ABSTRACT
Chromogranin B (CgB, also known as CHGB) is abundantly expressed in dense core secretory granules of multiple endocrine tissues and has been suggested to regulate granule biogenesis in some cell types, including the pancreatic islet β-cell, though the mechanisms are poorly understood. Here, we demonstrate a critical role for CgB in regulating secretory granule trafficking in the β-cell. Loss of CgB impairs glucose-stimulated insulin secretion, impedes proinsulin processing to yield increased proinsulin content, and alters the density of insulin-containing granules. Using an in situ fluorescent pulse-chase strategy to track nascent proinsulin, we show that loss of CgB impairs Golgi budding of proinsulin-containing secretory granules, resulting in a substantial delay in trafficking of nascent granules to the plasma membrane with an overall decrease in total plasma membrane-associated granules. These studies demonstrate that CgB is necessary for efficient trafficking of secretory proteins into the budding granule, which impacts the availability of insulin-containing secretory granules for exocytic release.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Chromogranin B, Secretory granule biogenesis, Insulin secretion, Granule trafficking, β-cell function
Summary: A novel fluorescent pulse-chase technology is used to demonstrate that CgB regulates granule budding in the islet β-cell.
INTRODUCTION
Pancreatic islet β-cells perform a critical role in maintaining whole-animal fuel homeostasis through the nutrient-dependent release of insulin, which enhances glucose uptake in peripheral tissues such as liver, adipose and skeletal muscle. Insulin insufficiency due to loss of islet β-cell mass and function, coupled with increasing peripheral (fat, muscle, liver) insulin resistance, leads to sustained hyperglycemia and ultimately, the development of type 2 diabetes (T2D). Compelling evidence highlights the central role of β-cell decompensation in this transition (Halban et al., 2014; Ward et al., 1984). In T2D, islet dysfunction presents with elevated basal insulin secretion, absent or delayed first phase (stimulated) secretion, and attenuated second phase secretion (Ward et al., 1984; Gerich, 2002). This decline in function coincides with degranulation of the β-cell, reduced insulin content and inefficient hormone processing, leading to increased release of proinsulin (Masini et al., 2012; Like and Chick, 1970; Alarcon et al., 2016). Despite the reduction in dense core granules observed in T2D islets, rodent models demonstrate that proinsulin synthesis is not limiting in T2D, but increased (Alarcon et al., 2016), suggesting that alterations in insulin granule formation and/or storage are contributing factors to the pathogenesis of T2D. Potentially, defects within the secretory pathway may directly impact granule formation and thereby contribute to β-cell secretory dysfunction in T2D.
Our understanding of granule protein sorting, either in the Golgi (sorting by entry) or in the immature granule (sorting by retention), in both healthy and disease contexts is limited, in part, because the regulatory mechanisms are not well defined (Dikeakos and Reudelhuber, 2007; Arvan and Castle, 1998; Chung et al., 1989). Aside from (pro)insulin, the most abundant proteins in the granule lumen are granin proteins, including chromogranin A (CgA, also known as CHGA), chromogranin B (CgB, also known as CHGB) and VGF (non-acronymic, unrelated to VEGF) (Helle, 2000; Bartolomucci et al., 2011; Suckale and Solimena, 2010). Granin proteins have been proposed to stimulate granulogenesis by clustering at trans-Golgi network (TGN) subdomains through direct and indirect associations with cholesterol-rich lipid rafts (Hosaka et al., 2004). Additionally, granins may chaperone the sorting and/or deposition of secretory cargo, such as prohormones, within the budding granule (Natori and Huttner, 1996), though direct associations with prohormones have yet to be documented. Reduced expression of some granins, such as CgB, has been reported in human T2D islets (Bugliani et al., 2013); however, conflicting knockout studies have hampered clear delineation of its role(s) in granule biology (Díaz-Vera et al., 2012; Hendy et al., 2006; Mahapatra et al., 2005; Dominguez et al., 2018). Global deletion of CgB reduces islet hormone secretion (glucagon, insulin and somatostatin) (Obermüller et al., 2010); however, ultrastructrural analysis failed to reveal defects in granule structure and number, suggesting CgB is not required for dense core granule formation. Whether CgB functions in related processes of granule cargo selection and trafficking remains to be determined, but this possibility is supported by decreased granule content in CgB knockout (KO) chromaffin cells (Díaz-Vera et al., 2010).
We have recently shown that islet β-cell-specific KO of VGF results in defective insulin release due to a lag in granule replenishment (Stephens et al., 2017). Loss of VGF is accompanied by an accumulation of granule proteins in the TGN, including proinsulin, CgA and CgB. These data posit VGF as a potential regulator of early-stage granule protein trafficking in the islet β-cell. Studies in PC12 pheochromocytoma and adrenal chromaffin cells demonstrate that VGF and CgB directly interact, whereas no interaction has been reported for either VGF or CgB with CgA (Fargali et al., 2014). Given the similarities in the VGF and CgB loss-of-function phenotypes and their direct interaction in PC12 and chromaffin cells, we reasoned that CgB may also regulate granule protein trafficking from the TGN in islet β-cells in a similar manner to VGF.
In the current study, we demonstrate that β-cell deficiency of CgB results in loss of insulin secretory function due to a trafficking delay of proinsulin/insulin transit from the TGN to the plasma membrane. In the absence of CgB, impaired glucose-stimulated insulin secretion is accompanied by reductions in proinsulin processing, altered granule density and decreased numbers of plasma membrane-bound insulin-containing secretory granules. Using a fluorescent pulse-chase labeling strategy to track nascent proinsulin in situ, we demonstrate a striking delay in the exit of newly synthesized proinsulin from the TGN that coincides with a profound reduction in the appearance of nascent secretory granules at the plasma membrane. We propose that CgB acts as a central mediator in the early stages of granule protein trafficking from the TGN to the plasma membrane, which is necessary to maintain full secretory capacity of the pancreatic islet β-cell.
RESULTS
Chromogranin B suppression impairs glucose-stimulated insulin secretion
Our previous work demonstrated that the granin protein VGF regulates granule protein trafficking in the islet β-cell (Stephens et al., 2017). In adrenal medullary chromaffin cells, VGF has been shown to directly interact with another granin protein, CgB, but not with CgA (Fargali et al., 2014). Here, we examined whether these interactions were conserved in pancreatic β-cells. As shown in Fig. 1A, VGF co-immunoprecipitated with CgB using antisera directed against either CgB or VGF in robustly glucose-responsive 832/3 insulinoma cells. In contrast, we could not detect VGF following CgA immunoprecipitation, nor could we detect CgA following VGF immunoprecipitation. These studies suggest that VGF and CgB may have related roles in regulating islet β-cell function; therefore, we further investigated the impact of CgB loss on islet β-cell function.
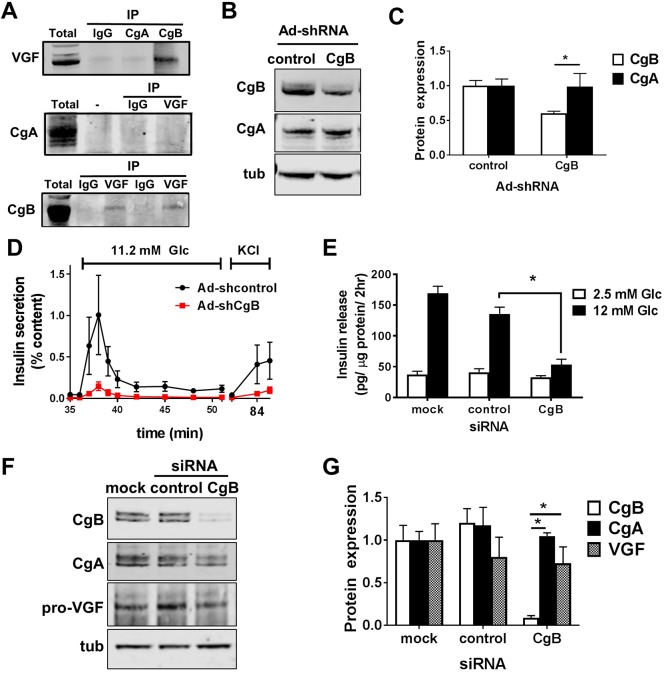
Fig. 1.
Loss of CgB impairs β-cell function. (A) Whole-cell lysates from 832/3 cells were immunoprecipitated with CgA, CgB, VGF (2 different antibodies), isotype control IgG antibodies or beads alone (−) and probed with VGF, CgA, CgB antibodies as indicated. (B–D) Islets isolated from C57Bl/6NJ mice (n=4–6 per group; 10–14 weeks of age) were treated with recombinant adenoviruses expressing either a control non-targeting (control) shRNA or CgB shRNA and assayed 96 h post-infection. Representative immunoblots from islet cell lysates (B) and quantification of relative protein expression (C). (D) Glucose-stimulated insulin secretion was determined by perifusion in media containing 2.5 mM Glc, 11.2 mM Glc or 35 mM KCl, sequentially, as indicated. (E–G) 832/3 insulinoma cells were transfected with pooled siRNA duplexes targeting CgB, a non-targeting control, or mock (no siRNA). (E) Glucose-stimulated insulin secretion was measured by static incubation in media containing 2.5 mM Glc followed by 12 mM Glc for 1 h each (n=3 independent experiments). Representative immunoblots are shown (F) and relative protein expression was quantified (G) from 832/3 whole cell lysates. (C,D,E,G) Data represent the mean±s.e.m. *P<0.05 by two-tailed, unpaired, t-test (C), two-way ANOVA with a Tukey post-test compared to control shRNA or siRNA treatments at similar glucose concentrations (D,E,G).
