Introduction
Harry Angelman, an English pediatrician, reported three cases of “Puppet Children” in 1965 (Angelman, 1965). These individuals displayed severe intellectual disability, ataxia, absent speech, jerky arm movements and bouts of inappropriate laughter. More cases were described as “Happy Puppet” syndrome (Bower and Jeavons, 1967), and additional consensus diagnostic criteria for what is now recognized as Angelman syndrome (AS) include microcephaly, seizures, EEG abnormalities, and hyperactivity (Williams et al., 2006). The current understanding of the clinical characteristics of AS children and adults has been summarized previously (Clayton-Smith and Laan, 2003; Dan, 2009; Van Buggenhout and Fryns, 2009).
Mapping of the AS genetic locus to the long arm of chromosome 15 between bands q11 and q13 (15q11-q13) was first reported in 1987 (Magenis et al., 1987) and confirmed in additional patients displaying chromosome 15q11-q13 deletions (Knoll et al., 1989). AS was only observed when the deletion occurred on the copy of chromosome 15 that was inherited from the mother (Knoll et al., 1989). This observation was remarkable because it had been previously found that a deletion of 15q11-q13 of the chromosome inherited from the father causes Prader-Willi syndrome (PWS), a disorder characterized by hyperphagia and obesity (Butler and Palmer, 1983). The intriguing observation that the 15q11-q13 deletion caused an entirely different disease depending on whether it was transmitted by the mother or the father was explained the discovery in 1989 that chromosome 15q11-q13 is subject to genomic imprinting (Nicholls et al., 1989), an epigenetic process whereby expression is allele-specific, i.e., one of the two parental copies of a gene is silenced.
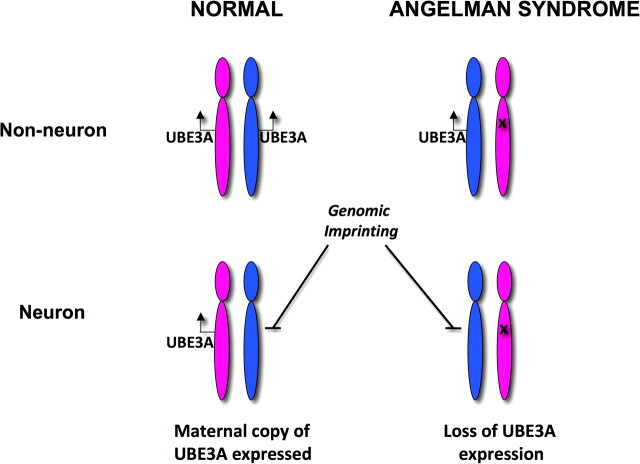
The specific gene in 15q11-q13 that causes AS encodes ubiquitin protein ligase E3A (UBE3A). UBE3A is also known as E6-associated protein (E6-AP) because it was originally discovered as a binding partner of the E6 protein of the human papilloma virus (Huibregtse et al., 1991, 1993). The relationship between genomic imprinting and UBE3A is illustrated in Figure 1. In non-neuronal tissues of normal individuals, UBE3A is expressed from both the chromosome 15 that is inherited from the father (blue) and the chromosome inherited from the mother (magenta). In neurons, the paternal copy (allele) of UBE3A is silenced by the process of genomic imprinting. As a result, only the maternal UBE3A allele is expressed in neurons from normal individuals (Albrecht et al., 1997; Rougeulle et al., 1997; Vu and Hoffman, 1997) (Fig. 1). In AS, the maternal UBE3A allele is absent or inactivated and, as a result, there is no active copy of UBE3A in neurons.
Figure 1.
In normal individuals, UBE3A is expressed from both the paternal (blue) and maternal (magenta) chromosomes 15 in most tissues. In neurons, the paternal UBE3A allele is silenced by the process of genomic imprinting. As a result, only the maternal UBE3A allele is expressed in neurons from normal individuals. In the case of AS, the maternal UBE3A allele is absent (deletion) or inactivated (mutation) and only the paternal allele is expressed in most tissues. UBE3A is not expressed in AS brain.
