Abstract
The marsupial inactive X chromosome expresses a long noncoding RNA (lncRNA) called Rsx that has been proposed to be the functional analog of eutherian Xist. Despite the possibility that Xist and Rsx encode related functions, the two lncRNAs harbor no linear sequence similarity. However, both lncRNAs harbor domains of tandemly repeated sequence. In Xist, these repeat domains are known to be critical for function. Using k-mer based comparison, we show that the repeat domains of Xist and Rsx unexpectedly partition into two major clusters that each harbor substantial levels of nonlinear sequence similarity. Xist Repeats B, C, and D were most similar to each other and to Rsx Repeat 1, whereas Xist Repeats A and E were most similar to each other and to Rsx Repeats 2, 3, and 4. Similarities at the level of k-mers corresponded to domain-specific enrichment of protein-binding motifs. Within individual domains, protein-binding motifs were often enriched to extreme levels. Our data support the hypothesis that Xist and Rsx encode similar functions through different spatial arrangements of functionally analogous protein-binding domains. We propose that the two clusters of repeat domains in Xist and Rsx function in part to cooperatively recruit PRC1 and PRC2 to chromatin. The physical manner in which these domains engage with protein cofactors may be just as critical to the function of the domains as the protein cofactors themselves. The general approaches we outline in this report should prove useful in the study of any set of RNAs.
Keywords: Polycomb, epigenetics, Xist, Rsx, lncRNAs
INTRODUCTION
The sex chromosomes of therian (eutherian and metatherian) mammals evolved from a pair of identical autosomes after the split of therian and monotreme mammals from their most recent common ancestor. Since that divergence, the Y chromosome has lost the large majority of its protein coding genes, creating a gene dosage imbalance between XY males and XX females. Part of the system that compensates for this imbalance is a process known as X-chromosome inactivation (XCI). Initiated early during female development, XCI results in the transcriptional silencing of one X chromosome in each somatic cell in female mammals. In eutherians, XCI is mediated by a long noncoding RNA (lncRNA) called Xist (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018).
The silencing function of Xist is thought to be mediated by the concerted action of several domains of tandemly repeated sequence that are interspersed throughout its length. These repeat domains harbor binding sites for distinct subsets of proteins that, through incompletely understood mechanisms, help Xist achieve different aspects of its function. “Repeat A” has been proposed to bind a number of proteins, including Polycomb repressive complex 2 (PRC2), SPEN, and RBM15, and is required for the stabilization of spliced Xist RNA, and for Xist to silence actively transcribed regions of the X chromosome (Wutz et al. 2002; Zhao et al. 2008; Hoki et al. 2009; Royce-Tolland et al. 2010; Engreitz et al. 2013; Cifuentes-Rojas et al. 2014; Chu et al. 2015; McHugh et al. 2015; Moindrot et al. 2015; Monfort et al. 2015; Patil et al. 2016). “Repeat B,” and at least a portion of “Repeat C,” bind HNRNPK to recruit the Polycomb repressive complex 1 (PRC1) to the inactive X chromosome (Almeida et al. 2017; Pintacuda et al. 2017; Colognori et al. 2019). “Repeat E” binds many proteins, including CIZ1, and is required for the stable association of Xist with X-linked chromatin and for the sustained recruitment of Polycomb repressive complex 2 (PRC2) to the inactive X (Smola et al. 2016; Ridings-Figueroa et al. 2017; Sunwoo et al. 2017).
Intriguingly, metatherians (marsupials) may have convergently evolved their own lncRNA, Rsx, to mediate XCI in XX females (Grant et al. 2012). Rsx shares no linear sequence similarity with Xist and is located in a different syntenic block on the marsupial X. Nevertheless, Rsx shares a number of surprising similarities with Xist. Both Xist and Rsx are expressed exclusively from the inactive X in females and are retained in the nucleus, forming what has been described as a “cloud-like” structure around their chromosome of synthesis. Moreover, both lncRNAs are spliced yet unusually long in their final processed form, and their expression correlates with the accumulation of histone modifications deposited by the PRCs on the inactive X (Grant et al. 2012; Wang et al. 2014).
Studies performed over the last three decades indicate that Xist is required for normal XCI in eutherians (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018). Given the similarities between Rsx and Xist, it has been proposed that the marsupial Rsx is the functional analog of Xist (Grant et al. 2012). While this hypothesis has yet to be directly tested, expression of an Rsx transgene on a mouse autosome does, to a certain extent, induce local gene silencing, supporting the notion that Rsx harbors Xist-like function (Grant et al. 2012).
Despite their lack of linear sequence similarity, Xist and Rsx both harbor long, internal domains of tandemly repeated sequence (Grant et al. 2012; Johnson et al. 2018). We recently discovered that evolutionarily unrelated lncRNAs that encode similar functions often harbor nonlinear sequence similarity in the form of k-mer content, where a k-mer is defined as all possible combinations of a nucleotide substring of a given length k (Kirk et al. 2018). Below, we describe our use of k-mer based methods to investigate the possibility that the repeat domains in Xist and Rsx harbor nonlinear sequence similarity that might be suggestive of shared function.
RESULTS
Lack of linear sequence similarity between repeat domains in Xist and Rsx
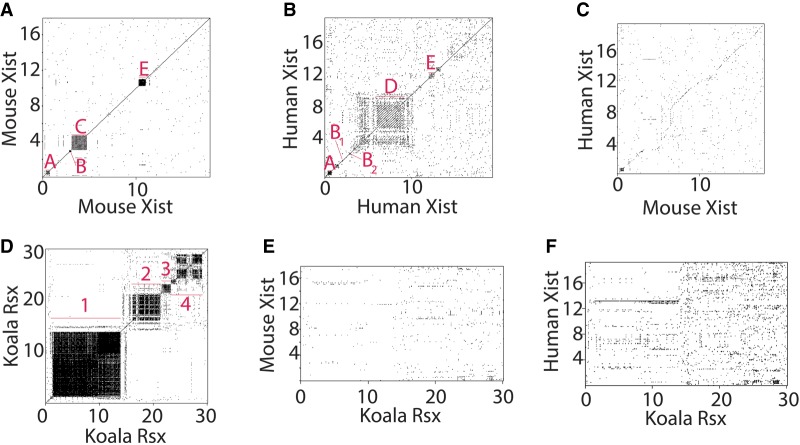
Xist and Rsx are both notable for their domains of highly repetitive sequence, which can be identified by aligning each lncRNA to itself and visualizing the alignment data as a dot plot (Supplemental File S1; Rice et al. 2000). In mouse Xist, the four major repetitive regions are referred to as Repeats A, B, C, and E (Fig. 1A; Brockdorff et al. 1992). Repeats A, B, and E are conserved in eutherian mammals, whereas Repeat C appears to be specific to murid rodents (Fig. 1C; Nesterova et al. 2001; Yen et al. 2007). In human Xist, the four major repetitive regions are referred to as Repeats A, B, D, and E (Fig. 1B; Brown et al. 1992). Relative to mouse, human Repeat B is comprised of two shorter Repeat B-like regions that appear to have been disrupted by insertion (Fig. 1B; Nesterova et al. 2001; Yen et al. 2007). Human Repeat D is comprised of eight core repeats flanked by several additional repeats that exhibit partial similarity to its core (Fig. 1B; Brown et al. 1992; Nesterova et al. 2001; Yen et al. 2007). While Repeat D is absent in murid rodents (Fig. 1C), Repeat D-like sequence appears in many other mammals (Supplemental Fig. S1; Supplemental File S2; Nesterova et al. 2001; Yen et al. 2007).
FIGURE 1.
Lack of linear sequence similarity between repeat domains in Xist and Rsx. (A–F) Dot plots comparing mouse and human Xist and Rsx to themselves and to each other. The location of repeat domains in all three lncRNA-to-self plots are marked with red bars and names/numbers.
In contrast to Xist, which is mostly comprised of nonrepetitive sequence, nearly all of the sequence in Rsx can be assigned to one of four repetitive domains (Fig. 1D; Johnson et al. 2018). Here, we refer to the repetitive domains in Rsx as Repeats 1 through 4. It has been suggested that Rsx Repeat 1 is functionally analogous to Xist Repeat A, because both repeats are the first to occur in each lncRNA, and because both repeats contain GC-rich elements (Grant et al. 2012; Johnson et al. 2018). Beyond this observation, little is known about the repetitive regions in Rsx and how they might relate to those in Xist. Hypothesizing that the repeat domains in Xist and Rsx recruit similar subsets of proteins, we expected that dot plots comparing the sequence of Xist to the sequence of Rsx would reveal regions of sequence similarity. However, this was not the case (Fig. 1E,F), nor was significant similarity between Xist and Rsx detected using BLASTN or the hidden-Markov based nhmmer (Altschul et al. 1990; Wheeler and Eddy 2013).
