Abstract
CRISPR-Cas systems are a class of adaptive immune systems in prokaryotes that use small CRISPR RNAs (crRNAs) in conjunction with CRISPR-associated (Cas) nucleases to recognize and degrade foreign nucleic acids. Recent studies have revealed that Type III CRISPR-Cas systems synthesize second messenger molecules previously unknown to exist in prokaryotes, cyclic oligoadenylates (cOA). These molecules activate the Csm6 nuclease to promote RNA degradation and may also coordinate additional cellular responses to foreign nucleic acids. Although cOA production has been reconstituted and characterized for a few bacterial and archaeal Type III systems, cOA generation and its regulation have not been explored for the Staphylococcus epidermidis Type III-A CRISPR-Cas system, a longstanding model for CRISPR-Cas function. Here, we demonstrate that this system performs Mg2+-dependent synthesis of 3–6 nt cOA. We show that activation of cOA synthesis is perturbed by single nucleotide mismatches between the crRNA and target RNA at discrete positions, and that synthesis is antagonized by Csm3-mediated target RNA cleavage. Altogether, our results establish the requirements for cOA production in a model Type III CRISPR-Cas system and suggest a natural mechanism to dampen immunity once the foreign RNA is destroyed.
Keywords: CRISPR-Cas, Cas10, crRNA, interference, oligoadenylate
INTRODUCTION
CRISPR (clustered regularly interspaced short palindromic repeats) and CRISPR-associated (Cas) proteins comprise a class of adaptive immune systems that protect prokaryotes against their viruses (known as phages) and other foreign nucleic acids (Barrangou et al. 2007; Brouns et al. 2008; Marraffini and Sontheimer 2008; Barrangou and Horvath 2017). CRISPR-Cas function requires the assembly of a ribonucleoprotein complex containing a CRISPR RNA (crRNA) able to base pair to an invading nucleic acid in conjunction with one or more Cas proteins that mediate a response to the detection of the foreign nucleic acid once base-pairing has occurred (Wright et al. 2016). Bioinformatic and functional investigations have revealed the existence of diverse CRISPR-Cas systems that can be grouped into two broad classes and six distinct Types (I–VI) based on their mechanisms and cas gene content (Makarova et al. 2015; Koonin et al. 2017). Class I systems possess multiprotein effector complexes and include Types I, III, and IV, whereas class II systems possess single protein effector complexes and include Types II, V, and VI. However, despite their architectural diversity, all CRISPR-Cas types exhibit the same three-step mechanism of immunity: adaptation, crRNA biogenesis, and interference (Klompe and Sternberg 2018). During adaptation, a short foreign nucleic acid (35–45 nucleotides [nt] in length) is acquired from the invader and incorporated into the CRISPR locus in between DNA repeats of similar length. These invader-derived sequences are termed spacers. During crRNA biogenesis, transcription of the repeat-spacer array generates a long precursor which is processed in one or more steps to yield mature crRNAs that each specify a single target for destruction. Mature crRNAs combine with one or more Cas proteins to form the effector complex, which during interference, can sense and destroy “protospacers,” the foreign targets bearing complementarity to the crRNA in the complex (Wright et al. 2016).
The effector complexes in Types I and III CRISPR-Cas systems are composed of an oligomer of a Cas7 homolog which binds the crRNA and additional proteins required for function (Wiedenheft et al. 2011a; Spilman et al. 2013; Staals et al. 2013, 2014; Makarova et al. 2015; Osawa et al. 2015). Despite the shared presence of the Cas7 homolog, Types I and III systems exhibit markedly different function. Type I CRISPR-Cas systems use a multisubunit complex exemplified by the well-characterized E. coli CASCADE complex, which mediates interference by base-pairing to the invading DNA and recruiting the Cas3 nuclease to carry out DNA cleavage (Brouns et al. 2008; Beloglazova et al. 2011; Sinkunas et al. 2011; Westra et al. 2012; Jackson et al. 2014; Mulepati et al. 2014; Zhao et al. 2014; Loeff et al. 2018). In contrast, Type III CRISPR-Cas systems, exemplified by the model system in Staphylococcus epidermidis, degrade invading DNA and RNA in a transcription-dependent manner (Marraffini and Sontheimer 2010; Deng et al. 2013; Goldberg et al. 2014; Samai et al. 2015) using the effector complex called Cas10-Csm (Hatoum-Aslan et al. 2013). In these systems, immunity is triggered when the crRNA base-pairs with the protospacer in the target RNA and initiates a cascade of events including RNA cleavage by the Cas7 homolog, and DNA cleavage by the Cas10 protein (Hale et al. 2009; Staals et al. 2013, 2014; Tamulaitis et al. 2014; Samai et al. 2015; Estrella et al. 2016; Liu et al. 2017).
Recently, a new biochemical activity for Type III systems was discovered: the ability of the Cas10 homolog, upon activation by crRNA-target RNA base-pairing, to synthesize ATP-derived second messengers via its Palm domain (Kazlauskiene et al. 2017; Niewoehner et al. 2017). The second messengers are oligomers of three to six adenosine monophosphate units linked in a 5′–3′ manner in a cyclic topology. These novel second messengers, termed cyclic oligoadenylates (cOA), bind to the CARF (CRISPR-associated Rossmann fold) domain of the Csm6 protein and cause allosteric activation of its HEPN (higher eukaryotes and prokaryotes nucleotide-binding) nuclease domain. In vivo experiments using the S. epidermidis Type III system overexpressed in Staphylococcus aureus cells demonstrated that the Csm6 CARF and HEPN domains in addition to the Cas10 Palm domain promote efficient immunity against a ΦNM1γ6 phage infection, thus implying that cOA production is required for immunity (Jiang et al. 2016; Niewoehner et al. 2017). However, cOA generation has not directly been tested in this system.
Building upon these reports, we have conducted an in-depth characterization of cOA synthesis by the Type III-A CRISPR-Cas system of S. epidermidis, an early and important model system for CRISPR biology (Marraffini and Sontheimer 2008). To date, the generation of cOA has not been reconstituted for this system, and their length and topology remain unknown. Additionally, many important questions regarding activation and deactivation of cOA synthesis have not been addressed. These questions are important because inadvertent activation or failure to shut off cOA synthesis could both be physiologically damaging (Makarova et al. 2014; Rostøl and Marraffini 2019). We report in vitro studies using purified S. epidermidis Cas10-Csm complex to determine the identity of the cOA species produced, establish a role for Csm3-mediated target RNA cleavage in deactivation of cOA synthesis, and identify how base-pairing interactions adjacent to the 5′-end of the crRNA affect cOA synthesis.
RESULTS
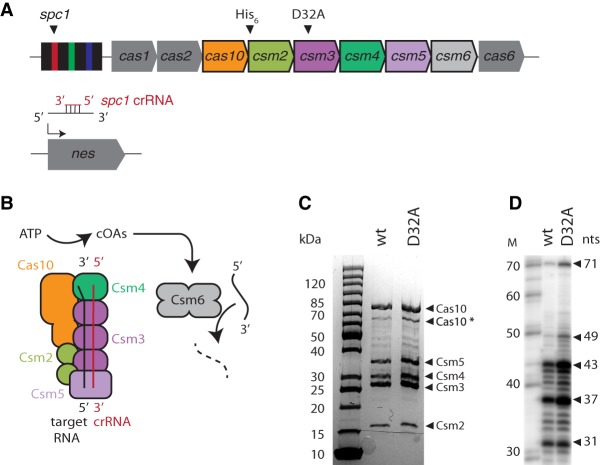
Detailed biochemical characterization of Cas10-Csm requires a homogeneous complex with high specific activity. Previous purification of Cas10-Csm was achieved by pull-down of the complex from S. epidermidis lysate with a biotinylated DNA oligonucleotide complementary to the crRNA or by recombinant expression in Escherichia coli followed by column chromatography (Hatoum-Aslan et al. 2013; Chou-Zheng and Hatoum-Aslan 2017). The former approach leads to an inactive complex because its crRNA is base-paired to DNA. The latter approach produces active complex but since the crRNA maturation takes place outside of its native complex, without the S. epidermidis nucleases associated with 3′-end maturation, the efficiency of maturation and thus the specific activity of the complex may be compromised (Walker et al. 2017). We isolated Cas10-Csm using the previously reported pcrispr expression plasmid to over-express the complex in the S. epidermidis LM1680 strain (Chou-Zheng and Hatoum-Aslan 2017). This construct contains a hexahistidine-tagged Csm2 subunit (Fig. 1A). Immobilized metal affinity chromatography was paired with ultracentrifugation over a sucrose gradient yielding an elution profile in the gradient consistent with the expected molecular weight of ∼300 kDa and an SDS-PAGE (polyacrylamide gel electrophoresis) banding pattern with all five Cas10-Csm proteins present (Fig. 1; Hatoum-Aslan et al. 2013). An additional substoichiometric band was observed by SDS-PAGE. Excision of this band from the gel followed by peptide sequencing by mass spectrometry identified Cas10 as the most abundant protein in the band as assessed by normalized spectral counts (Fig. 1C; Supplemental Table S1). For this reason, we believe the substoichiometric band is a truncated variant of Cas10. crRNAs were extracted from the complex and analyzed by PAGE revealing 37 and 43 nt species predominated with minor amounts of 31 and 49 nt species (Fig. 1D). Very little 71 nt species, a maturation intermediate, was present indicating efficient crRNA processing in the native S. epidermidis background. Purification and characterization of the resulting complex were carried out with wild-type complex and Csm3 D32A mutant complex, which is deficient in target RNA cleavage (Samai et al. 2015). When expressed on pcrispr, Cas10-Csm copurifies with crRNAs derived from the three spacers on the plasmid including spc1, which targets a region in the nes gene present on staphylococcal conjugative plasmids (Fig.1A; Marraffini and Sontheimer 2008; Hatoum-Aslan et al. 2011).