Prior studies from whole-body CgB KO mice demonstrated reduced glucose-stimulated insulin secretion (GSIS) and increased proinsulin content (Obermüller et al., 2010), similar to β-cell loss of VGF (Stephens et al., 2017); however, it is unclear whether the defects observed in the global CgB KO are driven by β-cell autonomous effects, developmental adaptions, or changes in central or peripheral cues from CgB loss in other endocrine tissues. To address this, we examined β-cell function in primary mouse islets treated with recombinant adenovirus expressing a CgB shRNA as compared to a non-targeting shRNA (control). Suppression of CgB (Fig. 1B,C) impaired both first and second phase glucose-stimulated insulin secretion as well as KCl-stimulated release (Fig. 1D) with no detectable effects on CgA expression (Fig. 1B,C). No change in the basal secretion response to 2.5 mM glucose (Glc) treatment was observed (data not shown). Similarly, siRNA-mediated suppression of CgB in 832/3 rat insulinoma cells also reduced glucose-stimulated insulin secretion as compared to non-targeting siRNA and mock-transfected controls (Fig. 1E) with no defects in basal insulin release detected. CgA and VGF expression were unchanged in response to reduced CgB expression (Fig. 1F,G). Furthermore, general constitutive secretion of proteins under non-stimulatory conditions (2.5 mM Glc) was not affected by CgB loss (Fig. S1A).
Loss of CgB impairs proinsulin processing
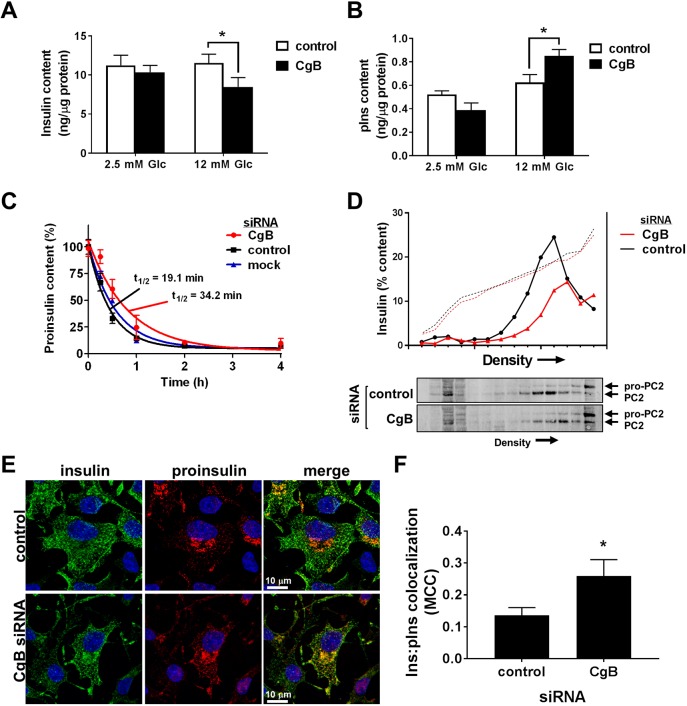
We next examined whether CgB loss was accompanied by changes in granule insulin content. As shown in Fig. 2A, we observed no change in insulin content in CgB knockdown cells at basal glucose as compared to control cells treated with non-targeting (control) siRNA duplexes. In contrast, 2 h glucose stimulation resulted in an ∼35% reduction in total insulin content in CgB-deficient cells (Fig. 2A) that was accompanied by a significant increase in proinsulin content (Fig. 2B), as observed in islets from CgB KO mice (Obermüller et al., 2010). Glucose stimulation is known to increase proinsulin biosynthesis (Guest et al., 1989), and thus the increase in proinsulin content observed in CgB knockdown cells may reflect a decrease in conversion to insulin. To address this, we examined proinsulin content following treatment with a protein synthesis inhibitor, cycloheximide, to halt additional proinsulin synthesis while allowing continued conversion of proinsulin to insulin (Haataja et al., 2013; Stephens et al., 2017). Using this method, we observed a striking increase in proinsulin half-life in CgB knockdown cells, suggesting a decrease in the proinsulin processing rate (Fig. 2C). Importantly, no change in proinsulin release was observed (data not shown). To further examine potential effects of CgB loss on secretory granules, we used density gradient sedimentation to resolve insulin-containing granule fractions and associated granule markers from other cellular organelles, such as ER, lysosomes and Golgi (Fig. S1B) (Stephens et al., 2017). As shown in Fig. 2D, we observed a significant depletion of the insulin granule fraction in CgB siRNA-treated cells at stimulatory glucose levels (12 mM Glc), as compared to control siRNA-treated cells, consistent with the decrease in total insulin content (Fig. 2A). Moreover, we also noted that insulin-containing fractions from CgB knockdown cells migrated to more dense regions of the gradient as compared to control cells (Fig. 2D). Immunoblot analysis of gradient fractions further revealed a sedimentation shift in the granule processing enzyme, prohormone convertase 2 (PC2, also known as PCSK2), similar to the observed shift of the insulin peak as measured by ELISA in CgB knockdown cells. Further interrogation of subcellular localization by confocal imaging revealed an increase in insulin staining (Fig. S2) associated with the TGN in CgB-deficient cells as compared to siRNA control-treated cells, with an increase in the overlap of proinsulin and insulin staining (Fig. 2E,F). The apparent discrepancy between sedimentation of insulin into denser fractions and the immunostaining in TGN-positive regions could be due to increased accumulation of proinsulin and other granule proteins within a Golgi subcompartment (Park et al., 2011) and/or additional changes in the granule membrane resulting in denser, rather than lighter, sedimentation. Taken together, these data demonstrate changes in proinsulin processing, granule protein composition and granule density in response to CgB loss in the islet β-cell.
Fig. 2.
Loss of CgB reduces proinsulin processing. 832/3 insulinoma cells were transfected with pooled siRNA duplexes targeting CgB, a non-targeting control siRNA, or mock (no siRNA) as indicated. (A,B) Glucose stimulation was performed by static incubation in media containing 2.5 mM Glc or 12 mM Glc for 2 h each. Insulin (A) and proinsulin (B) content were determined by ELISA from whole cell lysates and normalized to total cellular protein (n=3–4 independent experiments). (C) Cells were stimulated with Glc (12 mM) for 4 h while treated with the protein synthesis inhibitor cycloheximide (10 µM) at the indicated times to halt proinsulin synthesis. Proinsulin content was determined by ELISA from whole cell lysates. Nonlinear regression analysis (GraphPad) was used to determine curve fits and proinsulin half-life (n=3 independent experiments). (D) Cell lysates were resolved on 8–23% iodixanol gradients (representative traces shown of n=4). Solid line represents insulin as determined by ELISA from gradient fractions; dotted line represents gradient density (absorbance at 340 nm). Immunoblots of gradient fractions are shown. (E,F) Cells were immunostained for insulin (green), proinsulin (red) and counterstained with DAPI (blue). Representative images are shown (E) and colocalization determined (F) by Mander's correlation coefficient (MCC) from 37–38 cells from n=3 independent experiments. (A–C,F) Data represent the mean±s.e.m. *P<0.05 by two-way ANOVA with Tukey post-test analysis (A,B), or two-tailed, unpaired, t-test (F).
Pulse-chase labeling of nascent proinsulin/insulin-containing granules
Current methods for examining vesicle protein trafficking, such as fluorescent reporter fusion proteins and immunodetection, typically fail to temporally resolve granule proteins during granule maturation, owing, in part, to the prolonged half-life of proteins within the mature secretory granule, such as insulin (t½∼2.7 days), that are also highly abundant (Müller et al., 2017). To examine nascent granule protein trafficking from the Golgi, we utilized an in situ pulse-chase fluorescent labeling strategy based on the modified DNA repair enzyme, SNAP-tag, that self-labels by transfer of a synthetic (e.g. fluorescent) probe from a benzylguanidine-conjugated substrate (Ivanova et al., 2013). We inserted SNAP-tag within the C-peptide region of human preproinsulin, which should yield proCpepSNAP (proinsulin) and the mature processed fragments, insulin and CpepSNAP (C-peptide) (Fig. 3A) as performed previously with GFP (Haataja et al., 2013) and Gaussia lucerifase (Burns et al., 2015) to avoid potential protein folding and aggregation problems (Pouli et al., 1998). We used this construct to generate an insulinoma (832/3) cell line stably expressing proCpepSNAP and performed initial validation studies using an antibody directed against SNAP to evaluate (pro)CpepSNAP expression. Using density gradient sedimentation, we demonstrated that unprocessed proCpepSNAP, identified by both proinsulin and SNAP-directed antibodies, exclusively co-sedimented in the dense ER-containing fractions of an iodixanol gradient (Fig. 3C). In contrast, the proteolytically processed CpepSNAP fragment, identified by the SNAP antibody, but not the proinsulin-directed antibody, co-sedimented in the insulin-rich secretory granule (SG) fractions (Fig. 3C) as identified by insulin ELISA (data not shown), as well as denser fractions that may reflect accumulation in lysosomal vesicles. Furthermore, immunostaining of insulinoma cells showed strong colocalization of (pro)CpepSNAP with insulin throughout the cell body (Fig. S3A) with very few non-SNAP positive granules, indicating a high degree of incorporation of (pro)CpepSNAP within insulin granules. Finally, we used a cell-permeable, fluorescent-conjugated SNAP-tag substrate (TMR-SNAP) to verify that the labeling of (pro)CpepSNAP occurred in secretory granules in situ and confirmed co-localization of TMR-labeled (pro)CpepSNAP with CgB-positive (immunostained) granules (Fig. S3B).
