Abstract
The ability to detect harmful chemicals rapidly is essential for the survival of all animals. In Caenorhabditis elegans (C. elegans), repellents trigger an avoidance response, causing animals to move away from repellents. Dihydrocaffeic acid (DHCA) is a water-soluble repellent and nonflavonoid catecholic compound that can be found in plant products. Using a Xenopus laevis (X. laevis) oocyte expression system, we identified a candidate dihydrocaffeic acid receptor (DCAR), DCAR-1. DCAR-1 is a novel seven-transmembrane protein that is expressed in the ASH avoidance sensory neurons of C. elegans. dcar-1 mutant animals are defective in avoidance response to DHCA, and cell-specific expression of dcar-1 in the ASH neurons of dcar-1 mutant animals rescued the defect in avoidance response to DHCA. Our findings identify DCAR-1 as the first seven-transmembrane receptor required for avoidance of a water-soluble repellent, DHCA, in C. elegans.
Introduction
The detection of volatile or water-soluble chemicals, for example, odorants, pheromones, and tastants, is regulated by seven-transmembrane G-protein-coupled receptors (GPCRs) located in the olfactory epithelium, vomeronasal organ, and taste buds, respectively (Buck and Axel, 1991; Dulac and Axel, 1995; Adler et al., 2000). Although GPCRs are important for the detection of chemicals, many mammalian GPCRs are orphan receptors with no known ligand. In Caenorhabditis elegans (C. elegans), >1000 GPCRs distantly related to the rhodopsin superfamily are expressed in chemosensory neurons (Troemel et al., 1995; Robertson and Thomas, 2006), and most of them are orphan receptors. Genetic screens for chemosensory mutant animals have been unsuccessful in identifying receptors involved in the detection of chemicals, although they have led to the identification of downstream components of the GPCR signaling that are required for chemosensation. So far, the only chemosensory receptor in C. elegans whose ligand is known is the GPCR ODR-10 (Sengupta et al., 1996; Zhang et al., 1997). ODR-10 is expressed in the AWA olfactory neurons and has high affinity for the odorant diacetyl. Mutations in the odr-10 gene specifically affect the chemotaxis response to diacetyl, but not the responses to other compounds (Sengupta et al., 1996). Like ODR-10, other GPCRs have been identified that are specifically expressed in one or multiple kinds of chemosensory neurons, though their ligands are not known (Troemel et al., 1995).
To understand how animals sense chemicals in the environment, it is important to identify the ligands for individual chemosensory receptors. In this study, we identified a novel transmembrane receptor, dihydrocaffeic acid (DHCA) receptor (DCAR)-1, and its ligand, DHCA, using a Xenopus laevis (X. laevis) oocytes expression system. We show here that DCAR-1 is required for ASH-mediated water-soluble repellent, making it the first identified water-soluble taste receptor in these neurons. Our findings may lead to a better understanding of chemosensory signal transduction and its physiological relevance in the avoidance response in C. elegans.
Materials and Methods
X. laevis oocyte electrophysiology.
Electrophysiological analysis and expression cloning using X. laevis oocytes were performed as described previously (Miyamae et al., 1999). Ovarian lobes were obtained from mature female X. laevis. Ovarian tissue was removed and placed in OR2 solution (83 mm NaCl, 1.1 mm KCl, 0.49 mm MgCl2, 2.5 mm NaHCO3, 10 mm HEPES, pH 7.6). After treatment of the ovarian tissue with collagenase type I (3 mg/ml in OR2 solution; Sigma-Aldrich) to remove the follicular cells, the tissue was placed in Barth's solution (88 mm NaCl, 1 mm KCl, 0.33 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 2.4 mm NaHCO3, 10 mm HEPES, pH 7.4) supplemented with gentamycin (0.05 mg/ml) (Sigma-Aldrich). After 1 h, mature healthy oocytes were selected for cytoplasmic injections of 1.2 ng/nl (50 nl/oocyte) complementary RNA (cRNA) with a sharp pipette using a pressure injector (Narishige). Poly(A)+ RNA was extracted from C. elegans as described previously (Krause, 1995; Nakamura et al., 1999). Recombinant plasmid DNA (pSPORT-C06H5.7) was linearized with NotI, and in vitro transcription was performed using mMessage mMachine T7 transcription kit (Ambion). The oocytes injected with cRNA were again placed in fresh Barth's solution and incubated at 18°C. After 2–5 d, voltage-clamp recordings of injected oocytes were performed at a holding potential of −60 mV. Stimulants were dissolved in voltage-clamp solution (88 mm NaCl, 1 mm KCl, 0.33 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 2.4 mm NaHCO3, 15 mm HEPES, pH 7.6) and sequentially applied for 1 min with a 10 min interval left between the applications. Membrane potential was controlled, and membrane current was recorded by using a dual electrode voltage-clamp amplifier (Nihon Kohden).
Chemicals.
l-3,4-Dihydroxyphenylalanine (DOPA) was purchased from Nacalai Tesque. l-thereo-Dihydroxyphenylserine (DOPS) was kindly supplied by Sumitomo Pharmaceuticals. DHCA and 1-octanol were purchased from Wako Pure Chemical Industries. All the other chemicals were obtained from Sigma-Aldrich.
Cloning of DCAR-1.
With DOPS solution as a ligand, we isolated a positive clone as described as previously (Romero et al., 1998). The positive clone cDNA was the predicted open reading frame of C06H5.7 (dcar-1) in chromosome V (LG V), a putative seven-transmembrane protein (see Results, below).
Strains and genetics.
Nematodes were grown at 20°C and maintained using standard methods. Some strains were provided by the Caenorhabditis Genetics Center. Strains used in this study were as follows: N2 Bristol wild-type, dcar-1(nj66), dcar-1(tm2484), osm-9(ky10), ocr-2(ak47), odr-3(n2150), grk-2(rt97), osm-9(ky10);dcar-1(nj66), ocr-2(ak47);dcar-1(nj66), and odr-3(n2150) dcar-1(nj66).
Isolation of mutant alleles dcar-1(nj66) and dcar-1(tm2484).
A deletion allele dcar-1(nj66) was isolated in PCR screens of a deletion mutant library (∼24,000,000 genomes) generated by the trimethylpsoralen/ultraviolet method (Yandell et al., 1994; Gengyo-Ando and Mitani, 2000). Deletion allele dcar-1(tm2484) was kindly provided by the C. elegans National BioResource Project of Japan (http://shigen.lab.nig.ac.jp/c.elegans/index.jsp). dcar-1(nj66) and dcar-1(tm2484) mutant animals were backcrossed at least 10 times before behavioral testing. These mutant animals were viable and displayed normal locomotion and development.
RT-PCR.
Total RNA was prepared from mixed stage animals using RNeasy kit (Qiagen). First-strand cDNA was reverse-transcribed (SuperScript; Invitrogen) using an oligo (dT) primer according to the manufacturer's instructions. The cDNA was used for 30 cycles of PCR amplification with primers for dcar-1.
