Abstract
Posttranscriptional regulation plays a crucial role in shaping gene expression. During the maternal-to-zygotic transition (MZT), thousands of maternal transcripts are regulated. However, how different cis-elements and trans-factors are integrated to determine mRNA stability remains poorly understood. Here, we show that most transcripts are under combinatorial regulation by multiple decay pathways during zebrafish MZT. By using a massively parallel reporter assay, we identified cis-regulatory sequences in the 3′ UTR, including U-rich motifs that are associated with increased mRNA stability. In contrast, miR-430 target sequences, UAUUUAUU AU-rich elements (ARE), CCUC, and CUGC elements emerged as destabilizing motifs, with miR-430 and AREs causing mRNA deadenylation upon genome activation. We identified trans-factors by profiling RNA–protein interactions and found that poly(U)-binding proteins are preferentially associated with 3′ UTR sequences and stabilizing motifs. We show that this activity is antagonized by C-rich motifs and correlated with protein binding. Finally, we integrated these regulatory motifs into a machine learning model that predicts reporter mRNA stability in vivo.
Posttranscriptional regulation plays an essential role in shaping gene expression. 3′ UTRs represent a central regulatory hub that integrates multiple inputs to control mRNA translation, localization, stability, and polyadenylation status (Mayr 2017). In the cell, these inputs come from RNA-binding proteins (RBPs) and noncoding sequences such as microRNA binding sites and AU-rich elements (AREs), which function together with codon optimality and RNA modifications to regulate mRNA stability in the cell (Schoenberg and Maquat 2012; Gilbert et al. 2016; Mayr 2017; Hanson and Coller 2018). The search for noncoding regulatory elements has largely focused on the sequence of individual 3′ UTRs (Voeltz and Steitz 1998; Wirsing et al. 2011; Kristjánsdóttir et al. 2015). Transcriptome-wide analyses of mRNA stability, using pulse labeling (Miller et al. 2011) or blocking transcription (Geisberg et al. 2014), have led to the identification of potential regulatory sequences by searching for common motifs within mRNAs. Parallel reporter libraries (Oikonomou et al. 2014; Zhao et al. 2014; Wissink et al. 2016; Rabani et al. 2017) have been used to find the regulatory sequences within large sets of 3′ UTR sequences. However, these approaches have been limited by the diversity and length of the sequences tested. Short sequences provide limited information on how the sequence context influences regulation by individual motifs, whereas longer sequences lack resolution to pinpoint distinct segments driving regulation. To address these issues, we recently developed a high-throughput RNA-element selection assay (RESA) to measure the regulatory activities of mRNA sequences in vivo (Yartseva et al. 2017). RESA uses endogenous RNA fragments in a parallel reporter assay that allows for high sequence complexity and high-density coverage of the transcriptome, or targeted regions of interest, providing near nucleotide resolution of the regulatory activity of RNA sequences.
RNA regulatory elements are recognized by trans-factors, including miRNAs and RBPs (Glisovic et al. 2008). Once bound, RBPs can regulate the processing, stability, and translation of their target mRNAs (Gerstberger et al. 2014). Interactome capture has revealed a set of proteins in intimate contact with mRNAs across different eukaryotic systems (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Sysoev et al. 2016; Wessels et al. 2016; Despic et al. 2017). In vitro affinity selection methods, such as SELEX (Blackwell and Weintraub 1990; Ellington and Szostak 1990; Tuerk and Gold 1990), RNA affinity profiling (Tome et al. 2014), and RNAcompete (Ray et al. 2013), have been complemented with UV crosslinking and immunoprecipitation (CLIP) to provide the set of targets and the binding motifs in vivo for a number of RBPs (Ule et al. 2003; van der Brug et al. 2008; Chi et al. 2009; Hafner et al. 2010; König et al. 2010; Chan et al. 2014; Hansen et al. 2015; Murn et al. 2015; Sugimoto et al. 2015; Galloway et al. 2016; Scheckel et al. 2016; Rot et al. 2017). However, the presence of a specific sequence motif is not always indicative of regulation in vivo, suggesting that additional sequences, or combinatorial interactions between RBPs, influence the regulatory output on an mRNA. Current efforts have not yet linked the RNA regulatory maps with the RBP binding profiles to define the posttranscriptional regulatory network in vivo.
Identifying functional regulatory sequences together with RBP trans-factors is an essential step toward understanding mRNA posttranscriptional regulation. This regulation is of particular importance during the early stages of animal development, which are instructed by maternally provided mRNAs (Walser and Lipshitz 2011). During the maternal-to-zygotic transition (MZT), mRNAs deposited in the oocyte undergo coordinated remodeling. Individual pathways have been implicated in the regulation of maternal mRNAs (for review, see Lee et al. 2014; Yartseva and Giraldez 2015). For example, in Drosophila, the RBP SMAUG destabilizes maternal mRNAs (Dahanukar et al. 1999; Tadros et al. 2007). In Xenopus, AREs within the 3′ UTRs of maternal mRNAs trigger their deadenylation after egg activation and decay after the mid-blastula transition (MBT) (Audic et al. 1997; Voeltz and Steitz 1998). In zebrafish, zygotic transcription of microRNA miR-430 regulates ∼20% of destabilized maternal transcripts (Giraldez et al. 2006). Codon usage in the coding sequence of mRNAs influences differential mRNA stability during the MZT across several vertebrates (Bazzini et al. 2016; Mishima and Tomari 2016), and it influences mRNA half-life in yeast (Presnyak et al. 2015). mRNA methylation has been implicated in shaping mRNA stability during ES cell differentiation (Batista et al. 2014) and in MZT (Zhao et al. 2017). Despite these singular discoveries and the recent effort to develop models based on 3′ UTR sequence elements (Rabani et al. 2017), there is a paucity of quantitative models that integrate the various elements to predict mRNA deadenylation, degradation, and RBP binding. Thus, it is still poorly understood how sequence composition influences mRNA stability, how the sequence context affects the regulatory potential of each motif, what are the relative activities of different elements, and which RBPs mediate this regulation.
Here, we identified the RBPs bound to mRNAs during MZT, their preferential binding sequences, and the 3′ UTR cis-regulatory sequences shaping mRNA stability during zebrafish embryogenesis, which we have integrated into a quantitative model that explains the regulatory activity encoded within individual 3′ UTRs.
Results
Distinct pathways regulate maternal mRNA decay
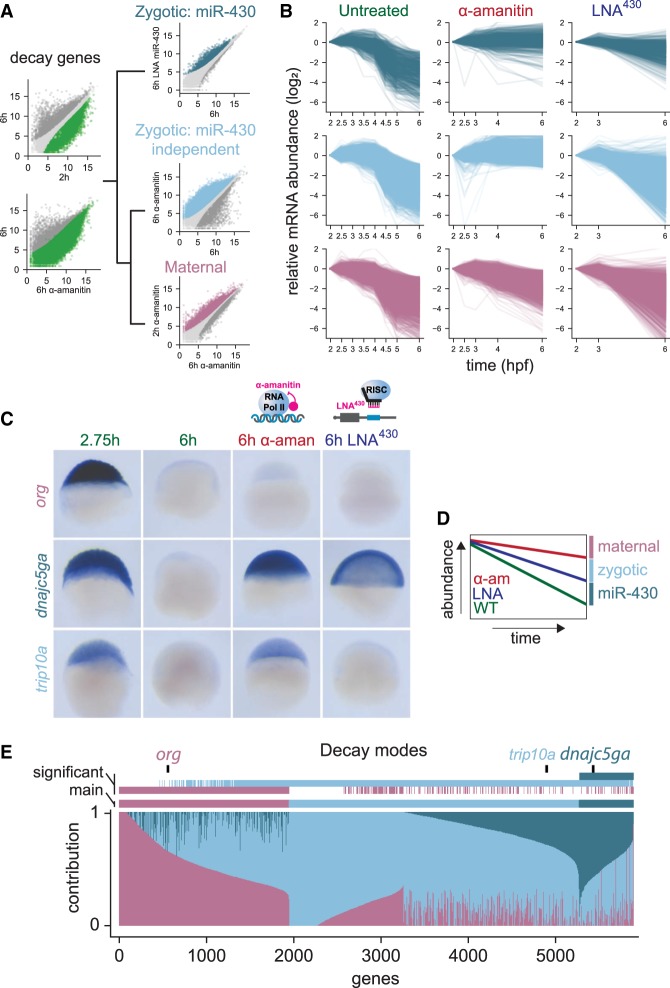
Fertilization triggers remodeling of the transcriptome required for the first steps of embryonic development. Maternal and zygotic posttranscriptional pathways regulate the dynamics of mRNA stability during MZT (Yartseva and Giraldez 2015). To identify which pathways regulate each mRNA, we analyzed transcript levels during the first 8 h postfertilization (hpf), at 30- to 60-min intervals in wild-type zebrafish embryos using mRNA-seq (Supplemental Table S1). We identified mRNAs whose decay was dependent on zygotic transcription or specifically on miR-430. These transcripts were significantly stabilized when zygotic transcription was inhibited with the RNA Pol II inhibitor, α-amanitin (Lindell et al. 1970; Kane et al. 1996), or when miR-430 was inhibited using an antisense tiny-LNA complementary to miR-430 (LNA430) (Staton et al. 2013). The remaining transcripts that underwent decay when zygotic transcription was inhibited were classified as regulated by the maternal mode. To normalize RNA expression across stages, we used exogenous yeast spike-in RNA, allowing us to quantify global changes in mRNA levels (Fig. 1A; Supplemental Table S2). Comparing early (2 hpf) to late (6 hpf) developmental stages, we defined the main regulatory mode for 5847 mRNAs undergoing decay: 3909 were regulated by the zygotic mode (67%), of which 616 were primarily dependent on miR-430 (11% of total unstable) and 1938 were primarily regulated by the maternal mode (33%) (Fig. 1B). In situ hybridization analysis of endogenous transcripts selected from each mode displays stabilization patterns in the absence of zygotic transcription (α-amanitin) and/or miR-430 function (LNA430) that were consistent with behaviors observed in the global RNA-seq analyses (Fig. 1C).
