ABSTRACT
The expression of transcription factors that initiate the specification of a unique cellular identity in multicellular organisms is often maintained throughout the life of the respective cell type via an autoregulatory mechanism. It is generally assumed that such autoregulation serves to maintain the differentiated state of a cell. To experimentally test this assumption, we used CRISPR/Cas9-mediated genome engineering to delete a transcriptional autoregulatory, cis-acting motif in the che-1 zinc-finger transcription factor locus, a terminal selector required to specify the identity of the ASE neuron pair during embryonic development of the nematode Caenorhabditis elegans. We show that che-1 autoregulation is indeed required to maintain the differentiated state of the ASE neurons but that it is also required to amplify che-1 expression during embryonic development to reach an apparent minimal threshold to initiate the ASE differentiation program. We conclude that transcriptional autoregulation fulfills two intrinsically linked purposes: one in proper initiation, the other in proper maintenance of terminal differentiation programs.
This article has an associated ‘The people behind the papers’ interview.
KEY WORDS: C. elegans, Autoregulation, Neuronal identity, Transcriptional control
Highlighted Article: Mutation of an autoregulatory motif of che-1, a cell identity-specifying transcription factor, shows that autoregulation is required for both acquisition and maintenance of cellular identity in C. elegans.
INTRODUCTION
Autoregulation is a feature common to many prokaryotic and eukaryotic transcriptional regulatory systems (Bateman, 1998; Crews and Pearson, 2009; Ptashne et al., 1976). It was first discovered in phage lambda where cI protein, a DNA binding transcription factor, represses the lytic stage of a bacteriophage infection, thereby promoting the lysogenic state. To maintain this specific regulatory state, cI binds to its own promoter to ensure its own continuous expression (Ptashne et al., 1976). The maintenance of cellular states by autoregulating transcription factors has subsequently been observed in a wide variety of cellular contexts in prokaryotic, eukaryotic and multicellular organisms. For example, in multicellular organisms, cell type-specific transcription factors define the identity of specialized, differentiated cell types via the activation of cell type-specific effector gene batteries (Bateman, 1998; Davidson et al., 2003; Hobert, 2016; Levine and Tjian, 2003) and such transcription factors are often also continuously expressed throughout the postembryonic life of a differentiated cell. In many cases, such maintained expression is ensured via autoregulation, in which the respective transcription factor binds to cognate cis-regulatory sites in its own locus (Bateman, 1998; Crews and Pearson, 2009; Schier and Gehring, 1992).
Even though many studies documented the existence of autoregulation (Bateman, 1998; Crews and Pearson, 2009), there have been few genetic loss-of-function studies that formally demonstrated that continuous expression of identity-specifying transcription factors is indeed required to maintain the differentiated state (e.g. Deneris and Hobert, 2014; Etchberger et al., 2009; Kadkhodaei et al., 2009; Kratsios et al., 2011; Liu et al., 2010; Serrano-Saiz et al., 2018, 2013; Song et al., 2011). In these studies, the identity-specifying and often autoregulating transcription factors were removed post-developmentally, i.e. after the differentiated state has been reached, and an ensuing loss of cellular identity was observed. However, formal proof of the importance of autoregulation should leave the transcription factor intact and only remove its autoregulatory aspect, ideally via mutating the cis-regulatory motif that is predicted to confer autoregulation. We provide here such formal proof in the context of neuronal cell type specification in the nematode Caenorhabditis elegans.
In this simple model system, a host of transcription factors, called terminal selectors, have been identified as key regulators of neuronal cell type differentiation (Hobert, 2008, 2016). Neuron type-specific terminal selectors are activated around the time of neuronal birth and control large batteries of effectors genes that impose specific phenotypic features onto individual neuron types. All known terminal selectors are continuously expressed throughout the life of the respective neuron type (Hobert, 2016). Although many terminal selector indeed autoregulate (Baumeister et al., 1996; Hobert et al., 1997; Masoudi et al., 2018; Way and Chalfie, 1989), the functional necessity of the autoregulatory phenomenon per se has not yet been examined.
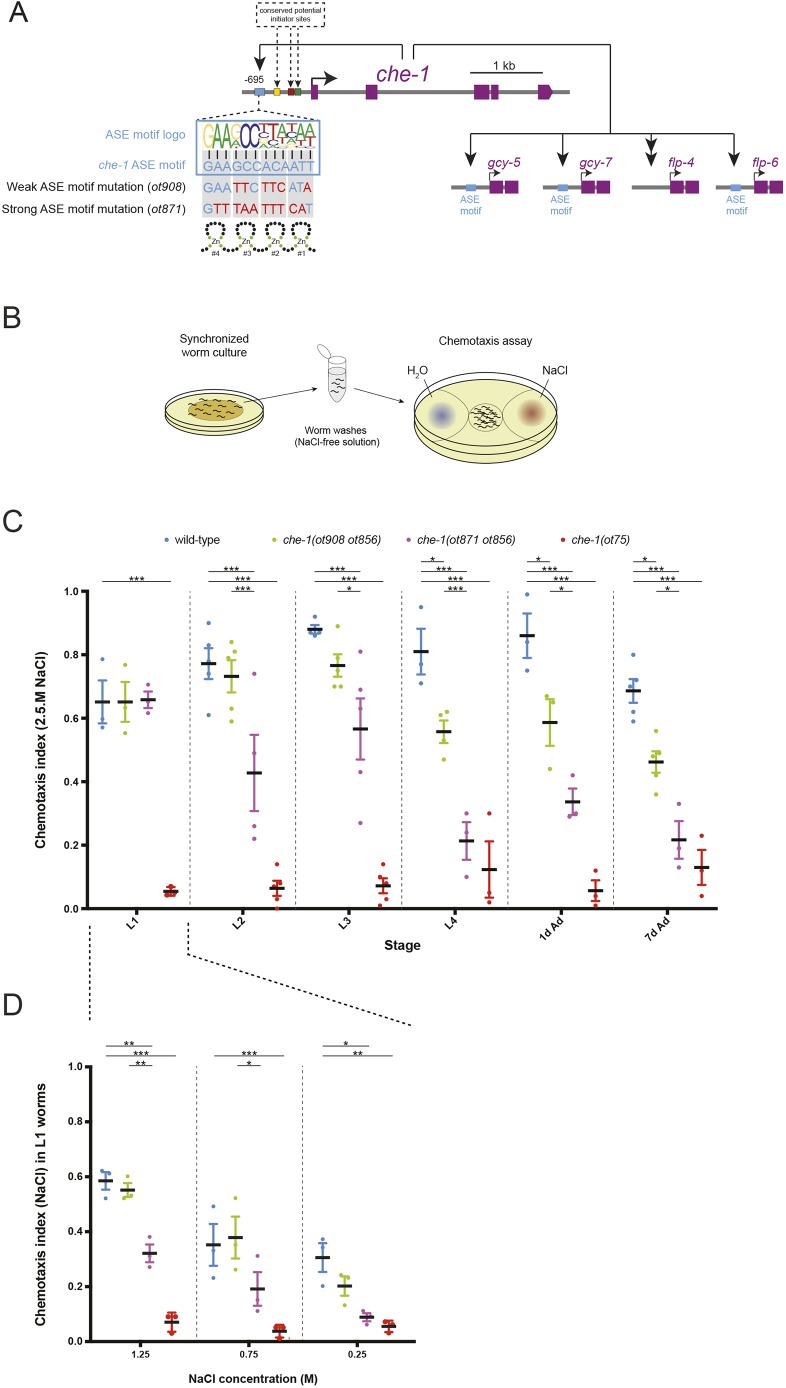
