Abstract
Behavioral circadian rhythms disintegrate progressively in the R6/2 mouse model of Huntington's disease (HD), recapitulating the sleep–wake disturbance seen in HD patients. Here we show that disturbances in circadian pacemaking are not restricted to the brain, but also encompass peripheral metabolic pathways in R6/2 mice. Notably, circadian rhythms of clock-driven genes that are key metabolic outputs in the liver are abolished in vivo. This deficiency is accompanied by arrhythmic expression of the clock genes Cry1 and Dbp, and a phase-advanced Per2 cycle. Compromised circadian metabolic cycles are not, however, a consequence of deficient pacemaking intrinsic to the liver, because when cultured in vitro, R6/2 liver slices exhibit self-sustained circadian bioluminescence rhythms. We therefore propose that compromised metabolic cycles arise from an internal desynchronization secondary to altered feeding patterns and impaired circadian signaling from the central pacemaker of the suprachiasmatic nucleus (SCN). Importantly, the SCN-independent food-entrainable oscillator remains intact in R6/2 mice and, when invoked, can restore daily behavioral cycles and reverse some of the metabolic abnormalities seen in the liver. Disturbances of metabolism have long been thought to be an important feature of HD. Uncoupling liver metabolism from circadian drives will reduce metabolic efficiency and cause imbalances in metabolites known to be deleterious to brain function. Thus, even subtle imbalances in liver function may exacerbate symptoms of HD, where neurological function is already compromised.
Introduction
HD is a neurodegenerative disorder caused by a CAG repeat expansion in the HTT gene (Bates et al., 2002). Although characterized by progressive striatal degeneration and motor symptoms, HD has a complexity of symptoms encompassing motor, cognitive, and psychiatric domains. HD patients also exhibit perturbed sleep and circadian rhythms (Morton et al., 2005; Arnulf et al., 2008; Aziz et al., 2009; Cuturic et al., 2009).
The R6/2 transgenic mouse model of HD carries the human HD mutation. We showed previously that circadian rhythms of rest and activity, as well as circadian gene expression in the central hypothalamic pacemaker of the suprachiasmatic nuclei (SCN), are disrupted in R6/2 mice (Morton et al., 2005), although the intrinsic molecular machinery for generating circadian rhythms in the SCN remains intact (Pallier et al., 2007). Furthermore, in vivo pharmacological regularization of the sleep–wake disturbance can ameliorate both motor and cognitive dysfunction, and restore some features of SCN gene expression (Pallier et al., 2007; Pallier and Morton, 2009).
Circadian pacemakers are distributed widely in the brain and peripheral organs (Yamazaki et al., 2000; Reddy and O'Neill, 2010), where they are synchronized to both solar time and each other by SCN-dependent cues (Yoo et al., 2004). Importantly, peripheral clocks regulate the local cycles of gene expression that direct physiological and metabolic rhythms (Kornmann et al., 2007). Thus, circadian disturbance arising from the HD mutation could adversely affect not only behavior but also metabolism.
Here we characterized the effects of the HD mutation on local circadian pacemakers in the brain and a range of peripheral organs, and examined the consequences of the mutation on circadian expression of metabolically relevant genes in R6/2 mice. In SCN-deficient wild-type (WT) mice, temporally restricted daily feeding (RF) can activate a food-entrainable oscillator (FEO) and thereby reimpose daily patterning to rest and activity (for review, see Mistlberger, 2009). We showed previously that pharmacological manipulations could restore daily patterning to the behavior of R6/2 mice (Pallier and Morton, 2009). Here, by subjecting R6/2 mice to RF, we tested whether the FEO could be activated and used to control the rest/activity cycle in the context of HD. If circadian disorganization can influence metabolically relevant stimuli, this would provide a novel perspective on the metabolic disturbances of unknown origin that are characteristic of HD [for references, see Bates et al. (2002)].
Materials and Methods
Animals.