Figure 1.
Distinct pathways regulate maternal mRNA decay. (A) Hierarchical protocol followed to separate genes into their predominant mode of decay. Among decaying genes (green) determined as stable early (2 hpf) or higher in absence of zygotic transcription (α-amanitin treatment), miR-430 decaying genes were first characterized using LNA430 treatment (dark blue). Remaining genes, as indicated by the black tree, were split into maternal (purple) and zygotic (light blue) decaying genes based on their dependence on zygotic transcription. Transcripts with nonsignificant expression changes are colored in light gray (see DESeq2 analysis in Methods). (B) Biplots representing mRNA expression levels over developmental time separated by decay pathways. Time-course is shown in WT, α-amanitin, and LNA430 conditions. Each line represents expression level of individual mRNA. (C) In situ hybridization illustrating three genes from the maternal, zygotic, and miR-430–dependent decay modes (n = 20/20). (D,E) Relative contribution of the three modes to RNA decay. Each gene total decay was assessed using fold-change between untreated early and late stages calculated in A. LNA430 and α-amanitin conditions were used to attribute the contribution of the miR-430 and zygotic modes, respectively. Remaining decay was attributed to the maternal mode. This normalization was taking into account that α-amanitin treatment is also blocking miR-430 transcription. For example, if a transcript's decay profile was not affected by blocking miR-430 or zygotic transcription, then that transcript's degradation was wholly attributed to maternal decay mode. In contrast, decay that was prevented by blocking miR-430 or zygotic transcription was attributed to miR-430 or zygotic decay pathways, respectively. As multiple comparisons, used in A to determine each gene mode of decay, can be significant, the main (lowest P) and significant (all significant P) modes of decay are shown (E).
Although we were able to identify the predominant mode of decay for each unstable mRNA, the stability of any individual mRNA is likely dictated by multiple regulatory mechanisms operating within the embryo. We identified multiple regulatory modes for numerous mRNAs. To dissect potential combinatorial regulation, we calculated the degree of stabilization conferred on individual mRNAs by the loss of miR-430 regulation, or zygotic transcription (Fig. 1D; Supplemental Table S3). We find that the majority of unstable transcripts were degraded by the combinatorial regulation of multiple pathways (Fig. 1E). For example, we observe that up to 3688 mRNAs (63%) are partially stabilized after blocking miR-430 function. Together, these results define three regulatory modes of maternal mRNA turnover and their relative contributions to shaping posttranscriptional regulation across the zebrafish transcriptome after fertilization.
RESA identifies regulatory RNA elements
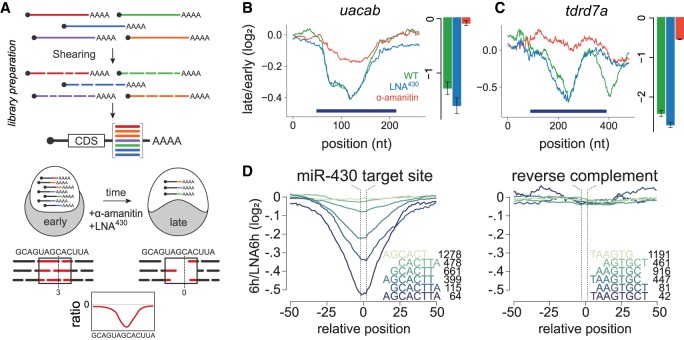
We hypothesized that the combinatorial regulation observed for maternal transcripts might be encoded in discrete elements within the mRNA. To identify these elements, we used RESA (Fig. 2A; Yartseva et al. 2017), which assesses the ability of sequences to regulate mRNA stability in vivo when placed in the 3′ UTR of a reporter mRNA library. We generated two RNA reporter libraries (Supplemental Table S1), one composed of random fragments spanning the entire embryonic transcriptome (∼30 nt length; transcriptome library) and a high-density library enriched for 3′ UTRs from 434 genes regulated during the MZT (∼100-nt length; targeted library). To identify regions that mediate differential mRNA stability, we injected each RNA reporter library into one-cell-stage embryos and quantified the abundance of each reporter within the RESA library using RNA-seq before and after zygotic transcription (2 and 6 hpf) in wild-type, α-amanitin, or LNA430-injected zebrafish embryos. Depletion or enrichment of sequences over developmental time revealed 1404 destabilizing and 295 stabilizing regions, respectively, across 3456 genes. Destabilizing regions were modulated by the maternal (593), zygotic (184), or miR-430 (627) modes of mRNA decay (Supplemental Fig. S1A). During early embryogenesis, deadenylation and decay are uncoupled (Voeltz and Steitz 1998), allowing us to distinguish between elements that predominantly cause deadenylation or decay. By inferring the relative poly(A) tail length from capture efficiency (i.e., depletion of reads from poly(A) selection over time relative to total reads indicates deadenylation) (Beilharz and Preiss 2007; Bazzini et al. 2012), we identified several hundred regions predominantly causing deadenylation (Supplemental Fig. S1A).
Figure 2.
RESA identifies regulatory RNA elements. (A) Schematic of RESA method. Sequence fragments derived from endogenous transcripts are placed within the 3′ UTR of reporter mRNAs. Regulatory activity is inferred from relative depletion or enrichment postinjection. Treatment with α-amanitin or LNA430 delineates sequences under zygotic and/or miR-430 regulatory pathways, respectively. (B,C) RESA identifies sequence regions within uacab and tdrd7a transcripts that are responsive to zygotic regulatory mode. Bar graphs display independent validation by qRT-PCR with reporters containing sequence inserts spanning regulated regions (dark blue bars): late/early fold-change in untreated (green), LNA430-treated (blue), α-amanitin treated (red). (D) Mean destabilization across all loci (minimum coverage of five counts per million at 2 hpf) centered on miR-430 target site variant, with number of represented loci indicated for each variant. Right panel shows mean destabilization for loci possessing the reverse complement for each miR-430 target sequence.
To test the regulatory activity of these sequences, we validated two reporter mRNAs containing 3′ UTR sequences identified by RESA using qRT-PCR (Fig. 2B,C). Each reporter was destabilized in wild-type embryos, and this effect was blocked when inhibiting zygotic transcription, consistent with the specific regulation of these regions by the zygotic mode. We observed that the mean destabilization across miR-430 target sites corresponded to the predicted microRNA target site strength (8-mer > 7-mer > 6-mer), whereas inserts containing reverse complement miR-430 target sites were not depleted (Fig. 2D), showing that RESA can accurately quantify regulatory strength across target sites. Together, these results indicate that RESA can identify several hundred regions that promote mRNA deadenylation and decay across the transcriptome.
Identifying destabilizing and stabilizing regulatory motifs
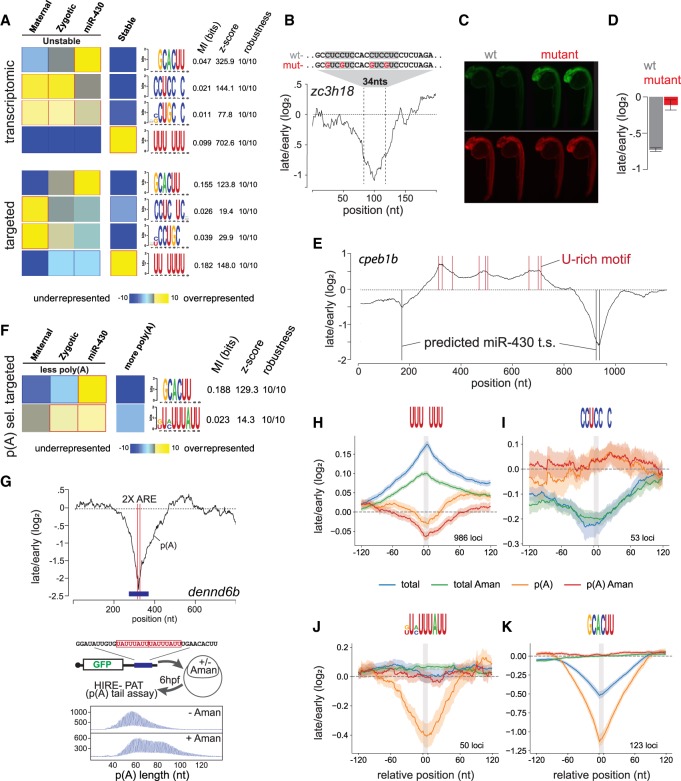
To identify short-linear motifs enriched in coregulated sequences, we used Finding Informative Regulatory Elements (FIRE) (Elemento et al. 2007; Oikonomou et al. 2014). This method analyzes all possible 7-mers to then optimize these seeds into sequence logos by maximizing mutual information (Elemento et al. 2007; Oikonomou et al. 2014). We identified motifs associated with destabilization and stabilization in the three decay modes (confidence cutoff Z-score >20 and >10 for the transcriptome and targeted libraries respectively, see Methods) (Fig. 3A; Supplemental Fig. S2). These motifs were consistent with those identified across mRNAs with different kinetics of decay (Rabani et al. 2017). We find that miR-430 seed target sequences were specifically enriched in unstable sequences that were stabilized by the loss of miR-430 function, confirming that our strategy identified known motifs in the expected mode of regulation. Independent of miR-430, the motifs most strongly associated with unstable sequences were CCUCCNC and CUGCNC (Z-score 144.1 and 77.8). To validate these findings independently from RESA, we analyzed the expression of a reporter mRNA containing multiple copies of the CCUC motif derived from the gene zc3h18 (that was in common between the transcriptomic and targeted libraries). We observed decreased stability for CCUC motifs compared with a reporter in which those motifs were mutated to CGUC, resulting in lower protein expression of a GFP reporter mRNA compared with a dsRed control (Fig. 3B–D). These results suggest that CCUC motifs are responsible for sequence-specific decay elements in the early zebrafish embryo. FIRE also identified U-rich motifs such as UUUNUUU, UUUUANA, or AANUAUU overrepresented in stable regions with UUUNUUU displaying the strongest activity (Supplemental Fig. S2). Stabilization at multiple U-rich sites was consistently observed within individual transcripts (Fig. 3E). In contrast, RESA samples prepared with additional poly(A) selection revealed that deadenylated sequences were enriched in miR-430 sites and U[CA]UUUAUU, an ARE known to be involved in regulating poly(A) tail length (Fig. 3F; Supplemental Fig. S2; Audic et al. 1997; Voeltz and Steitz 1998; Rabani et al. 2017; Yartseva et al. 2017). To validate this motif, we analyzed the poly(A) tail length of a reporter mRNA containing tandem copies of UAUUUAUU derived from the gene dennd6b (also known as fam116b). This analysis revealed that poly(A) shortening by AREs was dependent on activation of the zygotic genome, as it was abolished in α-amanitin–treated embryos (Fig. 3G).