Nonlinear similarity between repeat domains in Xist and Rsx
We hypothesized that sequence similarity between Xist and Rsx might become apparent using an algorithm we recently developed to detect sequence similarity between evolutionarily unrelated lncRNAs (Kirk et al. 2018). In our algorithm, called SEEKR (Sequence evaluation through k-mer representation), groups of lncRNAs are compared to each other by counting the number of occurrences of each k-mer of a given length k in each lncRNA, then normalizing k-mer counts by the length of the lncRNA in question, and finally calculating a z-score for each k-mer in each lncRNA. The list of z-scores for each k-mer in a lncRNA is referred to as its “k-mer profile” and represents the abundance of each k-mer in the lncRNA relative to the abundance of each k-mer in the other lncRNAs that were analyzed as part of the group. In SEEKR, k-mer profiles from lncRNAs of interest are compared to each other using Pearson's correlation. We previously demonstrated that SEEKR can be used to quantify the similarity between any number of lncRNAs regardless of their evolutionary relationships or differences in their lengths, and that similarities in k-mer profiles correlated with lncRNA protein binding potential, subcellular localization, and Xist-like repressive activity. A major strength of SEEKR is that it ignores positional information in similarity calculations, allowing it to quantify nonlinear sequence relationships (Kirk et al. 2018).
In order to compare Xist and Rsx via SEEKR, we calculated the k-mer profile at k = 4 of individual repeat domains in mouse Xist and koala Rsx, using all mouse lncRNAs from GENCODE as a background set to derive the mean and standard deviation of the counts for each k-mer (Derrien et al. 2012). The mechanisms through which Xist functions have been most extensively studied in mouse (Sahakyan et al. 2018). For this reason, we primarily used the repetitive regions from mouse Xist as search features in this work. However, because of the conservation of Repeat D-like domains in nonmurid eutherian mammals (Supplemental Fig. S1; Nesterova et al. 2001; Yen et al. 2007), we also included the sequence of human Xist Repeat D in our analyses. We used the sequence from koala Rsx as our exemplar, owing to the high quality of the koala genome build relative to builds from other marsupials (Johnson et al. 2018).
In our previous work, we found that SEEKR performed best when the length of the lncRNA or lncRNA fragment being studied was similar to 4k, i.e., the total number of possible k-mers at k-mer length k. In tests of Xist-like repressive activity, we found that comparisons of lncRNAs using k-mer lengths of k ≥ 7 underperformed relative to comparisons using smaller k-mer lengths, owing to the fact that most annotated lncRNAs are much less than 47 (16,384) nucleotides long, and k-mer profiles of individual lncRNAs at k ≥ 7 (≥16,384 possible k-mers) are dominated by “0” values (Kirk et al. 2018). Based on this observation, and because Repeats A and B, two essential repetitive regions within Xist (Wutz et al. 2002; Hoki et al. 2009; Royce-Tolland et al. 2010; Almeida et al. 2017; Pintacuda et al. 2017), are each about 44 (256) nucleotides in length, we reasoned that k-mer profiles at k = 4 (44 = 256 possible k-mers) would provide a reasonable estimate of sequence complexity for the repeats without being dominated by “0” values.
We also noted that relative to most lncRNAs, k-mer content in the repetitive regions of Xist and Rsx was skewed (Supplemental Fig. S2A). We therefore elected to log2-transform z-scores in k-mer profiles prior to comparison via Pearson's correlation, recognizing that this transformation would reduce skew and allow us to evaluate similarity in the context of a log-linear scale (Supplemental Fig. S2B).
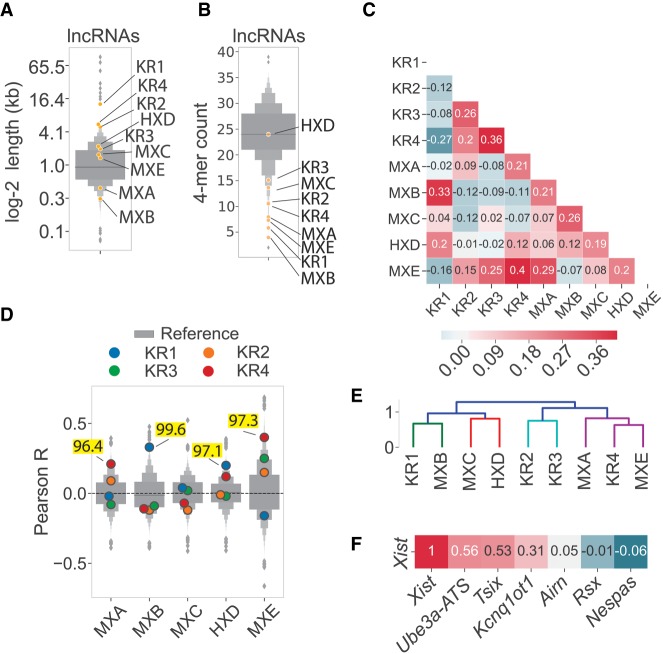
The individual repeat domains in Xist and Rsx vary substantially in terms of their length and sequence complexity. Xist repeats tend to be shorter and lower in overall complexity than repeats in Rsx (Fig. 2A,B). Despite these differences, using SEEKR, we identified substantial levels of similarity between the repeat domains of Xist and Rsx. The Repeat A region of Xist was most similar to Rsx Repeat 4, exhibited a weak positive correlation with Repeat 2, and had negative correlations with Rsx Repeats 1 and 3 (Pearson's r of 0.21, for Repeat A vs. Repeat 4, respectively, and r of −0.02, 0.09, and −0.08 for Repeats 1, 2, and 3, respectively; Fig. 2C). In contrast, Xist Repeat B was most similar to Rsx Repeat 1 and had negative correlations with Rsx Repeats 2 through 4 (Pearson's r of 0.33 for Repeat B vs. Repeat 1; Fig. 2C). Repeat C, which is specific to murid rodents (i.e., it is not found in other eutherians), had no appreciable correlation with any Rsx repeat, whereas human Repeat D had positive correlations with Rsx Repeat 1 and 4 (r of 0.20, 0.12, respectively; Fig. 2C). The k-mer profile of Xist Repeat E had positive correlations that increased progressively in Rsx Repeats 2, 3, and 4 (Pearson's r of 0.15, 0.25, 0.40, respectively; Fig. 2C).
FIGURE 2.
Nonlinear similarity between repeat domains in Xist and Rsx. (A) Length of Xist and Rsx repeat domains and (B) sequence complexity estimated by the number of unique 4-mers that constitute 25% of the total 4-mer counts in a transcript, each relative to all other GENCODE M18 lncRNA transcripts. For panels A–E: “M,” “H,” and “K” signify mouse, human, and koala, respectively, “X” and “R” signify Xist and Rsx, respectively, and the final letter or number in each abbreviation signifies the repeat domain in question. (C) Correlation matrix displaying the Pearson's r value derived from comparing k-mer profiles at k = 4 of each of the four repeat domains in koala Rsx (Repeats 1–4), the major repeats in mouse Xist (Repeats A,B,C,E), and human Xist Repeat D. The set of mouse lncRNAs from GENCODE was used to derive mean and standard deviation values for length-normalized abundance of each k-mer. (D) Similarity of repeat domains in Xist and Rsx relative to all lncRNA transcripts in the mouse GENCODE M18 database (Derrien et al. 2012). Each subplot shows the distribution of Pearson's r values describing the similarity between the Xist repeat in question and the set of GENCODE lncRNA transcripts. Similarities between Xist and Rsx that are above the 95th percentile of similarity for all mouse lncRNAs are highlighted in yellow. (E) The correlation matrix in A subject to hierarchical clustering. Colors represent clusters for all descendent links beneath the first node in the dendrogram with distance <70% of the largest distance between all clusters. (F) SEEKR-derived similarity (in the form of Pearson's r; Kirk et al. 2018) between full-length Xist, other cis-repressive lncRNAs in mouse, and koala Rsx (Johnson et al. 2018).
We sought to quantify the strength of the similarity between repeat domains in Xist and Rsx relative to other mouse lncRNAs. To do this, we used Pearson's correlation to compare the k-mer profile of each Xist repeat domain to the k-mer profiles of the set of spliced GENCODE M18 mouse lncRNAs (Derrien et al. 2012). We compared this distribution of Pearson's r values to the r value obtained when comparing each Xist repeat to each Rsx repeat.
This analysis revealed striking similarities between the repeat domains of Xist and Rsx. Xist Repeat B was more similar to Rsx Repeat 1 than it was similar to 99.6% of all lncRNAs (similarity ranked 65th out of 17,523 comparisons), despite the fact that the two repeats differ in length by ∼50-fold (Fig. 2A,D; Supplemental Tables S1, S2). Xist Repeat A was more similar to Rsx Repeat 4 than it was similar to 96.4% of all other lncRNAs (its similarity ranked 626th out of 17,523 comparisons), Xist Repeat D was more similar to Rsx Repeat 1 than it was similar to 97.1% of all other lncRNAs (its similarity ranked 515th out of 17,523 comparisons), and Xist Repeat E was more similar to Rsx Repeat 4 than it was similar to 97.3% of all other lncRNAs (its similarity ranked 467th out of 17,523 comparisons; Fig. 2D; Supplemental Tables S1, S2). No other repeat domains in Xist and Rsx fell above the 95th percentile in terms of their similarity to each other. Similar trends were observed when we used k-mer lengths k = 4, 5, and 6 for this analysis (Supplemental Fig. S3A).