Studies were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act (1986). The R6/2 line was maintained by backcrossing onto C57BL/6 × CBA F1 female WT mice. Genotyping of biopsies by Laragen confirmed that CAG repeat lengths of the R6/2 mice were 250 ± 2 repeats (n = 72). After weaning, mice were housed under a 12:12 light/dark (LD) cycle [lights on 7:00 A.M., Zeitgeber time 0 (ZT0)] in mixed-genotype, single-sex cages with ad libitum access to food and water (provided from a readily accessible waterspout). For circadian recording, mice (8–9 weeks old) were caged individually within light-tight ventilated cabinets. Activity was monitored by infrared movement detectors and a computerized recording system (ClockLab; ActiMetrics). Cages were changed at least once every 5 weeks and water twice every 15 d. Circadian behavior was analyzed with ClockLab (ActiMetrics) and Actiwatch (Cambridge Neurotechnology) software. For bioluminescence studies, R6/2 mice were crossed with mPer2::luciferase (mPER2::LUC) circadian reporter mice (Yoo et al., 2004). Prescreening confirmed disruption of circadian behavior in R6/2 mice before harvesting brain, SCN and peripheral tissues. For the food entrainment study, 3 d after the daily activity rhythm of R6/2 mice (n = 18) had disintegrated (i.e., there was no significant 24 h rhythm by periodogram), they and age-matched WT littermates (n = 18) were subjected to a daily 5 h window of restricted feeding by placing food in the hopper between ZT6 and ZT11. The time of feeding (circadian day) was deliberately selected to be different from the normal spontaneous peak (circadian night) so that any restoration of feeding behavior, its associated anticipatory behavior, and liver gene expression can be attributed directly to the RF schedule and not to an unmasking of a previously suppressed circadian drive. We weighed the animals twice a week, but there was no change in body weight other than the expected disease-related decline (data not shown). Tissue from R6/2 mice was harvested after 12 ± 2 d of RF.
In situ hybridization.
Frozen brain sections (12 μm) were hybridized overnight at 58°C against antisense riboprobes to mPer2 labeled with [35S] UTP (final activity of 1.5 × 107 cpm/ml) and apposed to Kodak film (Biomax MR) for 7–10 d (Morton et al., 2005). Sense probes generated no specific image.
Explant cultures.
Organotypic tissue explants were prepared from adult mice as described previously (Yoo et al., 2004) (Pallier et al., 2007). Total emitted bioluminescence was measured with Hamamatsu photomultiplier tube assemblies housed in a light-tight 37°C incubator for at least 5 d. Rhythmicity and period were assessed with BRASS software (Prof. A. Millar, University of Edinburgh, Edinburgh, UK). Rayleigh tests for significant phase-clustering between explants were conducted in Oriana software (Kovach), with the bioluminescence peak occurring between 12 and 36 h in culture as the phase reference.
Real-time PCR (qPCR).
RNA was extracted using TRIzol reagent (Invitrogen) and purified using RNeasy Mini Kits (Qiagen). One microgram of total RNA was reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For control reactions, mouse β-actin mRNA was amplified from the same RNA samples. Real-time PCR was performed on ABI 7900HT Sequence Detection System (Applied Biosystems) using standard protocols and the Taqman Gene Expression Assays listed below. The relative levels of each mRNA were calculated by the 2−ΔΔCt method and normalized to the corresponding β-actin mRNA levels. Each Ct value used for these calculations is the mean of at least four biological replicates of the same reaction; each PCR was done in duplicate.
Statistics.
Data are shown as mean ± SEM. Significant differences were assessed using unpaired Student's t test or by one- or two-way ANOVA with post hoc Bonferroni t test, where applicable.
Results
Dysregulation of circadian gene expression across the brain in vivo is rescued in vitro
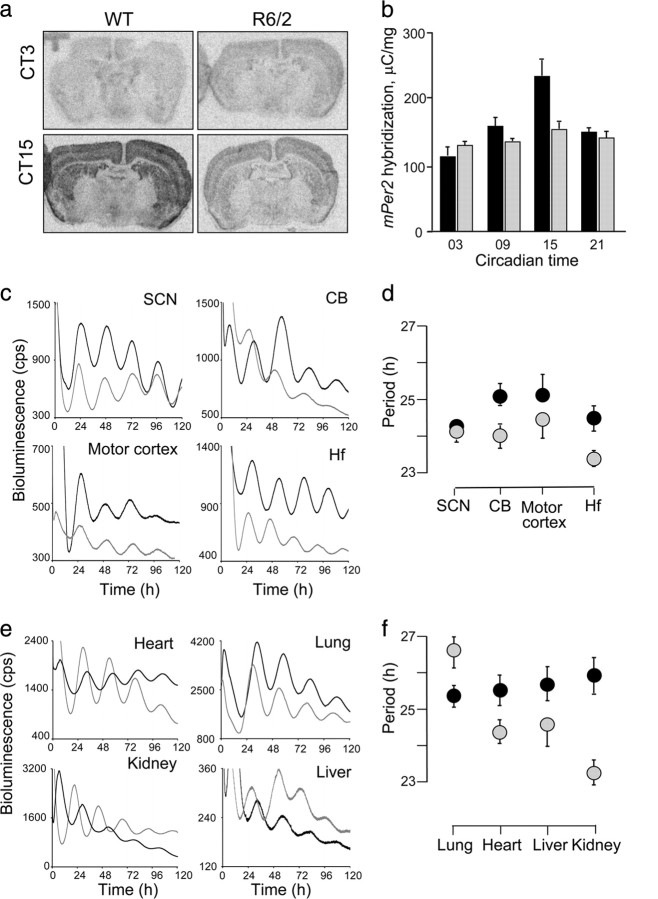
