Parkinson's disease (PD) is the second most common neurodegenerative disease affecting the Western world. The disease in humans is characterized by the selective loss of dopaminergic neurons in the substantia nigra pars compacta; however, modeling this disease in transgenic animals has proven exceptionally difficult. Until recently, knock-out murine models for the PD-associated genes LRRK2, α-synuclein, Parkin, PINK1, and DJ-1 have typically failed to generate a comparable pathological setting to aid basic and therapeutic research into PD pathogenesis. Researchers have therefore used genetic manipulation in Drosophila to gain insight into the signaling pathways affected in PD. Typically, these fly models consist of either RNA-interference- or P-element-generated knock-outs and/or overexpression of the mutated genes associated with PD. One of the more recent proteins to be assessed in Drosophila is LRRK2, mutations of which can induce dominantly inherited, late-onset PD.
LRRK2 is a formidable protein, which can be difficult to express due to its large size (280 kDa) and is likely to influence a number of cellular functions due to its multidomain structure including a Roc (Ras of complex), COR (C terminal of Roc), serine/threonine kinase domain, and several WD40 repeat protein–protein interaction domains. Although the PD-associated mutations in LRRK2 are located throughout the protein, several studies have reported that an increase in kinase activity is associated with pathogenesis. Consequently, the identification of LRRK2 substrates and inhibitors has been a key focus of recent investigations (Nichols et al., 2009).
The recent publication by Ng et al. (2009) in The Journal of Neuroscience assessed the effects of overexpressing LRRK2 wild-type (wt) and PD-associated mutations on age-dependent dopaminergic (DA), tyrosine hydroxylase-positive (TH+) neuron loss, altered dopamine levels, and climbing defects in flies. Unlike most previous studies, in which the Drosophila LRRK2 homolog (dLRRK) was overexpressed or knocked out, Ng et al. (2009) specifically expressed the human wt, G2019S, Y1699C, and G2385R LRRK2 variant proteins in the wt fly eye and brain. The location of these mutations within the kinase, COR, and WD40 domains, respectively, permit the authors to determine whether mutations within the different protein domains of LRRK2 can induce similar phenotypes. Notably, Ng et al. (2009) are the first group to assess the functional consequences of mutations within the WD40 domains of LRRK2 in Drosophila.
Consistent with most studies (Lee et al., 2007; Imai et al., 2008), Ng et al. (2009) found no eye abnormalities in any of their LRRK2 wt or PD-mutant overexpressing flies. However, they observed a significant age-dependent loss of TH+ neurons specifically in the PPM (protocerebral posterior medial) 1 and 3 neural clusters in flies expressing PD-associated forms of LRRK2. This neuronal loss was associated with reduced lifespan, which was particularly prominent in the G2019S lines. Climbing deficits were also observed in the G2019S and Y1699C lines, but surprisingly, despite the loss of TH+ neurons, no locomotor abnormalities were identified in the G2385R mutant line.
Additional differences between the LRRK2 mutant lines were observed when the authors assessed the susceptibility of the flies' DA neurons to degeneration by administering the PD-associated toxin rotenone. Although, under normal conditions, all the LRRK2 mutant fly lines showed similar TH+ neuron loss in the PPM1 and PPM3 clusters with age, rotenone exposure accelerated TH+ neuron loss in the PPM3 cluster in both the G2019S and G2385R lines. Furthermore, rotenone treatment induced a novel set of TH+ neurons to die in the PPM2 cluster in both the G2019S and G2385R lines, and neuron loss was observed in the PPL1 and PAL clusters of the G2385R mutant. Strikingly, rotenone treatment had no effect on flies overexpressing Y1966C.
Finally, the authors demonstrated that co-overexpression of human Parkin (a neuroprotective E3 ubiquitin ligase associated with early-onset recessive PD) in either the aged or rotenone-treated G2019S flies rescued the PPM1/3 and PPM2/3 TH+ neuronal death, respectively. The authors conclude that rotenone aggravates, whereas Parkin overexpression alleviates, the TH+ neuronal death induced by LRRK2 G2019S overexpression.
The finding that rotenone (a mitochondrial complex I inhibitor) accelerates TH+ neuron loss in a LRRK2 mutant model suggests that both environmental factors and genetics contribute to pathogenesis. The authors propose that these results, together with their previous work suggesting a direct interaction between LRRK2 and Parkin (Smith et al., 2005), indicate that interactions between LRRK2, Parkin, and mitochondria could have both therapeutic and mechanistic implications for PD.
