Abstract
To initiate X-Chromosome inactivation (XCI), the long noncoding RNA Xist mediates chromosome-wide gene silencing of one X Chromosome in female mammals to equalize gene dosage between the sexes. The efficiency of gene silencing is highly variable across genes, with some genes even escaping XCI in somatic cells. A gene's susceptibility to Xist-mediated silencing appears to be determined by a complex interplay of epigenetic and genomic features; however, the underlying rules remain poorly understood. We have quantified chromosome-wide gene silencing kinetics at the level of the nascent transcriptome using allele-specific Precision nuclear Run-On sequencing (PRO-seq). We have developed a Random Forest machine-learning model that can predict the measured silencing dynamics based on a large set of epigenetic and genomic features and tested its predictive power experimentally. The genomic distance to the Xist locus, followed by gene density and distance to LINE elements, are the prime determinants of the speed of gene silencing. Moreover, we find two distinct gene classes associated with different silencing pathways: a class that requires Xist-repeat A for silencing, which is known to activate the SPEN pathway, and a second class in which genes are premarked by Polycomb complexes and tend to rely on the B repeat in Xist for silencing, known to recruit Polycomb complexes during XCI. Moreover, a series of features associated with active transcriptional elongation and chromatin 3D structure are enriched at rapidly silenced genes. Our machine-learning approach can thus uncover the complex combinatorial rules underlying gene silencing during X inactivation.
X-Chromosome inactivation (XCI) is a developmental process in mammals that ensures equal gene dosage of X-linked genes between XX and XY individuals by transcriptional inactivation of one of the two X Chromosomes in female cells. In placental mammals XCI is triggered by the long noncoding RNA (lncRNA) Xist, which is up-regulated in a monoallelic fashion and coats the future inactive X Chromosome in cis, leading to the recruitment of several factors involved in transcriptional inactivation and eventually converting the entire X Chromosome into silent heterochromatin (Escamilla-Del-Arenal et al. 2011; Gendrel and Heard 2014; Galupa and Heard 2015).
Early events following Xist coating include the depletion of RNA polymerase II (RNAPII) from the Xist RNA domain and loss of active histone marks (Chaumeil et al. 2002, 2006) as well as gain of repressive chromatin modifications, such as H2AK119ub1 and H3K27me3, deposited by the Polycomb repressive complexes (PRC) 1 and 2, respectively (Plath et al. 2003, 2004; Silva et al. 2003; de Napoles et al. 2004). Subsequently, additional chromatin modifications are gained such as the histone variant macroH2A and DNA methylation of gene promoters (Escamilla-Del-Arenal et al. 2011). In recent years, progress has been made in identifying proteins that mediate Xist’s functions as well as the domains within the Xist RNA that recruit these proteins. Xist contains multiple conserved repeats, among which the A repeat mediates gene silencing through activation of SPEN and other factors including RBM15 (Chu et al. 2015; McHugh et al. 2015), and the B repeat recruits PRC indirectly through HNRNPK (Wutz et al. 2002; Brockdorff 2017; Pintacuda et al. 2017). The dynamics of Xist-mediated silencing are highly variable across genes (Chow et al. 2010; Borensztein et al. 2017; Żylicz et al. 2019), with a subset of so-called escapees remaining active even in somatic cells (Berletch et al. 2011). However, the determinants of susceptibility to XCI remain poorly understood. Because XCI is a multistep process, local interference with any step, such as Xist coating or access to the silencing machinery of one or several silencing pathways, could delay or prevent silencing of a certain gene. Defining the features that underlie differential susceptibility to XCI remains an important question, particularly because genes that are not fully silenced are implicated in diseases, such as autoimmune syndromes (Bianchi et al. 2012).
Xist RNA spreading occurs by proximity transfer to sites close to the Xist locus genomically or in 3D space (“Xist entry sites”) (Engreitz et al. 2013). From there, Xist has been proposed to move first into gene-dense regions and then spread to intergenic domains of the X Chromosome (Engreitz et al. 2013; Simon et al. 2013). In differentiated cells, Xist covers the entire X Chromosome but is reduced at escape genes (Engreitz et al. 2013; Simon et al. 2013). Xist coating is positively correlated with gene density and with PRC2 enrichment and negatively correlated with the density of LINE elements (Engreitz et al. 2013; Simon et al. 2013). Similarly, gene silencing tends to be slower for genes further from the Xist locus (Marks et al. 2015; Borensztein et al. 2017) and from LINE elements (Chow et al. 2010; Loda et al. 2017). Moreover, efficiently silenced genes tend to be enriched for Polycomb complexes (RING1B, H3K27me3) and depleted for CTCF and active marks such as H3K4me3 and H3K27ac prior to Xist-induced silencing (Kelsey et al. 2015; Loda et al. 2017). Thus, a variety of genetic and epigenetic features have been implicated in controlling gene-specific silencing efficiency. However, none of these features alone can predict whether and to what extent a gene will be silenced during XCI, and the associations with measured silencing efficiencies are generally weak. Because no predictive pattern of features has so far been identified, the susceptibility of genes to Xist-mediated silencing is likely to be controlled by a complex combination of different features.
In this study we set out to identify the genetic and chromatin features that predispose genes on the X Chromosome to be efficiently silenced or escape XCI. We measured chromosome-wide silencing dynamics of X-linked genes following induction of Xist expression, using allele-specific Precision nuclear Run-On sequencing (PRO-seq) (Kwak et al. 2013). We then trained two Random Forest machine-learning models to predict from 77 genomic and epigenetic features (1) whether a gene is subject to XCI, and (2) whether it will be silenced with fast or slow kinetics. Through forest-guided gene clustering, we identified feature sets that determine the silencing dynamics of subgroups of genes, indicating that variable silencing efficiencies might be associated with distinct silencing pathways. We have thus developed a framework to comprehensively assess the relative contribution of genetic and epigenetic factors to transcriptional silencing of the X Chromosome in an unbiased and quantitative manner.