Figure 3.
Identifying destabilizing and stabilizing regulatory motifs. (A) FIRE top selected motifs. In the heatmap, overrepresentation (yellow) and underrepresentation (blue) patterns are shown for each discovered motif in the corresponding category. Also shown are the mutual information values, Z-scores associated with a randomization-based statistical test and robustness scores from a threefold jackknifing test. (B) RESA transcriptome coverage ratio for the zc3h18 locus zoomed in on peak containing multiple copies of the CCUC motif. (C) Zebrafish 24-h embryos injected with WT (CCUC motifs) or mutant (CGUC motifs) reporters. (n = 50) (D) Bar graphs displaying independent validation by RT-PCR (three replicates, 25 embryos each). (E) RESA-targeted coverage ratio for the cpeb1b (also known as zorba) locus. U-rich and miR-430 target sites are marked with red and black bars, respectively. (F) Same as A for poly(A) selected RESA-targeted library. (G) RESA targeted with poly(A) selection coverage ratio plot showing the two ARE sites (red lines) within the dennd6b locus (top). HIRE-PAT assay measuring impact of dennd6b ARE sites on poly(A) tail length reporter. Shortening of poly(A) tail length is zygotic transcription dependent as shown by longer poly(A) tail after α-amanitin treatment (bottom). (H–K) Motif-centered metaplots for U-rich (H), CCUCCNC (I), ARE (J), and miR-430 (K) motifs. Targeted RESA libraries coverage ratio were averaged over windows centered on RBP motif (RESA minimum coverage >0.01 CPM). Motif is represented with gray bar. SEM of RESA is shown by shaded outlines.
To measure the regulatory activity of each identified motif, we analyzed the average decay across all the regions containing that motif in the RESA-targeted assay before (2 hpf) and after MZT (6–8 hpf). We performed two different assays: (1) We compared their effect in the total and the poly(A) selected library to distinguish effects on stability and deadenylation, and (2) we analyzed their dependency on the zygotic mode of decay by determining whether their regulatory activity was blocked in the absence of zygotic transcription (α-amanitin–treated samples). The U-rich motif showed a stabilizing effect (mean stabilization of 0.17 log2 fold-change (logFC) at 2 vs. 6 hpf) (Fig. 3H), whereas CCUCCNC had a destabilizing effect (mean destabilization −0.22 logFC 6/2 hpf) (Fig. 3I). Regulation by these elements was mainly controlled through the maternal mode on the total mRNA as it was still observed in α-amanitin–treated samples but had a weaker effect on the poly(A) libraries. Analysis of lower Z-score FIRE motifs CCUGC and UUAUU and Pumilio motifs (UGUA[AU]AUA) (Gamberi et al. 2002; Gerber et al. 2006) revealed weak regulation acting on total and poly(A) libraries (Supplemental Fig. S1B–D). As a control, meta-analysis over 123 miR-430 seeds (Fig. 3K) showed a strong zygotic-dependent regulatory effect on stability (−0.5 logFC 6/2 hpf) and deadenylation (−1.15 logFC 8/2 hpf), with an effect that was more than 2.5 times stronger than U-rich and CCUCCNC motifs. Consistent with the reporter analysis, meta-analysis of the U[CA]UUUAU ARE motif over 50 loci revealed that this motif causes zygotic-dependent deadenylation (−0.4 logFC 8/2 hpf) with a robust depletion of polyadenylated fragments, without a significant effect on the total mRNA abundance within the time frame analyzed (Fig. 3J). Relative regulatory activity of these elements revealed that within this context, miR-430 provides stronger regulation than ARE motif UAUUUAUU, with a rapid coupling of deadenylation and decay that is not observed in AREs. Together, these results identify sequence motifs that regulate reporter mRNA stability and deadenylation during the MZT.
Mapping RNA-interacting proteins in the embryo
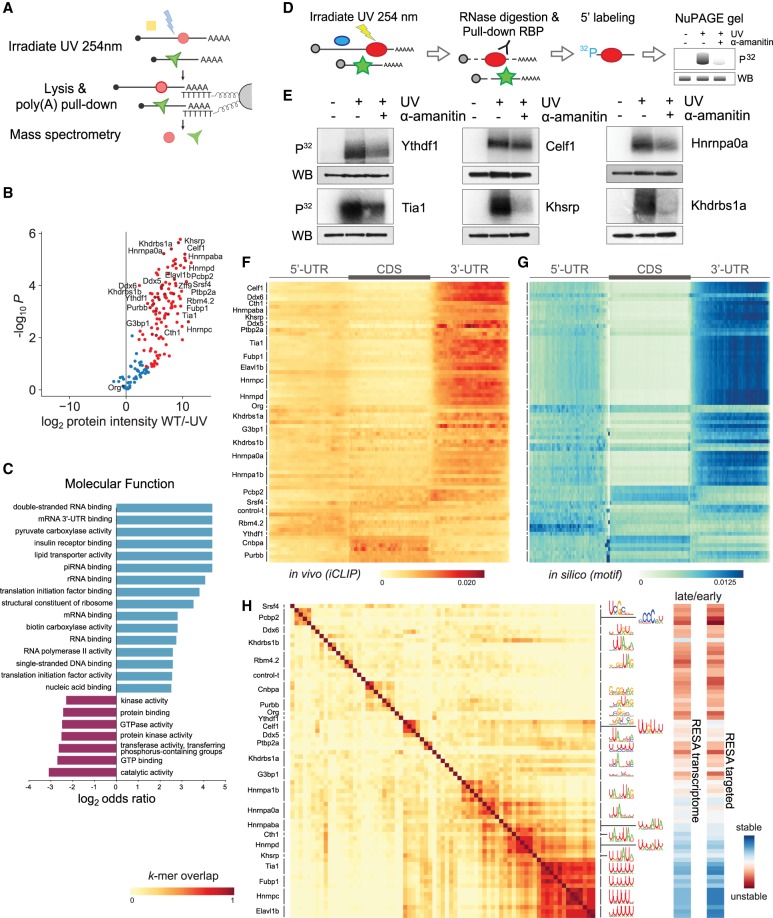
To identify the trans-factors binding mRNAs during the MZT, we adapted the interactome capture technique to zebrafish embryos (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Sysoev et al. 2016; Wessels et al. 2016; Despic et al. 2017). This method uses UV to crosslink protein–mRNA interactions and then poly(A) purification followed by mass spectrometry to identify the proteins bound to mRNA (Fig. 4A; Supplemental Fig. S3). We analyzed the interactome across three independent biological replicates with or without UV crosslinking. We queried the interactome at 4 hpf, a timepoint that precedes the widespread changes in maternal mRNA stability and deadenylation characterized here (Bazzini et al. 2012). By using label-free quantitative mass spectrometry, we identified 160 proteins with two or more peptides with at least one unique peptide. Of the 160 identified proteins, 112 were significantly enriched in the UV-crosslinked sample compared with controls (-UV) (Fig. 4B). From this core set of 112 proteins, 90 had also been identified in previous mRNA interactomes (Supplemental Table S4; Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Liao et al. 2016; Despic et al. 2017). We observed a significant enrichment of RBPs in the interactome, as 67.8% of the identified proteins had annotated RNA-binding domains compared with 8.2% of proteins detected in the input. Further, our analysis revealed an enrichment for Gene Ontology (GO) terms associated with RBPs (Fig. 4C) and a selective enrichment for bona fide RNA–protein interactions (Supplemental Fig. S4). These included RBPs involved in RNA processing and splicing such as Xrn2, Srsf2, or Celf1; mRNA translation such as Eif4a1a, Eif2a, and Eif4enif1; and RNA stability such as Pum1, Piwil1, Hnrnpd, Khsrp, and Stau2 among others. To validate their ability to bind RNA, we immunoprecipitated six of the identified proteins after UV crosslinking and radiolabeled the bound RNA for detection (Fig. 4D,E). Although all the proteins interacted with RNA, a subset of these interactions was also dependent on the activation of zygotic transcription (Ythdf1, Khsrp, Khdrbs1), because the levels of RNA pulled down were reduced in α-amanitin–treated embryos, without affecting the RBP levels. Taken together, the zebrafish interactome identified a large set of RBPs that participate in a wide range of RNA processing pathways during MZT, providing an entry point to identify the effector proteins that regulate mRNA stability and decay.
