Abstract
Calcium is a key mediator controlling essential neuronal functions depending on electrical activity. Altered neuronal calcium homeostasis affects metabolism of amyloid precursor protein (APP), leading to increased production of β-amyloid (Aβ), and contributing to the initiation of Alzheimer's disease (AD). A linkage between excessive glutamate receptor activation and neuronal Aβ release was established, and recent reports suggest that synaptic and extrasynaptic NMDA receptor (NMDAR) activation may have distinct consequences in plasticity, gene regulation, and neuronal death. Here, we report for the first time that prolonged activation of extrasynaptic NMDAR, but not synaptic NMDAR, dramatically increased the neuronal production of Aβ. This effect was preceded by a shift from APP695 to Kunitz protease inhibitory domain (KPI) containing APPs (KPI-APPs), isoforms exhibiting an important amyloidogenic potential. Conversely, after synaptic NMDAR activation, we failed to detect any KPI-APP expression and neuronal Aβ production was not modified. Calcium imaging data showed that intracellular calcium concentration after extrasynaptic NMDAR stimulation was lower than after synaptic activation. This suggests distinct signaling pathways for each pool of receptors. We found that modification of neuronal APP expression pattern triggered by extrasynaptic NMDAR activation was regulated at an alternative splicing level involving calcium-/calmodulin-dependent protein kinase IV, but overall APP expression remained identical. Finally, memantine dose-dependently inhibited extrasynaptic NMDAR-induced KPI-APPs expression as well as neuronal Aβ release. Altogether, these data suggest that a chronic activation of extrasynaptic NMDAR promotes amyloidogenic KPI-APP expression leading to neuronal Aβ release, representing a causal risk factor for developing AD.
Introduction
A hallmark of Alzheimer's disease (AD) is an extracellular deposit of insoluble amyloid-β (Aβ) in senile plaques (Selkoe, 2001). Elevated levels of oligomeric forms of Aβ in hippocampus and cortex may be responsible for early cognitive decline through the disruption of excitatory glutamatergic synapses (Klyubin et al., 2004; Lacor et al., 2007). Among the hypotheses proposed to explain this increased production of Aβ, dysregulation of neuronal calcium homeostasis was postulated early (Khachaturian, 1987; Green et al., 2007), and NMDA receptors (NMDARs) were shown to be especially involved because of their high Ca2+ permeability (Waxman and Lynch, 2005). Similarly, other authors reported a link between Aβ levels and excitatory synaptic activity (Buckner et al., 2005; Cirrito et al., 2005, 2008).
Production of Aβ is the result of the processing of amyloid-β precursor protein (APP) by β- and γ-secretase. In a non-Aβ-forming pathway, APP is cleaved by α- and γ-secretase to generate p3 peptide (Wilquet and De Strooper, 2004). The app gene can be alternatively spliced to produce isoforms varying in size from 695 to 770 aa (Selkoe, 2001). APP770 and APP751 contain a serine protease inhibitory domain encoded by exon 7 called Kunitz protease inhibitor (KPI) (Mahdi et al., 1995). In the brain, both are expressed in glial cells, whereas APP695 is abundant in neurons (LeBlanc et al., 1991). Neuronal expression of KPI-APP exhibits an important amyloidogenic potential (Ho et al., 1996), and APP770 mRNA is significantly elevated in the brain of AD patients (Rockenstein et al., 1995; Moir et al., 1998; Preece et al., 2004). Thus, a shift from APP695 to KPI-APP neuronal expression may result in a higher production of Aβ in the brain parenchyma.
Understanding the molecular mechanisms that regulate APP expression pattern and processing may provide new insights into AD pathogenesis. Several studies explored these mechanisms in neuronal cultures and highlighted the role of NMDAR activation in Aβ production. Although synaptic NMDAR activation was shown to promote nonamyloidogenic pathway (Marcello et al., 2007; Hoey et al., 2009), we have reported that chronic NMDAR activation increases Aβ production in neurons (Lesné et al., 2005).
NMDAR activation has opposite consequences on neuronal fate, according to their cellular localization (Hardingham and Bading, 2003; Léveillé et al., 2008). Stimulation of synaptic NMDARs induces prosurvival events through the activation of cAMP response element-binding protein (CREB) (Hardingham et al., 2002) and the extracellular signal-regulated kinase (ERK) cascade (Ivanov et al., 2006). Conversely, calcium flux through extrasynaptic NMDARs overrides these functions, causing mitochondrial dysfunction and cell death (Wittmann et al., 2004; Léveillé et al., 2008; Stanika et al., 2009). Thus, a distinct NMDAR activation signaling pathway was postulated, depending on their localization.
In the present work, we studied the influence of a selective stimulation of both populations of NMDARs on APP metabolism and neuronal Aβ production. We evaluated the therapeutic potential of memantine, a clinically approved NMDAR antagonist for the treatment of dementia, to counteract neuronal Aβ production. We report that extrasynaptic, but not synaptic, NMDAR activity stimulates neuronal amyloidogenic β-secretase-mediated APP processing and increases Aβ production.
Materials and Methods
Biological and chemical compounds
DMEM, poly-d-lysine, l-glutamine, cytosine β-d-arabino-furanoside (AraC), and protease and phosphatase inhibitor cocktails (P8340 and P2850) were from Sigma.
NMDA, (-)bicuculline methochloride (Bic), 4-aminopyridine (4-AP), (+)5-methyl-10, 11-dihydro-5H-dibenzo(a,d)cyclohepten-5,10-imine maleate (MK-801), d-2-amino-5-phosphonopentanoic acid (AP-5), CNQX, nimodipine, tetrodotoxin (TTX), BAPTA-acetoxymethyl ester (AM), STO-609 acetate, and memantine hydrochloride were purchased from Tocris Bioscience. KN-93 was from Merck Chemicals. Laminin, horse serum, and fetal bovine serum (FBS) were from Invitrogen. Mouse anti-amyloid-β precursor protein, KPI domain monoclonal antibody (MAB5354) and the monoclonal mouse antibody 22C11 (MAB348) were purchased from Millipore; rabbit anti-actin antibody and horseradish peroxidase-conjugated secondary antibodies were from Sigma.
Animals
Male Swiss mice of 6 weeks (25–30 g; Janvier) were housed in a temperature-controlled room on a 12 h light/dark cycle with food and water ad libitum. Experiments with animals were performed in accordance with French ethical laws (act no. 87–848; Ministère de l'Agriculture et de la Forêt) and European Communities Council Directives of November 24, 1986 (86/609/EEC) guidelines for the care and use of laboratory animals.
Primary cultures of cortical neurons
Mouse cortical cultures of neurons were prepared from 14- to 15-d-old embryos as described previously (Rose et al., 1993). Cerebral cortices were dissected, dissociated, and cultured on 24-well plates coated with 0.1 mg/ml poly-d-lysine (Sigma) and 0.02 mg/ml laminin (Invitrogen) in DMEM (Sigma) containing 5% FBS, 5% horse serum, and 2 mm glutamine (all from Sigma). Cultures were kept at 37°C in a humidified atmosphere containing 5% CO2. After 3–4 d in vitro (DIV), AraC (10 μm) was added to inhibit proliferation of non-neuronal cells. Treatments were performed on neuronal cultures after 12 DIV. Under these conditions, ∼98% of the cells were considered as neuronal, as judged by microtubule-associated protein-2 staining.
Neuronal culture treatments
Synaptic NMDAR activation (“synaptic protocol”).
At DIV 12, culture media were replaced with serum-free DMEM supplemented with 10 μm glycine. The day after, neurons were exposed to fresh 50 μm Bic and 2.5 mm 4-AP. The treatment was stopped after 1, 3, or 6 h for RNA analysis and after 12 or 24 h for protein analysis. Non-NMDAR contributions in Bic/4-AP treatment were studied by adding in the medium 10 μm CNQX as an AMPA/kainate receptor antagonist or 10 μm nimodipine as a voltage-sensitive calcium channel (VSCC) blocker. Brain-derived neurotrophic factor (BDNF) mRNA expression was thereafter measured as an indication of synaptic activity.
Extrasynaptic NMDAR activation (“extrasynaptic protocol”).
One day after serum removal (DIV 12), cortical neurons were exposed for 2 min to 50 μm Bic/2.5 mm 4-AP and consecutively to Bic/4-AP with MK-801 (10 μm), an open NMDA channel blocker, for 3 min, to irreversibly block activated synaptic NMDAR. After three extensive washings to remove unbound MK-801, cultures were incubated at 37°C for 30 min before the addition of 30 μm NMDA. Cells were thereafter incubated for 1, 3, or 6 h for total RNA extraction, and 12 or 24 h for protein extraction.
Quantification of neuronal cell death: lactate dehydrogenase release
Primary cortical neurons cultured in 24-well plates for 10 or 12 DIV were treated for varying times with 30 μm NMDA or according to the synaptic or extrasynaptic protocol, as described above. At the end of each time treatment (1 to 6 h for RNA analysis; 12 and 24 h for protein analysis), 50 μl of neuronal culture medium was removed, and neurotoxicity was quantified by measuring the amount of lactate dehydrogenase (LDH) released by dead and dying cells. LDH activity was determined by measuring the decrease in absorbance at 340 nm corresponding to oxidation of nicotinamide adenine dinucleotide (NADH) in NAD+. The LDH level corresponding to 100% neuronal death was determined in associated cultures exposed to 0.1% Triton X-100 for 10 min. Background LDH release was measured in cultures subjected to control wash and was subtracted from the experimental values. Results were expressed as the percentage of neuronal cell death on a scale between 0 and 100%.
Quantification of neuronal cell apoptosis
To evaluate neuronal apoptosis triggered by selective NMDAR activation, we used a double-stain apoptosis kit containing Hoechst 33342, a blue-fluorescence dye, and propidium iodide (PI), a red-fluorescence dye, when bound to DNA (Ref. No. V13244, Invitrogen). Hoechst reagent stains the condensed chromatin in apoptotic cells brightly, and PI is only permeant to dead cells. The kit was used according to manufacturer's instructions.
Morphological analysis of GFP-labeled cortical neurons
Primary cortical neuron cultures (10 DIV) were transfected with enhanced green fluorescent protein (eGFP) coding plasmid using the calcium phosphate coprecipitation method. To prevent excitotoxic cell death, 1 h before transfection neurons were incubated in the presence of kynurenic acid, a reversible glutamate antagonist with a preferential action at the glycine site of the NMDAR. One microgram of eGFP was mixed with 120 mm calcium phosphate in HBS buffer (20 mm HEPES, 150 mm NaCl, 1.5 mm Na2HPO4, pH 7.4). After mild vortexing, the mixture was incubated at room temperature for 20 min before being added dropwise to neuron cultures. Cells were exposed to the DNA/Ca2+ phosphate suspension for 1 h at 37°C with 5% CO2 before being washed and cultured again in DMEM. Under these conditions, transfection efficiency is evaluated at 3 to 5%. After 48 h of incubation, neurons were exposed to synaptic or extrasynaptic protocols, as described before. Quantification of cell death and varicosity-containing cells was performed on the whole-glass coverslip after 12 and 24 h of treatment in four independent experiments using Nikon EZ-C1 software at a resolution of 512 × 512 pixels.
Calcium imaging
Cultures on glass-bottom dishes (No. 0, coverglass 0.085–0.13 mm; Mattek Corporation) were loaded for 45 min with 10 μm fura-2/AM (F-1201, Invitrogen) and 0.2% pluronic acid and incubated for an additional 15 min at room temperature in HEPES and bicarbonate buffer containing the following (in mm): 116 NaCl; 5.4 KCl; 1.8 CaCl2; 0.8 MgSO4; 1.3 NaH2PO4; 12 HEPES; 5.5 glucose; 25 bicarbonate; and 10 μm glycine at pH 7.45. Experiments were performed at room temperature with continuous perfusion at 2 ml/min with a peristaltic pump, on the stage of a Nikon Eclipse inverted microscope equipped with a quartz light pathway, a 100 W Xenon lamp, and oil-immersion objective (Nikon CFI 40× NA 1.3 optimized for fluorescence imaging). An intensified CCD camera (Coolsnap EZ) recorded the fluorescence emissions (>510 nm) evoked by excitation through narrow bandpass filters (340 ± 5 nm/380 ± 6.5 nm) housed in a computer-controlled filter wheel. Fluorescence images were acquired on a PC-based computer at a maximal time resolution of 2 s per successive measurement and using Metafluor 6.3 (Universal Imaging Corporation). Images were background subtracted using a frame taken at each excitation wavelength from an area of the coverslip devoid of neurons. Measurements were obtained simultaneously from 30 to 40 neurons per randomly selected field. Calibration of the Fura-2 signal was achieved by determining both the maximum 340/380 ratio and the minimum 340/380 ratio of Fura-2. The maximum Fura-2 ratio was achieved with saturating levels of Ca2+, and the minimum Fura-2 ratio was obtained in the presence of the Ca2+ chelator EGTA with no added Ca2+, both in the presence of ionomycin. Ca2+ concentrations measured at intermediate ratios were calculated according to the relationship described for Fura-2: [Ca2+]i = K d[(R − R min)/(R max − R)] F 0/F s, where R is the measured ratio of 340/380 fluorescence, R min is the ratio measured in a Ca2+-free solution, R max is the ratio measured in a saturated Ca2+ solution, K d = 135 nm (the dissociation constant for Fura-2), and F 0 and F s are the fluorescence intensities measured, respectively, in a Ca2+-free solution at 380 nm or in a saturated Ca2+ solution at 380 nm (Grynkiewicz et al., 1985). We assessed intracellular calcium increase by measuring the area under the curve for 2 min after the beginning of the stimulation. Due to the uncertainties inherent in calibration procedures, presented concentrations should be considered as “estimated.”
Extraction of total RNA
Total RNAs were extracted from cultured neurons by using the NucleoSpin RNA II kit from Macherey-Nagel. Cells were lysed in a buffer containing 1% β-mercaptoethanol. After filtration of the lysate on Nucleospin filter units, total RNAs were purified on Nucleospin RNA II columns according to the manufacturer's instruction and eluted with RNase-free water. Contaminating genomic DNA was removed by rDNase digestion directly on the column. RNA concentration and quality were checked on a NanoDrop ND-1000 spectrophotometer (Labtech).