Results
Quantification of gene-specific silencing dynamics by PRO-seq
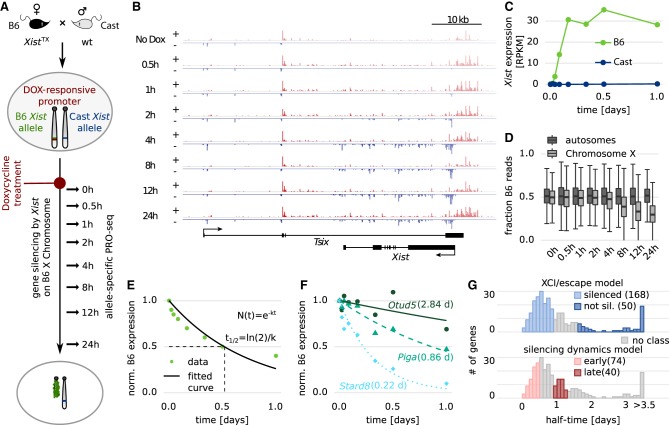
To quantify gene silencing dynamics during XCI in a chromosome-wide manner, we performed allele-specific PRO-seq during ectopic Xist induction in female murine embryonic stem cells (mESCs). Using an inducible system allowed us to overcome the asynchronous nature of XCI, which normally limits the temporal resolution of population measurements in differentiating mESCs, the classic model system to study XCI (Chow et al. 2010). We used the female TX1072 mESC line (Schulz et al. 2014), which was derived from a cross between two different mouse strains (C57BL/6 × CAST/EiJ) and in which Xist up-regulation from the endogenous locus on the B6 X Chromosome can be induced by doxycycline (Dox) treatment in undifferentiated cells. For a direct readout of gene silencing, we measured the nascent transcriptome by allele-specific PRO-seq (Kwak et al. 2013) at different time points up to 24 h of Dox treatment (Fig. 1A). Xist started to be up-regulated from the B6 chromosome about 1 h after Dox treatment and reached a plateau after 4 h (Fig. 1B,C; Supplemental Fig. S1), whereas global expression of the B6 X Chromosome was gradually reduced over time due to X inactivation, starting at 4 h of treatment (Fig. 1D).
Figure 1.
Measuring gene silencing dynamics. (A) Schematic of the experimental setup used in B–F. Using a hybrid female mESC line (B6 × CAST) carrying a Dox-responsive promoter in front of the endogenous Xist gene on the B6 allele, RNAPII activity was measured by allele-specific PRO-seq over a 24-h time course of Dox treatment. (B) Strand-specific read density at the Tsix-Xist locus. Plus-strand is shown in red, minus strand in blue; the y-axis indicates reads per million. (C) Xist expression from the two alleles. (D) Distribution of the fraction of B6 reads for autosomal and X-linked genes over time. (E,F) Schematic (E) and three examples (F) of how gene silencing half-times (in parentheses) were estimated from the allele-specific PRO-seq time course data through fitting an exponential decay function. (G) Distribution of estimated half-times for 280 X-linked genes with an assigned active transcription start site (TSS). The half-time ranges used to define the model classes and the number of genes falling in each category are indicated.
To quantify silencing dynamics, we fitted an exponential decay function to each gene and estimated gene-specific silencing half-times, that is, the time point when transcription on the B6 X Chromosome is reduced by 50% compared to the uninduced control (Fig. 1E and examples in 1F). After several filtering steps, we had estimated reliable half-times for 280 genes, which were used for all subsequent analyses and ranged from several hours up to several days (Fig. 1G; Supplemental Tables S1, S2).
Silencing dynamics are comparable in vitro and in vivo
To ensure that the relative silencing dynamics across genes, when XCI is induced in undifferentiated mESCs, are comparable to those in the cellular context where XCI occurs endogenously, we generated two additional data sets, in which mRNA-seq was performed at different time points of Dox treatment in undifferentiated and differentiating mESCs (Fig. 2A). The computed half-times were comparable between these two data sets (Pearson correlation coefficient: r = 0.75) (Fig. 2B), suggesting that the differentiation process only has a minor impact on relative gene silencing dynamics. When comparing half-times estimated from the two different data types (mRNA-seq vs. PRO-seq), correlation was generally a bit lower, independent of the cellular context (Pearson correlation coefficient r = 0.52 and r = 0.51) (Fig. 2C,D), which would be expected given that PRO-seq measures the direct transcription dynamics, whereas mRNA-seq kinetics are modulated by transcription, RNA-processing, and degradation. We also compared the estimated half-times to a previous study (Marks et al. 2015) that had used a different, Dox-independent strategy to make XCI nonrandom by inserting a stop-cassette in Xist’s repressive antisense transcript Tsix. The silencing classes defined in Marks et al. (2015) (early, intermediate, late, escapee) are in good agreement with the half-times estimated from the PRO-seq data (Fig. 2E), suggesting that Dox-induced XCI recapitulates endogenous gene silencing dynamics.
Figure 2.
Comparison of PRO-seq-based silencing half-times to other data sets. (A) Comparison of PRO-seq (undifferentiated mESC, upper) and mRNA-seq data (undifferentiated mESCs, middle; differentiated mESCs, lower). Fraction of B6 reads are shown for all genes covered in all three data sets, ordered by genomic position. (B–D) Comparison of estimated half-times (in days) between the data sets shown in A (replicate B only for mRNA-seq undiff.) with fitted regression lines (red). Pearson correlation coefficients are indicated. (E–G) Distribution of half-times within silencing classes defined previously in mESCs (E) (Marks et al. 2015), in preimplantation mouse embryos (F) (Borensztein et al. 2017), and the classes used for Random Forest modeling (G): (blue) XCI/escape model; (red) silencing dynamics model. (H) Estimated half-times (black circles) for all genes in the PRO-seq data set along the X Chromosome. A fitted smooth curve of the half-times is displayed as a blue line, and the Xist locus is marked with a gray line. (I) Cumulative distribution of half-times of genes silenced (independent, light gray) or not silenced (dependent, dark gray) by Xist lacking the repeat A element (Sakata et al. 2017).
Finally, we compared our Xist-induced gene silencing half-times in mESCs to the dynamics of (imprinted) XCI measured in preimplantation mouse embryos in vivo through single-cell RNA-seq (Borensztein et al. 2017). The gene classification in that study (early: 16-cell stage; intermediate: 32-cell stage; late: blastocyst stage or escapee) was once more in good agreement with the silencing half-times estimated from the PRO-seq data (Fig. 2F). Based on the PRO-seq derived silencing half-times, we classified all genes according to whether they are subject to XCI or escape (Figs. 1G, 2G, silenced/not silenced) and whether they are silenced with slow or fast kinetics (Figs. 1G, 2G, early/late). Genes with intermediate half-times between the classes were excluded from the analysis (see gap between groups in Fig. 1G). The resulting classes largely agree with those previously defined in differentiating mESCs and in preimplantation embryos (compare Fig. 2G with Fig. 2E,F). Moreover, the “not silenced” class contains 37 of 50 (74%) known escapees annotated from different cell types and is strongly enriched for escapees compared to the “silenced” class when considering only (high confidence) escapees identified in at least two different studies (e.g., Ddx3x, Taf1, Pdbc1, Kdm6a, Usp9x, Hcfc1, Hdac6, Mgmt1, Ftx, Nkap, and Uba1, odd ratio = 6.7, P = 2.6×10−5, Fisher's exact test) (Supplemental Table S2; Yang et al. 2010; Berletch et al. 2011; Splinter et al. 2011; Calabrese et al. 2012; Wu et al. 2014; Marks et al. 2015; Andergassen et al. 2017).
Identifying determinants of gene silencing dynamics with Random Forest modeling
We noted that genes close to the Xist locus tended to be silenced earlier than distal genes (Fig. 2H), in agreement with a previous study (Marks et al. 2015). However, many genes did not follow this trend. To uncover additional factors that potentially determine the susceptibility to Xist-mediated silencing, we developed a machine-learning model to predict silencing dynamics based on genomic and epigenetic features.