Loss of the active maternal UBE3A allele in AS is caused, in the majority of cases (∼75%), by maternal deletion of the chromosome 15q11-q13 region that contains UBE3A (Lossie et al., 2001). Another 20% of AS cases result from mutations of the maternal UBE3A allele (Kishino et al., 1997; Matsuura et al., 1997). The remainder result from uniparental disomy (the inheritance of two paternal and no maternal chromosomes 15) or from imprinting defects (see below). There are also cases of AS for which no molecular abnormality has been detected.
Genomic imprinting and Angelman syndrome
Genomic imprinting refers to a process whereby the maternal copy of a gene can be marked or “imprinted” differently than the paternal copy of the same gene (Reik and Walter, 2001). The imprinting process marks a relatively small number (100–200) of human genes and occurs in the germline. During the early stages of oogenesis, the paternal mark/imprint on a gene (coming from the father's chromosome) is erased and a maternal imprint is then placed on the gene. In this way, both gene copies in the egg are properly marked as maternal. Conversely, in spermatogenesis, the maternal imprint of a gene on the mother's chromosome is erased and then imprinted with a paternal mark such that both gene copies in sperm carry the appropriate paternal mark.
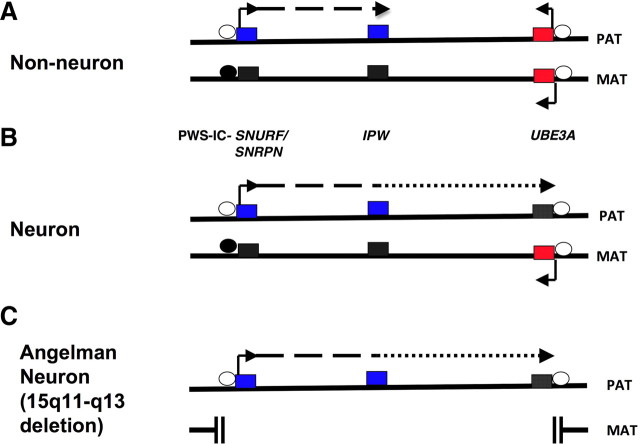
Imprinting marks are created by covalently attaching methyl groups to DNA, within specific segments that are rich in cytosine-guanine dinucleotides (CG-rich regions). In the germline, methyl groups are attached to or removed from the CG-rich imprinting region in a paternal- or maternal-specific pattern. The CG-rich imprinting region in chromosome 15q11-q13 is a 300 bp segment (Zeschnigk et al., 1997) that is called the Prader-Willi syndrome imprinting center (PWS-IC) (Fig. 2). The maternal copy of the PWS-IC is methylated during oogenesis and thus a methylated PWS-IC is the maternal imprint mark for 15q11-q13. Absence of methylation at the PWS-IC is the imprint of the paternal 15q11-q13. Since methylation is a characteristic of transcriptionally inactive chromatin, the methylation of the maternal copy of PWS-IC represses gene expression along the maternal chromosome (i.e., in cis-). As a result, the maternal alleles of genes within 15q11-q13 (Fig. 2A,B) are silenced. The paternal alleles of these genes are regulated in cis- by the paternal active (unmethylated) PWS-IC, and are expressed (Fig. 2A,B).
Figure 2.
