Abstract
The Vav proteins are guanine exchange factors (GEFs) that trigger the activation of the Rho GTPases in general and the Rac family in particular. While the role of the mammalian vav genes has been extensively studied in the hematopoietic system and the immune response, there is little information regarding the role of vav outside of these systems. Here, we report that the single Drosophila vav homolog is ubiquitously expressed during development, although it is enriched along the embryonic ventral midline and in the larval eye discs and brain. We have analyzed the role that vav plays during development by generating Drosophila null mutant alleles. Our results indicate that vav is required during embryogenesis to prevent longitudinal axons from crossing the midline. Later on, during larval development, vav is required within the axons to regulate photoreceptor axon targeting to the optic lobe. Finally, we demonstrate that adult vav mutant escapers, which exhibit coordination problems, display axon growth defects in the ellipsoid body, a brain area associated with locomotion control. In addition, we show that vav interacts with other GEFs known to act downstream of guidance receptors. Thus, we propose that vav acts in coordination with other GEFs to regulate axon growth and guidance during development by linking guidance signals to the cytoskeleton via the modulation of Rac activity.
Introduction
Vav proto-oncogenes act as guanine exchange factors (GEFs) for the Rho/Rac GTPase family. Vav proteins are activated by phosphorylation in response to stimulation by transmembrane receptors. Once activated, they convert the Rho GTPases from the inactive to the active state by facilitating the exchange of GDP for GTP (Bustelo, 2000). The main consequence of the Rho GTPase activation is the reorganization of the actin cytoskeleton, which is crucial for many biological processes such as cell division, adhesion, migration, and axon guidance. In addition to the Dbl homology and pleckstrin homology domains that are characteristic of the GEFs, Vav members consist of a calponin homology domain, an acidic region, a zinc finger region, and the structural hallmark of signal transduction proteins, the Src homology 2 (SH2) and SH3 domains. These different domains allow Vav proteins to combine catalytic and adaptor functions and make them unique among the GEF family. Thus, Vav proteins act as signaling molecules at the crossroads of many different pathways (Bustelo, 2000).
In mammals, vav1 is mainly expressed in the hematopoietic system, although vav2 and vav3 are expressed more broadly. Consequently, attention has been focused mainly on the role of vav in lymphocyte development and the immune response (Turner and Billadeau, 2002; Fujikawa et al., 2003). In addition, the three mammalian vav genes are also expressed in some areas of the CNS (Movilla and Bustelo, 1999; Betz et al., 2003; Chauvet et al., 2003). Furthermore, several cell culture studies have implicated vav2 and vav3 members in neuronal migration (Schmid et al., 2004), neurite outgrowth (Aoki et al., 2005), and the formation of dendrites and axons (Murata et al., 2006). However, there is very little evidence of a role for vav in axogenesis in a developing organism. In fact, only one study conducted using postnatal rat brains and primary cultured neurons reported the involvement of vav2 and vav3 during axon guidance (Cowan et al., 2005).
Here, we decided to take advantage of the Drosophila model system to further analyze the contribution of vav in axogenesis during development. The single Drosophila vav ortholog possesses the same catalytic specificity as its mammalian counterparts, suggesting that the main Vav activities and mechanisms are well conserved between species (Couceiro et al., 2005). Although ubiquitously expressed during Drosophila development, we show that vav displays higher levels of expression in the embryonic and larval CNSs. We have isolated mutants in the vav gene, and their phenotypic analysis reveals that vav is required at least at three different stages during development. During embryogenesis, vav participates in preventing longitudinal axons from crossing the midline. At larval stages, vav is required within the larval photoreceptors to trigger axon targeting of the optic lobe. Finally, in the adults, vav mutant brains display axon growth defects. Altogether, our results demonstrate that vav is used reiteratively throughout development to control axogenesis. Thus, our findings highlight the importance of the Vav proteins as key regulators of axon development.
Materials and Methods
Fly strains.