Figure 4.
Mapping RNA-interacting factors in the embryo and identifying putative regulatory factors driving mRNA stability. (A) Diagram summarizing the interactome capture protocol. After UV crosslinking and poly(A) mRNA pull-down, RBPs are identified using mass spectrometry. (B) Volcano plot showing the RBPs significantly enriched over background by interactome capture. (C) GO term enrichment analysis characterizing the molecular functions of the captured proteins. (D) Cartoon depicting the rationale behind a label-transfer experiment to validate RNA–protein interactions. P32 autoradiograph indicates the amount of RNA, whereas while FLAG western blot indicates RBP levels. (E) Validation of the RNA-binding activity during zebrafish development of representative RPBs identified in the interactome capture. (F) Heatmap representing iCLIP metaplots of RBP binding within protein-coding transcripts. The UTRs and CDS of each transcript were split into 50 bins to normalize their length. Metaplots averaged over each RBP were clustered to group similar binding profiles. (G) Heatmap representing in silico binding profiles obtained by scanning for RBP binding motifs within protein-coding transcripts. Motifs are represented in H. (H) Heatmap illustrating the overlap among RBP binding motifs. Motifs were characterized using the top 20 6-mers most bound normalized by iCLIP control. For example, two proteins with the same top 6-mers received a 1.0 overlap score and represented in dark red (left). RESA averaged coverage ratio for each motif (right).
Identifying putative regulatory factors driving mRNA stability
To identify potential effector proteins that mediate the regulatory activity observed in RESA, we performed iCLIP experiments on 24 RBPs identified in the zebrafish interactome and analyzed their target sequences (Supplemental Fig. S5; Supplemental Tables S1, S5). We reasoned that having a common tag would allow us to compare the signal between different proteins to identify specific binding events for each protein and distinguish them from background common to all samples. Thus, we analyzed FLAG-tagged versions of each protein expressed from an injected mRNA. To ascertain that this approach captures bone fide binding sites, we compared iCLIP signal of one candidate, Khsrp, from the pull down of the FLAG-tagged protein versus the endogenous protein using an endogenous antibody. We observed a similar enrichment of both the FLAG-tagged protein and the endogenous Khsrp in the 3′ UTR of endogenous mRNAs and similar motifs sharing the core UUUAU (Supplemental Fig. S6), suggesting that the FLAG-epitope does not significantly alter the binding motif of Khsrp. Next, we characterized the binding pattern and the sequence motif preferentially bound by each RBP across replicates (Supplemental Table S6). Cumulative count of the iCLIP reads within the 5′, 3′ UTRs and CDS revealed that the majority of RBPs displayed strong occupancy within the 3′ UTR (Fig. 4F). Within this class, we observed variable accumulation of reads in different regions of the 3′ UTR. Among these, Celf1, Elavl1b (also known as Hug), and multiple hnRNPs displayed preferential occupancy toward the distal end of the 3′ UTR. In contrast, Pcbp2 binding was more frequently observed directly proximal to and downstream from the annotated stop codon. iCLIP reads from cnbpa (also known as zff9) and purbb were distributed throughout the CDS. In particular, khsrp was preferentially enriched across 3′ UTRs in the transcriptome and was preferentially excluded from coding sequences and 5′ UTRs. At the exon junctions, most RBP binding was observed within exons and close to acceptor and donor sites similar to control profiles (Supplemental Fig. S7A). Known splicing factors such as Srsf4 had similar binding profiles as observed by Änkö et al. (2012), whereas Khsrp (Supplemental Fig. S7B) and Hnrnpd displayed high intron binding.
To assess the RNA sequence specificity of each RBP, we searched for the top 10 enriched hexamers bound by each RBP compared with a negative control lacking a FLAG epitope (Fig. 4H). For most proteins, the identified motifs were similar and partially overlapping to those previously identified in vitro (Supplemental Fig. S8; Ray et al. 2013). The iCLIP binding pattern for each RBP resembled the distribution of the top identified motifs (hereafter in silico binding) (Fig. 4G), suggesting that for most proteins, the presence of the binding motifs explains the binding distributions observed in vivo. However, we observed (1) higher density of in silico binding in the 5′ UTR than observed in vivo for several RBPs and (2) higher density of in silico binding to the CDS for pcbp2 despite similar in vivo and in vitro profiles for pcbp2 within the 3′ UTR. These differences raise the possibility that additional factors, such as the ribosome, might contribute to the observed occupancy profiles in vivo (Supplemental Fig. S9), as it has been suggested for UPF1 (Zünd et al. 2013).
We found that several of the RBP binding motifs possess regulatory activity as measured by RESA (Fig. 4H; Fig. 5D; right panels). For example, Elavl1b, Fubp1, Hnrnpc, and Tia1 were preferentially bound to poly(U) associated in RESA with stabilizing motifs. In contrast, RBPs such as Pcbp2, Ptbp2a, or Cnbpa were preferentially bound to destabilizing motifs. Taken together, these data correlate the regulatory activity of specific RNA motifs to the recognition by specific RBPs during zebrafish embryogenesis.
Figure 5.
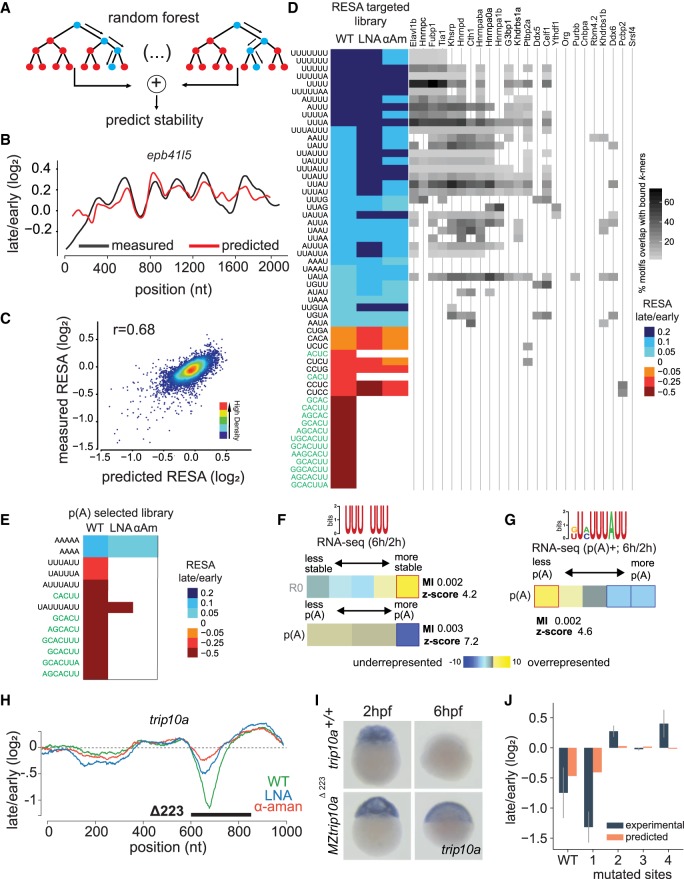
Modeling the effect of sequence on mRNA stability in vivo. (A) Scheme of the procedure for building the random forest model on RESA-targeted profiles. Data generated from window-sliding across the RESA profiles are used to train a random forest model. (B) Example of predicted (red) and RESA-targeted experimental (black) profiles for epb41l5 gene. (C) Model performance per window using fivefold cross-validation; model achieved 0.68 Pearson correlation between predicted stability and measured according to RESA-targeted library. (D, left) Top selected motifs according to the random forest model trained on RESA-targeted library (motifs in green text represent the miR-430 target sites). Columns represent WT, LNA430, and α-amanitin treatment. Color intensity represents the RESA fold-change difference between windows that do or do not contain each motif. Blue represents stabilizing motifs, and red represents destabilizing motifs (all motifs have P below 4.6 × 10−56 (Mann–Whitney U test followed by Bonferroni multiple testing adjustment). (Right) Heatmap representing motif enrichment in the top 50 hexamers enriched in iCLIP experiments. (E) Same as D with random forest model trained on the RESA targeted with poly(A) selection library. (F,G) U-rich (F) and ARE (G) motifs’ enrichment between 2 and 6 hpf within total and/or poly(A) selected RNA-seq. In heatmap, overrepresentation (yellow) and underrepresentation (blue) patterns are shown. Also shown are the mutual information values, Z-scores associated with a randomization-based statistical test, and robustness scores from a threefold jackknifing test. (H) RESA-targeted profile from the trip10a locus. Genetic deletion of a sequence spanning regulated region (black line) results in stabilization of the trip10a transcript as assessed by in situ hybridization (n = 20/20; I). (J) Random forest model validation. Barplot comparing experimental (RNA level by high-throughput sequencing) and predicted average stability of trip10a decay peak and four mutated sites (Spearman's correlation coefficient r = 0.60).