Current models suggest that the tandem repeats in Xist have distinct functions (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018). Thus, we were surprised to find that the repeat domains within Xist also exhibited high levels of similarity to each other (Supplemental Tables S1, S2). Repeat A was more similar to Repeat E than it was similar to 99.6% of all lncRNAs. Likewise, Repeats B and C were more similar to each other than they were similar to 97.8% and 99.6% of all other lncRNAs, respectively. Finally, Repeats C and D were more similar to each other than they were similar to 97.0% and 96.2% of all other lncRNAs, respectively (Supplemental Tables S1, S2).
The similarities between specific domains of Xist and Rsx were also evident in an unsupervised hierarchical cluster of the matrix from Figure 2C. Xist Repeat B and Rsx Repeat 1 formed a basal cluster which joined with a second basal cluster comprising Xist Repeat C and Xist Repeat D. Rsx Repeat 4 and Xist Repeat E formed a basal cluster that joined with Xist Repeat A. This multilevel cluster (Rsx Repeat 4, Xist Repeat E, and Xist Repeat A) joined with another basal cluster comprising Rsx Repeats 2 and 3 (Fig. 2E).
At k-mer length k = 4, Pearson's correlation with and without log-transformation of k-mer z-scores, as well as Spearman's correlation of nontransformed z-scores, detected similar relationships between Xist and Rsx repeat domains (Supplemental Fig. S4). While the similarities between individual domains were still evident at higher k-mer lengths, particularly when using Pearson's correlation of log-transformed k-mer counts, the clustering patterns that we observed at k-mer length k = 4 began to dissolve (Supplemental Fig. S4). At high k-mer lengths, Spearman's correlation was the least informative method of comparison, owing to the large number of “zero” values that populate k-mer profiles at these lengths (Supplemental Fig. S4). Thus, to a certain extent, the similarities in the repeat domains of Xist and Rsx are detectable regardless of prior assumptions about log-linear, linear, and monotonic relationships between k-mer profiles. However, the most robust similarities are detected using Pearson's correlation of log-transformed k-mer counts (Supplemental Fig. S4).
We observed that the similarities between Xist and Rsx were obscured when the k-mer profiles of the full-length lncRNAs were compared to each other (Pearson's r of −0.01 for the comparison of full-length Xist to full-length Rsx; Fig. 2F). This loss of similarity highlights the utility of domain-based similarity searches, particularly for lncRNAs whose functional domains may comprise a fraction of their overall length. The dissimilarity between k-mer profiles of full-length Xist and full-length Rsx likely stems from the fact that virtually all of Rsx is comprised of repetitive sequence domains that harbor limited k-mer diversity relative to the nonrepetitive sequence of Xist (Fig. 2B and compare Fig. 1A–C to 1D).
Sequence properties of Xist and Rsx repeat domains
Qualitative similarities between Xist and Rsx repeat domains were also revealed using MEME to visualize motifs that were enriched within individual domains (Bailey et al. 2009). In Xist Repeat A, MEME identified one motif comprised of short runs of G and C nucleotides and one motif most notable for runs of T nucleotides (Fig. 3A). Similar patterns were seen in the motifs enriched in Rsx Repeat 4 (Fig. 3B). The single motif from Repeat B was almost exclusively comprised of two tandemly arranged “GCCCC” motifs, and motifs containing runs of “G” and “C” nucleotides could be seen in Rsx Repeat 1 (Fig. 3A,B). The pyrimidine-rich runs that were characteristic of Xist Repeat E were also observed in Rsx Repeat 4 (Fig. 3A,B). Rsx Repeat 2 was unique in its enrichment of AAAG and GAAA motifs (Fig. 3B).
FIGURE 3.
Motifs enriched in Xist and Rsx repeat domains. (A–C) The top three de novo motifs identified by MEME in Xist repeats (panel A: all repeats from mouse Xist except for Repeat D, which is the human sequence), and in the four repeats in koala (B) and opossum (C) Rsx. The length in nucleotides of each repeat is shown in parentheses below the repeat name. The number of matches to each motif, as well as the expectation value for that number, is shown below each motif logo. Some repeats had less than three motifs detected by MEME.
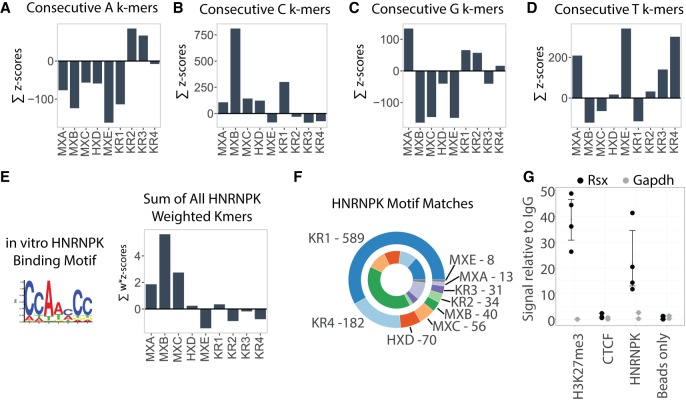
Several of the repeat domains in Xist and Rsx could be distinguished by the presence of k-mers comprised of runs of individual nucleotides that extended for two or more consecutive positions (such as AA, CC, GG, or TT; Supplemental File S1). Similar to enriched motifs, k-mers containing mononucleotide runs may function to recruit different subsets of RNA-binding proteins (Ray et al. 2013; Dominguez et al. 2018). We therefore sought to quantify the enrichment of k-mers containing mononucleotide runs in the repeat domains of Xist and Rsx, reasoning that this analysis might provide insight into function.
Similar to what we observed in our motif analysis (Fig. 3), Rsx Repeat 2 had the highest length-normalized abundance of poly(A) k-mers, followed closely by Rsx Repeat 3 (Fig. 4A). Repeat B, which is only ∼250 nucleotides (nt) long and is almost entirely comprised of poly(C) sequence, had the highest length-normalized abundance of poly(C) k-mers, followed by Rsx Repeat 1, and Xist Repeats C, D, and A (Fig. 4B). Mouse Repeat A had the highest length-normalized abundance of poly(G) k-mers, followed by Rsx Repeats 1, 2, and 4 (Fig. 4C). Xist Repeats A and E, as well as Rsx Repeats 3 and 4 had the highest length-normalized abundance of poly(T) k-mers, reflecting the high degree of SEEKR-detected similarity between these regions (Fig. 4D). Similar trends were detected when we used k-mer lengths k = 4, 5, and 6 for this analysis (Supplemental Fig. S3B). Thus, certain Xist and Rsx repeat domains share similarity in their overall k-mer profiles (Fig. 2) in their enriched motifs (Fig. 3), and in their enrichment in subsets of low-complexity k-mers that are comprised of mononucleotide runs (Fig. 4). The repeat domains also harbor differences in sequence composition that are consistent with their lack of alignment via methods designed to detect linear sequence similarity (Fig. 1).
FIGURE 4.
Mononucleotide runs and HNRNPK-binding motifs enriched in specific Xist and Rsx repeats. (A–D) The sum of z-scores in each repeat for k-mers containing consecutive (A) A, (B) C, (C) G, and (D) T nucleotides. For this analysis we defined “consecutive” as at least two consecutive nucleotides of the specified identity and used k-mer length k = 5 (Materials and Methods). Repeat abbreviations as in Figure 2. (E) The sum of z-scores for all k-mers in each Xist and Rsx repeat domain after weighting the k-mers by the likelihood with which they fit the consensus HNRNPK-binding motif. Motif logo that describes the consensus HNRNPK-binding motif obtained from Ray et al. (2013) is also shown. (F) Component arcs of outer circle indicate the proportion and number of HNRNPK-binding motif matches detected by FIMO (P < 0.01) in each Xist and Rsx repeat domain. Component arcs of inner circle indicate the proportion of motif matches in each repeat domain normalized for domain length. Repeat abbreviations as in Figure 2. (G) Rsx enrichment relative to IgG control after RNA IP-qPCR in cultured fibroblasts from female M. domestica. For each antibody, left (black) is enrichment of Rsx, right (gray) is enrichment of Gapdh. The histone modification H3K27me3 is enriched on the inactive X in marsupials, so an association with Rsx was expected. CTCF has nanomolar affinity for RNA and along with “bead only”/no-antibody IP serves as a negative control demonstrating IP specificity (Kung et al. 2015). Dots represent values from replicate RNA IP experiments; error bars represent bootstrap 95% CI.
HNRNPK-binding motifs are enriched in specific Xist and Rsx repeats
Xist Repeat B is known to bind a protein called HNRNPK, and this binding activity is essential for Xist to recruit PRC1 to the inactive X chromosome (Pintacuda et al. 2017). Given the quantitative and qualitative sequence similarities between Xist Repeat B and Rsx Repeat 1 (Figs. 2–4), we sought to compare HNRNPK-binding potential between the two repeats using two conceptually distinct approaches. First, we weighted z-scores of individual k-mers in all Xist and Rsx repeat domains by the probability that the k-mer would occur in the position-weight-matrix (PWM) describing the HNRNPK-binding motif from Ray et al. (2013) (see PWM in Fig. 4E). We then summed HNRNPK-scaled z-scores over each repeat, and plotted the results in a manner similar to Figure 4A–D. In this analysis, a positive sum indicates that k-mers matching the HNRNPK PWM occur more frequently in the domain in question than they occur in other lncRNAs in the GENCODE database.