Genomic imprinting of chromosome 15q11-q13 and epigenetic silencing of UBE3A in neurons. A, In non-neuronal tissues, both parental UBE3A alleles are expressed (red-filled boxes). The paternally expressed alleles (blue-filled boxes) of SNURF/SNRPN and IPW loci and the corresponding silent maternal alleles (black-filled boxes) are under the control of the PWS-IC. The PWS-IC, located within the SNURF/SNRPN promoter, is a CG-rich cluster that is differentially methylated: the paternal chromosome is not methylated (open circle), while the maternal copy is fully methylated (black-filled circle). The CG-rich UBE3A promoter is not methylated on either parental chromosome. The 3′-end of the paternal SNURF/SNRPN transcript is a long noncoding RNA of several hundred kilobases and incorporates IPW. B, In neurons, the paternal long noncoding RNA extends to and overlaps UBE3A as an antisense (UBE3A-ATS) with concomitant silencing of the paternal UBE3A allele (black-filled box). The transcriptional silencing of the paternal UBE3A allele in neurons is not associated with DNA methylation of the UBE3A promoter but results from as yet undiscovered mechanisms. C, In neurons from AS 15q11-q13 deletion patients, there is no active copy of UBE3A due loss of the maternal allele. Only a few representative genes from the 15q11-q13 imprinted region are illustrated here.
The PWS-IC is located in the upstream promoter region of the SNURF/SNRPN transcript (Sutcliffe et al., 1994; Zeschnigk et al., 1997; Bielinska et al., 2000; El-Maarri et al., 2001). The PWS-IC is one part of a bipartite imprinting regulatory element; the second component is located 35–40 kb upstream of PWS-IC and is referred to as the AS imprinting center (AS-IC) (Dittrich et al., 1996). The AS-IC appears to participate, by a yet undetermined mechanism, in the establishment of DNA methylation at the PWS-IC during oogenesis (Buiting et al., 2003). The AS-IC segment is deleted in a small number of AS cases that are termed imprinting defects. In these patients, the maternal chromosome 15 carries the paternal imprint, i.e., there is no DNA methylation of the maternal copy of the PWS-IC.
Although the PWS-IC is the master regulator of imprinting of the chromosome 15q11-q13 region, transcriptional repression of the paternal UBE3A allele involves additional brain-specific molecular mechanisms that remain to be conclusively elucidated (Fig. 2B). Brain-specific UBE3A imprinting does not result from parental-specific DNA methylation of the UBE3A promoter since both the maternal and paternal copy of the CpG-rich UBE3A promoter are unmethylated (Fig. 2) in all human and mouse tissues that have been examined, including human postmortem brain (Lossie et al., 2001). The observation that a noncoding antisense RNA (UBE3A-ATS) is paternally expressed in human brain (Fig. 2B) has led to the hypothesis that the paternal UBE3A allele is silenced by this cis-acting antisense transcript (Rougeulle et al., 1998; Chamberlain and Brannan, 2001; Runte et al., 2001; Yamasaki et al., 2003). The hypothesis that UBE3A imprinting is regulated by an antisense transcript is consistent with the finding that the imprinting of several other genes is mediated by antisense transcription (Lalande and Calciano, 2007).
UBE3A-ATS is a large (>600 kb) RNA that initiates in the PWS-IC (Fig. 2). The expression and/or processing of UBE3A-ATS differ between brain and other tissues. While UBE3A-ATS transcripts that include the IPW locus are detected in all tissues, it is only in neurons that UBE3A-ATS extends distally to overlap UBE3A (Fig. 2). In the case of AS, the maternal UBE3A allele is either absent or defective and UBE3A activity is lost in brain upon paternal UBE3A-ATS expression and concomitant epigenetic silencing of the normal paternal UBE3A allele (Fig. 2C).
There are several proposed mechanisms by which UBE3A-ATS could mediate the epigenetic silencing of UBE3A in neurons (Rougeulle and Heard, 2002; Shibata and Lee, 2004; Lalande and Calciano, 2007). These include transcriptional interference resulting from the simultaneous occupancy of RNA polymerase complexes on the positive and negative strands, and RNA interference induced by double-stranded RNA formed between sense and antisense RNAs. The antisense silencing mechanism could also involve epigenetic alterations of the sense transcript, such as the antisense-transcription-mediated repressive chromatin conformational changes that occur at the X chromosome inactivation locus (Navarro et al., 2005).