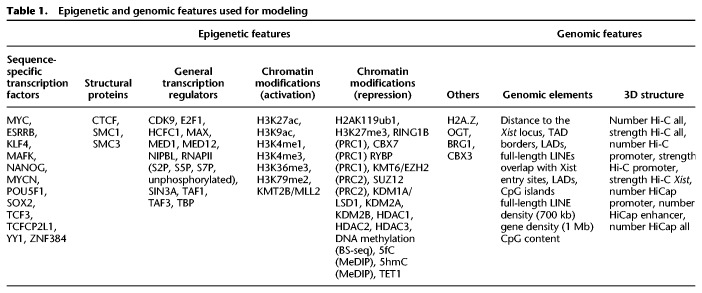
We collected 138 publicly available high-throughput data sets (ChIP-seq and bisulfite-seq) measuring chromatin modifications and DNA-binding factors, mostly in male mESCs. Because these data sets had been generated in undifferentiated mESCs, they correspond to the chromatin state before Xist induction. After stringent filtering on data quality, we computed the enrichment for 59 of these epigenomic features at promoters or gene body regions as appropriate (Table 1; Supplemental Text S1). In addition, we included a series of genomic and structural features, such as gene density, the frequency of 3D chromatin interactions with different genomic elements, and the linear distance to other genomic features, such as the distance to the Xist locus, the next TAD boundary, lamin-associated domain (LAD), or full-length LINE element (Table 1; Supplemental Text S1).
Table 1.
Epigenetic and genomic features used for modeling
A linear model to predict a gene's susceptibility to Xist-mediated silencing from the collected epigenetic and genomic features had little predictive power. This indicates that no single linear combination of features or rules could be defined to discriminate, for example, silenced from not silenced genes. The different functional domains of Xist might recruit distinct silencing complexes (e.g., PRC1 and SPEN/HDAC3), and susceptibility to each silencing pathway might be determined by distinct feature patterns. We expected to identify such feature patterns from our data set, because genes that require the Xist A repeat for silencing, which activates SPEN, exhibit longer silencing half-times compared to A repeat–independent genes that might be preferentially targeted by Polycomb complexes (Kolmogorov-Smirnov [KS] test, P = 2.2×10−6) (Fig. 2I; Sakata et al. 2017).
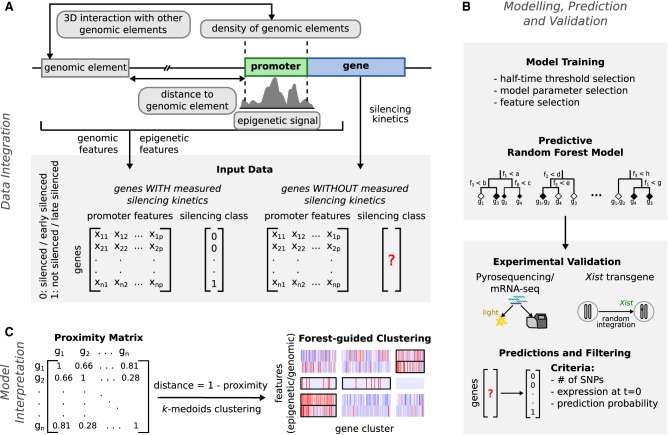
To identify combinatorial rule sets that could predict silencing susceptibility, we used Random Forest, a nonparametric machine-learning method that combines an ensemble of single classification trees, which successively split the feature input space in a nonlinear fashion, to predict the value of a discrete binary variable (Fig. 3A,B). We built two binary classification models to predict from a total of 77 epigenetic and genomic features whether a gene would be silenced or not (XCI/escape model), and whether it would be silenced early or late (silencing dynamics model) using the classification described above (Table 1; Fig. 1G). The half-time thresholds were selected such that they maximize the model classification accuracy (Supplemental Table S3; Supplemental Text S2). The XCI/escape model would identify combinations of factors, that are important for silencing in general, and the silencing dynamics model would find those that influence the kinetics of gene silencing.
Figure 3.
Schematic overview of our modeling approach. (A) Epigenetic and genomic input data for the model are collected, and feature matrices are computed for all X-linked genes with estimated half-times (labeled) and without estimated half-times (unlabeled). (B) After model training, the XCI/escape model is then used to predict the silencing class of all unlabeled X-linked genes given the same set of input features. The predictions are validated by comparing them to measured half-times from undifferentiated mRNA-seq data, with pyrosequencing experiments (few selected genes) and with measured silencing dynamics of genes in six transgenic mESCs clones. (C) A forest-guided clustering approach was developed for model interpretation. A proximity matrix between genes is computed from the trained model and converted into a distance matrix. Clusters of genes and their most significant associated features are displayed as a heatmap.
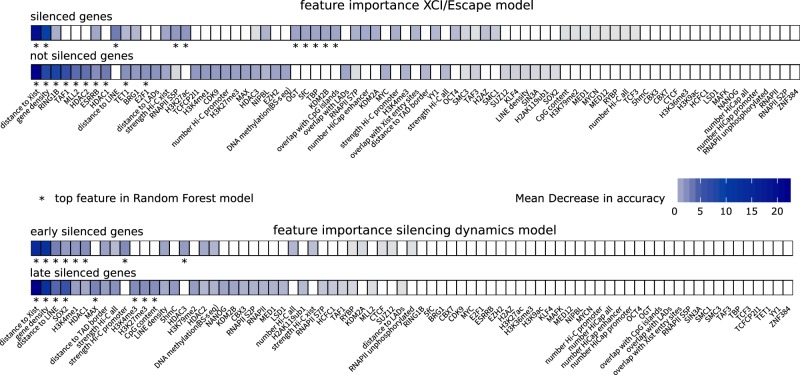
The two Random Forest models predict gene silencing dynamics with error rates of 28%–29% meaning that 71%–72% of genes are classified correctly. We assessed the individual contribution of each feature to the classification accuracy via Random Forest variable importance analysis for each class in the two models by the Mean Decrease in Accuracy (MDA), which indicates the importance of the feature for the classification performance (Fig. 4; Supplemental Fig. S2; Supplemental Text S2). We also trained our models on a set of only 10 (XCI/escape model) and 8 (silencing dynamics model) top features, which greatly improved the prediction error rate to 22.5% and 21.5%, respectively (Supplemental Fig. S3). The most important feature associated with silencing in both models was close genomic proximity of a gene's transcriptional start site (TSS) to the Xist locus (MDA 15%–16% in the XCI/escape model, MDA 14%–22% in the silencing dynamic model) (Fig. 4; Supplemental Figs. S2, S4, S5). Also a close proximity to LINE elements, low gene density, and enrichment for PRC1 (RING1B, H2AK119ub1, RYBP) and PRC2 (EZH2, H3K27me3) are associated with (early-) silenced genes in both models, with PRC1 playing a more prominent role in the XCI/escape model (MDA 8% for the not silenced class). (Early-) silenced genes are enriched for histone deacetylases (HDAC), involved in gene repression (MDA 1.6%–2.3% for the early-silenced class and 3.1%–3.8% for the not silenced class) and depleted for features associated with active transcription, such as H3K27ac and RNAPII S5P (MDA 1.8%/1.7% for the silenced class) and the general transcription factor TAF1 (MDA 6.3% for the not silenced class). Moreover, sequence-specific transcription factors, such as ESRRB (MDA 2.2% for the not silenced class) and SOX2 (MDA 1.6%–5.5% in the silencing dynamics model) contribute to model performance. Among the top features specific for the XCI/escape model we found binding of TET1, implicated in DNA demethylation, enriched at silenced genes (MDA 2.1% for the not silenced class). In the silencing dynamics model, in contrast, several features related to 3D chromosome organization seem to be important. Although genes located in close proximity to a TAD border tend to be silenced late, genes that are close to a LAD or highly connected to other genomic regions based on Hi-C/HiCap data tend to be silenced earlier. In summary, we have identified different feature sets that appear to influence whether or not a gene is subject to XCI and also whether silencing occurs with slow or fast dynamics.