As expected, at 16 weeks of age, the rest/activity cycles of R6/2 mice lost definition and coherence (Morton et al., 2005), with significant deterioration in the stability, consolidation, and peak/trough amplitude compared to R6/2 mice at 8 weeks of age and to age-matched WT mice (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These deficits were accompanied in vivo by globally downregulated and arrhythmic expression of the mPer2 gene across the forebrain (Fig. 1a,b). However, in vitro bioluminescence recordings of mPER2::LUC expression revealed that intrinsic pace-making mechanisms remained intact in the R6/2 SCN (Fig. 1c), consistent with results obtained using the mPer1 reporter (Pallier et al., 2007). Moreover, the motor cortex, hippocampus and cerebellum of mutant mice all exhibited clear circadian bioluminescence rhythms when isolated in culture (Fig. 1c,d). The peak phase of the SCN oscillation occurred around circadian time 12 from both WT and R6/2 tissues, with highly significant phase clustering between explants (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). The three other brain regions exhibited delayed peak phases relative to the SCN, consistent with previous work (Yoo et al., 2004). Interestingly, the phase of oscillation of the WT cerebellum was significantly different (by about 8 h) from that of WT motor cortex or hippocampus, whereas in R6/2 tissues, all three brain regions had peak phases of oscillation within 2 h of each other, indicating greater between-tissue synchronization in R6/2 than in WT explants. These expression data from R6/2 mice support the view that slice preparation releases tissue clocks from an inhibitory effect exerted in vivo, thereby resynchronizing or reactivating oscillations in individual cells across the brain.
Figure 1.
Real-time analysis of circadian expression of mPER2::LUC protein in the forebrain and periphery from WT and R6/2 animals. a, Representative autoradiographic images of mPer2 mRNA expression across forebrain of WT and R6/2 mutant mice sampled at circadian time 3 and 15. b, Circadian expression of the mPer2 mRNA across the brain in the R6/2 (gray bars) and WT mice (black bars). c, Representative bioluminescent measurements from explants of SCN, hippocampus (Hf), motor cortex, and cerebellum (CB) from 16-week-old WT (black lines) and R6/2 (gray lines) mice. d, Period values of mPER2::LUC bioluminescence rhythms from WT (black symbols) and R6/2 (gray symbols) in the SCN and other brain regions (symbols represent the mean ± SEM; n = 4–12/group). Overall there was genotype effect (2-way ANOVA, p < 0.05). Cerebellum and hippocampus have significant differences between R6/2 and WT (t test, p < 0.05). e, Representative bioluminescent measurements from static explant cultures of lung, heart, liver, and kidney from 16-week-old WT (black lines) and R6/2 (gray lines) mice. f, Period values of mPER2::LUC bioluminescence rhythms from WT (black symbols) and R6/2 (gray symbols) peripheral tissues described in e. Symbols represent mean ± SEM; n = 6–8/group.
We assessed the competence of circadian pacemaking in peripheral tissue explants from a second cohort of behaviorally arrhythmic R6/2 mice and rhythmic WT controls (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Both genotypes showed characteristic circadian cycles of mPER2::LUC bioluminescence (Fig. 1e), although tissues from R6/2 mice showed more variability in the mean period length between different peripheral organs than did WT explants (Fig. 1f). Explants from both genotypes showed significant phase clustering for all tissues (p < 0.01, Rayleigh test) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material) except the heart, where there was a wide range of phases in both WT (p < 0.05) and R6/2 (n.s., p = 0.29). Importantly, the R6/2 mutation was associated with a 4–8 h advance of the in vitro phase of oscillation of these peripheral clocks, compared to WT tissues. This was much greater than the changes in phase of oscillation seen in the brain slices, and varied markedly between different organs. This suggests that peripheral pacemakers are more susceptible to the in vivo arrhythmia/desynchronization associated with the R6/2 mutation than the brain pacemakers.