On a length-normalized basis, Xist Repeats B, C, A, and D, in descending order, had positive sums of HNRNPK-scaled z-scores. Repeat 1 was the only repeat in Rsx to have a positive sum, perhaps consistent with a role in recruiting HNRNPK to Rsx (Fig. 4E). The sum of HNRNPK-scaled z-scores in Rsx Repeat 1 was lower than the sums in Xist Repeats B, C, and A (Fig. 4E), which might be taken as evidence that on a length-normalized basis, Xist Repeats B, C, and A have a higher density of k-mers that are likely to bind HNRNPK than Rsx Repeat 1 or any other Rsx repeat. However, at 13 kb in length, Rsx Repeat 1 is ∼50 times longer than Xist Repeat B, and is over half the length of full-length Xist itself (Brockdorff et al. 1992; Brown et al. 1992; Johnson et al. 2018). Thus, we also counted the absolute number of matches to HNRNPK-binding motifs in Xist and Rsx repeats. Rsx Repeat 1 had 15 times more matches to HNRNPK-binding motifs than did Xist Repeat B (589 matches in Repeat 1 compared to 40 matches in Repeat B; Fig. 4F; Bailey et al. 2009). Rsx Repeat 4 also had a large number of matches to HNRNPK-binding motifs (182 matches), and human Repeat D and mouse Repeat C each had more HNRNPK-binding sites than Repeat B (70 and 56 matches, respectively, compared to 40 in Repeat B; Fig. 4F). CLIP performed in mouse and human cells supports a direct association between HNRNPK and Repeat C and Repeat D, respectively (Supplemental Fig. S5; Cirillo et al. 2016; Van Nostrand et al. 2016). Collectively, these data support the ideas that mouse Repeat C and human Repeat D cooperate with Repeat B in recruiting HNRNPK to Xist, and suggest that Rsx Repeat 1, and to a lesser extent, Rsx Repeat 4, could also recruit HNRNPK to Rsx.
We next used RNA immunoprecipitation (RNA IP) followed by RT-qPCR to determine whether we could detect evidence of HNRNPK association with Rsx. In fibroblast cells derived from a female gray short-tailed opossum, Monodelphis domestica, we found that HNRNPK IP enriched for Rsx 20-fold over IgG control IPs (Fig. 4G). This enrichment was similar to that seen for an IP using an antibody that detects histone H3-lysine27-trimethylation (H3K27me3), a modification known to be enriched on the opossum inactive X (Fig. 4G; Wang et al. 2014). Gapdh mRNA was not enriched by IP of HNRNPK or H3K27me3 (Fig. 4G). IP of CTCF, a protein that binds RNA with nanomolar affinity in a sequence nonspecific manner, showed neither Rsx nor Gapdh enrichment (Fig. 4G; Kung et al. 2015). Leaving out HNRNPK antibody prior to performing IP and qPCR also led to a loss of Rsx signal (“beads only” in Fig. 4G). DNase-treated input RNA (no reverse transcription control) did not yield signal in qPCR assays, indicating DNase digestion prior to cDNA synthesis and qPCR proceeded to completion (not shown). These data support our computational analyses and suggest that HNRNPK associates with Rsx in marsupial cells.
Conservation of repeat domains between koala and opossum Rsx
Considering that not all of the repeat domains in Xist exhibit conservation across eutherian mammals, we sought to determine whether or not the repeat domains in koala Rsx were conserved in another marsupial. Rsx was originally identified in opossum (Grant et al. 2012), but the most current assembly of the opossum genome (mondom5; Casper et al. 2018) harbors significant gaps within the sequence of Rsx.
To assemble a complete sequence of opossum Rsx for comparison to koala, we used Oxford Nanopore technology to sequence two bacterial artificial chromosomes (BACs) that encompassed the opossum Rsx locus (VMRC18-839J22 and VMRC18-303M7). De novo assembly and polishing of sequence reads identified a single 235,139 base contig aligning to chrX that had on average a 0.5% error rate with the mondom5 assembly (Supplemental File S3). Our assembly filled in 16,620 bases of unannotated sequence in the Rsx locus, 361 bases of which were a part of the spliced Rsx lncRNA annotation from Supplemental Table S3 and Grant et al. (2012).
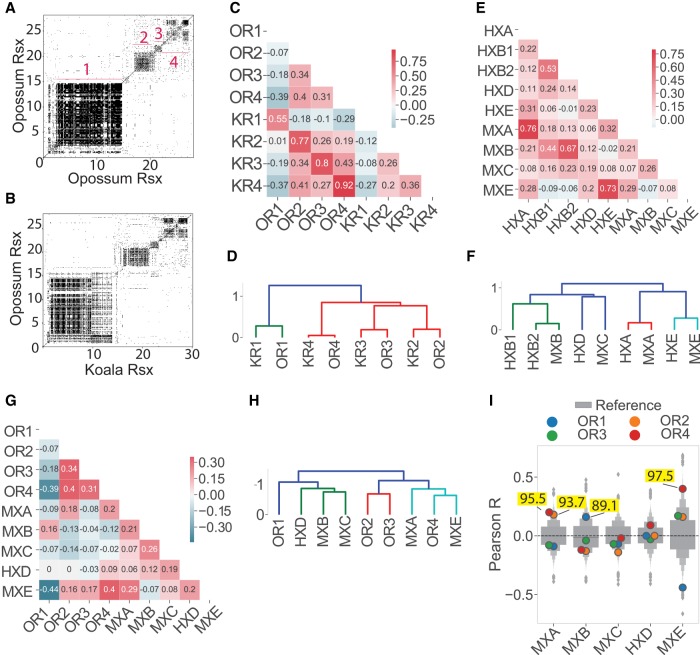
Alignment of this ungapped assembly of spliced opossum Rsx to koala Rsx revealed high levels of similarity between their repeat domains in a dot plot analysis (Fig. 5A,B). This similarity could also be seen at the level of k-mers (Fig. 5C,D), and by extraction of enriched motifs using MEME (Fig. 3B,C). Opossum and koala, which are members of distantly related American and Australian marsupial families, respectively, diverged approximately 82 million years ago (Kumar et al. 2017). By comparison, mouse and human are separated by approximately 90 million years of evolution (Kumar et al. 2017). Repeat domains 1 through 4 in opossum and koala Rsx exhibited levels of sequence similarity that approximated or exceeded the similarity found between the repeat domains in mouse and human Xist (with the exception of Repeat C/Repeat D; Fig. 5C–F). Thus, the repeat domains in Rsx appear to be at least as conserved between distantly related marsupials as the repeat domains in Xist are conserved among eutherians.
FIGURE 5.
Rsx repeat domains are conserved between koala and opossum. (A,B) Dot plots of opossum Rsx aligned to (A) itself or (B) koala Rsx. (C) Similarity between repeat domains in koala and opossum Rsx as calculated in Figure 2C. (D) Hierarchical cluster of similarity values from C. (E) Similarity between repeat domains in mouse and human Xist as calculated in Figure 2C. (F) Hierarchical clustering of similarity values from E. (G) Similarity between repeat domains in opossum Rsx and Xist repeat domains as calculated in Figure 2C. (H) Hierarchical cluster of similarity values from G. (I) Percentiles for Pearson's R for opossum Rsx repeat domains compared to each Xist repeat domain as in Figure 2D. Numbers mentioned in the body of the manuscript are highlighted in yellow.
Next, we compared the k-mer contents of repeat domains in Xist to the k-mer contents of repeat domains in opossum Rsx. We identified a level of similarity (Fig. 5G–I) that mirrored the similarity we found between repeat domains in Xist and koala Rsx (Fig. 2B,D,E). Xist Repeat A was most similar to opossum Repeats 2 and 4 (93.7th and 95.5th percentile relative to all other mouse lncRNAs, respectively); Xist Repeat B was most similar to opossum Repeat 1 (89.1st percentile relative to all other lncRNAs); and Xist Repeat E was most similar to opossum Repeat 4 (97.5th percentile relative to all other lncRNAs; Fig. 5I). Thus, the major repeat domains in Rsx are conserved between opossum and koala, and the repeat domains in Rsx from both marsupials harbor k-mer contents similar to those in repeat domains from mouse and human Xist.
