Chordoma is a primary bone cancer with no approved therapy1. The identification of therapeutic targets in this disease has been challenging due to the infrequent occurrence of clinically actionable somatic mutations in chordoma tumors2,3. Here we describe the discovery of therapeutically targetable chordoma dependencies via genome-scale CRISPR-Cas9 screening and focused small-molecule sensitivity profiling. These systematic approaches reveal that the developmental transcription factor T (brachyury; TBXT) is the top selectively essential gene in chordoma, and that transcriptional cyclin-dependent kinase (CDK) inhibitors targeting CDK7/12/13 and CDK9 potently suppress chordoma cell proliferation. In other cancer types, transcriptional CDK inhibitors have been observed to downregulate highly expressed, enhancer-associated oncogenic transcription factors (TFs)4,5. In chordoma, we find that T is associated with a 1.5-Mb region containing “super-enhancers” and is the most highly expressed super-enhancer-associated TF. Notably, transcriptional CDK inhibition leads to preferential and concentration-dependent downregulation of cellular brachyury protein levels in all models tested. In vivo, CDK7/12/13-inhibitor treatment substantially reduces tumor growth. Together, these data demonstrate small-molecule targeting of brachyury TF addiction in chordoma, identify a mechanism of T gene regulation that underlies this therapeutic strategy, and provide a blueprint for applying systematic genetic and chemical screening approaches to discover vulnerabilities in genomically quiet cancers.
Chordoma is a primary bone cancer that typically occurs in the skull-base, mobile spine, and sacrum6. Chordoma often manifests as a slow-growing but locally invasive malignancy, with a tendency to recur despite surgical and/or radiation therapy1,7. There are no approved targeted therapies, conventional cytotoxic chemotherapies, or immunotherapies for chordoma1. The lack of systemic treatment options, and an inadequate understanding of chordoma biology to guide the development of new therapies, contributes to poor prognoses for patients with advanced disease7.
Chordoma is hypothesized to originate from embryonic notochordal remnants8. Both cell types share high expression of the T-box-family TF brachyury (gene symbol: T; TBXT)9, an essential regulator of notochord development10 whose overexpression is characteristic of chordoma9. Beyond being a chordoma biomarker, brachyury appears to be critical for disease pathogenesis: germline T duplication confers susceptibility to familial chordoma11, a single-nucleotide polymorphism (SNP) in T is associated with chordoma12, some sporadic chordomas harbor somatic copy-number gains of T3,13, and T silencing inhibits growth of chordoma models13–15. Furthermore, brachyury is primarily expressed in the embryo and is absent from the majority of normal adult tissue9,10,16. These findings suggest that brachyury may act as an aberrantly activated developmental TF that is oncogenic and essential in a lineage-specific manner, akin to canonical “lineage-survival” oncogenes (e.g., MITF in melanoma)17.
Importantly, however, the full range of tumor dependencies in chordoma is not known. Few genes are recurrently mutated—and only at a modest frequency—in sporadic chordomas2,3; and nearly half of sporadic cases have no known driver mutation3. Furthermore, no systematic functional genomics studies have been conducted in chordoma models. Thus, it remains unclear if brachyury represents the central tumor dependency of chordoma, or whether there are critical dependencies left to be uncovered, and, if the former, whether brachyury overexpression can be targeted therapeutically. Like other TFs, brachyury is not readily inhibited pharmacologically18, and no small-molecule inhibitor of brachyury has been identified. It is also not known what underlies brachyury dysregulation in the majority of chordoma tumors, and whether any potential mediators of overexpression are therapeutically targetable. Somatic alterations in T occur in a minority of sporadic chordomas3 and cannot explain the nearly universal occurrence of brachyury expression. Therefore, a deeper understanding of essential genes in chordoma, including potential regulators of brachyury expression, is imperative for nominating candidate therapeutic targets.
Recent advances in systematic CRISPR-Cas9 screening and small-molecule sensitivity profiling approaches have enabled identification of tumor dependencies in multiple cancer types19. We integrated these complementary approaches to identify key tumor dependencies and candidate therapeutic targets in chordoma.
T is a selectively essential gene in chordoma
To identify genes essential for chordoma cell viability, we performed genome-scale pooled CRISPR-Cas9 loss-of-function screens in two chordoma cell lines (UM-Chor1, MUG-Chor1). We introduced a library of >74,000 single-guide RNAs (sgRNAs) targeting ~18,560 genes (Methods) into stably Cas9-expressing cells via lentiviral transduction, and after 21 days, quantified sgRNAs from the genomic DNA of surviving cells. Depleted sgRNAs, representing candidate essential genes, were identified by comparing these sgRNA abundances to those of the screening library. We ranked all sgRNAs by how much they reduced viability in chordoma cells relative to 125 non-chordoma cancer cell lines screened using the same sgRNA library (Broad Institute Project Achilles; https://depmap.org/portal/achilles/)20,, thus removing commonly essential genes to identify dependencies selective for chordoma.
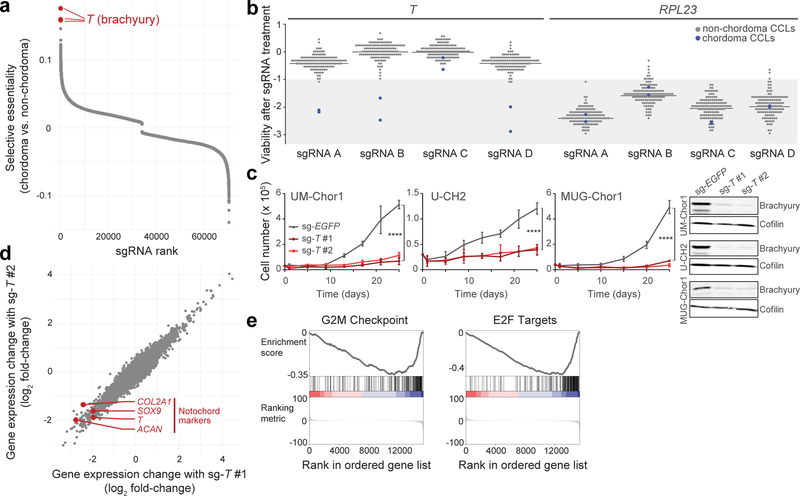
The top three selectively lethal sgRNAs, out of ~70,000 sgRNAs analyzed, all targeted the T gene (Fig. 1a). We confirmed that three of four T-targeted sgRNAs screened were lethal to both chordoma cell lines, but not to 125 non-chordoma cancer cell lines (Fig. 1b and Supplementary Table 1). Targeting T in chordoma cells induced a similar degree of viability reduction as targeting commonly essential ribosomal subunit genes RPL23, RPS11, and RPS19 (Fig. 1b, Extended Data Fig. 1, and Supplementary Table 1), indicating that chordoma cells are both exquisitely and selectively sensitive to loss of brachyury expression.
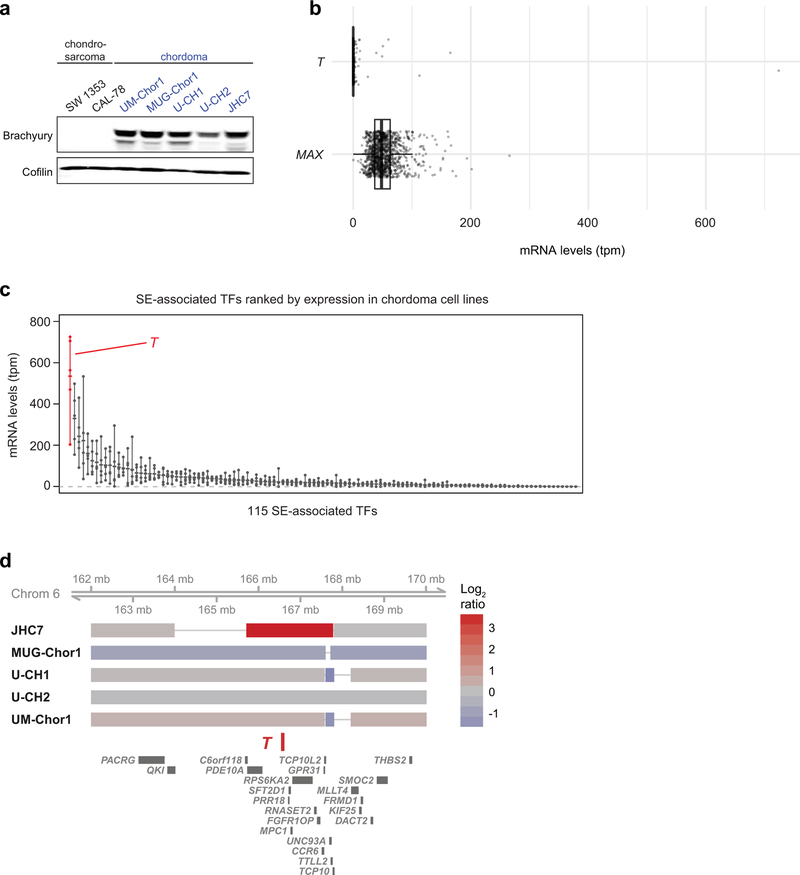
Figure 1. Genome-scale CRISPR-Cas9 screening identifies T (brachyury) as a selectively essential gene in chordoma cells.
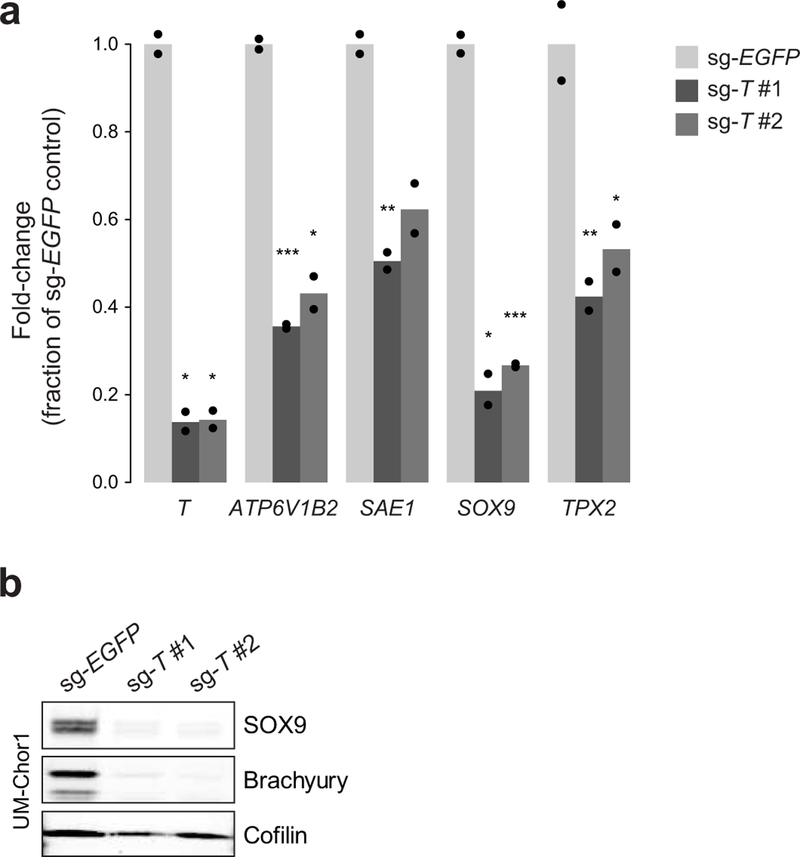
a) Comparative analysis of genome-scale CRISPR-Cas9 screening results generated in UM-Chor1 and MUG-Chor1 chordoma cell lines versus 125 non-chordoma cancer cell lines. SgRNAs were ranked by and plotted against selectively lethal effects (RNMI scores; see Methods) in chordoma versus non-chordoma cell lines. The top three sgRNAs (red circles) all target T. b) Viability after sgRNA treatment (represented by sgRNA-dependency scores; see Methods) corresponding to each of the four primary screening sgRNAs targeting either T or RPL23 across 127 cancer cell lines (CCLs), including 2 chordoma CCLs (blue circles). For each sgRNA, the median sgRNA-dependency score across 127 cell lines is indicated (gray line). Lower values indicate greater sgRNA depletion and thus essentiality of the target gene (shaded region). c) (Left) Proliferation of chordoma cell lines transduced with sgRNAs targeting T or a non-targeting sgRNA control. Points represent the mean ± SD (n = 3 biological samples measured in parallel). **** p < 0.0001, derived from a two-way ANOVA (p-values for the test comparing sg-EGFP and sg-T #1 are displayed). Exact p-values and effect sizes are reported in Supplementary Table 9. (Right) Immunoblot analysis of transduced cells confirming sgRNA-mediated protein repression. Proliferation experiments were performed twice with UM-Chor1 and U-CH2 cells (one representative experiment displayed for each) and once with MUG-Chor1 cells; immunoblots were performed once. d) Relative gene expression of UM-Chor1 cells transduced with one of two sgRNAs targeting T versus a non-targeting sgRNA control. Gene expression was measured with RNA sequencing. Data represent two biological replicates per condition. e) Gene-set enrichment analysis results show significant downregulation of genes associated with cell cycle progression, such as drivers of G2/M checkpoint progression and E2F targets.
We validated the screening results with two sgRNAs targeting independent sequences of T in three chordoma cell lines (UM-Chor1, MUG-Chor1, U-CH2) (Fig. 1c). The observed reduction in chordoma cell proliferation induced by sgRNA-mediated brachyury repression was consistent with previously reported effects of shRNA-mediated gene silencing of T in other chordoma cell lines13,14.
SgRNA-mediated T repression in UM-Chor1 cells led to downregulation of canonical notochord markers (SOX9, COL2A1, ACAN) (Fig. 1d), consistent with brachyury’s known role in notochord development21. Additionally, genes associated with the G2/M checkpoint and E2F targets were enriched among downregulated genes (Fig. 1e), suggestive of cell cycle arrest in sg-T-expressing cells. These data implicate T as an essential gene and regulator of notochord cell identity in chordoma.
Inhibitors of CDK7/12/13 and CDK9 suppress chordoma cell proliferation
In parallel to the genetic screens, we tested 459 small molecules for their ability to reduce proliferation or survival in four chordoma cell lines. Compounds were selected to collectively target many distinct nodes in cell circuitry, and included FDA-approved drugs, preclinical agents, and small-molecule probes (Supplementary Table 2). A largely overlapping small-molecule library was previously tested in ~800 non-chordoma cancer cell lines22, and we used these data to identify and prioritize compounds with antiproliferative effects selective for chordoma when possible (Fig. 2a; Methods).