Differential response of core clock genes and outputs to R6/2 mutation
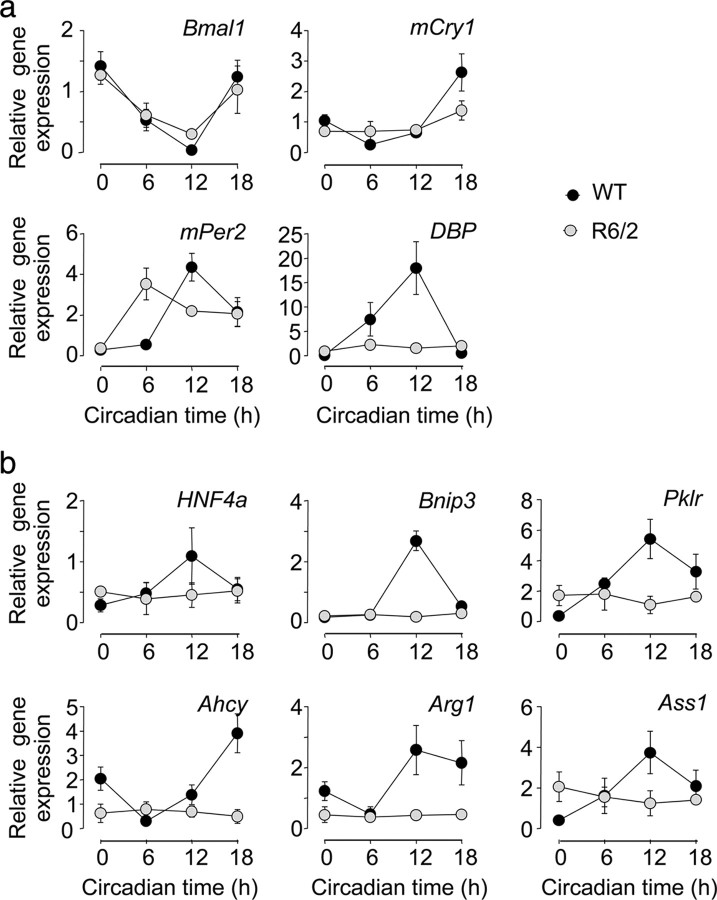
To assess the in vivo effect of the R6/2 mutation, we assayed core clock genes and output genes in the livers from WT and R6/2 mice. The expression of Bmal1 and mPer2 remained rhythmic in livers from R6/2 mice, although the peak expression of mPer2 was significantly phase advanced (Fig. 2a). Moreover, the rhythmic expression of mCry1 seen in WT was abolished in mutant liver, as was the rhythm of the canonical output gene albumin d-element-binding protein (Dbp). Normal maintenance and/or synchronization of various elements of the liver pacemaker were thus compromised in R6/2 mice in vivo.
Figure 2.
Compromised circadian expression of central clock genes and clock-controlled output genes in liver of R6/2 mice. a, Clock gene expression (mean ± SEM, n = 3–4 per group) in the livers of WT (black symbols) and R6/2 (gray symbols) mice. b, Expression patterns of clock-controlled (output) genes in livers of WT (black symbols) and R6/2 (gray symbols) mice (mean ± SEM, n = 3–4 per group). One-way ANOVA revealed significant change in expression over time for all genes in WT animals (p < 0.01). In livers from R6/2 mice, there was a significant rhythm in Bmal1 and mPer2 expression (ANOVA, p < 0.01) Expression of mCry1 and DBP and all of the downstream circadian genes was not rhythmic in the R6/2 mice.
We investigated hepatic circadian function further by assaying transcripts for key clock-driven metabolic outputs in the liver; hepatocyte nuclear factor 4 α (HNF4α), BCL2/adenovirus E1B 19 kDa protein-interacting protein (Bnip3), pyruvate kinase isozymes R/L (Pklr), adenosylhomocysteine hydrolase (Ahcy), arginase 1 (Arg1), and arginosuccinate synthase 1 (Ass1). As seen with DBP, these genes displayed arrhythmic profiles in the livers of R6/2 mice, with levels significantly and consistently below the peak of expression seen in WT mice (Fig. 2b). Thus, compromised expression of the core clock genes was accompanied by a severe loss of circadian drive to metabolically relevant output genes.
Consistent with findings in human cases (Sathasivam et al., 1999; Moffitt et al., 2009), the liver of R6/2 mice exhibited widespread ubiquitin- and huntingtin-positive nuclear inclusions, morphologically similar to those typically seen in brain tissue, but never seen in livers from WT mice (data not shown). Morphologically, the livers of R6/2 mice appeared normal, however, and did not stain with any markers indicative of a liver disorder per se. Thus, the compromise to circadian metabolic gene expression was not a result of a nonspecific or general hepatic pathology.