Modeling the effect of sequence on mRNA stability in vivo
Our analysis identifies several motifs that are enriched within regulated sequences in the mRNA. To determine whether sequence information can be used to model reporter mRNA regulation, we used machine learning and developed a random forest model (Breiman 2001). We reasoned that this model would allow us to capture the association between motif frequencies and their corresponding regulatory activities along RNA sequences (Fig. 5A). We analyzed the frequency of k-mers (1–8 nt) in 100-nt sliding windows across the RESA-targeted library. To build the model, we retained 387 k-mers out of 87,380 with an absolute Spearman's correlation coefficient above 0.1 between k-mer frequency and fold-change measured by RESA. Next, we used fivefold cross-validation to train and assess the performance of the random forest model. This model achieved a 0.68 Pearson correlation between the predicted change in mRNA abundance for each window and the corresponding change measured by RESA with asymptotic P = 0.0 (Fig. 5B,C). The random forest model selected 57 motifs with a dominant effect on RNA stability (Fig. 5D; left; Supplemental Fig. S11A).
To further analyze the importance of the motifs selected by the random forest model, we compared the differential stability between windows that contain or lack each motif. This analysis revealed that the top 25% stabilizing windows were significantly enriched in U-rich motifs, as well as UUAG and UGUA motifs (P < 1 × 10−146, 8 × 10−21, and 9 × 10−20, respectively, Mann–Whitney U test with Bonferroni multiple testing adjustment), confirming the trends observed above. On the other hand, the top 25% destabilizing windows were significantly enriched in miR-430 complementary sites and CCUC motifs (P < 2 × 10−96 and 6 × 10−95 respectively, Mann–Whitney U test with Bonferroni multiple testing adjustment) (Supplemental Fig. S10A,B). By intersecting these k-mers with the sequences identified in the iCLIP experiment, we identified potential RBPs that could act as trans-factors to regulate bound mRNAs (Fig. 5D, right) and revealed UUAG was significantly enriched among the top 50 k-mers bound by Hnrnpa1b (P < 2 × 10−73, χ2-test followed by Bonferroni multiple testing adjustment) and UUUU was significantly enriched in k-mers bound by Elavl1b and Hnrnpc (P < 1 × 10−174 and 2 × 10−92, respectively). On the other hand, CCUC was significantly enriched in k-mers bound by Pcbp2 (P < 2 × 10−23), and CUCU was significantly enriched in k-mers bound by Ptbp2a (P < 1 × 10−30). Together these analyses provided a model that predicts the regulatory information encoded in the 3′ UTR sequence to regulate reporter mRNA stability in vivo.
mRNAs can be controlled by maternally or zygotically encoded modes. To model the different modes of mRNA decay, we built additional random forest models in the absence of zygotic regulation (α-amanitin) or miR-430 regulation (LNA430). We observed a strong correlation between the predicted and the observed regulation in RESA targeted for both models (α-amanitin: r = 0.70 with asymptotic P = 0.0; LNA430: r = 0.74 with asymptotic P = 0.0) (Supplemental Fig. S11B,C). Consistent with the FIRE analysis, CCUC and U-rich motifs were identified as regulatory sequences independent of zygotic transcription as part of the maternal model, whereas miR-430 was identified as the main element regulating mRNA stability of the zygotic mode (Fig. 5D). In contrast, models built using the RESA-targeted libraries using poly(A) selection were less accurate (wild type: r = 0.53 with asymptotic P = 0.0; α-amanitin: r = 0.33 with asymptotic P = 1 × 10−248; LNA430: r = 0.31 with asymptotic P = 6 × 10−224) (Fig. 5E; Supplemental Fig. S12) and revealed that UAUUUAUU (AREs) motifs were the strongest zygotic-dependent regulator of the poly(A) tail after miR-430. The lower accuracy of the model when the zygotic mode is blocked suggests that regulation of the polyadenylation status by 3′ UTR sequence is mainly zygotic dependent. Based on these results, we conclude that our current models capture the importance of CCUC and U-rich motifs to maternally regulate RNA stability and of miR-430 and UAUUUAUU (AREs) to zygotically regulate mRNA polyadenylation.
Predicting stability of endogenous mRNAs
The random forest model captures the association between motif frequencies and stability. Thus, we applied it to predict differential stability of endogenous mRNAs, determined as the fold-change in mRNA levels between 2 and 6 hpf. To this end, we predicted the stability of endogenous mRNAs using the random forest model trained on the RESA-targeted library by averaging the predicted stability of all 100-nt sliding windows of the 3′ UTR for each endogenous mRNA. Our model achieved a 0.29 Pearson correlation between the predicted and measured mRNA fold-changes with asymptotic P = 1 × 10−158 (Supplemental Fig. S10D), indicating that a large fraction of mRNA regulation is not captured by the model developed on the 3′ UTR reporter mRNAs, consistent with the model developed by Rabani et al. (2017). This suggests that although the primary 3′ UTR sequence is responsible for about a third of endogenous mRNA stability, other factors not captured by RESA such as codon bias (Presnyak et al. 2015; Bazzini et al. 2016; Mishima and Tomari 2016) or RNA structure and modifications (Batista et al. 2014; Ke et al. 2017; Zhao et al. 2017; Beaudoin et al. 2018) could drive the remaining two-thirds (see Discussion).
Among the RESA enriched motifs, several were associated with differential stability or deadenylation of endogenous mRNAs. Endogenous mRNAs containing U-rich motifs were specifically stabilized in total RNA libraries (Fig. 5F). In contrast, we find that UAUUUAUU sequences (AREs) are enriched within endogenous mRNAs that are deadenylated, consistent with a role for AREs in poly(A) tail shortening (Fig. 5G). To test the regulatory activity of endogenous sequences identified by RESA and the motifs identified by the random forest model, we used CRISPR-Cas9 editing to mutate a potential destabilizing sequence element identified in the trip10a gene. We observed that a 223-nt deletion in the endogenous gene, targeting the region regulated in RESA, caused stabilization of the mutant mRNA compared with the wild type, without any apparent developmental phenotype. This shows that RESA can identify de novo functional regulatory elements in vivo (Fig. 5H,I). Sequence analysis of the regulatory region in trip10a 3′ UTR revealed multiple AUUUA, AUUA, and AAAUAAA, which, when disrupted, stabilized the mRNA as measured using high-throughput sequencing (Fig. 5J) and increased protein output (Supplemental Fig. S13B–D). We predicted the reporter RNA levels using our model and obtained a 0.60 Spearman's correlation coefficient with RNA levels measured experimentally (Fig. 5J; Supplemental Fig. S13A). We concluded that a significant part of the regulation is encoded in the 3′ UTR sequence and that it could be accurately predicted using machine learning.
Antagonistic effects of different RBP binding motifs
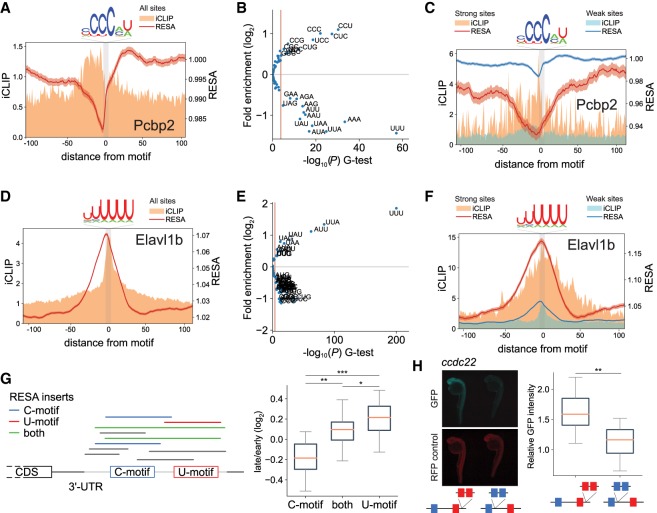
The regulatory effect of any particular motif can be expressed as the average activity across hundreds of loci with that motif. For example, loci containing the Pcbp2-binding motif displayed a mean destabilization of 0.985 across 10,305 loci (Fig. 6A). However, for most motifs, we observed a broad spectrum of regulation and RBP binding, suggesting that the mere presence of a given motif is not the unique determinant of regulatory activity or RPB binding. We hypothesized that the sequence context for each target site might explain differential regulation (RESA) and/or binding (iCLIP). To assess this, we ranked each locus possessing a Pcbp2, miR-430, or Elavl1b-binding motif according to its RESA activity. An analysis of flanking sequences revealed significant enrichment of specific 3-mers when comparing the most regulated versus least regulated loci. For Pcbp2, CCU, CUC, and CCC were significantly enriched within flanking sequences of the most destabilized sequences, which was associated with increased Pcbp2 binding as shown by cumulative iCLIP signal (Fig. 6B,C). In contrast U-rich sequences were significantly depleted within these sites. Conversely, the context of Elavl1b-binding sites was significantly enriched in UUU 3-mers within the most stabilized sequences, which were also among the most abundantly bound sequences by Elavl1b (Fig. 6D–F; Supplemental Fig. S14A–C). miR-430 binding sites did not appear to have a specific sequence bias associated with stronger regulation, consistent with the strength of the regulation being primarily modulated by the size of the seed (Supplemental Fig. S14D,E). Favorable sequence contexts for Pcbp2 and Elavl1b resemble the original binding site and have opposing nucleotide preferences. These results suggest that U-rich and poly(C)-binding proteins might antagonize each other's activity influencing the stability of the mRNA.