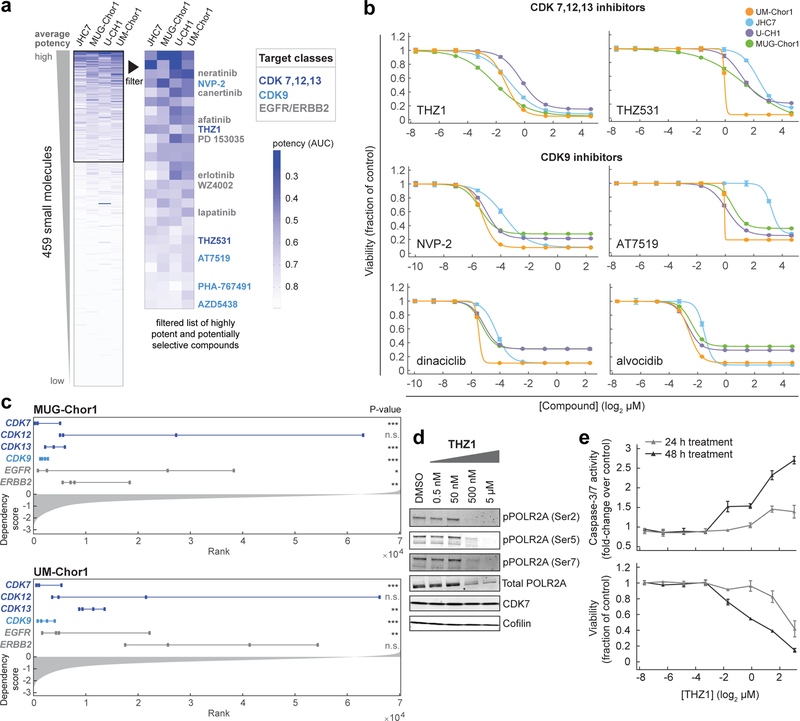
Figure 2. Small-molecule sensitivity profiling identifies inhibitors of CDK7/12/13 and CDK9 as potent antiproliferative agents in chordoma cells.
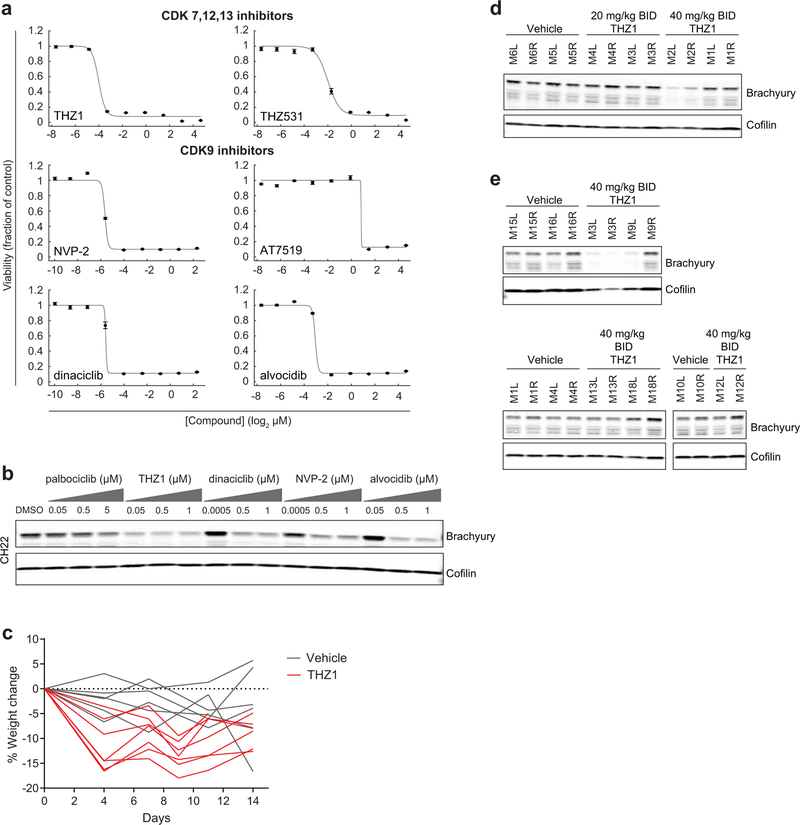
a) Profiling results for 459 small molecules tested in four chordoma cell lines. Data represent area-under-curve (AUC) values calculated from 8-point concentration-response curves generated in duplicate, and coloring in the heatmap is representative of smaller AUC values (blue) corresponding to more potent effects (AUC value of 1 = vehicle control; value of 0 = complete killing at all concentrations). Left heatmap, all compounds were ranked by average potency in chordoma cell lines; potent compounds (average AUC < 0.8, boxed) were then filtered to exclude those with cytotoxic effects non-selective for chordoma cell lines versus up to 891 non-chordoma cancer cell lines, when such data were available in CTRP. Compounds that were only tested in chordoma cell lines and were not available in CTRP were not filtered beyond the initial potency filter. Twenty-eight compounds (right heatmap) passed these criteria. b) Validation of primary screening hits and related compounds. Four chordoma cell lines were treated with indicated concentrations of candidate antiproliferative compounds and assayed for cell viability after 6 d with CellTiter-Glo. Response data are represented by a fitted curve to the mean fractional viability at each concentration relative to vehicle-treated cells; error bars represent the SEM (n = 4 biological samples measured in parallel). c) Rankings of sgRNA-level dependency scores (see Methods) for the indicated genes following genome-scale CRISPR-Cas9 screening (see Fig.1). Points represent each of four sgRNAs targeting a given gene that were present in the pooled CRISPR library used for screening. P-values were derived from a one-sided Mann-Whitney test. *, p < 0.05; **, p < 0.01; *** p < 0.001; n.s., not significant. The gray waterfall plot represents ranked dependency scores for all sgRNAs tested (median dependency score = −0.3257 for MUG-Chor1; median = −0.3283 for UM-Chor1). Median dependency scores for the four sgRNAs and exact Mann-Whitney p-values corresponding to each gene are reported in Supplementary Table 9. d) Immunoblot analysis of UM-Chor1 cells treated with indicated concentrations of THZ1 or DMSO for 24 h. Data are representative of two independent experiments. e) Caspase-3/7 activity (top) and cell viability (bottom) following THZ1 treatment of UM-Chor1 cells. Caspase-3/7 activity and cell viability were measured in parallel at the indicated time points using Caspase-Glo 3/7 and CellTiter-Glo reagents, respectively. Data are expressed as the fold-change of caspase-3/7 activity (top) or fraction of cell viability (bottom) relative to vehicle-treated cells and represent the mean ± SD (n = 4 biological samples measured in parallel).
Our analysis identified 28 potent antiproliferative compounds, including several inhibitors of either CDK7,12,13; CDK9; or EGFR/ERBB2 proteins (Fig. 2a and Supplementary Table 2). As CDK7 and CDK9 have related roles in regulating transcription23, we hypothesized that these inhibitor classes could have similar mechanistic effects.
The sensitivity of chordoma cells to compounds targeting CDK7/12/13 (THZ1)4; CDK12/13 (THZ531)24; CDK9 (NVP-2, AT7519, dinaciclib, alvocidib)23,25; and EGFR and/or ERBB2 (erlotinib, neratinib, canertinib, lapatinib, afatinib)26 was validated using multipoint concentration-response assays (Fig. 2b, Extended Data Fig. 2a, and Supplementary Table 3). Transcriptional CDK inhibitors including THZ1, dinaciclib, and alvocidib exhibit antiproliferative effects across a diversity of cancer cell lines4 (Extended Data Fig. 2b). Consistent with small-molecule sensitivity findings, sgRNAs targeting CDK7, CDK13, and CDK9, but not those targeting CDK12, were lethal to chordoma cells (Fig. 2c and Supplementary Table 1). SgRNAs targeting EGFR and ERBB2 were lethal to 2/2 and 1/2 chordoma cell lines, respectively (Fig. 2c and Supplementary Table 1). EGFR-inhibitor sensitivity in chordoma has been described previously27; thus, we focused on transcriptional CDK inhibitors, which are newly identified antiproliferative agents in chordoma.
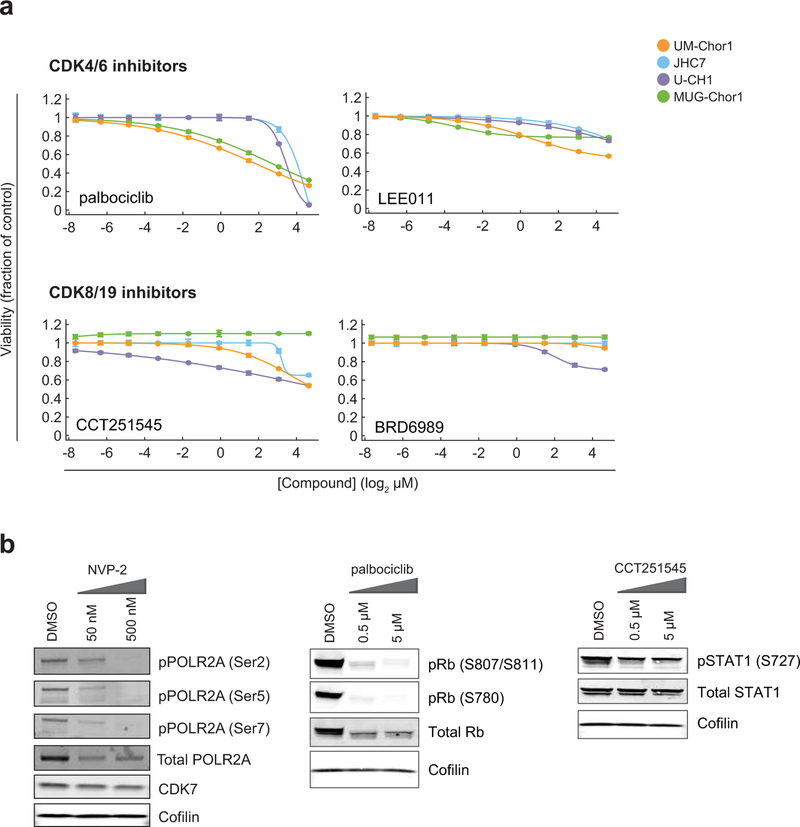
We confirmed on-target activity of the CDK7/12/13 inhibitor THZ1 by detecting reduced phosphorylation, particularly at serine 2, of the C-terminal domain (CTD) of RNA polymerase II (POLR2A) (Fig. 2d). Beyond having antiproliferative effects, THZ1 treatment induced apoptosis in chordoma cells, as measured by increased caspase-3/7 activity (Fig. 2e). We did not observe comparable sensitivity of chordoma cells to inhibitors of CDK proteins not implicated in direct phosphorylation of the POLR2A CTD, including CDK4/6 inhibitors (palbociclib/PD 0332991, LEE011)28; and CDK8/19 inhibitors (CCT251545, BRD6989)29,30 (Extended Data Fig. 3a and Supplementary Table 3). On-target activity was confirmed for a select number of CDK inhibitors tested (Extended Data Fig. 3b). Together, these data implicate inhibitors of CDK7/12/13 and CDK9 as potent antiproliferative agents in chordoma.
T is super-enhancer-associated in chordoma
Transcriptional CDK inhibitors are under clinical investigation ( NCT03134638) owing to their ability to downregulate preferentially oncogenic TFs in multiple cancer types5. Components of the transcriptional apparatus, including CDK7, can densely occupy super-enhancers—large clustered enhancers that drive genes required for tumor identity—often rendering the expression of super-enhancer-driven genes especially vulnerable to transcriptional inhibitors4,5,31,32. To investigate whether similar mechanisms connect the essentiality of T to the antiproliferative effects of transcriptional CDK inhibitors, we tested whether T is super-enhancer-associated in chordoma.
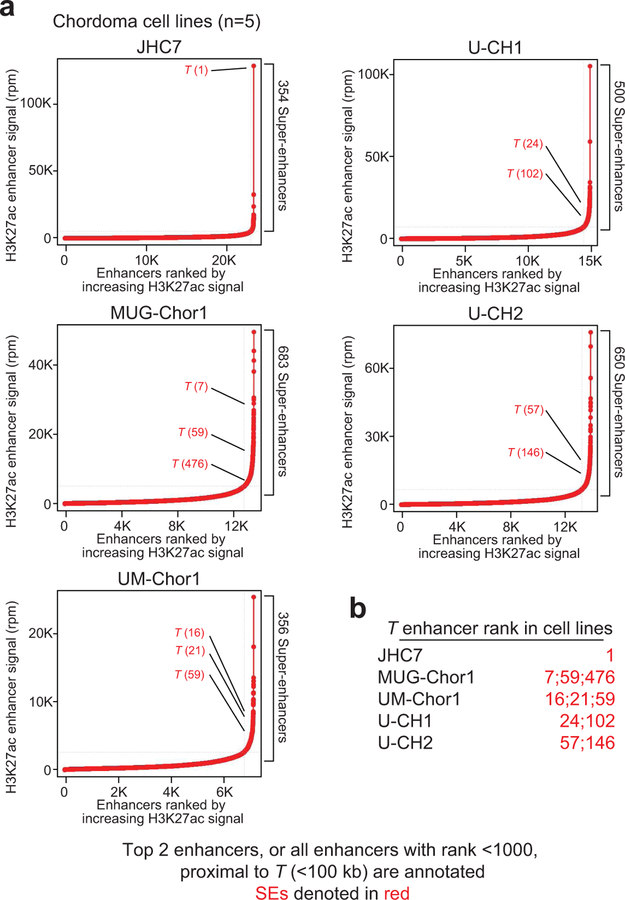
We mapped the enhancer landscape in five chordoma cell lines using chromatin immunoprecipitation sequencing (ChIP-seq) to detect histone H3 lysine 27 acetylation (H3K27ac), a chromatin modification associated with active enhancers32. We identified large enhancers overlapping and adjacent to the T locus in all five models (Fig. 3a, Extended Data Fig. 4, and Supplementary Table 4). T enhancers showed evidence of brachyury binding, as measured by brachyury ChIP-seq (using a published dataset33) (Fig. 3a), and potentially other TFs as suggested by an assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Fig. 3a), which measures chromatin accessibility and TF occupancy34. In contrast, super-enhancers and brachyury binding were not detected at the housekeeping TF MAX locus, despite the presence of other TF binding sites as inferred by ATAC-seq signal (Fig 3b and Supplementary Table 4).