The following fly stocks were used: UAS-vavwt and UAS-vavact (Couceiro et al., 2005); the X55 enhancer trap line; the ro-τlacZ line; the trio1, rac2, and mtl mutants; and the sos mutant [kind gifts from Dr. Christian Klämbt (Westfalische Wilhelms Universitat, Münster, Germany), Dr. Takashi Suzuki (Max Plank Institute of Medicine, Martinsried, Germany), Dr. Barry Dickson (Research Institute of Molecular Pathology, Vienna, Austria), and Dr. Long Yang (University of Pennsylvania School of Medicine, Philadelphia, PA), respectively]. Tubulin-Gal4, arm-Gal4, GMR-Gal4, sim-Gal4, UAS-trio, trio6A mutant flies, and the ovoD FRT101;flp38 stock that was used to produce the germline clones were obtained from the Bloomington Stock Center (Indiana University, Bloomington, IN). The vav11837FRT19A stock was generated by the University of California Los Angeles Undergraduate Research Consortium in Functional Genomics (Los Angeles, CA) and obtained from the Kyoto Stock Center (Kyoto Institute of Technology, Kyoto, Japan). The following double mutant stocks for vav and trio were made: vav2/FM7evelacZ;trio1/TM6UbxlacZ and vav1/FM7evelacZ;trio6A/TM6UbxlacZ.
For the rescue experiments, we made use of the Gal4/UAS system (Brand and Perrimon, 1993). For the rescue of lethality experiments, virgin females of the genotype vav/FMZ;tubulin-Gal4/TM6 were crossed to either FM7actGFP;UAS-vav59/TM6 or FM7actGFP;UAS-trio/TM3 (where GFP is green fluorescent protein) males to express ubiquitously vav and trio, respectively. For the rescue of the neuronal phenotype, the genotype used was vav/FMZ;arm-Gal4. Two different vav alleles, vav2 and vav3, were used in this study. Figure 2 D describes the adult male progeny obtained in all cases.
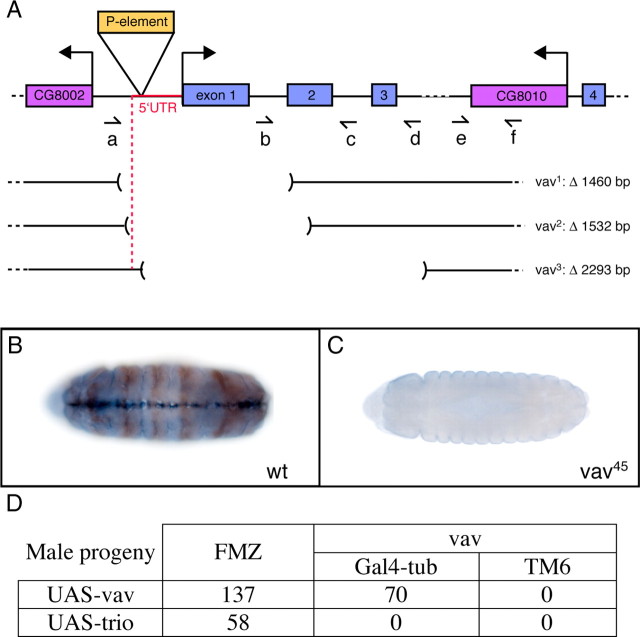
Figure 2.
Characterization of the vav deletion mutants. A, Schematic representation of the vav locus and mutations. Exons are represented by blue boxes, surrounding genes by purple boxes, and the P-element by an orange box. Shown in red is the vav 5′ untranslated region (UTR). Dashed lines indicate that the gene sequences continue but are not represented in entirety. Horizontal arrows designate transcription start sites for vav, for the gene located upstream and for the gene within the third exon of vav. Half-arrows (a–f) show the positions and directions (from 5′ to 3′) of the different primers used to determine the deletion endpoints of the three vav mutant alleles by PCR. vav1, vav2, and vav3 are the deletion mutants derived from y1P{SUPor-P}vavKG02022 by P-element imprecise excisions. The size of the deletions is indicated. For the three mutants, the vav locus is represented by a line and the region between brackets corresponds to the deletion. B, C, Vav expression is abolished in mutants. Anterior is to the left. Stage 15 embryos stained for β-gal (brown, immunostaining) and vav (blue, in situ hybridization). B, Vav2/FM7-eve-LacZ. The balancer chromosome gives positive β-gal staining and expresses the vav gene. Wt, Wild type. C, vav2 mutant shown by the absence of β-gal staining. The vav gene is not expressed. D, Table illustrating the rescue experiments. Crosses are described in Materials and Methods. The numbers of the adult males progeny are indicated in the table showing that the ubiquitous ectopic expression of vav but not trio can rescue the vav mutant lethality.