FIGURE 1.
Composition and purification of S. epidermidis Cas10-Csm complexes. (A) The pcrispr plasmid contains a repeat-spacer array and the cas operon facilitating purification of wt and mutant Cas10-Csm complexes from S. epidermidis. The crRNA encoded by spacer 1 (spc1) copurifies with Cas10-Csm and is complementary to the nes transcript present on conjugative plasmids. (B) Cas10-Csm complexes were recently reported to synthesize cOA from ATP when activated by a target RNA complementary to the crRNA. cOA binds to Csm6, stimulating its RNA cleavage activity. (C) SDS-PAGE analysis following purification confirms that the five proteins that constitute Cas10-Csm are present. (D) crRNAs extracted from the Cas10-Csm complex following purification have a distribution of lengths with 37 and 43 nt most prominent. A decade marker (M) was used to infer crRNA lengths.
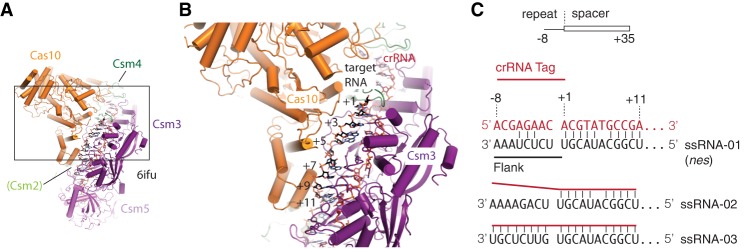
Recent studies revealing synthesis of cOA second messengers were performed with Streptococcus thermophilus, Enterococcus italicus, and Sulfolobus solfataricus Type III CRISPR-Cas complexes. However, cOA synthesis has not yet been reconstituted and characterized in the important model organism for CRISPR-Cas function, S. epidermidis (Marraffini and Sontheimer 2008; Kazlauskiene et al. 2017; Niewoehner et al. 2017; Rouillon et al. 2018). To address this gap, we first investigated the metal dependence and target RNA feature dependence of cOA synthesis by Cas10-Csm. Recent cryo-EM structures have revealed the architecture of two Cas10-Csm complexes at sufficient resolution for molecular modeling and have revealed the structural details of the 5′ crRNA tag–3′ target RNA flank interaction that regulates Type III-A interference (Fig. 2; Jia et al. 2018; You et al. 2019).
FIGURE 2.
The molecular environment of the crRNA–target RNA duplex. (A) An overview of the high-resolution cryo-EM structure of S. thermophilus Cas10-Csm bound to target RNA (6ifu). (B) The boxed region from A is magnified, showing that the +1 to +11 target RNA positions occupy a distinct environment, directly interacting with Cas10. The distal base pairs of the target RNA contact Csm2. (C) The 5′ tag region of crRNAs, in type Type III systems, is an 8 nt sequence derived from the repeat region of the CRISPR array. Base pairs between the crRNA and a foreign RNA target began downstream from the 5′ tag and are numbered starting with +1. Base pairs in the −1 to −8 region indicate a “self” nucleic acid, and this complementarity blocks Cas10-Csm–mediated interference. Three target RNAs that differ in the 5′ tag–3′ flank region of the duplex were used in this study. Csm3 occludes base-pairing at the +6 position.
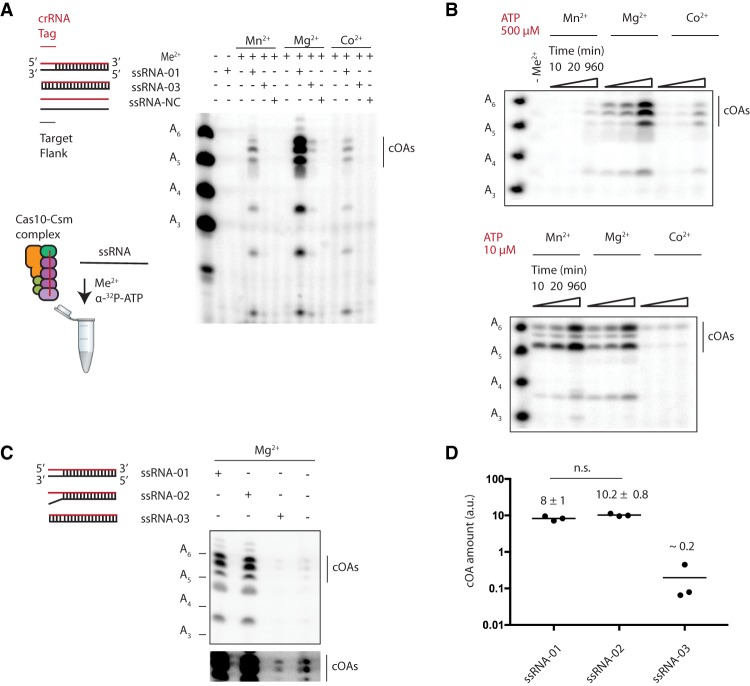
Enterococcus italicus Cas10-Csm cOA synthesis is reported to be Mg2+-dependent, while several divalent metals were shown to support cOA synthesis by S. thermophilus Cas10-Csm, including Co2+ (Kazlauskiene et al. 2017; Niewoehner et al. 2017). Incubations of Cas10-Csm with α-32P ATP in the absence of divalent metal or target RNA produced no oligoadenylates when reactions were analyzed by PAGE (Fig. 3A). However, the addition of 10 mM divalent metal and ssRNA-01 produced three to four prominent PAGE bands. ssRNA-01 is a synthetic oligo-containing sequence complementary to spc1 derived from the nes transcript, a bona-fide in vivo activator of Cas10-Csm (Fig. 1A; Marraffini and Sontheimer 2008). These results suggested that the Cas10-Csm-catalyzed reaction was target RNA-dependent, dependent on divalent metal and that the complex did not copurify with a divalent metal that supports cOA synthesis. An RNA lacking complementarity to crRNA did not activate cOA synthesis by Cas10-Csm while an RNA complementary to the 5′ tag region of the crRNA (the 8 nt on the 5′-end derived from the DNA repeat sequence) also failed to activate cOA synthesis (Fig. 3A). The latter observation agrees with previous reports that Cas10-Csm uses the 5′ tag of the crRNA to distinguish between foreign protospacers and self-nucleic acid (Marraffini and Sontheimer 2010; Rouillon et al. 2018).
FIGURE 3.
Divalent metal and target RNA requirements for oligoadenylates synthesis by Cas10-Csm. (A) A schematic showing the components of the target RNA-dependent cOA synthesis reaction. The Cas10-Csm complex with a crRNA (red) is incubated with either a single-stranded 5′ tag mismatch RNA (ssRNA-01), a 5′ tag complementary RNA (ssRNA-03), or a noncomplementary RNA (ssRNA-NC). α-32P-ATP is added to reactions analyzed by electrophoresis. Migration of the cOA species by PAGE is compared to a ruler of linear oligoadenylate species (A3–A6). (B) A time-resolved analysis of cOA synthesis with three different divalent metals reveals that Mg2+ supports efficient cOA production when ATP is abundant, while both Mn2+ and Mg2+ can efficiently produce cOA under low ATP conditions. (C) The ssRNA-01 target RNA contains three positions in the 5′ tag–3′ flank region capable of base-pairing, whereas ssRNA-02 contains no 5′ tag–3′ flank base pairs. The quantity of oligoadenylates produced by these target RNAs was compared to a crRNA-target RNA with complete complementarity in the 5′ tag–3′ flank region (ssRNA-03). Standard and overexposed (lower) images of a representative gel containing the oligoadenylates are shown. (D) Three replicate reactions described in C were analyzed by PAGE, and oligoadenylates were quantified by densitometry to produce the means and standard deviations given in arbitrary units (a.u.). The difference between the ssRNA-01 and ssRNA-02 reactions is not significant (n.s., P < 0.05).
Previous experiments demonstrating cOA synthesis by Type III CRISPR complexes used Co2+ or Mg2+ divalent metals (Kazlauskiene et al. 2017; Niewoehner et al. 2017). To determine how divalent metals affected cOA synthesis by Cas10-Csm, we performed experiments with the complex loaded with each of these divalent metals and also Mn2+. Experiments with 500 µM ATP, a concentration approaching physiological conditions, revealed that Mg2+ produced much more cOA product than the other divalent metals investigated and this was true for short or prolonged incubations (Fig. 3B). However, when we performed experiments with 10 µM ATP, we observed that Cas10-Csm with Mn2+ or Mg2+ produced similar amounts of cOA product (Fig. 3B). These results are consistent with the observation that multiple divalent metals can support cOA synthesis in vitro (Kazlauskiene et al. 2017).
The nes transcript (ssRNA-01) activates interference in vivo despite the fact that it can base-pair with spc1 crRNA at the –3 to –5 positions (Fig. 2C). We quantitated the amount of cOA produced by ssRNA-01 activation of Cas10-Csm versus a synthetic RNA (ssRNA-02) unable to base-pair with the 5′ tag of spc1 crRNA (Fig. 3C). This experiment revealed there was not a significantly different amount of cOA produced (P < 0.05) indicating the −3 to −5 base pairs are insufficient to affect licensing of cOA synthesis (Fig. 3D).