We used the C2H2 zinc-finger type terminal selector che-1, which is required for the differentiation of the gustatory ASE sensory neuron pair (Fig. 1A) (Chang et al., 2003; Etchberger et al., 2007; Uchida et al., 2003), as a paradigm for investigating the importance of autoregulation. Using a fosmid-based reporter transgene, we have previously shown that che-1 is exclusively expressed in the ASE sensory neuron pair, starting during embryogenesis when the ASE neurons are born, and being subsequently maintained through larval and adult stages (Sarin et al., 2009; Tursun et al., 2009). Initiation of che-1 expression in the embryo requires the orphan nuclear hormone receptor nhr-67/Tailless/Tlx (Sarin et al., 2009). nhr-67 is only transiently expressed in ASE until the first larval stage (Sarin et al., 2009), suggesting that nhr-67 cannot be responsible for the continuous expression of che-1 throughout larval and adult stages. We have also previously shown that a <700 bp region upstream of the che-1 locus, when fused to a fluorescent reporter, recapitulates the continuous ASE-specific expression observed with a fosmid reporter transgene (Etchberger et al., 2007). This upstream region behaves like many other cis-regulatory control regions of transcription factors: (1) it is turned off in ASE neurons in a che-1 mutant background, demonstrating that che-1 autoregulates its expression (Etchberger et al., 2007); and (2) in addition to putative initiator sites, it contains a single copy of a biochemically defined CHE-1 binding site (the ‘ASE motif’; Fig. 1A) (Etchberger et al., 2007), suggesting that autoregulation is direct.
Fig. 1.
The autoregulatory motif of the che-1 locus is required to maintain the functional state of ASE. (A) The che-1 paradigm. che-1 activates a battery of downstream terminal effectors and, additionally, activates its own expression through direct binding to its own promoter (autoregulation). The schematic of che-1 gene locus shows the ASE motif location as well as phylogenetically conserved motifs (colored boxes) that are likely required for initiation of che-1 expression (Etchberger, 2008) all located in a 700 bp upstream region that recapitulates che-1 expression and autoregulation (Etchberger et al., 2007). Another ASE motif that was not functionally validated is located in the second intron (not shown). ASE motif details and their binding to the four zinc fingers from CHE-1 are from Etchberger et al. (2007). Four che-1 targets, two chemoreceptor-encoding genes (gcy-5 and gcy-7) and two neuropeptide-encoding genes (flp-4 and flp-6) are used as ASE differentiation markers in this study. (B) Schematic of the salt chemotaxis assay. Synchronized worms are washed and placed on assay plates where chemotaxis to a sodium chloride gradient was tested. (C) Quantification of sodium chloride chemotaxis assays. Graphical representation of the chemotaxis index in wild type, che-1(ot908 ot856), che-1(ot871 ot856) and che-1(ot75) (null allele) mutants. Chemotaxis was assayed at different larval and adult stages: worm larvae at the first (L1), second (L2), third (L3) and fourth (L4) larval stages; 1-day-old adult (1d Ad) and 7-day-old adult (7d Ad) worms. 2.5 M NaCl was used to generate the gradients for all stages. (D) Chemotactic behavior of L1 stage worms at more dilute NaCl concentrations. The data in C,D are presented as individual values with each dot representing the value of each independent experiment with the mean±s.e.m. indicated. Two-way ANOVA followed by Tukey's multiple comparisons test; *P<0.05, **P<0.01, ***P<0.001. n≥3 independent experiments.
The advent of CRISPR/Cas9-based genome engineering technologies (Arribere et al., 2014; Doudna and Charpentier, 2014; Kim et al., 2014; Ward, 2014) has offered us the opportunity to examine the functional relevance of this autoregulatory cis-acting motif in the context of the endogenous che-1 locus. We demonstrate here that this autoregulatory motif is indeed required for maintaining ASE neuronal identity and function, and, in addition, we have uncovered evidence for a role of autoregulation in the initial amplification of che-1 expression.
RESULTS
Generation of cis-regulatory alleles of che-1
We used CRISPR/Cas9 to generate che-1(ot856[che-1::gfp]) animals in which we tagged the che-1 locus with gfp. We then used CRISPR/Cas9 again in these animals to generate two independent mutant alleles of the ASE motif in the 5′ promoter region of che-1, called che-1(ot908 ot856) and che-1(ot871 ot856) (Fig. 1A). The ASE motif is twelve nucleotides long, with each of the four Zn fingers of CHE-1 binding four adjacent sets of three nucleotides (Etchberger et al., 2007) (Fig. 1A). Based on previous extensive in vitro analysis of CHE-1 binding to ASE motifs (Etchberger et al., 2007), both CRISPR/Cas9-generated cis-regulatory alleles are predicted to affect CHE-1 binding. One of the two alleles still retains an invariant 3 nucleotide motif normally binding the fourth Zn finger (Etchberger et al., 2007) and could potentially display a weaker effect than the other allele, which disrupts almost all nucleotides known to be involved in CHE-1 binding (Fig. 1A).