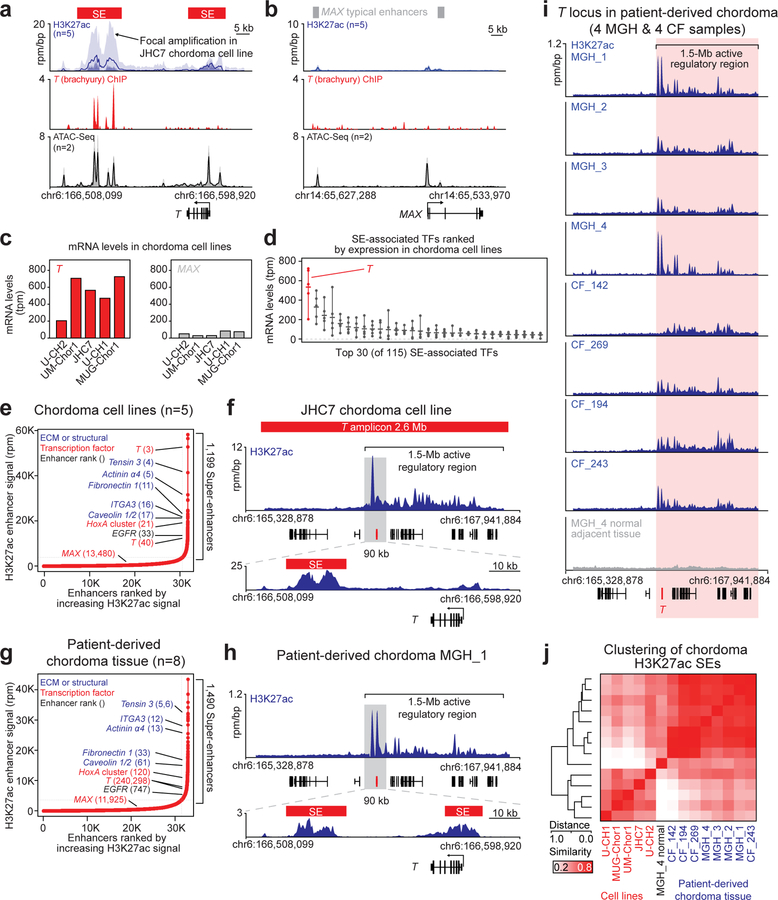
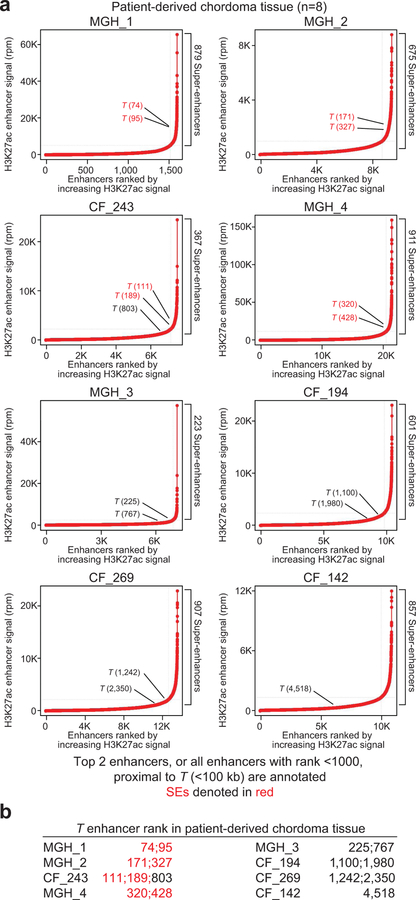
Figure 3. T (brachyury) is super-enhancer-associated and highly active across chordoma cell lines and in patient-derived chordoma tumors.
a,b) Gene tracks of H3K27ac, brachyury, and ATAC-seq signal (units of reads per million per base pair) at the T and MAX gene loci. For datasets with multiple samples (H3K27ac and ATAC-seq), signals for samples are plotted as a translucent shape and darker regions indicate regions with signal in more samples. An opaque line is plotted and gives the average signal across all samples in the group. H3K27Ac ChIP-seq was performed on 5 chordoma cell lines (UM-Chor1, MUG-Chor1, U-CH2, U-CH1, JHC7); brachyury ChIP-seq was performed on 1 chordoma cell line (U-CH1) as previously reported33; and ATAC-seq was performed on two chordoma cell lines (2 biological replicates for U-CH2 and 1 replicate for MUG-Chor1). c) Bar graphs of RNA-seq mRNA levels for T and MAX across 5 chordoma cell lines. Units are in transcripts per million (tpm). d) Gene expression levels of the top 30 (of 115) super-enhancer (SE)-associated transcription factors in 5 chordoma cell lines (points), ranked by mean expression (horizontal ticks). e,g) Enhancers in chordoma cell lines (UM-Chor1, MUG-Chor1, U-CH2, U-CH1, JHC7) or patient-derived chordoma tumor tissue ranked by average H3K27ac signal across samples. SEs and associated genes are annotated along the vertical axis and were determined by the inflection point of the plot. f,h) Gene tracks of H3K27ac signal at the T-amplified region in the JCH7 cell line and the corresponding region in the MGH_1 chordoma tumor. The amplicon and T-proximal SEs are shown (red boxes). i) Gene tracks of H3K27ac signal across eight patient-derived chordoma tumor samples and one matched normal adjacent muscle sample at the T locus. j) Clustergram of chordoma samples hierarchically clustered by similarity of H3K27ac signal at the union of all SE regions across samples.
All chordoma cell lines had high T (brachyury) expression, especially compared to non-super-enhancer-associated genes, such as MAX (Fig. 3b–c and Extended Data Fig. 5a); in contrast, T (brachyury) was not widely expressed in non-chordoma cancer cell lines (Extended Data Fig. 5a–b). Furthermore, T expression was highest among all super-enhancer-associated TFs in chordoma (Fig. 3d and Extended Data Fig. 5c), and when ranked against all other enhancers, the T-associated super-enhancer was among the largest in the chordoma genome (Fig 3e, Supplementary Table 5, and Extended Data Fig. 4). JHC7 cells had a large focal amplification at the T locus that encompassed adjacent T enhancers, as well as a 1.5-Mb upstream region with broad H3K27ac occupancy (Fig. 3f and Extended Data Fig. 5d).
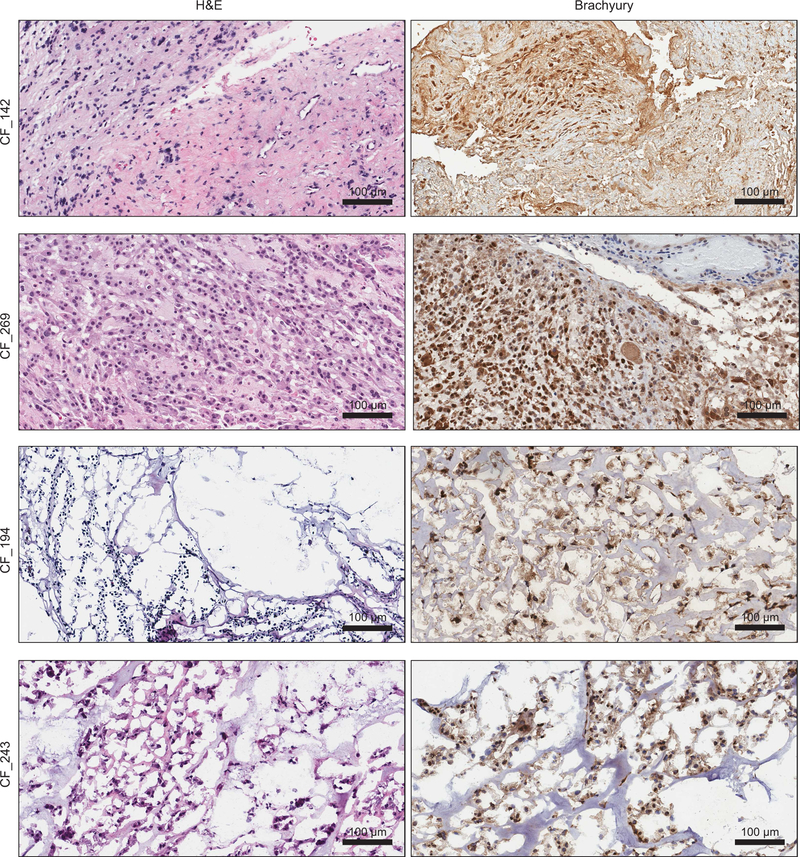
T-associated super-enhancers were similarly observed in patient-derived chordoma tumors (Supplementary Table 6), as detected by enhancer rank averaged across eight tumors (Fig. 3g and Supplementary Table 7) and in individual tumors (Fig. 3h, Extended Data Fig. 6, and Supplementary Table 4). Brachyury expression was confirmed for the subset of tumors tested (Extended Data Fig. 7). By enhancer ranking (Extended Data Fig. 6), 4/8 tumors had T-adjacent super-enhancers, 7/8 had T-adjacent enhancers ranking in the top 10% of all enhancers, and 8/8 were hyper-acetylated in the 1.5-Mb region found in JHC7 cells (Fig. 3i). In contrast, matched normal adjacent tissue lacked H3K27ac occupancy within this 1.5-Mb tumor hyper-acetylated region (Fig. 3i).
Enhancer patterns across tumors and cell lines showed grouping by in vivo or ex vivo state (Fig. 3j), similar to other cancer types35. Nonetheless, in both tumors and cell lines, super-enhancers associated with genes known to be highly expressed in chordoma: components of the extracellular matrix or its interactors (e.g., integrin subunit alpha 3, tensin 3, fibronectin 1, caveolin 1/2), mediators of de-differentiation (HoxA), and EGFR36–38. Thus, as in other cancers, super-enhancers associate with genes that define chordoma identity31,32, and regulation of T by super-enhancers is a dominant feature of the chordoma gene-regulatory landscape.
Transcriptional CDK inhibition downregulates T (brachyury) expression preferentially
The essentiality of T, together with its association with super-enhancers, led us to test whether the antiproliferative effects of transcriptional CDK inhibitors are mediated by preferential loss of brachyury expression.
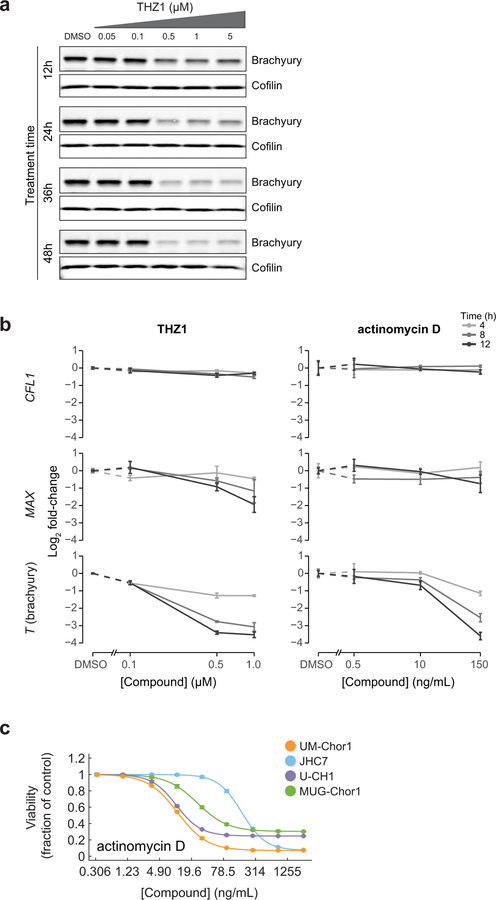
We found that THZ1 treatment reduced cellular brachyury protein levels in MUG-Chor1 and UM-Chor1 cells in a concentration- and time-dependent manner (Fig 4a and Extended Data Figure 8a). In contrast, brachyury protein expression was unchanged following treatment with compounds that are cytotoxic to chordoma cells but are not transcriptional CDK inhibitors (Supplementary Table 2), including inhibitors of EGFR (erlotinib), Src (dasatinib), and the BET bromodomain (JQ1) (Fig. 4a). Downregulation of brachyury expression was also observed following treatment with CDK9 inhibitors (NVP-2, dinaciclib, alvocidib), but not with the CDK4/6 inhibitor palbociclib, across multiple chordoma models (Fig. 4b).
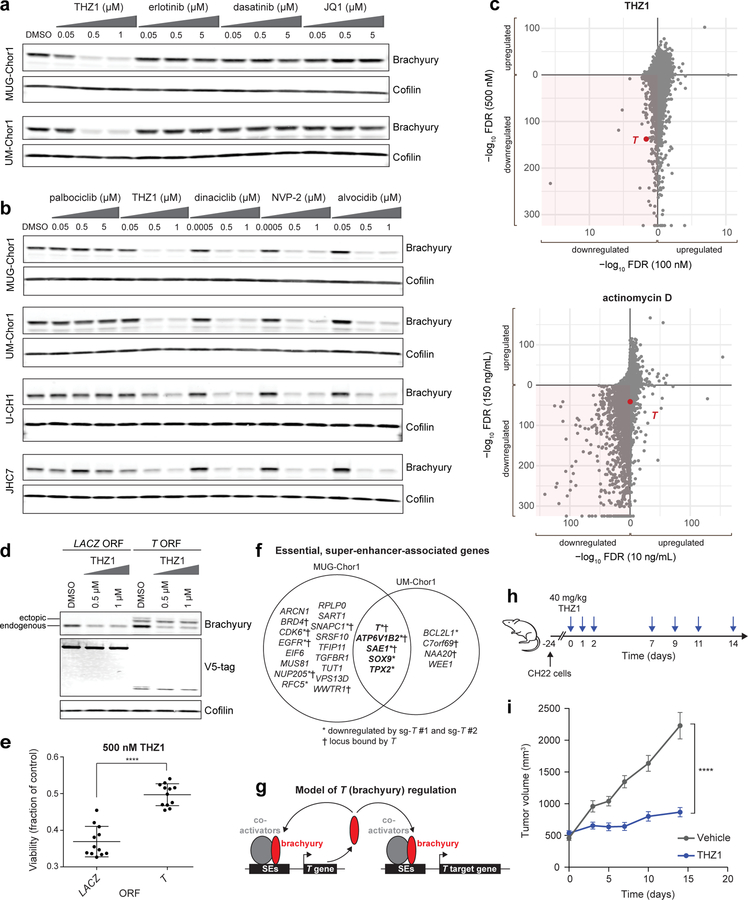
Figure 4. Inhibitors of CDK7/12/13 and CDK9 downregulate T (brachyury) expression, and THZ1 treatment reduces chordoma tumor proliferation in vivo.