Although the results reported by Ng et al. (2009) are intriguing and the mechanism underlying resistance to rotenone in their Y1699C line, is worthy of subsequent investigation, it is unfortunate that the experiments reported were not performed on all the LRRK2 mutant lines, to give a comprehensive assessment of all the phenotypes. Specifically, examination of dopamine levels in all fly lines, instead of just the G2019S line, may have provided an explanation for the lack of locomotor deficits, despite TH+ neuron loss, in the G2385R flies. These findings could open up an interesting possibility of compensatory mechanisms, perhaps similar to those occurring in patients, which could be investigated in this Drosophila model.
In addition, the increased sensitivity of the G2385R line to rotenone-induced TH+ neuronal death (compared to the G2019S line), particularly in neural clusters unaffected by aging, is worth noting. Previous studies have shown that the kinase activity of this LRRK2 mutant is not increased above wt levels, suggesting that pathogenesis is not induced by dysregulated kinase activity. However, expression of the G2385R protein induces cell death at comparable levels to the G2019S protein, which demonstrates increased kinase activity (West et al., 2007). Given these previous findings and the novel observations reported by Ng et al. (2009), showing that co-overexpression of Parkin can rescue TH+ neuron loss associated with age and rotenone treatment, it would have been interesting to assess the ability of Parkin co-overexpression to rescue the rotenone-induced G2385R phenotypes.
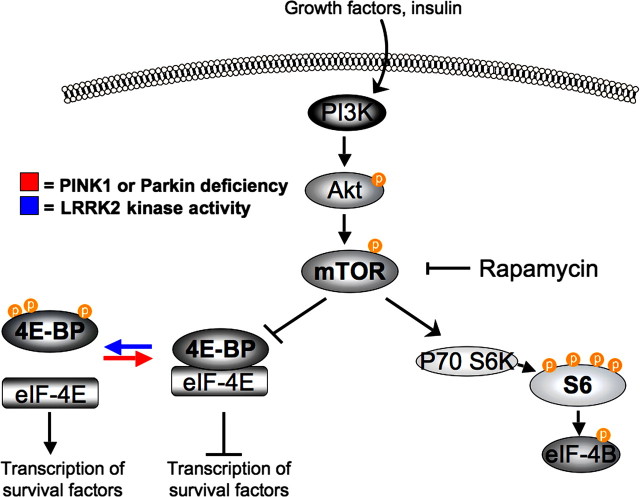
Another way Ng et al. (2009) could have gained insight into the differences in TH+ neuron loss in the aged or rotenone-treated fly lines would have been to assess 4E-BP phosphorylation levels. In 2008, it was shown that phosphorylation of 4E-BP on residues T37 and T46 are important for the pathogenic effects of mutant LRRK2 in Drosophila i.e., DA neuron loss, sensitivity to stress and reduced lifespan (Imai et al., 2008) (Fig. 1). Furthermore, a recent study by Tain et al. (2009) demonstrated that 4E-BP is hypophosphorylated in PINK1 and Parkin mutant fly lines and the pathogenesis observed in these lines can be rescued by either 4E-BP overexpression or rapamycin treatment (Fig. 1). The combination of these studies strongly suggests that 4E-BP phosphorylation levels are directly linked to the pathogenic phenotypes displayed in fly PD models and subsequent studies to dissect the involvement of the Akt/mTOR/4E-BP pathway in human pathogenesis should be an exciting area of future research. Given the differences in TH+ neuron loss, susceptibility to stress and increased mortality rates of their LRRK2 mutant lines, it is surprising that Ng et al. (2009) did not pursue this line of investigation, to gain mechanistic insights into their models. In particular, the group could have addressed (1) whether the prominent reduction in lifespan in the G2019S mutant flies was associated with increased phosphorylation of 4E-BP compared to the other fly lines, (2) whether the differential susceptibility to stress, observed in their fly lines, correlated with alterations in 4E-BP phosphorylation status, (3) whether the resistance to rotenone in their Y1699C line is associated with reduced 4E-BP phosphorylation levels, and finally (4) whether an alteration in 4E-BP phosphorylation state is induced in the rescued G2019S mutant line due to Parkin overexpression.
Figure 1.
Modulation of 4E-BP phosphorylation status by PD-associated proteins. 4E-BP phosphorylation levels are regulated by the mTOR kinase, which acts downstream of Akt. In its hypophosphorylated state, 4E-BP binds to the transcription factor eIF-4E and inhibits the subsequent transcription of survival factors. Upon phosphorylation, 4E-BP dissociates from eIF-4E and permits transcription. Recently, the active LRRK2 kinase was shown to phosphorylate 4E-BP, whereas loss of either PINK1 or Parkin function was shown to result in hypophosphorylated 4E-BP.