Effects of cis-regulatory alleles on ASE neuron function
To assess the functional consequence of mutating the che-1 autoregulatory motif, we first investigated the main animal behavioral output requiring ASE neuron function: salt chemotaxis (Fig. 1B). Sensory perception of sodium chloride induces ASE sensory neuron activity (Suzuki et al., 2008) and sodium chloride chemotaxis is abolished in animals in which either the ASE neuron is microsurgically removed (Bargmann and Horvitz, 1991) or which carry deletion or coding mutations of the che-1 locus (Uchida et al., 2003; Ward, 1973). We find that adult animals that carry either ASE cis-regulatory mutations fail to properly exhibit chemotaxis (Fig. 1C). In older stage animals, the effect of the strong cis-regulatory allele is almost as strong as a che-1 null allele, whereas the weaker cis-regulatory allele results in the retention of some chemotactic ability (Fig. 1C). Chemotaxis defects of the cis-regulatory alleles are less pronounced in earlier stage animals but are already detectable in first larval stage animals in more-challenging dilute NaCl gradients (Fig. 1C,D). Overall, the progressive worsening of the chemotactic defects during postembryonic maturation of the animal clearly demonstrate that the ASE motif and, hence, che-1 autoregulation, is required to maintain the functional properties of the ASE neurons. However, chemotaxis defects observed at earlier stages already suggest that ASE neurons may have actually never attained a fully functional state.
Effects of cis-regulatory alleles on molecular aspects of ASE neuron differentiation
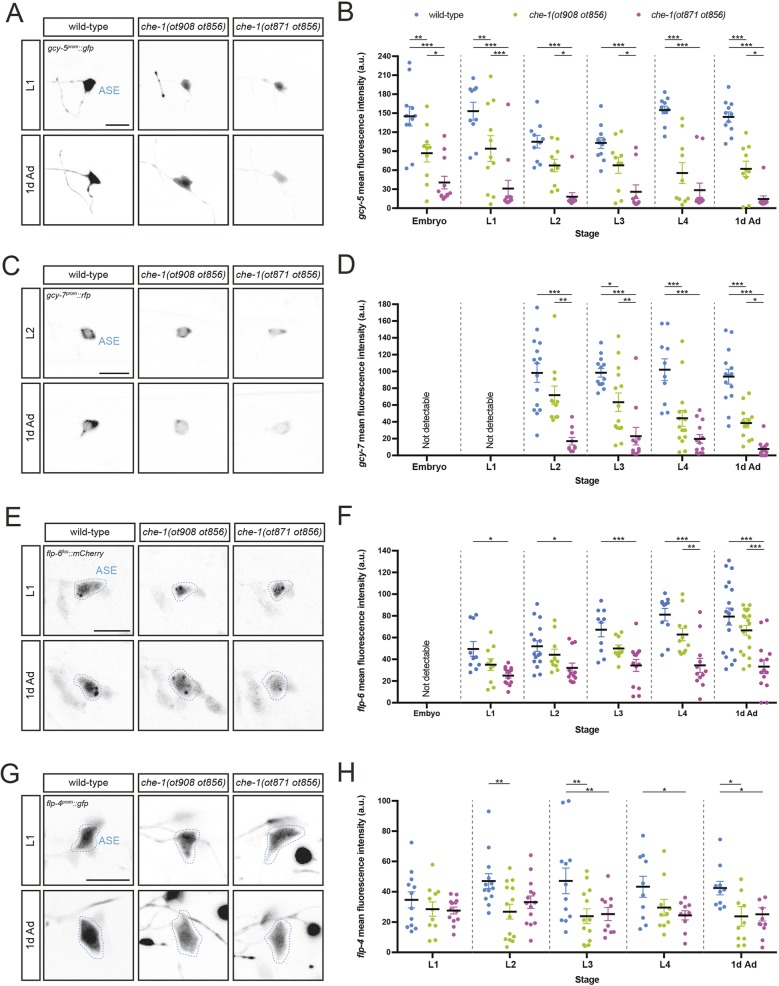
We also tested the effect of mutations in the autoregulatory motif of che-1 locus on the expression of four molecular markers of the ASE differentiated state: two chemoreceptor-encoding genes, the receptor-type guanylate cyclases gcy-5 and gcy-7 (both exclusively expressed in ASE; Yu et al., 1997); and two neuropeptide-encoding genes, the FMRFamides flp-4 and flp-6 (both expressed in ASE, as well as in other neurons; Kim and Li, 2004). All four genes are targets of the che-1 transcription factor, i.e. their expression is completely abrogated in che-1-null mutants, and three of them contain ASE motifs that have been shown to be required for expression in the ASE neurons (Chang et al., 2003; Etchberger et al., 2007; Uchida et al., 2003) (Fig. 1A). We found that the expression of gcy-5 in adult worms is significantly reduced in both cis-regulatory alleles, with close to undetectable expression levels in the stronger allele (Fig. 2A,B). Similar results were observed when gcy-7 and flp-6 expression was analyzed: a severe reduction in expression in the stronger allele and a milder defect in the weak cis-regulatory allele both in adult and larval stages (Fig. 2C-F). flp-4 expression is affected equally by both alleles (Fig. 2G,H).
Fig. 2.
che-1 autoregulation is required to adopt and to maintain the differentiated state, as determined by marker gene analysis. (A,C,E,G) Expression of reporters gcy-5prom::gfp [ntIs1] (A), gcy-7prom::rfp [otIs131] (C) flp-6fos::mCherry [otIs494] (E) and flp-4prom::gfp [ynIs30] (G) in wild-type (left), che-1(ot908 ot856) (middle column) or che-1(ot871 ot856) (right) at the L1 or L2 larval stage (for gcy-7prom::rfp, L2 images are shown as expression is not consistently detected at L1) (top) and 1-day-old adult stage (1d Ad) worms (bottom). Lateral views of the head are shown. Scale bars: 10 µm. (B,D,F,H) Quantification of gcy-5prom::gfp (B), gcy-7prom::rfp (D), flp-6fos::mCherry (F) and flp-4prom::gfp (H) fluorescence intensity in wild type, che-1(ot908 ot856) and che-1(ot871 ot856) mutants. Fluorescence intensity was analyzed at different embryonic, larval and adult stages: threefold embryos (embryo); worm larvae at the first (L1), second (L2), third (L3) and fourth (L4) larval stages; and 1-day-old adult (1d Ad) worms. The data in B,D,F,H are presented as individual values with each dot representing the expression level of one neuron with the mean±s.e.m. indicated. Two-way ANOVA followed by Tukey's multiple comparisons test; *P<0.05, **P<0.01, ***P<0.001. n≥10 for all genotypes. a.u., arbitrary units.