The food-entrainable oscillator is intact in R6/2 mice and can restore daily behavioral rhythms and hepatic gene expression
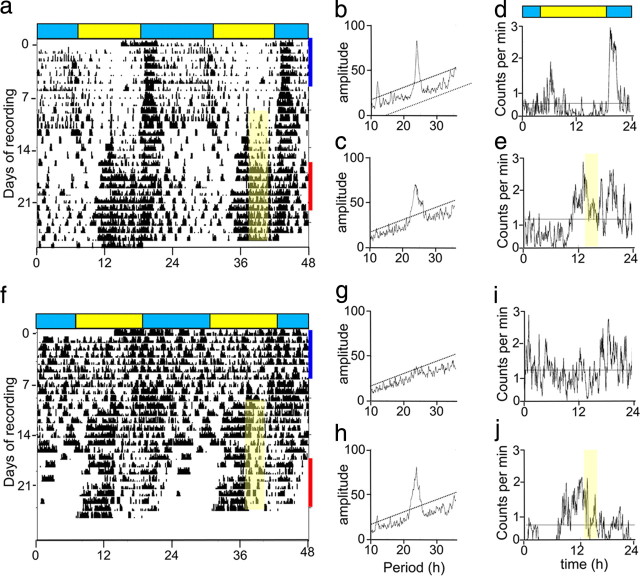
Given that central pacemaking is compromised in R6/2 mice, we tested whether their FEO (Mistlberger, 2009) remains intact and, if so, whether it might be used to restore daily patterning to behavior. R6/2 mice, whose circadian rhythms had already disintegrated, were subject to a RF regime (Fig. 3) and within a few days had regained a significant rhythm to their daily activity, with a peak onset of activity that anticipated the presentation of food. WT mice also shifted their peak onset of activity to anticipate presentation of food, although they also had an additional major peak of activity coincident with lights off, consistent with a functional, photically entrained SCN. Thus, although SCN-mediated control over daily behavior is compromised in R6/2 mice, the SCN-independent FEO remains functional and can be exploited to impose daily patterning over rest/activity behavior.
Figure 3.
Food entrainment restores circadian rhythmicity in R6/2 mice. a, f, Representative double-plotted actograms of continuous recordings of a WT (a) and an R6/2 (f) mouse monitored from 8 to 16 weeks of age, under a 12L:12D (LD) photoschedule. Only the last 4 weeks of recording are shown, over the period including the food entrainment (yellow shading). b, c, g, h, Representative periodogram analyses of the mice whose circadian behavior is illustrated in a and f. Dotted line denotes significant periodicity at p < 0.01. Note the highly significant peak in the WT but not in R6/2 mouse before food entrainment, and of both genotypes after initiation of food entrainment. d, e, i, j, Representative 24 h activity profiles (averaged counts over 7 d of recording under LD) of the individual WT (d, e) and R6/2 (i, j) mouse recorded at 13–14 weeks (blue bar in a and f) and 15.5–16.5 weeks (red bar in a and f), respectively. The food entrainment period is show as yellow shading.
We tested whether or not RF might be used to reverse compromised expression of circadian output genes in the livers of R6/2 mice. We elected to collect tissue at ZT12 and ZT18 when, under ad libitum feeding, expression levels for our genes of interest were consistently lower in R6/2 mice (Fig. 2b) and thus any increase in expression would be readily detectable. Under ad libitum feeding, expression in WT livers of Cry1 and Bmal1 significantly (p < 0.05, t test) increased between ZT12 to ZT 18, whereas Per2 and Dbp levels decreased (supplemental Table 1a, available at www.jneurosci.org as supplemental material). RF significantly altered these patterns (evidenced by main effect of feeding regime and/or interaction between time and regime in two-way ANOVA), in all cases consistent with a phase advance of the ad libitum pattern. In livers from ad libitum-fed R6/2 mice, there were no differences from WT in Cry1, Per2, and Bmal1 expression at ZT12 and ZT18, but a marked decline in Dbp levels at ZT12. RF again modified these profiles consistent with a phase advance (supplemental Table 1b, available at www.jneurosci.org as supplemental material), as anticipated.
Hepatic expression of the clock output genes Arg1, Ass1, and Ahcy was significantly lower in ad libitum fed R6/2 mice with disintegrated rhythms than in control mice (two-way ANOVA, genotype effect or genotype × time interaction). It did not, however, show any significant changes between ZT12 and ZT18 (post hoc t tests), and so data from these times were combined for analysis (Table 1). Importantly, relative to ad libitum feeding, RF increased the expression of all three clock-controlled genes in R6/2 mice. In the case of both Arg1 and Ahcy, values were comparable to those of WT tissues while expression of Ass1 in R6/2 tissue was still lower than in corresponding WT on RF. Thus RF reversed detrimental effects of the R6/2 mutation on both behavioral rhythmicity and hepatic metabolic gene expression.