Multiple protein-binding motifs are enriched to extreme levels in Xist and Rsx repeat domains
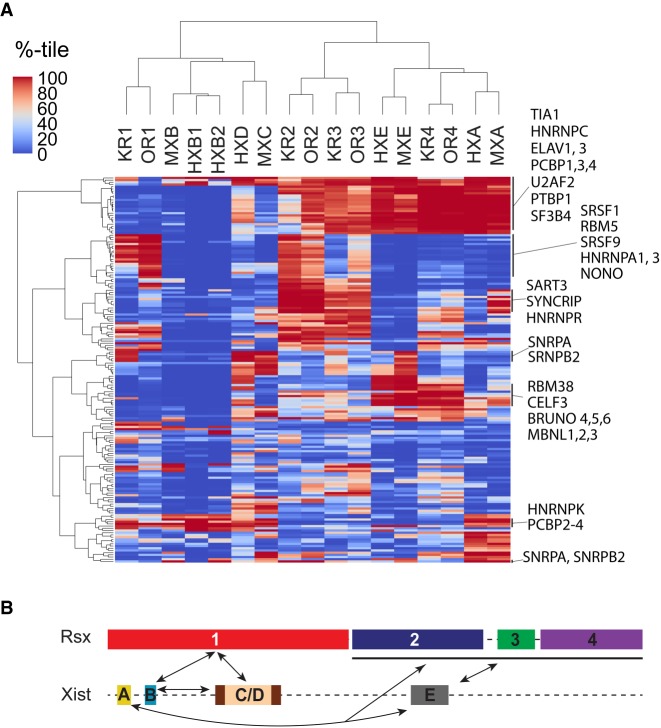
We examined the extent to which Xist and Rsx repeat domains were enriched for sequence motifs known to recruit RNA-binding proteins, hypothesizing that the patterns of enrichment might provide additional insight into similarities between the two lncRNAs. For this analysis, we downloaded PWMs for all mammalian RNA-binding proteins available in the CISBP-RNA database (Ray et al. 2013), and for each PWM in each repeat, we quantified enrichment by weighting k-mer z-scores by the probability that the k-mer matched the PWM, then calculating the sum of those weights, as we did for the HNRNPK PWM in Figure 4E. To gauge the extent of enrichment relative to other mouse lncRNAs, we determined the percentile rank of the sum for each PWM in each repeat relative to the sums generated from the same PWM-weighting procedure performed on all mouse lncRNAs. We then hierarchically clustered repeat domains from Xist and Rsx based on the percentile ranks of motif enrichment for each domain. The results of these analyses are shown in Figure 6A.
FIGURE 6.
Protein-binding motif enrichment in repeats of Xist and Rsx, and similarity model. (A) Hierarchically clustered heatmap of PWM weighted z-scores for each repeat in Xist and Rsx, expressed as a percentile relative to the set of all GENCODE M18 lncRNA annotations. (B) Regions predicted to have similar protein-binding functions in Xist and Rsx. Arrows connect domains in each lncRNA that have similar k-mer and motif contents.
Using ranked enrichment of protein-binding motifs as a metric for hierarchical clustering, we identified the same relationships between Xist and Rsx repeat domains as we did when we hierarchically clustered domains by their k-mer content alone (dendrogram in Fig. 6A compared to dendrograms in Figs. 2E, 5H). Via protein-binding motif enrichment, Xist Repeats B, C, and D formed a second order cluster that next joined with Rsx Repeat 1. This clustering order is the same as that detected using k-mer content alone (Fig. 6A vs. 2E and 5H). Likewise, protein-binding motif enrichment grouped Repeats A and E together with Rsx Repeat 4, while Rsx Repeats 2 and 3 formed a separate cluster that joined with the Repeat-A-E-4 cluster. Again, similar clustering patterns were obtained based purely on k-mer content (Fig. 6A vs. 2E and 5H). We note that the motifs used to create these clusters are limited in complexity and are capable of recruiting different proteins depending on cellular and sequence contexts (Ray et al. 2013; Dominguez et al. 2018). Thus, the enrichment of a particular protein-binding motif in an individual Xist or Rsx repeat domain does not provide direct evidence that the protein binds to the domain. Nevertheless, these results are consistent with the notions that lncRNA k-mer content encodes information about protein-binding potential (Kirk et al. 2018), and that the various repeats in Xist and Rsx encode function through the concerted recruitment of multiple RNA-binding proteins.
A closer inspection of the protein-binding motifs that were enriched in each repeat domain yielded several insights. First, our motif analysis uncovered relationships between repeat domains that were not obvious from direct k-mer comparisons. For example, both human and mouse Xist Repeat B were enriched in motifs that recruit poly(C)-binding proteins and little else (Fig. 6A; Supplemental Table S4). Rsx Repeat 1, which most closely resembles Xist Repeat B at the level of k-mers, was also enriched in poly(C)-binding motifs, in both koala and opossum (Fig. 6A; Supplemental Table S4). However, Repeat 1 from koala and opossum Rsx were also enriched in many motifs that were absent in Repeat B, such as motifs that bind the proteins SRSF1, SRSF9, and RBM5 (Fig. 6A; Supplemental Table S4). In addition, while both pure k-mer analysis and motif analysis identified similarities between Xist Repeats A and E and Rsx Repeats 2, 3, and 4, our motif analysis also identified similarities exclusive to pairs of domains within this group, such as similarities between Rsx Repeats 2 and 3 and similarities between Xist Repeat E and Rsx Repeat 4 (Fig. 6A; Supplemental Table S4).
Second, within individual repeat domains, many protein-binding motifs were enriched to extreme levels. Well over half of the motifs analyzed (101 out of 175) were in the 99th percentile in terms of their enrichment relative to other mouse lncRNAs in at least one Xist or Rsx repeat domain, and all repeat domains in Xist and Rsx harbored multiple protein-binding motifs that were enriched at the 99th percentile or greater (Fig. 6A; Supplemental Table S4). This extremity was notable considering that most Xist and Rsx repeats are greater in length than the average mouse lncRNA (Fig. 2A). For example, at 13 kb in length, Rsx Repeat 1 is longer than 99.97% of spliced mouse lncRNAs (Fig. 2A). Nevertheless, on a length-normalized basis, multiple protein-binding motifs were enriched in Repeat 1 at the 99th percentile, in both koala and opossum Rsx. Inasmuch as motif density is known to be an important driver of associations between proteins and RNA (Van Nostrand et al. 2016; Dominguez et al. 2018; Kirk et al. 2018), our data suggest that at the level of sequence composition, the repeat domains in Xist and Rsx each have the potential to serve as high-affinity binding platforms for multiple proteins.
Lastly, many of the most strongly enriched motifs in both Xist and Rsx are known to recruit near-ubiquitous RNA-binding proteins that play core roles in the process of splicing (Wahl and Lührmann 2015). These included PTBP1, RMB5, SF3B4, SNRPA, SNRPB2, U2AF2, multiple SR proteins, and multiple HNRNP proteins, including HNRNPA1, HNRNPC, and HNRNPK (Fig. 6A; Supplemental Table S4). We recognize that the motifs available for this analysis are biased toward RNA-binding proteins whose functions are best understood; overwhelmingly, these proteins are splicing factors (Ray et al. 2013; Dominguez et al. 2018). Nevertheless, it is possible that an extreme enrichment for a motif that recruits a ubiquitously expressed splicing factor may confer a function that a single binding motif would not. For example, we presume that the function of Repeat B could not be recapitulated by a single motif that binds HNRNPK (Pintacuda et al. 2017).
DISCUSSION
Xist has served as a paradigmatic regulatory lncRNA for more than 25 years (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018). Nevertheless, it has been challenging to apply the information gained from the study of Xist to other lncRNAs. This is because Xist has little linear sequence similarity to other RNAs, even to lncRNAs like Rsx, which seem likely to encode analogous functions (Grant et al. 2012; Wang et al. 2014). In the present study, we used a nonlinear method of sequence comparison called SEEKR (Kirk et al. 2018) to compare the repetitive regions of Xist and Rsx. Our data provide sequence-based evidence to support the hypothesis that Xist and Rsx are functional analogs that arose through convergent evolution, and provide insights into mechanisms through which their repeat domains may encode function.
Unexpectedly, at the level of k-mers, the repeat domains of Xist and Rsx partitioned into two major clusters. Xist Repeats B, C, and D were highly similar to each other and to Rsx Repeat 1, whereas Xist Repeats A and E were most similar to each other and to Rsx Repeats 2, 3, and 4. From prior analyses of sequence content, there is little that would have suggested that the repeats in these two lncRNAs would cluster together in such a manner. However, prior molecular analyses of Xist are consistent with such a clustering (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018).
Specifically, Xist Repeats B and C, through their ability to bind HNRNPK and possibly other proteins, are known to play important roles in recruiting PRC1 and tethering Xist to the inactive X (Pintacuda et al. 2017; Colognori et al. 2019). The similarity between Repeats B and C and Xist Repeat D and Rsx Repeat 1 suggests that the latter two repeats may also play roles in recruiting PRC1 and tethering Xist/Rsx to chromatin. Consistent with this possibility, we found that an antibody specific to HNRNPK robustly retrieved Rsx RNA in an IP. Moreover, eCLIP data show that Xist Repeat D is enriched for HNRNPK binding in human cells (Supplemental Fig. S5; Van Nostrand et al. 2016). Thus, even within Xist, murid and nonmurid mammals may have convergently evolved separate repeats that recruit PRC1 and simultaneously tether the lncRNA to chromatin, in the form of Repeats C and D, respectively.