Molecular biology and plasmid construction.
Subcloning and general DNA manipulations were performed using standard protocols (Sambrook et al., 1989). The dcar-1 (C06H5.7) genomic region was prepared using internal primers from cosmid E01B7. For the dcar-1::GFP fusion, 4 kb of sequence upstream of the dcar-1 gene as well as DNA encoding of the first 150 aa of the predicted DCAR-1 protein were amplified by PCR and subcloned into the pPD95.77 vector in-frame with GFP. To construct the sra-6p::dcar-1 plasmid, dcar-1 cDNA was inserted into the pBluescript SK vector between XbaI and SacII sites. Subsequently, 4 kb of the sra-6 promoter (sra-6p) region was amplified by PCR using primers with BamHI site and inserted at a SalI site in the pBluescript SK/C06H5.7 subclone 5′ of the dcar-1 cDNA. To construct the sra-6p::dcar-1::Venus plasmid, dcar-1 cDNA was inserted into the sra-6p::Venus between the SmaI and KpnI sites.
Transgenic strains.
Germline transformations were performed using standard microinjection methods (Mello et al., 1991). For rescue experiments, sra-6p::dcar-1 cDNA was injected at 2.5 ng/μl mixed with pKDK66 (ges-1p::NLS-GFP) as a coinjection marker.
Microscopy.
Animals expressing dcar-1::GFP and sra-6p::dcar-1 cDNA:: Venus were analyzed with an LSM 510 confocal scanning microscope (Carl Zeiss), which deconvolutes multiple sections of fluorescent micrographs to generate a projection and a three-dimensional reconstruction.
Behavioral assays.
The avoidance assay for water-soluble repellents was performed as described previously (Hilliard et al., 2002, 2004; Mellem et al., 2002). Briefly, a small drop of DHCA (1, 10, and 100 μm) in control buffer (30 mm Tris, pH 7.5, 100 mm NaCl, 10 mm KCl) was placed in the path of an animal as it moved forward on an agar plate. The response was recorded as the lag time from the initial contact with the solution to the initiation of backward movement. If animals did not reverse within 4 s, the reversal time was recorded as 4 s.
Cell ablation.
To identify the ASH neurons, we stained the wild-type animals with DiO (Invitrogen). Cells were ablated with a laser microbeam as described previously (Bargmann and Avery, 1995). Laser operations were performed at a late larval stage (L3–L4). Mock-ablated animals were treated identically except that there was no killing of cells. Ablated or mock-ablated animals were grown at 20°C for at least half a day after the operation. After the avoidance assay, we confirmed that the operated cells were damaged in >90% of the animals.
Statistical analysis.
Data are shown as mean ± SEM. The statistical significance of the results was analyzed using Student's t test and one-way ANOVA.
Results
dcar-1 encodes a predicted seven-transmembrane domain receptor
We previously proposed that DOPA, a precursor of dopamine, is a neurotransmitter (Misu and Goshima, 2006). In an attempt to clarify the receptor and/or receptor components in DOPA response pathway, we used the X. laevis oocyte expression system. We injected poly (A)+ RNA from C. elegans into the X. laevis oocytes and determined whether the application of DOPS, a synthetic precursor of noradrenaline, and DOPA receptor agonist (Yue et al., 1992; Miyamae et al., 1996) induced a current response in the oocytes (DOPS has DOPA-like activity and is more soluble than DOPA; therefore, we used DOPS as a ligand in our expression cloning in the oocytes). In the course of this study, we isolated a positive clone that was responsive to DOPS solution. The positive clone cDNA was the predicted open reading frame of C06H5.7 in chromosome V (LG V). Because this receptor was later shown to respond potently to DHCA (see Fig. 2), we designated the C06H5.7 gene DCAR-1.
Figure 2.
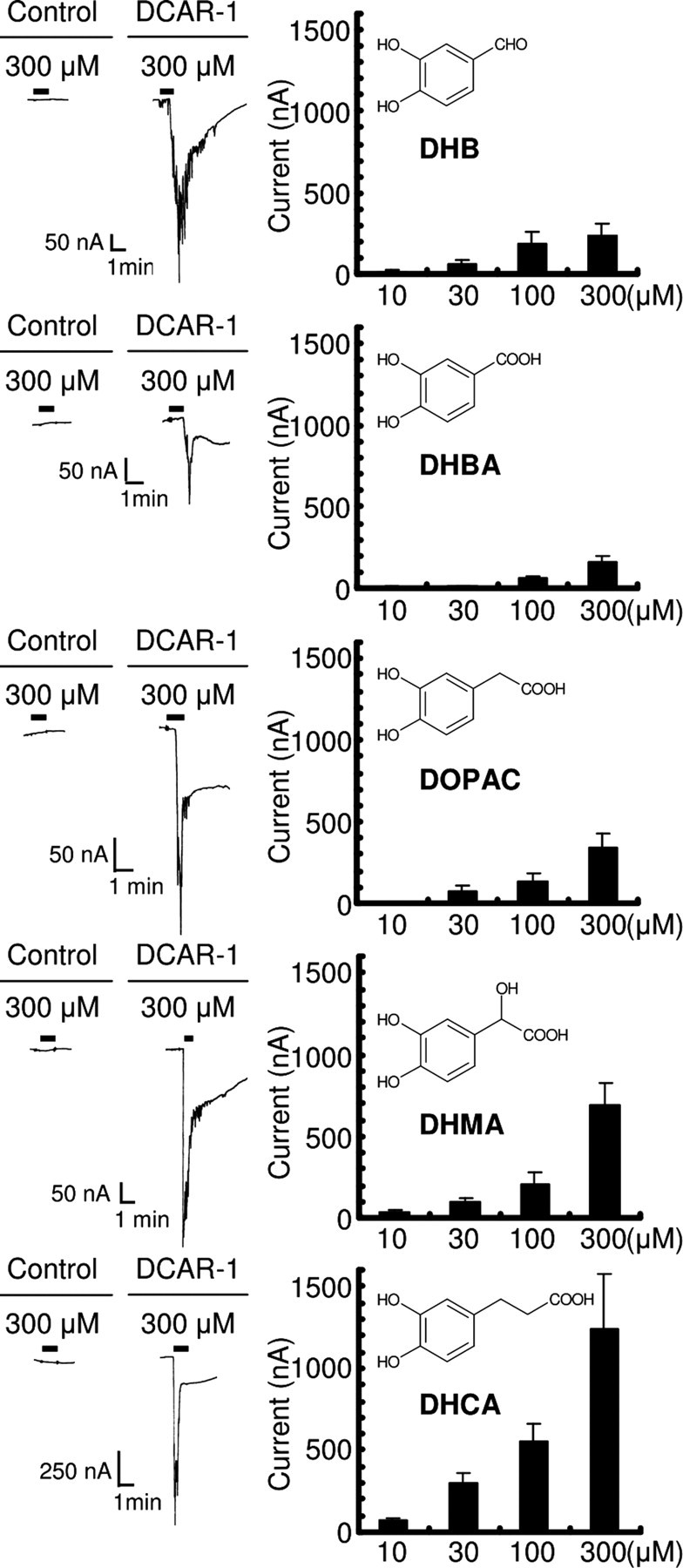
Summary of responses to active compounds by DCAR-1-expressing oocytes. 3,4-Dihydroxybenzaldehyde (DHB; 10 μm, n = 9; 30 μm, n = 7; 100 μm, n = 9; 300 μm, n = 7), 3,4-dihydroxybenzoic acid (DHBA; 10 μm, n = 6; 30 μm, n = 10; 100 μm, n = 9; 300 μm, n = 11), 3,4-dihydroxyphenylacetic acid (DOPAC; 10 μm, n = 7; 30 μm, n = 10; 100 μm, n = 11; 300 μm, n = 9), 3,4-dihydroxymandelic acid (DHMA; 10 μm, n = 6; 30 μm, n = 5; 100 μm, n = 7; 300 μm, n = 9), and dihydrocaffeic acid (DHCA; 10 μm, n = 5; 30 μm, n = 12; 100 μm, n = 7; 300 μm, n = 10) were all able to induce current responses in DCAR-1-expressing oocytes. DHB was identified as a candidate active substance in the solution of DOPS that induced a current response in oocytes injected with dcar-1 cRNA (data not shown).