Reverse transcription and real-time PCR
One microgram of total RNA from each sample was reverse-transcribed in cDNA at 37°C for 1 h using Moloney murine leukemia virus reverse transcriptase (200 U; Invitrogen) and 500 ng of poly(dT)12–18 oligonucleotides. Serial dilutions of the obtained cDNA were thereafter tested in amplification reactions with primer pairs designed for each gene. These preliminary amplifications allowed us to select the right cDNA dilution and to validate the primers. The specificity of the designed primers (Primer3, free available software or self-designed) was checked with the Blast database. The following primer sequences were used: APP695 (accession No. M18373), sense, 5′-GAT GAG GAT GTG GAG GAT GG-3′ (gene location 871–890), and antisense, 5′-GCT GCT GTC GTG GGA ACT C-3′ (gene location 1019–1001); APP770 (accession No. AY267348), sense, 5′-TGC TCT GAA CAA GCC GAG ACC-3′ (gene location 871–891), and antisense, 5′-CAT GCA GTA CTC TTC CGT GTC-3′ (gene location 1014–994); APP common part (AY267,348), sense, 5′-TGT GAT CTA CGA GCG CAT GAA CC-3′ (gene location 1605–1627), and antisense, 5′-AAG ACA TCG TCG GAG TAG TTC TGC-3′ (gene location 1730–1707); BDNF (accession No. NM_007,540), sense, 5′-GAT GCC GCA AAC ATG TCT ATG AGG-3′ (gene location 1019–1042), and antisense, 5′-ACT GTG ACC GTC CCG CCA-3′ (gene location 1164–1147); and cyclophilin as housekeeping gene (accession No. XM_913,899), sense, 5′-CAG GGT GGT GAC TTT ACA CGC-3′ (gene location 187–207), and antisense, 5′-TGT TTG GTC CAG CAT TTG CCA-3′ (gene location 319–299). Real-time PCR was performed on a Chromo 4 thermal cycler (Bio-Rad) using the SYBR Green containing MESA GREEN qPCR MasterMix Plus (Eurogentec). Twenty microliters of mix was added to 5 μl of diluted cDNA (1:50) in a 96-well plate with all negative controls. The amplification conditions were as follows: MeteorTaq activation, 95°C for 5 min; 40 cycles of PCR at 95°C for 15 s, and 60°C for 1 min. The expression level of interest gene was analyzed according to the ΔΔCt method (comparative Ct method) where Ct is the threshold cycle value, and cyclophilin the housekeeping gene. We previously checked that amplification efficiencies of both target gene and endogenous reference were approximately similar by looking at how ΔCt varies with template dilutions. Results were expressed using the following formula:
Immunoblotting
At the indicated times, neuronal cultures were harvested on ice in radioimmunoprecipitation assay (RIPA) buffer (Sigma) in the presence of a protease inhibitor mixture (1:100; Sigma). Cell lysates were centrifuged for 10 min (13,000 × g) at 4°C. Proteins in the supernatants were quantified using BCA protein assay (ThermoFisher Scientific) before being resolved on a 10% or gradient 4–12% NuPAGE Bis-Tris polyacrylamide gel (Invitrogen) in denaturing conditions. Proteins were transferred to a polyvinylidene difluoride membrane (PerkinElmer). Membranes were blocked with Tris-buffered saline (10 mm Tris, 200 mm NaCl, pH 7.4) containing 0.05% Tween-20, 5% nonfat dry milk for 1 h at room temperature. Blots were then incubated overnight at 4°C with the anti-amyloid-β precursor protein, KPI domain monoclonal antibody (1:1000) or 22C11 monoclonal antibody (Millipore Bioscience Research Reagents). Incubation with the secondary peroxidase-conjugated anti-mouse antibody (1:10,000) (Sigma) was performed for 1 h at room temperature. Specific proteins were visualized with an enhanced chemiluminescence ECL-Plus detection system (PerkinElmer). Blots were thereafter incubated in a stripping buffer before to be reprobed with an anti-actin antibody (1:2000) (Sigma) and revealed as described above.
Fluorescent immunocytochemistry
To detect neuronal KPI-APP proteins, primary neuron cultures were gently washed with PBS and fixed for 20 min with an ice-cold solution containing 4% paraformaldehyde and 4% sucrose. After three washings, cells were incubated consecutively in PBS containing 3% BSA and 0.1% Triton X-100 followed by an overnight incubation at 4°C with anti MAP-2 antibody (1/8000) and anti KPI-APP antibody (1/500). After three washings in PBS, neurons were incubated for 1 h at room temperature with the Alexa Fluor 488-conjugated anti-mouse (1/500) and Alexa Fluor 555-conjugated anti-chicken (1/500) (Invitrogen). Cultures were observed on a Nikon Eclipse (TE2000-E) inverted C1 confocal microscope equipped with an oil-immersion Nikon 60× objective, and images were acquired with Nikon EZC1 software.
CaM Kinase IV (CaMK IV) gene silencing
To determine transfection efficiency, we used the nonsilencing FAM-conjugated small interfering RNA (siRNA) sequence 5′-AAU UCU CCG AAC GUG UCA CGU-3′ (sense strand), 5′-ACG UGA CAC GUU CGG AGA AUU-3′ (antisense strand), with a 5′ fluorescein tag on the sense strand. Primary neuron cultures were transfected with this labeled siRNA (20 nm), as described below. Forty-eight hours later, neuron nuclei were counterstained with Hoechst 33342, and transfection efficiency was evaluated by counting under a fluorescence microscope.
Two pairs of siRNAs of 21 nt were used to target CaMK IV and were purchased from Sigma using the Mission siRNA program. The siRNAs were designed by Rosetta Inpharmatics and the sequences are as follows: forward, 5′-GUU GCU UAC CUG CAU GAA A(dT) (dT)-3′, reverse, 5′-UUU CAU GCA GGU AAG CAA C(dT) (dT)-3′ for the first pair; and forward, 5′-GGA GAA AGA UGC AGG UGU A(dT) (dT)-3′, reverse, 5′-UAC ACC UGC AUC UUU CUC C(dT) (dT)-3′ for the second pair. The two pairs target 795 nt and 1237 nt downstream from the start codon, respectively. Preliminary experiments showed that two pairs of siRNAs were much more potent than one pair to silence CaMK IV gene expression. A mixture of scrambled nonsilencing siRNA duplexes was used as negative control [forward, 5′-AAA UGG GUG GAG CUC UUG A(dT) (dT)-3′; reverse, 5′-UCA AGA GCU CCA CCC AUU U(dT) (dT)-3′]. Cortical neurons were cultured in 24-well plates for 9 d, and culture medium was replaced with serum-free DMEM. On the following day, cells were transfected in 500 μl of serum-free DMEM with 20 nm total siRNA (10 nm each) for CaMKIV or 20 nm scrambled control sequences using INTERFERin siRNA Transfection System (409-05; Polyplus Transfection). After 4 h at 37°C with 5% CO2, transfection medium was removed and neuron cultures were gently washed three times with serum-free medium. Neuron cultures were then incubated at 37°C for 48 h before specific treatments (synaptic or extrasynaptic NMDAR activation). At this time, the percentage of CamK IV gene knockdown was checked by real-time PCR and immunoblotting analysis.
Aβ(1-42) ELISA
Aβ(1-42) peptide release in the harvested conditioned media of cultured neurons was evaluated by ELISA (SIG-38952 BetaMark; Covance). This ELISA kit specifically detects rodent Aβ(1-42) with negligible cross-reactivity to Aβ(1-40). After indicated treatments, media were harvested in the presence of a protease inhibitor mixture, and were finally desalted and concentrated 25-fold using Microcon columns (Amicon; Millipore) with a nominal molecular weight limit of 3 kDa (YM-3). Aβ concentrations in samples were read in triplicate in a 96-well plate against a standard curve (0–250 pg/ml). Chemiluminescence signals were detected using a microplate reader (Sciencetec).
In vivo analysis of cortical KPI-APP expression pattern
Six-week-old male Swiss mice received an intraperitoneal injection of saline vehicle or memantine (1 or 30 mg/kg). Thirty minutes after this first injection, animals received a second intraperitoneal injection of PBS, pH 7.4, or NMDA at 120 mg/kg. After 6 h (for RNA extraction) or 24 h (for protein extraction), mice were deeply anesthetized with isoflurane 5% and perfused intraventricularly with 0.9% NaCl. The cortices were then rapidly collected and snap-frozen in liquid nitrogen. Samples were kept at −80°C until further processing. Total RNA was isolated by acidic phenol/chloroform extraction (Trizol, Invitrogen), and total proteins were extracted as described before.
Statistical analysis
Results are expressed as the mean ± SD from at least three independent biological samples. Statistical analyses were performed with Statview software (Abacus) by one-way ANOVA followed by Bonferroni–Dunn test or Student's t test.
Results
Characterization of calcium response induced by prolonged synaptic or extrasynaptic NMDAR stimulation
To study the implication of NMDAR activation on APP expression, we performed our experiments on murine cortical neuron cultures at DIV 12–14 and treated them with 10 μm AraC to prevent glial proliferation. We previously demonstrated that the time of culture (number of days) is a critical parameter directly affecting neuronal maturation, especially dendritic spine density and synaptic-mediated calcium response (Léveillé et al., 2008). To selectively activate synaptic or extrasynaptic NMDARs for long-lasting periods, we used protocols validated on both hippocampal (Tovar and Westbrook, 1999; Hardingham et al., 2002) and cortical (Léveillé et al., 2008) neurons. Intracellular calcium increase can be considered as an indirect measure of NMDAR activity as we showed that calcium response mainly relies on NMDAR current. By using CNQX (10 μm) or nimodipine (10 μm), we estimated the involvement of non-NMDAR response in the synaptic protocol. We found that VSCCs and AMPAR/kainate receptors were involved at 10 and 18%, respectively, in the Bic/4-AP-induced calcium response (data not shown). These results confirm previous reported data (Sattler et al., 1998; Léveillé et al., 2008). Selective synaptic NMDAR stimulation was achieved by blocking GABAA receptor function with 50 μm Bic in the presence of 4-AP (2.5 mm), a weak potassium channel blocker. For extrasynaptic NMDAR stimulation, we first evaluated calcium response evoked by successive bath application of increasing concentrations of NMDA (from 15 to 200 μm) (Fig. 1 A) after having blocked activated synaptic NMDAR with MK-801, an open channel blocker. To this aim, we measured the area under the curve for 2 min of treatment with each dose of NMDA. We showed a linear relationship between calcium increase for 2 min and NMDA concentration (Fig. 1 A,B). Efficiency of the blockade was controlled at the end of the experiment by the absence of Ca2+ response under Bic/4-AP treatment in the presence of CNQX (Fig. 1 A). This preliminary experiment allowed us to select an NMDA concentration (30 μm) leading to much lower intracellular calcium entry, when compared with that measured after a 2 min Bic/4-AP application (Fig. 1 B). Exposure of neurons to Bic/4-AP for 1 h produced continuous bursts of action potentials with transient intracellular Ca2+ increases (Fig. 1 C). We also checked that calcium entry in cells was fully blocked by TTX (0.5 μm), a sodium channel blocker (data not shown). After blocking, we thereafter stimulated extrasynaptic NMDAR for 1 h, and we monitored calcium response during this period (Fig. 1 D). We observed that the calcium entry profile was quite different from that obtained after Bic/4-AP exposure. NMDA (30 μm) application provokes a rapid initial increase in [Ca2+]i followed by a progressive return to a lower amount of Ca2+ entry. Measurement of the area under the curve in a time period of 1 h revealed a much lower intracellular Ca2+ accumulation (−54%; p < 0.01) evoked by extrasynaptic NMDAR stimulation when compared with Bic/4-AP treatment (Fig. 1 E). It should be mentioned that the NMDAR-induced calcium increase measured in either synaptic or extrasynaptic subpopulations may involve the release of calcium from intracellular pools in endoplasmic reticulum and/or mitochondria.
Figure 1.
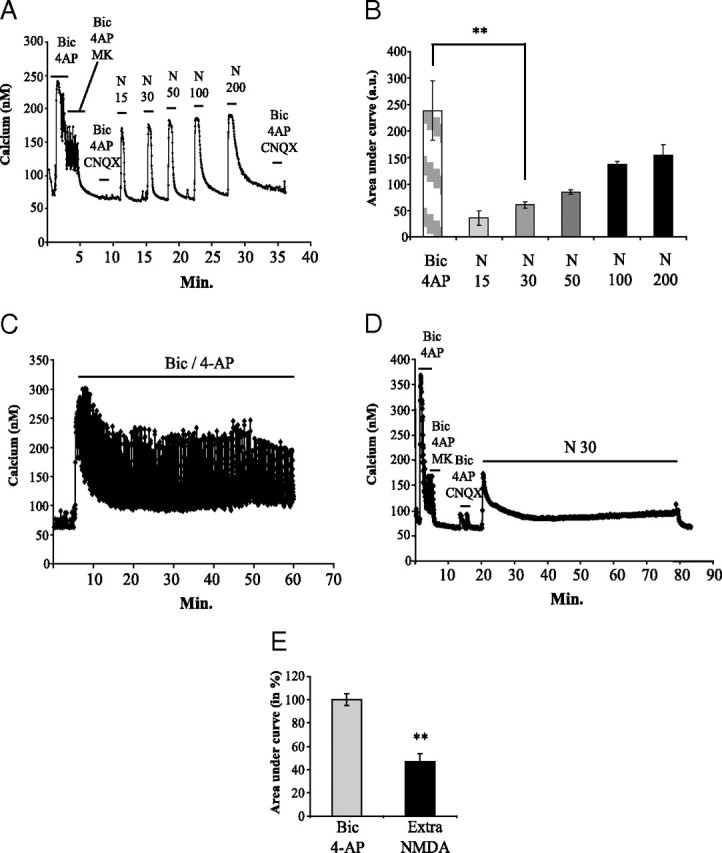
Characterization of prolonged synaptic and extrasynaptic NMDAR activation in cortical neuron cultures at DIV12. Intracellular Ca2+ concentrations were monitored by Fura-2 calcium imaging. A, Cortical neurons were exposed for 2 min to Bic/4-AP and consecutively to Bic/4-AP with MK-801 (10 μm), an open channel blocker, for 3 min to irreversibly block activated synaptic NMDAR. Efficiency of NMDAR blockade was controlled by the absence of Ca2+ response under Bic/4-AP treatment in the presence of CNQX (10 μm), to avoid any AMPA receptor-dependent response. After washing, a dose ranging of NMDA (from 15 to 200 μm for 2 min each) was applied to the culture, and Ca2+ entry in neurons was monitored. Blockade of synaptic NMDAR was finally checked by applying Bic/4-AP + CNQX. B, Calcium responses after Bic/4-AP or increasing doses of NMDA application were quantified by measuring area under curve and represented in histograms (n = 3; n = 137; **p < 0.01). C, Cortical neuron cultures were exposed for 1 h to 50 μm Bic/2.5 mm 4-AP, and cellular Ca2+ entry was monitored using Fura-2. D, After blockade of synaptic response as described in A, cortical neuron cultures were treated for 1 h with 30 μm NMDA to selectively activate extrasynaptic NMDAR. Calcium response was monitored using Fura-2. E, Intracellular calcium concentration was quantified by measuring the area under the curve during the whole treatment duration (1 h for both treatments) (C, D). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (Bic/4-AP: n = 3; n = 99; 30 μm NMDA after MK801 blockade: n = 3; n = 135; **p < 0.01).
