± 0.07, n = 8/5 mice) and WT mice (0.59 ± 0.07, n = 8/5 mice, p = 0.8) (data not shown). Together, these results are consistent with a decreased number of functional synapses impinging on CA1 pyramidal cells in Scrib1crc/+ mice, with a slight decrease of AMPAR content of the remaining ones, as well as an altered ability to undergo potentiation.
Figure 3.
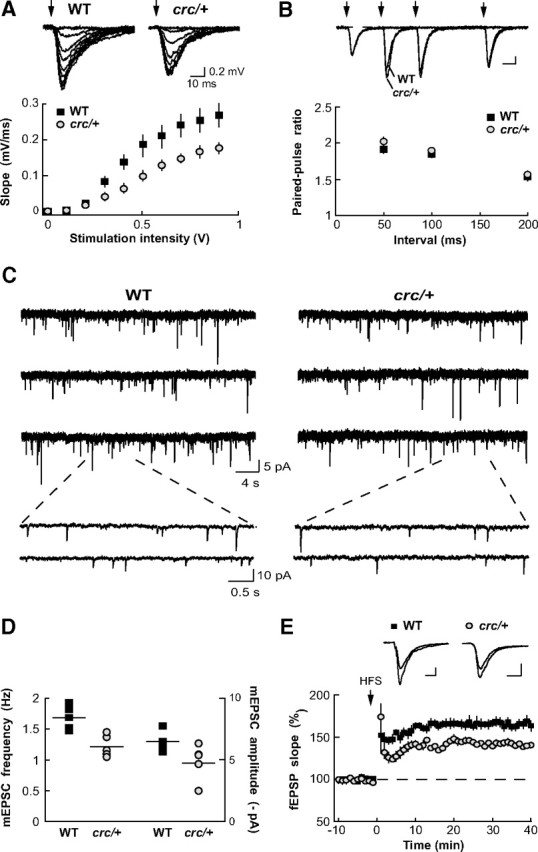
Reduced basal synaptic transmission and synaptic plasticity in the Scrib1crc/+ mice. A, Impaired input–output relationship. fEPSPs were recorded in response to increasing intensity of stimulation. The relation was modified in Scrib1crc/+ slices (n = 16/5) with respect to WT (n = 15/4), with a maximal slope (for maximal stimulation) ∼35% smaller, and a half-maximal response observed for ∼0.1 V higher stimulation intensity. B, Unaffected paired-pulse ratio. Two paired stimulations were applied within a 50, 100, or 200 ms interval. The paired-pulse ratio (fEPSPslope2/fEPSPslope1) was not statistically different for any of the three intervals (p > 0.05 for the three intervals; WT, n = 19/6 animals; Scrib1crc/+, n = 26/8 animals). Calibration: 0.2 mV, 20 ms. C, D, Miniature EPSCs. C, Miniature EPSCs were recorded using the whole-cell patch-clamp technique (V-clamp) in the presence of 1 μm TTX. D, A slight decrease in average amplitude appears significant in Scrib1crc/+ CA1 pyramidal cells (Scrib1crc/+, −4.8 ± 0.3 pA, n = 22/5; WT, −6.5 ± 0.4 pA, n = 16/5; p = 0.0012) concomitantly with an important (30%) decrease in the frequency of event occurrence (Scrib1crc/+, 1.2 ± 0.1 Hz n = 22/5; WT, 1.7 ± 0.1 Hz, n = 16/5; p = 0.002). E, Reduced long-term potentiation. After 20 min of fEPSP baseline recording (only 10 min shown here) with half-maximal intensity stimulation, a classic LTP protocol was applied (high-frequency stimulation: 3 × 1 s stimulation at 100 Hz). The potentiation observed in Scrib1crc/+ slices was smaller than in WT slices (WT, 165.4 ± 16.9% of baseline, n = 16/4; Scrib1crc/+, 140.5 ± 4.1% of baseline n = 24/7). Calibration: 0.2 mV, 20 ms.
Reduced Scrib1 levels improve learning and memory
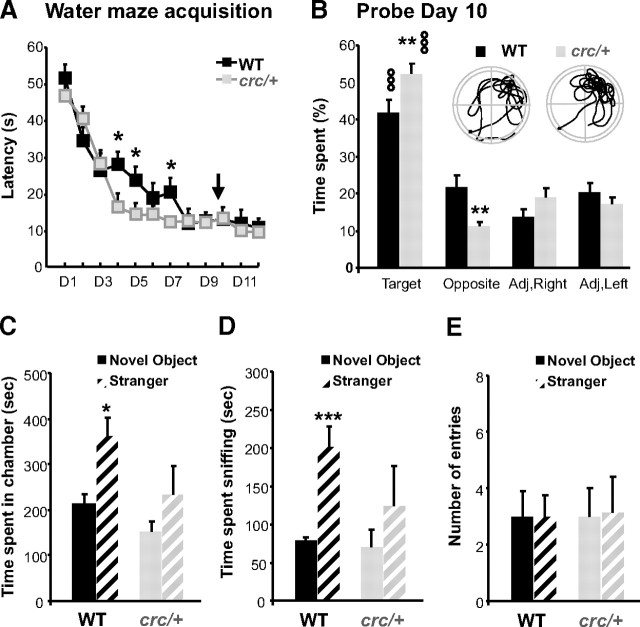
Then, to determine the functional consequence of the Scrib1crc/+ mutation, at a more integrated level, we investigated their spatial memory abilities. We first analyzed habituation to a novel environment, which is considered a simple form of spatial recognition. In response to novelty measured over 3 h, Scrib1crc/+ mice showed a decrease in exploratory activity over time as the context loses its novelty, and were not different from control mice (supplemental Fig. 5A, supplemental Table 1, available at www.jneurosci.org as supplemental material). These data show that this simple form of spatial recognition is preserved in Scrib1crc/+ mice. We next analyzed the effect of Scrib1 on spatial learning and memory in the Morris water maze. First, Scrib1crc/+ and control WT mice were trained to find a fixed hidden platform using distal cues (supplemental Fig. 5B, available at www.jneurosci.org as supplemental material). We found that Scrib1crc/+ mice could locate the platform faster, using as a parameter the latency to escape the platform (Fig. 4A) (genotype × time interaction, p = 0.02) or the distance traveled to find the platform (supplemental Fig. 5C, supplemental Table 2, available at www.jneurosci.org as supplemental material) (genotype × day interaction, p = 0.01), showing that spatial learning was accelerated in Scrib1crc/+ mice. These differences were not due to differences in swimming speed (supplemental Fig. 5D, available at www.jneurosci.org as supplemental material). To determine whether Scrib1crc/+ mice were better at learning this task, a probe test was performed on the 10th experimental day. In this condition, the platform is removed and the time spent in the target quadrant, as well as the numbers of crossings of target location are measured. As typically observed, WT mice spent significantly more time in the quadrant previously containing the platform (Fig. 4B) (t = 4.81, p < 0.001 against chance level). Scrib1crc/+ mice showed a higher preference for the platform-associated quadrant compared with WT mice, indicating an enhanced spatial memory (Fig. 4B) (p < 0.01). To measure long-term memory retention, an additional probe trial was performed 15 d after the last day of training. To prevent extinction due to the first probe test, we continued the training during 3 d after the first probe test (supplemental Fig. 5B, available at www.jneurosci.org as supplemental material). In this condition, both WT and Scrib1crc/+ mice retained the ability to identify the goal quadrant (supplemental Fig. 5E, available at www.jneurosci.org as supplemental material) (t = −2515, p = 0.02 for WT and t = 4.88, p < 0.001 for Scrib1crc/+ against the chance level). These results indicate that Scrib1crc/+ mice exhibited a better memory trace for the platform location and Scrib1crc mutation improves the formation of spatial memory, which are known to require important hippocampal function, whereas simpler forms of spatial recognition, such as habituation and novelty exploration, were not affected.