Imprecise excision of P-element to generate vav mutants.
The vav mutant alleles used in this study are deletions created by generating imprecise excision of the P-element insertion y1P{SUPor-P}vavKG02022 (Bloomington Stock Center) located within the vav 5′ untranslated region 427 bp upstream of the translational start site. y1P{SUPor-P}vavKG02022 flies were also recombined with yw flies to obtain y1wP{SUPor-P}vavKG02022 progeny. The P-element insertions were mobilized by introducing transposase on the third chromosome. Briefly, a total of 528 males of the genotype y1P{SUPor-P}vavKG02022;Δ2,3 Sb/+ and 228 males of the genotype y1wP{SUPor-P}vavKG02022;Δ2,3 Sb/+ were crossed individually to females carrying a FM7-ubiquitous-β-gal balancer. From the progeny, imprecise excisions of the P-element were isolated by selecting females that were yellow due to the loss of the y+ allele contained in the P-element. These females were individually crossed to males carrying an FM7-actin-GFP balancer, and their progeny was screened for lethality. Using a PCR-based screen, we examined 30 independent excised lines that were both lethal and yellow. Twenty of these mutants had a deletion in the vav gene (see Fig. 2 A, PCR with primers b and c). Among these candidates, only three mutants, named vav1, vav2, and vav3, also had intact neighboring genes (CG8010 and CG8002).
PCR and sequencing.
The imprecise excision of the P-element was verified by performing PCR over the excision breakpoint on genomic DNA extracted from different lethal lines recovered. Standard molecular biology protocols were used. The following primers were designed (primer locations within the vav locus are shown in Fig. 2 A) to map the deletion endpoints: a: 5′-TGCTTGCTTACTTCCTCTGAAAGG-3′; b: 5′-GAATGGGTGGCACACTCTAAACC-3′; c: 5′-TTGCTCGGACTGGCATCTATG-3′; d: 5′-ACGCACACGCACACAAAGCAACAAAC-3′; e: 5′-CGGAAAATCACTGTGGACGGTTG-3′; f: 5′-AGGAAACACCATCGCAAAGGTCT-3′. Sequencing was performed by STAB-VIDA, and sequences were determined on both strands.
In situ hybridization and immunohistochemistry.
Staining of embryos, imaginal discs, and larval CNS was performed according to standard procedure. Whole-mount in situ hybridization was done using an RNA probe corresponding to the full-length vav cDNA (Couceiro et al., 2005). The probe was labeled with a DIG labeling kit (Roche Applied Science). Staining was revealed with BCI/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) liquid substrate system or with Fast Red tablets (Roche Applied Science) in the case of fluorescent in situ hybridization. When required, in situ hybridization was followed by a standard immunostaining protocol. To identify unambiguously vav mutant embryos, the stocks bearing the different vav mutant alleles were balanced over FM7-eve-lacZ, a marked balancer. The following primary antibodies were used: rabbit anti-β-galactosidase (β-gal) at 1:2000 (Cappel); mouse anti-Fasciclin II (FasII; mAb 1D4) at 1:7 [Developmental Studies Hybridoma Bank (DSHB), University of Iowa, Iowa City, IA]; mouse anti-Wrapper at 1:10 (DSHB), mouse anti-Chaoptin (mAb 24B10) at 1:50 (DSHB), rabbit anti-Repo at 1:500 (kind gift from Dr. Benjamin Altenhein, University of Mainz, Mainz, Germany), and rabbit anti-GFP at 1/10000 (Invitrogen). Horseradish peroxidase with biotin-labeled secondary antibodies were used at 1/200 in combination with the Vectastain Elite ABC kit (Vector Laboratories) when staining was revealed with DAB. For fluorescent stainings, the secondary antibodies anti-rabbit labeled with Alexa Fluor 488 and anti-mouse labeled with Texas Red (Invitrogen) were used at 1:200. Stained embryos were mounted in glycerol, epon, or Vectashield (Vector Laboratories), and images were captured with an Axiophot microscope (Zeiss) using Nomarski optics and with a Leica TCS-SP2 confocal microscope.
Germline clones.
Germline clones were generated by x-ray-induced mitotic recombination. vav2/FM7 females were crossed to males containing the dominant female sterile mutation OvoD1 (Perrimon, 1984). The progeny was irradiated (Faxitron X-Ray model CP-160)with a dose of 1000 rad at the end of first larval instar and left to grow at 25°C. Adult females were crossed to FM7 males, and the resulting embryos lacking both vav maternal and zygotic products were examined by immunostaining.