Table 1.
Relative gene expression (mean ± SEM) in livers of WT and R6/2 mice kept on ad libitum or restricted feeding schedules
| Gene |
Ad libitum feeding |
Restricted feeding |
||
|---|---|---|---|---|
| WT (n = 11) | R6/2 (n = 12) | WT (n = 10) | R6/2 (n = 12) | |
| Per2 | 4.77 ± 0.46 | 4.21 ± 0.55 | 2.62 ± 0.78 | 2.26 ± 0.51 |
| Bmal1 | 3.90 ± 0.94 | 4.63 ± 0.84 | 6.95 ± 0.59 | 5.46 ± 0.48 |
| Cry1 | 4.57 ± 0.67 | 4.74 ± 0.43 | 5.15 ± 0.79 | 3.58 ± 0.45 |
| Ahcy | 5.96 ± 0.47 | 3.73 ± 0.22* | 4.74 ± 0.70 | 4.69 ± 0.31⋀ |
| Ass1 | 8.35 ± 0.81 | 4.62 ± 0.23* | 7.45 ± 0.70 | 5.92 ± 0.37⋀ |
| Arg1 | 8.38 ± 0.76 | 4.02 ± 0.35* | 6.50 ± 0.59 | 8.13 ± 0.50⋀ |
*p < 0.05 versus WT, ad libitum fed;
⋀p < 0.05 versus R6/2, ad libitum fed.
Discussion
Disrupted activity/rest cycles in R6/2 mice recapitulate sleep–wake disturbances observed in HD patients, and are accompanied by widespread downregulation of mPer2 expression across the forebrain (Morton et al., 2005), even though the machinery of the intrinsic brain pacemakers remains intact. Thus, it appears that circuitry factors (e.g., compromised interneuronal signaling and/or altered feedback from perturbed behavioral patterns) cause the dysrhythmic phenotype. Disrupted circadian function in vivo was also evident in the liver for both core clock genes (phase advanced mPer2 and arrhythmic mCry1 and Dbp) and metabolic output genes (for which circadian rhythms were abolished). Nevertheless, this was not a result of impaired local pacemaking, because in vitro liver tissue from R6/2 mice exhibited circadian mPER2::LUC bioluminescence rhythms. As with the brain, therefore, loss of direct or indirect synchronization from the SCN appears to underlie the in vivo deficits. Arrhythmic feeding (i.e., lack of daily metabolic stimulation) when food was available ad libitum likely causes this internal desynchrony, and consistent with this interpretation, activation of the SCN-independent FEO restored behavioral cycles and enhanced hepatic gene expression. These results provide a new context in which to understand the metabolic disturbances characteristic of R6/2 mice and HD patients.
Hepatic gene expression is differentially regulated such that the majority (∼90%) of circadian output genes are dependent upon the intrinsic hepatic pacemaker while ∼10% of genes, including Per2, are driven by rhythmic systemic cues (Kornmann et al., 2007). Consequently, loss of synchrony between systemic cycles and the local hepatic clock can initiate metabolic distress (Lamia et al., 2008). In R6/2 mice, the expression of particular metabolic output genes may be compromised because the sleep–wake and hence feeding cycles break down, even though other cues such as rhythmic hormone secretion, autonomic stimulation, and temperature cycles may remain sufficiently rhythmic to drive other circadian genes (Kornmann et al., 2007; Reddy and Maywood, 2007). We propose that such internal desynchronization is particularly relevant to HD patients, where food-entrainment and central cues may become uncoupled. In HD patients, for example, melatonin secretion remains rhythmic, albeit phase advanced (Aziz et al., 2009). Interestingly, livers taken from HD patients postmortem (but not early-stage disease) show significant atrophy (McCaughey, 1961; Bolt and Lewis, 1973; Chiu et al., 1975) and pathology consistent with mild abnormal liver function (Bolt and Lewis, 1973). The idea that liver and striatal pathology are linked is supported by the report that high indices for CAG repeat instability in a knock-in HD mouse model are restricted to the striatum and liver (Lee et al., 2010). Explicit liver enzyme abnormalities have not been reported in HD, but this is unsurprising. In R6/2 mice, circadian regulation is abnormal but genes continue to be expressed. Time of day of samples taken for hepatic function tests is rarely taken into account, and there is a wide range of “normal” liver function in humans that masks circadian fluctuations. Thus, deficits in circadian rhythms in liver function would not be observed unless multiple, timed samples were taken.