Animal models of Angelman syndrome
The high degree of evolutionary conservation of the UBE3A HECT domain amino acid sequence has allowed the generation of Drosophila models for AS (Wu et al., 2008; Lu et al., 2009). Null mutants display a movement disorder as well as deficits in long-term memory and circadian rhythms (Wu et al., 2008). Examination of dendritic morphogenesis and function in sensory neurons of Drosophila UBE3A-null mutants revealed defects in dendritic growth and branching and slower growth of fine terminal dendritic processes (Lu et al., 2009).
A mouse model engineered as a “knock-in” of a yellow fluorescent protein (YFP) marker into the Ube3a locus allowed visualization of the UBE3A distribution in brain (Dindot et al., 2008) and subcellularly in vitro. In cultured hippocampal neurons, the UBE3A:YFP fusion protein is found in the nucleus as well as the presynaptic and postsynaptic compartments (Dindot et al., 2008). Expression of maternally derived UBE3A:YFP fusion protein was observed in most brain regions, while the paternally inherited fusion protein was only faintly detected. These observations are consistent with other findings that genomic imprinting of Ube3a is widespread in brain (Landers et al., 2005) but are in contrast to previous reports that maternal-specific expression is observed predominantly in hippocampal neurons and cerebellar Purkinje cells (Albrecht et al., 1997; Jiang et al., 1998; Miura et al., 2002). The observation that both the paternal and maternal UBE3A:YFP fusion proteins are detected in glial fibrillary acidic protein (GFAP)-positive cells (Dindot et al., 2008) is a further indication that the UBE3A imprinting occurs in neurons and not in glial cells (Yamasaki et al., 2003).
Two mouse models of AS have been generated by targeted inactivation of Ube3a (Jiang et al., 1998; Miura et al., 2002). Upon inheritance of the mutation through the maternal but not the paternal germline, both mutant mouse models display several features characteristic of AS. These include microcephaly, deficits in motor function and long-term potentiation (LTP), abnormal context-dependent and spatial learning, seizures, abnormal hippocampal EEG, fast cerebellar oscillation associated with ataxia, and sleep disturbances (Jiang et al., 1998; Miura et al., 2002; Cheron et al., 2005; Colas et al., 2005; Heck et al., 2008). Defects in fluid consumption and licking observed in the mouse model (Heck et al., 2008) may recapitulate the swallowing problems and excessive drooling frequently associated with AS individuals (Williams et al., 2006). Mitochondrial dysfunction in CA1 hippocampal neurons (Su et al., 2009) and impaired adult neurogenesis (Mardirossian et al., 2009) are also observed in the mouse AS models.
In Ube3a knock-out mice (Jiang et al., 1998), abnormal dendritic spine development was found in cerebellar Purkinje cells as well as pyramidal neurons of the hippocampus and cortex (Dindot et al., 2008). Reduced dendritic spine length and density in neurons of the hippocampus and visual cortex were also observed in two other studies of Ube3a deficiency (Yashiro et al., 2009; Sato and Stryker, 2010). Moreover, Yashiro et al. (2009) found that this phenotype was only apparent in neurons from Ube3a-deficient mice that were subjected to sensory experience, correlating with loss of synaptic plasticity. The findings in both the Drosophila and mouse models thus strongly suggest that defects in dendritic spine growth and development contribute to AS neurological deficits.
Since Jiang et al. (1998) first reported a deficit in LTP, Weeber et al. (2003) reported that the deficits in Ube3a-deficient mice included both NMDA receptor-dependent and -independent LTP. They attributed this to an increased level of inhibitory autophosphorylation of the α subunit of calcium/calmodulin-dependent protein kinase II (αCaMKII), and hypothesized that decreased phosphatase activity could explain this phosphorylation defect. The introduction of an αCaMKII mutation that prevents inhibitory phosphorylation into maternal Ube3a-deficient mice rescued the LTP deficit in hippocampal neurons, corroborating the previous findings. This αCaMKII mutation also rescued many of the behavioral defects, including motor performance on an accelerating rotarod and contextual learning in the Morris water maze test (van Woerden et al., 2007). The changes in αCaMKII phosphorylation could, however, be regional- or developmental stage-specific since the increase in phosphorylation was not detected in visual cortex (Sato and Stryker, 2010).