Figure 4.
Feature importance for XCI/escape and silencing dynamics model. For each model, features are ranked class-wise according to their importance for the classification, quantified by the Mean Decrease in Accuracy (MDA) (Methods). (*) The top features of each class (10 for XCI/escape model; 8 for silencing dynamics model) that are used to build the final model. For more details, see Supplemental Figure S3. Similar results are obtained from the XCI/escape model trained on undifferentiated mRNA-seq data (Supplemental Fig. S20; Supplemental Text S8).
Forest-guided clustering of X-linked genes uncovers combinatorial rules of gene silencing
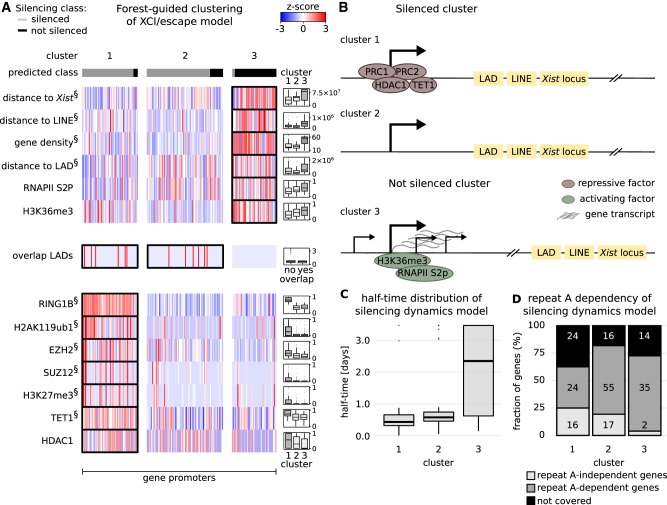
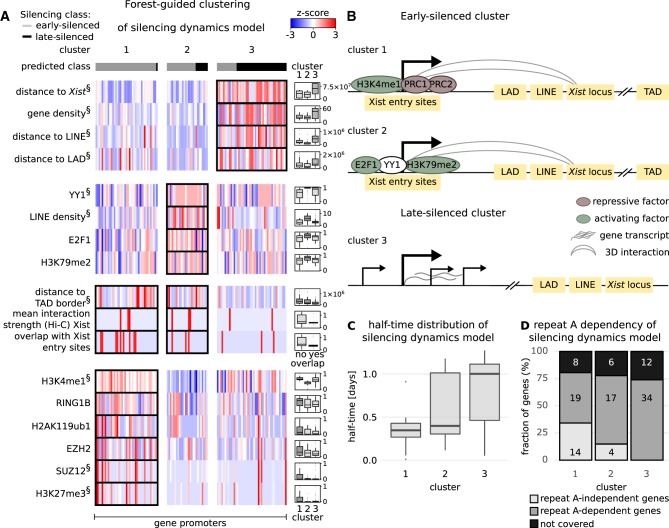
The preceding variable importance analysis pinpoints the individual contribution of each feature to the classification problem but cannot identify the role of correlated features and of feature combinations associated with different silencing pathways, which ultimately determine the silencing class of each gene. We therefore implemented a forest-guided clustering approach to stratify the genes into subgroups according to different combinations of rules. We used the proximity between genes within the Random Forest model to group genes that are regulated by the same set of genomic and epigenetic features. The number of clusters is chosen such that each cluster has a low degree of class mixture (containing mainly genes from one class and none or only a few genes from the other class) while maintaining a small number of clusters in total (Fig. 3C; Supplemental Fig. S6; Supplemental Text S3). The results are visualized in a heatmap showing the genes (columns), grouped by cluster, and a subset of features (rows) selected based on whether they were significantly different across clusters (P-value from an ANOVA test) (Figs. 5A, 6A).
Figure 5.
Classification rules for the XCI/escape model. (A) Results from the forest-guided clustering of the XCI/escape model visualized as a heatmap. Columns indicate the genes grouped by cluster; rows correspond to features with significant differences among clusters (ANOVA test). (§) The top 10 most significant features from the ANOVA test. Distributions of features in each cluster are shown in the box plots next to the heatmap, except for the feature "overlap LADs," where the number of genes in each category is shown. (B) Schematic view of the feature combinations promoting gene silencing (clusters 1 and 2) or escape (cluster 3). (C) Silencing half-time distribution in each cluster. (D) Proportion of genes in each cluster that undergo silencing in mouse trophoblasts, independent or dependent of the Xist-repeat A element (Sakata et al. 2017): (repeat A–dependent genes) genes with abrogated silencing in Xist-repeat A-mutant cells; (repeat A–independent genes) genes that still undergo silencing in the same cells; (not covered) our genes that were not covered in that data set. The numbers in each box indicate the number of genes that fall into each category. Similar results are obtained from the XCI/escape model trained on undifferentiated mRNA-seq data (Supplemental Fig. S20; Supplemental Text S8).
Figure 6.
Classification rules for the silencing dynamics model. (A) Results from the forest-guided clustering of the silencing dynamics model visualized as a heatmap. Columns indicate the genes grouped by cluster; rows correspond to features with significant differences among clusters (ANOVA test). (§) The top 10 most significant features from the ANOVA test. Differences in the distributions of features between clusters are highlighted in the box plots next to the heatmap, except for two features, where the number of genes in each category is shown. (B) Schematic view of the features associated with early (cluster 1 and 2) and late gene silencing (cluster 3). (C) Silencing half-time distribution for each cluster. (D) The proportion of genes which undergo silencing in mouse trophoblasts, independent or dependent of the Xist-repeat A element, is shown for each cluster, similar to Figure 5D. The numbers in each box indicate the number of genes that fall into each category.
For the XCI/escape model three clusters are found (Fig. 5A,B). Genes in clusters 1 and 2 are mainly predicted as silenced, and those in cluster 3 as not silenced (Fig. 5C). Genes tend to escape when they are far from the Xist locus, from LINE elements, and from LADs; they are found in gene-dense regions and are enriched for transcription elongation marks such as RNAPII S2P and H3K36me3 and never overlap with LADs. A cluster of silenced genes (cluster 1) is already marked by a repressive chromatin state (PRC1/2, HDAC1) and bound by TET1, whereas genes in the other silenced cluster (cluster 2) are depleted for those marks (Fig. 5A; Supplemental Figs. S7, S8).