Figure 6.
Antagonistic effects of different RBP binding motifs. (A,D) Motif-centered metaplots for Pcbp2 (A) and Elavl1b (D). RESA transcriptome coverage ratio and iCLIP signal were averaged over windows centered on RBP motif (RESA minimum coverage >0.05 CPM). The motif is represented with gray bar. SEM of RESA is shown by red shaded outline. (B,E) Sequence context of RBPs potential target sites. Volcano plot representing 3-mer enrichment 20 nt upstream of and downstream from the motif between the top 10% most destabilizing for Pcbp2 (B) and most stabilizing for Elavl1b (E) and the bottom 10%. P-values were calculated using a G-test. Red line indicates 1% significance cutoff after Bonferroni multiple test correction. (C,F) Motif-centered metaplots for Pcbp2 (C) and Elavl1b (F) comparing potential target sites with favorable context (red) or not (blue). Favorable context defined as top 10% of sites most enriched with significantly enriched 3-mers and least enriched in significantly depleted 3-mers as shown on panels B and E. Scale on y-axis was adjusted, showing higher regulation and wider iCLIP binding signal of sites with favorable context. (G) Schematic describing how to select RESA inserts to measure antagonistic effects of two motifs. Inserts from RESA-targeted overlapping C-motif only, U-motif only, and both motifs are colored in blue, red, and green, respectively. Inserts with a partial overlap in gray are ignored. To estimate stability of inserts, the same procedure is applied for early and late time-points, and the ratio of the number of inserts is calculated (left). Box-and-whisker plots showing stability of inserts overlapping C-rich and U-rich motifs separately and both together (right). (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, Wilcoxon signed-rank test. (H) Zebrafish 24-h embryos injected with [C-rich (blue box) and 3× U-rich motifs (red box)] or [3× C-rich and U-rich motifs] reporters (n = 18 per inserts). Reporter sequence is derived from 3′ UTR of ccdc22 gene. In G and H, boxes span first to last quartiles, and whiskers represent 1.5× the interquartile range. (**) P < 0.01, Mann–Whitney U test.
To test the antagonistic effect of U-rich and C-rich motifs on mRNA stability, we first searched for reporters in the RESA library overlapping strictly one or both motifs. Because RESA is composed of a large library of individual reporters, each reporter can be directly assessed instead of averaging the signal across reporters. We analyzed 18 loci with more than 20 RESA reporters overlapping a CCUCC motif, a UUUUUU motif, or both (Fig. 6G, left). We found that the reporters with both motifs had an intermediate stability compared with reporters with each individual motif (P = 2.14 × 10−3 and 4.29 × 10−2 for CCUCC and UUUUU compared with both motifs, respectively, Wilcoxon signed-rank test) (Fig. 6G; right). To validate the effect of these motifs independently from RESA, we analyzed the expression of reporters derived from the ccdc22 gene containing three copies of U-rich and one copy of C-rich motif and vice versa. U-rich reporters were more stable and resulted in higher protein expression compared with the C-rich reporter mRNA (P = 2.90 × 10−3, Mann–Whitney U test) (Fig. 6H). Together, these results show that RESA can measure the combined effects of multiple motifs and show that U-rich and C-rich motifs antagonize each other's activity during early embryogenesis.
Discussion
Posttranscriptional regulation plays a major role in shaping gene expression during cellular transitions in which mRNAs from the previous state undergo repression and decay. During the MZT, both maternal and zygotic modes regulate mRNA abundance. Here, we identified (1) cis-regulatory elements using RESA, (2) trans-factor RBPs using interactome capture, and (3) RBP target sequences using iCLIP. We integrated these regulatory sequences into a random forest prediction model, which largely recapitulated 3′ UTR–mediated regulation as measured by RESA. These findings integrate important aspects of the posttranscriptional regulation shaping mRNA stability in vivo.
Our implementation of RESA allowed us to measure the relative strength of regulatory sequences in vivo. Among the regions in the 3′ UTR, miR-430 targets were the strongest deadenylating and destabilizing elements. UAUUUAUU elements provided a strong deadenylating activity that was dependent on the activation of the zygotic genome, with a weaker effect on mRNA decay. On the same mRNA, RESA identified multiple sequence elements with antagonistic effects. For example, the cpeb1b (also known as zorba) mRNA contains adjacent stabilizing U-rich motifs and destabilizing miR-430 target sites. Combining these elements on the same mRNAs may provide differential temporal or spatial regulation, creating specific patterns of gene expression. We propose that these transcripts are first stabilized by maternally provided poly(U)-binding proteins and are later deadenylated and degraded by the zygotic mode of decay triggered by miR-430 or UAUUUAUU elements (Audic et al. 1997; Voeltz and Steitz 1998; Giraldez et al. 2006; Wu et al. 2006; Bazzini et al. 2012). Initial stabilization is likely coupled with active cytoplasmic polyadenylation, which would increase mRNA translation efficiency early in embryonic development (Subtelny et al. 2014). We find a strong correspondence between the motifs identified by RESA and those recently reported in zebrafish by Rabani et al. (2017). We further characterized the sequence contexts that influence the strength of the regulation associated with these elements. We identified an antagonistic activity between U-rich and CCUC sequences. Indeed, transcriptome-wide analysis of 3′ UTR mediated stability reveal a global tendency of 3′ UTR sequences (rich in U) to promote stabilization of the mRNA, with destabilizing islands that contain three main elements: miRNAs, CCUC domains, and UAUUUAUU motifs. Furthermore, the fact that deadenylation and decay are initially uncoupled during embryogenesis (Voeltz and Steitz 1998) allows us to define elements that preferentially affect mRNA stability (U-rich and CCUC) (Stoeckius et al. 2014), elements that mainly affect mRNA deadenylation (UAUUUAUU-ARE), and elements that induce both, such as miR-430. ELAV-like proteins (HuR) have been shown to stall decay following deadenylation mediated by AREs (Fan and Steitz 1998; Peng et al. 1998) and miRNAs (Kundu et al. 2012). The favorable sequence contexts for Elavl1b and Pcbp2 are similar to their binding motif and show a broader accumulation of iCLIP signal across the binding sites, suggesting that favorable context sites are bound by multiple proteins.
By using iCLIP, we characterized the binding motifs of multiple RBPs identified in the interactome capture. We observed that similar motifs are recognized by multiple RBPs, which are associated with similar regulatory activities. Although a high level of functional redundancy could be part of a robust developmental system, these RBPs might also recruit other proteins to achieve a specific regulatory response, as proteins with families containing the same binding domain are combined with different functional domains. Functional redundancy impairs our ability to genetically dissect their activity, an issue further complicated by the existence of multiple homologs for each RBP. Nevertheless, the genetic elimination of the regulatory elements identified in RESA clearly reveals the importance of these sites in the regulation of endogenous mRNA levels (Fig. 5H,I). These regulatory elements are enriched at the primary sequence level and associated with a high density of binding. Furthermore, we observed reduced RBP binding in the absence of zygotic transcription (Fig. 4E). This might be caused by lower levels of the target mRNAs in α-amanitin–treated embryos. Alternatively, RBP binding might be activated by the MZT. For instance, Khsrp shows differential binding to RNA in our interactome capture and in wild-type versus homozygous MK2 or MK3 kinase mutant cells (Boucas et al. 2015). The activation of MK2 is known to play a fundamental role during MZT (Holloway et al. 2009), which led us to hypothesize that the reduced binding of Khsrp to RNA might be caused by lack of posttranscriptional activation. This hypothesis opens the possibility that an additional layer of regulation is controlling the clearance of maternal RNAs, emphasizing the potential central role of the regulatory elements recognized by the RBPs in regulating RNA stability.
We integrated these regulatory activities into a random forest model. Our model was able to predict regulatory activity across the RESA-targeted library of reporters for maternal and zygotic signals that mediate mRNA stability. We also identified sequence motifs associated with lower polyadenylation status in the mRNA (i.e., miR-430 and UAUUUAUU) dependent on the zygotic mode. The stability of endogenous mRNAs was less accurately predicted (r = 0.29 with asymptotic P = 1 × 10−158) than the RESA 3′ UTR reporter library (per transcript Pearson's r = 0.82 with asymptotic P = 0.0) (Supplemental Fig. S10C,D). Our model is solely based on the primary sequence of the 3′ UTR, and its lower predictive power in endogenous transcripts is consistent with recent studies (Rabani et al. 2017). Generally, differences in the RESA and mRNA-seq assays could explain this difference in accuracy. Although using the same reporter backbone for all fragments tested in RESA allowed us to eliminate any bias owing to different coding sequences, we have yet to probe other factors regulating mRNA fate not assayed by RESA. Notably, the codon bias of the coding sequence of mRNAs influences their stability (Hanson and Coller 2018), specifically during the MZT (Bazzini et al. 2016; Mishima and Tomari 2016). Also, our RESA libraries are synthesized in vitro and therefore lack any RNA modifications. Modifications such as N6-methyladenosine (m6A) have been proposed to help the clearance of maternal mRNAs (Zhao et al. 2017), yet it is unclear whether m6A-containing mRNAs are specifically stabilized upon loss of m6A reader proteins (Kontur and Giraldez 2017). In addition, RNA structure analysis has identified long-range RNA interactions that are currently not being integrated in the RESA reporters owing to the limited size of the fragments tested (Aw et al. 2016; Lu et al. 2016; Sharma et al. 2016; Beaudoin et al. 2018). Finally, our model is restricted to linearly adding the effect of multiple motifs. This design was motivated by the additive effects of the U-rich and C-rich motifs we observed (Fig. 6G), but it excludes the cooperative effects of multiple sites (Hon and Zhang 2007; Saetrom et al. 2007; Lai et al. 2012). Including cooperative effects will require more sophisticated models capable of identifying the combined effects of all possible pairs of selected elements along with distance between each pair. This exponentially larger feature space will require a significantly larger training data set to avoid underfitting. Explaining the decay dynamics of all mRNAs will require the integration of the regulatory activities found in the 3′ UTRs with additional elements in the mRNA, namely, codons, RNA structure, and modifications to achieve a global prediction of mRNA dynamics.