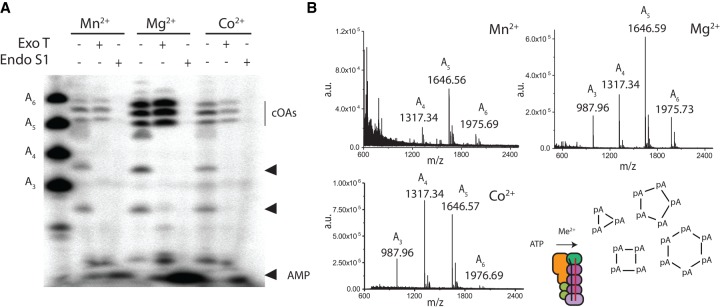
We next sought to identify the topology of the oligoadenylates that were visible by PAGE. We incubated the oligoadenylate products with either an endonuclease or exonuclease and polynucleotide kinase (PNK) and analyzed the resulting products by PAGE (Fig. 4A). PNK was included in the exonuclease reaction because it resolves 2′,3′-cyclic phosphates which can block exonuclease T activity. The major bands (in the vicinity of A5 and A6 of the ladder) collapsed to the bottom of the gel in the presence of endonuclease S1 but were unaltered by exonuclease T indicating the bands possess a cyclic topology. While exonuclease T did not digest the major bands, it did digest presumed synthesis intermediates in the lower half of the gel indicating the enzyme is active (Fig. 4A).
FIGURE 4.
Topology and length of Cas10-Csm oligoadenylates. (A) The α-32P-labeled products of each cOA synthesis reaction are incubated either with exonuclease T and PNK enzymes or endonuclease S1. 32P-labeled degradation products appear at the bottom of the gel in endonuclease reactions migrating in a manner consistent with AMP. Two arrows mark the position of presumed synthesis intermediates which are digested by exonuclease T. (B) MALDI mass spectra of cOA synthesis reaction products are shown along with a diagram depicting product formation.
Cyclic oligonucleotides are known to migrate more slowly in PAGE than their linear counterparts and cA3 was observed previously to migrate similar to a linear A6 (Kazlauskiene et al. 2017). Therefore, the prominent cyclic species observed by PAGE are most likely cA3–6 consistent with previous observations that Cas10-Csm produces a distribution of cOA lengths (Fig. 3A; Kazlauskiene et al. 2017; Niewoehner et al. 2017). Since nuclease digestion of the oligoadenylates did not change as a function of metal cofactor, this indicates the cofactor does not play a role in controlling the topology of the oligoadenylates (Fig. 4A).
To confirm our suspicion that the products observed by PAGE were cA3–6, we investigated the length of Cas10-Csm cOA products by matrix-assisted laser desorption ionization (MALDI) mass spectrometry (Fig. 4B). Under ideal conditions, four major bands were resolvable by PAGE and mass spectrometry revealed four major peaks with m/z values consistent with oligoadenylates of length A3–A6 (Fig. 4B). The m/z values observed are consistent with singly charged A3–A6 oligoadenylates with either a 2′,3′-cyclic phosphate terminus or a cyclic topology (M + H+: 988, 1317, 1647, 1976) created by a 3′-hydroxyl attack on the α-phosphate of the oligo's 5′-triphosphate. The inability of exonuclease to digest the oligoadenylates indicates they have a cyclic topology.
Revealing the interplay of the various biochemical activities of Cas10-Csm will be essential to understanding the biological functions of Type III systems. The interaction between target RNA cutting and the cOA synthesis activity of Cas10-Csm is a key example. We reasoned target RNA cleavage could be de-coupled from cOA activation by an oligonucleotide resistant to cleavage by Csm3. However, consistent with observations using S. thermophilus Cas10-Csm, a single-stranded DNA complementary to spc1 crRNA is unable to activate cOA synthesis (Fig. 5A; Kazlauskiene et al. 2017). We therefore sought a different means of blocking target RNA cutting so its role in cOA regulation could be better understood. While wt Cas10-Csm rapidly degrades target RNA, the point mutant, CsmCsm3D32A produces minimal RNA cleavage even after 120 min of incubation (Fig. 5B,C; Samai et al. 2015). We quantitatively followed cOA production over time in both wild-type and Cas10-CsmCsm3D32A and found that cOA levels rose abruptly in the wild-type reaction but increased very slowly after 5 min (Fig. 5D). In contrast, Cas10-CsmCsm3D32A cOA amounts continued to increase even at 120 min producing 7.4-fold more cOA than wild-type Cas10-Csm at the 60-min mark and 10-fold more cOA than wild-type at the 120-min mark (Fig. 5D). This result suggests that target RNA cleavage antagonizes cOA synthesis.
FIGURE 5.
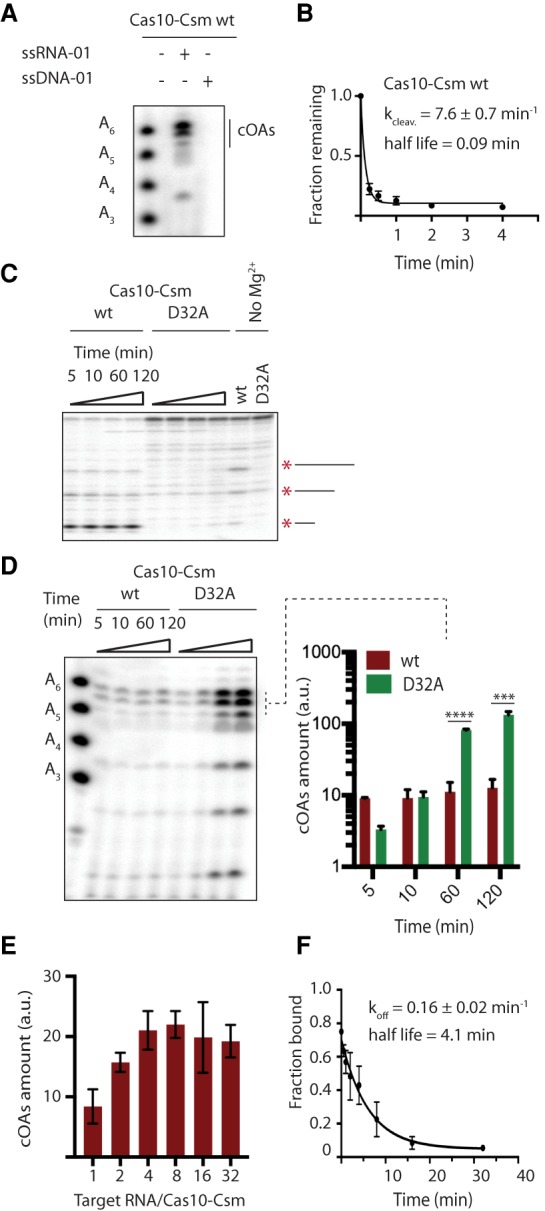
Cas10-Csm rapidly cleaves but slowly turns over target RNA. (A) PAGE analysis of a cOA synthesis reaction shows that a single-stranded DNA oligonucleotide complementary to spc1 crRNA is unable to activate cOA synthesis. (B) Three replicate reactions of target RNA cleavage (ssRNA-01) by wt Cas10-Csm were analyzed by PAGE to determine a rate constant for the process. (C) Cas10-Csm containing the Csm3 D32A mutant is unable to cleave target RNA, as shown by PAGE analysis of a reaction containing 32P-labeled ssRNA-01. An asterisk and line mark the location of RNA cleavage products. A control experiment incubating Cas10-Csm with ssRNA-01 for 120 min without Mg2+ is shown. (D) A time course of cOA synthesis analyzed by PAGE comparing the amount of cOA produced by wt and Cas10-CsmCsm3D32A. The reactions were performed in triplicate (one representative gel is shown), and a quantification of band intensities for three cOA species is shown. (***) P < 0.001; (****) P < 0.0001. (E) The amount cOA produced after 60 min was quantified for titrations of target RNA from a 1:1 mole ratio with Cas10-Csm up to a 32:1 mole ratio. (F) Dissociation of cleaved target RNA from its complex with crRNA bound Cas10-Csm was measured by a double filter binding assay to derive koff. Best-fit values for kcleav and koff are given along with standard error.
We sought to dissect mechanistically how target RNA cleavage might inhibit cOA synthesis considering two hypotheses: that once target RNA is cleaved it becomes unable to stimulate cOA synthesis by Cas10-Csm or that once target RNA is cleaved it rapidly dissociates from Cas10-Csm thereby causing the loss of activation. The latter hypothesis suggests that a large excess of target RNA would lead to sustained cOA synthesis over time. However, we measured cOA synthesis after 60 min in a series of reactions in which target RNA was titrated and found that no stimulation of cOA synthesis occurred for molar ratios of target RNA to Cas10-Csm above four (Fig. 5E). This observation leads us to measure the rate of target RNA dissociation in a pulse-chase experiment in which 32P-labeled target RNA cutting was initiated by Mg2+ and a chase of unlabeled target RNA allowed the rate of dissociation of cleaved RNA to be specifically followed in a double membrane-binding experiment (Fig. 5F). This experiment revealed that cleaved RNA dissociates from Cas10-Csm slowly, with a half-life of 4.1 min. This long half-life suggests a mechanism to explain the burst of cOA synthesis followed by repression of synthesis (Fig. 5D): Cleaved target RNA is unable to stimulate cOA synthesis and its slow dissociation rate ties up Cas10-Csm in an inactive state for extended periods of time.