Figure 4.
Scrib1crc/+ mice showed enhanced spatial memory and deficit in social behavior. A, Spatial learning in Scrib1crc/+ mice (gray line) and littermate WT control mice (black line) measured by latency to reach a submerged platform in the Morris water maze during 12 d of acquisition training. B, Probe test conducted 24 h after day 9 of training (probe day 10). The platform was removed, and the mean proportion of time spent in the target quadrant or one of the three other quadrants was measured. The dotted line represents chance performance. Representative swim-trace pattern for each probe trial, for WT and Scrib1crc/+ mice. Start position is marked by a black square, and a circle marks target location. C, Time spent in chamber with a stranger mouse versus empty chamber. D, Time spent sniffing with stranger mice versus empty cage. E, Number of entries into each side during the three-chambered choice test. ooop < 0.001 versus the chance level; *p < 0.05, **p < 0.01, ***p < 0.001 versus WT.
Reduced Scrib1 levels impair social interaction
A subset of mice carrying mutations in one of the proteins implicated in autism in human resumes some important aspects of human autism spectrum disorders (ASDs), including abnormal response to a novel environments or to a novel individual leading to limited social interaction, along with significant abnormalities in brain size and architecture, and enhanced learning capacities (Tabuchi et al., 2007; Hung et al., 2008; Jamain et al., 2008). For example, the Neuroligin 3 R451C mutant mouse, described by Südhof group as a model for studying autism-related behaviors, shows enhanced learning capacities associated with impaired social interaction (Tabuchi et al., 2007). For this reason, we decided to evaluate social interactions in the Scrib1crc/+ animals in a three-compartment chamber test (Moy et al., 2004). This task is designed to measure the preference for spending time with an unknown mouse enclosed in a wire cage (also called a “stranger”) versus an empty wire cage (also called a “novel object”). As expected, WT mice spent more time in the chamber containing the stranger mice than in the other chamber containing a novel object (Fig. 4C) (supplemental Table 3, available at www.jneurosci.org as supplemental material) (p = 0.012) (Moy et al., 2004; Nadler et al., 2004). In contrast, Scrib1crc/+ mice spent significantly less time in the chamber containing the stranger mice (Fig. 4C) (p = 0.03) compared with WT mice. Similar results were obtained when evaluating the time spent sniffing the stranger mice (Fig. 4D) (p = 0.02 for WT vs Scrib1crc/+ mice). Interestingly, during the habituation test, Scrib1crc/+ mice explore both compartments in a similar way, indicating that reduced interaction with the stranger mice was not due to deficit in the motivation to explore. The number of entries into both compartments was not significantly different between genotypes (Fig. 4E) (p > 0.05), confirming that exploratory tendencies and locomotor activity (as seen in Table 1) were not affected by the mutation. Altogether, the results indicated Scrib1crc/+ mice exhibited reduced social interactions.
Pyramidal neuron morphology is altered in Scrib1crc/+ mutant mice
It was possible that this clear reduction of plasticity in hippocampus during development could correlate with modifications of neuron morphology and spine number, and induced remodeling of the neuronal circuitries that are responsible for the behavioral outputs. We examined the dendritic structure of CA1 pyramidal neurons, the morphology and function of which have been extensively studied. We used the Golgi impregnation technique, which allows the visualization of the complete dendritic arborization of neurons, as well as dendritic spines (Fig. 5A–E). We found that the total length of basal dendrites was significantly increased in Scrib1crc/+ mice (WT, 663.68 ± 47.98 μm, n = 38/3 mice; Scrib1crc/+, 893.47 ± 62.89 μm, n = 31/4 mice; p < 0.05), whereas there was no significant difference in the total length of the apical dendrite (WT, 876.76 ± 48.55 μm, n = 35/3; Scrib1crc/+, 927.69 ± 55.13 μm, n = 39/4; p = 0.5) (Fig. 5B). Sholl analysis confirmed an increased complexity of hippocampal CA1 pyramidal neuron morphology in Scrib1crc/+ (Fig. 5C). In basal dendrites, the number of intersections increased significantly in Scrib1crc/+ mutant neurons. In apical dendrites, the number of intersections close to the cell body (<80 μm) was decreased, while the number of intersections distal to the cell body (>270 μm) was increased significantly (Fig. 5C). Since the total number of crossings remained the same, most of the changes seen on the apical dendritic tree suggest that branches tended to extend in the distal portion of the dendrite rather than to ramify close to the cell body.
Figure 5.