In line with the chemotaxis defects of young larvae, we also noted in the molecular marker analysis that removal of the autoregulatory motif affects expression of differentiation marker at early larval stages. Specifically, we observed that during embryonic and larval stages, gcy-5 expression levels in mutant worms never reached those observed in wild-type animals (Fig. 2B). Likewise, defects were already detected at the time when gcy-7 or flp-6 expression is activated (Fig. 2D,F). In summary, although effector gene expression defects observed upon disruption of che-1 autoregulation are generally more pronounced in the adult stage (corroborating the importance of autoregulation in maintaining the differentiated state of the ASE neurons), defects were already observed at much earlier developmental stages.
Mutation of autoregulatory elements reveals unexpected effects on expression of che-1
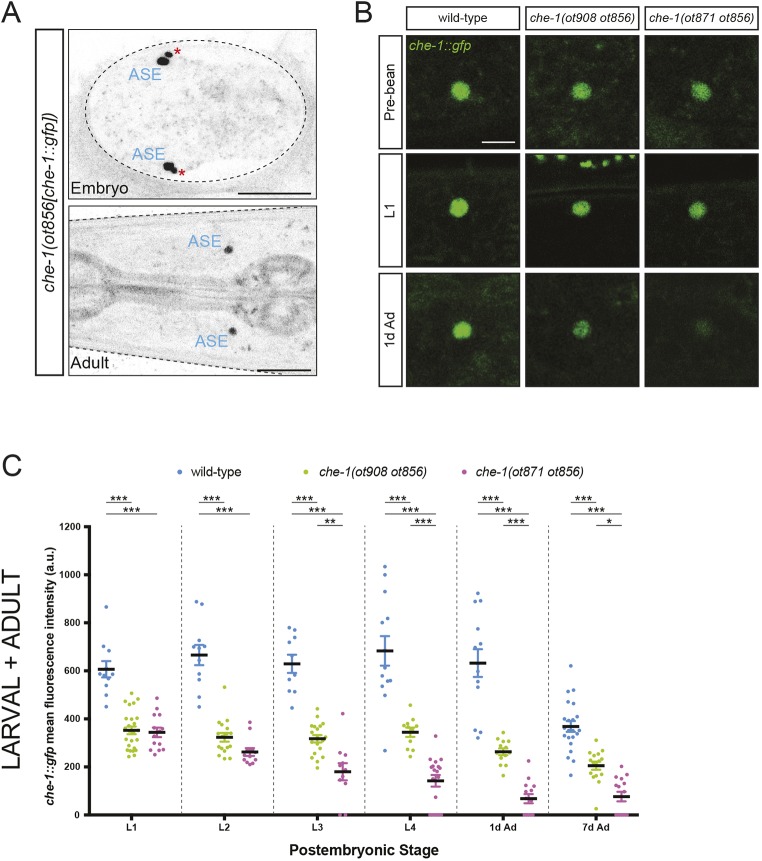
After characterizing the downstream effects of removing the che-1 autoregulatory motif, we examined che-1 expression, using the gfp-tagged che-1 locus (ot856 allele). We observed the same expression pattern as that found for the previously published fosmid-based reporter construct (Sarin et al., 2009). Specifically, expression was exclusively observed in the two ASE neurons, starting around the time of ASE cell birth in the embryo, and continuing throughout all ensuing embryonic, larval and adult stages (Fig. 3A-C). Examination of the expression of the gfp-tagged che-1 locus in which the ASE motif was mutated, che-1(ot908 ot856) and che-1(ot871 ot856), revealed an intriguing pattern, one part of it expected, another part unexpected. In larval and adult stages, we observed an expected reduction of che-1::gfp expression (Fig. 3B,C). Adult animals carrying the allele that disrupts the binding of all four zinc fingers exhibit a significantly greater decrease of che-1::gfp expression compared with the cis-regulatory allele in which one zinc finger of CHE-1 is still predicted to bind DNA. In larvae, we also observe a significant reduction in che-1::gfp expression in both cis-regulatory alleles compared with wild-type animals at all larval stages (Fig. 3B,C). This reduction is similar in the first (L1) and second (L2) larval stages, whereas the reduction in expression is significantly higher in the stronger allele starting at the L3 larval stage. Among all post-embryonic stages, animals carrying the weaker cis-regulatory allele always showed detectable che-1::gfp expression levels, possibly owing to residual binding of CHE-1 to the mutated ASE motif. In both alleles, che-1 expression defects are less pronounced at earlier stages, perhaps due to the perdurance of the embryonic initiators of che-1 expression, such as nhr-67, the expression of which fades postembryonically (Sarin et al., 2009). The overall pattern of decreases in che-1 expression correlates with the decreases in chemotactic behavior described above (Fig. 1), such that defects are generally less pronounced at early larval stages and more pronounced in late larval or adult stages. However, as the animals progresses through larval development, the precise timing of che-1 expression decreases is not well correlated with losses of chemotactic ability. For example, although animals carrying the stronger allele show a progressive reduction on che-1 expression levels, the chemotactic response drops abruptly in L2 larvae. As che-1 controls the expression of hundreds of target genes, many of which involved in controlling ASE function, the phenotypic consequence of partial losses of subsets of targets upon partial loss of che-1 expression is hard to predict.
Fig. 3.