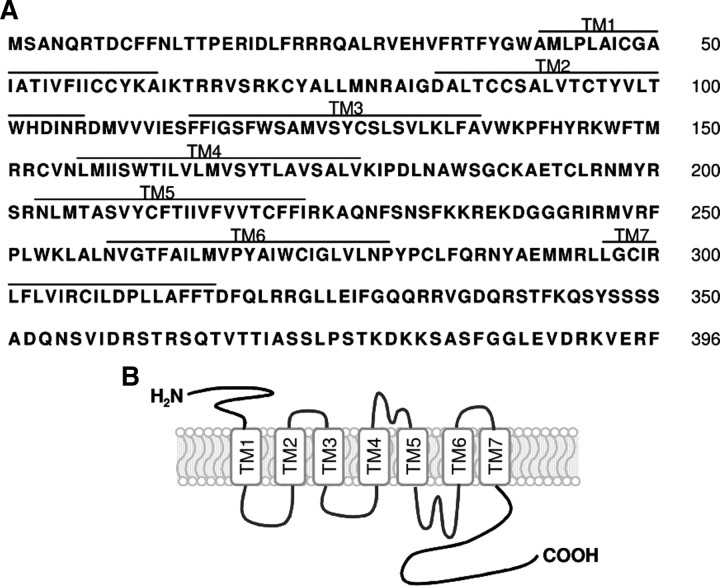
dcar-1 encodes a predicted protein of 396 aa (Fig. 1A). Hydrophobicity analysis of DCAR-1 revealed seven hydrophobic peaks that could correspond to membrane-spanning domains (Fig. 1), a topology characteristic of GPCRs (Bockaert and Pin, 1999). BLAST searches of GenBank and expressed sequence tag databases revealed a high degree of similarity in Caenorhabditis and Pristionchus species (C. brenneri, C. briggsae, C. remanei, C. japonica, and P. pacificus). Weak similarity was observed between dcar-1 and the srx family of candidate C. elegans chemosensory receptor genes (Robertson and Thomas, 2006).
Figure 1.
dcar-1 encodes a predicted seven-transmembrane domain receptor. A, Predicted amino acid sequence of dcar-1 cDNA. Amino acids are numbered beginning at the first methionine. Predicted transmembrane (TM) regions are numbered TM1 through TM7. The GenBank accession number for dcar-1 is BD314390. B, Predicted membrane topology of DCAR-1.
Identification of ligands that induce inward current response in DCAR-1-expressing X. laevis oocytes
In our experiments, DOPS was dissolved in ultra-pure water (10 mm stock solution). The solution was clear at first and subsequently turned brown during incubation at room temperature for 1 week. When the brown solution (diluted in voltage-clamp solution), but not the clear solution, was applied to the DCAR-1-expressing oocytes, it induced an inward current response (data not shown). No response was observed in the oocytes injected with vehicle control. This finding suggests that DCAR-1 could respond to an active derivative of DOPS in the brown solutions, but not to DOPS itself in the clear solutions. To identify the active derivatives, we performed high-performance liquid chromatography, NMR, and MS analysis on the brown sample (data not shown). Upon testing different compounds from the fractionation, we identified 3,4-dihydroxybenzaldehyde (DHB) as a candidate substance that could induce the current response in DCAR-1-expressing oocytes. We further observed that DHB induced the inward current responses in a dose-dependent manner (Fig. 2).
In an attempt to investigate the structure and activity relationship for the receptor, we tested the activity of several DHB-related compounds in DCAR-1-expressing oocytes. Several water-soluble compounds, including 3,4-dihydroxybenzoic acid, 3,4-dihydroxyphenylacetic acid, 3,4-dihydroxymandelic acid and DHCA, were able to induce current responses in the DCAR-1-expressing oocytes (Fig. 2). However, tyrosine, DOPA, dopamine, and noradrenaline did not induce current response in the DCAR-1-expressing oocytes (data not shown). Our data suggest that a relatively broad range of weakly acidic aromatic compounds, which possess a catechol moiety and a carboxyl group on the side chain and lack an amino group, act as DCAR-1 agonists.
dcar-1::GFP fusion genes are expressed in the ASH sensory neurons
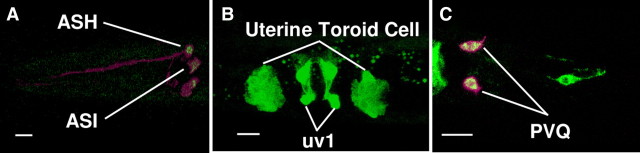
To determine where the dcar-1 gene is expressed in C. elegans, ∼4 kb of sequence upstream of the predicted translation start codon and ∼4 kb downstream of the dcar-1 coding region were used to generate a GFP reporter transgene. In transgenic animals, dcar-1::GFP was expressed in some sensory neurons and in some non-neuronal cells (Fig. 3). In sensory neurons, dcar-1::GFP was expressed in several amphid chemosensory neurons that have been shown to play an important role in chemical avoidance, including the ASH neurons (Fig. 3A). dcar-1::GFP was also expressed in the ASI sensory neurons (Fig. 3A), the uv1 neuroendocrine cells, the uterine toroid cells (Fig. 3B), and the PVQ interneurons (Fig. 3C). These data indicate that DCAR-1 is expressed in the C. elegans nervous system as well as in non-neuronal tissues.
Figure 3.