Effect of a selective and prolonged NMDAR activation on neuronal fate
To study the modifications of APP expression at transcriptional and translational levels, we performed long-lasting treatments on primary cortical neuron cultures (up to 24 h for protein analysis). First, we evaluated the neurotoxic impact triggered by prolonged NMDAR activation on cortical neuron cultures. To this end, we measured LDH release in the bath medium at 12 and 24 h after the onset of Bic/4-AP or extrasynaptic NMDAR stimulation protocol. We found no significant increase in LDH release after Bic/4-AP exposure (synaptic protocol), at 12 or 24 h (Fig. 2 A). This result is in accordance with previous published data revealing that Ca2+ overload through synaptic NMDARs is not neurotoxic (Léveillé et al., 2008). In the same way, we performed morphological analysis of eGFP-expressing neurons exposed to selective stimulation of synaptic NMDAR (Fig. 2 B). We quantified the relative abundance of varicosity-containing neurons, dead and living neurons transfected with an eGFP construct. We observed no modification of neuronal morphology at 12 and 24 h after Bic/4-AP exposure (Fig. 2 C). In contrast, selective activation of extrasynaptic NMDAR was shown to induce characteristic features of excitotoxicity (presence of varicosities, apoptosis), triggering neuronal death (Léveillé et al., 2008). We therefore chose a concentration of NMDA (30 μm) that never induced >25% neuronal death after 12 or 24 h of treatment. This toxic effect was fully reversed by the use of MK-801, indicating the involvement of extrasynaptic NMDAR (Fig. 2 A). Morphological analysis revealed that 23% of transfected neurons exposed to extrasynaptic protocol for 24 h were dead (Fig. 2 C, right) and 22% of cells contained dendritic varicosities (Fig. 2 B, right, arrows), whereas 55% of transfected cells were unaffected (68% after 12 h of treatment) (Fig. 2 C, left).
Figure 2.

Effect of selective and prolonged activation of NMDAR on cellular morphology and neuronal death. A, Primary cultured cortical neurons at DIV 12 were subjected to synaptic (50 μm Bic + 2.5 mm 4-AP) or extrasynaptic (50 μm Bic + 2.5 mm 4-AP + 10 μm MK801 followed by 30 μm NMDA application) NMDAR activation for 12 and 24 h. The level of LDH released in the neuronal cell culture medium was determined by measuring the decrease in absorbance at 340 nm resulting from the oxidation of NADH. Results are expressed as mean ± SD from five independent experiments. Histograms represent neuronal cell death normalized to maximum neuronal death (0.1% Triton X-100 for 10 min). Statistical analysis was performed by ANOVA followed by Bonferroni-Dunn's test. n = 5; **p < 0.01 vs corresponding control; ##p < 0.01 vs extrasynaptic NMDAR stimulation. B, Morphological analysis of eGFP-transfected neuronal cultures. eGFP-labeled cortical neurons (DIV 12; transfection efficiency of 3 to 6%) were treated according to the synaptic protocol or the extrasynaptic protocol described in A for 12 or 24 h. Images represent the projection of z-stack images acquired by confocal microscopy at the indicated times and are representative of the cultures. Scale bar, 20 μm. Arrows are pointing to typical dendritic varicosities. C, Quantification of living cells, dead cells, and varicosity-containing cells in cultures exposed or not to synaptic or extrasynaptic treatment during the indicated times (12 or 24 h). Quantification of the three cell populations was achieved on four independent cultures by counting eGFP-transfected neurons on the overall coverslip under confocal microscope. Histograms represent means ± SD, and statistical analysis was performed by ANOVA followed by Bonferroni–Dunn's test (n = 4; *p < 0.05). D, Quantification of living cells, necrotic cells, and apoptotic cells in cultures exposed or not to synaptic or extrasynaptic protocol during the indicated times (12 or 24 h). Quantification of the three cell populations was achieved on six independent cultures using a double-stain apoptosis detection kit containing the blue-fluorescent Hoechst 33342 dye and the red-fluorescent PI dye, which is permeant only to dead cells. Counting of living, apoptotic, and necrotic cell populations was performed under a fluorescence microscope. Histograms represent means ± SD and statistical analysis was performed by ANOVA followed by Bonferroni–Dunn's test (n = 6; *p < 0.05). E, The photo is an illustration of apoptotic and living cells stained with Hoechst 33342. Arrows are pointing to typical apoptotic cells exhibiting apoptotic bodies. Scale bar, 20 μm.
Next, by the use of a double-stain apoptosis kit, we evaluated the potential impact of both synaptic and extrasynaptic protocols in triggering neuron apoptosis. We found a nonsignificant increase in apoptosis at 12 and 24 h, whatever the protocol applied (Fig. 2 D,E).
Synaptic NMDAR activation with Bic/4-AP downregulates neuronal APP695 mRNA expression without inducing APP770/751 mRNA or protein expression
Calcium entry through synaptic NMDAR, but not extrasynaptic, was shown to induce CREB activity and BDNF mRNA expression (Hardingham et al., 2002). Thus, by real-time PCR analysis, we first checked the consequences of Bic/4-AP treatment on neuronal BDNF mRNA expression. We observed a significant increase in BDNF mRNA expression after 1 h (∼2-fold), 3 h (∼4-fold), and 6 h (∼5.5-fold) of Bic/4-AP application (Fig. 3 A). This increase was blocked by the coapplication of the competitive NMDAR antagonist AP-5 (100 μm). In contrast, coincubation with the L-type Ca2+ channel blocker nimodipine (10 μm) or with the AMPA receptor antagonist CNQX (10 μm) failed to inhibit BDNF induction. These results demonstrate that calcium influx through NMDAR is involved in the increase in BDNF mRNA expression triggered by Bic/4-AP. Concerning BDNF mRNA expression after extrasynaptic protocol, we observed a trend toward decreasing at 1, 3, and 6 h. However, the decrease in BDNF mRNA expression never reached statistical significance.
Figure 3.
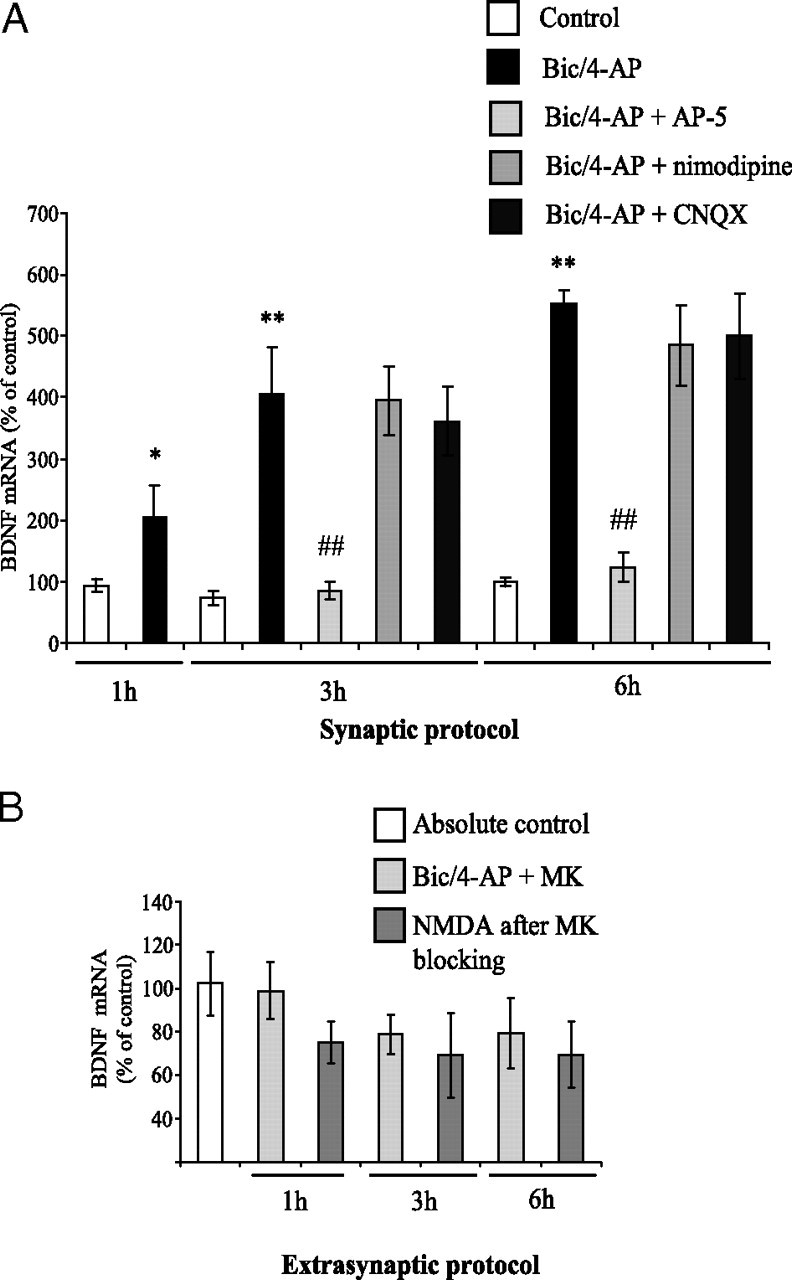
Synaptic, but not extrasynaptic, NMDAR activation induces BDNF mRNA expression in cortical neuron cultures. A, Primary cultured cortical neurons at 12 DIV were treated with 50 μm Bic + 2.5 mm 4-AP for 1, 3, or 6 h (synaptic protocol). Specificity of NMDAR in the effects of Bic/4-AP application was checked using AP-5 and nimodipine (to block VSCC), and CNQX (to block AMPA receptors). At the end of each time point, total RNA was extracted from neuron cultures and reverse transcribed in cDNA. Real-time PCR analysis was performed to quantify relative expression of BDNF mRNA in the different samples. The expression level of interest gene was analyzed according to the ΔΔCt method (comparative Ct method), where Ct is the threshold cycle value and cyclophilin the housekeeping gene. Histograms represent means ± SD, and statistical analysis was performed by ANOVA followed by Bonferroni-Dunn's test (n = 4; *p < 0.05, **p < 0.01 vs control; ##p < 0.05 vs Bic/4-AP treatment). B, Extrasynaptic NMDAR activation was performed by blocking activated NMDAR with 10 μm MK-801 for 3 min. After three extensive washings with culture medium, neurons were incubated at 37°C for 1 h before the addition of 30 μm NMDA for 1, 3, and 6 h. BDNF mRNA expression was measured as described above. Results are expressed as mean ± SD from five independent treatments.
Whereas APP770 and APP751 (KPI-APPs) are mainly expressed in the brain by astrocytes and microglia, APP695 (lacking the KPI domain) is considered as the main neuronal form of APP (LeBlanc et al., 1991). We and others previously showed that acute brain injuries (stroke and trauma) induced a shift in neuronal APP mRNA expression in favor of amyloidogenic KPI-APP expression (Kim et al., 1998; Lesné et al., 2005). This effect was linked to a calcium (Ca2+) influx resulting from a sublethal activation of NMDARs. Here, we therefore evaluated the effects of a selective long-lasting activation of synaptic NMDAR on APP expression pattern. We showed that synaptic NMDAR activation with Bic/4-AP downregulated APP695 mRNA expression (−50% at 1 h; ∼−70% at 3 and 6 h) (Fig. 4 A). However, this was not accompanied by an increase in KPI-APP mRNA expression (Fig. 4 B). Coincubation of neuron cultures with nimodipine (10 μm) or CNQX (10 μm) did not significantly modify Bic/4-AP-induced APP695 mRNA decrease. This shows that AMPA/kainate receptors and VSCCs are not involved in the modulations of APP695 mRNA expression. In the same way, Western blot analysis using a specific antibody directed against the KPI domain of APP revealed no induction of APP770/APP751 isoforms at the protein level (Fig. 4 E).
Figure 4.