To test whether these two clusters might be associated with different silencing pathways, we analyzed how they were affected in Xist mutants lacking either the A or the B and C repeats (Supplemental Text S4). We used data from a previous study that had analyzed both mutations in mESCs (Bousard et al. 2018) and from another study that had characterized the A-repeat mutant in trophoblasts in vivo (Sakata et al. 2017). Cluster 2 was enriched for genes that could still be silenced by a BC-repeat mutant compared to cluster 1 (repeat BC independent, odd ratio = 1.6, P = 0.19, Fisher's exact test), whereas cluster 1 was enriched for genes still silenced in the A-repeat mutant in either data set (mESC data: odd ratio = 2.3, P = 0.09 and trophoblast data: odds ratio = 2.76, P = 0.003, Fisher's exact test) (Fig. 5D; Supplemental Fig. S9). These findings are consistent with the idea that genes that require Polycomb for silencing (repeat BC dependent) are already premarked by PRC1/2 and the associated histone modifications (cluster 1), in contrast to genes that require that A repeat, which activates the SPEN/HDAC3 silencing pathway (cluster 2).
In the next step, we investigated the factors that would distinguish early- and late-silenced genes (silencing dynamics model). Again, the forest-guided clustering approach produced two early-silenced clusters (1, 2) with lower half-times and one late-silenced cluster (3) with higher half-times (Fig. 6A–C). Again, one early-silenced cluster (1) is premarked by Polycomb repressed chromatin (H2AK119ub1, RING1B, EZH2, SUZ12, H3K27me3) and also H3K4me1. The other early-silenced cluster (2) is mainly characterized by a preferential location of genes in LINE-dense regions, an enrichment of features related to transcriptional elongation, such as E2F1 subunit and H3K79me2 and the transcription factor YY1. Genes in both early-silenced clusters tend to be far away from TAD borders, to overlap with Xist entry sites, and to exhibit strong 3D contacts with the Xist locus. The late-silenced genes in cluster 3 are mainly characterized by genomic features; they are located in gene-dense regions, far from the Xist locus, from LINE elements, and from LADs (Fig. 6A; Supplemental Figs. S10, S11). We again analyzed the repeat A and repeat BC dependency in the two early-silenced clusters. Again, the Polycomb premarked cluster 1 tends to be enriched for repeat A–independent genes, albeit this effect was not statistically significant, and no difference was found for repeat BC dependent genes (Fig. 6D; Supplemental Fig. S9).
Experimental testing of model predictions
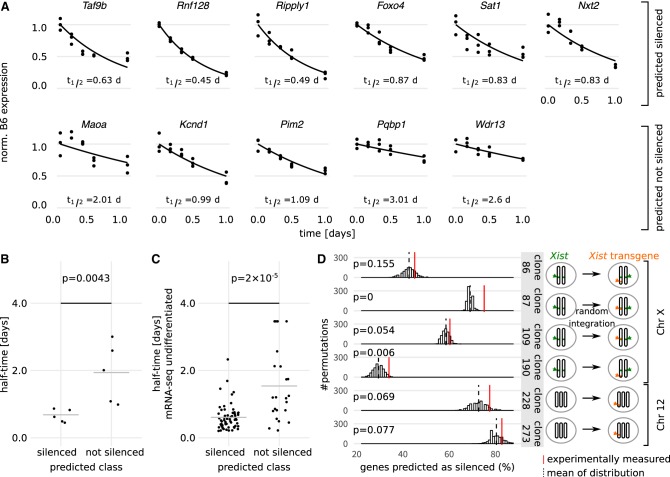
To validate our machine-learning model, we used the trained XCI/escape Random Forest model to predict the silencing class for X-Chromosomal genes that had not been measured in the PRO-seq experiment owing to insufficient coverage and had therefore not been used for model training (Fig. 3B; Supplemental Table S4). We performed an independent Dox induction time course experiment and used pyrosequencing to assess the allele-specific expression of six genes predicted to be silenced (Fig. 7A, top) and five genes predicted to be not silenced (Fig. 7A, bottom). The half-times of the silenced genes ranged from 0.45 to 0.87 d, and those of the not silenced genes lay between 0.99 and 3.01 d. The difference was highly significant (P = 0.0043, Wilcoxon rank-sum test) (Fig. 7B) and the half-time of all silenced genes fell in the silenced category (<0.9 d) (Fig. 1G), and three of five not silenced genes also fell in the respective category (>1.6 d) (Fig. 1G). To further validate the model, we compared all model predictions to the silencing half-times estimated from the mRNA-seq time course in undifferentiated mESCs (Fig. 2A, replicate A). Genes predicted as not silenced exhibited much longer silencing half-times than genes predicted as silenced (P = 1.7×10−5, Wilcoxon rank-sum test) (Fig. 7C). Genes that have not previously been reported as escapees, but are either measured or predicted from the PRO-seq model to be not silenced and are also measured as not silenced in the mRNA-seq data, are potentially novel escape genes, such as B630019k06Rik, Porcn, Ssr4, Gm14820, and Ppp1r3f (Supplemental Tables S2, S4).
Figure 7.
Experimental validation of model predictions. (A) Half-times of six candidate genes predicted as “silenced” (top) and five candidate genes predicted as “not silenced” (bottom) were estimated experimentally through allele-specific quantification by pyrosequencing at different time points during 24 h of doxycycline treatment in TX1072 cells in three independent experiments. Individual data points (dots), the fitted exponential decay function (line), and the estimated silencing half-times are shown. (B) Dot plot of the silencing half-times (t1/2) estimated in A. (C) Dot plot of undifferentiated mRNA-seq half-times for genes predicted as silenced and not silenced by our XCI/escape model. The gray line in B and C indicates the mean, and the P-value (Wilcoxon rank-sum test) indicates a significant difference between the mean of the two distributions. (D) Fraction of genes correctly predicted as silenced by the XCI/escape model (red lines) for six cell lines in which an inducible Xist transgene was integrated in different chromosomal locations (orange, cartoon on the right) (Supplemental Table S5). The background distributions of silenced predictions used to estimate empirical P-values is also shown (histogram, black dashed line represents the mean).
We next tested whether the model could predict silencing susceptibility to Xist transgenes located on an autosome. We used published allele-specific mRNA-seq data for a series of mESC clones that had integrated doxycycline-inducible Xist transgenes in different locations on the X Chromosome and on Chromosome 12 (Supplemental Table S5; Loda et al. 2017). The cells were trisomic for Chromosome 12 such that silencing of one copy should not affect cell viability. We adapted the model feature “distance to Xist” to account for the different locations of the transgene and calculated the fraction of genes predicted to be silenced by our model within all genes that were silenced, for each Xist transgene (Fig. 7D, red lines). These values varied considerably between clones depending on the size of the chromosomes and the location of the transgene. For five of six clones, the percentage predicted to be silenced was significantly higher than expected for a random sample, as estimated through a bootstrapping approach (Fig. 7D, compare red line to background distribution; Supplemental Text S5). Although potentially limited by the efficiency of the transgenes (Supplemental Fig. S12), this analysis shows that our model can to some extent be generalized even to other chromosomes. Taken together, these results confirm that our machine-learning model can predict X-Chromosomal gene silencing based solely on epigenetic and genomic features.