A dcar-1::GFP fusion gene is expressed in the ASH neurons. A–C, Confocal images of dcar-1::GFP expression in the head (A), midbody (B), and tail (C) of larvae. dcar-1::GFP is expressed in cell body of ASH and ASI amphid sensory neurons (A), uterine neuroendocrine cells (uv1) and uterine toroid cells (B), as well as PVQ interneurons (C). dcar-1::GFP merged with sra-6p::mCherry in the ASH, ASI, and PVQ neurons. Scale bars, 10 μm.
dcar-1(nj66) and dcar-1(tm2484) exhibit defects in avoidance response to the water-soluble DHCA
To elucidate the functions of DCAR-1 in C. elegans, we isolated a dcar-1 deletion allele, dcar-1(nj66), by PCR-based reverse genetics. In dcar-1(nj66) mutant animals, the deletion spans from intron 1 to intron 3 [32259–32967 on cosmid E01B7 (http://www.wormbase.org/): 709 bp], deleting exon 2 and exon 3 (Fig. 4A). We also used another deletion allele for our analysis, dcar-1(tm2484), which lacks part of exon 5 (34338–34561 on cosmid E01B7: 224 bp; obtained from the National Bioresource Project of Japan). When dcar-1 mRNA was examined in wild-type and mutant animals by RT-PCR, we observed abnormally short or scarce dcar-1 mRNAs in dcar-1(nj66) and dcar-1(tm2484) mutant animals (Fig. 4B). The dcar-1(nj66) and dcar-1(tm2484) gene products lacked the transmembrane regions known to be essential for proper binding of ligand (Bockaert and Pin, 1999). These data suggest that dcar-1(nj66) and dcar-1(tm2484) mutant animals are loss-of-function animals. The loss-of-function was confirmed by our finding that the brown solution of DOPS, a crude sample of DHB, induced the current response in oocytes injected with dcar-1 cRNA but not in oocytes injected with dcar-1(nj66) and dcar-1(tm2484) cRNAs (data not shown). Among the active compounds tested (see Identification of ligands that induce inward current response in DCAR-1-expressing X. laevis oocytes, above), we selected DHCA as a test compound in the behavioral assays because DHCA is easily dissolved in water and induced robust current responses in the DCAR-1-expressing oocytes (Fig. 2).
Figure 4.
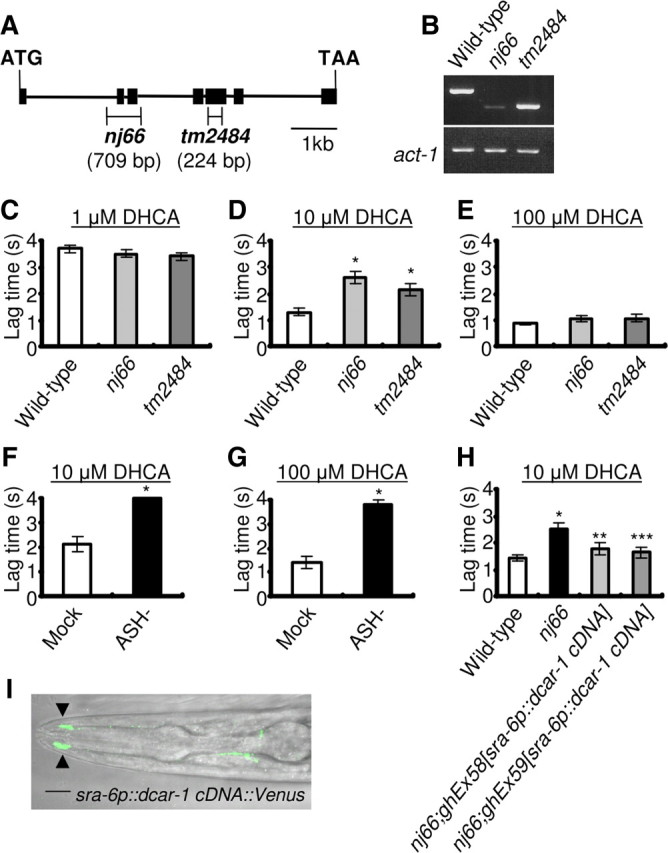
dcar-1 mutant animals show a defect in avoidance of DHCA. A, Structure of the dcar-1 gene. The locations and extent of dcar-1(nj66) and dcar-1(tm2484) deletions are indicated. Black boxes indicate exons; lines indicate introns. B, RT-PCR of total wild-type and dcar-1 mutant animals mRNA. act-1 messages were amplified in parallel as a positive control. C–E, Avoidance assays for DHCA. A small drop of DHCA buffer was placed in the path of an animal as it moved forward on agar plate. Average lag time for an animal to reverse direction after contacting with the DHCA was measured. If animals did not reverse within 4 s, the lag time was scored as 4 s. Data are shown as mean ± SEM (1 μm DHCA: wild-type, n = 31; nj66, n = 31; tm2484, n = 28; 10 μm DHCA: wild-type, n = 34; nj66, n = 35; tm2484, n = 32; 100 μm DHCA: wild-type, n = 33; nj66, n = 34; tm2484, n = 33). Asterisks indicate significant differences from wild-type animals (*p < 0.01 by one-way ANOVA). F, G, ASH-ablated animals showed a defect in the 10 and 100 μm DHCA avoidance. Data are shown as mean ± SEM (10 μm DHCA: mock-ablated, n = 9; ASH-ablated, n = 10; 100 μm DHCA: mock-ablated, n = 10; ASH-ablated, n = 11). Asterisks indicate significant differences from mock-ablated animals (*p < 0.001 by Student's t test). H, The 10 μm DHCA avoidance defect of dcar-1(nj66) mutant animals was rescued by the expression of dcar-1 cDNA in a subset of neurons including ASH neurons using the sra-6 promoter in two independent transgenic animals. Data are shown as mean ± SEM (wild-type, n = 46; nj66, n = 48; nj66;ghEx58[sra-6p::dcar-1 cDNA], n = 22; nj66;ghEx59[sra-6p::dcar-1 cDNA], n = 24). Asterisks indicate significant differences from wild-type animals (*p < 0.01 by one-way ANOVA) and dcar-1(nj66) mutant animals (**p < 0.05, ***p < 0.01 by one-way ANOVA). I, Confocal image of sra-6p::dcar-1::Venus expression in the head of larva. Black arrowheads indicate the sensory cilia of the head. Scale bar, 10 μm.