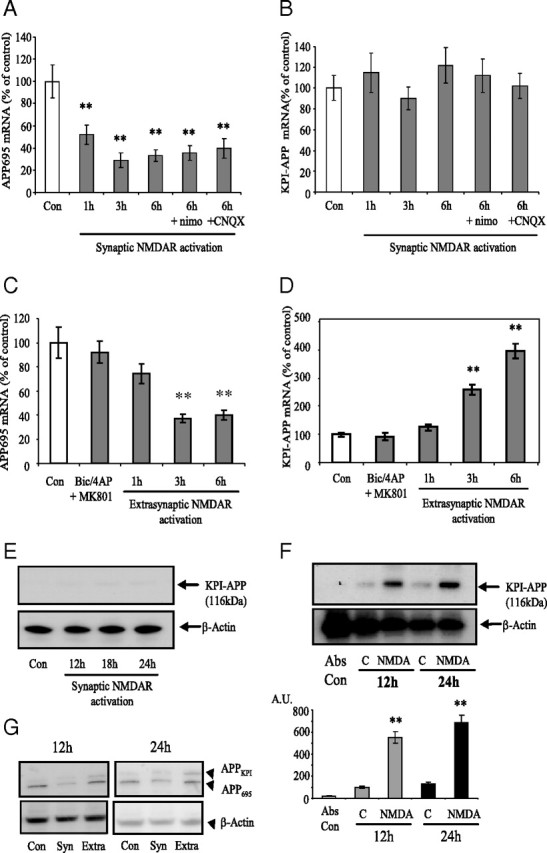
Synaptic and extrasynaptic NMDAR activity differently modulates APP expression in primary cultured neurons. A, Primary cultured cortical neurons at 12DIV were treated with 50 μm Bic + 2.5 mm 4-AP (synaptic protocol) for 1 h, 3, or 6 h. Real-time PCR analysis was performed to quantify relative expression of APP695 mRNA in the different samples. The expression level of interest gene was analyzed according to the ΔΔCt method (comparative Ct method) where Ct is the threshold cycle value and cyclophilin the housekeeping gene. Results are expressed as mean ± SD from five independent treatments. APP mRNA expression in control samples has been arbitrarily set at 100%. Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 5; **p < 0.01 to control). B, Neurons at DIV 12 were treated as before (in A), and real-time PCR analysis was performed to quantify relative expression of KPI-APP mRNA in the different samples with expression in control set at 100%. Results are expressed as mean ± SD from five independent treatments. Statistical analysis was realized by ANOVA followed by Bonferroni-Dunn's test (n = 5). C, Neurons at DIV 12 were exposed to 50 μm Bic/2.5 mm 4-AP for 2 min, and consecutively to Bic/4-AP with 10 μm MK801 for 3 min. After three extensive washings, cultures were incubated at 37°C for 1 h before the addition of 30 μm NMDA for 1, 3, and 6 h (extrasynaptic protocol). Total RNA was extracted, and real-time PCR analysis was performed to quantify relative expression of APP695 mRNA with 100% level arbitrarily set in the control. Statistical analysis was realized by ANOVA followed by Bonferroni-Dunn's test (n = 5; **p < 0.01 to control). D, KPI-APP mRNA expression was measured in cultured neurons exposed to extrasynaptic protocol, and results were analyzed as described in C (n = 5; **p < 0.01 to control). E, Representative immunoblot of KPI-APP after selective activation of synaptic NMDAR. Primary cultured cortical neurons at DIV12 were washed two times with serum-free DMEM before being treated with 50 μm Bic + 2.5 mm 4-AP for 12, 18, and 24 h (synaptic protocol). At each time point, cells were lysed in RIPA buffer, and immunoblots were performed with anti KPI-APP antibody. Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. F, Top, Representative immunoblot of KPI-APP after selective activation of extrasynaptic NMDAR. Primary cultured cortical neurons at DIV 12 were exposed to 50 μm Bic/2.5 mm 4-AP for 2 min, and consecutively to Bic/4-AP with 10 μm MK801 for 3 min. After three extensive washings, cultures were incubated at 37°C for 1 h before the addition of 30 μm NMDA for 12 and 24 h (extrasynaptic protocol). Cell cultures were lysed in RIPA buffer, and immunoblots were performed with anti KPI-APP antibody. Absolute controls (Abs Con) were only subjected to washings with DMEM. Controls at 12 and 24 h (C) were exposed to Bic/4-AP for 2 min, and consecutively to Bic/4-AP with MK801 for 3 min without further NMDA application. Bottom, Relative expression of KPI-APP protein compared with actin from immunoblot presented in the top. Densitometric analysis of the protein bands was performed with ImageJ software. Each column is the mean ± SD from three immunoblots (n = 3). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 3; **p < 0.01 vs respective control). G, Representative immunoblot of KPI-APPs versus APP695 protein expression in neurons subjected to synaptic or extrasynaptic protocol for 24 h. APP isoforms were revealed using the 22C11 monoclonal antibody. Protein loading was normalized using β-actin immunodetection.
Extrasynaptic NMDAR activation induces a shift from neuronal APP695 to KPI-APP expression
Selective activation of extrasynaptic NMDAR was performed as described in Materials and Methods (extrasynaptic protocol). After blocking of activated synaptic NMDAR with MK-801, we added 30 μm NMDA to the culture medium for 1, 3, and 6 h. In contrast to the synaptic NMDAR activation, stimulation of the extrasynaptic pool of NMDAR did not induce any increase in BDNF mRNA expression (Fig. 3 B). This result is in accordance with previous published data (Hardingham et al., 2002) and contributes to validating the protocols used for the selective activation of these receptor pools. Real-time PCR analysis revealed a time-dependent decrease in APP695 mRNA expression, the neuronal form of APP, with a significant inhibition 3 h (−62%) and 6 h (−60%) after the addition of 30 μm NMDA (Fig. 4 C). By using KPI domain-specific primers, we showed that extrasynaptic NMDAR activation induced a significant increase in APP770/APP751 mRNA after 3 h (+160%) and 6 h (+300%) of treatment (Fig. 4 D). These results were confirmed at the protein level by Western blot analysis. We observed an important increase in KPI-APP protein expression at 12 and 24 h (5.4-fold and 6.9-fold induction, respectively) after NMDA exposure (Fig. 4 F). By using the 22C11 monoclonal antibody recognizing all isoforms of APP (KPI-APPs and APP695), we also observed a strong increase in KPI-APP protein, 12 and 24 h after extrasynaptic NMDAR activation. This increase was not detected upon synaptic protocol (Fig. 4 G). Moreover, treatment of neurons with Bic/4-AP importantly reduced APP695 immunoreactivity at 12 h (−72%; p < 0.01) and 24 h (−56%, p < 0.01), confirming results at the mRNA level. APP695 protein decrease was more limited but significant upon extrasynaptic protocol (−28%; p < 0.05). In summary, if selective activation of each population of NMDARs significantly modifies APP expression pattern, only extrasynaptic NMDAR activation triggered expression of KPI-APP isoforms in neurons.
The calcium-/calmodulin-dependent kinase axis is required for the induction of neuronal KPI-APP protein expression after extrasynaptic NMDAR stimulation
Since NMDARs are gating high levels of calcium influx and are mainly involved in the activation protocols used here, we evaluated the consequences of intracellular chelation of calcium on APP expression following an extrasynaptic NMDAR activation. To this end, we pretreated cortical neuron cultures with the membrane-permeant Ca2+ chelator BAPTA-AM (10 μm) 30 min before NMDA application for 24 h in the extrasynaptic NMDAR activation protocol. BAPTA-AM (10 μm) did not significantly modify neuronal death 24 h later. In these conditions, we found that the extrasynaptic NMDAR-dependent increase in neuronal KPI-APP protein expression was reduced by 75% (p < 0.01) (Fig. 5 A,B), suggesting a calcium-dependent control of KPI-APPs protein expression in neurons. This crucial role exerted by calcium in this process led us to evaluate the implication of a Ca2+-activated enzyme associated with the NMDAR: the calmodulin (CaM)-dependent kinase (CaMK). To this aim, we pretreated neurone cultures with a CaMK inhibitor (KN-93) 30 min before NMDA application for 24 h. Our data show that KN-93 (10 μm) significantly decreased KPI-APP protein expression induced by extrasynaptic NMDAR activation (−63%, p < 0.01) (Fig. 5 A,B). These results demonstrate the involvement of the CaM/CaMK axis in the shift toward KPI-APP expression induced by Ca2+ entry in neurons through extrasynaptic NMDAR. Originally, the pharmacological reagents KN-62 and KN-93 were thought to be specific for blocking CaMKII activity (Tokumitsu et al., 1990). However, it is now well accepted that both of these reagents also suppress Ca2+/CaM activation of members of the CaM-dependent protein kinase kinase (CaMKK) cascade (CaMKI and CaMKIV) (Wayman et al., 2008). Thus, in light of the lack of specificity of KN-93, we considered this reagent in the present work as a global inhibitor of CaMK, all of them being Ca2+-activated enzymes. We therefore cotreated extrasynaptic NMDAR-activated neurons with STO-609 (1 μm), a selective and cell-permeable pharmacological inhibitor of Ca2+/CaM-KK (Tokumitsu et al., 2002). STO-609 at 1 μm was shown to have no significant inhibitory effect on CaMKII. Thus, its use allowed us to determine the involvement of the primary substrates of CaMKK (CaMKI and CaMKIV). Figure 5, C and D, shows that STO-609 at 1 μm significantly inhibited KPI-APP protein expression induced by extrasynaptic NMDAR activation (−76% at 12 h; −89% at 24 h). These results suggest that, in the calcium pathway, CaMKI and/or CaMKIV might be involved in the observed effect.
Figure 5.
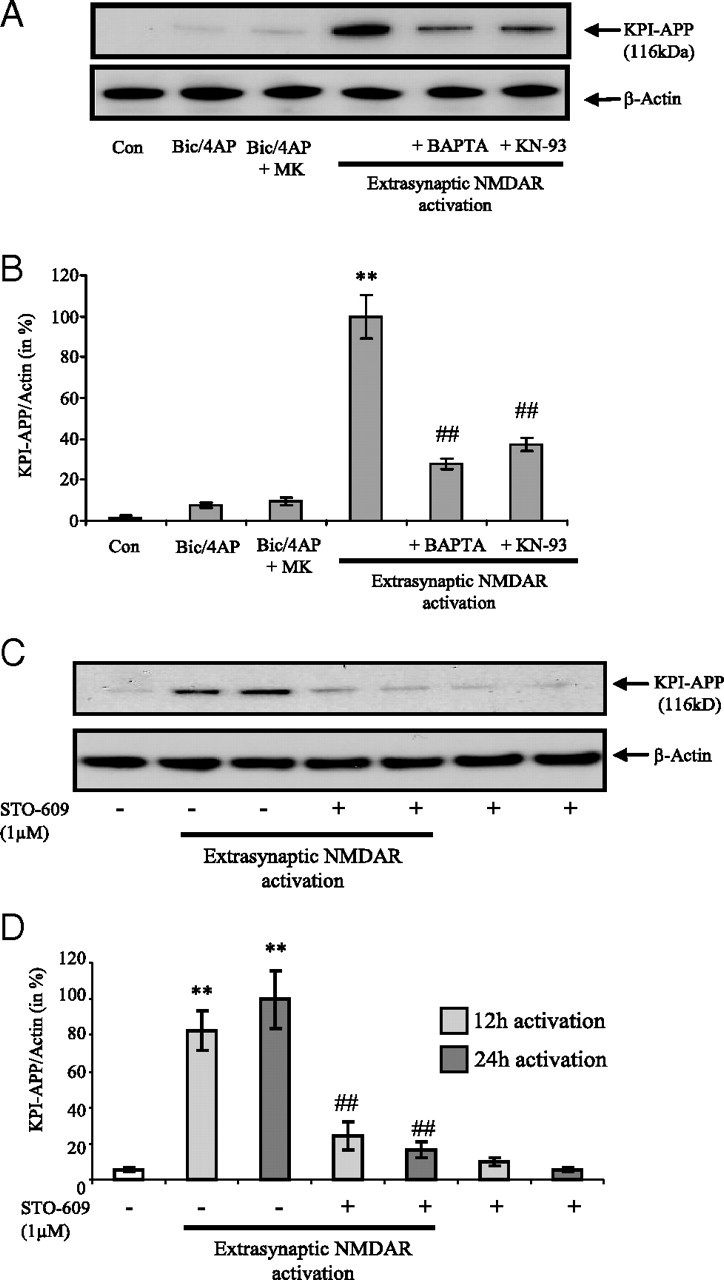
KPI-APP protein expression in cortical neurons exposed to extrasynaptic NMDAR activation is mediated by calcium signaling pathway. A, Immunoblotting analysis of KPI-APP protein in cortical neuron cultures 24 h after synaptic NMDAR activation with or without MK-801 blocking, or after extrasynaptic NMDAR activation (extrasynaptic protocol) in the presence or not of 10 μm BAPTA-AM, an intracellular calcium chelator, or 10 μm KN-93, an inhibitor of CaM kinases. Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. B, Relative expression of KPI-APP compared with actin from experiments presented in A. Densitometric analysis of the protein bands was performed with ImageJ software. Each column is the mean ± SD from three immunoblots (n = 3). Statistical analysis was realized by ANOVA followed by Bonferroni-Dunn's test (n = 3; **p < 0.01 vs control; ##p < 0.01 vs extrasynaptic NMDAR activation). C, Immunoblotting analysis of KPI-APP protein in cortical neuron cultures 12 and 24 h after extrasynaptic NMDAR activation in the presence or not of 1 μm STO-609, a selective Ca2+/calmodulin-dependent protein kinase kinase. Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. D, Relative expression of KPI-APP compared with actin from experiments presented in C. Densitometric analysis of the protein bands was performed with ImageJ software. Each column is the mean ± SD from three immunoblots (n = 3). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 3; **p < 0.01 vs respective control; ##p < 0.01 vs extrasynaptic NMDAR activation).
Extrasynaptic NMDAR activation regulates splicing of APP mRNA and modifies APP695/KPI-APP mRNA ratio in favor of KPI-APP, whereas synaptic activation decreases APP expression
The app gene contains 18 exons and is subjected to alternative pre-mRNA splicing that may give rise to several isoforms. The three main isoforms contain 695, 751, or 770 aa, respectively, and are termed APP695, APP751 and APP770. Our results showed that synaptic and extrasynaptic NMDAR activation modulated the expression of APP695 and KPI-containing APPs in different ways. Therefore, we investigated whether these modifications of APP isoforms abundance were due to changes in APP gene expression or to regulations at an mRNA splicing level. To this end, we designed a pair of primers directed against the common part of the three APP isoforms and performed real-time PCR analysis of the overall APP mRNA expression. Figure 6 A shows that synaptic NMDAR activation induced a significant decrease in total APP mRNA after Bic/4-AP application. This decrease was observed as soon as 1 h after the onset of the activation paradigm (−76%, p < 0.01) and remained pronounced at 3 h (−70%, p < 0.01) and at 6 h (−55%, p < 0.01) (Fig. 6 A).
Figure 6.