Relatedly, Xist Repeats A and E have been implicated in recruitment of PRC2 to the inactive X both via direct and cooperative means (Kohlmaier et al. 2004; Zhao et al. 2008; Cifuentes-Rojas et al. 2014; Davidovich et al. 2015; Almeida et al. 2017; Ridings-Figueroa et al. 2017; Sunwoo et al. 2017; Wang et al. 2017). The similarity between Xist Repeats A and E and Rsx Repeats 2, 3, and 4 suggests that the Rsx repeats could also play roles in recruiting PRC2. Indeed, PRC2 can bind Repeat A and other RNAs with high affinity (Cifuentes-Rojas et al. 2014; Davidovich et al. 2015), preferentially through G-quadruplex-like structures (Wang et al. 2017). G-quadruplexes can be encoded by repeated runs of as few as two consecutive G nucleotides separated by a few nucleotides (Wang et al. 2017). Xist Repeat A and Rsx Repeats 1, 2, and 4 were all enriched in k-mers containing runs of G nucleotides (Fig. 4C) and thus may be capable of binding PRC2. Moreover, a portion of the similarity between Xist Repeats A and E and Rsx Repeat 4 was driven by a shared enrichment for k-mers that had greater-than-average levels of poly-T(U) sequence (Fig. 4D). While the function of the poly-T segment of Repeat A is unknown (Brockdorff et al. 1992; Brown et al. 1992; Wutz et al. 2002; Minks et al. 2013; Kirk et al. 2018), polypyrimidine-rich sequence in Repeat E likely recruits several proteins, including CIZ1, that help stabilize Xist and PRC2 on chromatin (Smola et al. 2016; Ridings-Figueroa et al. 2017; Sunwoo et al. 2017; Stewart et al. 2019).
Based on these data, we propose that the two major clusters of repeats in Xist and Rsx function in part to cooperatively recruit PRC1 and PRC2 to chromatin. Within Xist, Repeat B plays a dominant role in recruiting PRC1 via its ability to bind HNRNPK; in turn, PRC1-induced chromatin modifications likely stimulate loading of PRC2 onto chromatin of the inactive X (Almeida et al. 2017; Pintacuda et al. 2017). Nevertheless, a PRC1-dominant model does not preclude other repeats in Xist or Rsx from functioning in PRC2 recruitment. Indeed, while there does not appear to be a single domain in Xist that is absolutely required to recruit PRC2 during the early stages of XCI (Wutz et al. 2002; Kohlmaier et al. 2004), it is possible that multiple domains in Xist recruit PRC2 duplicatively, such that deletion of any single domain alone does not cause complete loss in PRC2 recruitment. This hypothesis is supported by prior studies that link both Repeat A and E to recruitment of PRC2 (Kohlmaier et al. 2004; Zhao et al. 2008; Cifuentes-Rojas et al. 2014; Davidovich et al. 2015; Almeida et al. 2017; Ridings-Figueroa et al. 2017; Sunwoo et al. 2017; Wang et al. 2017), and by our own data that show Xist Repeats A and E and Rsx Repeats 2, 3, and 4 have similar k-mer profiles and motif contents. PRC1, PRC2, and related complexes function cooperatively in flies, mammals, and plants (Schuettengruber et al. 2014; Blackledge et al. 2015; Li et al. 2018). Considering this cooperativity, it is conceivable that the repeat domains in Xist and Rsx also cooperate to distribute PRC1 and PRC2 on chromatin.
Beyond recruiting PRCs, Xist evades nuclear export, it associates with transcribed regions of chromatin, and it induces Polycomb-independent gene silencing (da Rocha and Heard 2017; Balaton et al. 2018; Brockdorff 2018; Sahakyan et al. 2018). It is possible that Rsx carries out many, if not all of these actions, and that Rsx relies on sets of proteins similar to those used by Xist to achieve them (Grant et al. 2012; Wang et al. 2014). We found that all Xist and Rsx repeat domains harbored extreme levels of enrichment for multiple motifs known to recruit different subsets of RNA-binding proteins. Most of these proteins have been best characterized in the context of splicing, rather than epigenetic silencing.
In light of these data, we suggest that the repeat domains in Xist and Rsx may encode some of their functions not by recruiting a set of dedicated RNA silencing factors, but by engaging with ubiquitously expressed RNA-binding proteins in ways that are distinct from most other RNAs. Such a model was recently proposed (Brockdorff 2018), and agrees well with what is known about the specificity of RNA–protein interactions. Most RNA-binding proteins have limited sequence specificity, and are capable of binding many thousands of regions in hundreds to thousands of expressed RNAs (Ray et al. 2013; Van Nostrand et al. 2016; Dominguez et al. 2018). SPEN and HNRNPK are two RNA-binding proteins that are critical for Xist-induced silencing, yet they clearly associate with RNAs other than Xist (Cirillo et al. 2016; Van Nostrand et al. 2016). HNRNPK in particular is a ubiquitously expressed RNA-binding protein that functions in the cytoplasm and nucleus and its enrichment is not unique to Xist nor to Rsx (Bomsztyk et al. 2004; Huelga et al. 2012; Van Nostrand et al. 2016). Relatedly, many other proteins important for Xist-induced silencing play central roles in RNA splicing and nuclear export and, through these latter roles, likely associate with a large portion of the transcriptome (Moindrot et al. 2015). Thus, Xist and Rsx may distinguish themselves from other chromatin-associated transcripts not necessarily by the proteins to which they bind, but by the manner in which they bind these proteins.
That the related repeat domains were present in a different order in Xist and Rsx supports the notion that within a lncRNA, the order of functional domains is likely to be less important than the presence of the functional domains (Fig. 6B). This notion is consistent with a body of work that suggests lncRNAs encode regulatory function in a modular fashion, via discrete domains that recruit distinct subsets of effector proteins (Wutz et al. 2002; Tsai et al. 2010; Johnson and Guigo 2014; Kelley et al. 2014; Hezroni et al. 2015; Somarowthu et al. 2015; Hacisuleyman et al. 2016; Lu et al. 2016; Patil et al. 2016; Smola et al. 2016; Liu et al. 2017; Pintacuda et al. 2017; Kirk et al. 2018; Lubelsky and Ulitsky 2018).
From a methodological standpoint, our manuscript outlines approaches that should prove useful in the study of functional domains in other sets of RNAs. Intuitively, k-mer based comparisons like SEEKR seem most likely to succeed in identifying similarity when the domains of interest are repetitive. By nature, repetitive domains that share enrichments of similar subsets of k-mers will be more similar to each other than they will be similar to the average nonrepetitive region in the transcriptome.
Nevertheless, similarity between two repetitive domains, when observed, should be carefully considered, especially when the similarity occurs in lncRNAs such as Xist and Rsx, which are expressed at similar levels in equivalent subcellular compartments (Grant et al. 2012; Wang et al. 2014). Motif density is known to be a dominant factor driving protein-RNA interactions (Van Nostrand et al. 2016; Dominguez et al. 2018; Kirk et al. 2018). All other variables being equal, two lncRNAs that harbor domain-specific k-mer similarity should possess similar protein-binding profiles that could specify similar or analogous functions.
However, SEEKR is not limited to analysis of repetitive domains. It also has the ability to detect similarity between repetitive and nonrepetitive domains and between strictly nonrepetitive domains as well. In any given sequence, a set of k-mers can be arranged in repetitive or nonrepetitive ways, and SEEKR has no inherent preference for one over the other. As a contrived example, the sequence of Xist Repeat D can be shuffled in a way that eliminates its repeated monomers, yet entirely preserves its k-mer content (Supplemental File S1). By BLAST, this shuffled sequence has little internal similarity to itself or to Repeat D (Supplemental Fig. S6). Yet, by k-mer content, the shuffled sequence and Repeat D are literally identical (Supplemental Fig. S6). In a real-world example, the top five lncRNAs that SEEKR found to be the most similar to Repeat D are not nearly as repetitive as Repeat D itself (Supplemental Fig. S6). Of all Xist and Rsx repeats, Repeat D is the most complex (Fig. 2B). Nevertheless, these results demonstrate that k-mer based similarity searches performed with repetitive domains can identify nonrepetitive top hits.
With regard to nonrepetitive domains, our 2018 study showed that SEEKR rivaled BLAST-like alignment in its ability to detect lncRNA homologs in human and mouse (Kirk et al. 2018). The majority of homologs detected by SEEKR either lacked obvious repetitive elements, or were predominantly comprised of nonrepetitive sequence; the lncRNAs H19, Hottip, Malat1, Miat, and RMST being specific examples. We also found that SEEKR could identify Xist-like repressive activity in several synthetic and natural lncRNAs that lacked repetitive elements (Kirk et al. 2018). Thus, even in nonrepetitive regions of RNA, SEEKR should be capable of detecting meaningful similarities. However, functional domains comprised of high-complexity sequence elements will likely remain challenging to identify, regardless of the method in use.