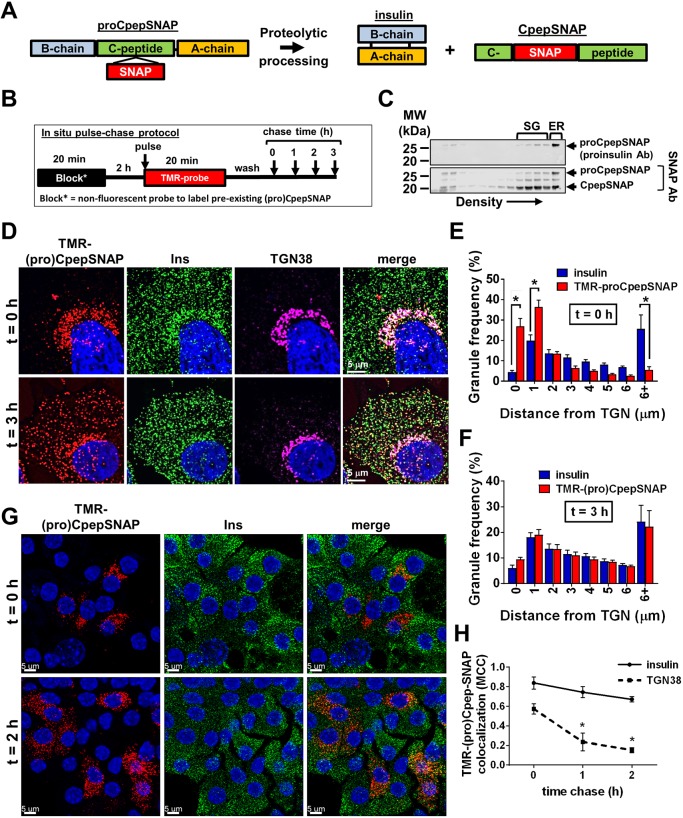
Fig. 3.
Expression, processing and trafficking of proCpepSNAP. (A) Schematic of the proCpepSNAP construct and processed peptides, insulin and CpepSNAP. (B) Schematic of fluorescent pulse-chase labeling of proCpepSNAP. (C–F,H) 832/3 insulinoma cells stably expressing proCpepSNAP were evaluated for protein expression. (C) Cell lysates were resolved on 8–23% iodixanol gradients. Immunoblots demonstrate proCpepSNAP detected by the proinsulin-specific antibody and CpepSNAP detected by the SNAP-specific antibody. (D–F,H) Cells were pulse-labeled with SNAP-TMR (red) for 20 min and chased for the indicated times before fixation. Labeled cells were immunostained for insulin (green), TGN38 (magenta), and counterstained with DAPI (blue). (D) Confocal images (maximum projection from five z-stacks) are shown. Gain and contrast are set independently for the t=0 h and t=3 h time points to account for signal intensity. Frequency distribution of binned granule distances from the TGN following t=0 h chase (E) and t=3 h chase (F). (H) Colocalization was quantified by Mander's correlation coefficient (MCC). (G) Mouse islets (C57Bl/6NJ; 10–14 weeks) treated with AdRIP-proCpepSNAP were dispersed into monolayers 72 h post-infection. Islet cells were pulse-labeled with SNAP-TMR (20 min) and chased for the indicated times prior to fixation. Islet cells were immunostained for insulin (green) and counterstained with DAPI (blue). Representative confocal images are shown (maximum projection from five z-stacks). (E,F,H) Data represent the mean±s.e.m. collected from 55–60 cells per time point (n=3 independent experiments). *P<0.05 by two-way ANOVA with Sidak post-test analysis (E,F) or one-way ANOVA with Bonferroni post-test (H).
SNAP-tag strategies have been previously used to examine the contribution of aged granules to insulin secretion (Ivanova et al., 2013; Müller et al., 2017). We postulated that the temporal control afforded by SNAP-tag pulse-chase labeling could enable the visualization of granule loading dynamics. Importantly, SNAP-tag labeling is a terminal event; once labeled, the reactive cysteine is not available for subsequent labeling. Taking advantage of this, proCpepSNAP-expressing 832/3 cells were pre-labeled with a non-fluorescent SNAP-tag probe to mask the existing pool of (pro)CpepSNAP (Fig. 3B). This was followed by a 2 h recovery time to allow de novo protein synthesis of proCpepSNAP so that our analysis could focus on the trafficking of newly synthesized proCpepSNAP. We then pulse-labeled (20 min) cells with a cell-permeable TMR-labeled SNAP-tag substrate and monitored protein-trafficking dynamics during the subsequent chase period. Using granule distance from the TGN as a measure of granule budding and trafficking, we demonstrated that initially ∼60% of TMR-labeled proCpepSNAP is within 1 µm of the TGN (t=0 h chase) and is distinct from the total insulin granule distribution present throughout the cell body (Fig. 3D,E). Additional analysis demonstrated the co-localization of CgB-positive, TMR-labeled granules adjacent to the TGN (Fig. S3C). Following a 3 h chase, TMR-labeled (pro)CpepSNAP precisely mirrored the insulin granule distribution (Fig. 3D,F). Correlation analysis confirmed a time-dependent loss of TMR-labeled (pro)CpepSNAP colocalization with TGN marker TGN38 (also known as TGOLN2), but not insulin, within a 2 h chase period (Fig. 3H). These data are consistent with the known processing kinetics of proinsulin to mature insulin (Rhodes and Halban, 1987; Zhu et al., 2002). We further confirmed that regulated secretion of TMR-CpepSNAP occurred in response to the secretagogues, glucose and KCl, in a similar manner to insulin (Fig. S3D). These data demonstrate that the trafficking and secretion of proCpepSNAP is consistent with the regulation of endogenous proinsulin/insulin.
To determine whether this pulse-chase labeling system is also amenable to the study of insulin granule trafficking in primary β-cells, we generated a recombinant adenovirus expressing proCpepSNAP under control of the rat insulin promoter (RIP). Isolated mouse islets were treated with AdRIP-proCpepSNAP and dispersed into monolayers prior to pulse-chase labeling to facilitate high-resolution imaging. Using the labeling scheme described for insulinoma cells (Fig. 3B), initial labeling in primary β-cells (t=0 h chase) occurred in the perinuclear region, consistent with Golgi localization (Fig. 3G). Following a 2 h chase, TMR-labeled granules are evenly distributed throughout the cell body, consistent with trafficking into the insulin granule storage pool. These data demonstrate the utility of this system to study insulin granule trafficking in primary β-cells.
CgB is necessary for efficient budding of nascent secretory granules from the TGN
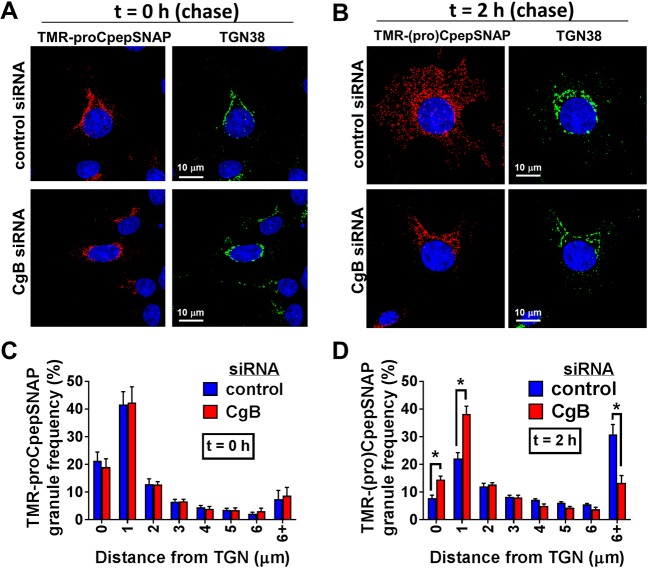
Using the SNAP-tag pulse-chase labeling strategy, we investigated the impact of CgB loss on proinsulin/insulin trafficking in 832/3 cells stably expressing proCpepSNAP. As shown in Fig. 4A, initial TMR labeling (t=0 h) of proCpepSNAP occurred in a perinuclear region of the cell, colocalizing with the TGN marker, TGN38, in both CgB siRNA- and control siRNA-treated cells, with ∼60% of the TMR-labeled proCpepSNAP within 1 µm of the TGN (Fig. 4C). CgB-negative cells were identified by immunostaining (data not shown). Following a 2 h chase, TMR-labeled (pro)CpepSNAP was well distributed throughout the cell body in control siRNA-treated cells, with a large fraction greater than 6 µm from the TGN (Fig. 4B,D), consistent with maturation and trafficking of nascent granules into the secretory granule pool. In contrast, loss of CgB resulted in the retention of nascent TMR-labeled (pro)CpepSNAP in close proximity (within 2 µm) to the TGN following the 2 h chase.
Fig. 4.
Loss of CgB delays early-stage granule trafficking from the TGN. 832/3 cells stably expressing proCpepSNAP were transfected with a pool of siRNA duplexes targeting CgB or a non-targeting control siRNA. Cells were pulse-labeled with SNAP-TMR (red) for 20 min and chased for the indicated times. (A,B) Cells were immunostained for TGN38 (green), and counterstained with DAPI (blue). Confocal images (maximum projection from five z-stacks) are shown from t=0 h chase (A) and t=2 h chase (B). Comparative images of the same time point were imaged and contrasted identically. (C,D) Frequency distribution of binned granule distances from the TGN following t=0 h chase (C) and t=2 h chase (D) are shown. Data represent the mean±s.e.m. of 21–37 imaged cells per condition from n=3 independent experiments. *P<0.05 by two-way ANOVA with Sidak post-test analysis.