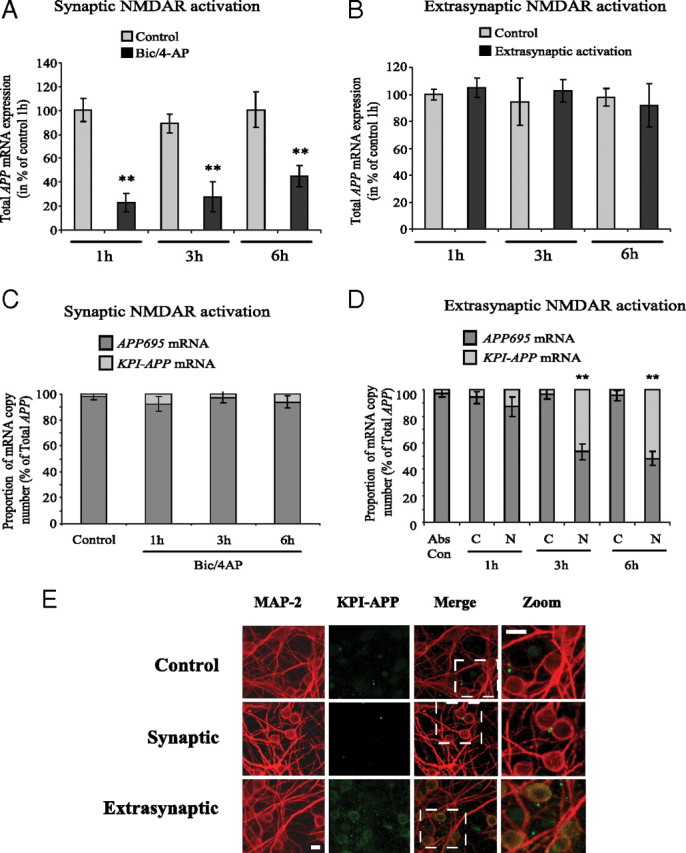
Synaptic NMDAR activation downregulates overall APP mRNA expression, whereas extrasynaptic NMDAR activation acts on APP isoforms mRNA ratio via an alternative splicing pathway. A, Real-time PCR analysis of total APP mRNA expression in cortical neurons exposed to Bic/4AP for 1, 3, or 6 h (synaptic protocol). A new pair of primers directed against the common part of the different APP isoforms was designed. At each time, APP mRNA expression in treated neurons was compared with its control. The expression level of APP was analyzed according to the ΔΔCt method (comparative Ct method) where Ct is the threshold cycle value and cyclophilin is the housekeeping gene. Results are representative of three independent experiments performed in triplicate. Statistical analysis was realized by ANOVA followed by Bonferroni-Dunn's test (n = 9; **p < 0.01 to respective control). B, Real-time PCR analysis of total APP mRNA expression in cortical neurons exposed to extrasynaptic NMDAR activation for 1, 3, or 6 h (extrasynaptic protocol). At each time, APP mRNA expression in treated neurons was compared with its control. Results are representative of three independent experiments performed in triplicate and expression level of APP was analyzed according to the ΔΔCt method. C, APP695/KPI-APP mRNA ratio in cortical neuron cultures treated with 50 μm Bic/2.5 mm 4-AP for 1, 3, and 6 h (synaptic protocol). Absolute quantification of APP695 and KPI-APP mRNA in each sample was performed by real-time PCR using known concentrations of DNA standard molecules (PCR products). Histograms represent the relative part of the two mRNA populations with the total of both mRNA arbitrarily set at 100%. D, APP695/KPI-APP mRNA ratio in cortical neuron cultures exposed to 50 μm Bic/2.5 mm 4-AP for 2 min, and consecutively to Bic/4-AP with 10 μm MK801 for 3 min. After three extensive washings, cultures were incubated at 37°C for 1 h before the addition of 30 μm NMDA for 1, 3, and 6 h (extrasynaptic protocol). Absolute quantification of APP695 and KPI-APP mRNA in each sample was performed by real-time PCR using known concentrations of DNA standard molecules (PCR products) as described in Materials and Methods. Histograms represent the relative part of the two mRNA populations with the total of both mRNA arbitrarily set at 100%. Statistical analysis was performed by ANOVA followed by Bonferroni-Dunn's test (n = 3; **p < 0.01 vs respective control). Abs Con, Absolute control; C, control (Bic/4-AP and MK-801 blocking); N, NMDA treatment after MK-801 blocking. E, Expression of KPI-APP isoforms is induced in neurons exposed to extrasynaptic but not to synaptic NMDAR activation. Immunocytochemical analysis of KPI-APP expression in cultured cortical neurons subjected to synaptic or extrasynaptic NMDAR activation for 24 h. Neurons were stained with an antibody raised against the neuronal marker MAP-2 (red), and KPI-APP protein expression was determined by using the monoclonal antibody directed against the KPI domain of APP (green). KPI-APP appeared colocalized (yellow) to neuronal cytosol and plasma membrane when images were overlaid. No expression of KPI-APP was detected in neurons exposed to Bic/4-AP for 24 h (synaptic protocol). Scale bars, 20 μm.
In contrast to synaptic treatment, no significant modification of total APP mRNA expression was measured after extrasynaptic NMDAR activation, whatever the time of NMDA application (1 to 6 h) after MK-801 blocking (Fig. 6 B). Overall, these results suggest that activation of both populations of NMDARs in cortical neuron cultures differently modulates APP mRNA processing. Synaptic NMDAR activation with Bic/4-AP downregulates total APP gene expression, whereas extrasynaptic NMDAR activation acts on APP mRNA splicing, giving rise to KPI-APP isoform expression.
To further evidence the specific effects of an extrasynaptic NMDAR activation on the abundance of APP isoforms, we used an absolute quantification strategy by real-time PCR. To this aim, we related the PCR signal to input copy number using a calibration curve based on known concentrations of DNA standard molecules, as detailed in Materials and Methods. For each gene, a linear regression was obtained between threshold values (Ct) and log (number of copies). As detailed in the legend of Figure 6, C and D, each column represents the relative part of both mRNA populations with the total arbitrarily set at 100%. This quantification assay showed that, in resting conditions, the APP695 mRNA isoform is predominant in primary cortical neuron cultures (Fig. 6 C). KPI-APP isoform represents only ∼2% of total APPs. This result is in accordance with previous published data arguing that KPI-containing APPs are weakly expressed in neuronal cells, whereas APP695 is abundant (LeBlanc et al., 1991). The detection of a low level of KPI-APP could be explained by the presence of astrocytes (2 to 5%) in our neuronal culture model, as determined previously (Rose et al., 1993).
Synaptic NMDAR activation for 1, 3, or 6 h did not significantly modify the APP695/KPI-APP ratio in neuron cultures, with KPI-APP isoforms remaining at <8% of total APPs (Fig. 6 C). These observations are in accordance with the results presented in Figure 4 B, showing no modification of KPI-APP mRNA expression after synaptic treatment.
By using the same absolute quantification method, we next evaluated the consequences of extrasynaptic NMDAR activation on neuronal APP695/KPI-APP mRNA ratio. Our results revealed significantly higher levels of KPI-APP mRNA, reaching 55% of total APP mRNA at 6 h (Fig. 6 D). These results corroborate the data presented previously in Figure 4, C and D, showing that a switch from APP695 to KPI-APP mRNA expression was detected 3 and 6 h after the onset of extrasynaptic NMDAR activation. Conversely, since KPI-APP is highly expressed in astrocytes, it is relevant to investigate cellular localization of KPI-APP expression following NMDAR activation. We performed immunocytochemical analysis to localize KPI-APP induction in primary cortical cultures. Neurons were treated for 24 h according to either synaptic or extrasynaptic protocol and incubated with an antibody raised against the neuronal marker MAP-2 (red), and a monoclonal antibody directed against the KPI domain of APP (green). We observed that a 24 h exposure to extrasynaptic protocol-induced KPI-APP immunoreactivity colocalized with MAP-2 immunostaining (Fig. 6 E). In contrast, no induction of neuronal KPI-APP was observed 24 h after synaptic NMDAR activation. We conclude that extrasynaptic, but not synaptic, NMDAR activation specifically induces neuronal KPI-APP expression.
CaMKIV, hnRNPA1, and SC-35 play a role in extrasynaptic NMDAR activation-induced KPI-APP expression in cortical neuron cultures
In neurons, many splicing events have been shown to be controlled by chronic depolarization or by treatments that stimulate calcium signaling pathways (Li et al., 2007). This mode of regulation affects very different neuronal transcripts of proteins such as those involved in synaptogenesis (neurexins and neuroligins), proteins involved in neurotransmitter release (SNAP25, syntaxin), PSD proteins (Homer 1), reeling receptor apolipoprotein E receptor 2 (APOER2), and especially ion channel receptor (e.g., NMDAR1, BK channel, calcium ATPase, GABA receptors) (An and Grabowski, 2007; Li et al., 2007). One of the major splicing mediators identified in neurons is CaMK IV, which is known to repress splicing through CaMK IV-responsive RNA elements located in the target exon. This mechanism has been well described for STREX exon of BK channel mRNA (Xie et al., 2005) and for E21 exon of NMDAR1 (Lee et al., 2007). Since we found that a splicing regulation pathway could be involved in extrasynaptic NMDAR-induced KPI-APP mRNA expression, we evaluated the role of CaMK IV in the retention of exon18 encoding for the KPI domain. To this end, we used a double siRNA transfection strategy to knock down CaMK IV 48 h before to perform extrasynaptic treatments on cortical neuron cultures for 1, 3, and 6 h (mRNA analysis) and 24 h (protein analysis). We first aimed to evaluate the siRNA transfection efficiency in primary cortical neuron cultures by using a nonsilencing 6-carboxyfluorescein (FAM)-conjugated siRNA (20 nm; see Materials and Methods). Counts indicated that 65.8 ± 4.4% of cells, identified by Hoechst staining, were transfected (n = 30; mean +/− SD) (Fig. 7 D). Preliminary experiments using only one siRNA directed against CaMKIV did not allow us to obtain sufficient gene knockdown (data not shown). Different conditions of siRNA concentration and time incubation were tested to find a good compromise between the absence of cell toxicity and gene knockdown. With the experimental conditions chosen (10 nm for each siRNA, 4 h of incubation), we obtained ∼70% of inhibition for CaMK IV mRNA expression (Fig. 7 A). This inhibition was confirmed at the protein level (Fig. 7 B). Real-time PCR analysis in neurons transfected with CaMK IV siRNAs showed that KPI-APP mRNA induction was 37.5% lower after 3 h of extrasynaptic treatment and 52% lower after 6 h of treatment, when compared with untransfected neuron cultures (Fig. 7 C). Western blot analysis on transfected neurons after extrasynaptic NMDAR activation also revealed a lower induction of KPI-APP protein expression (−73% at 12 h; −51% at 24 h) when compared with control neuron cultures (Fig. 7 E). These results strengthen the hypothesis of the involvement of CaMK IV in extrasynaptic NMDAR activation-induced KPI-APP mRNA expression through the regulation of alternative splicing of APP pre-mRNA. By siRNA knockdown experiments, it was recently shown that heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and SC35 are major factors involved in skipping of exon 7 (corresponding to KPI domain) of APP pre-mRNA (Donev et al., 2007). hnRNP proteins were initially described as a splicing repressor by directly antagonizing the recognition of splice sites (for review, see Martinez-Contreras et al., 2007). Conversely, a positive role for hnRNP proteins in pre-mRNA splicing was also described (Martinez-Contreras et al., 2006). In the study of Donev et al. (2007), this mechanism was investigated in transfected HEK293 cells. It was therefore interesting to check the modification of hnRNPA1 and SC35 mRNA expression in cortical neuron cultures, transfected or not with siRNA against CaMKIV, and submitted to extrasynaptic NMDAR activation. Figure 8, A and B, shows that hnRNPA1 and SC35 mRNA expression were significantly decreased in nontransfected primary neuron cultures after extrasynaptic NMDAR activation. This decrease reached 60% for both hnRNPA1 mRNA (at 6 h) and SC35 mRNA (at 1, 3, and 6 h). Conversely, no significant modification of expression was measured after synaptic NMDAR activation (Fig. 8 A,B). The same experiments performed on transfected neurons show that the decrease in hnRNPA1 and SC35 mRNA expression was reduced at all times of treatment with significant differences at 6 h for hnRNPA1 and at 3 and 6 h for SC35. These data suggest that CaMKIV regulated both splicing factors and confirmed their involvement in the skipping of exon 7 of the APP gene in cortical neurons. The reduced neuronal expression of these two splicing factors consecutive to extrasynaptic NMDAR stimulation could therefore contribute to the retention of exon 7 corresponding to the KPI domain of APP.
Figure 7.
