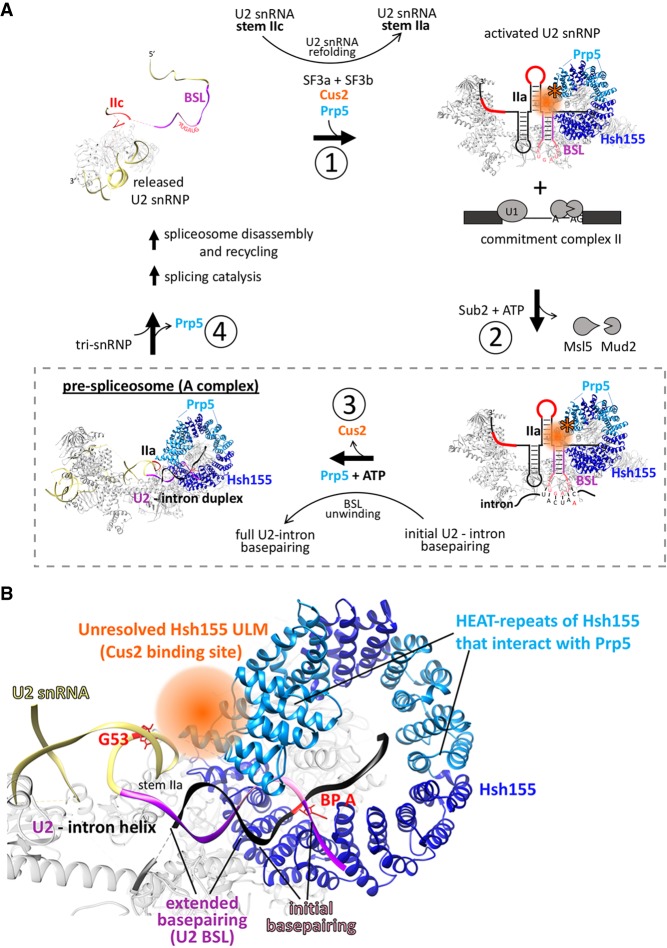
FIGURE 7.
A model of ATP-dependent pre-spliceosome assembly. (A) After a completed round of splicing and spliceosome disassembly, U2 snRNA is released from the spliceosome in the stem IIc (red) form and lacks the SF3a and SF3b subcomplexes. (1) Prior to another round of splicing, the BSL (purple) of U2 snRNA is reformed and Cus2 (orange) associates with both U2 snRNA and the ULM of Hsh155 to promote the transition from stem IIa to stem IIc. (2) The activated U2 snRNP then engages the intron BP in the commitment complex through a limited number of initial base pairs between the BP and the loop of the BSL (pink), possibly displacing the Msl5-Mud2 heterodimer from the commitment complex. (3) ATP hydrolysis by Prp5 disrupts the Cus2 UHM/Hsh155 ULM interaction to release Cus2 and unwinds the helix of the BSL to promote stable duplex formation between U2 snRNA and the intron BP generating the pre-spliceosome. (4) Stable duplex formation between U2 snRNA and the intron BP results in release of Prp5 and recruitment of the U4/U5/U6 tri-snRNP. Aspects of this model specific to this work are boxed with a dashed line. (B) Cryo-EM model (PDB 6G90) of the yeast pre-spliceosome showing the putative binding site of Cus2 relative to Hsh155, the U2/intron helix, and stem IIa of U2 snRNA. The unresolved ULM of Hsh155 and the space likely occupied by Cus2 is orange. HEAT repeats of Hsh155 previously shown to interact with Prp5 are light blue. Sequences of U2 snRNA that compose the BSL are purple (stem) and pink (loop). The G53A mutation in U2 snRNA that destabilizes stem IIa and is suppressed by Cus2 is indicated in red.