Finally, it would have been interesting if additional aspects of the rescue phenotypes in the G2019S-Parkin coexpressing fly lines had been investigated. For example, Parkin overexpression has been shown to upregulate PINK1 protein levels in Drosophila, and recently, PINK1 was shown to phosphorylate Parkin, resulting in Parkin's relocalization to mitochondria (Yang et al., 2006; Kim et al., 2008). Since PINK1 can protect against oxidative stress, alleviate mitochondrial damage induced by complex I inhibitors and is tightly linked to Parkin (Deas et al., 2009), it seems premature for the authors to attribute all of the protective effects observed exclusively to Parkin.
In summary, Ng et al. (2009) have provided novel insights into the effects of the LRRK2 G2385R mutant protein in vivo, a PD-associated mutation which has received relatively little attention in the literature compared to mutations in the Roc and COR domains. The location of this mutation in the protein–protein interaction WD40 domains, rather than an enzymatic domain of LRRK2, raises interesting questions about the mechanism by which mutant LRRK2 mediates PD pathogenesis. The generation of this particular fly model is therefore an important step toward advancing our overall understanding of LRRK2 function in PD. Furthermore, as the second comprehensive, detailed analysis of TH+ neuron loss in a combination of LRRK2 Drosophila models, these results finally allow a direct comparison between studies. The ability to compare studies is essential because several groups investigating the effects of PD-associated LRRK2 (or equivalent dLRRK) mutations on photoreceptor degeneration, TH+ neuron loss, muscle pathology, locomotor function, and lifespan have reported discrepancies in fly phenotypes (Lee et al., 2007; Imai et al., 2008; Liu et al., 2008; Vendarova et al., 2009). Specifically, LRRK2 wt and PD mutant fly lines range from having photoreceptor degeneration to having no abnormal eye phenotype, significant reductions in TH+ neuron numbers in all neuronal clusters to no apparent loss of TH+ neurons in any of the clusters, severe muscle pathology resulting in abnormal wing posture to no overt muscle phenotype, and either decreased, normal, or increased lifespan. On the surface, these discrepancies are confusing given that the authors all use the same GAL4 drivers, i.e., GMR (eye specific), elav (pan neuronal), Ddc (DA neuron specific), and TH (DA neuron specific), to selectively target LRRK2 wt or mutant protein expression to specific tissue regions. However, different phenotypes can be caused by insertion-site-specific effects on transgene expression levels or dosage effects caused by multiple transgene insertions in a single embryo (Spradling and Rubin, 1983). In addition, perhaps some of the discrepancy, specifically in measures of TH+ neuron loss, could be attributed to the fact that not all studies have assessed the same neural clusters. Moreover, in some cases, neurons assigned to different clusters by some groups have been combined and examined as a single cluster by others, and this may have resulted in the TH+ neuron loss dropping below the level of significance. However, before any of the phenotypes are potentially ignored (based on a consensus between studies), it is important to remember that the pathology of LRRK2 patients carrying the same pathogenic mutation is also varied (Giasson et al., 2006). Given these observations in patients, it is perhaps not surprising that a range of phenotypes have been observed in fly models recapitulating the disease. As a consequence, further assessments of LRRK2 pathogenic function in Drosophila will be required to address these issues.
Footnotes
We thank Dr. Helene Plun-Favreau and Abi Li for critical appraisal of the article and helpful discussions.
Editor's Note: These short, critical reviews of recent papers in the Journal, written exclusively by graduate students or postdoctoral fellows, are intended to summarize the important findings of the paper and provide additional insight and commentary. For more information on the format and purpose of the Journal Club, please see http://www.jneurosci.org/misc/ifa_features.shtml.
References
- Deas E, Plun-Favreau H, Wood NW. PINK1 function in health and disease. EMBO Mol Med. 2009;1:152–165. doi: 10.1002/emmm.200900024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM. Biochemical and pathological characterization of Lrrk2. Ann Neurol. 2006;59:315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim W, Lee S, Chung J. Loss of LRRK2/PARK8 induces degeneration of dopaminergic neurons in Drosophila. Biochem Biophys Res Commun. 2007;358:534–539. doi: 10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, Ross CA, Montell C, Smith WW. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, Bamborough P, Reith AD, Alessi DR. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson's disease. Biochem J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell. 1983;34:47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venderova K, Kabbach G, Abdel-Messih E, Zhang Y, Parks RJ, Imai Y, Gehrke S, Ngsee J, Lavoie MJ, Slack RS, Rao Y, Zhang Z, Lu B, Haque ME, Park DS. Leucine-rich repeat kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Hum Mol Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]