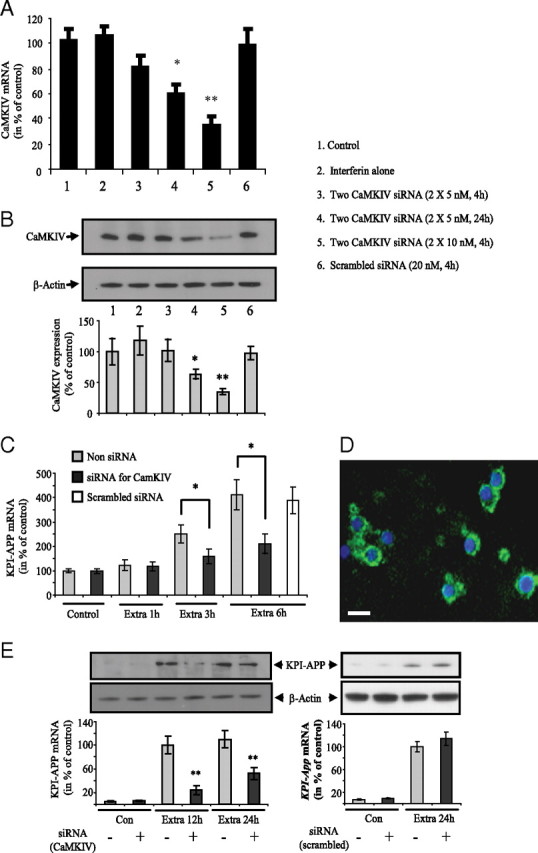
Knockdown of CaMKIV reduces extrasynaptic NMDAR activation-induced KPI-APP expression in cortical neuron cultures. A, Cortical neuron cultures at 9 DIV were transfected with a mixture of two siRNA directed against CaMK IV using INTERFERin as transfection reagent. Two concentrations of siRNA (5 and 10 nm each) and two incubation times with siRNA/INTERFERin complex (4 and 24 h) were tested. For the scramble siRNA control, a single concentration (20 nm) was used. CaMK IV mRNA expression was measured by real-time PCR 48 h after transfection. Each column is the mean ± SD of six independent transfections (n = 6), and statistical analysis was performed by ANOVA followed by Bonferroni–Dunn's test (*p < 0.05, **p < 0.01 vs control). B, Cortical neuron cultures at 9 DIV were transfected with a mixture of two siRNAs (10 nm each) directed against CaMK IV. Different conditions of transfection were tested as described in A. Forty-eight hours after transfection, CaMK IV protein expression was evaluated by Western blot analysis. Each column is the mean ± SD of three independent transfections (n = 3), and statistical analysis was performed by ANOVA followed by Bonferroni-Dunn's test (*p < 0.05, **p < 0.01 vs control). C, Following preliminary experiments (A, B), neurons at 9 DIV were transfected with two siRNAs directed against CaMKIV (10 nm for each siRNA, 4 h of incubation). After three washings, neuron cultures were incubated for 48 h at 37°C and exposed to extrasynaptic NMDAR activation for 1, 3, and 6 h (extrasynaptic protocol). For each time, extrasynaptic treatment was compared with its respective control (non-siRNA-transfected neuron cultures). For each end-point, KPI-APP mRNA expression was measured by real-time PCR as described before. Each column is the mean ± SD of six independent experiment (n = 6). Statistical analysis was performed by ANOVA followed by Bonferroni–Dunn's test (*p < 0.05 vs control). D, Transfection of 20 μm nonsilencing FAM-labeled siRNA (green) in cultured cortical neurons at DIV12. Cell nuclei were counterstained with Hoechst 33342 (blue). Scale bar, 25 μm. E, Neuron cultures at DIV9 were transfected with two siRNAs directed against CaMKIV (10 nm for each siRNA, 4 h of incubation) or with a scrambled siRNA (20 nm). Forty-eight hours later, the cultures were exposed to extrasynaptic NMDAR activation for 12 and 24 h. Proteins were extracted in RIPA buffer in the presence of protease inhibitors for Western blot analysis (top). Quantification of band intensities was performed with ImageJ software (bottom). Each column is the mean ± SD from three immunoblots (n = 3). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 3; **p < 0.01 vs control).
Figure 8.
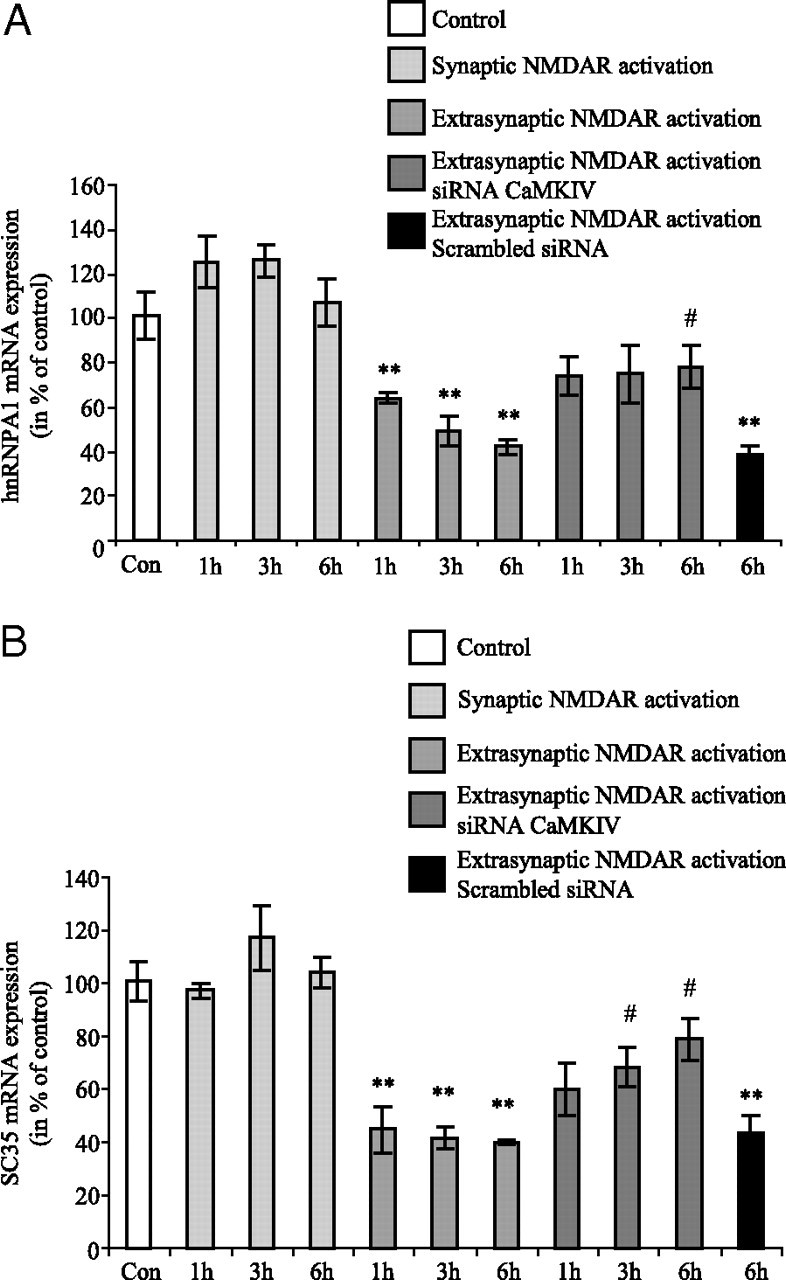
A, B, Extrasynaptic, but not synaptic, NMDAR activation downregulates the expression of two splicing factors, hnRNPA1 (A) and SC35 (B), at the mRNA level in cortical neuron cultures. Primary cultured cortical neurons at 12 DIV were treated according to the previously described synaptic protocol or extrasynaptic protocol for 1, 3, and 6 h. Extrasynaptic NMDAR activation was performed in neurons, transfected or not with siRNA directed against CaMKIV. At the end of each time point, total RNA was extracted from neurons and real-time PCR analysis was performed using primers specific for hnRNPA1 or SC35. Results are expressed as mean ± SD from three independent treatments with mRNA expression level in controls (Con) arbitrarily set at 100%. Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 3; **p < 0.01 to control; #p < 0.05 to respective time of extrasynaptic treatment).
Memantine dose-dependently modulates extrasynaptic NMDAR-induced KPI-APP protein expression in cortical neurons
Memantine is a low-affinity NMDAR antagonist that was clinically shown to counteract pathological consequences of NMDAR activation. This pharmacological compound was initially shown to preferentially block extrasynaptic NMDAR activation without disrupting synaptic activity, thus limiting neuronal injury (Chen et al., 1998). We and others presented further experimental data confirming these observations (Léveillé et al., 2008; Okamoto et al., 2009). It was therefore interesting to evaluate the ability of memantine to modulate extrasynaptic NMDAR-mediated KPI-APP protein expression. Figure 9, A and B, shows that both 7.5 μm NMDA bath application and extrasynaptic treatment (at 12 and 24 h), but not Bic/4-AP, induce KPI-APP protein expression. We observed that coapplication of memantine with 30 μm NMDA in the extrasynaptic protocol significantly inhibited KPI-APP expression at both mRNA and protein levels, when applied at 1 μm (−51% for mRNA; −59% for protein) and at 10 μm (−79% for mRNA; −88% for protein). In additional experiments, we also tested the effects of memantine, at the same concentrations, on 7.5 μm NMDA-treated neurons for 6 h (mRNA analysis) and 24 h (protein analysis). At 1 and 10 μm, we found the same levels of inhibition on KPI-APP mRNA and protein induction and no significant effect at 0.1 μm (data not shown). Together, these results demonstrate that memantine is efficient in decreasing the neuronal expression of amyloidogenic forms of APP induced by NMDA or extrasynaptic NMDAR activation. This open channel blocker may exert its effect by uncompetitive blockade of predominant NMDA subunits expressed in cortical neurons (NR2A and NR2B). However, we cannot rule out that some observed effect of memantine may rely on a blockade of NR2D receptors (Kotermanski and Johnson, 2009).
Figure 9.
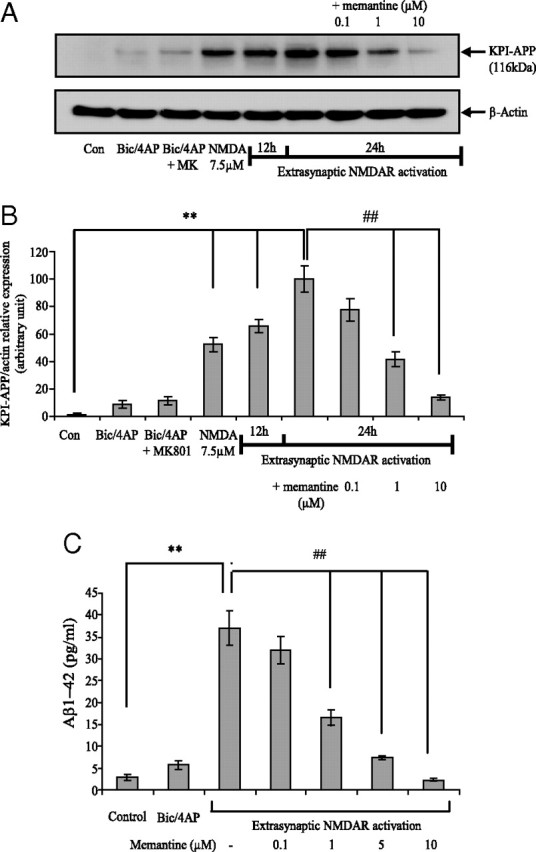
Memantine dose-dependently inhibits extrasynaptic NMDAR-induced KPI-APP protein expression and Aβ production from cortical neuron cultures. A, Immunoblotting analysis of KPI-APP protein in cortical neuron cultures exposed for 24 h to Bic/4-AP (synaptic protocol), to Bic/4AP + MK-801 followed by three washings, to 7.5 μm NMDA, and to extrasynaptic NMDAR activation for 12 or 24 h. For extrasynaptic activation, neuron cultures were incubated in the presence or not of increasing doses of memantine (0.1, 1, and 10 μm). Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. Con, Control. B, Relative quantification of KPI-APP protein expression compared with actin from experiments presented in A. Densitometric analysis of the protein bands was performed with ImageJ software. Each column is the mean ± SD from three immunoblots (n = 3). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 3; **p < 0.01 vs control; ##p < 0.01 vs extrasynaptic NMDAR activation). C, Primary cultured cortical neurons at 12 DIV were treated for 24 h with DMEM (control), with 50 μm Bic + 2.5 mm 4-AP (synaptic protocol) or according to extrasynaptic protocol in the presence of increasing concentrations of memantine. After 24 h, conditioned culture media were harvested in the presence of a protease inhibitor mixture and finally desalted and concentrated 25-fold using Microcon columns (Amicon; Millipore) with a nominal molecular weight limit of 3 kDa (YM-3). Media were processed for measurement of Aβ(1-42) levels using a mouse/rat Aβ(1-42) ELISA kit, as detailed in Materials and Methods. Each column is the mean ± SD of nine independent treatments (n = 9), and statistical analysis was performed by ANOVA followed by Bonferroni-Dunn's post-test (**p < 0.01 vs control; ##p < 0.01 vs extrasynaptic NMDAR activation).
Memantine inhibits neuronal Aβ production and release induced by extrasynaptic NMDAR activation
Our results showing that memantine inhibits extrasynaptic NMDAR-induced KPI-APP expression prompted us to evaluate its effect on neuronal Aβ production by measuring the level of Aβ(1-42) in the neuronal culture medium. To this end, we used an ELISA kit that specifically detects rodent Aβ(1-42) with negligible cross-reactivity to Aβ(1-40). To ensure that detected Aβ(1-42) is linked to the treatment, shortly before the onset of NMDAR activation, neuronal culture medium was replaced with fresh medium without serum. Aβ measurements were performed after 24 h of treatment. We first compared the amount of Aβ released after selective stimulation of each population of NMDAR. We showed that, if stimulation of synaptic NMDAR with Bic/4-AP for 24 h induced no significant modification of neuronal Aβ(1-42) secretion, extrasynaptic NMDAR activation caused an important increase in Aβ(1-42) release in the neuronal culture medium (from 4 to 74 pg/ml; p < 0.01) (Fig. 9 C). The coapplication of an irreversible NMDAR antagonist, MK-801 (10 μm), completely abolished Aβ(1-42) secretion (data not shown). This indicates the NMDAR specificity of the observed effects. Furthermore, we investigated the effects of different concentrations of memantine, ranging from 0.1 to 10 μm, on extrasynaptic NMDAR activation-induced Αβ secretion. We found a dose-dependent inhibition of neuronal Aβ release with memantine. We especially observed that at 1 μm, a clinically achievable concentration in the CSF, coincubation with memantine induced a 55% inhibition of Aβ release. In the presence of 10 μm memantine, a stronger inhibition rate was reached (93%).
Memantine inhibits cortical induction of KPI-APP expression in mice subjected to NMDA intraperitoneal injection
The adaptation of extrasynaptic protocol in an in vivo model is experimentally nonachievable. Several studies have described a rapid modification of cerebral gene expression (at both mRNA and protein levels) in mice subjected to intraperitoneal NMDA injection (Nagendra et al., 1997; Chung et al., 2000; Taniura et al., 2007). To validate our in vitro findings, we evaluated the expression of KPI-APP in cortex of NMDA-treated mice by a single intraperitoneal injection. Six hours after NMDA injection (120 mg/kg), we found a significant increase in KPI-APP mRNA expression (+ 120%; p < 0.05) (Fig. 10 A) confirmed at the protein level 24 h after NMDA administration (+ 97%; p < 0.05) (Fig. 10 B).
Figure 10.