a) Immunoblot analysis of chordoma cells treated with indicated concentrations of antiproliferative compounds or DMSO for 48 h. The experiment was performed four times with UM-Chor1 cells (one representative experiment displayed) and once with MUG-Chor1 cells. b) Immunoblot analysis of chordoma cells treated with indicated concentrations of inhibitors targeting CDK4/6 (palbociclib), CDK7/12/13 (THZ1), or CDK9 (dinaciclib, NVP-2, alvocidib) or DMSO for 48 h. The experiment was performed twice with MUG-Chor1, UM-Chor1, and U-CH1 cells (one representative experiment displayed for each) and once for JHC7 cells. c) Relative gene expression of UM-Chor1 cells treated with THZ1 (100 nM or 500 nM) (top), or actinomycin D (10 ng/mL or 150 ng/mL) (bottom), versus vehicle-treated cells. Cells were treated with compound or vehicle (DMSO) for 4 h. Gene-expression profiling was performed with RNA-seq, and genes downregulated following treatment with both concentrations of a given compound are indicated (shaded region). Ranked by significance, T has a rank of 128 and 139 out of 37,043 genes for 100 nM and 500 nM THZ1, respectively; and a rank of 11,607 and 2,539 out of 37,043 genes for 10 ng/mL and 150 ng/mL actinomycin D, respectively. Data represent two biological replicates per condition, and false-discovery-rate (FDR) values were derived from a two-sided Wald test, with a Benjamini-Hochberg correction. d) Immunoblot analysis of UM-Chor1 cells transduced with V5-tagged ORFs encoding T or a LACZ control gene and then treated with indicated concentrations of THZ1. Unlike endogenous brachyury expression, ectopic brachyury expression was not greatly reduced with THZ1 treatment. The experiment was performed twice (one representative experiment displayed). e) Cell viability of UM-Chor1 cells transduced with an ORF encoding either LACZ or T and then treated with 500 nM THZ1. Data are expressed as the fraction of cell viability relative to vehicle-treated cells and represent the mean ± SD (n = 12 biological samples measured in parallel). **** p < 0.0001, derived from a two-tailed, unpaired t test. Exact p-values and effect sizes are reported in Supplementary Table 9. f) Integration of super-enhancer-associated and essential genes in UM-Chor1 and MUG-Chor1 cell lines. Essential genes were identified for each cell line by applying the STARS algorithm (see Methods) to genome-scale CRISPR-Cas9 screening data described in Fig. 1. Asterisk, genes downregulated following sg-T-mediated T repression as measured by RNA-seq (experiment described in Fig. 1d). Cross, gene loci bound by brachyury as measured by brachyury ChIP-seq (using a previously reported dataset33; see Methods). g) Model of T (brachyury) gene control. h) Schematic of the dosing schedule for THZ1 treatment of a human xenograft mouse model of chordoma. i) Tumor proliferation in mice engrafted with CH22 cells and treated with vehicle or THZ1. Data represent the mean tumor volume ± SEM of 12 (vehicle-treated) or 14 (THZ1-treated) mouse tumors at the indicated time points. **** p < 0.0001, derived from a two-way ANOVA with repeated measures. Exact p-values and effect sizes are reported in Supplementary Table 9.
THZ1 treatment also substantially reduced mRNA expression of T, especially compared to MAX or the housekeeping gene CFL1 (Extended Data Fig. 8b). For comparison, we tested actinomycin D, a transcriptional inhibitor not thought to act through CDK inhibition39, and similarly observed mRNA downregulation of T, but not MAX or CFL1 (Extended Data Fig. 8b). However, compared to THZ1, actinomycin D induced higher cytotoxicity at concentrations leading to substantial T-mRNA downregulation (Extended Data Fig. 8b–c and Supplementary Table 3), suggesting that THZ1 and actinomycin D could differ in how selectively they induce T downregulation.
We confirmed this difference using transcriptome-wide gene-expression profiling of THZ1- and actinomycin D-treated cells (Fig. 4c). Only THZ1 treatment led to preferential downregulation of T expression (Fig. 4c), indicating that super-enhancer-driven T transcription is particularly vulnerable to CDK7/12/13 inhibition.
Furthermore, we tested whether THZ1’s antiproliferative effects are mediated by loss of super-enhancer-driven brachyury expression. To dissociate T from its endogenous regulatory elements, we transduced UM-Chor1 cells with an ORF encoding T under the control of an exogenous promoter. Unlike endogenous brachyury protein, ectopic brachyury expression levels were not greatly reduced with THZ1 treatment (Fig. 4d). Moreover, ectopic brachyury expression was sufficient to partially rescue loss of viability induced by 500 nM THZ1 treatment (Fig. 4e). Thus, downregulation of brachyury via its endogenous regulatory elements directly contributes to the antiproliferative effects of THZ1.
Besides T, only four genes were identified to be both super-enhancer-associated and essential in UM-Chor1 and MUG-Chor1 cells: SOX9, SAE1, TPX2, and ATP6V1B2 (Fig. 4f). These genes appear to be under direct and/or indirect transcriptional control by brachyury: sgRNA-mediated T repression significantly reduced expression levels of all four genes (Fig. 4f, Supplementary Table 8, and Extended Data Fig. 9); and brachyury bound genomic regions corresponding to two of them (SAE1 and ATP6V1B2; Fig. 4f and Supplementary Table 8).
These data support a model whereby T (brachyury) is expressed at high levels by a super-enhancer, is auto-regulated, and through direct binding to other super-enhancers and/or indirect mechanisms, can regulate other essential, super-enhancer-associated genes (Fig. 4g). As transcriptional CDKs including CDK7 and CDK9 are localized at enhancers4,31, these data implicate transcriptional CDK inhibition in both upstream and downstream targeting of T (brachyury) gene control.
Finally, THZ1 efficacy was tested in vivo. A xenograft mouse model was generated with the human chordoma cell line CH2240. Ex vivo, CH22 cells were sensitive to transcriptional CDK inhibitors (Extended Data Fig. 10a), and compound treatment induced downregulation of brachyury expression (Extended Data Fig. 10b). Mice were treated with 40 mg/kg THZ1 BID, on a pulse-dosing schedule (Fig. 4h). This dose was reasonably well tolerated (Extended Data Fig. 10c), and was empirically determined to downregulate brachyury expression in target engagement studies of mouse tumors (Extended Data Fig. 10d), although we note that detection of brachyury downregulation varied across experiments and mice (Extended Data Fig. 10e).
THZ1 treatment led to a significant tumor-suppressive effect (p = 1.1 × 10−5; Fig. 4i and Supplementary Table 9). These data provide evidence for the in vivo efficacy of THZ1 for the treatment of chordoma.
In conclusion, these findings nominate transcriptional CDK inhibition as a therapeutic strategy to target brachyury-driven transcriptional addiction in chordoma. Our results indicate that T (brachyury), a transcriptional regulator of notochord development, might drive chordoma tumorigenesis when dysregulated or persistently expressed. Thus, T resembles other “lineage-survival” oncogenes (e.g., MITF in melanoma), which arise from dysregulated expression of master genes that mediate normal lineage development17. More generally, these findings provide evidence for “transcriptional addiction” in chordoma, a state of exquisite dependence on specific regulators of gene expression5. As transcriptional dependencies are not typically discovered by tumor genome sequencing, these findings underscore the value of functional screening approaches, such as CRISPR-Cas9 and small-molecule sensitivity screening, to identify key vulnerabilities that may not manifest as mutations in the tumor genome.
TFs including brachyury are challenging to target directly with candidate drugs. Transcriptional CDK inhibition provides a therapeutic opportunity to downregulate preferentially T (brachyury). The recent entrance of next-generation transcriptional CDK inhibitors into the clinic may provide a much-needed therapeutic option for chordoma patients with refractory disease.
METHODS
Cell lines
UM-Chor1, MUG-Chor1, U-CH1, U-CH2, and JHC7 chordoma cell lines were obtained from the Chordoma Foundation. CH22 cells were obtained from the Chordoma Foundation and Massachusetts General Hospital and have been described previously40. UM-Chor1 cells were maintained in IMDM/RPMI (4:1) media + 10% fetal bovine serum (FBS) and 1X non-essential amino acids. MUG-Chor1, U-CH1, and U-CH2 cell lines were maintained in IMDM/RPMI (4:1) media + 10% FBS. JHC7 cells were maintained in DMEM/F12 (1:1) + 10% FBS. CH22 cells were maintained in RPMI media + 10% FBS. All chordoma cell lines were maintained on collagen I-coated plates. SW 1353 and CAL-78 chondrosarcoma cell lines were obtained from the Broad Institute Cancer Cell Line Encyclopedia project41. SW 1353 cells were maintained in DMEM/F12 (1:1) + 10% FBS media. CAL-78 cells were maintained in RPMI media + 20% FBS.
Genome-scale CRISPR-Cas9 screening
Cas9-expressing UM-Chor1 and MUG-Chor1 cells were generated as follows: each parental cell line was incubated with lentivirus corresponding to the pLX_311-Cas9 plasmid (Addgene plasmid #96924), encoding the Cas9 protein, in the presence of 4 μg/mL polybrene, dispensed in 12-well plates (1.5 × 106 cells/well), and spin-infected at 2,000 rpm for 2 h at 30°C. After spin-infection, 2 mL of standard growth media was added to each well, and cells were incubated at 37°C overnight. The following day, for each cell line, cells were trypsinized and expanded in selective media containing blasticidin (2–3 µg/mL for UM-Chor1 and 2 µg/mL for MUG-Chor1). Following selection for infected cells, Cas9 activity was confirmed in each transduced cell line using a previously described Cas9-activity assay42.
Screening was performed using a genome-scale library of 74,687 unique sgRNAs targeting ~18,560 genes (typically 4 sgRNAs per gene) and 1,000 non-targeting control sgRNAs (Broad Institute Avana sgRNA library)20. UM-Chor1-Cas9 and MUG-Chor1-Cas9 cells were each incubated with lentivirus corresponding to the pooled CRISPR library in the presence of 4 μg/mL polybrene, dispensed in 12-well plates (21 plates with 1.5 × 106 cells/well for UM-Chor1 and 15 plates with 3 × 106 cells/well for MUG-Chor1), and spin-infected at 2,000 rpm for 2 h at 30°C. Lentivirus was titered in each cell line to achieve a low MOI (<1), and infections were performed with a sufficient a number of cells to achieve a representation of >800 cells per sgRNA per replicate after selection for infected cells. After spin-infection, 2 mL of standard growth media was added to each well, and cells were incubated at 37°C overnight. The following day, for each cell line, cells were trypsinized, divided into three replicates, and expanded in selective media containing puromycin (3 µg/mL for UM-Chor1 and 6 µg/mL for MUG-Chor1) and blasticidin (3 µg/mL for UM-Chor1 and 2 µg/mL for MUG-Chor1). Cells were grown in culture for 21 d post-infection, with carryover of >40 × 106 cells at each passage. Cells were grown in selective media until 7 d (UM-Chor1) or 8 d (MUG-Chor1) post-infection, after which they were grown in standard growth media. At 21 d post-infection, cells were collected and stored at −20°C in PBS until genomic DNA isolation steps.
Genomic DNA was purified from cell pellets using the NucleoSpin Blood XL Kit (Macherey-Nagel). The sgRNA sequence was PCR-amplified with sufficient gDNA to maintain representation and then quantified using massively parallel sequencing43,44. For each cell line, primary screening was performed once with three replicates.
CRISPR-Cas9 screening data quality control
Cell lines with poor Cas9 activity (<50%) were excluded from screening. Quality-control measures were used to remove cell line replicate sample data where (1) the single-nucleotide polymorphism (SNP) genotype fingerprint failed to match the reference cell line as previously described43; (2) the reproducibility between replicates was less than 70%; (3) the strictly standardized mean difference (SSMD) using positive (proteasome, ribosome, and spliceosome components) and non-targeting controls of fold-change data was poor (>−0.75); and (4) principal component analysis showed a replicate or cell line to be a global outlier of the dataset.
CRISPR-Cas9 data processing and analysis
Data were processed in a reproducible GenePattern pipeline containing individual modules, all available from GParc (gparc.org). Normalized read counts per million were calculated and compared to the abundance of the initial DNA plasmid pool to calculate a log2 fold change per sgRNA after removing sgRNAs with low abundance in the initial plasmid pool (GP modules: FilterLowPert, PertFoldChange)43. The data were batch corrected using the ComBat algorithm to account for a known reagent change during screening (PertBatchCor)45. The median of non-targeting controls (n = 1,000) in the Avana library was subtracted from each sgRNA to generate a sgRNA-level score (“sgRNA-dependency score”) and plots (NormToNegCntrls, collapseReps). Guides that targeted more than one perfect match location were removed and normalization across cell lines was performed using LOWESS to enable cross-cell-line comparisons (gctRowKeep, NormLines). STARS analysis was performed on LOWESS-normalized sgRNA-level data. The comparison of chordoma and non-chordoma lines was performed using PARIS43 on LOWESS-normalized sgRNA-level data. The rescaled normalized mutual information (RNMI) score from PARIS was used to rank chordoma selectively essential sgRNAs.
The Mann-Whitney test reported in Fig. 2c was performed in MATLAB (MathWorks, Inc.).
Validation of CRISPR-Cas9 screens
UM-Chor1, MUG-Chor1, and U-CH2 cells were each incubated with lentivirus corresponding to the pXPR_001 plasmid, encoding either sg-T #1, sg-T #2, or sg-EGFP (sequences below), in the presence of 4 μg/mL polybrene, dispensed in 12-well plates (1.5 × 106 cells/well for UM-Chor1 and MUG-Chor1, 1.2 × 106 cells/well for U-CH2), and spin-infected at 2,000 rpm for 2 h at 30°C. After spin-infection, 2 mL of standard growth media was added to each well, and cells were incubated at 37°C overnight. The following day, for each cell line, cells were trypsinized and expanded in selective media containing puromycin (2 µg/mL for UM-Chor1, 1–2 µg/mL for MUG-Chor1, 3 µg/mL for U-CH2). Following selection for infected cells, cells were seeded in 24-well plates in selective media (30,000 cells/well for UM-Chor1 and U-CH2 and 40,000 cells/well for MUG-Chor1, with three replicates per timepoint), and counted at indicated intervals over a 25 d period. In parallel, selected cells were harvested (at least 9 d post-transduction) for immunoblotting to confirm sgRNA-mediated protein reduction. Proliferation experiments were performed twice for UM-Chor1 and U-CH2 with minor modifications between experiments and once for MUG-Chor1, with three replicates per experiment. Immunoblots were performed once. Statistical analysis was performed with GraphPad Prism v7.