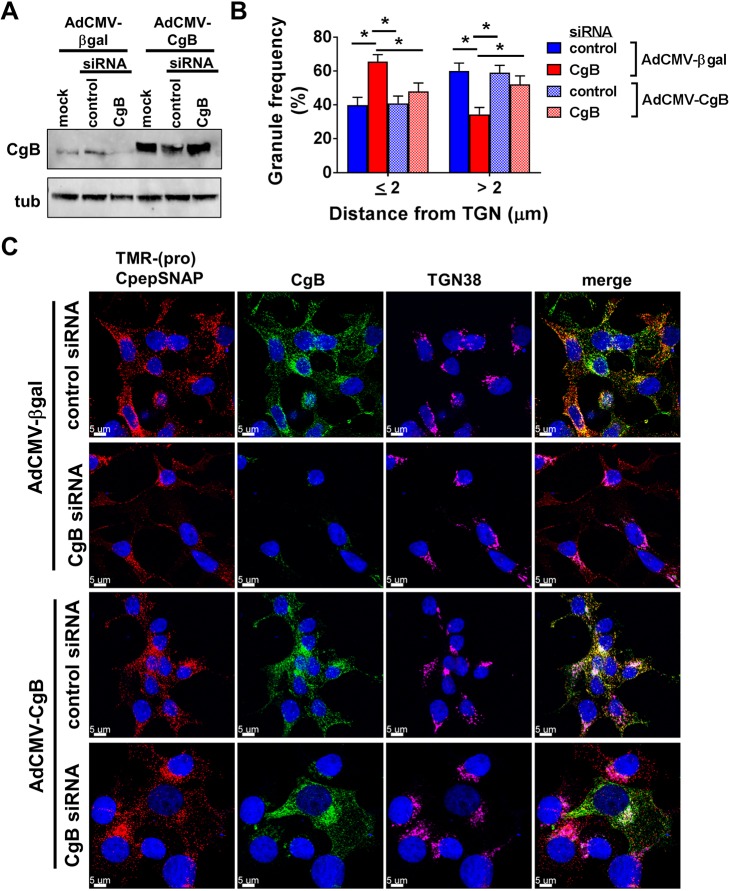
To determine whether the defects in granule trafficking were specific to CgB loss, we examined the ability of re-expression of CgB to rescue granule trafficking defects in CgB knockdown cells. Insulinoma 832/3 cells stably expressing proCpepSNAP were transfected with CgB or control siRNAs, and subsequently transduced with recombinant adenovirus expressing human CgB lacking siRNA targeting sequences or expressing a control transgene expressing β-galactosidase. CgB overexpression was evident by immunoblot (Fig. 5A) and CgB immunostaining was used to delineate CgB-expressing vs CgB-knockdown cells for microcopy (Fig. 5C). Using the pulse-chase strategy (Fig. 3B), we focused our attention on the 2 h chase period where we observed the strong (pro)CpepSNAP distribution phenotype (Fig. 4B,D). Consistent with our previous data, CgB suppression resulted in an increase in TMR-labeled granule association near the TGN (<2 µm) as compared to control siRNA-transfected cells co-treated with control virus, AdCMV-βgal (Fig. 5B,C). While no difference was observed in cells treated with AdCMV-CgB and control siRNA, re-expression of CgB (AdCMV-CgB) in CgB siRNA knockdown cells restored the granule distribution to control siRNA-treated levels. Notably, neighboring CgB-positive vs CgB-negative cells were distinct in their cellular distribution of TMR-labeled granules (Fig. 5C, lower panel, CgB siRNA, AdCMV-CgB treatment). CgB-negative cells had primarily Golgi-localized granules whereas CgB-positive cells displayed an even distribution of TMR-labeled granules in the cell body.
Fig. 5.
Re-expression of CgB rescues CgB siRNA-mediated defects in proCpepSNAP Golgi exit. 832/3 cells stably expressing proCpepSNAP were transfected with CgB siRNA or a non-targeting control siRNA duplex and treated with recombinant adenoviruses expressing human CgB (AdCMV-CgB) or β-galactosidase (AdCMV-βgal). (A) Representative immunoblot of CgB expression. (B,C) Cells were pulse-labeled with SNAP-TMR (red) for 20 min and chased for 2 h. Cells were immunostained for CgB (green) to confirm knockdown and overexpression, TGN38 (magenta), counterstained with DAPI (blue), and imaged by confocal microscopy. (B) Frequency distributions of binned granule distances from the TGN were determined. (C) Representative images are shown (maximum projection from five z-stacks). (B) Data represent the mean±s.e.m. of 25–31 imaged cells per condition from n=3 independent experiments. *P<0.05 by two-way ANOVA with Sidak post-test analysis.
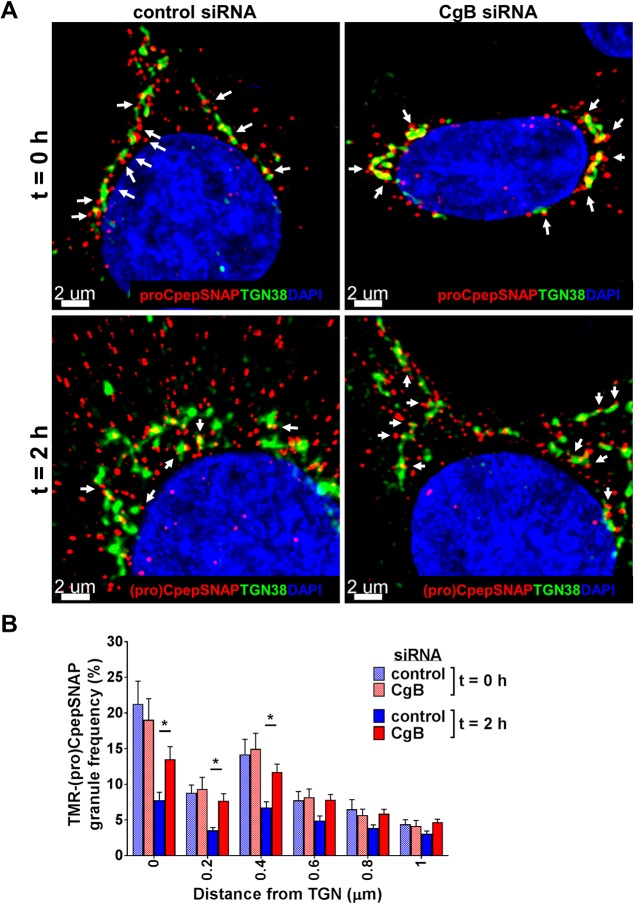
We next performed a more detailed examination of our pulse-chase labeling data (Fig. 4A–D) restricted to TMR-labeled granules at the TGN to delineate possible granule budding defects. As shown in Fig. 6A, numerous TMR-labeled granules are evident along terminal tips (white arrows) of the TGN at t=0 h of the chase period in both control and CgB siRNA-treated cells, which likely reflect budding sites for nascent granules. Notably, in control cells, these budding granules are highly enriched for CgB (Fig. S3C). Image analysis examining only TMR-labeled granules ≤1 µm from the TGN showed no differences in nascent granule frequency associated with the TGN in control and CgB siRNA-treated cells at the beginning of the chase period (Fig. 6B, t=0 h). Following the 2 h chase, TGN-associated granules are greatly reduced in control cells (Fig. 6A,B; t=2 h), consistent with efficient budding and trafficking into the cellular granule pool. In CgB knockdown cells, numerous TMR-labeled granules remain evident along TGN borders after the 2 h chase (Fig. 6A, t=2 h) with significant retention of nascent granules within 0.4 µm of the TGN (Fig. 6B, t=2 h) as compared to control cells. In follow-up studies with longer chase duration (t=18 h), TMR-labeled granules in CgB siRNA-treated cells were well-distributed throughout the cell body with no discernible differences from control cells (Fig. S4C). The co-localization of TMR-labeled CpepSNAP and insulin vs TGN38 was comparable to control cells (Fig. S4A), as was the cellular granule distribution relative to the TGN (Fig. S4B). Thus, it is unlikely that nascent granules are budding and being rapidly degraded in CgB knockdown cells. Taken together, these data suggest that CgB regulates efficient budding of proinsulin-containing secretory granules from the TGN in β-cells, but additional, redundant, albeit slower, pathways allow for granule budding and maturation to occur.
Fig. 6.
CgB is required for efficient budding of nascent granules from the TGN. 832/3 cells stably expressing proCpepSNAP were transfected with a pool of siRNA duplexes targeting CgB or a non-targeting control. Cells were pulse-labeled with SNAP-TMR (red) for 20 min and chased for the indicated times. Cells were immunostained for TGN38 (green), and counterstained with DAPI (blue). (A) Confocal images (3D view, Imaris) are shown from the indicated chase periods (t=0 h or t=2 h). White arrows denote TMR-labeled granules with close (<0.4 µm) apposition to the TGN. (B) Frequency distribution of binned granules ≤1 µm from the TGN are shown from the indicated chase periods (t=0 h or t=2 h). Data represent the mean±s.e.m. of 21–37 imaged cells per condition from n=3 independent experiments. *P<0.05 by two-way ANOVA with Sidak post-test analysis. (Note: this is the same data set as used in Fig. 4.)