The metabolic and neurological consequences of abolishing circadian oscillations of liver gene expression are unknown, but may contribute to diverse aspects of the HD phenotype. For example, there is considerable pathology in skeletal muscle of R6/2 mice (Ribchester et al., 2004), with cachexia similar to that seen in late-stage HD patients. Disruption of metabolic circadian rhythms would likely have deleterious effects on skeletal muscle, which after the brain and heart, has the highest metabolic demand of any tissue. Equally, the clock output genes we studied have been implicated in neurological disturbance. Ahcy encodes S-adenosylhomocysteine hydrolase, defects of which cause hypermethioninemia (Baric et al., 2004), and cognitive deficits in rats (Stefanello et al., 2007). Notably, HD patients are reported to have abnormal handling of homocysteine (Andrich et al., 2004), and mutant huntingtin influences homocysteine metabolism by modulating the activity of cystathionine-β-synthase (Boutell et al., 1998). Dysregulation of Ass1 and Arg1, key components of the urea cycle, could lead to the accumulation of toxic metabolites such as ammonia that could exacerbate HD neuropathology (Picker et al., 2003; Deignan et al., 2006). Interestingly, urea cycle abnormalities characterized by hyperammonemia, high blood citrulline, and suppression of urea cycle enzymes have been shown in two mouse models of HD (Chiang et al., 2009).
Importantly, our findings show that by exploiting the FEO it is possible to regularize circadian behavioral and metabolic disturbances in R6/2 mice. This has significant implications for therapy in HD patients. Our mice were phenotypically “late stage,” when we put them onto the RF regime, and were already beyond the point typically used for behavioral studies (although they would be expected to survive for another 4–6 weeks). Nevertheless, they responded positively to the RF regime, showing that the machinery of the FEO was functional, even though the control of the overall circadian behavior of the mice had disintegrated. Our data suggest, therefore, that up until the stage that neurons have degenerated, both behavioral and physiological dysfunctions (typified by the disintegration of circadian rhythms) are potentially reversible. That is, the R6/2 brain can sustain 24 h behavior, if provided with the appropriate stimulus. This offers hope that if brain circuitry in HD patients is similarly intact, the onset of disease in HD patients could be delayed. Restoration of metabolic output patterns may provide a way to slow down a diverse range of clinically relevant features of HD.
Footnotes
This work was supported by CHDI Foundation (A.J.M.), Medical Research Council (M.H.H. and E.S.M.), and the Wellcome Trust (A.B.R., WT083643MA). A.J.M. is a Royal Society Leverhulme Trust Senior Research Fellow; A.B.R. is a member of the MRC Centre for Obesity and Related Metabolic Diseases. We gratefully acknowledge the excellent technical assistance from Jo Chesham, Tracey Butcher, and Zhiguang Zheng.
References
- Andrich J, Saft C, Arz A, Schneider B, Agelink MW, Kraus PH, Kuhn W, Müller T. Hyperhomocysteinaemia in treated patients with Huntington's disease homocysteine in HD. Mov Disord. 2004;19:226–228. doi: 10.1002/mds.10629. [DOI] [PubMed] [Google Scholar]
- Arnulf I, Nielsen J, Lohmann E, Schiefer J, Wild E, Jennum P, Konofal E, Walker M, Oudiette D, Tabrizi S, Durr A. Rapid eye movement sleep disturbances in Huntington disease. Arch Neurol. 2008;65:482–488. doi: 10.1001/archneur.65.4.482. [DOI] [PubMed] [Google Scholar]
- Aziz NA, Pijl H, Frölich M, Schröder-van der Elst JP, van der Bent C, Roelfsema F, Roos RA. Delayed onset of the diurnal melatonin rise in patients with Huntington's disease. J Neurol. 2009;256:1961–1965. doi: 10.1007/s00415-009-5196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric I, Fumic K, Glenn B, Cuk M, Schulze A, Finkelstein JD, James SJ, Mejaski-Bosnjak V, Pazanin L, Pogribny IP, Rados M, Sarnavka V, Scukanec-Spoljar M, Allen RH, Stabler S, Uzelac L, Vugrek O, Wagner C, Zeisel S, Mudd SH. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc Natl Acad Sci U S A. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GP, Harper PS, Jones L. Huntington's disease. Ed 3. Oxford: Oxford UP; 2002. [Google Scholar]