Lentiviral vectors used for screening validation and functional characterization
The T-targeting sgRNAs used for low-throughput experiments were cloned in the pXPR_001 lentiviral vector, which encodes the Cas9 protein46. Spacer sequences for T-targeting sgRNAs were as follows: sg-T #1, CCCTGAGACCCAGTTCATAG; sg-T #2, TGGCTGGTGATCATGCGCTG. The pXPR_001 plasmid encoding sg-EGFP was provided by the laboratory of Feng Zhang (Broad Institute, Cambridge, MA) and has been described previously (“EGFP sgRNA 6”)46. Lentivirus was produced by transfection of 293T packaging cells with three plasmids [pXPR_001-sgRNA, Δ8.9 (gag, pol), and VSV-G]; and the FuGene6 transfection reagent (Roche). Virus-containing supernatant was harvested 3 d post-transfection.
Small-molecule sensitivity profiling
Chemical sensitivity tests similar to those reported in the Cancer Therapeutics Response Portal (CTRP: www.broadinstitute.org/ctrp/) were performed using 459 small molecules chosen collectively to target a diverse set of nodes in cell circuitry, and with a high degree of overlap to published informer sets (Supplementary Table 2)22, including 428 compounds published in CTRP. UM-Chor1, MUG-Chor1, U-CH1, and JHC7 cells were each seeded overnight in 384-well BioCoat Collagen I (Corning) microtiter plates at a density of 1200, 1800, 2000, and 1600 cells per well, respectively. The following day, compound or DMSO was added to wells (1:400 dilution) using a CyBio pinning instrument. Each compound was tested using 8 concentrations in duplicate. Cells were incubated at 37°C, and cell viability was assayed 6 d after the addition of compound or DMSO using the CellTiter-Glo reagent (Promega). For each cell line, primary screening was performed once with two replicates.
Computational analysis software for small-molecule sensitivity studies
Computational analyses and visualizations were performed in Microsoft Excel Professional Plus 2013, GraphPad Prism (v6 or v7), Pipeline Pilot (version 8.5) (Accelrys, Inc.), or MATLAB 8.4 (2014b) or MATLAB 9.4 (2018a) (MathWorks, Inc.), as described in the following specific Methods sections.
Identification of candidate antiproliferative small molecules in chordoma
The response of each chordoma cell line to a tested compound was quantified as follows: log-transformed duplicate data were averaged during normalization of luminescence values to vehicle (DMSO) treatment47, and at each compound concentration, an average percent-viability (PV) score was calculated. PV response-point measurements were computed across all cell lines, compounds, and concentrations. Curves were fit with nonlinear sigmoid functions, and the area-under-curve (AUC) for each CCL-compound pair was calculated by numerically integrating under the 8-point concentration-response curve essentially as described previously22. In our analysis, we differed from the published procedure in (1) the number of concentrations tested; (2) the use of fixed concentration limits of integration for AUCs across all compounds; (3) standardization of AUC values on the range from 1 (DMSO or no effect) to 0 (complete killing throughout the concentration range of integration); cf. https://github.com/remontoire-pac/ctrp-reference/tree/master/auc. Data pre-processing from instrument files through DMSO normalization was performed in Pipeline Pilot, and curve-fitting, numerical integration, and subsequent analysis steps were performed in MATLAB.
To identify candidate antiproliferative small molecules in chordoma, a threshold was applied to include only potent compounds (average AUC across four chordoma cell lines < 0.8). Next, to assess whether a compound had selectively cytotoxic effects in chordoma lines relative to other cell lines tested in CTRP, we computed the median AUC for 4 chordoma lines and z-scored this median with respect to the mean and standard deviation of all other cell lines tested with that compound. We applied this selectivity analysis to filter out potent compounds whose cytotoxic effects were non-selective for chordoma cell lines (z-score < −1), when those data were available. We note that some compounds were only tested in the chordoma lines, and therefore data about their performance across CTRP lines were not available; in such cases, we did not apply any additional filtering step beyond the initial potency filter. Twenty-eight compounds passed these criteria, and 431 compounds were filtered out. Compounds targeting proteins of interest (CDK 7,12,13,9 and EGFR/ERBB2) were carried forward for secondary screening.
Validation of small-molecule sensitivity screens
UM-Chor1, MUG-Chor1, U-CH1, JHC7, and CH22 cells were each seeded overnight in 384-well BioCoat Collagen I (Corning) microtiter plates at a density of 1200, 1800, 2000, 1600, and 1200 cells per well, respectively. The following day, compound or DMSO was added to wells using an HP D300 digital dispenser instrument. Each compound was tested using 9 concentrations, in quadruplicate (four wells treated in parallel). Cell viability was assayed 6 d after compound addition with the CellTiter-Glo reagent (Promega). Luminescence values were treated as with the primary screening data (vide supra). Data pre-processing from instrument files through DMSO normalization was performed in Pipeline Pilot for UM-Chor1, MUG-Chor1, U-CH1, JHC7, or Microsoft Excel for CH22; and curve-fitting, numerical integration, and subsequent analysis steps were performed in MATLAB. Experiments depicted in Fig. 2b, Extended Data Fig. 2a, and Extended Data Fig. 3a were performed at least twice in each cell line (except afatinib treatment in JHC7 cells, which was performed once); experiments depicted in Extended Data Fig. 8c were performed at least three times in UM-Chor1 cells and once for all other cell lines. Experiments depicted in Extended Fig. 10a were performed once. For experiments performed multiple times, compound addition was performed using either a digital dispenser instrument, as described above, or a Cybio pinning instrument.
Small-molecule reagents
Small molecules used for low-throughput ex vivo experiments were obtained from the following sources: THZ1, THZ531, and NVP-2 were provided by the laboratory of Nathanael Gray (Dana-Farber Cancer Institute; Boston, MA); JQ1 was provided by the laboratory of James Bradner (Dana-Farber Cancer Institute; Boston, MA); CCT251545 was provided by Paul Clarke and Aurelie Mallinger (The Institute of Cancer Research; London, UK); alvocidib, AT7519, dinaciclib, LEE011, and palbociclib were purchased from Selleck Chemicals; erlotinib and dasatinib were purchased from LC Laboratories; actinomycin D was purchased from Gibco; neratinib, canertinib, lapatinib, and afatinib were obtained from Broad Institute Compound Management and were originally purchased from Selleck Chemicals; BRD6989 was obtained from Broad Institute Compound Management and was originally purchased from Vitas-M Laboratory (cat# STL241555). Small molecules used for high-throughput screening were obtained from Broad Institute Compound Management (Broad Institute Compound IDs reported in Supplementary Table 2). THZ1 Hydrochloride used for in vivo studies was purchased from MedChemExpress.
RNA-sequencing
For RNA-sequencing experiments with sgRNA-treated cells: UM-Chor1 cells were incubated with lentivirus corresponding to the pXPR_001 plasmid, encoding either sg-T #1, sg-T #2, or sg-EGFP in the presence of 4 μg/mL polybrene, dispensed in 12-well plates (1.3 × 106 cells/well), and spin-infected at 2,000 rpm for 2 h at 30°C. After spin-infection, 2 mL of standard growth media was added to each well, and cells were incubated at 37°C overnight. The following day, cells were trypsinized and expanded in selective media containing puromycin (2 µg/mL). Following selection for infected cells, selective media was replaced with standard growth media, and cells were harvested 2 d later. Cell pellets were used for RNA extraction using an RNeasy Kit (Qiagen). Sequencing libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs), and libraries were sequenced using the Illumina HiSeq 2500 instrument (rapid-run mode) with paired-end 100 bp reads. Sequencing was performed once with two biological replicates for each condition.
For RNA-sequencing experiments with compound-treated cells: UM-Chor1 cells were seeded overnight in 6 cm collagen I-coated dishes at a density of 750,000 cells per dish. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Compound- and DMSO-treated cells were harvested 4 h after compound or DMSO addition. Cell pellets were used for RNA extraction using an RNeasy Kit (Qiagen). Sequencing libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina), and libraries were sequenced using the Illumina NextSeq 500 instrument with single-end, 75 bp reads. Sequencing was performed once with two biological replicates for each condition.
RNA-sequencing analysis
For all RNA-seq experiments, raw reads were processed with Trim Galore! (version 0.4.1) (Babraham Bioinformatics; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) to remove adapter sequences and aligned with STAR (version 2.4.2)48 to the human genome (GRCh38, primary assembly) using the GENCODE annotation (version 23). Differential expression analysis was performed in R (version 3.2.1) (http://www.R-project.org/) using the DESeq2 package (version 1.10.0)49.
Gene set enrichment analysis was performed with GSEA2 (version 2.2.0) and gene sets from MSigDB (version 5.0)50,51. We used the “preranked” algorithm to analyze gene lists ranked by the negative decadic logarithm of adjusted p-values obtained from the differential-expression analysis with DESeq2. To separate up- and down-regulated genes, we artificially assigned a negative sign to values for downregulated genes (thus using the decadic logarithm of adjusted p-values). We used the options –nperm 1000, -set_max 1500, and –set_min 5.
Copy number variation analysis
Whole-exome sequencing data for all chordoma cell lines were obtained from the Chordoma Foundation (available at www.cavatica.org) and processed according to the GATK Best Practices52. Briefly, starting with reads in an unaligned BAM file, we marked sequencing adapters with Picard (version 1.999) MarkIlluminaAdapters, aligned reads to the human genome (hg19) with BWA (version 0.7.12)53, and marked duplicates with Picard MarkDuplicates (Broad Institute Picard; https://github.com/broadinstitute/picard). We then used the GATK3 toolkit (version 2.2) to perform Indel Realignment and Base Recalibration (Broad Institute GATK; software.broadinstitute.org/gatk)54. Somatic copy number variations were determined using the GATK4 toolkit (Broad Institute GATK; https://gatkforums.broadinstitute.org/gatk/discussion/9143/how-to-call-somatic-copy-number-variants-using-gatk4-cnv).
RT-qPCR
RT-qPCR of compound-treated cells
UM-Chor1 cells were seeded overnight in 6-well collagen I-coated dishes at a density of 275,000 cells per well. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Compound- and DMSO-treated cells were harvested 4, 8, and 12 h after compound or DMSO addition, and cell pellets were used for RNA extraction using an RNeasy Kit (Qiagen). Three biological replicates were generated per condition. Total RNA was reverse transcribed to cDNA using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). cDNA was used for qPCR using the Taqman Gene Expression Master Mix and Taqman Gene Expression probes (Applied Biosystems). For each cDNA sample, four technical replicate reactions were performed using probes for the gene of interest, and, in parallel, for the GAPDH reference gene. The Taqman probes used in the study were as follows: T, hs00610080_m1; MAX, hs00811069_g1; CFL1, hs02621564_g1; GAPDH, hs02758991_g1. Amplification was performed using the QuantStudio 6 Flex instrument (Applied Biosystems). The experiment was performed twice with variations in cell seeding density and replicate numbers, with similar conclusions.
RT-qPCR of sgRNA-treated cells
UM-Chor1 cells were transduced with sgRNAs targeting T and EGFP and subjected to RNA extraction as described in the RNA sequencing methods section. Total RNA was used for cDNA synthesis and qPCR, with four technical replicates per qPCR reaction condition, as described above. The Taqman probes used in the study were as follows: T, hs00610080_m1; GAPDH, hs02758991_g1; ATP6V1B2, hs00156037_m1; SAE1, hs00271440_m1; SOX9, hs00165814_m1; TPX2, hs00201616_m1. The experiment was performed once with two biological replicates for each sgRNA.
Analysis of RT-qPCR data
RT-qPCR data were analyzed according to the instrument manufacturer’s instructions for relative quantity (RQ) calculations using the ΔΔCT method. For each sample, cycle threshold (CT) values were averaged across technical replicates. ΔCT values were then calculated by subtracting the average reference-gene (GAPDH) CT values from average target-gene CT values, for each biological replicate. We applied a one-sided Welch’s t-test comparing ΔCT values for treatment and reference samples to determine significant differences. Log2-fold-changes were calculated as ΔΔCT values by subtracting ΔCT values for the reference samples from ΔCT values for the treated samples. All calculations were done in R (version 3.4.4).
Caspase-3/7 activity assay
UM-Chor1 cells were seeded overnight in 384-well BioCoat Collagen I (Corning) microtiter plates at a density of 1200 cells per well. The following day, THZ1 or DMSO was added to wells using a HP D300 digital dispenser instrument. Cells were incubated at 37°C, and caspase-3/7 activity and cell viability were assayed in parallel 24 h and 48 h after the addition of compound or DMSO using the Caspase-Glo 3/7 and CellTiter-Glo reagents, respectively (Promega). Four replicate wells were treated per condition. Luminescence values corresponding to compound-treated cells were normalized to those of DMSO-treated cells to calculate relative caspase-3/7 activity or relative cell viability. The experiment was performed twice.
Compound or sgRNA treatment for immunoblot analysis
For Fig. 4a: MUG-Chor1 and UM-Chor1 cells were seeded overnight in 6 cm collagen I-coated dishes at a density of 750,000 cells per dish. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Cells were harvested 48 h after compound or DMSO addition and cell pellets were used for immunoblot analysis. The experiment was performed four times for UM-Chor1 with variations in compound concentration and incubation times, with similar conclusions, and once for MUG-Chor1.
For Fig. 4b and Extended Data Fig. 10b: MUG-Chor1, UM-Chor1, U-CH1, JHC7, and CH22 cells were each seeded overnight in 6 cm collagen I-coated dishes at a density of 750,000 cells per dish. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Cells were harvested 48 h after compound or DMSO addition, and cell pellets were used for immunoblot analysis. The experiment was performed two times for MUG-Chor1, UM-Chor1, and U-CH1, with variations in compound concentration and incubation times, with similar conclusions, and once for JHC7 and CH22.
For Extended Data Fig. 8a: UM-Chor1 cells were seeded overnight in 6 cm collagen I-coated dishes at a density of 750,000 cells per dish. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Cells were harvested 12 h, 24 h, 36 h, and 48 h after compound or DMSO addition and cell pellets were used for immunoblot analysis. The experiment was performed once.