Discussion
In this study we developed a machine-learning model that can predict a gene's susceptibility to Xist-mediated silencing from a combination of epigenetic and genomic features. To train the model, we measured silencing kinetics with high temporal resolution through allele-specific PRO-seq. Compared to previous studies (Marks et al. 2015; Borensztein et al. 2017), we assessed silencing dynamics by measuring nascent transcription, therefore observing instantaneous changes in transcription by transcriptionally engaged RNAPII allowing a more direct quantification of silencing compared to mRNA-seq, which was previously used. Moreover, the use of an inducible system allowed us to uncouple XCI from differentiation and to avoid the use of mutations to ensure nonrandom XCI. Unlike previous studies that focused on just a few genes and/or investigated a few selected promoter features that are potentially linked to the XCI (Kelsey et al. 2015; Marks et al. 2015; Loda et al. 2017), we set out to identify silencing determinants in an unbiased manner based on a large number of epigenetic and genomic features. To uncover the combinatorial rules that control silencing dynamics, we went one step beyond classical variable importance analysis in Random Forests and introduced a random forest-guided visualization scheme. The determinants of silencing for groups of clustered genes recapitulated previous observations but also shed light on novel players or combination of features potentially controlling a gene's susceptibility to Xist-mediated inactivation.
Linear distance and 3D interactions with the Xist locus (leading to fast Xist deposition at so-called “entry sites”) are thought to be the prime determinants of early Xist spreading (Engreitz et al. 2013). Our model found the same features to be highly predictive of gene silencing dynamics, an association that has been described previously (Marks et al. 2015; Borensztein et al. 2017) and suggests that efficient Xist coating is required for fast silencing. However, previous studies had shown that Xist RNA initially tends to spread to gene-dense and LINE-poor regions (Engreitz et al. 2013; Simon et al. 2013), but in our analysis, gene density was associated with reduced silencing and LINE elements were found in proximity to rapidly silenced genes. A similar association was reported previously (Chow et al. 2010; Loda et al. 2017), suggesting that Xist coating is not the only determinant of silencing.
The Xist RNA recruits several protein complexes that mediate gene silencing, such as SPEN, which binds directly to the repeat A element and PRC complexes, which are indirectly recruited by the repeat B (Wutz et al. 2002; Chu et al. 2015; Monfort et al. 2015; Brockdorff 2017). Our model identified groups of genes associated with each of these silencing pathways. Repeat B/PRC-associated genes are already enriched for PRC1 and PRC2 prior to the onset of XCI, suggesting that Polycomb premarking might promote and even accelerate gene silencing and/or reinforce Xist spreading, as suggested by a recent study (Colognori et al. 2019). A similar enrichment of PRC components was previously found at genes susceptible to ectopic silencing by Xist transgenes (Kelsey et al. 2015; Loda et al. 2017). Although we did not find a clear signature at repeat A–associated silenced genes in the XCI/escape model, early-silenced genes in the silencing dynamics model that are not enriched for PRC, are located in particularly LINE-dense regions, suggesting that LINE elements might promote silencing of Polycomb-independent genes.
Previous studies looking at post-XCI cells proposed a role of CTCF in XCI (Filippova et al. 2005; Berletch et al. 2015) and found a moderate enrichment of CTCF prior to XCI at promoters of escapees compared to promoters of silenced genes (Loda et al. 2017). Although CTCF was not one of the discriminating promoter features in our XCI/escape model, we observed a significant enrichment of CTCF signal at enhancers of not silenced X-linked genes (Supplemental Fig. S13; Supplemental Text S6), suggesting a potential role of CTCF in gene silencing mediated by chromatin looping between enhancers and promoters.
Finally, our analysis identified several structural features that appear to modulate the dynamics of silencing. A high “connectivity” of some genes, that is, how much the gene is involved in 3D interactions with other genomic elements, is associated with faster silencing, maybe because Xist RNA can spread more easily to these genes through proximity transfer. Moreover, early silencing preferentially occurs at genes that are close to LADs, which generally contain repressed genes (van Steensel and Belmont 2017), whereas genes close to TAD boundaries tend to be silenced late.
In conclusion, we developed two Random Forest models that can accurately predict silenced and not silenced/escape genes, but also classes of early- versus late-silenced genes, constituting the first chromosome-wide predictive models of gene silencing from a very large set of features. We confirmed the predictive nature of our models by experimental testing of model predictions. The Random Forest approach allowed us to quantify the relative contribution of several features that were previously associated with XCI (e.g., linear distance to Xist, LINE elements, enrichment for PRC1 and PRC2, etc.) and suggested new features, which can be tested in more detail in future studies, such as TET1, and some pluripotency factors, such as ESRRB and SOX2, which have recently been implicated in reactivation of the X Chromosome during reprogramming (Janiszewski et al. 2019). Additional features could be included in the model in the future to further improve our ability to predict silencing susceptibility, and a detailed experimental investigation of the different silencing pathways elicited by Xist will facilitate the interpretation of the features that predict silencing dynamics as well as escape from XCI.
Methods
ES cell culture
The female TX1072 mESC line was grown in serum-containing medium, supplemented with LIF and 2i, as previously described (Schulz et al. 2014). Details on the cell line and culture conditions are given in Supplemental Text S7.
PRO-seq
PRO-seq was performed as described previously with some modifications (Mahat et al. 2016). For details about the experiment and the allele-specific bioinformatics analysis, see Supplemental Text S7.
mRNA-seq and pyrosequencing
RNA was isolated and converted to cDNA using standard procedures. For mRNA-seq, libraries were prepared using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) and sequenced and analyzed using standard procedures (Supplemental Text S7). For pyrosequencing, an amplicon containing a SNP is amplified by PCR from cDNA using GoTaq G2 Flexi (Promega) with 2.5 mM MgCl2 or HotStarTaq (Qiagen) for 40 cycles. The PCR product was sequenced using the PyroMark Q24 system (Qiagen). Assay details are given in Supplemental Table S6.