Reduced experience-dependent maturation of synapses in the visual cortex was also observed in maternal Ube3a-deficient mice (Yashiro et al., 2009). Normal rearing and dark rearing paradigms were used to elicit sensory experience, and sensory deprivation, respectively. Ube3a-deficient mice subjected to normal rearing conditions lost synaptic plasticity, observed as defects in both LTP and long-term depression (LTD). This synaptic rigidity could be prevented and reversed by dark rearing. Another study using ocular dominance plasticity as an assay reported similar findings. However, this group also observed that repression of paternal Ube3a and a change in subcellular localization of Ube3a protein also occurs during the window in which experience-dependent plasticity is lost in Ube3a-deficient mice (Sato and Stryker, 2010).
Loss of UBE3A function and Angelman syndrome
UBE3A functions in the ubiquitin-proteasome system by ligating ubiquitin to target proteins and thereby initiating degradation of the target protein via the 26S proteasome complex. The first step in the UBE3A ubiquitination process involves activation of ubiquitin by an E1 enzyme, followed by transfer of ubiquitin via an E2 enzyme to UBE3A. UBE3A then attaches the activated ubiquitin to a target protein.
The great majority of UBE3A mutations that lead to AS are predicted to disrupt or delete the 350 aa C-terminal HECT (homologous to the E6-AP C terminus) domain (Lossie et al., 2001). This domain is prototypical of the HECT domain class of E3 ubiquitin-protein ligases (Scheffner et al., 1993). The terminal 100 aa region of the HECT domain is essential for ligation of the activated ubiquitin to the protein that is targeted for degradation by the ubiquitin-proteasome degradation system. Given the likelihood that the accumulation of such target proteins underlies the AS disease mechanism, there is great interest in identifying the protein targets that are ubiquitinated by UBE3A in neurons.
The first UBE3A target gene identified in brain, the Ras homolog-specific guanine exchange factor (Rho/GEF), pebble, was discovered in a screen of Drosophila heads using a human UBE3A transgene (Reiter et al., 2006). The mouse ortholog of pebble, is epithelial cell transforming sequence 2 (Ect2) and pebble/Ect2 is known to play an essential role cytokinesis, and to regulate neuronal outgrowth in postmitotic cells. Reiter et al. studied Ect2 expression by immunohistochemical staining in the brains of normal and Ube3a mutant mice. Expression of Ect2 was increased and mislocalized in the CA3 region of the hippocampus in Ube3a-null mice with levels of Ect2 reduced in the soma of Purkinje cells (Reiter et al., 2006).
The recent identification of activity-regulated, cytoskeletal-associated protein (ARC) as a target of ubiquitylation by UBE3A, provides a tantalizing prospect for the primary neuronal defect underlying AS (Greer et al., 2010). ARC participates in the activity-dependent trafficking of AMPA (α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate) receptors at synapses onto hippocampal neurons. Fewer AMPA receptors are present on the cell surface in stimulated neurons lacking Ube3a, which results in reduced AMPA receptor-mediated currents (Greer et al., 2010). These findings are consistent with the previously observed LTP and LTD deficits in Ube3a-deficient mice, since ARC is important for both LTP and LTD (Shepherd et al., 2006). This finding may also explain why a mutant form of αCaMKII that is incapable of inhibitory autophosphorylation could rescue the AS phenotype in mice (van Woerden et al., 2007): increased αCaMKII activity may increase AMPA receptor function at the synaptic surface (Rose et al., 2009). However, these results are inconsistent with those of Yashiro et al., which suggest that the ratio of AMPA to NMDA receptor currents is comparable between wild type and Ube3a-deficient mice in the visual cortex (Yashiro et al., 2009). The apparent discrepancy in AMPA:NMDA receptor ratios could reflect regional (e.g., hippocampus vs visual cortex) or temporal (e.g., P14 vs P28) differences between the two studies. It is also not clear whether decreased AMPA receptor currents can lead to the increased inhibitory phosphorylation of αCaMKII that was observed by Weeber et al. (2003). Additional experiments will need to be done to determine whether increased levels of postsynaptic ARC can explain the phenotypic manifestations of AS.