To further test the hypothesis that target RNA cleavage is the primary mechanism for inactivation of cOA synthesis, rather than RNA dissociation, we constructed a mathematical model able to simulate the cOA synthesis progress curve. The model considered the rate of nucleotide polymerization chemistry (Cas10-CsmCsm3D32A), the rate of RNA cleavage and the dissociation rate of cleaved RNA (Fig. 6A). Simulations in the case of excess target RNA showed that if target RNA cleavage inactivated cOA synthesis (Model 1), active Cas10-Csm would rapidly be depleted due to the slow dissociation rate of cleaved RNA while a large constant amount of active Cas10-Csm would be present if cOA synthesis was stimulated by bound RNA regardless of its cleavage state (Model 2) (Fig. 6B). Next, we simulated cOA synthesis progress curves and compared these results to experimental progress curves from wt Cas10-Csm and Cas10-CsmCsm3D32A (Fig. 6C,D). Simulations with Model 1 possessed a burst of cOA synthesis from 0 to 1 min followed by a very slow rate of additional synthesis after 1 min, whereas Model 2 simulations predicted a linear increase of cOA over time. A wt Cas10-Csm experimental cOA progress curve from 0 to 1 min was consistent with an exponential decay of active complex as predicted by Model 1 and resembled the predicted amounts of cOA accumulation over 60 min by Model 1. In the presence of excess target RNA, the Cas10-CsmCsm3D32A mutation requires this complex to behave similar to Model 2 in a near perpetual RNA-bound state. Cas10-CsmCsm3D32A resembled the linear Model 2 progress curve from 0 to 45 min. Beyond 45 min, the curve deviates slightly from linearity likely due to additional factors the mathematical model does not account for such as product inhibition. From the progress curves, we concluded that intact target RNA is required to stimulate cOA synthesis and that the combination of rapid RNA cleavage and its slow dissociation rate constrain cOA synthesis to primarily a brief burst following target RNA binding (Fig. 6C).
FIGURE 6.
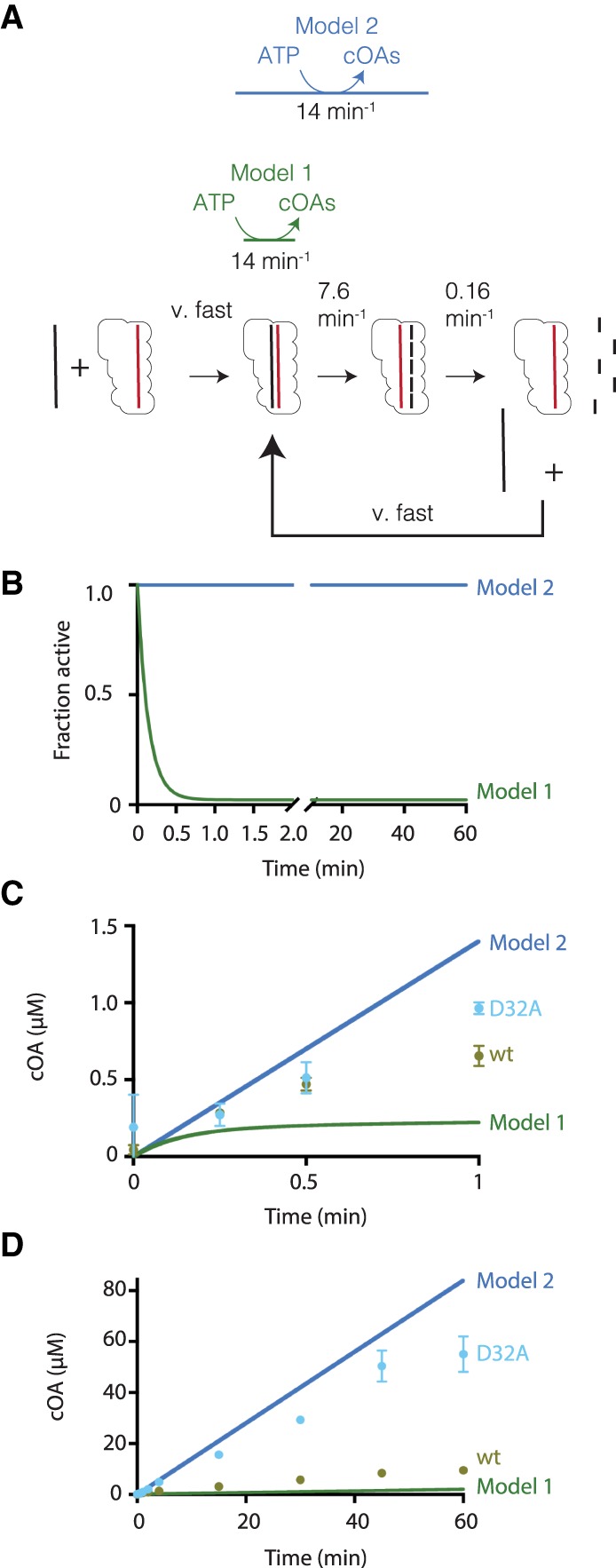
Target RNA cleavage inhibits cOA synthesis. (A) Two models for control of cOA synthesis are shown. In Model 1, Cas10-Csm is active in cOA synthesis only when bound to an intact target RNA. In Model 2, Cas10-Csm bound to target RNA is active in cOA synthesis regardless of whether the RNA is cleaved or not. (B) A mathematical model built from the rates in A predicts the fraction of Cas10-Csm active in cOA synthesis as a function of time in each model. (C) An experimental, wt cOA progress curve and a Cas10-CsmCsm3D32A, experimental cOA progress curve are compared to the model curves for 0–1 min. (D) Experimental and model progress curves are shown from 0 to 60 min.
For prokaryotes harboring a Type III-A CRISPR-Cas system, an inherent trade-off exists between specific and promiscuous RNA targeting. Stringent requirement of full complementarity between crRNA and target RNA allows a foreign transcript with a single mismatch to avoid targeting. However, too loose a requirement for complementarity could allow self-nucleic acid targeting and cell death. Recent in vivo studies have indicated that only a relaxed complementarity between crRNA and target RNA is required for Cas10-Csm defense against foreign genetic elements while in vitro cOA synthesis studies have revealed that some double mismatches block cOA production (Kazlauskiene et al. 2017; Pyenson et al. 2017; Rouillon et al. 2018).
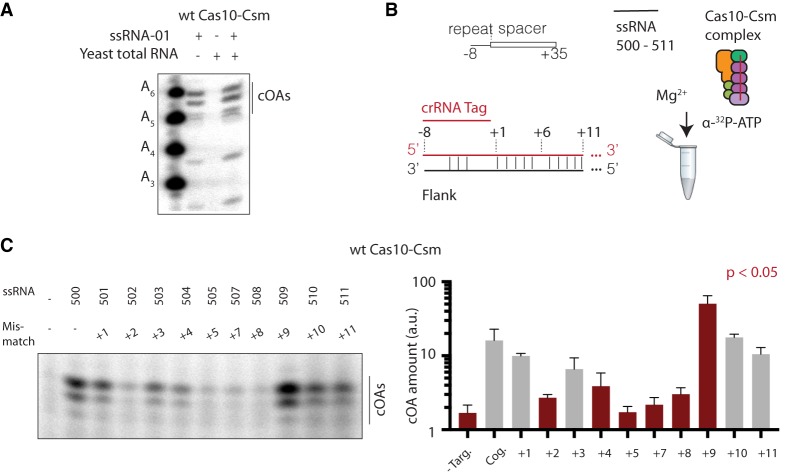
We began an investigation of target RNA specificity on cOA synthesis by asking if a complex mixture of total RNA extract would activate cOA synthesis. We found that 480 µg/mL total RNA from Saccharomyces cerevisiae, under our reaction conditions, did not lead to an observable signal for cOA synthesis (Fig. 7A). Next, we asked whether targeting of a complementary RNA (ssRNA-01) would be hindered by the complex mixture of RNA (S. cerevisiae total RNA) and found that Cas10-Csm still readily activated cOA synthesis (Fig. 7A). This observation indicates that Cas10-Csm avoids off-target activation in vitro and efficiently searches complex RNA mixtures in vitro reproducing two features required of the enzyme in vivo.
FIGURE 7.
Specificity of Cas10-Csm cOA synthesis activity. (A) Target RNA, yeast total RNA or both were incubated with Cas10-Csm and α-32P ATP. The cOA reaction products were separated by PAGE. (B) A panel of ssRNAs, each possessing a single base-pair mismatch with crRNA, was assayed for its effect on cOA synthesis. (C) The effect on cOA synthesis of introducing single mismatches in target RNA in the +1 to +11 base pairs was tested. The reactions were performed in triplicate (one representative gel is shown) and a quantification of band intensities for three cOA species is shown. cOA levels significantly different than cognate RNA (no mismatch) are shown with red bars.