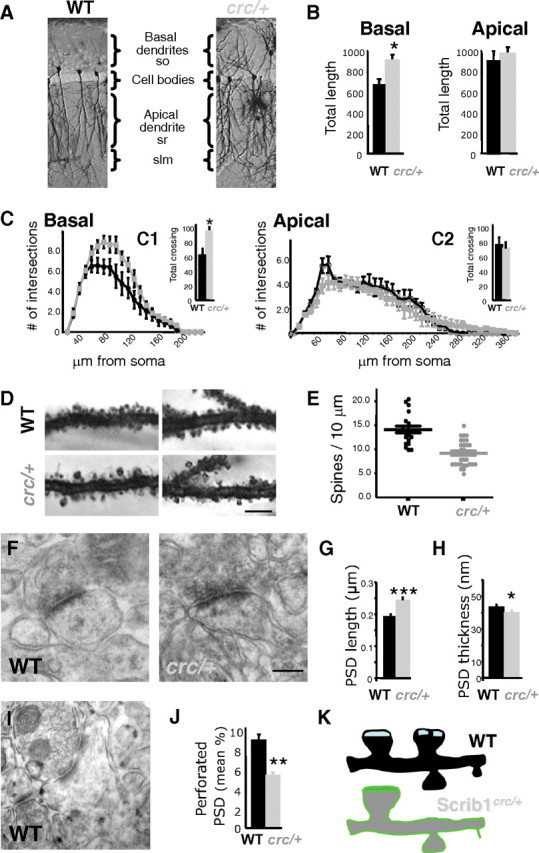
Neuronal architecture: fewer spines but bigger synapses are found in the Scrib1crc/+ mice. A, Representative photographs of Golgi-stained CA1 pyramidal neurons of WT and Scrib1crc/+ mice. Four to six sections (70 μm thick) through the whole hippocampus were used per animal and at least five neurons were analyzed for each section (WT, n = 6 mice; Scrib1crc/+, n = 6 mice) (slm, stratum lacunosum moleculare; so, stratum oriens; sr, stratum radiatum). B, Quantification of total length of apical and basal CA1 pyramidal cell dendrites using the Neurolucida system show increased arborization of the pyramidal neurons in Scrib1crc/+. C, Sholl analysis was performed on apical and basal dendrites to determine whether the three-dimensional organization of dendrites relative to the cell body was changed in the Scrib1crc/+ animals. In basal dendrites, a significant increase in intersections was observed both in dendrites close and distal to the cell body. In apical dendrites, the number of intersections was significantly increased only distally to the cell body in the Scrib1crc/+ compared with the WT. Total number of crossing shown for basal (C1) and apical (C2) dendrites. D, Representative high-magnification photographs of Golgi-stained CA1 pyramidal neuron dendrites bearing spines of WT and Scrib1crc/+ mice. Scale bar, 5 μm. E, The data show a significant decrease (32%) in spine density per micrometer of dendrite in the Scrib1crc/+ hippocampal neurons compared with WT mice (*p < 0.05). Only spines on apical dendrites in the stratum radiatum between 100 and 250 μm from the soma were counted. At least 500 spines were counted. F, Representative electron micrographs of hippocampal CA1 stratum radiatum spinous synapses from adult WT and Scrib1crc/+ mice. Complete synapses (PSDs apposed to presynaptic boutons with vesicles) were readily identified in the Scrib1crc/+. Scale bar, 250 nm. G, H, Quantitative analysis of the PSD length and thickness. The PSD length was increased while the PSD thickness was decreased in the Scrib1crc/+ mice. Differences are significant for both length and thickness. I, Representative electron micrograph of a perforated synapse in hippocampal CA1 stratum radiatum. J, Number of perforated PSDs in adult WT and Scrib1crc/+ mice CA1 hippocampus. K, Representative schema of spine distribution in WT vs Scrib1crc/+ hippocampus.
Since Scrib1 was present in dendritic spines (Fig. 1E–G), we investigated whether it could regulate spine number and morphology. Five-week-old Scrib1crc/+ CA1 pyramidal neurons showed 14% fewer spines per micrometer than WT mice (WT, 1.45 ± 0.08 spines/μm; Scrib1crc/+, 1.24 ± 0.02 spines/μm; p < 0.001) (data not shown), whereas 10-week-old Scrib1crc/+ CA1 pyramidal neurons showed a greater decrease, 32% fewer spines per micrometer than WT pyramidal neurons (WT, 1.41 ± 0.05 spines/μm; Scrib1crc/+, 0.96 ± 0.04 spines/μm; p < 0.001) (Fig. 5D,E). The mean spine length (WT, 1.22 ± 0.02 μm, n = 516; Scrib1crc/+, 1.30 ± 0.03 μm, n = 515; p < 0.05) and the percentage of spines > 1.6 μm long (WT, 19.77%; Scrib1crc/+, 32.33%) were significantly increased in Scrib1crc/+ neurons (data not shown). The mean width of spine heads increased in Scrib1crc/+ neurons (WT, 0.61 ± 0.01 μm, n = 516; Scrib1crc/+, 0.65 ± 0.01 μm, n = 515; p < 0.05), with a significantly increased percentage of spine heads with width > 0.7 μm (WT, 7.82%; Scrib1crc/+, 19.16%) (data not shown). We confirmed at the EM level that WT spine heads were smaller than in Scrib1crc/+ mice (WT, 0.116 ± 0.005 μm2, n = 155; Scrib1crc/+, 0.123 ± 0.005 μm2, n = 186; Kolmogorov–Smirnov test, p < 0.001, all values based on pooled data from 2 mice for each genotype). In general, spines appeared normal with a presynaptic bouton well aligned with the postsynaptic compartment. The presynaptic terminals appeared normal (Fig. 5F). Compared with synapses in WT mice, Scrib1crc/+ synapses were less numerous (mean number of spines: WT, 56.00 ± 2.68, number of spines = 3168, number of EM fields = 58, total area = 9521 μm2; Scrib1crc/+, 45.96 ± 2.57, number of spines = 2896, number of EM fields = 63, total area = 9741 μm2; t test, p < 0.01, all values based on pooled data from three mice for each genotype). These data are consistent with our electrophysiological data and show that Scrib1 is necessary for normal spine development.
Scrib1 regulates PSD morphology
We then analyzed the morphology of Scrib1crc/+ pyramidal neurons PSD, as it was reported in previous studies that an increase in spine size could be correlated to an increase in the PSD size (Bourne and Harris, 2008). Compared with synapses in WT mice, Scrib1crc/+ synapses had larger PSD (WT, 0.189 ± 0.005 μm, n = 150; Scrib1crc/+, 0.243 ± 0.006 μm, n = 183; Kolmogorov–Smirnov test, p < 0.001; all values based on pooled data from 3 mice for each genotype) (Fig. 5F,G). Also, Scrib1crc/+ PSDs were significantly thinner (WT, 43.17 ± 1.28 nm, n = 139; Scrib1crc/+, 40.05 ± 1.29 nm, n = 185; Kolmogorov–Smirnov test, p < 0.05; all values based on pooled data from 3 mice for each genotype) (Fig. 5H). These changes at the ultrastructural level are consistent with the decrease in spine density and increase in spine size observed with the Golgi impregnation technique (Fig. 5D,E).