Summary and future perspectives
UBE3A was identified as the gene responsible for AS in 1997 (Kishino et al., 1997; Matsuura et al., 1997). Since this discovery, animal models have been generated thus permitting insights into the AS disease mechanism. Ube3a-mutant mice share several of the phenotypic characteristics that are diagnostic of AS. These include ataxia and motor dysfunction, defects in learning and memory, abnormal EEG and sleep disturbances. The growth and development of dendritic spines are affected by loss of UBE3A in brains of both mouse and Drosophila. The recent discovery that ARC is targeted by UBE3A in dendritic spines of hippocampal neurons in an activity-dependent manner suggests that neurological defects in AS could be associated with the trafficking of AMPA receptors. It is likely that additional research will uncover other neurobiological and molecular abnormalities that underlie the AS phenotype and that, eventually, these might lead to therapies to alleviate the neurological problems of this disease. The advent of induced pluripotent stem cell reprogramming technologies should make available live human AS neurons to accelerate the discovery and validation of drug therapies (Chamberlain et al., 2008).
Normal neurological function requires appropriate regulation of UBE3A expression levels. While deficiency of UBE3A leads to AS, excess levels of UBE3A also result in neurological abnormalities. Over expression of UBE3A in Drosophila nervous system leads to abnormal locomotion and decreased dendritic branching in sensory neurons (Wu et al., 2008; Lu et al., 2009). In humans, duplication of the chromosome 15q11-q13 is associated with a clinically distinct syndrome characterized by delayed motor skills and language development, cognitive and learning disabilities and seizures (Battaglia, 2005). Chromosome 15q11-q13 duplication is also the most frequent cytogenetic abnormality observed in autism (Veenstra-Vanderweele et al., 2004). The increased risk of autism is associated with maternal but not paternal transmission of chromosome 15q11-q13 duplications suggesting that over expression of UBE3A in brain is a candidate mechanism in autism (Cook et al., 1997). In contrast, an autistic-like mouse phenotype results from paternal duplication of the homologous Ube3a chromosomal region (Nakatani et al., 2009). While the reason for these strikingly contradictory results remains to be resolved, it is evident that the identification molecular networks and pathways regulated by UBE3A will further our understanding of neurogenetic diseases in addition to AS.
Footnotes
Editor's Note: Disease Focus articles provide brief overviews of a neural disease or syndrome, emphasizing potential links to basic neural mechanisms. They are presented in the hope of helping researchers identify clinical implications of their research. For more information, see http://www.jneurosci.org/misc/ifa_minireviews.dtl.
We are grateful to Drs Richard Mains, Elizabeth Eipper and Eric Levine for their careful reading of the manuscript. Sources of additional information are as follows: Angelman syndrome Foundation (www.angelman.org/), National Institute of Neurological Disorders of the National Institutes of Health (www.ninds.nih.gov/disorders/angelman/angelman.htm), Department of Pediatrics, University of Florida (http://www.peds.ufl.edu/divisions/genetics/programs/angelman_syndrome.htm), and Genomic imprinting, Medical Research Council, Harwell, UK (http://www.har.mrc.ac.uk/research/genomic_imprinting/).