Silencing half-times
To normalize for sequencing depth, the reads mapping on the B6 genome were divided by the total number of allele-specific reads for each gene as follows:
| (1) |
was averaged across replicates (0, 24 h), which showed good replicate agreement (Supplemental Fig. S14), resulting in a total of eight time points (t = 0, 0.5, 1, 2, 4, 8, 12, 24 h). The allelic ratio was calculated as follows:
| (2) |
and normalized to the uninduced control (t = 0) to correct for basal skewing (different transcriptional activity at the two alleles in the absence of Dox) by the following:
| (3) |
To estimate gene-specific silencing half-times, an exponential decay function was used:
| (4) |
where k represents the silencing rate that was fitted to using the nls function (stats R package) and the half-time was calculated as
| (5) |
A maximum value of k = 5, corresponding to a half-time of 3.5 d was set, because higher half-times cannot be reliably estimated from our data as a result of the limited range of time points from 0 to 24 h. The goodness of fit was evaluated via the square root of the sum of squared residuals sqrtRSS defined as
| (6) |
After filtering out genes without SNPs, with fewer than 10 reads for at least one time point and strong allelic skewing ( or ) and sqrtRSS > 1.5, we obtained reliable half-times for 296 X-Chromosomal genes on mouse genome mm10 (Supplemental Tables S1, S2). Those genes were mapped to the mouse genome mm9 with the liftOver tool from the UCSC Genome Browser (Kuhn et al. 2007).
Gene half-times from both differentiated and undifferentiated mRNA-seq time series were computed in the same way as described above. For the undifferentiated mRNA-seq data set, we discarded replicate B because of insufficient read coverage and only used replicate A (Fig. 2A), which resulted in computing half-times for 346 genes. For the differentiated mRNA-seq data set, we averaged replicate A and B for each time point and computed half-times for 379 genes. For 233 genes, half-time could be estimated from all three data sets.
Definition of model features
The epigenetic and genomic features used for the Random Forest models are summarized in Table 1 and listed in detail in Supplemental Table S7. In total, 138 ChIP-seq libraries and one bisulfite sequencing experiment on undifferentiated male mESCs (only HDAC3 was assessed in female mESCs) were collected from various sources. After performing ChIP-seq library quality control with the deepTools package (Ramírez et al. 2014), 58 ChIP-seq libraries and the bisulfite sequencing experiment were used in the model (Supplemental Figs. S15, S16; Supplemental Table S8; Supplemental Text S1). Epigenetic features are defined as the average ChIP-seq signal in a predefined genomic region, normalized to the signal of a matched control experiment in the same region. Read counts of each feature were normalized to the control with the R package normR (Supplemental Fig. S17; Helmuth et al. 2016; Kinkley et al. 2016). Although for most features the signal is extracted in a genomic region around the active gene TSS, for broader features such as elongation marks H3K36me3, RNAPII S2P, and H3K79me2, the signal was averaged over the entire gene body (Supplemental Table S7). The active TSS for each gene was identified based on the PRO-seq data at time point t = 0 through the dREG method, which finds regions that harbor bidirectional transcription (Supplemental Fig. S18; Danko et al. 2015). In addition to epigenetic features, we defined 18 genomic features, including distance of each gene's TSS to the Xist locus, the next TAD border, or the next full-length LINE element; gene overlap with LADs, LINEs, and CpG islands; and strength and number of 3D chromatin interactions of gene promoters with other genomic elements. For details, see Supplemental Text S1.
Random Forest classification models
Two statistical models were developed to distinguish (1) “silenced” from “not silenced” genes, referred to as “XCI/escape model,” and (2) “early”- from “late”-silenced genes, referred to as “silencing dynamics model.” The continuous half-time values were therefore assigned to discrete classes in both models, according to fixed thresholds, which were chosen such that the error rate from the classification model (described below) would be minimized (Figs. 1G, 2G; Supplemental Table S3). Genes were defined as “silenced” for t1/2 < 0.9, as “not silenced” for t1/2 > 1.6, as “early-silenced” for t1/2 < 0.5, and as “late-silenced” for 0.9 < t1/2 < 1.3. The XCI/escape model was trained on 218 genes (168 from the “silenced” and 50 from the “not silenced” class) and the silencing dynamics model on 114 genes (74 from the “early”-silenced and 40 from the “late”-silenced class).
The two Random Forest classification models were implemented with the randomForest R package and use 77 predictor variables (epigenetic and genomic features), which show a variable degree of correlation between each other (Supplemental Fig. S19; R Core Team 2009). The error rate of the models is computed based on the out-of-bag (OOB) error, which is the mean prediction error over all the trees of the Random Forest. Importance of each feature is computed as “mean decrease in accuracy” (MDA) (Supplemental Text S2). Variables with large positive values of the MDA correspond to important features for the classification, whereas variables with MDA close to zero or negative correspond to noise.
Feature importance (MDA) and classification performance (OOB error) measures were further averaged over a collection of five hundred Random Forests to obtain stable results.
Simple feature selection was performed to improve the model performance by removing weaker or redundant features, which potentially introduce noise. We retained only the top 10 features from each class of the XCI/escape model and the top eight features from each class of the silencing dynamics model, which yield the model minimal error rate (Supplemental Text S2). For both models, the classification performance on the top features is reported as an average of 500 Random Forests.
Given the trained XCI/escape model, we predicted the silencing class of all X-linked genes, which were not included in the training set and chose few genes for experimental validation with pyrosequencing (Supplemental Text S2).
Forest-guided clustering for model interpretation
We can extract a proximity matrix from the trained Random Forest, which is a rough estimate of the distance between genes based on the proportion of times the genes are found in the same leaf node of a tree (Fig. 3C; Supplemental Text S3).
Based on this proximity matrix, genes were grouped into three clusters with the k-medoids algorithm for both models (Reynolds et al. 2006). Genes of the same silencing class (e.g., not silenced) are largely expected to cluster together according to a certain combination of epigenetic and genomic features. Given the nonlinear nature of the classification problem modeled here, we also expect, to some extent, genes from the same silencing class to be grouped in different clusters according to different combinations of features.
Similar to k-means clustering, k-medoids clustering requires setting in advance the number of clusters k (Supplemental Text S3). The results of the k-medoids clustering are visualized for both models as heatmaps, displaying the top 10 features that have a significant variation across clusters according to the P-value of an ANOVA test and few others which are also important for classification. Compared to classical Random Forest feature importance, the outcome of the forest-guided clustering enables an alternative interpretation of the Random Forest predictions in terms of combinatorial rules that determine the silencing state of groups of genes.
Statistical tests
Kolmogorov-Smirnov (KS) test was performed to test differences in silencing dynamics between A repeat–dependent and A repeat–independent genes. A Fisher's exact test was performed to test for enrichment of escapees in the silenced class and for A or BC repeat–dependent genes in the clusters of both models. A Wilcoxon rank-sum test was used to test for differences in half-times between silenced and not silenced genes from the pyrosequencing experiment and for comparison with mRNA-seq data. An analysis of variance (ANOVA) test was performed to find significantly different features across clusters in both models. All statistical tests were performed in R with the base statistical functions package.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE121144. The Random Forest pipeline is available as Supplemental Code, and the scripts for all additional analysis are on GitHub (https://github.com/marsicoLab/xist_mediated_gene_silencing).