We assessed the effects of DHCA at various concentrations in wild-type and dcar-1 mutant animals using the drop assay (Hilliard et al., 2002, 2004; Mellem et al., 2002). In this assay, a drop was placed on the agar a few millimeters ahead of an animal that was moving forward, so that it encountered the substance absorbed in the agar. Adult animals were tested on agar plates in the absence of bacteria. When tested with control buffer, almost all the wild-type and dcar-1 mutant animals moved forward without changing direction (data not shown). In contrast, when tested with 1 μm DHCA, a few wild-type and dcar-1 mutant animals stopped first then moved backward after a brief delay (Fig. 4C). Moreover, when tested with 10 or 100 μm DHCA, a large number of wild-type and dcar-1 mutant animals showed the avoidance behavior (Fig. 4D,E). For the 10 μm DHCA drop stimulus, the mean lag time for reversal of wild-type animals was 1.30 ± 0.16 s, while the lag time of dcar-1(nj66) and dcar-1(tm2484) mutant animals was 2.62 ± 0.23 s and 2.17 ± 0.23 s, respectively (Fig. 4D). The longer lag times observed in dcar-1(nj66) and dcar-1(tm2484) mutant animals indicates that these mutants are defective in the DHCA avoidance response.
DCAR-1 in ASH neurons is required for the avoidance response to DHCA
We observed that dcar-1::GFP is expressed in ASH neurons, which were known to respond to several nociceptive stimuli and have an important role in repellent avoidance behavior. If the expression of DCAR-1 in ASH neurons is required for the avoidance response to DHCA, animals lacking ASH neurons should be defective in DHCA avoidance. To test this hypothesis, we performed an avoidance assay with animals in which ASH neurons were killed by laser microsurgery. When compared with mock-ablated animals, the ASH-ablated animals were severely defective in avoidance response to 10 and 100 μm DHCA (Fig. 4F,G). These data suggest that ASH neurons are required for the DHCA avoidance response.
To confirm that the avoidance defects observed in dcar-1(nj66) mutant animals were due to loss of DCAR-1 function, we generated transgenic animals that expressed the wild-type dcar-1 cDNA in the ASH neurons using the sra-6 promoter, which shows strong expression in the ASH and PVQ neurons and weak expression in ASI neurons (Troemel et al., 1995). The mean lag time of wild-type animals was 1.43 ± 0.14 s, while that of nj66 mutant animals and nj66;ghEx58[sra-6p::dcar-1 cDNA] and nj66;ghEx59[sra-6p::dcar-1 cDNA] rescued animals were 2.53 ± 0.20 s, 1.77 ± 0.23 s, and 1.65 ± 0.20 s, respectively (Fig. 4H). These results suggest that expression of dcar-1 cDNA under the sra-6 promoter was sufficient to restore the avoidance response to 10 μm DHCA in two independent transgenic lines (Fig. 4H). In addition, we observed that Venus-tagged DCAR-1 specifically expressed in ASH neurons under the sra-6 promoter was localized in the sensory cilia (Fig. 4I), suggesting a role in sensory transduction. These results suggest that DCAR-1 expressed in the ASH neurons is necessary for the DHCA avoidance response and that DCAR-1 is a receptor for DHCA.
In C. elegans, calcium imaging has been used as an indicator of neuronal activity in ASH neurons (Hilliard et al., 2005). To record the responses from ASH neurons when they are exposed to ligands in vivo, we analyzed the intracellular Ca2+ transients in ASH neurons following stimulation with DHCA in wild-type animals using the Ca2+ indicator protein cameleon (Hilliard et al., 2005). We stimulated animals with 10 and 100 μm DHCA using the olfactory microfluidic chip (Chronis et al., 2007). However, we could not detect any significant rises in the cytosolic Ca2+ concentration in ASH neurons when exposed to DHCA, whereas 10 mm CuCl2 induced a significant rise in the Ca2+ concentration using the same protocol (data not shown). DHCA at 1 mm tended to increase the Ca2+ concentration in ASH neurons, although this effect was not significant. One possibility is that the trimeric G-proteins and intracellular signal transduction machinery coupled with DCAR-1 in X. laevis oocytes may differ from those in ASH neurons in wild-type worms. Indeed, signal transduction mechanisms of quinine, glycerol, and nose touch responses, in which ASH neurons are involved, remain obscure (Tobin et al., 2002; Hilliard et al., 2004; Kahn-Kirby et al., 2004). Another possibility is that DHCA stimulation in wild-type animals induced a Ca2+ response in ASH neurons, but it was below the detectable threshold in our assay system.
We also examined the avoidance behavior of several other water-soluble repellents (quinine, glycerol, copper sulfate, acetic acid, SDS). Using the drop assay, we tested the behavioral responses to chemorepellents that are also detected by the ASH neurons (Hilliard et al., 2002, 2004). Wild-type, dcar-1(nj66), and dcar-1(tm2484) mutant animals exhibited similar avoidance response to these water-soluble repellents (Table 1).
Table 1.
Avoidance assays for volatile and water-soluble repellents
| Volatile repellents | Genotype | Seconds |
|---|---|---|
| Benzaldehyde | Wild-type | 2.79 ± 0.26 (n = 30) |
| dcar-1(nj66) | 5.61 ± 0.57** (n = 30) | |
| dcar-1(tm2484) | 4.84 ± 0.63* (n = 31) | |
| nj66;ghEx58[sra-6p::dcar-1 cDNA] | 3.36 ± 0.51*** (n = 32) | |
| 1-Octanol | Wild-type | 1.19 ± 0.11 (n = 30) |
| dcar-1(nj66) | 1.27 ± 0.14 (n = 30) | |
| dcar-1(tm2484) | 1.60 ± 0.12 (n = 31) |
| Water-soluble repellents | Genotype | Seconds |
|---|---|---|
| Quinine (10 mm) | Wild-type | 1.80 ± 0.19 (n = 15) |
| dcar-1(nj66) | 1.59 ± 0.17 (n = 15) | |
| dcar-1(tm2484) | 1.65 ± 0.10 (n = 15) | |
| Glycerol (1 m) | Wild-type | 1.13 ± 0.16 (n = 20) |
| dcar-1(nj66) | 0.90 ± 0.13 (n = 21) | |
| dcar-1(tm2484) | 1.39 ± 0.20 (n = 16) | |
| Copper sulfate (0.1 m) | Wild-type | 0.39 ± 0.04 (n = 19) |
| dcar-1(nj66) | 0.40 ± 0.03 (n = 20) | |
| dcar-1(tm2484) | 0.38 ± 0.03 (n = 20) | |
| Acetic acid (0.1 m) | Wild-type | 1.73 ± 0.12 (n = 20) |
| dcar-1(nj66) | 1.61 ± 0.16 (n = 20) | |
| dcar-1(tm2484) | 1.65 ± 0.20 (n = 18) | |
| Sodium dodecyl sulfate (0.1%) | Wild-type | 0.99 ± 0.12 (n = 21) |
| dcar-1(nj66) | 0.98 ± 0.12 (n = 21) | |
| dcar-1(tm2484) | 0.99 ± 0.09 (n = 17) |
Volatile repellents: Animals were confronted with a microcapillary containing 5 μl of undiluted benzaldehyde or 1-octanol. Each animal was assayed once for each volatile repellent, with at least 10 min interval between trials. If animals did not reverse within 20 s, the reversal time was scored as 20 s. The benzaldehyde avoidance defect of dcar-1(nj66) mutant animals was rescued by the expression of dcar-1 cDNA in a subset of neurons including ASH neurons using the sra-6 promoter. Asterisks indicate significant differences from wild-type animals (*p < 0.05, **p < 0.01 by one-way ANOVA) or from dcar-1(nj66) mutant animals (***p < 0.01 by one-way ANOVA). Data are shown as mean ± SEM.