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Angelman H. “Puppet Children.” A report of three cases. Dev Med Child Neurol. 1965;7:681–688. [Google Scholar]
- Battaglia A. The inv dup(15) or idic(15) syndrome: a clinically recognisable neurogenetic disorder. Brain Dev. 2005;27:365–369. doi: 10.1016/j.braindev.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- Bower BD, Jeavons PM. The “happy puppet” syndrome. Arch Dis Child. 1967;42:298–302. doi: 10.1136/adc.42.223.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, el-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Palmer CG. Parental origin of chromosome 15 deletion in Prader-Willi syndrome. Lancet. 1983;1:1285–1286. doi: 10.1016/s0140-6736(83)92745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Li XJ, Lalande M. Induced pluripotent stem (iPS) cells as in vitro models of human neurogenetic disorders. Neurogenetics. 2008;9:227–235. doi: 10.1007/s10048-008-0147-z. [DOI] [PubMed] [Google Scholar]
- Cheron G, Servais L, Wagstaff J, Dan B. Fast cerebellar oscillation associated with ataxia in a mouse model of Angelman syndrome. Neuroscience. 2005;130:631–637. doi: 10.1016/j.neuroscience.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis. 2005;20:471–478. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- Dan B. Angelman syndrome: current understanding and research prospects. Epilepsia. 2009;50:2331–2339. doi: 10.1111/j.1528-1167.2009.02311.x. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, Walter J, Horsthemke B. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck DH, Zhao Y, Roy S, LeDoux MS, Reiter LT. Analysis of cerebellar function in Ube3a-deficient mice reveals novel genotype-specific behaviors. Hum Mol Genet. 2008;17:2181–2189. doi: 10.1093/hmg/ddn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Graham JM, Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell Mol Life Sci. 2007;64:947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers M, Calciano MA, Colosi D, Glatt-Deeley H, Wagstaff J, Lalande M. Maternal disruption of Ube3a leads to increased expression of Ube3a-ATS in trans. Nucleic Acids Res. 2005;33:3976–3984. doi: 10.1093/nar/gki705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, Theriaque D, Hutson A, Nicholls RD, Zori RT, Williams CA, Driscoll DJ. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18:454–462. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenis RE, Brown MG, Lacy DA, Budden S, LaFranchi S. Is Angelman syndrome an alternate result of del(15)(q11q13)? Am J Med Genet. 1987;28:829–838. doi: 10.1002/ajmg.1320280407. [DOI] [PubMed] [Google Scholar]
- Mardirossian S, Rampon C, Salvert D, Fort P, Sarda N. Impaired hippocampal plasticity and altered neurogenesis in adult Ube3a maternal deficient mouse model for Angelman syndrome. Exp Neurol. 2009;220:341–348. doi: 10.1016/j.expneurol.2009.08.035. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, Wagstaff J. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, Iwamoto K, Kato T, Okazawa M, Yamauchi K, Tanda K, Takao K, Miyakawa T, Bradley A, Takumi T. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature. 1989;342:281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J, Jin SX, Craig AM. Heterosynaptic molecular dynamics: locally induced propagating synaptic accumulation of CaM kinase II. Neuron. 2009;61:351–358. doi: 10.1016/j.neuron.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Heard E. Antisense RNA in imprinting: spreading silence through air. Trends Genet. 2002;18:434–437. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso C, Fontés M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- Runte M, Hüttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci U S A. 2010;107:5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Lee JT. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol. 2004;14:1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Su H, Fan W, Coskun PE, Vesa J, Gold JA, Jiang YH, Potluri P, Procaccio V, Acab A, Weiss JH, Wallace DC, Kimonis VE Advance online publication. Mitochondrial dysfunction in CA1 hippocampal neurons of the UBE3A deficient mouse model for Angelman syndrome. Neurosci Lett. 2009 doi: 10.1016/j.neulett.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Van Buggenhout G, Fryns JP. Angelman syndrome (AS, MIM 105830) Eur J Hum Genet. 2009;17:1367–1373. doi: 10.1038/ejhg.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, Jiang YH, Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, Laan LA, Magenis RE, Moncla A, Schinzel AA, Summers JA, Wagstaff J. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, Fischer JA. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci U S A. 2008;105:12399–12404. doi: 10.1073/pnas.0805291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M, Schmitz B, Dittrich B, Buiting K, Horsthemke B, Doerfler W. Imprinted segments in the human genome: different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum Mol Genet. 1997;6:387–395. doi: 10.1093/hmg/6.3.387. [DOI] [PubMed] [Google Scholar]