MARCM analysis.
MARCM (mosaic analysis with a repressible cell marker) was carried out as described by Wu and Luo (2006). Larvae were obtained by crossing FRT19A,tubGAL80,hsFLP,w;UAS-mCD8::GFP males (Bloomington Stock Center) to our vav mutant stocks. Briefly, 0–24 h posthatching larvae of the following genotypes were heat-shocked for 1.5 h at 37°C, raised at 25°C, and dissected at the late third instar larval stage: (1) generation of wild-type control clones: FRT19A/FRT19A,tub-Gal80,hsFLP,w; UAS-mCD8::GFP/+; tubulinGal4/+ or FRT19A/FRT19A, tub-Gal80,hsFLP,w; UAS-mCD8::GFP/eyelessGal4; (2) generation of vav mutant clones: vav3FRT19A/FRT19A,tub-Gal80,hsFLP,w; UAS-mCD8:: GFP/+; tubulinGal4/+ or vav3FRT19A/FRT19A,tub-Gal80,hsFLP,w; UAS-mCD8::GFP/eyeGal4.
Adult brain paraffin sections.
Paraffin embedding and sectioning were performed according to Boquet et al. (2000a). Briefly, adult flies of the genotype yw, y1P{SUPor-P}vavKG02022 (control flies) or from the vav2 mutant line were placed in collars, fixed, dehydrated, and embedded in paraffin at 60°C overnight. Seven micrometer frontal sections were performed using a microtome. Paraffin was next eliminated by sequential washes in xylol and ethanol. Brain sections were subjected to immunostaining directly on the slide according to normal procedures.
Results
Vav is highly expressed in the ventral midline during embryonic development
The only Drosophila vav gene has been reported as being ubiquitously expressed during embryonic development (Dekel et al., 2000). A more detailed analysis of vav expression shows that although early in embryogenesis vav mRNA displays a ubiquitous distribution (Fig. 1 A), from stage 11 transcripts start to accumulate in more restricted regions of the embryo such as the CNS and the invaginating midgut (Fig. 1 B). At stage 13, vav is still detected in the CNS and midgut with a prominent expression in the midline and a central region of the midgut (Fig. 1 C). The high expression of vav in the midline persists until the end of embryogenesis (Fig. 1 D).
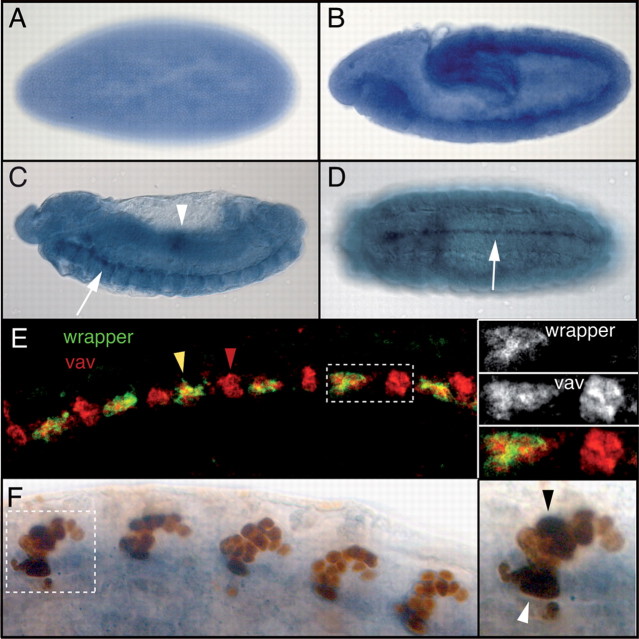
Figure 1.