During this analysis, we also noticed that perforated spines were less numerous in the Scrib1crc/+ mutant. We observed a 34.72% reduction in the mutant compared with WT mice (WT, 8.62 ± 0.77%, n = 3, perforated/total spines = 265/3168, 58 fields, total area = 9521 μm2; Scrib1crc/+, 5.62 ± 0.76%, n = 3, perforated/total spines = 148/2896, 63 fields, total area = 9741 μm2; Mann–Whitney rank sum test, p < 0.001) (Fig. 5I,J). These data suggest that Scrib1 is necessary for normal spine splitting, and a defect in this process could result in the larger spines and PSD that we see in the Scribcrc/+ mice (Fig. 5K).
A tight regulation of Scrib1 levels is necessary to form and maintain spines
As Scrib1 is enriched in dendrites and spines of pyramidal neurons (Fig. 1), we examined whether Scrib1 modifies spinogenesis and synaptogenesis in 14–18 DIV hippocampal neurons. We used GFP- or mRFP-β-actin with and without expression plasmids to quantify changes in spine density and morphology (Brünig et al., 2004; Calabrese and Halpain, 2005; Saneyoshi et al., 2008). Tagged Scrib1 overexpressed in neurons was targeted to dendrites and dendritic spines as identified by GFP or HA expression, and colocalized with native PSD-95 and Bassoon (data not shown). Neurons overexpressing full-length Scrib1 together with RFP-β-actin had more dendritic spines than control neurons, and these spines were smaller on average (Fig. 6A,B,D–F). On the contrary, in neurons overexpressing the Scribcrc form, we observed a clear decrease in spine number and an increase in spine size (Fig. 6A,C–F). We also analyzed the impact of a reduction of Scrib1 expression using shRNAs. The specificity and efficacy of three shRNAs for Scrib1 were tested in HEK cells with transfected Scrib1 cDNA. We selected shRNA#1 and shRNA#3 for our loss-of-function studies as they decreased Scrib1 expression by at least 70% (supplemental Fig. 6A, available at www.jneurosci.org as supplemental material). Transfection of shRNA#1 and shRNA#3 in hippocampal neurons resulted in a 70.05% and 80.45% reduction, respectively, of endogenous Scrib1 staining (red) within the cell body, compared with that of untransfected neurons or neurons transfected with control shRNA (supplemental Fig. 6C–F, available at www.jneurosci.org as supplemental material). The number of spines was reduced in these neurons, whereas the number of filopodia increased significantly in Scrib1 shRNA-expressing neurons (Fig. 6G–I,L,M), and this phenotype was rescued by overexpression of a shRNA-resistant human form of Scrib1 (Fig. 6J,L,M) (supplemental Fig. 6B, available at www.jneurosci.org as supplemental material). The Scrib1crc form had a partial rescued effect, suggesting that the shorter form retained some of the Scrib1 functions (Fig. 6K–M). These data strongly support the critical role of Scrib1 in the efficient formation of dendritic spines.
Figure 6.
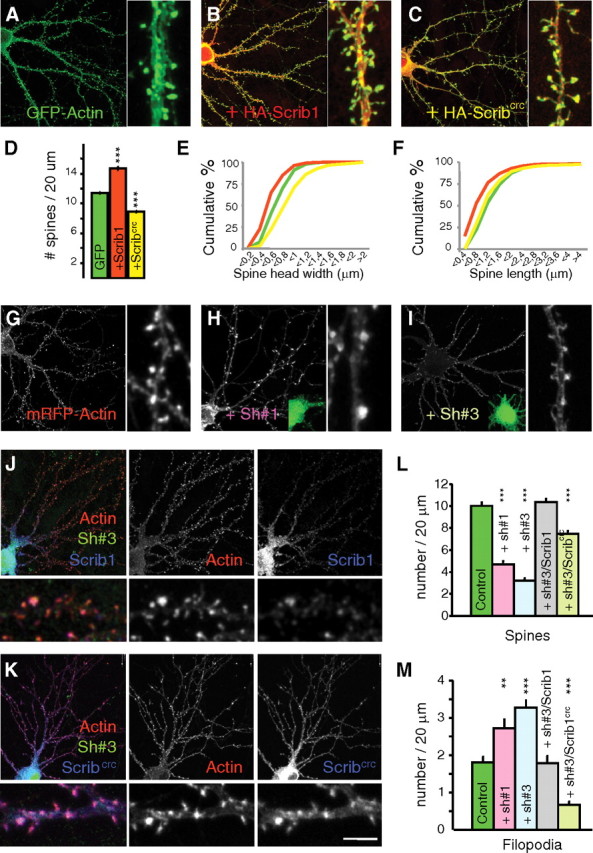
Effects of Scrib1 overexpression or downregulation in hippocampal neurons. A–C, Neuron cotransfected with GFP-actin (A), GFP-actin and HA-Scrib1 (B), or GFP-actin and HA-Scrib1crc (C). Scrib1 is enriched in dendritic spines. Right panels: high-magnification image of a dendrite. D, Quantification of spine density (number of spines/20 μm of dendrite length) in neurons as defined with GFP expression (mean ± SEM). The number of spines increased in Scrib1- (red) and decreased in Scrib1crc- (yellow) transfected neurons (13.95 ± 0.42 spines/20 μm for Scrib1 neurons, 9.25 ± 0.21 spines/20 μm for Scrib1crc neurons, vs 11.53 ± 0.20 spines/20 μm for GFP-actin neurons; p < 0.001). The data are the means ± SEM. E, Cumulative distribution of spine head width in transfected neurons with GFP-actin (green), GFP-actin + HA-Scrib1 (red), or GFP-actin + HA-Scrib1crc (yellow). The mean width of spine heads decreased in HA-Scrib1-transfected neurons (0.57 ± 0.006 μm; p < 0.001) but increased with HA-Scrib1crc (0.87 ± 0.01 μm) compared with the control (0.69 ± 0.005 μm); p < 0.001 for both. F, Cumulative distribution of spine length in transfected neurons with GFP-actin (green), GFP-actin + HA-Scrib1 (red), or GFP-actin + HA-Scrib1crc (yellow). The mean length of spines decreased in HA-Scrib1-transfected neurons (0.88 ± 0.17 μm, p < 0.001) and decreased less, but significantly, with HA-Scrib1crc (1.13 ± 0.02 μm) compared with the control (1.25 ± 0.02 μm; p < 0.001) for both. G–I, Neuron cotransfected with mRFP-actin (G), mRFP-actin + Sh#1 (H), or mRFP-actin + Sh#3 (I). Right panel: high-magnification images of a dendrite. J, Rescue experiment: neuron transfected with HA-Scrib1, Sh#3, and mRFP-actin. K, Scrib1crc effect on a Scrib1 KO background: neuron transfected with HA-Scrib1crc, Sh#3, and mRFP-actin. L, M, Quantification of spine (L) and filopodia (M) density (number of spines or filopodia/20 μm of dendrite length) in neurons as defined with mRFP expression (mean ± SEM). The number of spines decreased with Scrib1 shRNA#1 (pink) and Scrib1 ShRNA#3 (light blue) transfected neurons (4.61 ± 0.27 spines/20 μm for Sh#1 neurons, 3.23 ± 0.22 spines/20 μm for Sh#3 neurons vs 10.31 ± 0.32 spines/20 μm for control neurons; p < 0.001). In neurons transfected with mRFP-Actin + Sh#3 and HA-Scrib1, Scrib1 restored the formation of spines (10.46 ± 0.33 spines/20 μm, p > 0.05), but not Scrib1crc (7.47 ± 0.23 spines/20 μm; p < 0.001). On the contrary, the number of filopodia increased in Scrib1 shRNA#1 (pink) and Scrib1 ShRNA#3 (light blue) transfected neurons (2.71 ± 0.24 filopodia/20 μm for Sh#1 neurons, 3.27 ± 0.19 filopodia/20 μm for Sh#3 neurons, versus 1.91 ± 0.16 filopodia/20 μm for control neurons; p < 0.01 and p < 0.001 respectively). In neurons transfected with mRFP-Actin+Sh#3 and HA-Scrib1, Scrib1 restored the formation of filopodia (2.05 ± 0.22 filopodia/20 μm; p > 0.05), but not Scrib1crc (0.67 ± 0.08 spines/20 μm, p < 0.001). Scale bar: K, 10 μm.