For Fig. 2d and Extended Data Fig. 3b: UM-Chor1 cells were seeded overnight in 6 cm collagen I-coated dishes at a density of 750,000 cells per dish. The following day, media was replaced with standard growth media containing compound at the final concentration or DMSO. Cells were harvested 24 h after compound or DMSO addition, and cell pellets were used for immunoblot analysis. The experiment was performed 2 times for THZ1, with minor variations in compound concentrations, with similar conclusions, and CCT251545; and once for other compounds.
For Extended Data Fig. 9b, UM-Chor1 cells were transduced with sgRNAs targeting T and EGFP and used for immunoblotting as described in the methods section describing validation of CRISPR-Cas9 screens. Transductions were performed once; immunoblots were performed twice.
Immunoblotting
Cell pellets were resuspended in lysis buffer (50 mM Tris pH 7.4, 2.5 mM EDTA pH 8, 150 mM NaCl, 1% Triton X-100, 0.25% IGEPAL CA-630) containing protease inhibitors (Roche) and Phosphatase Inhibitor Mixtures I and II (Calbiochem). Lysates were incubated on ice for >2 min, then centrifuged for 2 min at 15,700 x g. Supernatants were quantified using a BCA Protein Assay Kit (Pierce), normalized, reduced, and denatured. Protein samples were then resolved using Tris-Glycine gels (Novex), and resolved protein was transferred to iBlot Transfer Stack nitrocellulose membranes (Novex). Membranes were probed with primary antibodies at 4°C overnight. The antibodies against brachyury (clone N-19, #sc-17743; 1:1,000) and CDK7 (clone C-4, #sc-7344, 1:1,000) were purchased from Santa Cruz Biotechnology. Antibodies against cofilin (clone D3F9, #5175; 1:10,000), total Rb (clone 4H1, #9309; 1:2,000), phospho-Rb S807/S811 (clone D20B12, #8516; 1:1,000), phospho-Rb S780 (clone D59B7, #8180; 1:1,000), SOX9 (clone D8G8H, #82630, 1:1,000), total STAT1 (clone 9H2, #9176; 1:1,000), and phospho-STAT1 S727 (clone D3B7, #8826; 1:1,000) were purchased from Cell Signaling Technology. Antibodies against phospho-POLR2A Ser2 (clone 3E1, #04–1571-I; 1: 1,000), phospho-POLR2A Ser5 (clone 3E8, #04–1572-I; 1:1,000), and phospho-POLR2A Ser7 (clone 4E12, #04–1570-I; 1:1,000) were purchased from Millipore. The antibody against total POLR2A (#A300–653A; 1:400) was purchased from Bethyl Laboratories. The antibody against the V5 tag (#46–0705, 1:5,000) was purchased from Invitrogen. Membranes were incubated with IRDye secondary antibodies (1:10,000; LI-COR Biosciences) and detected with the Odyssey Imaging System (LI-COR Biosciences). Immunoblots were quantified by densitometry, typically using the predominant band species, with ImageJ software (NIH). Quantification results appear in Supplementary Table 10.
Patient-derived chordoma tumor and normal adjacent tissue samples
Patient-derived tumor and normal adjacent tissue specimens from Massachusetts General Hospital (MGH) were obtained from the clinical archives of Dr. Francis Hornicek (Department of Orthopaedic Surgery, Massachusetts General Hospital). Institutional review board (IRB) approval was obtained to study these tumor samples via Dana-Farber/Harvard Cancer Center protocol number 03–344. Patients were consented according to protocol. Following resection, tissue was flash frozen and stored at −80°C until later use.
Patient-derived tumor specimens from the Chordoma Foundation (CF) Biobank were collected under the CF’s IRB-approved protocol QR26112/1. Patients gave their written informed consent for scientific study of their tumor cells. Following resection, tissue was flash frozen and stored in liquid nitrogen or −80°C until later use for ChIP-seq. Immunohistochemistry was performed as described below.
Studies were in compliance with all relevant ethical regulations.
Immunohistochemical staining for brachyury expression
Staining was performed by the Histology Core Facility at Ohio State University. Slides were cut from FFPE blocks of patient tissue and were heated at 75°C for 15 min, then dewaxed in xylene and graded alcohols as follows: xylene, 5 min (2X); 100% ethanol, 5 min (2X); 95% ethanol, 2 min; 70% ethanol, 2 min; 50% ethanol, 2 min; distilled water, 2 min. Antigen retrieval was performed by steaming in citric acid pH 6 (10 mM) for 15 min. Slides were cooled for 15 min, then washed with water for 5 min. Slides were blocked with hydrogen peroxide 3% for 15 min at RT then washed with water for 5 min, followed by a quick PBS wash. Slides were incubated with blocking buffer for 30 min at RT (Dako, #X0909), followed by incubation with the brachyury antibody (Santa Cruz Biotechnology, #sc-17743, 1:150) overnight at 4°C. Slides were washed with PBS 3 X 5 min, then incubated with secondary antibody (Cell IDx, #GH-015) for 1h at 37°C. Slides were washed with PBS 3 X 5 min. Stains were developed using DAB, then washed with PBS 3 X 5 min. Counterstaining was performed with hematoxylin. Tissue sections were dehydrated with graded alcohols as follows: 50% ethanol, 2 min; 70% ethanol, 2 min; 95% ethanol, 2 min; 100% ethanol, 2 min (2X); xylene, 5 min (2X). Slides were mounted using Cytoseal (Richard-Allan Scientific).
Images were converted to high-resolution digital images using Leica ScanScope XT. Images were visualized for figures using Aperio ImageScope software.
Chromatin immunoprecipitation (ChIP)
For H3K27ac ChIP of patient-derived tissue:
approximately 100 mg of flash-frozen tissue was minced into 1–2 mm pieces and incubated in 1% formaldehyde for 15 minutes, followed by quenching with glycine (125 mM final concentration). Fixed tissue pieces were homogenized with a Tissue Tearor rotor stator homogenizer (Biospec) set to 30,000 RPM for 60 seconds. Homogenate was washed with ice-cold PBS containing 1X HALT protease inhibitor. Homogenized pellets were then resuspended in cytosolic lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% IGEPAL, 0.25% Triton X-100, 1X HALT protease inhibitor) and the protocol was continued as described below.
For H3K27ac ChIP of chordoma cell lines:
Chordoma cell lines were grown in 175 cm collagen I-coated plates (3 plates per cell line) and cross-linked by adding 1/10 of the cell culture volume of 11% formaldehyde solution (1M HEPES-KOH pH 7.5, 0.5 M EDTA pH 8.0, 0.5 M EGTA pH 8.0, 5 M NaCl, 37% formaldehyde) for 10 minutes, followed by quenching (125 mM glycine). Cells were washed in PBS and harvested by cell scraper in PBS. Cells were centrifuged at 1,350 X g for 5 min at 4°C, washed with PBS, and centrifuged again at 1,350 X g for 5 min at 4°C. Cell pellets were flash frozen and stored at −80°C until further processing.
For both cell line and tissue ChIP-seq:
samples were subsequently rotated end-over-end for 10 minutes at 4º C in cytosolic lysis buffer and then collected by spinning at 1,350 x g for 5 minutes. Samples were resuspended in nuclear lysis buffer (10 mM Tris-HCl pH 8, 200 mM NaCl, 1 mM EDTA, 0.5mM EGTA, 1X HALT protease inhibitor) and rotated end-over-end for 10 minutes at 4º C and spun at 1,350 x g for 5 minutes. Samples were resuspended in 1 mL sonication buffer ( 0.1% Na-Deoxycholate, 50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1X HALT protease inhibitor) with SDS added to a final concentration of 0.5%. Chromatin was sheared by sonication using a Bioruptor water bath sonicator (Diagenode) for 25 cycles. Samples were clarified by centrifuging 10 minutes at 20,000 x g at 4º C. A 50 uL aliquot of supernatant was set aside as non-IP input control. The remaining sheared chromatin was diluted 1:5 in sonication buffer and each sample was incubated overnight at 4º C with 100 uL of Protein G Dynabeads (Life Technologies) bound with 10 µg of anti-H3K27ac antibody (abcam, #ab4729). Beads were washed while rotating end-over-end at 4º C with the following buffers: twice in sonication buffer, once in sonication buffer with NaCl added to a final concentration of 500 mM, once in LiCl wash buffer (20 mM Tris pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% IGEPAL, 0.5% Na-Deoxycholate), and once in TE-NaCl buffer (50 mM Tris-HCl pH 8, 50 mM NaCl). Chromatin was eluted by resuspending beads in 200 uL elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS) and heating for 15 minutes at 65º C. Beads were pelleted by centrifuging at 20,000 G for 30 seconds and supernatant was collected. Elution buffer (150 uL) was also added to input chromatin samples. Samples were incubated at 65º C overnight to reverse crosslinks. Samples were then incubated with RNAse A (Thermo Scientific) for 2 hours at 37º C, followed by incubation with Proteinase K (Ambion) for 30 minutes at 55º C. DNA isolation was performed via phenol-chloroform extraction and concentrated by ethanol precipitation. Sequencing libraries were generated with the Thruplex DNAseq Single Index Kit (Rubicon) and sequenced on an Illumina NextSeq 500 with single-end, 75 bp reads. ChIP-seq was performed once for each sample.
Assay for transposase-accessible chromatin (ATAC)
For each cell line (MUG-Chor1 and U-CH2), 50,000 cells were lysed for 10 minutes at 4°C in lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-360). After lysis, the pellets were subject to a transposition reaction (37°C, 60 minutes) using the 2X TD buffer and transposase enzyme (Illumina Nextera DNA preparation kit, FC-121–1030). The transposition mixture was purified using a Qiagen MinElute PCR purification kit. Library amplification was performed using custom Nextera primers and the number of total cycles determined by running a SYBR-dye based qPCR reaction and calculating the cycle number that corresponds to ¼ the maximum. Amplified libraries were purified using a Qiagen PCR purification kit and sequenced with paired-end 100 bp reads on an Illumina HiSeq 2000. ATAC-seq was performed once for each cell line, with two biological replicates for U-CH2 and one replicate for MUG-Chor1.
ChIP-seq data analysis
Genomic coordinates and gene annotation.
All coordinates and gene annotations in this study were based on human reference genome assembly hg19, GRCh37 (ncbi.nlm.nih.gov/assembly/2758/) and RefSeq genes.
ChIP-seq data processing.
All datasets were aligned using Bowtie2 (version 2.2.1) to build version NCBI37/HG1955. Alignments were performed using all default parameters except for –N 1.
Calculating read density.
We calculated the normalized read density of a ChIP-seq dataset in any genomic region using the Bamliquidator (version 1.0) read density calculator (https://github.com/BradnerLab/pipeline/wiki/bamliquidator). Briefly, ChIP-seq reads aligning to the region were extended to 200 bp and the density of reads per base pair (bp) was calculated. The density of reads in each region was normalized to the total number of million mapped reads producing read density in units of reads per million mapped reads per bp (rpm/bp).
Plotting composite representations of ChIP-seq signal at individual loci.
To compactly display ChIP-seq signal from multiple cell line or patient-derived chordoma samples at individual genomic loci samples, we developed a simple meta representation as previously described35. For all samples within a group, ChIP-seq signal is smoothed using a simple spline function and plotted as a translucent shape in units of rpm per bp. Darker regions indicate regions with signal in more samples. An opaque line is plotted and gives the average signal across all samples in a group.
Identifying enriched regions.
We used the MACS version 1.4.2 (model-based analysis of ChIP-seq) peak finding algorithm to identify regions of ChIP-seq enrichment over background56. A p-value threshold of enrichment of 1e-9 was used for all datasets.
Defining active genes.
Across chordoma cell lines, active genes were defined as those with an enriched H3K27ac region in at least one cell line in the +/− 1kb region flanking the transcription start site (TSS).
Mapping typical enhancers and super-enhancers using H3K27ac enhancer definitions in individual samples.
H3K27ac super-enhancers (SEs) and typical enhancers (TEs) in individual chordoma samples were mapped using the ROSE2 software package described previously57 and available at https://github.com/BradnerLab/pipeline - ROSE2_main.py. For each dataset, a stitching parameter was determined that consolidated proximal peaks while optimizing the enriched fraction of stitched peaks. Peaks contained within the +/− 2.5 kb flanking the transcription start site of genes were excluded (-t 2500 parameter). To assign enhancers to the T or MAX genes, a 100 kb proximity window was used (Extended Data Figs. 4 and 6).
Mapping typical enhancers and super-enhancers using H3K27ac enhancer definitions across composites of cell lines or patient-derived chordoma samples.
H3K27ac super-enhancers (SEs) and typical enhancers (TEs) composite landscapes in either chordoma cell lines or patient-derived samples were mapped using the ROSE2 software package described previously57 and available at https://github.com/BradnerLab/pipeline - ROSE2_META.py. Briefly, for a given cohort of samples (e.g. cell lines), the union of all discrete H3K27ac enriched regions was determined. For each cohort of samples, a stitching parameter was determined that consolidated proximal peaks while optimizing the enriched fraction of stitched peaks. The average background subtracted signal for each sample across a cohort was averaged and used to rank enhancers. To assign enhancers to all genes, a 50 kb proximity window was used. Across chordoma cell lines, we identify a total of 1,199 SEs (Fig. 3e and Supplementary Table 5). Across patient-derived chordoma samples, we identified 1,490 SEs (Fig. 3g and Supplementary Table 7). Locations, ranks, proximal and overlapping genes for composite chordoma cell line and patient-derived sample SEs are provided in Supplementary Tables 5 and 7.
Quantifying expression of SE-associated transcription factors.
Cell line expression data were provided by the Chordoma Foundation (available at www.cavatica.org). Briefly, reads were aligned with STAR aligner and quantified with Sailfish to produce expression measurements in units of transcripts per million (tpm). We selected all genes that were (1) associated with SEs in chordoma cell lines, (2) expressed (>1 tpm in at least 1 sample), and (3) annotated as transcription factors in UniProt, resulting in 115 genes. Gene-expression levels for the top 30 SE-associated TFs, ranked by mean expression, are shown in Fig. 3d; levels for all 115 genes are shown in Extended Data Fig. 5c.
Clustering ChIP-seq samples.
To cluster ChIP-seq samples by similarity of H3K27ac enhancer profiles, we first defined the union of all enhancer regions present in any individual chordoma sample. ChIP-seq signal was calculated at all regions, background subtracted, and then median normalized. Samples were then clustered using hierarchical clustering of cross sample Pearson correlation (Fig. 3j).