Pyenson and coworkers, measuring the efficiency of foreign DNA transformation, found that crRNA-target RNA base-pairing in the Cas10 proximal region (see Figs. 2, 7B) is the most sensitive to mismatches (Pyenson et al. 2017). Additionally, recent high-resolution cryo-EM structures of Cas10-Csm complexes indicate Cas10 makes direct contacts to crRNA-target RNA base pairs in the vicinity of +1 to +11, however, does not make direct contacts to the distal base pairs of the duplex (Fig. 2; Jia et al. 2018; You et al. 2019). Therefore, we investigated the effect of mismatches at the +1 to +11 positions on in vitro synthesis of cOA. We generated a panel of target RNAs each containing a single mismatch generated by mutating a nucleotide to its base-pair partner (e.g., A–U at position +1 becomes A–A) (Fig. 2C). Because target RNA cleavage antagonizes cOA synthesis and the combined effect of mismatches on RNA cleavage and cOA synthesis could be complex, we performed cOA synthesis experiments using the panel and wild-type Cas10-Csm or Cas10-CsmCsm3D32A.
Wild-type Cas10-Csm produced cOA as expected when activated by the target RNA mimicking the nes transcript sequence (ssRNA-500) (Fig. 7C). As shown in Figure 5D, the cOA amount produced by wild-type Cas10-Csm with ssRNA-500 is lower than that produced by Cas10-CsmCsm3D32A (Figs. 7C, 8A). For wild-type Cas10-Csm, a mismatch at +2, +4, +5, +7, or +8 significantly reduced cOA synthesis while a mismatch at +1, +3, +10, and +11 did not significantly affect cOA amounts (Fig. 7C). One mismatch, at the +9 position, produced slightly higher amounts of cOA (Fig. 7C). We suspected the cOA production decreases of the mismatched RNAs are due to a failure to activate Cas10. However, another possible explanation is that less cOA was produced because the mismatches were somehow accelerating target RNA cleavage (Fig. 5D). Therefore, we performed cOA synthesis experiments with Cas10-CsmCsm3D32A. When provided mismatched target RNA, Cas10-CsmCsm3D32A produced significantly less cOA product for all mismatches tested except +9 and +10 (Fig. 8A). As with wild-type Cas10-Csm, a mismatch at +2, +5, +7, or + 8 elicited the greatest defect in cOA production (Figs. 7C, 8A). We investigated whether a change in the affinity of the mismatched RNAs for Cas10-Csm might explain the cOA synthesis defect. To address this possibility, we titrated Cas10-CsmCsm3D32A against target RNAs containing a mismatch at +2, +5, +7, or + 8 and found that 125 nM Cas10 complex is sufficient to completely bind each target RNA, a concentration comparable to the 100 nM Cas10 complex used in the cOA synthesis assays (Fig. 8B). Taken together, the experiments with mismatched target RNAs indicate that a single mismatch in particular positions of a nes transcript reduces cOA synthesis similar to background levels observed when no target RNA is present.
FIGURE 8.
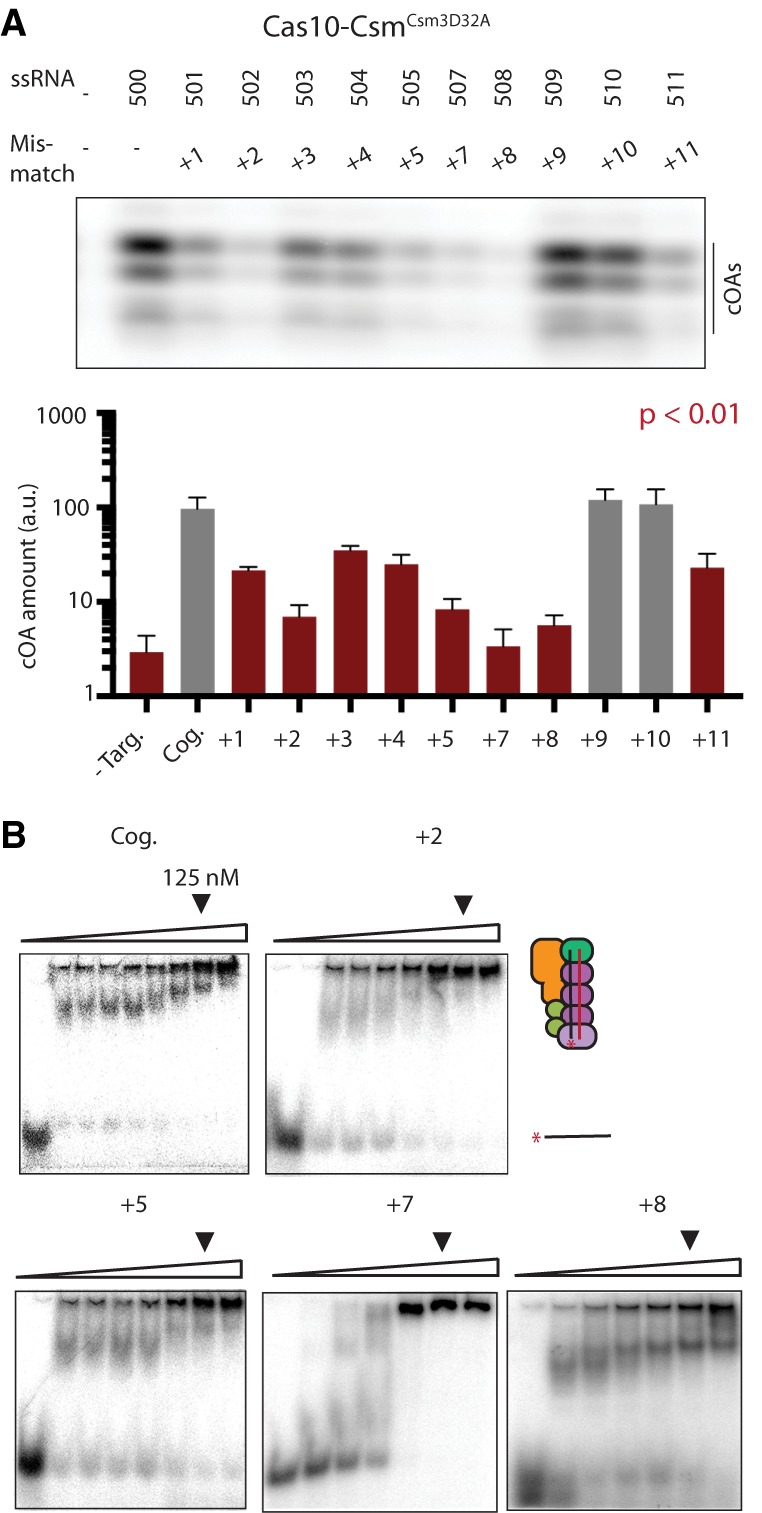
The effect of single base-pair mismatches on cOA synthesis in Cas10-Csm deficient in RNA cleavage. (A) The effect of single base-pair mismatches on cOA synthesis by Cas10-CsmCsm3D32A assayed by PAGE. Reactions were performed in triplicate and a representative gel is shown. Quantitation of cOA amounts is shown as a bar chart. Red bars indicate cOA amounts significantly different from the amount produced in the presence of cognate target RNA with P < 0.01. (B) Electrophoretic mobility shift assays show that single base-pair mismatched target RNAs are fully bound to Cas10-Csm at the 125 nM concentration of complexes used in cOA synthesis assays.
DISCUSSION
Here we report, to our knowledge, the first in vitro characterization of cOA production by the Cas10-Csm complex in S. epidermidis, which comprises a well-established model Type III-A CRISPR-Cas system. In vitro assays of cOA production by Cas10-Csm required the development of a new purification approach because the previous approach of purifying Cas10-Csm from S. epidermidis relied on capturing the complex with a 5′-biotinylated oligo complementary to the crRNA (Chou-Zheng and Hatoum-Aslan 2017). The resulting crRNA-DNA duplex was an obstacle to downstream activity assays. Therefore we coupled IMAC to the size-based selection approach, ultracentrifugation, to obtain high purity Cas10-Csm.
Divalent metal effects on cOA synthesis
Recent studies have demonstrated that the Cas10 Palm domain is responsible for cOA synthesis in S. thermophilus, E. italicus, and S. solfataricus (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Rouillon et al. 2018). Additionally, genetic evidence implicates the Palm domain of Cas10 as the site of cOA synthesis in S. epidermidis (Niewoehner et al. 2017). The Palm domains in Cas10 homologs contain a GGDD motif similar to the Palm polymerase domains of nucleotide cyclases and DNA polymerases (Makarova et al. 2002). In the latter, the conserved aspartates coordinate divalent cations to facilitate polynucleotide chain elongation. Mg2+, Mn2+, and Co2+ have all been shown to support activity in many DNA polymerases, although Mg2+ is believed to be the physiological metal (Vashishtha et al. 2016). For this reason, we investigated the effect of the three metals on cOA production by Cas10-Csm. In DNA polymerases, Mn2+ dramatically elevates the levels of incorporation of nucleotides not matching the DNA template, thus causing a loss of specificity (Vashishtha et al. 2016). While a strictly analogous mechanism could not be in play during cOA synthesis, we speculated that different metals may have some effect on the regulation of cOA synthesis or the nature of the products synthesized. Interestingly, we observed that the efficiency with which Mn2+ supports cOA synthesis depends on ATP concentration: Under low ATP conditions, Mn2+ produced cOA amounts similar to Mg2+ (Fig. 2B). This cannot be explained by titration of divalent metal concentrations by ATP, as each divalent metal was included at 10 mM.