Water-soluble repellents: A small drop of quinine (10 mm), glycerol (1 m), copper sulfate (0.1 m), acetic acid (0.1 m), and sodium dodecyl sulfate (0.1%) in control buffer (30 mm Tris, pH 7.5, 100 mm NaCl, 10 mm KCl) was placed in the path of an animal as it moved forward on an agar plate. Average lag time was measured for an animal to reverse direction after contacting with the repellents. Data are shown as mean ± SEM.
dcar-1(nj66) and dcar-1(tm2484) show defects in avoidance response to the volatile repellent benzaldehyde
We next examined the avoidance behavior to volatile repellents in C. elegans. It has been reported that the repellent benzaldehyde is detected in undiluted form by the ASH neurons and undiluted 1-octanol by the ASH and ADL neurons (Troemel et al., 1995). A microcapillary tube containing undiluted benzaldehyde was placed in front of a free-moving animal, and the lag time before initiation of a reversal response was measured. Wild-type animals responded to undiluted benzaldehyde and 1-octanol within a few seconds (Table 1). However, both dcar-1(nj66) and dcar-1(tm2484) mutant animals were defective in avoiding benzaldehyde (Table 1). Both mutant animals responded to undiluted 1-octanol similarly to wild-type animals (Table 1).
We examined whether the expression of DCAR-1 in ASH neurons could rescue the phenotypic defects in dcar-1(nj66) mutant animals. We observed that ASH expression of DCAR-1 under the sra-6 promoter rescued the benzaldehyde avoidance defect in transgenic animals (Table 1). These results together suggest that DCAR-1 is required in the ASH neurons for the avoidance response to undiluted benzaldehyde. In contrast to DHCA, however, benzaldehyde at 100 pm to 1 mm was essentially unable to induce the current response in DCAR-1-expressing oocytes.
Other sensory signaling mutants that are defective in DHCA avoidance
In C. elegans, several sensory signaling molecules are expressed and function in the ASH neurons (Colbert et al., 1997; Roayaie et al., 1998; Jansen et al., 1999; Tobin et al., 2002; Fukuto et al., 2004; Kahn-Kirby et al., 2004, Bargmann, 2006). Among these, the Gi/o-like protein ODR-3 is necessary for generating the response to osmotic strength, nose touch, and high salt concentrations (Roayaie et al., 1998). OSM-9 and OCR-2 are members of the vanilloid subfamily of transient receptor potential channel TRPV proteins that regulate the avoidance of osmotic strength and nose touch (Colbert et al., 1997; Tobin et al., 2002). Finally, GRK-2 is a GPCR kinase (GRK) protein that plays a crucial role in G-protein signaling (Fukuto et al., 2004). We investigated whether these mutant animals were defective in the avoidance response to DHCA. In osm-9(ky10), ocr-2(ak47), and odr-3(n2150) mutant animals, the avoidance response to 10 μm DHCA was defective (Fig. 5A). In addition, grk-2(rt97) mutant animals were severely defective in the avoidance response to 10 μm DHCA. Moreover, several double-mutant animals (osm-9(ky10);dcar-1(nj66), ocr-2(ak47);dcar-1(nj66), and odr-3(n2150) dcar-1(nj66)) respond to 10 μm DHCA similarly to the single mutant animals. These data suggest that these signaling proteins could function in the same signaling pathway with DCAR-1.
Figure 5.
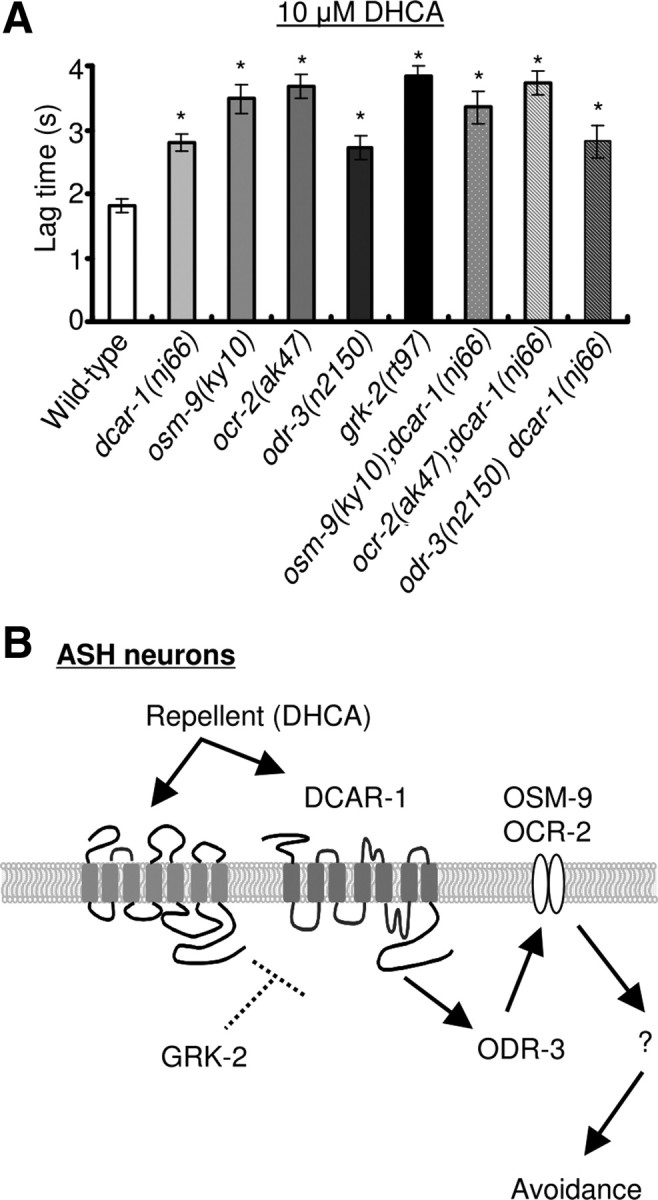
osm-9, ocr-2, and odr-3 mutant animals show defects in avoidance of DHCA. A, Avoidance responses to 10 μm DHCA in osm-9(ky10), ocr-2(ak47), and odr-3(n2150) mutant animals were defective. No significant additive defects were observed between single- and double-mutant animals. Data are shown as mean ± SEM (wild-type, n = 98; dcar-1(nj66), n = 97; osm-9(ky10), n = 20; ocr-2(ak47), n = 20; odr-3(n2150), n = 50; grk-2(rt97), n = 11; osm-9(ky10);dcar-1(nj66), n = 21; ocr-2(ak47);dcar-1(nj66), n = 19; odr-3(n2150) dcar-1(nj66), n = 27). Asterisk indicates significant differences from wild-type animals (*p < 0.01 by one-way ANOVA). B, Sensory signaling model for the avoidance response to DHCA.