ATAC-seq data analysis
All paired-end datasets were aligned using Bowtie2 (version 2.2.1) to build version NCBI37/HG19 with the following parameters: -k 1. Aligned bams were filtered and sorted using samtools (0.1.19) by removing chrM and duplicate reads, and filtering against the ENCODE blacklist (https://sites.google.com/site/anshulkundaje/projects/blacklists). Peaks were called using MACS version 1.4.2 with a p-value cutoff of 1e-9.
ORF rescue experiments
For cell viability experiments, UM-Chor1 cells were seeded overnight in 384-well BioCoat Collagen I (Corning) microtiter plates at a density of 1000 cells per well. The following day, cells were incubated with lentivirus corresponding to the pLX-Blast-V5 expression vector58 encoding either the LACZ or T (p.Gly177Asp variant)12 ORF in the presence of 4 μg/mL polybrene, spin-infected at 1,126 × g for 30 min at 30 °C, then incubated at 37 °C for an additional 4.5 – 5.5 h before replacing media with standard growth media. Three days post-infection, cells were treated with 500 nM THZ1 (12 replicate wells treated in parallel) or DMSO (8 replicate wells treated in parallel) using a using an HP D300 digital dispenser instrument. Cell viability was assayed 6 d after the addition of THZ1/DMSO using the CellTiter-Glo reagent (Promega). Infection efficiency was assayed in a parallel experiment by adding 3 μg/mL blasticidin to infected wells one day post-infection. The experiment was performed three times. Statistical analysis was performed with GraphPad Prism v7.
For the immunoblot experiment depicted in Fig. 4d, UM-Chor1 cells were seeded overnight in 6-well collagen I-coated plates at a density of 300,000 cells per well. The following day, cells were treated with lentivirus corresponding to the pLX-Blast-V5 expression vector encoding either the LACZ or T (p.Gly177Asp variant) ORF in the presence of 8 μg/mL polybrene, and incubated at 37 °C for ~19–20 h before replacing media with standard growth media. The following day, media was replaced with selective media containing blasticidin (4 μg/mL). Following selection for infected cells (at least 7 d later), media was replaced with standard growth media containing compound at the final concentration or DMSO. Cells were harvested 48 h after compound or DMSO addition, and cell pellets were used for immunoblot analysis. The experiment was performed twice.
STARS and integrative analyses
STARS, an algorithm designed to rank genes in genetic perturbation screens (https://portals.broadinstitute.org/gpp/public/software/stars ), was used to identify significantly essential genes in each chordoma line screened. sgRNAs were ranked by the sgRNA-dependency score in each line, then STARS was run at a 5% threshold to ensure that at least 2 independent sgRNAs scored for each gene in the top 5% of all sgRNAs screened. Genes scored as essential in each line, if they passed these criteria. For Fig. 4f, those genes scoring in each chordoma cell line were then intersected with the list of expressed, super-enhancer-associated genes in the same cell line. Gene-expression levels in each cell line were assessed using RNA-sequencing data, which was obtained from the Chordoma Foundation (available at www.cavatica.org), and processed as described in the RNA-sequencing analysis methods. H3K27ac super-enhancers (SEs) in individual chordoma cell lines were mapped as described in the ChIP-seq data analysis methods section, except that we used a (higher confidence) 50 kb proximity window to assign enhancers to genes. For a gene to be considered downregulated by sg-T #1 and sg-T #2 in Fig. 4f, the relative gene expression (as measured by RNA-sequencing) of cells transduced with sg-T versus a non-targeting control must have had a log2-fold change < 0 and an adjusted p-value < 0.01. Brachyury ChIP-seq analysis was performed as described in the associated methods section.
Brachyury ChIP-seq analysis
We obtained FASTQ files for a previously reported brachyury ChIP-seq experiment33. We aligned ChIP-seq reads to the human reference genome (hg19) using Bowtie 2 (version 2.2.1)55 and converted the aligned BAM files to BED files using BEDtools (version 2.26.0)59. Peaks were then called with MACS (version 1.4.2)56 and annotated with nearby genes using HOMER (version 4.9.1) (http://homer.ucsd.edu/homer/)60. To be demarcated with a cross in Fig. 4f, a gene must have been associated with a peak passing the significance threshold (p < 0.05) in all three ChIP-seq replicates.
In vivo treatment studies
All breeding, mouse husbandry, and in vivo experiments were performed with the approval of the NYU Langone Medical Center (NY, NY) Animal Care and Use Committee.
SCID Hairless Outbred (SHO), all female, 6–8 week-old mice were purchased from Charles River Laboratories International Inc. (#474). CH22 cells were cultured in RPMI media + 10% FBS on collagen I-coated 175 cm2 flasks (Corning). Cells were resuspended in serum-free medium mixed with an equal amount of Matrigel (Corning, #354234). For the study depicted in Fig. 4h–i and Extended Data Fig. 10e (bottom panel): mice were injected with 8 million cells per shot and 2 locations per mouse in the flanks. The mice were randomly grouped. Each cohort included 10 mice. Treatment was started when the mean tumor size per group reached approximately 500 mm3. Three mice of each group were taken out on Day 3 for target engagement analysis. One mouse in the vehicle group did not have tumor engrafted and was therefore excluded from the study. Tumor sizes were monitored weekly, and volumes were calculated with the following formula: (mm3) = length × width × width × 0.5.
THZ1 was dissolved in DMSO:5% dextrose (1:10) and dosed as 40 mg/kg twice daily via the intraperitoneal route. Vehicle-group mice received DMSO:5% dextrose (1:10) via the same route. The dosing schedule for the first week was continuous dosing for the first three days; for subsequent weeks, animals were dosed three times a week, on alternating days, as depicted in Fig. 4h. Statistical analysis was performed with GraphPad Prism v7. The efficacy study was performed once.
For the experiments depicted in Extended Data Fig. 10d and Extended Data Fig. 10e (top panel), injections and compound treatment were performed in a similar fashion as the target engagement study described above, except that the experiment depicted in Extended Data Fig. 10d was performed for 5 d and had an additional treatment arm of 20 mg/kg THZ1 twice daily. Each of the three target engagement studies reported was performed once.
For immunoblotting of tumor tissue, tumor fragments were homogenized in lysis buffer (recipe described in the immunoblotting methods section) using the Precellys Evolution (Bertin Technologies) instrument (3 × 7500 rpm for 20 s, pausing for 15 s between rounds). Homogenized lysates were centrifuged for 2 min at 15,700 x g. Supernatants were quantified and used for immunoblotting as described in the immunoblotting methods section. Immunoblots were performed once.
Studies were in compliance with all relevant ethical regulations.
Code availability
Computational code used for ChIP-seq analysis can be found at github.com/linlabcode/chordoma_code. Code for CRISPR-Cas9 screening analysis is available on GParc (www.gparc.org). Code for small-molecule sensitivity profiling analysis; and DNA and RNA sequencing analysis are available upon reasonable request.
Data availability
CRISPR-Cas9 screening data for two chordoma cell lines (pertains to Figs. 1, 2, 4, and Extended Data Fig. 1) are available at Figshare (DOI: 10.6084/m9.figshare.7302515). CRISPR-Cas9 screening data for all other cancer cell lines (pertains to Figs. 1, 2, 4, and Extended Data Fig. 1) were generated as part of Project Achilles (Broad Institute Project Achilles; https://depmap.org/portal/achilles/). All RNA-sequencing data (pertains to Figs. 1 and 4) are available at Gene Expression Omnibus (GEO) (accession number: GSE121846). Small-molecule sensitivity data generated using non-chordoma cell lines and used for comparative analyses (pertains to Fig. 2) are available at the National Cancer Institute’s CTD2 Data Portal (https://ocg.cancer.gov/programs/ctd2/data-portal) and the CTRP (www.broadinstitute.org/ctrp/). The analysis of new small-molecule primary screening data generated using chordoma cell lines (pertains to Fig. 2) was performed as described previously22, except as noted in the Methods, and the resulting AUC values are provided in Supplementary Table 2. Raw plate-reader data files and accompanying Pipeline Pilot and MATLAB scripts for small-molecule primary screening and low-throughput compound sensitivity analysis (pertains to Fig. 2, Extended Data Figs. 2a, 3a, 8c, and 10a) are available upon reasonable request. Chromatin profiling data (pertains to Figs. 3, 4, and Extended Data Figs. 4 and 6) are available at GEO (accession number: GSE109794).
Extended Data
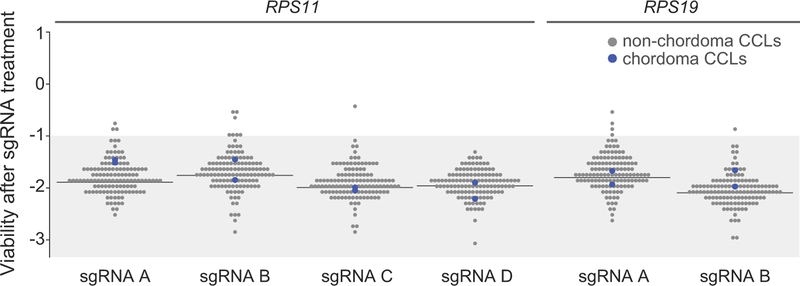
Extended Data Figure 1. Dependency scores for sgRNAs targeting commonly essential genes.
Viability after sgRNA treatment (represented by sgRNA-dependency scores; see Methods) corresponding to each of the primary screening sgRNAs targeting either RPS11 or RPS19 across 127 cancer cell lines (CCLs), including 2 chordoma CCLs (blue circles). For a given sgRNA, the median sgRNA-dependency score across 127 cell lines is indicated (gray line). Lower values indicate greater sgRNA depletion and thus essentiality of the target gene (shaded region).
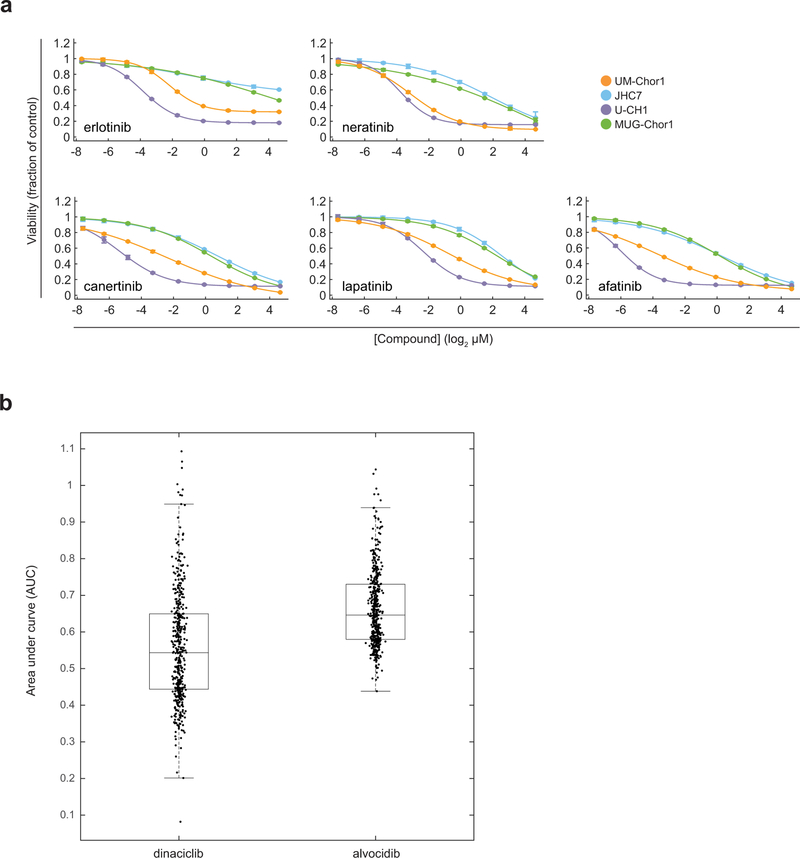
Extended Data Figure 2. Sensitivity of chordoma cells to EGFR and/or ERBB2 inhibitors and of non-chordoma cells to CDK9 inhibitors.
a) Validation of primary screening hit compounds targeting EGFR and/or ERBB2. Four chordoma cell lines were treated with indicated concentrations of candidate antiproliferative compounds and assayed for cell viability after 6 d with CellTiter-Glo. Response data are represented by a fitted curve to the mean fractional viability at each concentration relative to vehicle-treated cells; error bars represent the SEM (n = 4 biological samples measured in parallel). b) Dinaciclib and alvocidib have antiproliferative effects across a wide range of cancer cell lines. Area-under-curve (AUC) values corresponding to cell lines in CTRP treated with either dinaciclib or alvocidib. Each point represents a cancer cell line in CTRP treated with the indicated compound. Boxplots depict the inner quartiles (boxes) and median value (horizontal line) with whiskers representing 1.5 × the interquartile range of 445 (dinaciclib-treated) or 440 (alvocidib-treated) cell lines. AUCs were computed as described in the Methods and at https://github.com/remontoire-pac/ctrp-reference/tree/master/auc.
Extended Data Figure 3. Chordoma cells are less sensitive to CDK4/6 and CDK8/19 inhibitors.
a) Response of chordoma cells to compounds targeting CDK4/6 and CDK8/19 proteins. Four chordoma cell lines were treated with indicated concentrations of compounds and assayed for cell viability after 6 d with CellTiter-Glo. Response data are represented by a fitted curve to the mean fractional viability at each concentration relative to vehicle-treated cells; error bars represent the SEM (n = 4 biological samples measured in parallel). b) Immunoblot analysis of UM-Chor1 cells treated with indicated concentrations of inhibitors or DMSO for 24 h. The experiment was performed twice for CCT251545 (one representative experiment displayed) and once for other compounds.
Extended Data Figure 4. T is super-enhancer-associated across chordoma cell lines.
a) Enhancers in 5 chordoma cell lines ranked by H3K27ac signal in each sample. Enhancers proximal (within 100 kb) to the T gene start site are annotated, as described in the figure. Super-enhancers (SEs) were determined by the inflection point of the plot. b) Table showing the ranks of top T-associated enhancers in each chordoma sample.