The autoregulatory CHE-1 binding site is required to maintain che-1 expression in larval and adult stages. (A) che-1(ot856[che-1::gfp]) expression in embryo (top, dorso-ventral view, bean stage embryo) and adult stage (bottom, head dorso-ventral view) animals. Red asterisks indicate the ASE sister cell, which eventually undergoes apoptosis and is consumed by the ASE cell. Scale bars: 20 µm. (B) che-1::gfp expression is reduced in che-1(ot908 ot856) (middle column) and che-1(ot871 ot856) (right) compared with wild-type (left) animals, at the pre-bean embryonic stage (top, dorso-ventral images), L1 larval stage (middle row, lateral images) and 1-day-old adult stage (1d Ad) worms (bottom, lateral images). Scale bar: 5 µm. (C) Quantification of che-1(ot856[che-1::gfp]) fluorescence intensity in wild type, che-1(ot908 ot856) and che-1(ot871 ot856) mutants. Fluorescence intensity was analyzed at different larval and adult stages: worm larvae at the first (L1), second (L2), third (L3) and fourth (L4) larval stages; and 1-day-old adult (1d Ad) and 7-day-old adult (7d Ad) worms. The data are presented as individual values with each dot representing the expression level of one neuron with the mean±s.e.m. indicated. Two-way ANOVA followed by Tukey's multiple comparisons test; *P<0.05, **P<0.01, ***P<0.001. n≥10 for all genotypes. a.u., arbitrary units.
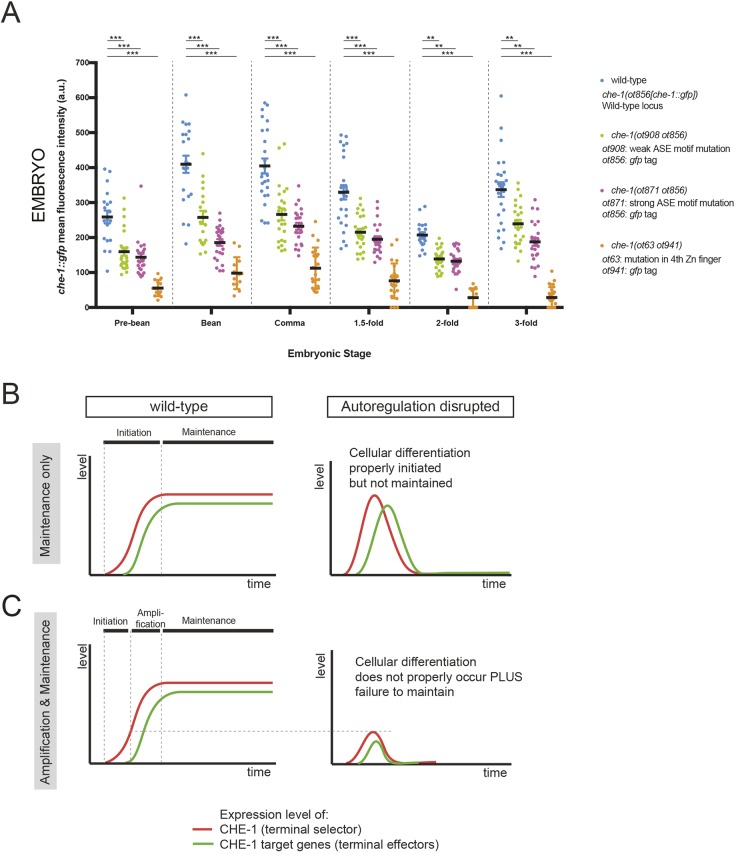
The unexpected part of the che-1::gfp expression pattern was revealed during embryogenesis. We found that in the two ASE motif-mutant strains, che-1::gfp expression never reaches wild-type levels even at those earlier embryonic stages at which che-1 expression first becomes detectable (Fig. 3B and 4A). Specifically, we find that, in wild-type animals, che-1::gfp expression is normally initiated at the pre-bean stage, and amplified from this stage to the bean stage. The observed onset of CHE-1 expression at the late pre-bean stage agrees with previous reports in which the onset was determined by fosmid reporter expression (Sarin et al., 2009) and by smFISH (Cochella et al., 2012), suggesting that our analysis captures the very onset of CHE-1 expression. Transiently expressed initiators of che-1 expression, such as nhr-67, trigger this initiation (Sarin et al., 2009) but, as we show here, this initial amplification is defective in both cis-regulatory alleles (Fig. 4A). These results indicate that che-1 autoregulation is not only required to maintain wild-type che-1 expression levels at larval and adult stages but that it may also be required to amplify che-1 expression during initial specification of ASE neuron identity.
Fig. 4.
che-1 is required to amplify its own expression in the embryo. (A) Quantification of che-1(ot856[che-1::gfp]) fluorescence intensity in wild type, che-1(ot908 ot856), che-1(ot871 ot856) and che-1(ot63 ot941) mutants. Fluorescence intensity was analyzed at different embryonic stages: pre-bean, bean, comma, 1.5-fold, 2-fold and 3-fold. The data are presented as individual values with each dot representing the expression level of one neuron with the mean±s.e.m indicated. Two-way ANOVA followed by Tukey's multiple comparisons test; **P<0.01, ***P<0.001. n≥13 for all genotypes. a.u., arbitrary units. (B,C) Conceptualization of the function of autoregulation. (B) In this scenario, autoregulation is required only for maintenance of the cellular state. The initiation phase of che-1 induction triggers sufficiently high levels of che-1, effector gene activation and cellular differentiation occurs normally, and autoregulation is exclusively required to maintain the differentiated state. (C) In this scenario, found to apply here to che-1, autoregulation is also required for the boosting of initial transcription factor expression, ensuring initial activation of the effector battery and, later, maintaining transcription factor expression to ensure continuous effector gene expression.
One caveat of this interpretation is that the ASE motif may not operate as an autoregulatory motif at the stage of initiation of che-1 expression, but that it may rather serve as a binding site for distinct upstream factors that initiate che-1 expression. To test this possibility, we used CRISPR/Cas9 genome editing to engineer gfp into the che-1(ot63) loss-of-function allele that carries a missense mutation in the fourth zinc-finger domain of CHE-1 (Cys255Tyr) (Chang et al., 2003). This fourth zinc finger is essential for CHE-1 binding to the ASE motif in vitro (Etchberger et al., 2007) and che-1 gene function in vivo (Chang et al., 2003). We reasoned that if che-1 had no role in the initial establishment of che-1 expression, we would observe normal initiation of che-1 expression, followed by a failure to maintain its expression; if, in contrast, che-1 had a role in the initial establishment of che-1 expression, we should observe little to no che-1 expression. We indeed found that che-1(ot63 ot941) animals show a substantially reduced level of gfp expression in the embryo, failing to reach che-1 expression levels detected in wild-type animals (Fig. 4A). The effect of disabling CHE-1 protein function on the initiation of che-1 gene expression (i.e. initial amplification) is even stronger than mutation of the ASE motif (Fig. 4A). We ascribe this effect to the presence of an additional ASE motif in the second intron in the che-1 locus, which may contribute to CHE-1 autoregulation. Thus, disabling CHE-1 protein function may completely eliminate the ability to properly amplify che-1 expression through either of the two ASE motifs. We conclude that che-1 is indeed required to amplify its own expression, likely pursuant to a weak initial regulatory input.