Whole-mount in situ hybridization to detect vav expression in Drosophila embryos. Anterior is toward the left in all panels. A, Vav is ubiquitously expressed in the cellular blastoderm. B, Lateral view of a stage 11 embryo when the germ band is extended, showing vav expression in the midline primordium and the head. C, Stage 13 germ band-retracted embryo. Lateral view shows vav expression in the embryonic midline (white arrow) and in the midgut (a region with higher levels of vav expression is indicated with a white arrowhead). D, Ventral view of stage 15 embryo highlighting vav expression in the midline (white arrow). E, F, Colocalization of vav mRNA and midline marker proteins in stage 15 embryos. E, Left, Confocal views of vav fluorescent in situ hybridization. Vav transcripts (red) colocalize with wrapper proteins (green), showing that vav is expressed in anterior and medial midline glia (yellow arrowhead) plus other cells where Wrapper expression is excluded (red arrowhead). Right, High magnification of region shown in the dashed rectangle. Wrapper expression alone is shown at the top, vav expression in the middle, and merged staining at the bottom. F, Left, Distribution of vav transcripts (blue) with regard to X55 enhancer-trap expression (brown) driven in midline neurons and posterior midline glia (MGP). Vav is expressed in the MGP (white arrowhead) and a subset of neurons (black arrowhead). Right, High magnification of the segment in the dashed box showing vav expression in both glial and neuronal midline cells.
We next examined whether vav expression was restricted to a specific midline cell type by using markers expressed in glia and neurons. Midline glia include three pairs of cells, the anterior (MGA), medial (MGM), and posterior (MGP) midline glia. Wrapper is a marker for midline glia that is predominantly expressed at the surface of MGA and MGM (Noordermeer et al., 1998). By using combined in situ and antibody staining, we could detect vav transcripts in all Wrapper-expressing cells. Vav expression is also observed in additional cells in which Wrapper is not detected (Fig. 1 E). To identify these cells, we used the X55 enhancer trap that drives LacZ expression in several types of midline neurons and in the MGP (Klämbt et al., 1991). By stage 15, when midline cells are differentiated, vav expression is detected in the most posterior cluster of X55-expressing cells, which corresponds to the MGP. In addition, vav and β-gal also colocalize in a subset of neuronal cells, which can be seen as a darker brown signal (Fig. 1 F). Together, these results indicate that vav is expressed in all midline glial cells and in some midline neurons, showing that vav expression is not restricted to a specific cell type in the embryonic midline.
Generation of vav loss-of-function mutations
The fact that vav is specifically expressed in the midline and its transcripts can be found in both glial and neuronal cell populations prompted us to analyze a possible role for vav in axogenesis or guidance. For this, we generated vav null mutants by imprecise excision of a P-element inserted upstream of the vav start codon y1P{SUPor-P}vavKG02022 (see Materials and Methods). Given the importance of the vav genes in mammals, it seemed likely that vav functions would be essential for viability and its deletion would be lethal. Among the 30 independent lethal lines recovered from the mutagenesis, we isolated three mutants for vav named vav1, vav2, and vav3. Sequence analysis of these mutants revealed that the deletions removed 1460, 1532, and 2293 nt, respectively, of the genomic vav locus as illustrated in Figure 2 A. The three vav alleles caused deletions both upstream and downstream of the transcription start site. We confirmed by in situ hybridization that they were null alleles, as they did not produce detectable transcripts (Fig. 2 B,C).
The lethality of the vav mutant chromosomes is due to the loss of vav function, as it could be rescued by the ectopic expression of a vav cDNA (UAS-vav driven by tubulin-Gal4 or arm-Gal4) in vav2 and vav3 mutant backgrounds (Fig. 2 D). Significantly, since the ubiquitous ectopic expression of another GEF called trio was not able to rescue the vav mutant lethality, these results demonstrate that the mutants generated affect specifically the vav gene.
Although the majority of homozygous vav mutants die before eclosion, we found occasional male escapers, indicating that the mutations are semilethal. These wild-type-looking escapers display strong locomotion defects and die shortly after birth glued to the food and unable to walk or fly away. If such flies are rescued from the food, they show impairment of spontaneous locomotor activity and display a “shaking” phenotype. When we dissected out late pupae that failed to eclose, we observed that they contained fully developed adults also displaying strong locomotion defects. This suggests that many mutants die as pharate adults trapped in the pupae case, as they fail to achieve the normal movements needed for eclosion. The fact that vav mutants normally survive until the pupal stages allows us to address vav function at different times during development.