Discussion
Our present study identifies DCAR-1 as the first seven-transmembrane receptor shown to be involved in the avoidance response to a water-soluble repellent. Indeed, when compared with wild-type animals, both dcar-1(nj66) and dcar-1(tm2484) mutant animals showed delayed responsiveness to DHCA. The introduction of sra-6p::dcar-1 cDNA in the dcar-1(nj66) mutant animals restored the defect in the avoidance response (Fig. 4H). In addition, dcar-1 cDNA::Venus driven by sra-6 promoter was localized in the sensory cilia (Fig. 4I). Since the cilia are believed to be the compartment in which sensory transduction occurs, the localization of DCAR-1 to the sensory cilia suggests a sensory function for this protein. These results provide evidence that DCAR-1 expressed in ASH neurons is involved in the physiological response to DHCA, a water-soluble repellent.
In addition to DCAR-1, several unknown GPCRs may also participate in the avoidance response to DHCA, since the avoidance response is attenuated but not abolished in dcar-1(nj66) and dcar-1(tm2484) mutant animals. In contrast, the laser ablation of ASH neurons almost completely abolished the avoidance response to DHCA, suggesting other DHCA receptors might be present in these neurons (Fig. 4F,G). It is also possible that DCAR-1 functions as a component of a receptor complex, which specifically detects the repellent ligands at lower concentrations, since the avoidance defect for both dcar-1(nj66) and dcar-1(tm2484) mutant animals was seen at a concentration of 10 μm but not 100 μm DHCA (Fig. 4D,E).
In our studies, odr-3, osm-9, ocr-2, and grk-2 mutant animals were all defective in the avoidance response to DHCA (Fig. 5A). ODR-3 is a Gi/o-like α-subunit protein that is essential for sensing stimuli such as osmotic strength, nose touch, and high salt concentrations in ASH neurons. The ODR-3 protein is localized in the cilia of ASH neuron (Roayaie et al., 1998), suggesting that it is directly involved in the transduction of chemical stimuli. OSM-9 and OCR-2 are members of the vanilloid subfamily of TRPV proteins that regulate the avoidance of osmotic strength and nose touch in ASH neurons (Colbert et al., 1997; Tobin et al., 2002). These channel proteins are similar to the Drosophila phototransduction channel, TRP, as well as the vertebrate capsaicin receptor, VR1, which is required for detecting painful stimuli such as capsaicin and noxious heat (Phillips et al., 1992; Caterina et al., 1997). GRK-2 is a GRK protein that plays a role in the avoidance of repellents in ASH neurons (Fukuto et al., 2004). In vertebrates, GRKs are involved in desensitization of receptor signaling (Premont and Gainetdinov, 2007). Thus, all these genes encode molecules that might act downstream of DCAR-1 in chemosensory signaling (Fig. 5B). Consistent with this possibility, several double-mutant combinations responded to 10 μm DHCA similarly to the constituent single mutant animals. We observed robust DHCA responses in X. laevis oocytes expressing ectopic DCAR-1 protein. The plasma membrane of X. laevis oocyte expresses calcium-dependent chloride channels, which are activated by several G-proteins (Gαi, Gαo, Gαq, Gαs, Gβγ) and phospholipase C-β (PLC-β) pathway (Kasahara and Sugiyama, 1994; Nakamura et al., 1994; Guttridge et al., 1995; de la Peña et al., 1995). Conceivably, DCAR-1 couples to the endogenous G-proteins (Gαi or Gαo) and then activates the PLC-β to induce the chloride currents in the X. laevis oocytes.
DHCA is classed as a polyphenol, a generic name for a group of phytochemical pigments and defensive components. The polyphenols are known to possess antioxidant properties and are very astringent or bitter. Therefore, DCAR-1 might be involved in detecting the sensation of bitter taste in C. elegans. Considering that the avoidance response is one of the biological warning systems in living organisms, it is possible that chemorepellent receptors such as DCAR-1 show a lower specificity in recognizing repellent molecules than receptors for chemoattractants such as ODR-10. ODR-10, which is involved in the chemoattractive response to diacetyl, indeed shows high ligand specificity for diacetyl (Sengupta et al., 1996); however, further work is needed to categorize the ligand selectivity of DCAR-1.
In C. elegans, the receptor for benzaldehyde has not yet been determined. The present behavioral analysis clearly indicates that the mutations in dcar-1 lead to a selective loss in the ability to respond to undiluted benzaldehyde as a repulsive volatile odorant. Both dcar-1(nj66) and dcar-1(tm2484) mutant animals showed significantly prolonged latency times in the avoidance response to undiluted benzaldehyde when compared with wild-type animals (Table 1). In contrast, these mutants showed a similar 1-octanol avoidance response to wild-type animals (Table 1). These defects suggest that DCAR-1 is involved in the recognition of undiluted benzaldehyde but not 1-octanol, and that the recognition pathway of benzaldehyde may be distinct from that of 1-octanol. In addition, it has been reported that low concentration of benzaldehyde is detected by AWC neurons (Bargmann et al., 1993). dcar-1(nj66) mutants showed normal chemotaxis to 100 mm benzaldehyde (data not shown), suggesting that other receptors could be involved in benzaldehyde sensation.
Although undiluted benzaldehyde induced an avoidance response in C. elegans, it did not induce a current response in DCAR-1-expressing X. laevis oocytes. Interestingly, numerous genes encoding soluble odorant-binding proteins (OBPs) have been identified in vertebrates (Hekmat-Scafe et al., 2002). These OBPs are suggested to transport chemical compounds to their receptors on dendrite membranes (Pelosi et al., 2006). It is possible that certain receptors can only recognize certain repellents (possibly including benzaldehyde) that are bound to the OBPs in vivo. Alternatively, benzaldehyde absorbed into nematodes could be converted to other active compounds, and these converted substances might induce the avoidance response in C. elegans. In fact, benzaldehyde and its metabolic intermediates are effectively degraded by some species of fungi (Kamada et al., 2002). The oxidation and reduction occur simultaneously, forming benzyl alcohol and benzoic acid as major products. Hydroxylation reactions occur on benzaldehyde to form corresponding 4-hydroxyl derivatives, resulting in the formation of DHB. This may reflect the capability of C. elegans to recognize some environmental substances and their related substances through these enzymatic reactions, reminiscent of the formation of diacetyl from pyruvate, both of which are ligands for ODR-10, by bacterial species through the diacetyl synthesis pathway (Zhang et al., 1997).