Key variables to decide upon when using SEEKR are the k-mer length and the appropriate set of RNAs that define the background k-mer frequency; that is, the set of RNAs used to define the means and standard deviations from which k-mer z-scores are calculated. At present, data regarding functional domains in lncRNAs are too limited to arrive at conclusive recommendations for either variable. We favor using a k-mer length at which 4k most closely resembles the length of the shortest domain being analyzed. This approach minimizes the number of k-mers that yield counts of zero in the domain. Data from the present study as well as our prior work suggest that this minimization increases discriminatory power (Supplemental Fig. S4; Kirk et al. 2018).
In terms of the set of RNAs that should be used to define the background k-mer frequency, it is worth noting that SEEKR measures relative, not absolute, similarity. Pearson's r values returned by SEEKR reflect the similarity between two sequences relative to the k-mer frequency present in the background set of RNAs. We have found that using a background set of all lncRNAs in a genome provides a convenient way to identify trends. For example, in the present study, we used all known spliced lncRNAs in the mouse as a background set. Accordingly, we were able to identify properties in the repeat domains of Xist and Rsx that were distinct from the average spliced lncRNA annotated by GENCODE (Derrien et al. 2012).
In our initial description of SEEKR, we used k-mer contents of full-length lncRNAs as search features; we did not examine k-mer contents at the level of individual domains (Kirk et al. 2018). The domain-centric approaches outlined in the present study may be better suited for lncRNAs such as Xist and Rsx, which have multiple functions that are likely to be distributed among multiple domains. Indeed, at the level of k-mers, full-length Xist and Rsx were negatively correlated to each other. Similarities between the two lncRNAs emerged only when we took a domain-centric approach. Other eutherian lncRNAs known to harbor an Xist-like silencing function, such as Kcnq1ot1 and Airn, are exceptionally long—each on the order of 90 kb. Extrapolating from our findings above, we would expect these lncRNAs to harbor the greatest levels of similarity to each other not at the level of their full-length transcripts, but at the level of specific domains.
MATERIALS AND METHODS
Dot plots
Dot plots were generated using EMBOSS dotmatcher (Rice et al. 2000). For clarity, different visualization thresholds were used to generate the different dot plots shown in the manuscript. Figures 1A,C,D, and 5A,B, used a window size 10 and a threshold of 40. Figure 1B,E,F used window size 10 and threshold 45. Supplemental Figure S1 used a window size 20 and a threshold of 50.
Definition of repeat domains in Xist and Rsx, and nonhuman/nonmouse Xist sequences
The sequence of all Xist and Rsx repeat domains used in this work can be found in Supplemental File S1. The sequences of all full-length Xist and Rsx lncRNAs used in this work can be found in Supplemental File S2. The spliced mouse Xist sequence was sourced from the mm10 build of the mouse genome and annotations for the tandem repeats were sourced from (Brockdorff et al. 1992). The spliced human Xist sequence was sourced from the hg38 build of the human genome and the annotations for the tandem repeats were sourced from (Brown et al. 1992; Yen et al. 2007).
The sequences of spliced Xist used to generate the dot plots in Supplemental Figure S1 were obtained directly from annotations in the UCSC Genome Browser (Tyner et al. 2017), or, for genomes in which full annotations were unavailable, were reconstructed from partial annotations by UCSC and RNA-seq data from Hezroni et al. (2015). In the case of the vole Microtus rossiaemeridionalis, Xist sequence was obtained directly from Nesterova et al. (2001).
Spliced koala Rsx was obtained from Johnson et al. (2018). To identify repeat domains, Rsx was aligned to itself using EMBOSS dotmatcher with a 10 bp window and a 40% threshold (Rice et al. 2000). Start and stop positions of each repeat were defined by visual inspection of the dot plot. We considered separating the fourth major repeat in Rsx into two repeat domains, one 500 bp and the other 5000 bp in length (see Fig. 1D); however, analysis of the shorter sub-repeat within Repeat 4 revealed its k-mer content to be highly similar to the larger sub-repeat (not shown). Thus, to simplify our analyses and to clarify our presentation, we elected to merge the sub-repeats. Repeat domains in opossum Rsx (after filling in the gaps in assembly; see below) were defined in the identical manner.
K-mer based comparisons (i.e., SEEKR)
SEEKR was performed essentially as described in Kirk et al. (2018), with minor modifications. As a reference for normalization, we first calculated the mean and standard deviation for all k-mers at k = 4 in the GENCODE M18 lncRNA annotation file. We then generated length-normalized counts of all k-mers at k = 4 for each repeat domain in Xist and Rsx and calculated z-scores for each k-mer by subtracting the mean and dividing by the standard deviation for each k-mer from our reference set of GENCODE lncRNAs. Prior to performing Pearson's correlation, z-scores were log2 transformed.
To generate the distributions of Pearson's values in Figures 2B and 5I, we calculated the k-mer profile for each repeat domain and each GENCODE M18 lncRNA using the mean and standard deviation values from the full-length GENCODE M18 lncRNA annotation file, as described above. We then log2-transformed the z-scores and used Pearson's correlation to compare all lncRNAs to the Xist repeat in question.
Hierarchical clustering
Hierarchical clustering was performed using the SciPy hierarchy package in Python 3.6 (Jones et al. 2001), with distance defined as d = 1 − r, where r is defined as the Pearson correlation, using complete linkage.
De novo motif analysis
Motifs in each Xist and Rsx repeat domain were detected with MEME (version 5.0.2; Bailey et al. 2009), and run using the following options: -mod anr -dna -bfile bkg.meme -nmotifs 100 -minw 4 -maxw 12 -maxsites 1000, where the “bkg.meme” file specified a background frequency of 0.25 for all four nucleotides.
Consecutive k-mer analyses
To calculate the sums of z-scores for k-mers containing matches to mononucleotide runs in Figure 4A–D, we used the following approach. A mononucleotide run was defined as at least two consecutive occurrences of the nucleotide in question. For each nucleotide [A|C|G|T], we multiplied the z-score for each k-mer that contained a run by (the nucleotide length of the run minus 1). The sum of these products for each repeat domain at k-mer length k = 5 is plotted in Figure 4A–D. Identical trends were seen using k-mer lengths k = 4, 5, and 6 (Supplemental Fig. S3). K-mer length k = 5 was chosen for plotting in Figure 4 to emphasize trends that were present but less pronounced when using k-mer length k = 4. The set of mouse lncRNAs from GENCODE M18 was used to derive z-scores that described the length-normalized abundance of each k-mer in each repeat domain.
Weighting k-mer z-scores by likelihood of matching the HNRNPK-binding motif
To weight the sums of z-scores by the HNRNPK PWM in Figure 4E we performed the following calculation. For all k-mers at k = 5 we calculated the probability of a given k-mer's sequence occurring in the PWM for HNRNPK. The probability was defined as the independent probability of each letter in the k-mer occurring at the corresponding location within the PWM for each possible frame within the PWM. The HNRNPK motif is 8 nt long, therefore there were three possible frames for a 5-mer to fall within. The z-score for the k-mer in question was then weighted by taking the sum of the product between the z-score and each probability. The height of the bars in Figure 4E represent the sum of weighted z-scores for each Xist and Rsx repeat domain. The set of mouse lncRNAs from GENCODE M18 was used to derive z-scores that described the length-normalized abundance of each k-mer in each repeat domain.
Detecting HNRNPK-binding motif matches
Motifs occurrences in each Xist and Rsx repeat domain were detected with FIMO (version 5.0.2; Bailey et al. 2009), run using the following nondefault option: −thresh 0.01.
RNA immunoprecipitation
Cultured female M. domestica fibroblast cells were harvested at 70% confluency by scraping, then aliquoted into 1 × 107 cells, pelleted by centrifugation at 200g, then snap-frozen and stored at −80°C until used. RIPs from noncrosslinked cells were performed essentially as described in Zhao et al. (2010), using the following antibodies from Abcam: H3K27me3 (ab6002), CTCF (ab70303), HNRNPK (ab39975), and mouse IgG (ab18413). Briefly, cell pellets were gently resuspended in 1 mL of ice-cold RIPA buffer supplemented with 1× EDTA-free Proteinase Inhibitor Cocktail (Thermo Scientific) and lysed for 15 min at 4°C. Samples were sonicated at 4°C (Qsonica Q700 with cup horn accessory) at 12% amplitude for fifteen 30 sec intervals, with 30 sec resting steps between intervals. Cell debris was removed by centrifugation (at 6000g for 5 min), and samples were subsequently diluted to 1 mg of protein per ml with ice-cold RIPA buffer. Lysates with 1 mg of total protein (i.e., 500 µL) were incubated with the appropriate antibody coupled to Protein G beads (Life Technologies), overnight at 4°C with end-over-end rotation. Beads with no antibodies (mock IP) were used as background control. Beads were removed from lysate using a magnetic stand and were resuspended in 1 mL of ice cold NP-40 buffer (50 mM Tris at pH 7.5, 50 mM NaCl, 10 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) and washed for 15 min at 4°C with end-over-end rotation, repeated twice, followed by three washes with RIPA buffer. Following the last wash, beads were collected and resuspended in 1 mL of TRIzol (Life Technologies) for RNA extraction. A total of 10% of the input lysate (i.e., 50 µL) was processed in parallel. RNA was cleaned using RNeasy spin columns (Qiagen), following the manufacturer's “RNA Cleanup” protocol, with on-column RNase-free DNase Set (Qiagen) treatment. cDNA was synthesized using input and immunoprecipitated RNA with SuperScript III reverse transcriptase (Life Technologies) and random hexamer priming. Rsx was detected by RT-qPCR (in technical triplicate) with primer pair L2 from Grant et al. (2012). Cycle threshold (Ct) values were normalized to input and relative to the IgG. Fold enrichment was determined by relative quantification, which was calculated using the 2e(−ΔΔCt) method. The level of Gapdh mRNA enrichment was used as an internal nontarget index in the qPCR analysis.