Embryonic midline guidance defects in vav mutants
Many genes expressed in the midline are involved in axon guidance, controlling which axons should cross the midline and which should not. In wild-type late embryos, as it is the case in y1P{SUPor-P}vavKG02022 embryos (Fig. 3 A), the anti-FasII antibody labels three bundles of axons running longitudinally along each side of the midline (Grenningloh et al., 1991). While we could not detect any defects in vav mutant embryos at stages 13 and 14 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), we found that ∼14% of stage 17 vav mutant embryos (selected by the absence of β-gal staining) displayed misrouting of a subset of longitudinal axons across the midline (Fig. 3 B). These embryos showed two ectopic crosses on average (Fig. 3 H). This phenotype was observed in the three different vav alleles isolated. However, as these three alleles are derived from a single parental chromosome, we also used an unrelated allele (vav11837FRT19A) containing a P-element insertion disrupting the vav gene. The same phenotype was observed with the vav11837FRT19A allele (Fig. 3 G), confirming that the defects were indeed due to the absence of vav. To see whether other axons were misrouted in vav mutant embryos, we used the BP102 monoclonal antibody (Seeger et al., 1993) that labels both axons crossing the midline (commissural axons) and axons on each side of the midline (longitudinal axons) and allows us to visualize the overall CNS axon pattern. Vav mutant embryos stained with BP102 did not display any obvious phenotype (data not shown).
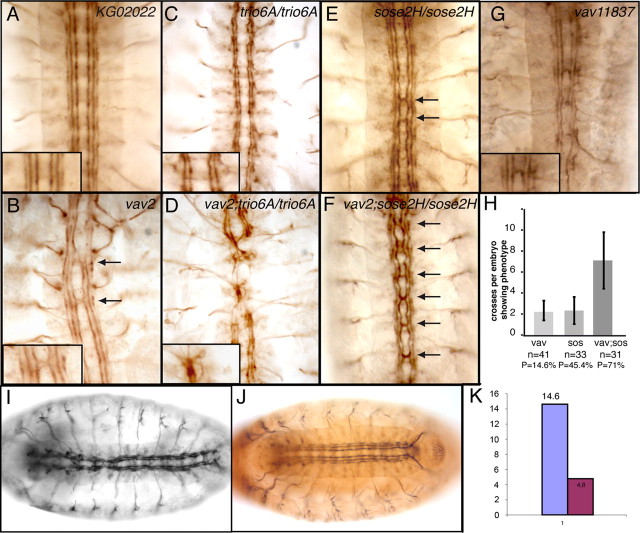
Figure 3.
Vav is involved in axon formation during embryonic development and interacts genetically with other GEFs. Embryos were stained with the monoclonal anti-FasII (or 1D4) antibody to visualize a subset of CNS axons. Stage 17 embryos were filleted (B–D) or mounted in epon (A, E–G). Anterior is up. Higher magnification is shown in insets. A, KG02022 embryo showing the normal organization of the longitudinal axons in three fascicles at each lateral side of the ventral midline. B, Vav2 mutant embryos showing axon formation and guidance defects. Mutants display midline crossing defects where axon bundles abnormally cross the midline (arrows). C, Trio mutants (trio6A/trio6A) display interrupted axon tracts. D, In vav;trio double mutants (vav2;trio6A/trio6A), axon tracts are more severely disrupted and axons fascicles are joined together at the midline. E, Sos embryos (sose2H/sose2H) display midline crossing defects (arrows). F, In vav;sos double mutants (vav2;sose2H/sose2H), midline crossing defects (arrows) are much stronger than in the single mutants. G, The vav 11837FRT19A mutant embryos display the same defects as the vav alleles isolated here (compare with B). H, Quantification of the midline crossing defects observed in vav, sos, and vav;sos mutants. Partial genotypes are indicated on the x-axis with the number of embryos analyzed (n) and the penetrance (P) of the phenotype in each case. The average number of ectopic crosses per embryo displaying at least one cross (expressivity) is indicated on the y-axis. Error bars represent standard deviations. I, Vav2 mutant embryos show axon defects. J, Ubiquitous expression of a wild-type version of vav, UAS-vav-HA, tagged with a HA epitope (brown), can partially rescue the embryonic CNS defects. K, Quantification of the rescue of the neuronal phenotype. The purple bar represents the percentage of vav embryos showing midline crossing defects, and the pink bar represents the rescue of this phenotype by ubiquitous expression of a wild-type version of vav.