Scrib1 regulates fast actin dynamics and reorganization
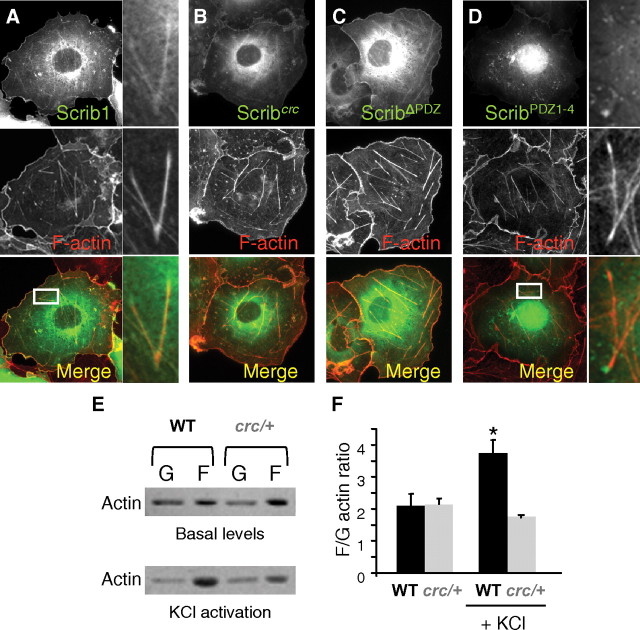
Scrib1 is known for its involvement in cytoskeleton rearrangement (Montcouquiol et al., 2003; Humbert et al., 2006). Because dendritic spine formation is dependent on actin remodeling, and induction of LTP also involves rapid polymerization of actin in spines, we analyzed the relationship between actin and Scrib1 (Bramham, 2008). In COS-7 cells, transfected Scrib1 colocalized with F-actin (Fig. 7A). The crc mutation does not disrupt this localization, nor a deletion of all PDZ domains (Fig. 7B,C), but a truncation of the N-terminal domain of Scrib1 did strongly reduce the colocalization (Fig. 7D). We next measured the F-actin/G-actin ratio in hippocampus of WT and Scrib1crc/+ animals. No significant differences were seen in F-actin/G-actin ratios between WT and mutant animal in basal condition. However, upon stimulation with 20 mm KCl, the F/G-actin ratio significantly increased in the control, indicating the quick change of G-actin to a polymerized F-actin state. In hippocampus from Scrib1 mutants, the levels did not change, suggesting an impairment or reduction in quick actin polymerization in neurons where Scrib1 levels were reduced (Fig. 7E,F).
Figure 7.
Scrib1 associates with F-actin. A–D, Immunofluorescence microscopy of COS-7 cells transiently transfected with the full length of Scrib1 (Scrib1), the 960stop mutation of Scrib1 (Scrib1crc), a truncated Scrib1 form of PDZ domain (Δ PDZ), and the Scrib1 form lacking the LRR domain (PDZ 1-4). A, Merge image showing that the full length of Scrib1 (green) colocalizes with actin filament (phalloidin staining, red). Right panels: high-magnification images of the region in the inset. B–D, Scrib1crc (B) and Δ PDZ (C) do not disrupt this colocalization, while this colocalization is altered with the PDZ1-4 form (D). Right panels: high-magnification images of the region in the inset. E, Immunoblot analysis of G-actin (G) and F-actin (F) fraction from WT and Scrib1crc/+ extracts from hippocampus in basal condition and after treatment with 20 mm KCl. G-actin and F-actin are probed with anti-actin antibody. F, Quantitative F-actin/G-actin ratios in hippocampus were measured with actin polymerization in vivo assay kit. Ratios between genotypes were not significantly different in basal condition. After 60 s in vitro stimulation with 20 mm KCl, the F-actin/G-actin ratio increased in the WT (n = 4), while no significant increase was seen in the mutants (n = 4). Ratios of F-actin/G actin were determined from the blots by optical density measurements. *p < 0.05, Student's t test.
Molecular correlates of spine and PSD hypertrophy
We next quantified the level of expression of Scrib1 downstream signaling proteins in the PSD. By immunoblotting, we confirmed that Scrib1 was present in the PSD, and that Scrib1crc/+ animals displayed an ∼50% reduction of Scrib1 in the PSD (Fig. 8B), similar to what was observed in the whole brain at E18.5 (Fig. 2E) or the total hippocampus at 10 weeks (supplemental Fig. 4B, available at www.jneurosci.org as supplemental material). Using quantitative Western blotting, we found no significant changes in the level of expression of PDZ proteins, such as the MAGUK (PSD-95 and SAP102), SHANK, and Erbin, a member of the LAP family (Fig. 8B). Since Scrib1 forms a tight complex with βPIX/GIT1 that is also known to affect Rac1 localization and activation (Audebert et al., 2004; Nola et al., 2008; Richier et al., 2010) (Fig. 8A), we investigated whether the crc mutation could affect the Scrib1/βPIX complex. We did a GST pull-down with the different PDZ domains of Scrib1 and showed that all PDZ domains, except for PDZ4, can bind to βPIX (supplemental Fig. 7A, available at www.jneurosci.org as supplemental material). We also coimmunoprecipitated βPIX with Scrib1wt or Scrib1crc and showed that the βPIX interaction was reduced with Scrib1crc compared with WT (supplemental Fig. 7B, available at www.jneurosci.org as supplemental material). We then analyzed the levels of these proteins in the PSD. There was a significant reduction of βPIX, PAK, and GIT1 (∼40% for βPIX and ∼20% for PAK and GIT1) (Fig. 8C). Interestingly, CaMKII (Fig. 8D) was also reduced in mutant PSDs. Using a GST-PAK binding assay, we measured the level of activated Rac1, Rac1-GTP, in WT versus Scrib1crc/+ mice hippocampus. We found that Rac1-GTP levels were increased in Scrib1crc/+ mice when compared with WT (Fig. 8E) and were unchanged after KCl stimulation (data not shown). Thus, the abilities of the Scrib1crc form to reduce spine number and increase their size is due to the reduced amount of βPIX and PAK in the PSD and is not due to an inefficient activation of Rac1, which is quite high even at basal level.