Methods
Early zebrafish embryo transcriptome
For the large developmental gene expression time-course, RNA-seq libraries were prepared after collecting developing embryos between 0 and 8 hpf. Yeast total RNA was spiked to allow for appropriate normalization of fold-changes. After extraction, RNA was subjected to poly(A)-selected RNA-seq library preparation and ribosomal RNA-depleted total RNA-seq library preparation. Sequenced reads were then mapped onto zebrafish genome Zv9 using STAR (Dobin et al. 2013) and gene annotation from Ensembl r78 (Aken et al. 2017). Significantly over- and underexpressed genes were determined using DESeq2 (Love et al. 2014). Decay mode of genes (Maternal, Zygotic: miR-430–dependent and Zygotic: miR-430–independent) and their contribution to gene decay were determined based on significant DESeq2 tests, which are described in the Supplemental Methods.
In situ hybridization for the org, trip10a, and dnajc5ga genes were performed as by Thisse and Thisse (2008).
RESA
The transcriptome-based reporter library was generated by overlap-extension PCR with primers mapping to the SP6 promoter and downstream from the SV40 polyadenylation site (Yartseva et al. 2017). Libraries were injected into one-cell zebrafish embryos. After library preparation (Yartseva et al. 2017) and sequencing, RESA profiles were obtained for each transcript (transcriptomic library) or UTR (targeted library). They represented positional read coverage normalized to count per million (CPM) using total counts of all transcript/UTR profiles per sample. RESA peaks in UACA and tdrd7a loci were validated using qRT-PCR. For trip10a, CRISPR-mediated mutagenesis (Moreno-Mateos et al. 2015) was used to obtain a 223-nt deletion. Stabilization of the trip10a transcript was assessed by in situ hybridization.
To evaluate the regulatory effect of short RNA elements, RESA profiles were split into fixed-length sequence segments (30 and 100 nt long for the transcriptomic and targeted libraries, respectively) with 33% overlaps (Oikonomou et al. 2014). For each segment, a score was computed and differences between scores (early and late and between conditions) were transformed to Z-scores. The scores were used to categorize segments to destabilizing and stabilizing modes of regulation: maternal, zygotic/miR-430 dependent, and zygotic/miR-430 independent. Within each category, segments were grouped based on their category-specific P. Then, FIRE, a computational framework for the discovery of regulatory elements (Elemento et al. 2007), was used for de novo discovery of short motifs that are significantly informative of the different modes of regulation. To this end, FIRE was extended to include N-fold cross-validation, a restriction on the average degeneracy of the elements, and zebrafish-specific options.
Interactome capture
Wild-type zebrafish embryos were irradiated at 4 hpf with UV at 254 nm for 4 min and then collected. Interactome capture was conducted as described by Castello et al. (2012) with oligo(dT) magnetic beads. The –UV control group of embryos were not UV-irradiated before collection. After peptides were eluted, LC-MS/MS was performed to identify them. Detailed protocol is included in the Supplemental Methods. Finally, enrichment in the molecular function category of identified proteins was tested.
RBP binding: CLIP
FLAG-tagged RBPs genes were first cloned and in vitro transcribed to capped mRNA. UV crosslinking and pull-down was conducted on 4 hpf embryos. The iCLIP protocol described by Huppertz et al. (2014) adapted to zebrafish was performed. Khsrp was iCLIPed using endogenous antibody. After read mapping using STAR (Dobin et al. 2013), RBP binding profiles were computed. To summarize RBP binding within protein-coding transcripts, all UTR and CDS iCLIP binding profiles were transformed into metagene profiles using 50-nt binning. To define RBP motifs, enrichment of 6-mer sequences at RBP target sites was calculated while taking into account the iCLIP experimental background. Logo representation of the top-10 motifs was built by aligning them using MAFFT (Katoh and Standley 2013). Finally, to simultaneously analyze binding and regulatory activity, sequences matching RBP motifs were searched within the transcriptome. Within the 100-nt window, iCLIP (5′ end) and RESA (coverage ratio) windows were averaged.
Modeling the effect of sequence on mRNA stability
The frequency of all k-mers (1–8 nt) within the 100-nt sliding windows (10-nt step) of the RESA profiles was used to train a random forest model with 500 trees, aiming to build a set of decision trees able to capture the association between k-mer frequencies and their correlation with RESA stability values. To make the learning process more efficient, a preprocessing, unbiased, and fast k-mer filtering step was implemented. After filtering rare, not correlated, and not important (using feature importance function) k-mers, 57 k-mers with the most significant contribution were selected. Trained models were validated using fivefold cross-validation for each library (RESA targeted and poly(A) selected). For each of these two libraries, a separate model was trained and validated for each condition (WT, LNA-430, and α-amanitin). Finally, the stability score per each transcript in the RESA-targeted library was calculated as the average of all the sliding windows predicted scores across the 3′ UTR of that transcript.
Stability of endogenous transcripts was predicted using the random forest model trained on RESA-targeted library by averaging the predictions obtained using the same sliding window approach.
Independent validation using targeted mutations on the trip10a locus was performed comparing (1) GFP expression compared with the control dsRed expression using fluorescence microscopy on 24 hpf zebrafish embryos and (2) stability measured by high-throughput sequencing of wild-type versus mutated trip10a reporter RNAs.
Data access
The sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra). The developmental time-course mRNA-seq data are available in SRP189512, in addition to the previously published SRP072296 (Bazzini et al. 2016) and SRP149556 (Beaudoin et al. 2018). RESA sequencing is available in SRP189389, in addition to the previously published SRP090954 (Yartseva et al. 2017). Demultiplexed and raw reads for iCLIP data are available in SRP189499. SRP149368 contains demultiplexed and raw reads for endogenous Khsrp iCLIP (Beaudoin et al. 2018). For iCLIP multiplexed data, a column barcode in the SRA annotation indicates the internal barcode used to multiplex replicates (see Methods). To facilitate data download, internal to laboratory (AGR) and SRA (SRR), run IDs are listed in Supplemental Table S1 and at https://data.giraldezlab.org. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD009514. Source codes of the FIRE software (Supplemental Code S1) and the RESA random forest model (Supplemental Code S2) are also available at https://data.giraldezlab.org. Updated gene counts, data sets, and genome tracks are available at https://data.giraldezlab.org.
Supplementary Material
Acknowledgments
We thank Karen Bishop for technical help, and Ariel Bazzini, Miguel Moreno-Mateos and all the members of the Giraldez laboratory for intellectual and technical support, Dr. Matthias Hentze and Dr. Alfredo Castello for providing the original interactome capture protocol, and Cassandra Kontur and Dr. Valerie Tornini for manuscript editing. The Swiss National Science Foundation (grant P2GEP3_148600 to C.E.V.), NIH grants NHGRI (2R01HG003219) and NHGRI (1R01HG009065; P.O. and S.T.), the Deutsche Forschungsgemeinschaft (DFG; M.S. fellowship), the Eunice Kennedy Shriver National Institute of Child Health and Human Development–NIH grant K99HD071968 (D.C.), HHMI Faculty Scholars program (A.J.G.), and NIH grants R21 HD073768, R01 HD074078, R01 GM102251, and R35 GM122580 (A.J.G.) supported this work. The Yale Scholars Program and Whitman fellowship funds were provided by E.E. Just, Lucy B. Lemann, Evelyn and Melvin Spiegel, The H. Keffer Hartline and Edward F. MacNichol Jr. of the Marine Biological Laboratory in Woods Hole, MA, to A.J.G. The research of A.J.G. was supported in part by a faculty scholar grant from the Howard Hughes Medical Institute and the Simons Foundation.
Author contributions: C.M.T., V.Y., C.E.V., D.C., M.A.M., and A.J.G. conceived the project. C.M.T., V.Y., D.C., S.L., R.C., M.S., and H.D.-C. performed the experiments. C.E.V., M.A.M., P.O., and R.C. performed computational analysis of the data. C.M.T., C.E.V., V.Y., and M.T.L. performed RESA analysis. D.C., S.L., and R.C. performed interactome capture. M.A.M. performed random forest analysis, model-based motifs identification, and reporter mRNA predictions. D.C., S.L., and C.E.V. performed iCLIP and data analysis. C.M.T., V.Y., and M.S. performed reporter analysis. P.O. performed FIRE analysis. M.A.M., J.-D.B., H.D.-C., and C.E.V. performed trip10a reporter analysis. D.M. and C.E.V. performed ccdc22 reporter analysis. C.M.T., V.Y., D.C., C.E.V., M.A.M., P.O., M.S., R.C., and A.J.G. interpreted the results. A.J.G. supervised the project, with the contribution of S.T. and T.C.W. C.E.V., C.M.T., and V.Y. drafted the manuscript. C.E.V., M.A.M., and A.J.G. wrote the manuscript with input from the other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.245159.118.