Islet β-cell loss of CgB impairs nascent granule trafficking
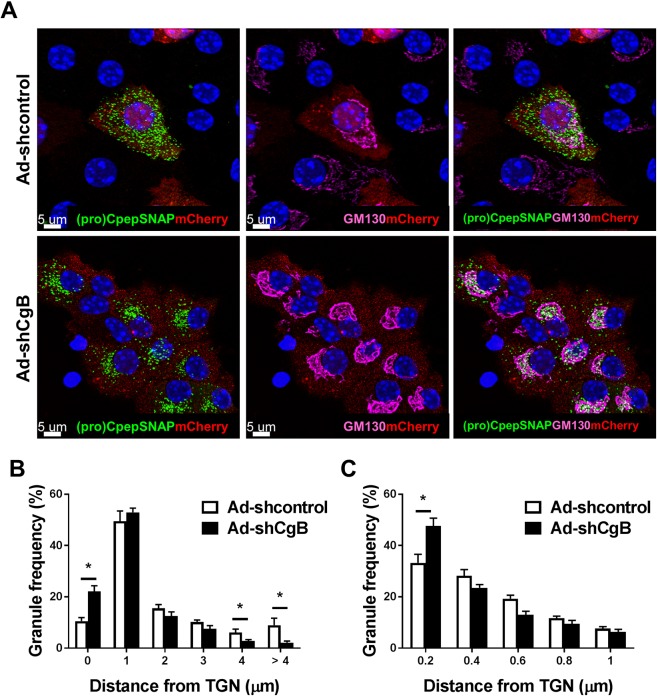
To determine whether the impairment in insulin granule trafficking could also be observed in primary β-cells, we utilized our pulse-chase labeling strategy in isolated mouse islets. In these studies, islets were treated with recombinant adenovirus expressing either a shRNA targeting CgB or a control, non-targeting shRNA as shown in Fig. 1B–D. Notably, this viral backbone co-expresses mCherry from a separate RNA polymerase II promoter (PGK) allowing the specific identification of transduced (CgB or control shRNA) islet cells. Islets were co-treated with recombinant adenovirus expressing proCpepSNAP under control of the rat insulin promoter (RIP) to ensure β-cell specific transgene expression, such that mCherry-positive proCpepSNAP-positive cells correspond to CgB knockdown (or transduced control) β-cells. Due to spectral overlap of the mCherry reporter and the TMR-labeled SNAP substrate, we used a green fluorescent SNAP substrate, 505-Star. Here, we focused on the primary observation at the 2 h chase time observed in insulinoma cells. As shown in Fig. 7A, nascent (505-Star-labeled) granules in control islet β-cells were noticeably punctate, and evenly distributed throughout the cell body, consistent with the trafficking and maturation of insulin granules. In contrast, nascent (505-Star-labeled) granules in CgB shRNA (mCherry-positive) β-cells remained predominantly adjacent to the Golgi (labeled for marker GM130) and failed to distribute into the cell body during the 2 h chase period, consistent with our data in insulinoma cells (Figs 4 and 6). By examining granule distributions, we demonstrate a clear increase in granules within 1 µm to the Golgi in CgB knockdown β-cells and a corresponding decrease in labeled granules distal (≥4 µm) to the Golgi (Fig. 7B). Further analysis of the subset of granules in close proximity to the Golgi (≥1 µm) revealed a strong increase in labeled granules in CgB shRNA-treated β-cells that were 0.2 µm or less from the Golgi (Fig. 7C), consistent with a delay in Golgi budding. These data suggest that impairments in proinsulin/insulin trafficking kinetics occur in CgB-deficient primary β-cells.
Fig. 7.
Loss of CgB impairs granule protein trafficking in islet β-cells. Mouse islets (C57Bl/6NJ; 10–14 weeks) were treated with recombinant adenoviruses (AdU6-shRNA-PGK-mCherry) targeting CgB or non-targeting (control) and co-treated with AdRIP-proCpepSNAP. 72 h post-infection, islets were dispersed onto coverslips and cultured overnight. Islet cells were pulse-labeled with SNAP-505-Star (20 min) and chased for 2 h prior to fixation. (A) Islet cells were imaged by confocal microscopy and representative images of 505-Star-labeled (pro)CpepSNAP (green), mCherry (red), Golgi (GM130, magenta) and DAPI (blue) are shown (3D projection from five z-stacks, Imaris). (B,C) Frequency distribution of binned granule distances (µm) from the TGN are shown. Data represent the mean±s.e.m. of 22–25 imaged cells per condition from n=3 mice per condition. *P<0.05 by two-way ANOVA with Sidak post-test analysis.
Delayed appearance of nascent granules at the plasma membrane in CgB-deficient cells
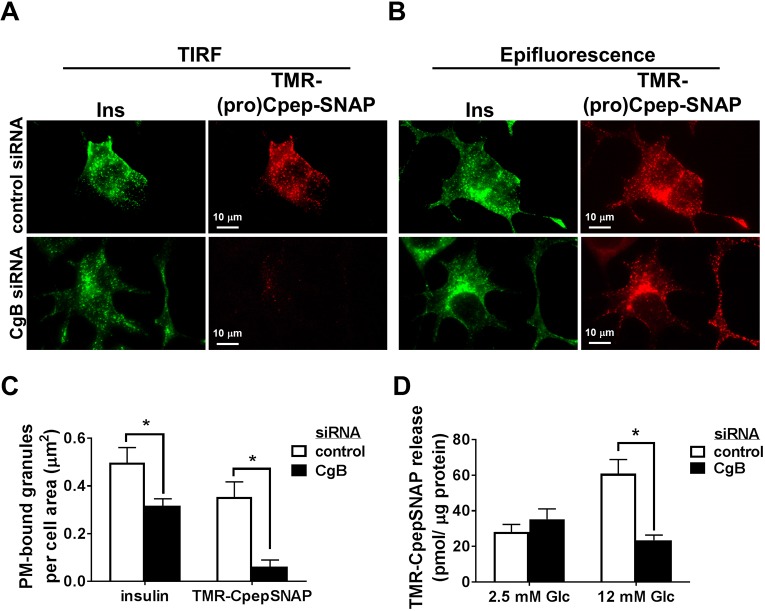
A strong preference exists for secretion of newly formed, rather than aged, insulin granules (Straub et al., 2004; Rhodes and Halban, 1987; Rorsman and Renstrom, 2003; Hao et al., 2005). We hypothesized that the decrease in β-cell function observed in CgB deficiency resulted from the reduced appearance of newly-synthesized insulin-containing granules at the plasma membrane available for exocytosis. To address this, we investigated the kinetics of nascent granule protein appearance at the plasma membrane using both total internal reflection fluorescent (TIRF) and epifluorescence microscopy, which allows the direct identification of granules adjacent (i.e. within 100–150 nm) to the plasma membrane (Fig. 8A, TIRF) that are distinct from the total granule pool present throughout the cell body (Fig. 8B, epifluorescence). Following a 4 h chase, fluorescent pulse-labeled (pro)CpepSNAP was readily evident at the plasma membrane in control cells (Fig. 8A), corresponding to ∼70% of the total insulin staining [compare TMR-(pro)CpepSNAP to insulin (Ins)]. In CgB knockdown cells, we observed a ∼40% decrease in total insulin-containing granules at the plasma membrane per cell area (Fig. 8A,C), consistent with the decrease in total insulin content (Fig. 2A,C). Although TMR-labeled-(pro)CpepSNAP granules were readily detected in CgB knockdown cells (Fig. 8B, epifluorescence), we observed a dramatic reduction in the appearance of TMR-labeled granules at the plasma membrane as observed by TIRF microscopy (Fig. 8A,C). Consistent with this, glucose stimulation revealed a striking absence of TMR-labeled CpepSNAP release in CgB knockdown cells as compared to siRNA control cells (Fig. 8D). Taken together, these data demonstrate a significant delay in the arrival of newly synthesized insulin-containing granules at the plasma membrane in CgB-deficient cells that corresponds to the loss of stimulated secretion.
Fig. 8.
CgB deficiency impairs trafficking of nascent granules to the plasma membrane. 832/3 cells stably expressing proCpepSNAP were transfected with CgB siRNA or a non-targeting control siRNA duplex. Cells were pulse-labeled with SNAP-TMR (red) for 20 min and chased for 4 h. (A–C) Cells were immunostained for insulin (green). Images captured in TIRF mode at a penetration depth of 150 nm (A) and epifluorescent mode (B) are shown. (C) The total proportion of insulin-positive and TMR-positive granules identified from TIRF microscopy images relative to the total cell area is shown (150–167 cells per condition). (D) Glucose-stimulated TMR-CpepSNAP secretion was determined by static incubation in media containing 2.5 mM Glc or 12 mM Glc for 1 h. (C,D) Data represent the mean±s.e.m. from n=3 independent experiments. *P<0.05 by two-way ANOVA with Sidak post-test analysis.
DISCUSSION
In this report, we show that loss of CgB in the pancreatic islet β-cell has significant effects on glucose-stimulated insulin secretion directly related to its role in granule protein trafficking. Previous studies have demonstrated that loss of CgB negatively impacts hormone release in some endocrine tissues, such as pancreatic islets (Obermüller et al., 2010) and adrenal chromaffin cells (Díaz-Vera et al., 2012; Dominguez et al., 2018), but is dispensable in other cell types including hippocampal neurons (Dominguez et al., 2018). This may reflect specific dependencies for granins in the regulation of some cargo types such as proinsulin that poorly oligomerize in the TGN (Quinn et al., 1991; Kuliawat and Arvan, 1994), whereas other vesicle cargo, such as neuropeptide Y (NPY), may have inherent structural features that negate the need for granins in cargo storage and release. Prior studies from CgB KO mice revealed increased numbers of immature secretory granules in pancreatic β-cells, increased proinsulin content, and reduced glucose-stimulated insulin release (Obermüller et al., 2010). Consistent with this, we show that the rise in proinsulin in CgB-depleted β-cells following sustained glucose stimulation is likely due to a decreased rate of proinsulin processing. Although no changes in ultrastructural features of mature dense core granules were previously observed in CgB KO mice (Obermüller et al., 2010), our studies reveal changes in granule density and alterations in insulin and proinsulin immunostaining, suggesting that hormone packaging may be negatively impacted in the absence of CgB.
Using a fluorescent probe-based pulse-chase procedure to label nascent proinsulin (proCpepSNAP), we suggest that loss of CgB impairs TGN budding of secretory granules and subsequently delays their appearance into the cellular pool of mature insulin-containing granules. Furthermore, we observed a striking delay in the appearance of nascent granules at the plasma membrane in CgB-deficient cells that were available for secretion. Importantly, we demonstrate that proCpepSNAP is processed, trafficked and released in a manner consistent with the kinetics of native insulin and C-peptide (Zhu et al., 2002; Rhodes and Halban, 1987). Several studies have demonstrated that newly-formed insulin is preferentially secreted from β-cells in both healthy (Straub et al., 2004; Rhodes and Halban, 1987; Rorsman and Renstrom, 2003; Hao et al., 2005) and T2D islets (Gandasi et al., 2018). This may reflect the directed movement of nascent granules from microtubule organizing centers on the Golgi membrane toward delivery sites at the plasma membrane (Efimov et al., 2007; Hoboth et al., 2015). Thus, impediments or inefficiencies early in the secretory pathway, such as those arising from loss of CgB function as shown here, can lead to dramatic reductions in β-cell secretory output by reducing the availability of the preferred, nascent granule pool for directed exocytosis. Nevertheless, total granule number may be minimally impacted owing to the balance of reduced secretion with reduced secretory granule budding.