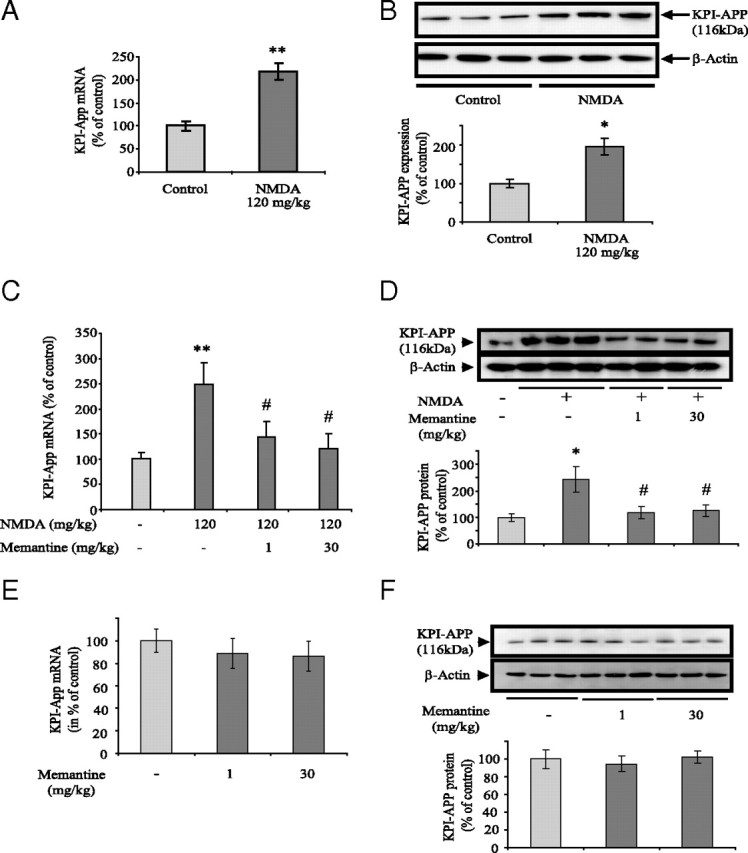
Memantine at low or high doses inhibits the increase in KPI-APP expression in cortex of mice injected intraperitoneally with NMDA. Swiss mice received an intraperitoneal injection of saline vehicle or memantine (1 or 30 mg/kg). Thirty minutes after this first injection, animals received a second intraperitoneal injection of PBS, pH 7.4, or NMDA in a dose of 120 mg/kg. Mice were exposed to this treatment for 6 h (for RNA extraction) or 24 h (for protein extraction). A, Real-time PCR analysis of KPI-APP mRNA expression in cortical tissue of mice injected with 120 mg/kg NMDA. Total RNA was isolated from cortices by acidic phenol/chloroform extraction before to be reverse transcribed in cDNA. The expression level of APP was analyzed according to the ΔΔCt method with cyclophilin as the housekeeping gene. Results are representative of six independent experiments. Statistical analysis was realized by ANOVA followed by Bonferroni-Dunn's test (n = 6; **p < 0.01 vs control). B, Immunoblotting analysis of KPI-APP protein expression in cortical tissue of mice injected or not with 120 mg/kg NMDA. Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. Relative expression of KPI-APP compared with actin is presented in histogram below the blot. Each column is the mean ± SD from six immunoblots (n = 6). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 6; *p < 0.05 vs control). C, Real-time PCR analysis of KPI-APP mRNA expression in cortical tissue of mice 6 h after 120 mg/kg NMDA injection, with or without memantine (1 or 30 mg/kg). (n = 6; **p < 0.01 vs control; # p < 0.05 vs NMDA alone). D, Immunoblotting analysis of KPI-APP protein expression in cortical tissue of NMDA-injected mice with or without memantine (1 or 30 mg/kg). Relative expression of KPI-APP normalized with actin is presented in histogram below the blot. Each column is the mean ± SD from six immunoblots (n = 6). Statistical analysis was realized by ANOVA followed by Bonferroni–Dunn's test (n = 6; *p < 0.05 vs control; #p < 0.05 vs NMDA alone). E, Real-time PCR analysis of KPI-APP mRNA expression in cortical tissue of mice 6 h after injection of saline or memantine alone (1 or 30 mg/kg). F, Top, Immunoblotting analysis of KPI-APP protein expression in cortical tissue of mice 24 h after injection of saline or memantine alone (1 or 30 mg/kg). Blots were rehybridized with an anti-actin antibody to estimate the total amount of proteins loaded. Bottom, Relative quantification of KPI-APP protein expression compared with actin from experiments presented above. Densitometric analysis of the protein bands was performed with ImageJ software. Each column is the mean ± SD from three immunoblots (n = 3).
In a second set of experiments, we evaluated the effect of an injection of memantine on this KPI-APP expression increase. According to two recent studies, we selected two doses of memantine (1 and 30 mg/kg, i.p.) injected 30 min before a single NMDA administration (120 mg/kg). Real-time PCR and Western blot analysis revealed that both doses of memantine significantly inhibited the NMDA-induced KPI-APP expression (Fig. 10 C,D). It is important to note that memantine alone (at both doses) did not modify the level of KPI-APP mRNA or protein in mice brain (Fig. 10 E,F). This would indicate that memantine does not modify physiological neurotransmission. Since memantine at 1 mg/kg was described to block predominantly extrasynaptic NMDAR (Okamoto et al., 2009; Milnerwood et al., 2010), our results reinforce the idea that extrasynaptic NMDAR activation triggers KPI-APP induction in vivo, confirming our observations obtained in primary cortical neuron cultures.
Discussion
Recent reports suggested that even mild deregulation of the glutamatergic neurotransmission may represent a causal risk for developing AD by modifying the metabolism of pathological proteins (Aβ and protein tau) (Green et al., 2007; Green and LaFerla, 2008). A major finding of the present work is that activation of NMDAR induced a very different neuronal APP expression pattern leading to different levels of Aβ release, depending on their cellular location. While activation of synaptic NMDAR induced a downregulation of APP mRNA expression, extrasynaptic NMDAR activation induced a shift from APP695 (neuronal form) toward KPI-APPs, and consequently led to important production of amyloid peptide. These findings suggest that chronic activation of each pool of receptors regulates APP expression through distinct intracellular signaling pathways. We first aimed to determine whether these effects could be explained by different levels of intracellular calcium concentration reached during these experimental conditions. Data in the literature suggest that only 20 to 36% of NMDARs in mature neuron cultures are extrasynaptic (Rosenmund et al., 1995; Tovar and Westbrook, 1999; Thomas et al., 2006; Harris and Pettit, 2007), contributing only moderately to calcium entry. Conversely, in a recent report, extrasynaptic NMDARs were shown to induce larger intracellular Ca2+ elevations than synaptic NMDARs (Stanika et al., 2009). By using calcium imaging experiments, we showed that intracellular calcium levels reached after extrasynaptic treatment are much lower than those obtained after synaptic activation. Calcium levels measured here are certainly not the result of a maximal synaptic or extrasynaptic NMDAR activation, and other channel types may be involved. However, evaluation of calcium response allows us to verify that, in our experimental conditions, the induction of KPI-APP expression cannot be explained by higher intracellular calcium concentrations obtained following selective activation of extrasynaptic NMDARs.
Second, we showed that extrasynaptic NMDAR activation modifies APP isoform abundance by regulating alternative splicing of its mRNA without affecting its overall expression. It has been recently reported that neuronal alternative pre-mRNA splicing is dynamically regulated by calcium signaling pathways and that CaMK IV is a crucial factor involved in the repression of exon splicing of different neuronal genes (Lee et al., 2007; Li et al., 2007). The use of pharmacological compounds revealed that KPI-APP expression was mediated by Ca2+-dependent signaling pathways activated by extrasynaptic NMDAR. The use of STO-609 to inhibit CaMKK further indicated that CaMK I and/or CaMK IV (the two main substrates of CaMKK) are involved in these effects. In addition, our experiments using siRNA directed against CaMK IV strengthened the hypothesis of an alternative splicing mechanism in which CaMK IV plays a major role. Interestingly, a recent publication demonstrated the fundamental role of two neuronal splicing factors, hnRNPA1 and SC35, in the regulation of exon 7 (containing KPI domain) skipping during APP pre-mRNA maturation (Donev et al., 2007). By lowering the expression of one or both factors, they reported an increase in KPI-APP expression with a subsequent elevated Aβ production in neuronal cells. Since hnRNPA1, among other splicing regulators, was reported to be regulated by CaMK IV following cell depolarization (An and Grabowski, 2007; Li et al., 2007), we investigated the mRNA expression of both splicing factors in our model of cortical neuron cultures subjected to selective activation of NMDAR pools. We found a significant decrease in both hnRNPA1 and SC35 mRNA expression after extrasynaptic NMDAR activation, whereas no change was observed after synaptic NMDAR stimulation. This suggests that a prolonged extrasynaptic NMDAR activation modulates alternative splicing of the APP pre-mRNA, shifting the balance of the APP isoforms toward KPI-containing APPs. This splicing pathway involves CaMK IV activation and the downregulation of hnRNPA1 and SC35.
Previous reports indicated a link between the induction of KPI-APP isoforms and glutamate exposure (Willoughby et al., 1995) or NMDA treatment (Lesné et al., 2005) in neurons. However, to our knowledge, no published data are available describing a possible link between extrasynaptic NMDAR activation and amyloidogenesis. Many studies have established that synaptic NMDAR activation induced an upregulation of key pathways (ERK, CaMK), transcription factors (CREB), or prosurvival genes (BDNF). Conversely, activation of extrasynaptic NMDAR was shown to cause a loss of mitochondrial membrane potential (Hardingham et al., 2002; Léveillé et al., 2008; Gouix et al., 2009), and to have opposite effects on synaptic NMDAR-activated CREB function (Hardingham et al., 2002; Soriano and Hardingham, 2007; Mulholland et al., 2008), ERK pathway (Ivanov et al., 2006; Gouix et al., 2009), or BDNF expression (Vanhoutte and Bading, 2003). Very recently, extrasynaptic NMDARs were also shown to be implicated in the pathogenesis of Huntington's disease (Okamoto et al., 2009; Milnerwood et al., 2010). Here, we provide new insights into the potential involvement of the extrasynaptic pool of NMDAR in a different area of neuronal pathophysiology: APP expression pattern and amyloidogenesis. We observed that induction of KPI-APP expression is clearly associated with subsequent elevated amount of neuronal Aβ(1-42) production. We previously evidenced a cellular mechanism linking neuronal expression of KPI-APP proteins to increased Aβ secretion by revealing a shift from α-secretase to β-secretase (Lesné et al., 2005). According to our present findings, we can assume that inhibiting extrasynaptic NMDAR-mediated neuronal KPI-APP expression would also reduce subsequent Aβ production. Following this idea, memantine, the only clinically approved NMDAR antagonist for the treatment of AD, has been shown to preferentially block excessive NMDAR activity without disrupting normal synaptic activity (Chen et al., 1998; Lipton, 2004, 2007; Parsons et al., 2007). It was reported that memantine relatively spared synaptic NMDAR-mediated activity while blocking extrasynaptic activity, thus predominantly accounting for the drug's efficiency and clinically tolerated action (Lipton, 2004; Zhao et al., 2006). In the present work, memantine dose-dependently inhibited extrasynaptic NMDAR-induced KPI-APP protein expression as well as neuronal Aβ(1-42) release. It is interesting to note that these effects were observed at a clinically achievable concentration (1 μm) after prolonged activation of extrasynaptic NMDAR (24 h), thus reflecting a pathophysiological situation. This is in accordance with previous observations reporting that memantine only acts under pathological conditions without much affecting normal functions, and so relatively spares normal excitatory synaptic function (Lipton, 2006 and 2007). Furthermore, our in vivo data validate our in vitro results. NMDA intraperitoneal injection increased cortical KPI-APP expression in mice, while low-dose memantine, described to block extrasynaptic NMDAR (Okamoto et al., 2009; Milnerwood et al., 2010), significantly inhibited KPI-APP induction.
A controversy still exists about the role of neuronal activity in the determination of extracellular Aβ levels. It was reported that synaptic activity increases extracellular Aβ levels through the involvement of a change in APP processing (Kamenetz et al., 2003). This increase has also been proposed to be due to the direct stimulation of a readily releasable pool of Aβ present at nerve terminal (Cirrito et al., 2005, 2008). Alternatively, other studies reported a NMDAR-mediated inhibition of neuronal Aβ release resulting from increased α-secretase-mediated cleavage of APP (Marcello et al., 2007, 2008; Hoey et al., 2009). However, most of these studies aimed to understand how APP processing is dynamically regulated by electrical activity, under physiological conditions. Thus, these authors mainly focused on the measurement of α-, β-, and γ-secretase activities in short periods of time to determine the induction of Aβ or non-Aβ-forming pathways. In the present work, we aimed rather to mimic a pathophysiological situation on a neuronal culture model. Since KPI-APP isoforms are known to be significantly elevated in AD brains, we investigated transcriptional and translational changes in the APP isoforms expressed in neurons after prolonged treatments.
It is unlikely that blocked synaptic receptors remained unavailable during all of the time course of extrasynaptic NMDAR stimulation (24 h). For such long periods of time, NMDAR turnover as well as lateral mobility may occur between both receptor pools (Tovar and Westbrook, 2002; Triller and Choquet, 2005). However, others strongly suggested an absence of significant extrasynaptic NMDAR lateral mobility (Harris and Pettit, 2007; Bengtson et al., 2008). In our experiments, only extrasynaptic NMDAR activation led to the induction of neuronal KPI-APP expression. Conversely, we never observed any induction of KPI-APPs after Bic/4-AP application. Thus, despite a possible partial recovery of synaptic NMDAR activity during prolonged times of treatment, this is not sufficient to modify the consequences of extrasynaptic NMDAR activation on APP expression pattern.
In summary, the results of the present study reveal a direct linkage between prolonged extrasynaptic NMDAR activation and neuronal Aβ production. The release of Aβ is preceded by the neuronal induction of KPI-APPs, isoforms exhibiting an important amyloidogenic potential. The change in APP expression pattern is the consequence of its pre-mRNA splicing regulation, involving CaMK IV without affecting the overall APP expression. Finally, memantine was shown to efficiently counteract this process by preferentially blocking extrasynaptic NMDARs, confirming the interest of this NMDAR antagonist in the treatment of AD.