References
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, et al. 2017. Ensembl 2017. Nucleic Acids Res 45: D635–D642. 10.1093/nar/gkw1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. 2012. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Omilli F, Osborne HB. 1997. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol 17: 209–218. 10.1128/MCB.17.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, Tapsin S, Chan YS, Tan CP, Sim AY, et al. 2016. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol Cell 62: 603–617. 10.1016/j.molcel.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. 2014. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. 10.15252/embj.201694699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin JD, Novoa EM, Vejnar CE, Yartseva V, Takacs CM, Kellis M, Giraldez AJ. 2018. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat Struct Mol Biol 25: 677–686. 10.1038/s41594-018-0091-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T. 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997. 10.1261/rna.569407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110. 10.1126/science.2174572 [DOI] [PubMed] [Google Scholar]
- Boucas J, Fritz C, Schmitt A, Riabinska A, Thelen L, Peifer M, Leeser U, Nuernberg P, Altmueller J, Gaestel M, et al. 2015. Label-free protein–RNA interactome analysis identifies Khsrp signaling downstream of the p38/Mk2 kinase complex as a critical modulator of cell cycle progression. PLoS One 10: e0125745 10.1371/journal.pone.0125745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Machine Learning 45: 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486. 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. 1999. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell 4: 209–218. 10.1016/S1097-2765(00)80368-8 [DOI] [PubMed] [Google Scholar]
- Despic V, Dejung M, Gu M, Krishnan J, Zhang J, Herzel L, Straube K, Gerstein MB, Butter F, Neugebauer KM. 2017. Dynamic RNA–protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res 27: 1184–1194. 10.1101/gr.215954.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. 2007. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350. 10.1016/j.molcel.2007.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. 1998. Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460. 10.1093/emboj/17.12.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway A, Saveliev A, Łukasiak S, Hodson DJ, Bolland D, Balmanno K, Ahlfors H, Monzón-Casanova E, Mannurita SC, Bell LS, et al. 2016. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 352: 453–459. 10.1126/science.aad5978 [DOI] [PubMed] [Google Scholar]
- Gamberi C, Peterson DS, He L, Gottlieb E. 2002. An anterior function for the Drosophila posterior determinant Pumilio. Development 129: 2699–2710. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. 2014. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156: 812–824. 10.1016/j.cell.2013.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492. 10.1073/pnas.0509260103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA, Schaening C. 2016. Messenger RNA modifications: form, distribution, and function. Science 352: 1408–1412. 10.1126/science.aad8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986. 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HT, Rasmussen SH, Adolph SK, Plass M, Krogh A, Sanford J, Nielsen FC, Christiansen J. 2015. Drosophila Imp iCLIP identifies an RNA assemblage coordinating F-actin formation. Genome Biol 16: 123 10.1186/s13059-015-0687-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19: 20–30. 10.1038/nrm.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BA, Gomez de la Torre Canny S, Ye Y, Slusarski DC, Freisinger CM, Dosch R, Chou MM, Wagner DS, Mullins MC. 2009. A novel role for MAPKAPK2 in morphogenesis during zebrafish development. PLoS Genet 5: e1000413 10.1371/journal.pgen.1000413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon LS, Zhang Z. 2007. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 8: R166 10.1186/gb-2007-8-8-r166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. 10.1016/j.ymeth.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Hammerschmidt M, Mullins MC, Maischein HM, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, et al. 1996. The zebrafish epiboly mutants. Development 123: 47–55. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, et al. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontur C, Giraldez A. 2017. RNA methylation clears the way. Dev Cell 40: 427–428. 10.1016/j.devcel.2017.02.024 [DOI] [PubMed] [Google Scholar]
- Kristjánsdóttir K, Fogarty EA, Grimson A. 2015. Systematic analysis of the Hmga2 3′ UTR identifies many independent regulatory sequences and a novel interaction between distal sites. RNA 21: 1346–1360. 10.1261/rna.051177.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. 2012. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 40: 5088–5100. 10.1093/nar/gks148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Lai X, Schmitz U, Gupta SK, Bhattacharya A, Kunz M, Wolkenhauer O, Vera J. 2012. Computational analysis of target hub gene repression regulated by multiple and cooperative miRNAs. Nucleic Acids Res 40: 8818–8834. 10.1093/nar/gks657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Castello A, Fischer B, Leicht S, Föehr S, Frese CK, Ragan C, Kurscheid S, Pagler E, Yang H, et al. 2016. The cardiomyocyte RNA-binding proteome: links to intermediary metabolism and heart disease. Cell Rep 16: 1456–1469. 10.1016/j.celrep.2016.06.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. 1970. Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science 170: 447–449. 10.1126/science.170.3956.447 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2017. Regulation by 3′-untranslated regions. Annu Rev Genet 51: 171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L, et al. 2011. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7: 458 10.1038/msb.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y. 2016. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885. 10.1016/j.molcel.2016.02.027 [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12: 982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murn J, Zarnack K, Yang YJ, Durak O, Murphy EA, Cheloufi S, Gonzalez DM, Teplova M, Curk T, Zuber J, et al. 2015. Control of a neuronal morphology program by an RNA-binding zinc finger protein, Unkempt. Genes Dev 29: 501–512. 10.1101/gad.258483.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou P, Goodarzi H, Tavazoie S. 2014. Systematic identification of regulatory elements in conserved 3′ UTRs of human transcripts. Cell Rep 7: 281–292. 10.1016/j.celrep.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J 17: 3461–3470. 10.1093/emboj/17.12.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Pieper L, Chew GL, Schier AF. 2017. A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Mol Cell 68: 1083–1094.e5. 10.1016/j.molcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rot G, Wang Z, Huppertz I, Modic M, Lenče T, Hallegger M, Haberman N, Curk T, von Mering C, Ule J. 2017. High-resolution RNA maps suggest common principles of splicing and polyadenylation regulation by TDP-43. Cell Rep 19: 1056–1067. 10.1016/j.celrep.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P, Heale BSE, Snøve O, Aagaard L, Alluin J, Rossi JJ. 2007. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res 35: 2333–2342. 10.1093/nar/gkm133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckel C, Drapeau E, Frias MA, Park CY, Fak J, Zucker-Scharff I, Kou Y, Haroutunian V, Ma'ayan A, Buxbaum JD, et al. 2016. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife 5: e10421 10.7554/eLife.10421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. 2012. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259. 10.1038/nrg3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. 2016. Global mapping of human RNA–RNA interactions. Mol Cell 62: 618–626. 10.1016/j.molcel.2016.04.030 [DOI] [PubMed] [Google Scholar]
- Staton AA, Knaut H, Giraldez AJ. 2013. Reply to: “On the robustness of germ cell migration and microRNA-mediated regulation of chemokine signaling”. Nat Genet 45: 1266–1267. 10.1038/ng.2812 [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Grün D, Kirchner M, Ayoub S, Torti F, Piano F, Herzog M, Selbach M, Rajewsky N. 2014. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO J 33: 1751–1766. 10.15252/embj.201488769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. 2015. HiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. 10.1038/nature14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoev VO, Fischer B, Frese CK, Gupta I, Krijgsveld J, Hentze MW, Castello A, Ephrussi A. 2016. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun 7: 12128 10.1038/ncomms12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155. 10.1016/j.devcel.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3: 59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Tome JM, Ozer A, Pagano JM, Gheba D, Schroth GP, Lis JT. 2014. Comprehensive analysis of RNA–protein interactions by high-throughput sequencing-RNA affinity profiling. Nat Methods 11: 683–688. 10.1038/nmeth.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215. 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, Martindale J, Xie C, Ahmad R, Thomas KJ, et al. 2008. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci 105: 10244–10249. 10.1073/pnas.0708518105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Steitz JA. 1998. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol 18: 7537–7545. 10.1128/MCB.18.12.7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD. 2011. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev 21: 431–443. 10.1016/j.gde.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Wessels HH, Imami K, Baltz AG, Kolinski M, Beldovskaya A, Selbach M, Small S, Ohler U, Landthaler M. 2016. The mRNA-bound proteome of the early fly embryo. Genome Res 26: 1000–1009. 10.1101/gr.200386.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsing A, Senkel S, Klein-Hitpass L, Ryffel GU. 2011. A systematic analysis of the 3′UTR of HNF4A mRNA reveals an interplay of regulatory elements including miRNA target sites. PLoS One 6: e27438 10.1371/journal.pone.0027438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink EM, Fogarty EA, Grimson A. 2016. High-throughput discovery of post-transcriptional cis-regulatory elements. BMC Genomics 17: 177 10.1186/s12864-016-2479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci 103: 4034–4039. 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yartseva V, Giraldez AJ. 2015. The maternal-to-zygotic transition during vertebrate development: a model for reprogramming. Curr Top Dev Biol 113: 191–232. 10.1016/bs.ctdb.2015.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yartseva V, Takacs CM, Vejnar CE, Lee MT, Giraldez AJ. 2017. RESA identifies mRNA-regulatory sequences at high resolution. Nat Methods 14: 201–207. 10.1038/nmeth.4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. 2014. Massively parallel functional annotation of 3′ untranslated regions. Nat Biotechnol 32: 387–391. 10.1038/nbt.2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. 2017. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542: 475–478. 10.1038/nature21355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zünd D, Gruber AR, Zavolan M, Mühlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943. 10.1038/nsmb.2635 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.