Granule lumenal proteins arrive at the cisternal Golgi face via anterograde COP II-coated transport vesicles. As these proteins mature in the Golgi stacks, they are thought to coalesce at terminal sites of vesicle budding, partially segregated from constitutively (non-regulated) secreted proteins by their aggregation in the Golgi lumen (Chanat and Huttner, 1991; Jain et al., 2002); however, it is unclear whether additional inclusion and/or exclusion criteria in the TGN further promote protein selectivity in the budding granule. Our data, demonstrating delayed vesicle budding in CgB-deficient β-cells, would suggest that certain events or criteria need to be satisfied for efficient budding of nascent secretory granules from the TGN. In adrenal chromaffin cells, CgA and CgB, as well as other granule proteins such as tissue-type plasminogen activator, can regulate exocytic fusion pore lifetimes (Abbineni et al., 2018). Though our studies cannot rule out this effect on exocytosis by CgB loss in β-cells, it is interesting to speculate that in the budding granule, a structural role for granins may also serve to maintain access for partially aggregated TGN lumenal cargo. Thus, in the absence of CgB, slow delivery of key cargo may delay necessary signals to stimulate granule budding. In support of this model, amperometry measurements from CgB KO chromaffin cells show up to 30% decrease in granule lumen content (Díaz-Vera et al., 2010), consistent with abnormalities in granule cargo loading.
Previously, our lab demonstrated that another granin, VGF, also regulates early-stage secretory function in the β-cell (Stephens et al., 2017). Given the direct interaction of VGF and CgB observed in β-cells (this report) and other endocrine cell models (Fargali et al., 2014), it is possible that VGF and CgB are involved in regulating hierarchical assemblies of the granule proteome necessary for the efficient delivery and trafficking of proinsulin and requisite maturation factors (i.e. prohormone convertases) into the budding granule. Whether CgA also participates in this complex seems unlikely given the lack of detectable association; however, future studies are needed to further delineate these roles. Notably, granins are expressed throughout all endocrine tissues. Thus, it is unlikely that granins bind specific cargo such as proinsulin, given the diversity of endocrine hormones. Alternatively, granins may serve to define the relevant cellular contexts for proper loading and assembly that are necessary to signal the timely budding of the nascent granule from the TGN. In addition, it is important to note that functional redundancies likely exist between granins, which allows the endocrine cell secretory system to continue to operate in the absence of a specific granin, albeit at reduced efficiency.
In T2D, the impairment in nutrient-regulated insulin release from the islet β-cell is associated with distinct changes in secretory function including a reduction in the number of mature, dense core secretory granules (Masini et al., 2012; Like and Chick, 1970; Alarcon et al., 2016) and deficiencies in proinsulin processing (Kahn et al., 2009; Kahn and Halban, 1997) despite elevated proinsulin biosynthesis (Alarcon et al., 2016). A culmination of factors, including genetic predisposition and environmental stresses (ER and oxidative stress, inflammation) likely contribute to β-cell decline in T2D, yet how these events directly impact secretory capacity is less clear (Halban et al., 2014). Importantly, the disconnect between proinsulin biosynthesis and insulin storage in dense core granules may reflect alterations or defects in terminal stages of the secretory pathway, such as granule formation and protein trafficking, that directly contribute to the overall decline of islet β-cell function in T2D. The data described in this current study on CgB, as well as our past work on VGF (Stephens et al., 2017), show that an impediment in trafficking can significantly impair insulin secretion and glucose homeostasis, though their role(s) in the pathology of T2D remains to be determined. Although it is not known what specific changes in the granule content occur in the context of T2D, alterations, particularly in regulatory proteins, could directly impact the trafficking and fate of other granule proteins and the overall ability of β-cells to maintain mature insulin stores for nutrient-dependent release. We should note that the distinct effects of VGF and CgB loss on granule density suggest that separate effects may drive their respective phenotypes. Thus, understanding the molecular mechanisms regulating secretory granule formation in the islet β-cell remains a critical area toward understanding the pathology of islet dysfunction in diabetes.
MATERIALS AND METHODS
Cell culture, islet isolation and reagents
Rat insulinoma 832/3 cells (a kind gift from Dr Christopher Newgard, Duke University, Durham, NC, USA) were cultured as previously described (Hohmeier et al., 2000). SMARTpool (Dharmacon) siRNAs targeting rat CgB were transfected using Dharmafect I (Dharmacon) and cells assayed 72 h post-transfection. Cells were compared against those transfected with a non-targeted siRNA control duplex or mock-transfected (no duplex). Cell culture reagents were from Thermo Life Technologies unless specified otherwise. Chemical reagents were from Sigma-Aldrich unless specified otherwise. Mouse islets were isolated via collagenase V (Sigma-Aldrich) digestion and purified using Histopaque 1077 and 1119 (Sigma-Aldrich). Islets were cultured in RPMI supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin and maintained at 37°C in 5% CO2. Pools of islets were transduced with ∼2×107 infectious units/ml adenovirus (MOI ∼100–200) for 18 h and assayed 72–96 h post-treatment. All animal protocols were approved by the Institutional Animal Use and Care Committee at the University of Iowa.
Plasmids, viruses and stable cell line
SNAP-tag was inserted near the ApaI site of human preproinsulin via gBlock synthesis (IDT) and recombined into pDONR221 P4r-P3r via BP Clonase II (Thermo Life). Rat insulin promoter (RIP)-driven proCpepSNAP-hGH polyA was assembled into a modified pAd-PL/DEST via multi-site Gateway cloning using LR Clonase II plus (Haldeman et al., 2018). Plasmid containing a U6 promoter-driven mouse-specific CgB shRNA targeting sequence or a non-targeting control (siSAFE) and a PGK promoter-expressing mCherry reporter in an adenoviral backbone was obtained from Vector Builder. Human CgB cDNA (DNASU) was PCR subcloned into pDONR221 using BP Clonase II (Thermo Life Technologies) and recombined into pAd-PL/DEST/CMV via LR Clonase recombination. Recombinant adenoviruses were generated in HEK293 cells and purified using a cesium chloride gradient. All recombinant viruses were determined to be E1A-deficient using a quantitative PCR screen (Haldeman et al., 2018). EF1α-proCpepSNAP-hGH polyA was assembled into JN302 via multi-site Gateway cloning using LR Clonase II plus (Haldeman et al., 2018) and co-transfected with pCMV(CAT)T7-SB100 into low-passage 832/3 cells. pCMV(CAT)T7-SB100 was Addgene plasmid #34879 (Mátés et al., 2009), deposited by Zsuzsanna Izsvak. Polyclonal cells were selected with puromycin resistance (0.75 μg/ml) and expanded. All sequences were verified by the Iowa Institute of Human Genetics, University of Iowa.
Glucose-stimulated insulin secretion
Insulin secretion was measured by means of static incubation of 832/3 cells as previously described (Hohmeier et al., 2000), in secretion assay buffer (SAB) containing 2.5 mM glucose (Glc, basal) for 1 h at 37°C followed by incubation in SAB containing 12 mM Glc (stimulatory) for 1 h. Cells were lysed in RIPA buffer and total protein determined using BCA (Pierce). For perifusion studies, approximately 40 islets were perifused at 37°C using a BioRep Perifusion system with a flow rate of 100 µl/min. Perifusate was collected at 1–2 min intervals. Following stabilization of insulin release under basal (2.5 mM) glucose conditions (up to 32 min), islets were stimulated with 11.2 mM glucose for up to 32 min, returned to basal (2.5 mM) Glc for 16 min and stimulated with 35 mM KCl at basal Glc for 16 min. Islets were collected from perifusion chambers and lysed in RIPA buffer. Insulin (secreted and content) and proinsulin (content) were measured using the appropriate ELISA, rodent 80-INSMR-CH10 (ALPCO) and PINSRT-E01 (ALPCO), respectively. Proinsulin content was determined as the percentage of proinsulin relative to insulin+proinsulin. The insulin ELISAs have very little cross-reactivity to pro-insulin (<18%) and negligible reactivity with C-peptide; proinsulin ELISAs have no detectable cross-reactivity with insulin or C-peptide. Secreted TMR-CpepSNAP was detected from media using a SpectraMax i3 fluorescent plate reader (Molecular Dynamics) at 540/580 nm (excitation/emission).
Immunoblot and immunoprecipitation analysis
Clarified cell lysates were resolved on 4–12% NuPAGE gels (Thermo Life Technologies) and transferred to supported nitrocellulose membranes (Bio-Rad). Membranes were probed with diluted antibodies raised against chromogranin A (goat; Santa Cruz Biotechnology, C-20; 1:1000), chromogranin B (goat; Santa Cruz Biotechnology, C-19; 1:1000), cation-dependent M6PR (rabbit; Sigma-Aldrich, GW21444; 1:1000), GRP94 (rabbit; kind gift from Dr Christopher Nicchitta, Duke University, USA; 1:5000), PC2 (rabbit; Thermo Scientific, PA1-058; 1:1000), proinsulin (mouse monoclonal; Developmental Studies Hybridoma Bank, University of Iowa, USA, GS-9A8; 1:500), SNAP-tag (rabbit; GenScript, A00684-40; 1:1000), and VGF (rabbit antiserum; kind gift from Dr Stephen Salton, Icahn School of Medicine at Mount Sinai, USA; 1:1000). Donkey anti-mouse, anti-rabbit, or anti-goat antibodies coupled to IR-dye 680 or 800 (Licor, 926-68072, 926-32214, 926-32213; 1:10,000) were used to detect primary antibodies. Blots were developed using an Odyssey CLx Licor Instrument.