- Bolt JM, Lewis GP. Huntington's chorea. A study of liver function and histology. Q J Med. 1973;42:151–174. [PubMed] [Google Scholar]
- Boutell JM, Wood JD, Harper PS, Jones AL. Huntingtin interacts with cystathionine beta-synthase. Hum Mol Genet. 1998;7:371–378. doi: 10.1093/hmg/7.3.371. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Chen HM, Lai HL, Chen HW, Chou SY, Chen CM, Tsai FJ, Chern Y. The A2A adenosine receptor rescues the urea cycle deficiency of Huntington's disease by enhancing the activity of the ubiquitin-proteasome system. Hum Mol Genet. 2009;18:2929–2942. doi: 10.1093/hmg/ddp230. [DOI] [PubMed] [Google Scholar]
- Chiu E, Mackay IR, Bhathal PB. Hepatic morphology in Huntington's chorea. J Neurol Neurosurg Psychiatry. 1975;38:1000–1002. doi: 10.1136/jnnp.38.10.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuturic M, Abramson RK, Vallini D, Frank EM, Shamsnia M. Sleep patterns in patients with Huntington's disease and their unaffected first-degree relatives: a brief report. Behav Sleep Med. 2009;7:245–254. doi: 10.1080/15402000903190215. [DOI] [PubMed] [Google Scholar]
- Deignan JL, Livesay JC, Yoo PK, Goodman SI, O'Brien WE, Iyer RK, Cederbaum SD, Grody WW. Ornithine deficiency in the arginase double knockout mouse. Mol Genet Metab. 2006;89:87–96. doi: 10.1016/j.ymgme.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-M, Zhang J, Su AI, Walker JR, Wiltshire T, Kang K, Dragileva E, Gillis T, Lopez ET, Boily M-J, Cyr M, Kohane I, Gusella JF, MacDonald ME, Wheeler VC. A novel approach to investigating tissue-specific trinucleotide repeat instability. BMC Syst Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey WTE. The pathological spectrum of Huntington's chorea. J Nerv Ment Dis. 1961;133:91–103. [Google Scholar]
- Mistlberger RE. Food-anticipatory circadian rhythms: concepts and methods. Eur J Neurosci. 2009;30:1718–1729. doi: 10.1111/j.1460-9568.2009.06965.x. [DOI] [PubMed] [Google Scholar]
- Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington's disease. PLoS One. 2009;4:e8025. doi: 10.1371/journal.pone.0008025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. Disintegration of the sleep–wake cycle and circadian timing in Huntington's disease. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallier PN, Morton AJ. Management of sleep/wake cycles improves cognitive function in a transgenic mouse model of Huntington's disease. Brain Res. 2009;1279:90–98. doi: 10.1016/j.brainres.2009.03.072. [DOI] [PubMed] [Google Scholar]
- Pallier PN, Maywood ES, Zheng Z, Chesham JE, Inyushkin AN, Dyball R, Hastings MH, Morton AJ. Pharmacological imposition of sleep slows cognitive decline and reverses dysregulation of circadian gene expression in a transgenic mouse model of Huntington's disease. J Neurosci. 2007;27:7869–7878. doi: 10.1523/JNEUROSCI.0649-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker JD, Puga AC, Levy HL, Marsden D, Shih VE, Degirolami U, Ligon KL, Cederbaum SD, Kern RM, Cox GF. Arginase deficiency with lethal neonatal expression: evidence for the glutamine hypothesis of cerebral edema. J Pediatr. 2003;142:349–352. doi: 10.1067/mpd.2003.97. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Maywood ES. Circadian rhythms: per2bations in the liver clock. Curr Biol. 2007;17:R292–R4. doi: 10.1016/j.cub.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Reddy AB, O'Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribchester RR, Thomson D, Wood NI, Hinks T, Gillingwater TH, Wishart TM, Court FA, Morton AJ. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington's disease mutation. Eur J Neurosci. 2004;20:3092–3114. doi: 10.1111/j.1460-9568.2004.03783.x. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Hobbs C, Turmaine M, Mangiarini L, Mahal A, Bertaux F, Wanker EE, Doherty P, Davies SW, Bates GP. Formation of polyglutamine inclusions in non-CNS tissue. Hum Mol Genet. 1999;8:813–822. doi: 10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- Stefanello FM, Monteiro SC, Matté C, Scherer EB, Netto CA, Wyse AT. Hypermethioninemia increases cerebral acetylcholinesterase activity and impairs memory in rats. Neurochem Res. 2007;32:1868–1874. doi: 10.1007/s11064-007-9464-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]