DISCUSSION
Although transcription factor autoregulation is a pervasive gene regulatory phenomenon, the functional relevance of autoregulation in cellular differentiation has not been extensively probed through the genetic removal of autoregulatory, cis-acting transcription factor binding motifs. The only instance that we are aware of is the removal of a large cis-regulatory element with several Krox20-binding sites from the mouse and zebrafish Krox20 loci, which implicates Krox20 autoregulation in rhombomere specification (Bouchoucha et al., 2013; Torbey et al., 2018). We have undertaken a more fine-grained analysis in a distinct cellular and organismal context, addressing the importance of autoregulation during the execution of a terminal differentiation program in the nervous system. Through constitutive or post-developmental removal of che-1 gene activity, this transcription factor locus was previously known to be required to initiate and maintain the differentiated state of a single chemosensory neuron in C. elegans (Etchberger et al., 2009). If maintenance of expression were the only function of autoregulation, mutation of the autoregulatory motif would be predicted to result in effects schematized in Fig. 4B. Transiently expressed factors (such as nhr-67)(Sarin et al., 2009) initiate che-1 expression and levels of CHE-1 are sufficient to initially trigger the onset of expression of che-1 target genes and, hence, ASE differentiation. Subsequently, owing to the lack of autoregulation, che-1 expression, as well as effector gene expression, would fade out. Although we clearly do observe a progressive worsening of the loss of che-1 and effector gene expression, as well as neuron function (corroborating the previously described maintenance role of che-1; Etchberger et al., 2009), we also observed effects that we interpret as schematized in Fig. 4C. In the absence of the autoregulatory motif, che-1 expression never reaches normal wild-type levels of expression, even early in the embryo when che-1 expression is initiated. Hence, the fully functional differentiated state is never attained. We interpret this to mean that the factor(s) that initiate che-1 expression (such as nhr-67) (Sarin et al., 2009) may be weak activators that are alone not sufficient to produce the amounts of che-1 expression required to activate che-1 target genes. Autoregulation of che-1 seems to ensure that che-1 expression is amplified to reach the critical threshold for activation of downstream target genes. This mechanism contrasts with other autoregulation cases that follow the more conventional model shown in Fig. 4B, where the autoregulation phase is clearly separated from the initiation phase. For example, in the tunicate Ciona intestinalis, maintenance of Ebf expression in atrial siphon muscle founder cells relies on autoregulation, while the initial amplification depends on an initiation event that requires MAPK signaling (Razy-Krajka et al., 2018). In contrast, in mouse, Krox20 autoregulation is required for the conversion of a transient input into a stable fate commitment (Bouchoucha et al., 2013). However, Krox20 expression is transient and its function is not required for the maintenance of the differentiated state. We describe here a case in which transcription factor autoregulation is required both to maintain the differentiated neuronal state in adult animals and to amplify transcription factor expression during embryonic development to initially specify neuronal identity.
In conclusion, the function of transcriptional autoregulation goes beyond a mere maintenance function by ‘boosting’ transcription factor expression beyond a critical threshold required to activate its downstream effectors. This auto-amplifying, boosting mechanism enables the detection of weak and transient regulatory inputs to eventually lock them into a stably maintained, terminal state. Considering the previously reported case of vertebrate Krox20, for which a similar auto-amplification has been reported in the context of rhombomere development (Bouchoucha et al., 2013), we propose that the functional duality of transcriptional autoregulation, initial amplification and ensuing maintenance, constitutes a widely used gene regulatory principle during animal development.
MATERIAL AND METHODS
Strains
The C. elegans strains used in this study were: OH14130, che-1(ot856[che-1::gfp]) (Leyva-Diaz et al., 2017); OH15579, che-1(ot908 ot856) (generated in this study); OH15683, che-1(ot871 ot856) (generated in this study); OH15815, che-1(ot63 ot941) (generated in this study); OH610, che-1(ot63) (Chang et al., 2003); and OH13098, che-1(ot75) (Chang et al., 2003). The reporter transgenes were: OH3192, ntIs1[gcy-5prom::gfp] (Chang et al., 2003); OH1092, otIs131[gcy-7prom::rfp] (Chang et al., 2003); OH12372, otIs494[flp-6fosmid::sl2::1xNLS::mChOpti]; and NY2030 ynIs30[flp-4prom::gfp] (Kim and Li, 2004). Animals were maintained at 20°C with abundant E. coli OP50 as food following standard conditions (Brenner, 1974).
Targeted genome modification in C. elegans
The CRISPR/Cas9-mediated modification of the genome to mutate the ASE motif on the che-1 promoter was performed using a Co-CRISPR screening strategy (Arribere et al., 2014; Kim et al., 2014; Ward, 2014). This screening strategy uses a visible phenotype at one locus to help identify edits at a second locus of interest. Briefly, che-1(ot856[che-1::gfp]) adult hermaphrodites were used for gonad injection of a DNA injection mix containing: eft-3::Cas9 plasmid (50 ng/µl), che-1 sgRNA plasmid (70 ng/µl), che-1 homology directed repair (HDR) plasmid (40 ng/µl), Co-CRISPR sgRNA plasmid (40 ng/µl) and myo-2::mCherry plasmid (3 ng/µl). We used a guide RNA that targets a sequence 108 bp downstream the ASE motif on the che-1 promoter (target sequence: ATCACAAAAATAAAGAGGG). The che-1 HDR plasmid contains a 2.2 kb homology region centered around the ASE motif on the che-1 promoter (−1486, +758). In this plasmid, the ASE motif was substituted into an EcoRI restriction site for the che-1(ot908) mutation and into a PmeI site for the che-1(ot871) mutation, while the PAM site was mutated to prevent Cas9 from cutting the repair template (TGG to TAG). che-1(ot908) was generated using unc-22 mutations for Co-CRISPR, while che-1(ot871) was generated using a gain-of-function dpy-10(cn64) Co-CRISPR approach. When dpy-10(cn64) Co-CRISPR was used, a dpy-10 repair oligo (Arribere et al., 2014) (14 ng/µl) was included in the injection mix instead of myo-2::mCherry. For the che-1(ot908 ot856) mutation, transformed F1 progeny of the injected animals were identified based on the myo-2::mCherry co-injection marker and singled into independent plates. F1 worms that produced F2 progeny with a ‘twitching’ phenotype (unc-22 phenotype) were screened by PCR to identify animals carrying the desired modification. For the che-1(ot871 ot856) mutation, che-1(ot908 ot856) and che-1(ot871 ot856) mutations were outcrossed at least five times. CRISPR/Cas9-mediated gfp tagging of the che-1(ot63) locus was carried out with the same gfp cassette present in che-1(ot856[che-1::gfp]), using a previously described protocol (Dokshin et al., 2018). che-1(ot63) adult hermaphrodites were used for gonad injection of a mix containing S. pyogenes Cas9 protein (250 ng/µl, IDT), tracrRNA (100 ng/µl, IDT), che-1 crRNA (56 ng/µl, IDT), dsDNA donor cocktail (200 ng/µl) and PRF4::rol-6(su1006) plasmid (40 ng/µl). We used a crRNA that targets a sequence 21 bp upstream the che-1 STOP codon (target sequence: CACAGAGTGGGAACTTGCAT).