For immunoprecipitations, cells were lysed in 50 mM MES pH 5.5, 150 mM NaCl, 10 mM CaCl2, 0.1% NP-40, halt protease inhibitor (Thermo Life Technologies). Antisera or control IgG (Dako) and Protein A/G agarose resin (Thermo Life Technologies) was added to clarified lysates and rotated end over end overnight at 4°C. Collected resin was washed with lysis buffer and proteins eluted in LDS sample buffer (Thermo Life Technologies). Samples were prepared as described for immunoblot analysis.
In situ SNAP-tag labeling, immunofluorescence and microscopy
832/3 cells (parental or stably expressing proCpepSNAP) were plated on HTB9-coated coverslips 48 h post-siRNA transfections at low density and cultured overnight as previously described (Hayes et al., 2017; Stephens et al., 2017). Isolated islets were dispersed using Accutase (Sigma-Aldrich) and plated onto HTB9-coated coverslips. For SNAP-tag labeling, cells were initially incubated with SNAPcell block (10 µM; NEB) diluted in culture media for 20 min, washed three times for 10 min each, and cultured for an additional 2 h. For pulse-labeling, cells were cultured with SNAPcell-TMR (1 µM; NEB) or SNAPcell-505-Star (10 μM) for 20 min in media, washed three times for 10 min each in culture media with reduced glucose (5 mM) and chased as indicated. Following treatments, cells were fixed in 10% neutral-buffered formalin. For immunostaining, cells were incubated overnight with antibodies raised against insulin (guinea pig; Dako, Cat. no. A056401-2; 1:200; partial reactivity with proinsulin), chromogranin B (rabbit; Proteintech, 14968-1-AP; 1:200), GM130 (mouse monoclonal; BD Transduction, 610822; 1:200), proinsulin (mouse monoclonal; Developmental Studies Hybridoma Bank, University of Iowa, USA, GS-9A8; 1:50) and TGN38 (mouse monoclonal; Novus Biologicals, 2F7.1; 1:100) as indicated. Highly cross-adsorbed fluorescent dye-conjugated secondary antibodies donkey anti-guinea pig-Alexa Fluor 488 (Cat. no. 706-545-148) donkey anti-rabbit Rhodamine Red-X (Cat. no. 711-296-152) donkey anti-mouse Alexa Fluor 647 (Cat. no. 715-606-150) (all from Jackson ImmunoResearch) were used for detection. For confocal studies, cells were counterstained with DAPI (Sigma-Aldrich) and mounted using Fluorosave (Calbiochem). Images were captured on a Leica SP8 confocal microscope 40× (NA=1.4) oil objective with 5× zoom as z-stacks (∼5–7 per set, 0.25 μm step, 0.88 μm optical section) and deconvolved (Huygen's Professional). Granule distance measurements from the TGN were determined using a distance transformation module in Imaris (Bitplane) from spot-rendered granules (insulin immunostained or SNAP-TMR labeled) and surface rendering of the TGN identified through TGN38 immunostaining. Granule distances were binned (MS Excel) and expressed as a percentage of the total to normalize between cells. Co-localization was determined as the Mander's correlation coefficient using the JACoP plugin in Fiji/ImageJ from individual slices. For plasma membrane detection, immunostained (fixed) cells were mounted in aqueous media containing 0.1% n-propyl gallate and imaged using a Leica TIRF AM microscope via a 100× oil objective in epifluorescent and TIRF modes with a penetration depth of 110–150 nm. Granule numbers from SNAP labeling and insulin immunostaining, and cell area were determined using Fiji/ImageJ plugins.
Density gradient isolation of secretory granules
Cells were collected, washed in ice-cold PBS, and disrupted using 15 strokes in a pre-chilled ballbearing cell homogenizer (Isobiotec) with a 16 µm clearance in 10 mM MES pH 6.5, 1 mM MgSO4, 1 mM EDTA, 0.3 M sucrose. Post-nuclear supernatants were layered atop 8–23% linear iodixanol (Optiprep, Sigma-Aldrich) gradients and resolved at 110,000 g in an SW41 for 16–18 h. Fractions were manually collected by tube puncture. Iodixanol gradients were verified by measurement of absorbance at 340 nm. Insulin content was determined by ELISA (ALPCO) and normalized to the total insulin content from post-nuclear supernatant. For immunoblots, total protein was TCA-precipitated, resuspended in LDS sample buffer (Thermo Life Technologies) and prepared as described above.
Statistical analysis
Data are presented as the mean±s.e.m. For statistical significance determinations, data were analyzed by two-tailed unpaired, Student's t-test or by one- or two-way ANOVA with post-hoc analysis for multiple group comparisons as indicated (GraphPad Prism). Nonlinear regression analysis was used to compare curve fits for proinsulin turnover studies (GraphPad Prism).
Supplementary Material
Acknowledgements
The authors would like to acknowledge use of the University of Iowa Central Microscopy Research Facility, a core resource supported by the Vice President for Research and Economic Development, the Holden Comprehensive Cancer Center and the Carver College of Medicine. The acquisition of the Leica SP8 Laser Scanning Confocal microscope with STED capability was made possible by a generous grant from the Roy J. Carver Charitable Trust. Additional funding was provided by the University's Office of the Vice President for Research and Economic Development, the Carver College of Medicine, and the College of Liberal Arts and Sciences. Additional technical support was provided by Thomas Moninger and Jian Shao. We would like to thank Dr D. Thomas Rutkowski and Dr Andrew Norris for careful reading of this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.C.B., C.J.B., S.B.S.; Methodology: S.C.B., M.B., J.M.H., S.B.S.; Validation: S.C.B., C.J.B., M.B., J.M.H., S.S., S.B.S.; Formal analysis: S.C.B., C.J.B., S.S., S.B.S.; Investigation: S.C.B., C.J.B., S.B.S.; Resources: M.B.; Writing - original draft: S.B.S.; Writing - review & editing: S.B.S.; Supervision: S.B.S.; Project administration: S.B.S.; Funding acquisition: S.B.S.
Funding
This work was supported in part by a K01 award from the National Institute of Diabetes and Digestive and Kidney Diseases (DK099294) to S.B.S., pilot grant from the Office of the Vice Provost, University of Iowa to S.B.S., and startup funds provided by the Fraternal Order of Eagles Diabetes Research Center, University of Iowa to S.B.S. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.231373.supplemental
References
- Abbineni P. S., Bittner M. A., Axelrod D. and Holz R. W. (2018). Chromogranin A, the major lumenal protein in chromaffin granules, controls fusion pore expansion. J. Gen. Physiol. 116, 524A 10.1016/j.bpj.2018.11.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C., Boland B. B., Uchizono Y., Moore P. C., Peterson B., Rajan S., Rhodes O. S., Noske A. B., Haataja L., Arvan P. et al. (2016). Pancreatic beta-cell adaptive plasticity in obesity increases insulin production but adversely affects secretory function. Diabetes 65, 438-450. 10.2337/db15-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P. and Castle D. (1998). Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem. J. 332, 593-610. 10.1042/bj3320593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomucci A., Possenti R., Mahata S. K., Fischer-Colbrie R., Loh Y. P. and Salton S. R. J. (2011). The extended granin family: structure, function, and biomedical implications. Endocr. Rev. 32, 755-797. 10.1210/er.2010-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugliani M., Liechti R., Cheon H., Suleiman M., Marselli L., Kirkpatrick C., Filipponi F., Boggi U., Xenarios I., Syed F. et al. (2013). Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin-proteasome system in pancreatic beta cell dysfunction. Mol. Cell. Endocrinol. 367, 1-10. 10.1016/j.mce.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Burns S. M., Vetere A., Walpita D., Dančík V., Khodier C., Perez J., Clemons P. A., Wagner B. K. and Altshuler D. (2015). High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic beta-cell function. Cell Metab. 21, 126-137. 10.1016/j.cmet.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Chanat E. and Huttner W. B. (1991). Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 115, 1505-1519. 10.1083/jcb.115.6.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. N., Walter P., Aponte G. W. and Moore H. P. (1989). Molecular sorting in the secretory pathway. Science 243, 192-197. 10.1126/science.2911732 [DOI] [PubMed] [Google Scholar]
- Díaz-Vera J., Morales Y. G., Hernandez-Fernaud J. R., Camacho M., Montesinos M. S., Calegari F., Huttner W. B., Borges R. and Machado J. D. (2010). Chromogranin B gene ablation reduces the catecholamine cargo and decelerates exocytosis in chromaffin secretory vesicles. J. Neurosci. 30, 950-957. 10.1523/JNEUROSCI.2894-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Vera J., Camacho M., Machado J. D., Domínguez N., Montesinos M. S., Hernández-Fernaud J. R., Luján R. and Borges R. (2012). Chromogranins A and B are key proteins in amine accumulation, but the catecholamine secretory pathway is conserved without them. FASEB J. 26, 430-438. 10.1096/fj.11-181941 [DOI] [PubMed] [Google Scholar]