Extended Data Figure 5. Brachyury is highly expressed in chordoma cell lines.
a) Immunoblot analysis of chordoma and chondrosarcoma cell lines. Chordoma cell lines selectively express high levels of the brachyury protein. The experiment was performed once. b) Expression of T and MAX, as measured by RNA-sequencing, across 935 non-chordoma cancer cell lines derived from diverse tumor types. Data were generated as part of the Broad Institute Cancer Cell Line Encyclopedia (quantified data obtained from: https://ocg.cancer.gov/ctd2-data-project/translational-genomics-research-institute-quantified-cancer-cell-line-encyclopedia). Boxplots depict the inner quartiles (boxes) and median value (horizontal line) with whiskers representing 1.5 × the interquartile range. c) Gene expression levels of 115 super-enhancer (SE)-associated transcription factors in 5 chordoma cell lines (points), ranked by mean expression (horizontal ticks). d) T is amplified in the JHC7 chordoma cell line. Genomic copy-number alterations, inferred from whole-exome sequencing data, in five chordoma cell lines. A region of 2.06 Mb around the T locus on chromosome 6 shows 26-fold amplification in JHC7. This finding is consistent with the 2.6-Mb amplicon inferred from ChIP-seq whole-cell extract.
Extended Data Figure 6. T is super-enhancer-associated in patient-derived chordoma tumors.
a) Enhancers in chordoma tumors ranked by H3K27ac signal in each sample. Super-enhancers (SEs, red) and typical enhancers (black) proximal (within 100 kb) to the T gene start site are annotated. SEs were determined by the inflection point of the plot. b) Table showing the ranks of top T-associated SEs or typical enhancers in each chordoma sample.
Extended Data Figure 7. Patient-derived chordoma tumors express brachyury.
Immunohistochemical staining of patient-derived chordoma tumors for brachyury expression. H&E, hematoxylin and eosin. The experiment was performed once.
Extended Data Figure 8. THZ1 and actinomycin D reduce expression of T (brachyury) in a concentration- and time-dependent fashion.
a) Immunoblot analysis of UM-Chor1 cells treated with indicated concentrations of THZ1 or DMSO for 12, 24, 36, or 48 h. The experiment was performed once. b) UM-Chor1 cells were treated with indicated concentrations of compound or DMSO for 4, 8, or 12 h and subjected to RT-qPCR. Data are expressed as the log2 fold-change of transcript levels relative to vehicle-treated cells, normalized to GAPDH levels, and represent the mean ± SD (n = 3 biological samples measured in parallel). Results of statistical analyses of RT-qPCR data, derived from a one-sided Welch’s t-test, are reported in Supplementary Table 9. c) Response of chordoma cells to treatment with actinomycin D. Four chordoma cell lines were treated with indicated concentrations of compound and assayed for cell viability after 6 d with CellTiter-Glo. Response data are represented by a fitted curve to the mean fractional viability at each concentration relative to vehicle-treated cells; error bars represent the SEM (n = 4 biological samples measured in parallel).
Extended Data Figure 9. Expression of ATP6V1B2, SAE1, SOX9, and TPX2 is downregulated following sgRNA-mediated T (brachyury) repression.
a) UM-Chor1 cells were transduced with sgRNAs targeting T or a non-targeting sgRNA control and subjected to RT-qPCR. Data are expressed as the fold-change of transcript levels relative to sgRNA control-treated cells, normalized to GAPDH levels, and represent the mean (n = 2 biological samples measured in parallel, represented by black points). * p < 0.05; ** p < 0.01; *** p < 0.001; p-values were derived from a one-sided Welch’s t-test. Exact p-values and effects sizes are reported in Supplementary Table 9. b) Immunoblot analysis of UM-Chor1 cells transduced with sgRNAs targeting T or a non-targeting sgRNA control. SgRNA treatment was performed once and immunoblotting was performed twice (one representative experiment displayed).
Extended Data Figure 10. THZ1 treatment can reduce brachyury expression in CH22 cells ex vivo and in vivo.
a) CH22 chordoma cells were treated with indicated concentrations of transcriptional CDK inhibitors and assayed for cell viability after 6 d with CellTiter-Glo. Response data are represented by a fitted curve to the mean fractional viability at each concentration relative to vehicle-treated cells; error bars represent the SEM (n = 4 biological samples measured in parallel). b) Immunoblot analysis of CH22 cells treated with indicated concentrations of inhibitors targeting CDK4/6 (palbociclib), CDK7/12/13 (THZ1), or CDK9 (dinaciclib, NVP-2, alvocidib) or DMSO for 48 h. The experiment was performed once. c) Weight change of mice treated with THZ1 or vehicle for the study depicted in Fig. 4h–i. d) THZ1 can downregulate brachyury expression in vivo. Immunoblot analysis of CH22 xenograft tumors following treatment with indicated doses of THZ1 or vehicle twice daily for 5 d. The experiment was performed once. e) Immunoblot analysis of CH22 xenograft tumors following treatment with THZ1 or vehicle twice daily for 3 d. Top and bottom panels represent two independent studies (bottom panel corresponds to the study depicted in Fig. 4h–i).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Vrcic, K. Hartland, S. Figueroa-Lazu, C. Biasetti, and J. Santos for assistance with small-molecule sensitivity experiments; members of the Broad Institute Genetic Perturbation Platform for sgRNA library construction and technical assistance with DNA processing after pooled screening; the Histology Core Facility at Ohio State University for immunohistochemistry studies; X. Zhang for guidance with RNA sequencing; L. Gechijian and P. Veeraraghavan for technical and analytical assistance; R. Meyers and A. Cherniack for guidance with copy-number analysis; and R. Belizaire, Y. Zou, N. Kwiatkowski, J. Dempster, S. Gill, and T. Sundberg for helpful discussions. This work was generously supported by the Chordoma Foundation, the Roye family, the Fuchs family, and the NCI’s Cancer Target Discovery and Development (CTD2) Network (grant number U01CA176152/U01CA217848 awarded to S.L.S. and U01CA176058 awarded to W.C.H.). In addition to being supported by the Chordoma Foundation, C.Y.L is supported by the Cancer Prevention Research Institute of Texas (RR150093) and by the NIH and NCI (1R01CA215452–01), and is a Pew-Stewart Scholar for Cancer Research (Alexander and Margaret Stewart Trust). The Institute of Cancer Research authors were funded by Cancer Research UK (Programme Grant number C2739/A22897). P.W. is a Cancer Research UK Life Fellow. S.L.S. is an investigator at the Howard Hughes Medical Institute. This paper is dedicated to the memory of Todd Fuchs, whose spirit and perseverance were an inspiration for all involved.
Footnotes
COMPETING INTERESTS
T.S. is a consultant for Jnana Therapeutics. N.S.G. is equity holder and scientific advisor for Syros, Gatekeeper, Soltego, C4, Petra and Aduro companies. Syros has licensed intellectual property from Dana-Farber Cancer Institute covering THZ1. P.A. Clarke, J.B., and P.W. are current employees of The Institute of Cancer Research, which has a Rewards to Inventors scheme and has a commercial interest in the development of inhibitors of the WNT pathway, CDK8/19 and other CDKs, with intellectual property licensed to Merck and Cyclacel Pharmaceuticals. PW is a consultant for Astex Pharmaceuticals, CV6 Therapeutics, Nextechinvest, Nuevolution and Storm Therapeutics and holds equity in Chroma Therapeutics, Nextech and Storm. D.E.R. receives research funding from members of the Functional Genomics Consortium (Abbvie, Jannsen, Merck, Vir), and is a director of Addgene, Inc. W.C.H. is a consultant for Thermo Fisher, Paraxel, AjuIB, MPM Capital and KSQ Therapeutics, is a founder of KSQ Therapeutics and receives research support from Deerfield. J.E.B. is now an executive and shareholder of Novartis AG, and has been a founder and shareholder of SHAPE (acquired by Medivir), Acetylon (acquired by Celgene), Tensha (acquired by Roche), Syros, Regency and C4 Therapeutics. K.K.W. is a founder and equity holder of G1 Therapeutics and he has consulting/sponsored research agreements with AstraZeneca, Janssen, Pfizer, Array, Novartis, Merck, Takeda, Ono, Targimmune and BMS. C.Y.L. is a consultant for Jnana Therapeutics and is a shareholder of and inventor of intellectual property licensed to Syros Pharmaceuticals. J.D.K. is a founder, executive, and shareholder of Jnana Therapeutics. S.L.S. is a member of the Board of Directors of the Genomics Institute of the Novartis Research Foundation (“GNF”); a shareholder and member of the Board of Directors of Jnana Therapeutics; a shareholder of Forma Therapeutics; a shareholder of and adviser to Decibel Therapeutics; an adviser to Eisai, Inc., the Ono Pharma Foundation, and F-Prime Capital Partners; and a Novartis Faculty Scholar.
All other authors declare no competing interests.
REFERENCES
- 1.Stacchiotti S, Sommer J & Chordoma Global Consensus Group. Building a global consensus approach to chordoma: a position paper from the medical and patient community. Lancet Oncol 16, e71–83 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Choy E, et al. Genotyping cancer-associated genes in chordoma identifies mutations in oncogenes and areas of chromosomal loss involving CDKN2A, PTEN, and SMARCB1. PLoS One 9, e101283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarpey PS, et al. The driver landscape of sporadic chordoma. Nature communications 8, 890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradner JE, Hnisz D & Young RA Transcriptional Addiction in Cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMaster ML, Goldstein AM, Bromley CM, Ishibe N & Parry DM Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer causes & control : CCC 12, 1–11 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Barry JJ, et al. The next step: innovative molecular targeted therapies for treatment of intracranial chordoma patients. Neurosurgery 68, 231–240; discussion 240–231 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Salisbury JR The pathology of the human notochord. The Journal of pathology 171, 253–255 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Vujovic S, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. The Journal of pathology 209, 157–165 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Showell C, Binder O & Conlon FL T-box genes in early embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists 229, 201–218 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XR, et al. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet 41, 1176–1178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillay N, et al. A common single-nucleotide variant in T is strongly associated with chordoma. Nat Genet 44, 1185–1187 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Presneau N, et al. Role of the transcription factor T (brachyury) in the pathogenesis of sporadic chordoma: a genetic and functional-based study. The Journal of pathology 223, 327–335 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hsu W, et al. Generation of chordoma cell line JHC7 and the identification of Brachyury as a novel molecular target. Journal of neurosurgery 115, 760–769 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah SR, et al. Brachyury-YAP Regulatory Axis Drives Stemness and Growth in Cancer. Cell reports 21, 495–507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palena C, et al. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin Cancer Res 13, 2471–2478 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Garraway LA & Sellers WR Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 6, 593–602 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Berg T Inhibition of transcription factors with small organic molecules. Current opinion in chemical biology 12, 464–471 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Sharifnia T, Hong AL, Painter CA & Boehm JS Emerging Opportunities for Target Discovery in Rare Cancers. Cell chemical biology 24, 1075–1091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyers RM, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet (2017). [DOI] [PMC free article] [PubMed]
- 21.Barrionuevo F, Taketo MM, Scherer G & Kispert A Sox9 is required for notochord maintenance in mice. Developmental biology 295, 128–140 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Seashore-Ludlow B, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov 5, 1210–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittaker SR, Mallinger A, Workman P & Clarke PA Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacology & therapeutics 173, 83–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol 12, 876–884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galli GG, et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell 60, 328–337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modjtahedi H, Cho BC, Michel MC & Solca F A comprehensive review of the preclinical efficacy profile of the ErbB family blocker afatinib in cancer. Naunyn-Schmiedeberg’s archives of pharmacology 387, 505–521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheipl S, et al. EGFR inhibitors identified as a potential treatment for chordoma in a focused compound screen. The Journal of pathology 239, 320–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathy D, Bardia A & Sellers WR Ribociclib (LEE011): Mechanism of Action and Clinical Impact of This Selective Cyclin-Dependent Kinase 4/6 Inhibitor in Various Solid Tumors. Clin Cancer Res 23, 3251–3262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dale T, et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat Chem Biol 11, 973–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannessen L, et al. Small-molecule studies identify CDK8 as a regulator of IL-10 in myeloid cells. Nat Chem Biol (2017). [DOI] [PMC free article] [PubMed]
- 31.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson AC, et al. An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. The Journal of pathology 228, 274–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin CY, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwab JH, et al. Chordoma and chondrosarcoma gene profile: implications for immunotherapy. Cancer immunology, immunotherapy : CII 58, 339–349 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jager D, et al. HOXA7, HOXA9, and HOXA10 are differentially expressed in clival and sacral chordomas. Scientific reports 7, 2032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalaby A, et al. The role of epidermal growth factor receptor in chordoma pathogenesis: a potential therapeutic target. The Journal of pathology 223, 336–346 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Trask DK & Muller MT Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc Natl Acad Sci U S A 85, 1417–1421 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, et al. Establishment and characterization of a novel chordoma cell line: CH22. Journal of orthopaedic research : official publication of the Orthopaedic Research Society 30, 1666–1673 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Scientific data 1, 140035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguirre AJ, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov 6, 914–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dancik V, et al. Connecting Small Molecules with Similar Assay Performance Profiles Leads to New Biological Hypotheses. Journal of biomolecular screening 19, 771–781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current protocols in bioinformatics 43, 11–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown JD, et al. NF-κB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Molecular cell 56, 219–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CRISPR-Cas9 screening data for two chordoma cell lines (pertains to Figs. 1, 2, 4, and Extended Data Fig. 1) are available at Figshare (DOI: 10.6084/m9.figshare.7302515). CRISPR-Cas9 screening data for all other cancer cell lines (pertains to Figs. 1, 2, 4, and Extended Data Fig. 1) were generated as part of Project Achilles (Broad Institute Project Achilles; https://depmap.org/portal/achilles/). All RNA-sequencing data (pertains to Figs. 1 and 4) are available at Gene Expression Omnibus (GEO) (accession number: GSE121846). Small-molecule sensitivity data generated using non-chordoma cell lines and used for comparative analyses (pertains to Fig. 2) are available at the National Cancer Institute’s CTD2 Data Portal (https://ocg.cancer.gov/programs/ctd2/data-portal) and the CTRP (www.broadinstitute.org/ctrp/). The analysis of new small-molecule primary screening data generated using chordoma cell lines (pertains to Fig. 2) was performed as described previously22, except as noted in the Methods, and the resulting AUC values are provided in Supplementary Table 2. Raw plate-reader data files and accompanying Pipeline Pilot and MATLAB scripts for small-molecule primary screening and low-throughput compound sensitivity analysis (pertains to Fig. 2, Extended Data Figs. 2a, 3a, 8c, and 10a) are available upon reasonable request. Chromatin profiling data (pertains to Figs. 3, 4, and Extended Data Figs. 4 and 6) are available at GEO (accession number: GSE109794).