A role for target RNA cleavage in cOA regulation
By performing assays with a Cas10-Csm carrying the Csm3 D32A mutant, which is defective in target RNA cleavage, we have shown that target RNA cleavage inhibits cOA synthesis (Fig. 5D). This could be either because the cleaved RNA is unable to activate cOA synthesis or because the RNA diffuses away from Cas10-Csm in the cleaved form. Recently, Rouillon and coworkers made a similar observation to ours. In an archaeal Type III-D system, they showed that cOA synthesis persisted when CRISPR-Cas was bound to an RNA modified to be cleavage resistant, but cOA synthesis was abruptly terminated coincident with RNA cleavage (Rouillon et al. 2018). In this study, it was speculated that cOA synthesis was inhibited by the dissociation of cleaved target RNA from the CRISPR-Cas complex (Rouillon et al. 2018). We designed experiments to distinguish between inhibition of cOA synthesis by target RNA cutting, Model 1, and inhibition of cOA synthesis by target RNA dissociation, Model 2 (Figs. 5D–F, 6). We observed that molar ratios of target RNA to Cas10-Csm greater than four did not lead to increased cOA synthesis, a finding that disagrees with Model 2. Additionally, we observed that dissociation of cleaved RNA occurred slowly, on the time scale of minutes, explaining why excess target RNA failed to increase cOA synthesis (Fig. 5F). Because the ultimate rate of cOA synthesis is affected by the intrinsic rate of enzyme-mediated chemistry, the RNA cleavage rate and the dissociation rate of cleaved RNA, we generated a mathematical model to assist in visualizing the interplay of these factors. Simulations of cOA synthesis progress curves under Model 1 and Model 2 reveal that the experimentally observed wild-type curve agrees much better with Model 1. We therefore concluded that target RNA cutting inactivates cOA synthesis by Cas10-Csm, presumably by reversing a conformational change required for activation. The difference between the behavior of the mesophilic Cas10-Csm and the S. solfataricus Type III-D system which appeared to be inhibited in cOA synthesis by target RNA dissociation may be related to the fact that S. solfataricus is a thermophile and nucleic acid dynamics are accelerated at its growth temperature. An interesting consequence of Model 1 is that it suggests cOA are primarily produced in a rapid burst within seconds to minutes after target RNA recognition and that the amount of cOA produced is weakly dependent on the abundance of the target transcript (Figs. 5E, 6).
Target RNA cleavage was the first activity identified for Type III CRISPR-Cas and was long thought to be integral for interference (Hale et al. 2009). However, accumulating data indicate the primary role of Csm3/Cmr4-mediated target RNA cleavage is the regulation of interference by antagonizing cOA synthesis and Cas10-mediated DNA cleavage (Estrella et al. 2016; Kazlauskiene et al. 2016; Rouillon et al. 2018). Regulation of cOA levels appears to be important in vivo as it was recently shown that the S. solfataricus Type III-D system contains ring nucleases dedicated to degrading cOA (Athukoralage et al. 2018). Additionally, it was shown that many CARF domain proteins exist which could act as proteases or transcription regulators in response to cOA (Shah et al. 2018; Shmakov et al. 2018). Therefore, a broad reprogramming of cellular pathways could result in cOA synthesis which would be detrimental to fitness if sustained under the wrong conditions. As a prime example, hyper-activating Csm6 could be detrimental to cell fitness since it appears to cut RNA indiscriminately, favoring only A-rich or C-rich sequences (Jiang et al. 2016; Foster et al. 2018; Gootenberg et al. 2018), and such persistent activation is hypothesized to lead to cell death (Makarova et al. 2014; Rostøl and Marraffini 2019). Studies on the role of Csm3 RNA cutting during bacteriophage infection showed that Csm3's RNA cleavage activity was dispensable for immunity (Jiang et al. 2016). However, a double mutant defective in Csm3 and Csm6 activity did not efficiently clear bacteriophage when targeting a late-expressed transcript (Jiang et al. 2016). Additionally, it has been shown that target RNA cleavage antagonizes Type III DNA cleavage providing spatial and temporal control of this activity (Estrella et al. 2016; Kazlauskiene et al. 2016). Our findings provide further evidence for the recently advanced model, in which Csm3 target RNA cleavage plays a minor role in Cas10-Csm–mediated interference, but a crucial regulatory role in shutting off interference once the foreign nucleic acid has been eliminated (Estrella et al. 2016; Kazlauskiene et al. 2016; Rouillon et al. 2018).
The effect of crRNA-target RNA mismatches on cOA synthesis and CRISPR immunity
The earliest studies of CRISPR-Cas function showed that single-nucleotide polymorphisms between a crRNA and a bacteriophage protospacer or regions adjacent to the protospacer could block CRISPR-Cas–mediated immunity (Barrangou et al. 2007; Deveau et al. 2008; Mojica et al. 2009). Multiple genetic studies in Type I and Type II systems showed that single-nucleotide polymorphisms affected immunity in multiple sequence contexts and later studies elucidated the structural basis for this phenomenon (Deveau et al. 2008; Semenova et al. 2011; Wiedenheft et al. 2011a,b). It was found in both Type I and II CRISPR-Cas systems that a seed region exists in the crRNA-protospacer duplex wherein full complementarity of the duplex is required for cleavage of foreign DNA (Semenova et al. 2011; Gorski et al. 2017). Additionally, a protospacer adjacent motif (PAM) is required to license interference in these systems (Mojica et al. 2009; Gasiunas et al. 2012; Jinek et al. 2012; Jiang et al. 2013). In contrast, Type III CRISPR-Cas systems do not rely upon a PAM and it has been argued do not use a seed sequence in the crRNA-protospacer duplex (Marraffini and Sontheimer 2010; Maniv et al. 2016; Pyenson et al. 2017). However, we wondered if spacer-protospacer mismatches near the 5′-tag might have a weighted impact on cOA production. Indeed, we have shown that single mismatches at several positions of the crRNA-target RNA duplex are sufficient to abrogate cOA production by Cas10-Csm (Fig. 7C). Our data taken together with previous studies (Kazlauskiene et al. 2017; Rouillon et al. 2018) show that in several sequence contexts subtle crRNA-target RNA mismatches can interfere with cOA synthesis which is crucial for interference (discussed further below).
Recent high-resolution cryo-EM structures of Type III-A CRISPR-Cas bound to a target RNA reveal that some base pairs are directly contacted by Cas10 (Fig. 2B; Jia et al. 2018; You et al. 2019). In Thermococcus onnurineus Cas10-Csm, these are base pairs +1 and +4 to +7, while in S. thermophilus Cas10-Csm, these are base pairs +3 to +13 (Jia et al. 2018; You et al. 2019). Cas10-Csm interference is inefficient against a target with multiple mismatches in the +1 to +10 or + 11 to +20 regions but is efficient despite multiple mismatches in the +26 to +35 region (Pyenson et al. 2017). This observation combined with the structural data may indicate the base pairs in direct contact with Cas10 are more important for interference than the distal base pairs. By measuring the effect of mismatches within the +1 to +11 region we have identified specific base pairs, which in the context of the spc1 crRNA-target RNA duplex, have an outsized effect on cOA activation.
It has been shown that cOA switch on the RNase activity of Csm6 and that this activity is required for defense by Cas10-Csm when targeting late-expressed bacteriophage transcripts (Niewoehner et al. 2017). In vivo experiments showed that all protein components of the cOA pathway are required for a wild-type level of defense: the Cas10 Palm domain which synthesizes cOA, the Csm6 CARF domain which binds cOA and the Csm6 HEPN domain nuclease activity (Niewoehner et al. 2017). Interestingly, genetic studies have indicated both Csm6 and its archaeal homolog Csx1 are required for anti-plasmid immunity also (Deng et al. 2013; Hatoum-Aslan et al. 2014). Taken together, these results indicate that the RNase activity of Csm6 plays an important role in Type III-A CRISPR-Cas–mediated immunity. The requirement for Csm6 in Type III CRISPR-Cas function implies that crRNA-target RNA mismatches that block cOA synthesis, in the right context, should affect CRISPR-Cas–mediated immunity. Additional investigations will be needed to test this hypothesis.
Type III CRISPR-Cas systems are capable of mounting a sophisticated and multifaceted response to foreign nucleic acids. Currently much is unknown concerning the specific activation and regulation of Type III CRISPR-Cas systems. Since specificity and regulation of CRISPR-Cas impinge strongly on bacterial fitness, these domains are crucial to understanding CRISPR biology. Identifying the molecular mechanisms controlling specific activation of cOA production and the in vivo consequences of the dysregulation of cOA production appear to be salient future directions.
MATERIALS AND METHODS
Strains and growth conditions
Staphylococcus epidermidis LM1680 (Hatoum-Aslan et al. 2013) was propagated in Brain Heart Infusion (BHI) broth (Difco), and S. aureus RN4220 (Kreiswirth et al. 1983) was grown in Tryptic Soy Broth (TSB, Difco). For S. epidermidis strains, media were supplemented with 10 µg/ml chloramphenicol (for selection of pcrispr-based plasmids) and 15 µg/mL neomycin (for selection of S. epidermidis).