- Dikeakos J. D. and Reudelhuber T. L. (2007). Sending proteins to dense core secretory granules: still a lot to sort out. J. Cell Biol. 177, 191-196. 10.1083/jcb.200701024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez N., van Weering J. R. T., Borges R., Toonen R. F. G. and Verhage M. (2018). Dense-core vesicle biogenesis and exocytosis in neurons lacking chromogranins A and B. J. Neurochem. 144, 241-254. 10.1111/jnc.14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P. M., Andreyeva N., Gleeson P., Galjart N., Maia A. R. R., Mcleod I. X. et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12, 917-930. 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargali S., Garcia A. L., Sadahiro M., Jiang C., Janssen W. G., Lin W.-J., Cogliani V., Elste A., Mortillo S., Cero C. et al. (2014). The granin VGF promotes genesis of secretory vesicles, and regulates circulating catecholamine levels and blood pressure. FASEB J. 28, 2120-2133. 10.1096/fj.13-239509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandasi N. R., Yin P., Omar-Hmeadi M., Ottosson Laakso E., Vikman P. and Barg S. (2018). Glucose-dependent granule docking limits insulin secretion and is decreased in human type 2 diabetes. Cell Metab. 27, 470-478.e4. 10.1016/j.cmet.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Gerich J. E. (2002). Is reduced first-phase insulin release the earliest detectable abnormality in individuals destined to develop type 2 diabetes? Diabetes 51 Suppl. 1, S117-S121. 10.2337/diabetes.51.2007.S117 [DOI] [PubMed] [Google Scholar]
- Guest P. C., Rhodes C. J. and Hutton J. C. (1989). Regulation of the biosynthesis of insulin-secretory-granule proteins. Co-ordinate translational control is exerted on some, but not all, granule matrix constituents. Biochem. J. 257, 431-437. 10.1042/bj2570431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haataja L., Snapp E., Wright J., Liu M., Hardy A. B., Wheeler M. B., Markwardt M. L., Rizzo M. and Arvan P. (2013). Proinsulin intermolecular interactions during secretory trafficking in pancreatic beta cells. J. Biol. Chem. 288, 1896-1906. 10.1074/jbc.M112.420018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halban P. A., Polonsky K. S., Bowden D. W., Hawkins M. A., Ling C., Mather K. J., Powers A. C., Rhodes C. J., Sussel L. and Weir G. C. (2014). Beta-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care 37, 1751-1758. 10.2337/dc14-0396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldeman J. M., Conway A. E., Arlotto M. E., Slentz D. H., Muoio D. M., Becker T. C. and Newgard C. B. (2018). Creation of versatile cloning platforms for transgene expression and dCas9-based epigenome editing. Nucleic Acids Res. 47, e23 10.1093/nar/gky1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M., Li X., Rizzo M. A., Rocheleau J. V., Dawant B. M. and Piston D. W. (2005). Regulation of two insulin granule populations within the reserve pool by distinct calcium sources. J. Cell Sci. 118, 5873-5884. 10.1242/jcs.02684 [DOI] [PubMed] [Google Scholar]
- Hayes H. L., Peterson B. S., Haldeman J. M., Newgard C. B., Hohmeier H. E. and Stephens S. B. (2017). Delayed apoptosis allows islet beta-cells to implement an autophagic mechanism to promote cell survival. PLoS One 12, e0172567 10.1371/journal.pone.0172567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle K. B. (2000). The chromogranins. Historical perspectives. Adv. Exp. Med. Biol. 482, 3-20. 10.1007/0-306-46837-9_1 [DOI] [PubMed] [Google Scholar]
- Hendy G. N., Li T., Girard M., Feldstein R. C., Mulay S., Desjardins R., Day R., Karaplis A. C., Tremblay M. L. and Canaff L. (2006). Targeted ablation of the chromogranin a (Chga) gene: normal neuroendocrine dense-core secretory granules and increased expression of other granins. Mol. Endocrinol. 20, 1935-1947. 10.1210/me.2005-0398 [DOI] [PubMed] [Google Scholar]
- Hoboth P., Müller A., Ivanova A., Mziaut H., Dehghany J., Sönmez A., Lachnit M., Meyer-Hermann M., Kalaidzidis Y. and Solimena M. (2015). Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies. Proc. Natl. Acad. Sci. USA 112, E667-E676. 10.1073/pnas.1409542112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmeier H. E., Mulder H., Chen G., Henkel-Rieger R., Prentki M. and Newgard C. B. (2000). Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49, 424-430. 10.2337/diabetes.49.3.424 [DOI] [PubMed] [Google Scholar]
- Hosaka M., Suda M., Sakai Y., Izumi T., Watanabe T. and Takeuchi T. (2004). Secretogranin III binds to cholesterol in the secretory granule membrane as an adapter for chromogranin A. J. Biol. Chem. 279, 3627-3634. 10.1074/jbc.M310104200 [DOI] [PubMed] [Google Scholar]
- Ivanova A., Kalaidzidis Y., Dirkx R., Sarov M., Gerlach M., Schroth-Diez B., Muller A., Liu Y., Andree C., Mulligan B. et al. (2013). Age-dependent labeling and imaging of insulin secretory granules. Diabetes 62, 3687-3696. 10.2337/db12-1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Chang W. T., Geetha C., Joyce P. B. and Gorr S.-U. (2002). In vitro aggregation of the regulated secretory protein chromogranin A. Biochem. J. 368, 605-610. 10.1042/bj20021195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn S. E. and Halban P. A. (1997). Release of incompletely processed proinsulin is the cause of the disproportionate proinsulinemia of NIDDM. Diabetes 46, 1725-1732. 10.2337/diab.46.11.1725 [DOI] [PubMed] [Google Scholar]
- Kahn S. E., Zraika S., Utzschneider K. M. and Hull R. L. (2009). The beta cell lesion in type 2 diabetes: there has to be a primary functional abnormality. Diabetologia 52, 1003-1012. 10.1007/s00125-009-1321-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliawat R. and Arvan P. (1994). Distinct molecular mechanisms for protein sorting within immature secretory granules of pancreatic beta-cells. J. Cell Biol. 126, 77-86. 10.1083/jcb.126.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Like A. A. and Chick W. L. (1970). Studies in the diabetic mutant mouse: II. Electron microscopy of pancreatic islets. Diabetologia 6, 216-242. 10.1007/BF01212232 [DOI] [PubMed] [Google Scholar]
- Mahapatra N. R., O'connor D. T., Vaingankar S. M., Hikim A. P. S., Mahata M., Ray S., Staite E., Wu H., Gu Y., Dalton N. et al. (2005). Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J. Clin. Invest. 115, 1942-1952. 10.1172/JCI24354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini M., Marselli L., Bugliani M., Martino L., Masiello P., Marchetti P. and DE Tata V. (2012). Ultrastructural morphometric analysis of insulin secretory granules in human type 2 diabetes. Acta Diabetol. 49 Suppl. 1, S247-S252. 10.1007/s00592-012-0446-6 [DOI] [PubMed] [Google Scholar]
- Mátés L., Chuah M. K. L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D. P., Schmitt A., Becker K., Matrai J. et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753-761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- Müller A., Neukam M., Ivanova A., Sönmez A., Münster C., Kretschmar S., Kalaidzidis Y., Kurth T., Verbavatz J.-M. and Solimena M. (2017). A global approach for quantitative super resolution and electron microscopy on cryo and epoxy sections using self-labeling protein tags. Sci. Rep. 7, 23 10.1038/s41598-017-00033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natori S. and Huttner W. B. (1996). Chromogranin B (secretogranin I) promotes sorting to the regulated secretory pathway of processing intermediates derived from a peptide hormone precursor. Proc. Natl. Acad. Sci. USA 93, 4431-4436. 10.1073/pnas.93.9.4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermüller S., Calegari F., King A., Lindqvist A., Lundquist I., Salehi A., Francolini M., Rosa P., Rorsman P., Huttner W. B. et al. (2010). Defective secretion of islet hormones in chromogranin-B deficient mice. PLoS ONE 5, e8936 10.1371/journal.pone.0008936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. J., Gondre-Lewis M. C., Eiden L. E. and Loh Y. P. (2011). A distinct trans-Golgi network subcompartment for sorting of synaptic and granule proteins in neurons and neuroendocrine cells. J. Cell Sci. 124, 735-744. 10.1242/jcs.076372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouli A. E., Kennedy H. J., Schofield J. G. and Rutter G. A. (1998). Insulin targeting to the regulated secretory pathway after fusion with green fluorescent protein and firefly luciferase. Biochem. J. 331, 669-675. 10.1042/bj3310669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn D., Orci L., Ravazzola M. and Moore H. P. (1991). Intracellular transport and sorting of mutant human proinsulins that fail to form hexamers. J. Cell Biol. 113, 987-996. 10.1083/jcb.113.5.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes C. J. and Halban P. A. (1987). Newly synthesized proinsulin/insulin and stored insulin are released from pancreatic B cells predominantly via a regulated, rather than a constitutive, pathway. J. Cell Biol. 105, 145-153. 10.1083/jcb.105.1.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P. and Renstrom E. (2003). Insulin granule dynamics in pancreatic beta cells. Diabetologia 46, 1029-1045. 10.1007/s00125-003-1153-1 [DOI] [PubMed] [Google Scholar]
- Stephens S. B., Edwards R. J., Sadahiro M., Lin W.-J., Jiang C., Salton S. R. and Newgard C. B. (2017). The prohormone VGF regulates beta cell function via insulin secretory granule biogenesis. Cell Rep. 20, 2480-2489. 10.1016/j.celrep.2017.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. G., Shanmugam G. and Sharp G. W. G. (2004). Stimulation of insulin release by glucose is associated with an increase in the number of docked granules in the beta-cells of rat pancreatic islets. Diabetes 53, 3179-3183. 10.2337/diabetes.53.12.3179 [DOI] [PubMed] [Google Scholar]
- Suckale J. and Solimena M. (2010). The insulin secretory granule as a signaling hub. Trends Endocrinol. Metab. 21, 599-609. 10.1016/j.tem.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Ward W. K., Bolgiano D. C., Mcknight B., Halter J. B. and Porte D. Jr (1984). Diminished B cell secretory capacity in patients with noninsulin-dependent diabetes mellitus. J. Clin. Invest. 74, 1318-1328. 10.1172/JCI111542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Orci L., Carroll R., Norrbom C., Ravazzola M. and Steiner D. F. (2002). Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc. Natl. Acad. Sci. USA 99, 10299-10304. 10.1073/pnas.162352799 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.