Footnotes
This work was supported by the National Research Agency (SEST 06), France-Alzheimer Foundation, and Region Basse Normandie. We thank Dr. F. Léveillé and Dr. B. Haelewyn for helpful discussions.
References
- An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson CP, Dick O, Bading H. A quantitative method to assess extrasynaptic NMDA receptor function in the protective effect of synaptic activity against neurotoxicity. BMC Neurosci. 2008;9:11. doi: 10.1186/1471-2202-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationalship between default activity, amyloid and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Wang YF, Rayudu PV, Edgecomb P, Neill JC, Segal MM, Lipton SA, Jensen FE. Neuroprotective concentrations of the N-methyl-D-aspartate open-channel blocker memantine are effective without cytoplasmic vacuolation following post-ischemic administration and do not block maze learning or long-term potentiation. Neuroscience. 1998;86:1121–1132. doi: 10.1016/s0306-4522(98)00163-8. [DOI] [PubMed] [Google Scholar]
- Chung KC, Shin SW, Yoo M, Lee MY, Lee HW, Choe BK, Ahn YS. A systemic administration of NMDA induces immediate early gene pip92 in the hippocampus. J Neurochem. 2000;75:9–17. doi: 10.1046/j.1471-4159.2000.0750009.x. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Yamada KA, Finn MB, Sloviter RS, Bales KR, May PC, Schoepp DD, Paul SM, Mennerick S, Holtzman DM. Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Kang JE, Lee J, Stewart FR, Verges DK, Silverio LM, Bu G, Mennerick S, Holtzman DM. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donev R, Newall A, Thome J, Sheer D. A role for SC35 and hnRNPA1 in the determination of amyloid precursor protein isoforms. Mol Psychiatry. 2007;12:681–690. doi: 10.1038/sj.mp.4001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouix E, Léveillé F, Nicole O, Melon C, Had-Aissouni L, Buisson A. Reverse glial glutamate uptake triggers neuronal cell death through extrasynaptic NMDA receptor activation. Mol Cell Neurosci. 2009;40:463–473. doi: 10.1016/j.mcn.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Green KN, LaFerla FM. Linking calcium to Aβ and Alzheimer's Disease. Neuron. 2008;59:190–194. doi: 10.1016/j.neuron.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Green KN, Smith IF, Laferla FM. Role of calcium in the pathogenesis of Alzheimer's disease and transgenic models. Subcell Biochem. 2007;45:507–521. doi: 10.1007/978-1-4020-6191-2_19. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hardingham GE, Bading H. The Yin and Yang of NMDA receptor signalling. Trends Neurosci. 2003;26:81–89. doi: 10.1016/S0166-2236(02)00040-1. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL. Extrasynaptic and synaptic NMDA receptors form stable and uniform pools in rat hippocampal slices. J Physiol. 2007;584:509–519. doi: 10.1113/jphysiol.2007.137679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Fukuchi K, Younkin SG. The alternatively spliced Kunitz protease inhibitor domain alters amyloid β protein precursor processing and amyloid β protein production in cultured cells. J Biol Chem. 1996;271:30929–30934. doi: 10.1074/jbc.271.48.30929. [DOI] [PubMed] [Google Scholar]
- Hoey SE, Williams RJ, Perkinton MS. Synaptic NMDA receptor activation stimulates α-secretase amyloid precursor protein processing and inhibits amyloid-β production. J Neurosci. 2009;29:4442–4460. doi: 10.1523/JNEUROSCI.6017-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Pellegrino C, Rama S, Dumalska I, Salyha Y, Ben-Ari Y, Medina I. Opposing role of synaptic and extrasynaptic NMDA receptors in regulation of the extracellular signal-regulated kinases (ERK) activity in cultured rat hippocampal neurons. J Physiol. 2006;572:789–798. doi: 10.1113/jphysiol.2006.105510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Khachaturian ZS. Hypothesis on the regulation of cytosol calcium concentration and the aging brain. Neurobiol Aging. 1987;8:345–346. doi: 10.1016/0197-4580(87)90073-x. [DOI] [PubMed] [Google Scholar]
- Kim HS, Lee SH, Kim SS, Kim YK, Jeong SJ, Ma J, Han DH, Cho BK, Suh YH. Post-ischemic changes in the expression of Alzheimer's APP isoforms in rat cerebral cortex. Neuroreport. 1998;9:533–537. [PubMed] [Google Scholar]
- Klyubin I, Walsh DM, Cullen WK, Fadeeva JV, Anwyl R, Selkoe DJ, Rowan MJ. Soluble arctic amyloid beta protein inhibits hippocampal long-term potentiation in vivo. Eur J Neurosci. 2004;19:2839–2846. doi: 10.1111/j.1460-9568.2004.03389.x. [DOI] [PubMed] [Google Scholar]
- Kotermanski SE, Johnson JW. Mg2+ imparts NMDA receptor subtype selectivity to the Alzheimer's drug memantine. J Neurosci. 2009;29:2774–2779. doi: 10.1523/JNEUROSCI.3703-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, Viola KL, Klein WL. Aβ oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer's disease. J Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc AC, Chen HY, Autilio-Gambetti L, Gambetti P. Differential APP gene expression in rat cerebral cortex, meninges, and primary astroglial, microglial and neuronal cultures. FEBS Lett. 1991;292:171–178. doi: 10.1016/0014-5793(91)80861-v. [DOI] [PubMed] [Google Scholar]
- Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S, Ali C, Gabriel C, Croci N, MacKenzie ET, Glabe CG, Plotkine M, Marchand-Verrecchia C, Vivien D, Buisson A. NMDA receptor activation inhibits α-secretase and promotes neuronal amyloid-β production. J Neurosci. 2005;25:9367–9377. doi: 10.1523/JNEUROSCI.0849-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé F, El Gaamouch F, Gouix E, Lecocq M, Lobner D, Nicole O, Buisson A. Neuronal viability is controlled by a functional relation between synaptic and extrasynaptic NMDA receptors. FASEB J. 2008;22:4258–4271. doi: 10.1096/fj.08-107268. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in NMDA receptor antagonist drug development: molecular mechanism of uncompetitive inhibition by memantine in the treatment of Alzheimer's disease and other neurologic disorders. J Alzheimer Dis. 2004;6([6 Suppl]):61–74. doi: 10.3233/jad-2004-6s610. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Pathologically activated therapeutics for neuroprotection. Nat Rev Neurosci. 2007;8:803–808. doi: 10.1038/nrn2229. [DOI] [PubMed] [Google Scholar]
- Mahdi F, Van Nostrand WE, Schmaier AH. Protease nexin-2/amyloid beta-protein precursor inhibits factor Xa in the prothrombinase complex. J Biol Chem. 1995;270:23468–23474. doi: 10.1074/jbc.270.40.23468. [DOI] [PubMed] [Google Scholar]
- Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, Borroni B, Cattabeni F, Sala C, Padovani A, Di Luca M. Synapse-associated protein-97 mediates α-secretase ADAM10 trafficking and promotes its activity. J Neurosci. 2007;27:1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello E, Epis R, Di Luca M. Amyloid flirting with synaptic failure: towards a comprehensive view of Alzheimer's disease pathogenesis. Eur J Pharmacol. 2008;585:109–118. doi: 10.1016/j.ejphar.2007.11.083. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RW, Vasuta OC, Graham RK, Hayden MR, Murphy TH, Raymond LA. Early increase in extrasynaptic NMDA receptor signalling and expression contributes to phenotype onset in Huntington's disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Moir RD, Lynch T, Bush AI, Whyte S, Henry A, Portbury S, Multhaup G, Small DH, Tanzi RE, Beyreuther K, Masters CL. Relative increase in Alzheimer's Disease of soluble forms of cerebral Aβ amyloid protein precursor containing the Kunitz protease inhibitory domain. J Biol Chem. 1998;273:5013–5019. doi: 10.1074/jbc.273.9.5013. [DOI] [PubMed] [Google Scholar]
- Mulholland PJ, Luong NT, Woodward JJ, Chandler LJ. Brain-derived neurotrophic factor activation of extracellular signal-regulated kinase is autonomous from the dominant extrasynaptic NMDA receptor extracellular signal-regulated kinas shutoff pathway. Neuroscience. 2008;151:419–427. doi: 10.1016/j.neuroscience.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendra SN, Faiman MD, Davis K, Wu JY, Newby X, Schloss JV. Carbamoylation of brain glutamate receptors by a disulfiram metabolite. J Biol Chem. 1997;272:24247–24251. doi: 10.1074/jbc.272.39.24247. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, Zaidi R, Clemente A, Kaul M, Graham RK, Zhang D, Vincent Chen HS, Tong G, Hayden MR, Lipton SA. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CG, Stöffler A, Danysz W. Memantine: a NMDA receptor antagonist that improves memory by restoration of homeostasis in the glutamatergic system—too little activation is bad, too much is even worse. Neuropharmacology. 2007;53:699–723. doi: 10.1016/j.neuropharm.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Preece P, Virley DJ, Costandi M, Coombes R, Moss SJ, Mudge AW, Jazin E, Cairns NJ. Amyloid precursor protein mRNA levels in Alzheimer's disease brain. Mol Brain Res. 2004;122:1–9. doi: 10.1016/j.molbrainres.2003.08.022. [DOI] [PubMed] [Google Scholar]
- Rockenstein EM, McConlogue L, Tan H, Power M, Masliah E, Mucke L. Levels and alternative splicing of amyloid beta protein precursor (APP) transcripts in brains of APP transgenic mice and humans with Alzheimer's disease. J Biol Chem. 1995;270:28257–28267. doi: 10.1074/jbc.270.47.28257. [DOI] [PubMed] [Google Scholar]
- Rose K, Goldberg MP, Choi DW. Cytotoxicity in murine cortical cell culture. In: Tyson CA, Frazier JM, editors. In vitro biological methods. Methods in toxicology. San Diego: Academic; 1993. pp. 46–60. [Google Scholar]
- Rosenmund C, Feltz A, Westbrook GL. Synaptic NMDA receptor channels have a low open probability. J Neurosci. 1995;15:2788–2795. doi: 10.1523/JNEUROSCI.15-04-02788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler R, Charlton MP, Hafner M, Tymianski M. Distinct influx pathways, not calcium load, determine neuronal vulnerability to calcium neurotoxicity. J Neurochem. 1998;71:2349–2364. doi: 10.1046/j.1471-4159.1998.71062349.x. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Soriano FX, Hardingham GE. Compartimentalized NMDA receptor signalling to survival and death. J Physiol. 2007;584:381–387. doi: 10.1113/jphysiol.2007.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanika RI, Pivovarova NB, Brantner CA, Watts CA, Winters CA, Andrews SB. Coupling diverse routes of calcium entry to mitochondrial dysfunction and glutamate excitotoxicity. Proc Natl Acad Sci U S A. 2009;106:9854–9859. doi: 10.1073/pnas.0903546106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H, Iijima S, Kambe Y, Georgiev D, Yoneda Y. Tex261 modulates the excitotoxic cell death induced by N-methyl-D-aspartate (NMDA) receptor activation. Biochem Biophys Res Commun. 2007;362:1096–1100. doi: 10.1016/j.bbrc.2007.08.126. [DOI] [PubMed] [Google Scholar]
- Thomas CG, Miller AJ, Westbrook GL. Synaptic and extrasynaptic NMDA receptor NR2 subunits in cultured hippocampal neurons. J Neurophysiol. 2006;95:1727–1734. doi: 10.1152/jn.00771.2005. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Chijiwa T, Hagiwara M, Mizutani A, Terasawa M, Hidaka H. KN-62, 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-L-tyrosyl]-4-phenylpiperazine, a specific inhibitor of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 1990;265:4315–4320. [PubMed] [Google Scholar]
- Tokumitsu H, Inuzuka H, Ishikawa Y, Ikeda M, Saji I, Kobayashi R. STO-609, a specific inhibitor of the Ca2+/calmodulin dependent kinase kinase. J Biol Chem. 2002;277:15813–15818. doi: 10.1074/jbc.M201075200. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro . J Neurosci. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. Mobile NMDA receptors at hippocampal synapses. Neuron. 2002;34:255–264. doi: 10.1016/s0896-6273(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Triller A, Choquet D. Surface trafficking of receptors between synaptic and extrasynaptic membranes: and yet they do move! Trends Neurosci. 2005;28:133–139. doi: 10.1016/j.tins.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Vanhoutte P, Bading H. Opposing roles of synaptic and extrasynaptic NMDA receptors in neuronal calcium signalling and BDNF gene regulation. Curr Opin Neurobiol. 2003;13:366–371. doi: 10.1016/s0959-4388(03)00073-4. [DOI] [PubMed] [Google Scholar]
- Waxman EA, Lynch DR. N-methyl-D-aspartate receptors subtypes: Multiple roles in excitotoxicity and neurological disease. Neuroscientist. 2005;11:37–49. doi: 10.1177/1073858404269012. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Silva A, Soderling TR. Calmodulin kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby DA, Rozovsky I, Lo AC, Finch CE. Beta-amyloid precursor protein (APP) and APP-RNA are rapidly affected by glutamate in cultured neurons: selective increase of mRNAs encoding a Kunitz protease inhibitor domain. J Mol Neurosci. 1995;6:257–276. doi: 10.1007/BF02736785. [DOI] [PubMed] [Google Scholar]
- Wilquet V, De Strooper B. Amyloid-β precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Bengtson CP, Bading H. Extrasynaptic NMDA receptors: mediators of excitotoxic cell death. In: Krieglstein J, Klumpp S, editors. Pharmacology of cerebral ischemia. Stuttgart, Germany: Medpharm Scientific Publishers; 2004. pp. 253–266. [Google Scholar]
- Xie J, Jan C, Stoilov P, Park J, Black DL. A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA. 2005;11:1825–1834. doi: 10.1261/rna.2171205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Marszalec W, Toth PT, Huang J, Yeh JZ, Narahashi T. In vitro galantamine-memantine co-application: mechanism of beneficial action. Neuropharmacology. 2006;51:1181–1191. doi: 10.1016/j.neuropharm.2006.08.007. [DOI] [PubMed] [Google Scholar]