C. elegans microscopy
Worms were anesthetized using 100 mM sodium azide (NaN3) and mounted on 5% agarose pads on glass slides. Images were acquired using a Zeiss confocal microscope (LSM880). Several z-stack images (each ∼0.7 µm thick) were acquired with the ZEN software. Images were reconstructed via maximum intensity z-projection of 2-10 µm z-stacks using the ImageJ software (Schneider et al., 2012). Representative images are shown following orthogonal projection of 2-10 µm z-stacks.
For fluorescence intensity quantification, reporter expression levels were analyzed using the mean gray value of ImageJ version 1.51. Fluorescence intensity was measured in the focal plane with the strongest ASE expression within the z-stack. For each fluorescent reporter, the ASE neuron area was delineated (region of interest, ROI). For each image, the same ROI was also used to measure the background next to ASE in the same focal plane, and this value was then subtracted from the reporter fluorescence intensity value in ASE. For all images used for fluorescence intensity quantification, the acquisition parameters were maintained constant among all samples (same pixel size, laser intensity, PMT voltage, pinhole, etc.) for each of the fluorescent reporters. The fluorescence intensity of the gcy-5prom::gfp and flp-4prom::gfp reporters is much higher than that of che-1::gfp. We tested that che-1::gfp fluorescence was not detectable with the acquisition settings used for gcy-5prom::gfp or flp-4prom::gfp so that che-1::gfp fluorescence did not interfere with the reporters fluorescence intensity acquisition.
Chemotaxis assays
The response to NaCl gradients was assayed as previously described (Bargmann and Horvitz, 1991). Briefly, 10 ml of buffered agar (20 g/l agar, 1 mM CaCl2, 1 mM MgSO4 and 5 mM KPO4) was poured into 10 cm diameter petri dishes. To establish the chemical gradient, we applied 10 μl of NaCl solution to the attractant spot and 10 μl of double-distilled H2O to the control spot. The NaCl was allowed to diffuse for 14-16 h at room temperature before the assay. To increase the steepness of the gradient, another 4 μl of NaCl solution or water was added to the same spots 4 h before the assay. We applied a 1 μl drop of 1 M sodium azide to both attractant and control spots 10 min before the assay to immobilize worms that reached these areas. Synchronized animals were washed three times with CTX solution (1 mM CaCl2, 1 mM MgSO4 and 5 mM KPO4) and 100-200 animals were placed in the center of the assay plate in a minimal volume of buffer. Animals were allowed to move about the agar surface for 1 h, after which assay plates were placed at 4°C overnight. For worms assayed at all stages in Fig. 1C, a 2.5 M NaCl solution was applied to the attractant spot. Additionally, for L1 worms, different salt dilutions (1.25 M, 0.75 M and 0.25 M NaCl) were used in Fig. 1D.
The distribution of animals across the plate was then determined and a chemotaxis index was calculated as the number of animals at the NaCl spot minus the number of animals at the control spot, divided by the total number of animals. Animals that did not leave the initial inner circle were not included in the count of total number of animals, as these animals were dead or had movement defects. Chemotaxis assays were performed on at least three independent days with at least three assays per day.
Statistical analysis
For quantification data shown in graphs of all figures, dot symbols represent individual values, the black line indicates the mean value and error bars represent the s.e.m. Statistical analyses were carried out using a two-way ANOVA followed by Tukey's multiple comparisons test. Wild type versus weak, wild type versus strong, wild type versus null and weak versus strong allele comparisons are shown if significant (Figs 1–3); only comparisons with wild type are shown in Fig. 4; *P<0.05, **P<0.01, ***P<0.001. The Shapiro-Wilk normality test was performed on all data to test whether the values follow a normal distribution. All statistical analyses were performed using GraphPad Prism 7.0d for Mac OS X.
Acknowledgements
We thank Chi Chen for the C. elegans injections, Dylan Rahe for providing the GFP-tagged che-1(ot856) allele, Tulsi Patel for providing the flp-6 fosmid reporter, and Mark Ptashne, Steve Crews and members of the Hobert lab for comments on this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.L.-D., O.H.; Formal analysis: E.L.-D.; Investigation: E.L.-D.; Writing - review & editing: E.L.-D.; Visualization: E.L.-D.; Supervision: O.H.; Project administration: O.H.