In conclusion, our findings suggest that DCAR-1 is a key mediator in avoidance response for water-soluble repellent in the chemosensory system of C. elegans. Further analysis may provide insights into the nature of the signal transduction in ASH neurons, its physiological role in the avoidance response, and its relevance to bitter taste perception.
Footnotes
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sport, Science, and Technology of Japan; the Naito Foundation; and Japanese Smoking Research Foundation (Y.G.). R.A. was supported by a fellowship from the 21 COE program (the Ministry of Education, Culture, Sport, Science, and Technology of Japan). We are grateful for the expertise of Kazushi Yoshida in laser ablation of ASH neurons. We thank Dr. Jon Audhya for the mCherry clone, Dr. Jim McGhee for the ges-1::GFP clone, Dr. Andrew Fire for the C. elegans expression vectors, and Dr. Cornelia I. Bargmann, Dr. Naoya Yamashita, and Sandy Chen for critical reading of our manuscript. We also thank Drs. Isao Katsura, Takeshi Ishihara, Toru Sakakibara, Shouhei Mitani, and Keiko Gengyo-Ando for helpful discussions, constructive input, and encouragement; and Takayo Ueda, Yasuhiro Murota, and Rie Katayama for their excellent technical assistance. Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C. elegans (October 25, 2006), WormBook, ed. The C. elegans Research Community, WormBook. 2006. doi/ 10.1895/wormbook.1.123.1, http://www.wormbook.org.
- Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña P, del Camino D, Pardo LA, Domínguez P, Barros F. Gs couples thyrotropin-releasing hormone receptors expressed in Xenopus oocytes to phospholipase C. J Biol Chem. 1995;270:3554–3559. doi: 10.1074/jbc.270.8.3554. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Fukuto HS, Ferkey DM, Apicella AJ, Lans H, Sharmeen T, Chen W, Lefkowitz RJ, Jansen G, Schafer WR, Hart AC. G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron. 2004;42:581–593. doi: 10.1016/s0896-6273(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- Guttridge KL, Smith LD, Miledi R. Xenopus Gqα subunit activates the phosphatidylinositol pathway in Xenopus oocytes but does not consistently induce oocyte maturation. Proc Natl Acad Sci U S A. 1995;92:1297–1301. doi: 10.1073/pnas.92.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI, Bazzicalupo P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol. 2002;12:730–734. doi: 10.1016/s0960-9822(02)00813-8. [DOI] [PubMed] [Google Scholar]
- Hilliard MA, Bergamasco C, Arbucci S, Plasterk RH, Bazzicalupo P. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 2004;23:1101–1111. doi: 10.1038/sj.emboj.7600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, Schafer WR. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Kahn-Kirby AH, Dantzker JL, Apicella AJ, Schafer WR, Browse J, Bargmann CI, Watts JL. Specific polyunsaturated fatty acids drive TRPV-dependent sensory signaling in vivo. Cell. 2004;119:889–900. doi: 10.1016/j.cell.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Kamada F, Abe S, Hiratsuka N, Wariishi H, Tanaka H. Mineralization of aromatic compounds by brown-rot basidiomycetes: mechanisms involved in initial attack on the aromatic ring. Microbiology. 2002;148:1939–1946. doi: 10.1099/00221287-148-6-1939. [DOI] [PubMed] [Google Scholar]
- Kasahara J, Sugiyama H. Inositol phospholipid metabolism in Xenopus oocytes mediated by endogenous G(o) and Gi proteins. FEBS Lett. 1994;355:41–44. doi: 10.1016/0014-5793(94)01170-2. [DOI] [PubMed] [Google Scholar]
- Krause M. Techniques for analyzing transcription and translation. Methods Cell Biol. 1995;48:513–529. doi: 10.1016/s0091-679x(08)61401-6. [DOI] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV. Decoding of polymodal sensory stimuli by postsynaptic glutamate receptors in C. elegans. Neuron. 2002;36:933–944. doi: 10.1016/s0896-6273(02)01088-7. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu Y, Goshima Y. Neurobiology of DOPA as a neurotransmitter. Boca Raton, FL: CRC; 2006. [Google Scholar]
- Miyamae T, Yue JL, Goshima Y, Misu Y. Depressor action of l-threo-dihydroxyphenyl serine in the rat nucleus tractus solitarii. Eur J Pharmacol. 1996;300:105–108. doi: 10.1016/0014-2999(95)00875-6. [DOI] [PubMed] [Google Scholar]
- Miyamae T, Goshima Y, Shimizu M, Shibata T, Kawashima K, Ohshima E, Suzuki F, Misu Y. Some interactions of l-DOPA and its related compounds with glutamate receptors. Life Sci. 1999;64:1045–1054. doi: 10.1016/s0024-3205(99)00031-4. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Goshima Y, Strittmatter SM, Kawamoto S. Isolation of receptor clones by expression screening in Xenopus oocytes. Methods Mol Biol. 1999;128:1–18. doi: 10.1385/1-59259-683-5:1. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Nukada T, Haga T, Sugiyama H. G protein-mediated inhibition of phosphoinositide metabolism evoked by metabotropic glutamate receptors in frog oocytes. J Physiol. 1994;474:35–41. doi: 10.1113/jphysiol.1994.sp020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AM, Bull A, Kelly LE. Identification of a Drosophila gene encoding a calmodulin-binding protein with homology to the trp phototransduction gene. Neuron. 1992;8:631–642. doi: 10.1016/0896-6273(92)90085-r. [DOI] [PubMed] [Google Scholar]
- Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinase and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Thomas JH. The putative chemosensory receptor families of C. elegans (January 06, 2006), WormBook, ed. The C. elegans Research Community, WormBook. 2006. doi/10.1895/wormbook.1.66.1, http://www.wormbook.org.
- Romero MF, Kanai Y, Gunshin H, Hediger MA. Expression cloning using Xenopus laevis oocytes. Methods in Enzymology. 1998;296:17–52. doi: 10.1016/s0076-6879(98)96004-9. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. Odr-10 encodes a seven-transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C. Combinatorial expression of TRPV channel proteins defines their sensoryfunctions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven-transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Yandell MD, Edgar LG, Wood WB. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1994;91:1381–1385. doi: 10.1073/pnas.91.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JL, Goshima Y, Nakamura S, Misu Y. L-DOPA-like regulatory actions of l-threo-3,4-dihydroxyphenylserine on the release of endogenous noradrenaline via presynaptic receptors in rat hypothalamic slices. J Pharm Pharmacol. 1992;44:990–995. doi: 10.1111/j.2042-7158.1992.tb07080.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chou JH, Bradley J, Bargmann CI, Zinn K. The Caenorhabditis elegans seven-transmembrane protein ODR-10 functions as an odorant receptor in mammalian cells. Proc Natl Acad Sci U S A. 1997;94:12162–12167. doi: 10.1073/pnas.94.22.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]