Discussion
Here, we show that Scrib1 has a new role in addition to regulating cell migration and polarity. Scrib1 is capable of influencing the morphology and function of synapses. We show that the Scrib1crc/+ mutant mice possess enhanced learning and memory capacity, and decreased sociability, which are positively associated with increased numbers of enlarged spines, and negatively with LTP. These results show that Scrib1 plays a crucial role in regulating higher brain functions that might be relevant to ASD.
Involvement of Scrib1 in polarity
Our analysis of homozygote Scrib1crc/crc mouse brains show a consistent protrusion of tissue through an opening in the upper part of the skull of the embryo, referred to as “encephalocele” in previous work (Rachel et al., 2000). The hippocampal structure is also greatly affected, as are the cortical layers and the other main areas of the brain in the embryo. The lateral and third ventricles are larger in size, an enlargement also observed in other mutants for planar cell polarity (PCP) proteins, such as Fzd3 and Celsr3 (Tissir et al., 2005; Wang and Nathans, 2007; Zhou et al., 2008). This enlargement of the ventricles was also present in adult Scrib1crc/+ mice but seemed to only affect the thickness of visual cortex, leaving unchanged the other brain regions. Analysis of adult Scrib1crc/+ hippocampal tissue showed a 50% reduction of Scrib1, and a complete disappearance of the protein in homozygote Scrib1crc/crc tissues, similar to that observed in murine embryonic fibroblasts (MEFs) of the same mutant (Nola et al., 2008), confirming the nature of the loss of function of the mutation. Our current knowledge of the various roles of Scrib1 in apicobasal polarity (Assémat et al., 2008), migration (Nola et al., 2008), or proliferation (Dow and Humbert, 2007) suggests a disruption in any of these events or any combination of them could be responsible for such dramatic neuronal phenotype. We cannot exclude, however, that part of this phenotype is due to the presence of the putative truncated protein of 960 aa containing only the LRR and the first two PDZ domains of Scrib1, and resulting from the frame-shift mutation in Scrib1crc/crc (Murdoch et al., 2003).
Involvement of Scrib1 in dendrite development
Our data show that Scrib1 participates in normal maturation of neurons, a property that is shared with other identified PCP genes (Montcouquiol et al., 2006b; Wang and Nathans, 2007; Goodrich, 2008; Chacon-Heszele and Chen, 2009). We did not find indications of abnormal polarization of pyramidal neurons in Scrib1crc/+ mice; they had an apical dendrite, three to five basal dendrites, and one axon. We, however, observed an increased total length and branching complexity of the basal dendrites compared with their WT counterparts. Since only half of the Scrib1 is present in the mutants, our data suggest that Scrib1 inhibits an overgrowth of dendritic arborization in WT animals, through the regulation of the cytoskeleton. These results are consistent with the role of PCP proteins in controlling changes in the cytoskeleton in various tissues and species, both locally and globally. For instance, the large atypical cadherin Flamingo is required for controlling the extension and/or guidance of dendrites and axons of multiple types of neurons during neural development, but also controls the orientation of actin-rich hair covering Drosophila wings (Usui et al., 1999; Tissir et al., 2005; Shima et al., 2007; Goodrich, 2008; Zhou et al., 2008). It is interesting to note that only the length of basal dendrites was affected, whereas apical dendrites had normal length but increased intersections/branching only in their distal portion. Further analysis will be necessary to evaluate whether this is due to asymmetric protein accumulation or increase in specific pathways.
Involvement of Scrib1 in dendritic spines formation and maintenance
Synaptic elimination, which is a normal developmental mechanism necessary to avoid too much conflicting information arriving on a dendrite, appeared exacerbated with the loss of Scrib1 with development/aging. Along with this overall decrease in spine density, we observed an increase in the number of spines with larger heads in Scrib1crc/+ mice. These observations were recapitulated by in vitro manipulation of Scrib1 levels in hippocampal neurons. Overexpression of full-length Scrib1 led to an increased number of spines that were smaller on average, whereas overexpression of the truncated form of Scrib1 (Scrib1crc), or downregulation with shRNAs, led to an overall decrease in spine number, with an increase in the number of larger spines. These data show that Scrib1 is required for normal spine development and maturation, but that it also affects spine stability, having an overall negative effect on spine morphing. Although many spines can persist for days to months, the “morphing” of spine shape is described as the ability of spines to change size and shape continually on a time scale of seconds to minutes. Studies have shown that most of the mutations in synaptic proteins result in a decrease in spine number and/or size, and only a few mutants present an increase in size of the PSD and/or spine number. The Scrib1crc/+ mutant displays a combination of both phenotypes. Interestingly, a transgenic mouse expressing a dominant-negative PAK, a downstream target of Scrib1, also has fewer spines and an increased proportion of larger ones in the cortex (Hayashi et al., 2004). PAK and two of its known interactors, βPIX and GIT1, are significantly reduced in Scrib1crc/+ mice PSDs, but not in homogenates, suggesting that they could participate in the phenotype of the Scrib1crc/+ mice.