Construction of S. epidermidis LM1680 bearing pcrispr/csm3D32A
pcrispr/csm3D32A was constructed from pcrispr-cas (Hatoum-Aslan et al. 2013) using Gibson assembly (Gibson et al. 2009). Briefly, PCR products using primers F027/F014 and F028/F016 (listed in Supplemental Table S2) were purified (EZNA Cycle Pure Kit [Omega]) and assembled. The assembled plasmid was first electroporated into S. aureus RN4220. The mutation was verified in three independent colonies using sequencing of a colony PCR product with primers A426/F004. The plasmid was purified and sequenced (using primers A414-A426) from one mutant to confirm the absence of unintended mutations within the cas genes. This plasmid was then electroporated into S. epidermidis LM1680 for use in further experiments.
Purification of Cas10-Csm
Cas10-Csm expression and immobilized metal affinity chromatography were performed as reported by Chou-Zheng and Hatoum-Aslan (2017). Briefly, overnight cultures of S. epidermidis LM1680 carrying the plasmids pcrispr or pcrispr/csm3D32A were used to inoculate 1 L of brain heart infusion media containing appropriate antibiotics. Growth was continued at 37°C until OD600 ∼2.0 was reached, and cells were harvested by centrifugation. Lysis was achieved by sonication and the addition of lysostaphin to 28 µg/ml, and cells were clarified by centrifugation at 19,000g for 30 min. The lysate was added to 1 mL of Ni2+-NTA resin, and nonspecifically bound protein was removed with wash buffer (100 mM NaH2PO4 pH 8.0, 600 mM NaCl, 40 mM imidazole). Cas10-Csm was eluted with wash buffer containing 250 mM imidazole. Cas10-Csm enriched fractions were pooled and layered onto a 5%–20% (w/v) sucrose gradient composed of 50 mM Tris HCl pH 8.0, 150 mM NaCl, and 5% (v/v) glycerol. Ultracentrifugation was performed for 41 h at 118,000g on a Beckman SW-32TI rotor. Fractions containing intact Cas10-Csm complex were identified by A280 measurements and appeared near the bottom of the gradient, consistent with a molecular weight of ∼ 300 kDa. Sample purity was analyzed by 4%–20% SDS-PAGE and purified Cas10-Csm was flash-frozen and stored at −80°C.
Purification and visualization of crRNAs
crRNAs were extracted from the protein complex with 1:1 (v/v) ratio of phenol-chloroform-isoamyl alcohol (25:24:1) twice, followed by one extraction with 1 vol chloroform. CrRNAs were 32P-labeled by incubation with γ-32P-ATP (3000 Ci/mmol) and T4 PNK (NEB) at 37°C for 1 h. Unreacted γ-32P-ATP was removed by passing the reaction over a G-25 spin column. Extracted crRNAs were mixed 1:1 with formamide dye (5 mM EDTA pH 8.0, 95% formamide v/v, 0.025% w/v bromophenol blue, 0.025% w/v xylene cyanol) and heated at 70°C for 2 min before loading to a 12%, 8 M urea gel. Invitrogen Decade Markers were added to the gel to infer crRNA sizes. Electrophoresis was carried out at 50 W for approximately 90 min. Phosphorimaging was performed with storage phosphor screens and a Typhoon FLA 7000 both from GE Healthcare.
cOA synthesis
All reactions were carried out in 50 mM Tris HCl pH 8.0, 150 mM NH4Cl, 5% (v/v) glycerol, 500 µM ATP and 10 mM Mg2+ (TNG buffer) at 37°C except where differences in divalent metal or ATP concentrations are noted. Reactions analyzed by PAGE were supplemented with 30 nM α-32P-ATP 3000 Ci/mmol. Reaction conditions for Figures 3 and 4 included 200 nM Cas10-CsmCsm3D32A, 200 nM target RNA in TNG buffer for 4-h incubation. cOA synthesis was performed overnight to generate the products for mass spectrometry (Fig. 4B). Exonuclease treatment to determine cOA topology was performed as follows: 5 µL cOA reaction product (Fig. 4A) was incubated with 20 units T4 PNK for 1 h at 37°C in a total volume of 10 µL in PNK buffer, T4 PNK was then inactivated at 65°C for 2 min. A total of 2.5 µL NEB Buffer 4 and 10 units exonuclease T (NEB) were added to the reaction, which was then incubated at room temperature for 1 h. Endonuclease treatment of cOA products was performed as follows: 5 µL of cOA reaction product (Fig. 4A) was incubated with 160 units endonuclease S1 in S1 nuclease buffer (Promega) in 10 µL total volume for 1 h at 37°C.
The reactions in Figure 5A were performed with Cas10-Csm 200 nM, 200 nM target RNA or DNA as indicated in TNG buffer for 2-h incubation. Reactions in Figure 5D were performed with 200 nM Cas10-Csm or Cas10-CsmCsm3D32A, 200 nM ssRNA-01 in TNG buffer. Figure 5E reactions were carried out with 100 nM Cas10-Csm and 100–3200 nM ssRNA-01 in TNG buffer for 60 min. cOA synthesis progress curves (Fig. 6C,D) were obtained using 100 nM Cas10-Csm and 3000 nM ssRNA-01 or Cas10-CsmCsm3D32A and 3000 nM ssRNA-01 incubated in TNG buffer. cOA synthesis in the presence of S. cerevisiae total RNA was performed with Cas10-Csm 200 nM in TNG buffer for 2-h incubation. ssRNA-01 was present at 200 nM where indicated. S. cerevisiae total RNA used in Figure 5A was obtained using the Ambion RiboPure Yeast kit and added to reactions to 480 µg/mL as indicated. For reactions measuring the effect of mismatches on cOA synthesis (Figs. 7, 8), reactions were carried out with 100 nM Cas10-Csm or Cas10-CsmCsm3D32A, 400 nM target RNA in TNG buffer for 12 min.
PAGE analysis of cOA reaction products was carried out with 24% w/v acrylamide (19:1), 8 M urea, tris-borate-EDTA gels. Prior to gel loading, cOA synthesis reactions were quenched by adding 1:1 formamide dye and incubating at 70°C for 2 min. Electrophoresis was performed at 2900 V for approximately 3 h.
Target RNAs
Target RNAs used in cOA synthesis assays are given in Supplemental Table S3. All RNAs are derived from the sequence of the nickase (nes) gene of the pG0400 conjugative plasmid that can be targeted by the first spacer (spc1) crRNA of pcrispr (Marraffini and Sontheimer 2008; Hatoum-Aslan et al. 2013). ssRNA-01 is identical to 43 nt of the nes transcript. ssRNA-02 is modified from the nes transcript to possess no base pairs in the tag-flank region while the ssRNA-03 has been altered to fully base-pair with the 5′ tag of spc1 crRNA. ssRNA-NC was designed by randomly shuffling the sequence of ssRNA-01 with EMBOSS (Rice et al. 2000). Synthetic RNAs were made and PAGE purified by Dharmacon. DNA templates for in vitro transcription of target RNAs were generated by overlap extension PCR using Phusion polymerase (NEB) according to the manufacturer's instructions using the primers shown in Supplemental Table S4. In vitro transcription was performed using the RiboMax (Promega) kit. Cleanup of the in vitro transcription products was performed by DNase I digestion, phenol-chloroform extraction, and ethanol precipitation according to the manufacturer's instructions, except phenol-chloroform-isoamyl alcohol 25:24:1 was used for extractions. An additional PAGE purification was performed on the in vitro transcription products. The quality of the purified RNAs was checked by a 12% acrylamide urea-PAGE run at 300 V for 30 min, stained with SYBR green II (Lonza), and imaged on a Typhoon FLA 7000 imager.
Mass spectrometry
The presence of a ∼70 kDa truncated Cas10 variant in the Cas10-Csm complex was identified by excision of the band following 10% acrylamide SDS-PAGE. The band was digested overnight with Trypsin Gold, Mass Spectrometry Grade (Promega, cat. # V5280) following the manufacturer's instructions. Peptide extracts were reconstituted in 0.1% formic acid/ddH2O at 0.1 µg/µL. Electrospray ionization tandem mass spectrometry was carried out, and the data were processed, searched, filtered, grouped, and quantified, as previously reported in detail (Ludwig et al. 2016). Mass spectrometry was performed by the UAB Cancer Center Mass Spectrometry and Proteomics Shared Facility.
For mass spectrometry analysis of cOA reactions, products were desalted with C18 ziptips. Trifluoroacetic acid (TFA) was added to the products to 0.6% v/v and the products were adsorbed to the C18 matrix, washed with 0.1% TFA and eluted with 0.1% TFA and 50% acetonitrile. One microliter of desalted cOA products was mixed with 1 µL of matrix (25 mM ammonium citrate, saturated THAP in 50% v/v acetonitrile) and subjected to matrix-assisted laser desorption ionization (MALDI) mass spectrometry with a Bruker rapifleX MALDI TOF/TOF.
RNA cleavage
ssRNA-01 was 32P-labeled with T4 PNK and γ-32P-ATP as previously described for extracted crRNAs. The rate of target RNA cleavage (Fig. 5B) was measured in a single-turnover assay with 1000 nM Cas10-Csm and 50 nM ssRNA-01. RNA cleavage was initiated with Mg2+ and reactions were quenched with formamide dye and subjected to 12% urea-PAGE at 50 W for 120 min followed by phosphorimaging. Target RNA cleavage in Figure 5C was measured with 200 nM Cas10-Csm or Cas10-CsmCsm3D32A, 200 nM ssRNA-01, 10 mM Mg2+ in TNG buffer. Reactions were quenched and analyzed by PAGE as above.