Supplementary Material
Acknowledgments
We thank the NGS platform of the Curie Institute for sequencing, J. Helmuth and Q. Deletang for help with data analysis, and S. Rocha and A. Bousard for providing RNA-seq data of Xist mutants associated with a recent preprint. This work was supported by NIH grant GM025332 to J.T.L.; Center for Vertebrate Genomics (Cornell University) stipend to J.T.L. and I.J.; Helen Hay Whitney Postdoctoral Fellowship and Rosalind Franklin Fellowship (University of Groningen) to I.J.; ERC Advanced Investigator award (ERC-2014-AdG no. 671027), Labelisation La Ligue, FRM (grant DEI20151234398), ANR DoseX 2017, Labex DEEP (ANR-11-LBX-0044), part of the IDEX Idex PSL (ANR-10-IDEX-0001-02 PSL), and ABS4NGS (ANR-11-BINF-0001) funding to E.H.; ARC Foundation for Cancer Research (France; post-doctoral fellowship and grant R17016KK) and the ATIP-AVENIR program (INSERM / CNRS, France; grant C16002KS) to J.C.; IMPRS for Computational Biology and Scientific Computing funding to L.B.A.S.; and HFSP long-term fellowship (LT000597/2010-L), Max-Planck Research Group Leader program and E:bio Module III—Xnet grant (BMBF 031L0072) to E.G.S.
Author contributions: J.T.L. and E.H. conceived the PRO-seq experiment and strategy; A.M., E.G.S., and E.H. conceived the main idea of the computational analysis; E.G.S., I.J., C.-J.C., and L.S. performed the PRO-seq experiment and preprocessing of the data; L.B.A.S. and A.M. performed statistical analysis and data modeling; J.C., C.P., and L.S. generated and processed the mRNA-seq data, I.D. and B.F. performed the pyrosequencing experiment. L.B.A.S., A.M., and E.G.S. wrote the original draft; I.J., J.T.L., and E.H. reviewed and edited the paper; J.T.L., E.H., E.G.S., and A.M. financed the study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.245027.118.
Freely available online through the Genome Research Open Access option.
References
- Andergassen D, Dotter CP, Wenzel D, Sigl V, Bammer PC, Muckenhuber M, Mayer D, Kulinski TM, Theussl HC, Penninger JM, et al. 2017. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. eLife 6: e25125 10.7554/eLife.25125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. 2011. Genes that escape from X inactivation. Hum Genet 130: 237–245. 10.1007/s00439-011-1011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X. 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet 11: e1005079 10.1371/journal.pgen.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi I, Lleo A, Gershwin ME, Invernizzi P. 2012. The X chromosome and immune associated genes. J Autoimmun 38: J187–J192. 10.1016/j.jaut.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N, et al. 2017. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 24: 226–233. 10.1038/nsmb.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousard A, Raposo AC, Żylicz JJ, Picard C, Pires VB, Qi Y, Syx L, Chang HY, Heard E, da Rocha ST. 2018. Exploring the role of Polycomb recruitment in Xist-mediated silencing of the X chromosome in ES cells. bioRxiv 10.1101/495739 [DOI] [PMC free article] [PubMed]
- Brockdorff N. 2017. Polycomb complexes in X chromosome inactivation. Philos Trans R Soc Lond B Biol Sci 372: 20170021 10.1098/rstb.2017.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963. 10.1016/j.cell.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. 2002. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res 99: 75–84. 10.1159/000071577 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237. 10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969. 10.1016/j.cell.2010.04.042 [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT. 2019. Xist deletional analysis reveals an interdependency between Xist RNA and Polycomb complexes for spreading along the inactive X. Mol Cell 74: 101–117.e10. 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A. 2015. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods 12: 433–438. 10.1038/nmeth.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Del-Arenal M, da Rocha ST, Heard E. 2011. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Hum Genet 130: 307–327. 10.1007/s00439-011-1029-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8: 31–42. 10.1016/j.devcel.2004.10.018 [DOI] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2015. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev 31: 57–66. 10.1016/j.gde.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. 2014. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol 30: 561–580. 10.1146/annurev-cellbio-101512-122415 [DOI] [PubMed] [Google Scholar]
- Helmuth J, Li N, Arrigoni L, Gianmoena K, Cadenas C, Gasparoni G, Sinha A, Rosenstiel P, Walter J, Hengstler JG, et al. 2016. normR: regime enrichment calling for ChIP-seq data. bioRxiv 10.1101/082263 [DOI]
- Janiszewski A, Talon I, Song J, De Geest N, To SK, Bervoets G, Marine JC, Rambow F, Pasque V. 2019. Dynamic erasure of random X-chromosome inactivation during iPSC reprogramming. bioRxiv 10.1101/545558 [DOI] [PMC free article] [PubMed]
- Kelsey AD, Yang C, Leung D, Minks J, Dixon-McDougall T, Baldry SEL, Bogutz AB, Lefebvre L, Brown CJ. 2015. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol 16: 208 10.1186/s13059-015-0774-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkley S, Helmuth J, Polansky JK, Dunkel I, Gasparoni G, Fröhler S, Chen W, Walter J, Hamann A, Chung H-R. 2016. reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T cells. Nat Commun 7: 12514 10.1038/ncomms12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. 2007. The UCSC genome browser database: update 2007. Nucleic Acids Res 35: D668–D673. 10.1093/nar/gkl928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda A, Brandsma JH, Vassilev I, Servant N, Loos F, Amirnasr A, Splinter E, Barillot E, Poot RA, Heard E, et al. 2017. Genetic and epigenetic features direct differential efficiency of Xist-mediated silencing at X-chromosomal and autosomal locations. Nat Commun 8: 690 10.1038/s41467-017-00528-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. 2016. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11: 1455–1476. 10.1038/nprot.2016.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kerstens HH, Barakat TS, Splinter E, Dirks RA, van Mierlo G, Joshi O, Wang SY, Babak T, Albers CA, et al. 2015. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol 16: 149 10.1186/s13059-015-0698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep 12: 554–561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. 2017. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish Polycomb-mediated chromosomal silencing. Mol Cell 68: 955–969.e10. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. 2004. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 167: 1025–1035. 10.1083/jcb.200409026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org/. [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith VJ. 2006. Clustering rules: a comparison of partitioning and hierarchical clustering algorithms. J Math Model Algor 5: 475–504. 10.1007/s10852-005-9022-1 [DOI] [Google Scholar]
- Sakata Y, Nagao K, Hoki Y, Sasaki H, Obuse C, Sado T. 2017. Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development 144: 2784–2797. 10.1242/dev.149138 [DOI] [PubMed] [Google Scholar]
- Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Blüthgen N, Heard E. 2014. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14: 203–216. 10.1016/j.stem.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AHFM, Jenuwein T, Otte AP, Brockdorff N. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 Polycomb group complexes. Dev Cell 4: 481–495. 10.1016/S1534-5807(03)00068-6 [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504: 465–469. 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25: 1371–1383. 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Luo J, Yu H, Rattner A, Mo A, Wang Y, Smallwood PM, Erlanger B, Wheelan SJ, Nathans J. 2014. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 81: 103–119. 10.1016/j.neuron.2013.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622. 10.1101/gr.103200.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, da Rocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176: 182–197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.