The apparent contradiction in the number of spines and their size might be explained by the fact that synapse “consolidation” involves a series of alternating events, such as making and breaking of adhesions, and extension and retraction of actin, which, by necessity, must involve alternating positive and negative influences. Accordingly, we find that Scrib1 colocalizes with actin filaments and that actin fast polymerization was perturbated in the Scrib1crc/+ mutant. Scrib1 might function as a key hub protein linking membrane receptors and associated βPIX-based Rac1 signaling complex to actin filaments for spine dynamics. In fact, Scrib1 forms a tight complex with βPIX/GIT1, which is known to promote Rac1 activation through PAK, but βPIX can also interact directly with Rac1 (Audebert et al., 2004). Our data show that Scrib1 disruption leads to an increase in activated Rac1 levels in hippocampus. Similar results were obtained by another group with MEFcrc/crc cultures and cancer epithelial cell lines (Nola et al., 2008), but another group reported that depletion of Scrib1 or βPIX induced a significant decrease in activated Rac1 in carcinoma (Zhan et al., 2008). These discrepancies possibly reflect tissue-/stage-specific dependence on various GEFs. In fact, in PAK1−/− cells, the activity of Rac1 is also increased (ten Klooster et al., 2006), suggesting that PAK and Scrib1 act in the same downstream pathway. Furthermore, Tashiro and Yuste (2004) reported that a subset of spine heads in constitutively active Rac-transfected neurons was larger than control. Altogether, we believe that Scrib1 regulates structural and molecular organization in the spine linking membrane receptors to the actin cytoskeleton.
Improved learning and memory and impaired social interactions
We found that Scrib1crc/+ animals had better spatial learning abilities and impaired social behaviors. These behavioral phenotypes could result from the alteration of CA3-CA1 hippocampal synaptic connectivity, a critical element for the amplification and processing of information. However, synaptic changes occurring within other hippocampal or brain areas might also be involved.
Mixed cognitive phenotypes in humans primarily affecting social interactions, deficits in communication, and repetitive behaviors, are typical of ASD (Bourgeron, 2009). Still, no human mutation has yet been found in the Scrib1 gene. Interestingly, enlargement of the ventricle and modified dendritic spines has also been reported in mouse models of severe neuropsychiatric disorders including autism and schizophrenia (Hikida et al., 2007; Hung et al., 2008; Gant et al., 2009) and in human patients (Bienvenu and Chelly, 2006; Wu et al., 2007; Mata et al., 2009). Moreover, in mouse models for autism, the synaptic imbalance resulting from excess or reduced connectivity in vivo has also been implicated (Bourgeron, 2009). For instance, inhibition is increased in Mecp2-mutant mice or NL2 and NL3 models (Dani et al., 2005; Tabuchi et al., 2007; Hines et al., 2008). We can hypothesize that, in our case, decreasing excitatory transmission might have the same effect as enhancing inhibitory drive. Many mutated genes implicated in autism, including NRXN/neurexin 1, NL3–4, or SHANK3, alter synaptic function and belong to a pathway upstream of the βPIX/GIT1 complex (Bourgeron, 2009). Interestingly KO mice for SHANK1 and NL3 KI mice display enhanced memory, and NL3 mutants also display deficits in social behaviors (Tabuchi et al., 2007; Hung et al., 2008). Thus, Scrib1crc mutation, which leads to a similar phenotype, functionally implicates Scrib1 as a gene belonging to this pathway and regulating synaptic functions. It will be of interest to determine whether these behavioral changes in the Scrib1crc/+ mice result from changes occurring during the developmental period or to the network ability to adapt to ongoing behavior. These findings make a key step forward in the dissection of the multiple roles of Scrib1 in the developing and adult brain, and will lead to a better understanding of its role in the pathophysiology of behavior.
Footnotes
Supported by grants from INSERM AVENIR (N.S. and M.M.), Conseil Regional d'Aquitaine (N.S., M.M.), La Fondation pour la Recherche Medicale (N.S., M.M.), ANR Projet ANR-07-NEUR-031–01 (N.S.), ANR fellowship (M.M.M.), FRM fellowship (C.D.), and the European Commission Coordination Action ENINET (contract number LSHM-CT-2005-19063; N.S., M.M.). The sequencing was performed at the Sequencing Facility of Bordeaux (Conseil Régional d'Aquitaine Grants 20030304002FA and 20040305003FA, and European Union, FEDER Grant 2003227). We thank R. Rachel for the circletail mice. We thank J. Murdock for providing the Scrib ISH probe, I. Macara for the GFP-Scrib constructs, P.O. Humbert for the IRES–Scrib1 construct, J. P. Borg for the Scrib, Lano, and Erbin antibodies and the HA–Scrib1 and GST–PAK constructs, C. Gauthier-Rouvière for the mRFP–actin construct, V. Vasioukhin for the Lgl1 antibody, and R. J. Wenthold for the MAGUK and GluR antibodies. We also thank N. Aubailly and her team for excellent technical assistance with mutant animals, and D. Gonzales and D. Arma for genotyping and sequencing. The confocal microscopy was done at the Bordeaux Imaging Center of the Neurosciences Institute of the University of Bordeaux II. We thank C. Funk for the help with behavioral tests, A. Fabre for the Neurolucida training, M. C. Donat for excellent technical assistance, and M. Warchol and R. J. Wenthold for helpful comments and discussion. This work is dedicated to Robert J. Wenthold, an inspiring mentor.
References
- Assémat E, Bazellières E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lécine P, Bellaiche Y, Dupont JL, Premont RT, Sempéré C, Strub JM, Van Dorsselaer A, Vitale N, Borg JP. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 2004;14:987–995. doi: 10.1016/j.cub.2004.05.051. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Brünig I, Kaech S, Brinkhaus H, Oertner TG, Matus A. Influx of extracellular calcium regulates actin-dependent morphological plasticity in dendritic spines. Neuropharmacology. 2004;47:669–676. doi: 10.1016/j.neuropharm.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Calabrese B, Halpain S. Essential role for the PKC target MARCKS in maintaining dendritic spine morphology. Neuron. 2005;48:77–90. doi: 10.1016/j.neuron.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Chacon-Heszele MF, Chen P. Mouse models for dissecting vertebrate planar cell polarity signaling in the inner ear. Brain Res. 2009;1277:130–140. doi: 10.1016/j.brainres.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Humbert PO. Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int Rev Cytol. 2007;262:253–302. doi: 10.1016/S0074-7696(07)62006-3. [DOI] [PubMed] [Google Scholar]
- Driver JE, Racca C, Cunningham MO, Towers SK, Davies CH, Whittington MA, LeBeau FE. Impairment of hippocampal gamma-frequency oscillations in vitro in mice overexpressing human amyloid precursor protein (APP) Eur J Neurosci. 2007;26:1280–1288. doi: 10.1111/j.1460-9568.2007.05705.x. [DOI] [PubMed] [Google Scholar]
- Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17:343–352. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Gant JC, Thibault O, Blalock EM, Yang J, Bachstetter A, Kotick J, Schauwecker PE, Hauser KF, Smith GM, Mervis R, Li Y, Barnes GN. Decreased number of interneurons and increased seizures in neuropilin 2 deficient mice: implications for autism and epilepsy. Epilepsia. 2009;50:629–645. doi: 10.1111/j.1528-1167.2008.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV. The plane facts of PCP in the CNS. Neuron. 2008;60:9–16. doi: 10.1016/j.neuron.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, Chattarji S, Kirkwood A, Tonegawa S. Altered cortical synaptic morphology and impaired memory consolidation in forebrain-specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–787. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andradé M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines RM, Wu L, Hines DJ, Steenland H, Mansour S, Dahlhaus R, Singaraja RR, Cao X, Sammler E, Hormuzdi SG, Zhuo M, El-Husseini A. Synaptic imbalance, stereotypies, and impaired social interactions in mice with altered neuroligin 2 expression. J Neurosci. 2008;28:6055–6067. doi: 10.1523/JNEUROSCI.0032-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert PO, Dow LE, Russell SM. The Scribble and Par complexes in polarity and migration: friends or foes? Trends Cell Biol. 2006;16:622–630. doi: 10.1016/j.tcb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Frahm J, Fischer J, Bourgeron T, Ehrenreich H, Brose N. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci U S A. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- Lahuna O, Quellari M, Achard C, Nola S, Méduri G, Navarro C, Vitale N, Borg JP, Misrahi M. Thyrotropin receptor trafficking relies on the hScrib-betaPIX-GIT1-ARF6 pathway. EMBO J. 2005;24:1364–1374. doi: 10.1038/sj.emboj.7600616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Mata I, Perez-Iglesias R, Roiz-Santiañez R, Tordesillas-Gutierrez D, Gonzalez-Mandly A, Vazquez-Barquero JL, Crespo-Facorro B. A neuregulin 1 variant is associated with increased lateral ventricle volume in patients with first-episode schizophrenia. Biol Psychiatry. 2009;65:535–540. doi: 10.1016/j.biopsych.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. Identification of Vangl2 and Scrib1 as planar polarity genes in mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Crenshaw EB, 3rd, Kelley MW. Noncanonical Wnt signaling and neural polarity. Annu Rev Neurosci. 2006a;29:363–386. doi: 10.1146/annurev.neuro.29.051605.112933. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Sans N, Huss D, Kach J, Dickman JD, Forge A, Rachel RA, Copeland NG, Jenkins NA, Bogani D, Murdoch J, Warchol ME, Wenthold RJ, Kelley MW. Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J Neurosci. 2006b;26:5265–5275. doi: 10.1523/JNEUROSCI.4680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Murdoch JN, Henderson DJ, Doudney K, Gaston-Massuet C, Phillips HM, Paternotte C, Arkell R, Stanier P, Copp AJ. Disruption of scribble (Scrib1) causes severe neural tube defects in the circletail mouse. Hum Mol Genet. 2003;12:87–98. doi: 10.1093/hmg/ddg014. [DOI] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, Crawley JN. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Navarro C, Nola S, Audebert S, Santoni MJ, Arsanto JP, Ginestier C, Marchetto S, Jacquemier J, Isnardon D, Le Bivic A, Birnbaum D, Borg JP. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 2005;24:4330–4339. doi: 10.1038/sj.onc.1208632. [DOI] [PubMed] [Google Scholar]
- Nola S, Sebbagh M, Marchetto S, Osmani N, Nourry C, Audebert S, Navarro C, Rachel R, Montcouquiol M, Sans N, Etienne-Manneville S, Borg JP, Santoni MJ. Scrib regulates PAK activity during the cell migration process. Hum Mol Genet. 2008;17:3552–3565. doi: 10.1093/hmg/ddn248. [DOI] [PubMed] [Google Scholar]
- Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel RA, Murdoch JN, Beermann F, Copp AJ, Mason CA. Retinal axon misrouting at the optic chiasm in mice with neural tube closure defects. Genesis. 2000;27:32–47. doi: 10.1002/1526-968x(200005)27:1<32::aid-gene50>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Richier L, Williton K, Clattenburg L, Colwill K, O'Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, Krueger SR, Pawson T, Fawcett JP. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci. 2010;30:4796–4805. doi: 10.1523/JNEUROSCI.3726-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ME. Redistribution of synaptic AMPA receptors at glutamatergic synapses in the dorsal cochlear nucleus as an early response to cochlear ablation in rats. Hear Res. 2006;216–217:154–167. doi: 10.1016/j.heares.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Sala C, Cambianica I, Rossi F. Molecular mechanisms of dendritic spine development and maintenance. Acta Neurobiol Exp (Wars) 2008;68:289–304. doi: 10.55782/ane-2008-1696. [DOI] [PubMed] [Google Scholar]
- Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron. 2008;57:94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol. 2003;5:520–530. doi: 10.1038/ncb990. [DOI] [PubMed] [Google Scholar]
- Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- Sfakianos MK, Eisman A, Gourley SL, Bradley WD, Scheetz AJ, Settleman J, Taylor JR, Greer CA, Williamson A, Koleske AJ. Inhibition of Rho via Arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J Neurosci. 2007;27:10982–10992. doi: 10.1523/JNEUROSCI.0793-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi SY, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci. 2007;10:963–969. doi: 10.1038/nn1933. [DOI] [PubMed] [Google Scholar]
- Sun Y, Aiga M, Yoshida E, Humbert PO, Bamji SX. Scribble interacts with beta-catenin to localize synaptic vesicles to synapses. Mol Biol Cell. 2009;20:3390–3400. doi: 10.1091/mbc.E08-12-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol Cell Neurosci. 2004;26:429–440. doi: 10.1016/j.mcn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol. 2006;172:759–769. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Bar I, Jossin Y, De Backer O, Goffinet AM. Protocadherin Celsr3 is crucial in axonal tract development. Nat Neurosci. 2005;8:451–457. doi: 10.1038/nn1428. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nathans J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 2007;134:647–658. doi: 10.1242/dev.02772. [DOI] [PubMed] [Google Scholar]
- Wenthold RJ, Prybylowski K, Standley S, Sans N, Petralia RS. Trafficking of NMDA receptors. Annu Rev Pharmacol Toxicol. 2003;43:335–358. doi: 10.1146/annurev.pharmtox.43.100901.135803. [DOI] [PubMed] [Google Scholar]
- Wu S, Yue W, Jia M, Ruan Y, Lu T, Gong X, Shuang M, Liu J, Yang X, Zhang D. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:492–495. doi: 10.1002/ajmg.b.30495. [DOI] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Bar I, Achouri Y, Campbell K, De Backer O, Hebert JM, Jones K, Kessaris N, de Rouvroit CL, O'Leary D, Richardson WD, Goffinet AM, Tissir F. Early forebrain wiring: genetic dissection using conditional Celsr3 mutant mice. Science. 2008;320:946–949. doi: 10.1126/science.1155244. [DOI] [PMC free article] [PubMed] [Google Scholar]