Measuring koff of cleaved target RNA
The dissociation rate of cleaved target RNA was measured with a pulse-chase strategy. A total of 100 nM Cas10-Csm was incubated with 1 nM 32P-labeled ssRNA-01 for 5 min at 37°C with 1 mM EDTA present. A total of 10 nM ssRNA-NC was also present to prevent nonspecific binding. RNA cleavage was initiated with 10 mM Mg2+, and simultaneously a 100 nM unlabeled ssRNA-01 chase was added. Timepoints were collected by adding 20 µL reaction volumes to a BioDot apparatus containing a nitrocellulose-Hybond membrane sandwich followed by vacuum filtering. A 100 µL wash was applied to the membrane sandwich after filtering. Dried membranes were exposed to a phosphorimaging screen to measure the fraction of 32P-labeled RNA associated with each membrane.
Electrophoretic mobility shift assays (EMSA)
Varying concentrations of Cas10-CsmCsm3D32A (0, 1, 5, 10, 50, 125, 250 nM) were incubated with 1 nM 32P-radiolabeled ssRNA at 37°C for 30 min. The binding buffer contained 50 mM Tris HCl, pH 8.0, 10 mM Mg2+, 5% (v/v) glycerol, 0.1 mg/mL BSA, and 0.5 mM EDTA. The binding reactions were analyzed by a 6% native PAGE gel at 25°C followed by phosphorimaging.
Data analysis and modeling cOA synthesis progress curves
32P-labeled cOA or target RNAs were quantitated by phosphorimaging and analysis in GE ImageQuant TL 8.1. Significance tests and curve fitting were performed using GraphPad Prism 7. Significance tests were performed as two-tailed t-tests. Where absolute amounts, rather than relative amounts, of cOA were reported, cOA amounts were calculated using a 32P-labeled oligonucleotide of known specific activity included in the PAGE gel as an internal standard. A rate constant for target RNA cleavage (kcleav) was obtained by fitting data from a single turnover assay to a one-phase exponential decay. A rate constant for dissociation of cleaved target RNA (kdis) from Cas10-Scm was obtained by fitting to a one-phase exponential decay. A kcat ≃ 14 min−1 for cOA synthesis was obtained by linear regression of initial velocity data with Cas10-CsmCsm3D32A complex to decouple kcat from RNA cleavage.
Modeling cOA synthesis progress curves was performed using MatLab. A system of differential equations, dI/dt = −kcleav • [I] + kon • [U], dC/dt = −kdis • [C] + kcleav • [I], dU/dt = −kon • [U] + kdis • [C], where I represents Cas10-Csm complex with a bound and intact target RNA, C represents complex with a bound and cleaved target RNA, and U represents complex unbound to target RNA, was solved with the dsolve function producing expressions of I, C, and U as a function of time. The initial conditions were set to [I] = 0, [C] = 0, and [U] = 1 fraction of the total Cas10-Csm. The rate constant kon, which describes target RNA binding to Cas10-Csm, was assumed to be very rapid (109 M−1s−1). In Model 1, only species I is active in cOA synthesis, whereas in Model 2, species I and C are active in cOA synthesis. The cOA progress curves were simulated for Model 1 by integrating [I] • kcat • [Cas10-Csm] or for Model 2 by integrating [I + C] • kcat • [Cas10-Csm] from time = 0 to t.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The National Science Foundation (CHE-1726812) provided support for the acquisition of the Bruker rapifleX MALDI TOF/TOF mass spectrometer. The UAB Cancer Center Mass Spectrometry/Proteomics (MSP) Shared Facility is supported by the National Institutes of Health (P30CA013148). The UAB Cancer Center MSP Shared Facility is also supported by the UAB Institutional Core Funding Mechanism. National Institutes of Health (1R15GM129671-01) and University of Alabama startup funds provided funding for publication charges.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070417.119.
REFERENCES
- Athukoralage JS, Rouillon C, Graham S, Grüschow S, White MF. 2018. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562: 277–280. 10.1038/s41586-018-0557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Horvath P. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol 2: 17092 10.1038/nmicrobiol.2017.92 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. 2011. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J 30: 4616–4627. 10.1038/emboj.2011.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou-Zheng L, Hatoum-Aslan A. 2017. Expression and purification of the Cas10-Csm complex from Staphylococci. Bio Protoc 7: e2353 10.21769/BioProtoc.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87: 1088–1099. 10.1111/mmi.12152 [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo FT, Bailey S. 2016. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev 30: 460–470. 10.1101/gad.273722.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Kalter J, Woodside W, Terns RM, Terns MP. 2018. The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol 16: 449–460. 10.1080/15476286.2018.1493334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. 2014. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514: 633–637. 10.1038/nature13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360: 439–444. 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Vogel J, Doudna JA. 2017. RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol 18: 215–228. 10.1038/nrm.2016.174 [DOI] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956. 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. 2011. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci 108: 21218–21222. 10.1073/pnas.1112832108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. 2013. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem 288: 27888–27897. 10.1074/jbc.M113.499244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. 2014. Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J Bacteriol 196: 310–317. 10.1128/JB.01130-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. 2014. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345: 1473–1479. 10.1126/science.1256328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Mo CY, Wang C, Eng ET, Marraffini LA, Patel DJ. 2018. Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol Cell 73: 264–277.e5. 10.1016/j.molcel.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Samai P, Marraffini LA. 2016. Degradation of phage transcripts by CRISPR-associated RNases enables Type III CRISPR-Cas immunity. Cell 164: 710–721. 10.1016/j.cell.2015.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C, Siksnys V. 2016. Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62: 295–306. 10.1016/j.molcel.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V. 2017. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357: 605–609. 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- Klompe SE, Sternberg SH. 2018. Harnessing “a billion years of experimentation”: the ongoing exploration and exploitation of CRISPR–Cas immune systems. CRISPR J 1: 141–158. 10.1089/crispr.2018.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37: 67–78. 10.1016/j.mib.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305: 709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- Liu TY, Iavarone AT, Doudna JA. 2017. RNA and DNA targeting by a reconstituted Thermus thermophilus type III-A CRISPR-Cas system. PLoS One 12: e0170552 10.1371/journal.pone.0170552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeff L, Brouns SJJ, Joo C. 2018. Repetitive DNA reeling by the Cascade-Cas3 complex in nucleotide unwinding steps. Mol Cell 70: 385–394.e3. 10.1016/j.molcel.2018.03.031 [DOI] [PubMed] [Google Scholar]
- Ludwig MR, Kojima K, Bowersock GJ, Chen D, Jhala NC, Buchsbaum DJ, Grizzle WE, Klug CA, Mobley JA. 2016. Surveying the serologic proteome in a tissue-specific kras(G12D) knockin mouse model of pancreatic cancer. Proteomics 16: 516–531. 10.1002/pmic.201500133 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. 2002. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res 30: 482–496. 10.1093/nar/30.2.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. 2014. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet 5: 102 10.3389/fgene.2014.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13: 722–736. 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniv I, Jiang W, Bikard D, Marraffini LA. 2016. Impact of different target sequences on type III CRISPR-Cas immunity. J Bacteriol 198: 941–950. 10.1128/JB.00897-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463: 568–571. 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. 2014. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345: 1479–1484. 10.1126/science.1256996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. 2017. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548: 543–548. 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Sato C, Numata T. 2015. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell 58: 418–430. 10.1016/j.molcel.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Pyenson NC, Gayvert K, Varble A, Elemento O, Marraffini LA. 2017. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22: 343–353.e3. 10.1016/j.chom.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Rostøl JT, Marraffini LA. 2019. Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat Microbiol 4: 656–662. 10.1038/s41564-018-0353-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S, Grüschow S, White MF. 2018. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 7: e36734 10.7554/eLife.36734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. 2015. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161: 1164–1174. 10.1016/j.cell.2015.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci 108: 10098–10103. 10.1073/pnas.1104144108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Alkhnbashi OS, Behler J, Han W, She Q, Hess WR, Garrett RA, Backofen R. 2018. Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol 16: 530–542. 10.1080/15476286.2018.1483685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov SA, Makarova KS, Wolf YI, Severinov KV, Koonin EV. 2018. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci 115: E5307–E5316. 10.1073/pnas.1803440115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342. 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. 2013. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 52: 146–152. 10.1016/j.molcel.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. 2013. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52: 135–145. 10.1016/j.molcel.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. 2014. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 56: 518–530. 10.1016/j.molcel.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. 2014. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell 56: 506–517. 10.1016/j.molcel.2014.09.027 [DOI] [PubMed] [Google Scholar]
- Vashishtha AK, Wang J, Konigsberg WH. 2016. Different divalent cations alter the kinetics and fidelity of DNA polymerases. J Biol Chem 291: 20869–20875. 10.1074/jbc.R116.742494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker FC, Chou-Zheng L, Dunkle JA, Hatoum-Aslan A. 2017. Molecular determinants for CRISPR RNA maturation in the Cas10-Csm complex and roles for non-Cas nucleases. Nucleic Acids Res 45: 2112–2123. 10.1093/nar/gkw891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605. 10.1016/j.molcel.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. 2011a. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477: 486–489. 10.1038/nature10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. 2011b. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108: 10092–10097. 10.1073/pnas.1102716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. 2016. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 164: 29–44. 10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- You L, Ma J, Wang J, Artamonova D, Wang M, Liu L, Xiang H, Severinov K, Zhang X, Wang Y. 2019. Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176: 239–253.e16. 10.1016/j.cell.2018.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. 2014. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515: 147–150. 10.1038/nature13733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.