Nanopore sequencing and annotation of opossum Rsx
High molecular weight DNA from VMRC18-839J22 and VMRC18-303M7 BACs was prepared using the NucleoBond BAC 100 kit (Machery Nagel). DNA from the two BAC preparations was pooled, sheared to an average length of 20 kb using a g-TUBE (Covaris), and then sequenced on the Oxford Nanopore Technologies (ONT) MinION using an R9.4 flow cell (FLO-MIN106) following the 1D ligation protocol (SQK-LSK109).
Reads were base-called with Albacore 2.3.1 (ONT), then assembled using Flye 2.3.5b (Kolmogorov et al. 2019). The six resulting scaffolds were aligned to E. coli K12 (NC_000913.3), opossum chromosome X (MonDom5, NC_008809.1), and the pCC1BAC cloning vector (EU140750.1). Scaffolds consisting entirely of E. coli or cloning vector DNA were removed. Three scaffolds aligned to adjacent regions of the MonDom5 X chromosome. These were merged together into a single candidate assembly sequence that was then polished iteratively with Racon 1.3.2 four times (Vaser et al. 2017), followed by Nanopolish 0.10.1 (Loman et al. 2015), to produce a final complete assembly of 235,139 nt (Supplemental File S3).
This polished assembly sequence was aligned again to MonDom5 using BLASTN to establish start and end coordinates to use as a reference when replacing the gaps in MonDom5 with the completed sequence in our assembly. The final sequence of opossum Rsx used in this work (Supplemental File S2) was generated using splice annotations from Grant et al. (2012), and replacing the N's in mondom5 with the corresponding sequence from our polished assembly (nucleotide substitutions are listed in Supplemental Table S3). Raw sequencing reads were deposited in NCBI's SRA, under accession number PRJNA522427.
Shuffling of Repeat D sequence
The sequence of Repeat D was shuffled using ushuffle (Jiang et al. 2008).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank UNC colleagues for discussions. This work was supported by National Institutes of Health (NIH) grant GM121806 and Basil O'Connor Award #5100683 from the March of Dimes Foundation (to J.M.C.), NIH Grant R014214 (to P.B.S.), and the Australian Research Council grant DP180100931 (to P.D.W.). D.S. was supported in part by an NIH training grant in pharmacology (T32 GM007040). J.R.W. was supported by the University Cancer Research Fund.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069815.118.
REFERENCES
- Almeida M, Pintacuda G, Masui O, Koseki Y, Gdula M, Cerase A, Brown D, Mould A, Innocent C, Nakayama M, et al. 2017. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356: 1081–1084. 10.1126/science.aal2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaton BP, Dixon-McDougall T, Peeters SB, Brown CJ. 2018. The eXceptional nature of the X chromosome. Hum Mol Genet 27: R242–R249. 10.1093/hmg/ddy148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J. 2004. hnRNP K: one protein multiple processes. Bioessays 26: 629–638. 10.1002/bies.20048 [DOI] [PubMed] [Google Scholar]
- Brockdorff N. 2018. Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Noncoding RNA 4: E28 10.3390/ncrna4040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. 10.1016/0092-8674(92)90519-I [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46: D762–D769. 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. 2014. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55: 171–185. 10.1016/j.molcel.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo D, Blanco M, Armaos A, Buness A, Avner P, Guttman M, Cerase A, Tartaglia GG. 2016. Quantitative predictions of protein interactions with long noncoding RNAs. Nat Methods 14: 5–6. 10.1038/nmeth.4100 [DOI] [PubMed] [Google Scholar]
- Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT. 2019. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol Cell 74: 101–117 e110. 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Heard E. 2017. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol 24: 197–204. 10.1038/nsmb.3370 [DOI] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. 2015. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 57: 552–558. 10.1016/j.molcel.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867 e859. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487: 254–258. 10.1038/nature11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Shukla CJ, Weiner CL, Rinn JL. 2016. Function and evolution of local repeats in the Firre locus. Nat Commun 7: 11021 10.1038/ncomms11021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. 2015. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11: 1110–1122. 10.1016/j.celrep.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. 2009. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136: 139–146. 10.1242/dev.026427 [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178. 10.1016/j.celrep.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Anderson J, Gillespie J, Mayne M. 2008. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinformatics 9: 192 10.1186/1471-2105-9-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Guigo R. 2014. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA 20: 959–976. 10.1261/rna.044560.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RN, O'Meally D, Chen Z, Etherington GJ, Ho SYW, Nash WJ, Grueber CE, Cheng Y, Whittington CM, Dennison S, et al. 2018. Adaptation and conservation insights from the koala genome. Nat Genet 50: 1102–1111. 10.1038/s41588-018-0153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P. 2001. SciPy: open source scientific tools for Python. http://www.scipy.org/
- Kelley DR, Hendrickson DG, Tenen D, Rinn JL. 2014. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol 15: 537 10.1186/s13059-014-0537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JM, Kim SO, Inoue K, Smola MJ, Lee DM, Schertzer MD, Wooten JS, Baker AR, Sprague D, Collins DW, et al. 2018. Functional classification of long non-coding RNAs by k-mer content. Nat Genet 50: 1474–1482. 10.1038/s41588-018-0207-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171 10.1371/journal.pbio.0020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34: 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, Colognori D, Jeon Y, Szanto A, del Rosario BC, Pinter SF, et al. 2015. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 57: 361–375. 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Fu X, Wang Y, Liu R, He Y. 2018. Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat Genet 50: 1254–1261. 10.1038/s41588-018-0190-0 [DOI] [PubMed] [Google Scholar]
- Liu F, Somarowthu S, Pyle AM. 2017. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol 13: 282–289. 10.1038/nchembio.2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Ulitsky I. 2018. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555: 107–111. 10.1038/nature25757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks J, Baldry SE, Yang C, Cotton AM, Brown CJ. 2013. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin 6: 23 10.1186/1756-8935-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep 12: 562–572. 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep 12: 554–561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. 2001. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res 11: 833–849. 10.1101/gr.174901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. 2017. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell 68: 955–969 e910. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Ridings-Figueroa R, Stewart ER, Nesterova TB, Coker H, Pintacuda G, Godwin J, Wilson R, Haslam A, Lilley F, Ruigrok R, et al. 2017. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev 31: 876–888. 10.1101/gad.295907.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce-Tolland ME, Andersen AA, Koyfman HR, Talbot DJ, Wutz A, Tonks ID, Kay GF, Panning B. 2010. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat Struct Mol Biol 17: 948–954. 10.1038/nsmb.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A, Yang Y, Plath K. 2018. The role of Xist in X-chromosome dosage compensation. Trends Cell Biol 28: 999–1013. 10.1016/j.tcb.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Oded Elkayam N, Sexton T, Entrevan M, Stern S, Thomas A, Yaffe E, Parrinello H, Tanay A, Cavalli G. 2014. Cooperativity, specificity, and evolutionary stability of Polycomb targeting in Drosophila. Cell Rep 9: 219–233. 10.1016/j.celrep.2014.08.072 [DOI] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113: 10322–10327. 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary structure. Mol Cell 58: 353–361. 10.1016/j.molcel.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart ER, Turner RML, Newling K, Ridings-Figueroa R, Scott V, Ashton PD, Ainscough JFX, Coverley D. 2019. Maintenance of epigenetic landscape requires CIZ1 and is corrupted in differentiated fibroblasts in long-term culture. Nat Commun 10: 460 10.1038/s41467-018-08072-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Colognori D, Froberg JE, Jeon Y, Lee JT. 2017. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc Natl Acad Sci 114: 10654–10659. 10.1073/pnas.1711206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, Fischer CM, Gibson D, Gonzalez JN, Guruvadoo L, et al. 2017. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45: D626–D634. 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27: 737–746. 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Lührmann R. 2015. SnapShot: spliceosome dynamics I. Cell 161: 1474–e1471. 10.1016/j.cell.2015.05.050 [DOI] [PubMed] [Google Scholar]
- Wang X, Douglas KC, Vandeberg JL, Clark AG, Samollow PB. 2014. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res 24: 70–83. 10.1101/gr.161919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, Youmans DT, Cech TR, Davidovich C. 2017. Targeting of Polycomb repressive Complex 2 to RNA by short repeats of consecutive guanines. Mol Cell 65: 1056–1067 e1055. 10.1016/j.molcel.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29: 2487–2489. 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Yen ZC, Meyer IM, Karalic S, Brown CJ. 2007. A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 90: 453–463. 10.1016/j.ygeno.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953. 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.