Funding
E.L.-D. was supported by a European Molecular Biology Organization long-term fellowship. O.H. is an Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
References
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S. and Fire A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I. and Horvitz H. R. (1991). Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729-742. 10.1016/0896-6273(91)90276-6 [DOI] [PubMed] [Google Scholar]
- Bateman E. (1998). Autoregulation of eukaryotic transcription factors. Prog. Nucleic Acid Res. Mol. Biol. 60, 133-168. 10.1016/S0079-6603(08)60892-2 [DOI] [PubMed] [Google Scholar]
- Baumeister R., Liu Y. and Ruvkun G. (1996). Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes Dev. 10, 1395-1410. 10.1101/gad.10.11.1395 [DOI] [PubMed] [Google Scholar]
- Bouchoucha Y. X., Reingruber J., Labalette C., Wassef M. A., Thierion E., Desmarquet-Trin Dinh C., Holcman D., Gilardi-Hebenstreit P. and Charnay P. (2013). Dissection of a Krox20 positive feedback loop driving cell fate choices in hindbrain patterning. Mol. Syst. Biol. 9, 690 10.1038/msb.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R. J. Jr. and Hobert O. (2003). A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17, 2123-2137. 10.1101/gad.1117903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L. and Hobert O. (2012). Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151, 1229-1242. 10.1016/j.cell.2012.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S. T. and Pearson J. C. (2009). Transcriptional autoregulation in development. Curr. Biol. 19, R241-R246. 10.1016/j.cub.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., McClay D. R. and Hood L. (2003). Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. USA 100, 1475-1480. 10.1073/pnas.0437746100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris E. S. and Hobert O. (2014). Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899-907. 10.1038/nn.3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin G. A., Ghanta K. S., Piscopo K. M. and Mello C. C. (2018). Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210, 781-787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A. and Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Etchberger J. F. (2008). The cis-regulatory logic of gustatory neuron development in Caenorhabditis elegans. PhD thesis, Department of Biochemistry and Molecular Biophysics. New York: Columbia University. [Google Scholar]
- Etchberger J. F., Lorch A., Sleumer M. C., Zapf R., Jones S. J., Marra M. A., Holt R. A., Moerman D. G. and Hobert O. (2007). The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21, 1653-1674. 10.1101/gad.1560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J. F., Flowers E. B., Poole R. J., Bashllari E. and Hobert O. (2009). Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development 136, 147-160. 10.1101/gad.1560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2008). Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. USA 105, 20067-20071. 10.1073/pnas.0806070105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2016). Terminal selectors of neuronal identity. Curr. Top. Dev. Biol. 116, 455-475. 10.1016/bs.ctdb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Hobert O., Mori I., Yamashita Y., Honda H., Ohshima Y., Liu Y. and Ruvkun G. (1997). Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron 19, 345-357. 10.1016/S0896-6273(00)80944-7 [DOI] [PubMed] [Google Scholar]
- Kadkhodaei B., Ito T., Joodmardi E., Mattsson B., Rouillard C., Carta M., Muramatsu S.-I., Sumi-Ichinose C., Nomura T., Metzger D. et al. (2009). Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J. Neurosci. 29, 15923-15932. 10.1523/JNEUROSCI.3910-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. and Li C. (2004). Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475, 540-550. 10.1002/cne.20189 [DOI] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D. Jr., Shirayama M. and Mello C. C. (2014). A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069-1080. 10.1534/genetics.114.166389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M. and Hobert O. (2011). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15, 205-214. 10.1038/nn.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. and Tjian R. (2003). Transcription regulation and animal diversity. Nature 424, 147-151. 10.1038/nature01763 [DOI] [PubMed] [Google Scholar]
- Leyva-Diaz E., Stefanakis N., Carrera I., Glenwinkel L., Wang G., Driscoll M. and Hobert O. (2017). Silencing of repetitive DNA is controlled by a member of an unusual Caenorhabditis elegans gene family. Genetics 207, 529-545. 10.1534/genetics.117.300134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Maejima T., Wyler S. C., Casadesus G., Herlitze S. and Deneris E. S. (2010). Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 13, 1190-1198. 10.1038/nn.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudi N., Tavazoie S., Glenwinkel L., Ryu L., Kim K. and Hobert O. (2018). Unconventional function of an Achaete-Scute homolog as a terminal selector of nociceptive neuron identity. PLoS Biol. 16, e2004979 10.1371/journal.pbio.2004979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara M. M., Zhang F. and Hobert O. (2010). Maintenance of neuronal laterality in Caenorhabditis elegans through MYST histone acetyltransferase complex components LSY-12, LSY-13 and LIN-49. Genetics 186, 1497-1502. 10.1534/genetics.110.123661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B. and Sauer R. T. (1976). Autoregulation and function of a repressor in bacteriophage lambda. Science 194, 156-161. 10.1126/science.959843 [DOI] [PubMed] [Google Scholar]
- Razy-Krajka F., Gravez B., Kaplan N., Racioppi C., Wang W. and Christiaen L. (2018). An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. eLife 7, e29656 10.7554/eLife.29656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Antonio C., Tursun B. and Hobert O. (2009). The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development 136, 2933-2944. 10.1242/dev.040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A. F. and Gehring W. J. (1992). Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature 356, 804-807. 10.1038/356804a0 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R. J., Felton T., Zhang F., de la Cruz E. D. and Hobert O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659-673. 10.1016/j.cell.2013.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Leyva-Diaz E., De La Cruz E. and Hobert O. (2018). BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr. Biol. 28, 2813-2823.e2812. 10.1016/j.cub.2018.06.045 [DOI] [PubMed] [Google Scholar]
- Song N.-N., Xiu J.-B., Huang Y., Chen J.-Y., Zhang L., Gutknecht L., Lesch K. P., Li H. and Ding Y.-Q. (2011). Adult raphe-specific deletion of lmx1b leads to central serotonin deficiency. PLoS ONE 6, e15998 10.1371/journal.pone.0015998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Thiele T. R., Faumont S., Ezcurra M., Lockery S. R. and Schafer W. R. (2008). Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454, 114-117. 10.1038/nature06927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torbey P., Thierion E., Collombet S., de Cian A., Desmarquet-Trin-Dinh C., Dura M., Concordet J. P., Charnay P. and Gilardi-Hebenstreit P. (2018). Cooperation, cis-interactions, versatility and evolutionary plasticity of multiple cis-acting elements underlie krox20 hindbrain regulation. PLoS Genet. 14, e1007581 10.1371/journal.pgen.1007581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I. and Hobert O. (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4, e4625 10.1371/journal.pone.0004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida O., Nakano H., Koga M. and Ohshima Y. (2003). The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130, 1215-1224. 10.1242/dev.00341 [DOI] [PubMed] [Google Scholar]
- Ward S. (1973). Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. USA 70, 817-821. 10.1073/pnas.70.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D. (2014). Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199, 363-377. 10.1534/genetics.114.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C. and Chalfie M. (1989). The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 3, 1823-1833. 10.1101/gad.3.12a.1823 [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E. and Garbers D. L. (1997). Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94, 3384-3387. 10.1073/pnas.94.7.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]