As it has been reported that when midline glia cells are absent some CNS axons cross the midline inappropriately (Stemerdink and Jacobs, 1997), we decided to test whether the midline glia was present in vav mutant embryos. By using an antibody directed against the Wrapper protein, which specifically labels midline glia, we found that midline glia correctly expressed Wrapper in vav mutant embryos (data not shown). In addition, as decreased levels of the repulsive cue Slit, such as those found in lola mutant embryos (Crowner et al., 2002), also result in midline crossing defects, we tested whether Slit expression was altered in vav mutants and found it was not (data not shown). Thus, although we cannot exclude a possible additional role for vav in the midline glia, our results indicate that the midline crossing phenotype that we observed in vav mutants cannot be ascribed to an absence of midline glial cells nor to gross alterations in the levels of the secreted ligand Slit.
Because the penetrance of the phenotype was only 14%, we tested whether vav maternal product could compensate for the lack of zygotic vav, even though RNA transcripts were not detected from early stages in vav mutant embryos. Thus, we generated embryos in which both the maternal and the zygotic products were absent. We could not detect stronger midline crossing defects than those already observed (data not shown), suggesting the Vav maternal contribution, if any, is not important. In conclusion, vav contributes at the end of embryogenesis to prevent misrouting of axons at the midline.
Genetic interactions between vav and other GEFs
One of the best described functions of Vav is its activity as a GEF for the Rho GTPase family, particularly for the Rac proteins in both mammals and Drosophila (Turner and Billadeau, 2002; Hornstein et al., 2003; Couceiro et al., 2005). The three Drosophila rac members have been shown to play a crucial role in axon growth and guidance during development (Hakeda-Suzuki et al., 2002; Ng et al., 2002). However, while some rac mutants display midline crossing phenotypes that resemble those observed in vav mutants, the double and triple rac mutants show additional defects that are not found in vav mutants, such as axon growth defects. We therefore hypothesized that vav could be cooperating with other GEFs to mediate distinct aspects of Rac function during embryonic axon growth and guidance. In fact, another Rac GEF, trio, is required for proper axon growth in the embryo (Awasaki et al., 2000; Bateman et al., 2000). Interestingly, mutations in trio recapitulate only some aspects of the rac phenotype, as is the case for vav. For instance, trio mutants display discontinuous longitudinal axon tracts but do not show midline crossing defects (Awasaki et al., 2000; Bateman et al., 2000). Hence, we decided to test for genetic interaction between vav and trio. Strikingly, most vav; trio double mutants display severe patterning defects, making axogenesis difficult to analyze in these mutants (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Those mutant embryos that do not show gross morphological defects exhibit stronger and more frequent CNS phenotypes than the single mutants alone (Fig. 3 D). In fact, axon projection defects, both along the longitudinal tracts and across the midline, are enhanced in the double mutants. Interestingly, the axon defects found in vav;trio double mutants are reminiscent of those observed in triple loss-of-function Rac mutants (Hakeda-Suzuki et al., 2002).
Sos is another GEF that have been implicated in axon guidance during embryonic CNS formation through the activation of Rac (Yang and Bashaw, 2006). Removal of sos results in midline crossing defects similar to those described here for vav (Fritz and VanBerkum, 2000) and (Fig. 3 E). Notably, double vav;sos embryos display a midline crossing phenotype with a much higher expressivity (seven crosses per embryo on average) than the single mutants (Fig. 3 F,H). However, these embryos do not show any other defects within and outside of the CNS (data not shown), as opposed to vav;trio double mutants.
These results suggest that vav, trio, and sos cooperate to regulate Rac activities in the embryonic CNS during development.
Photoreceptor axon targeting defects in vav mutants
Next, we decided to test whether vav function during axogenesis was restricted to embryonic stages or whether, on the contrary, vav was also required to regulate axon guidance at other developmental stages. Another good model to assess defects in axon guidance in Drosophila is the targeting of photoreceptor axons from the eye imaginal disc to the optic lobe of the brain during larval development. We analyzed vav expression in the developing eye by performing whole-mount in situ hybridization on third instar larvae. In the eye imaginal disc, vav is expressed ubiquitously with higher levels in two stripes around the morphogenetic furrow (Fig. 4 A). In the brain, transcripts are detected almost ubiquitously in the optic lobes, whereas in the ventral nerve cord they are restricted to the midline as well as two lateral stripes (Fig. 4 B). This expression is vav specific, as no transcripts were detected in vav mutant eye discs (Fig. 4 A′) and larval brains (data not shown).
Figure 4.
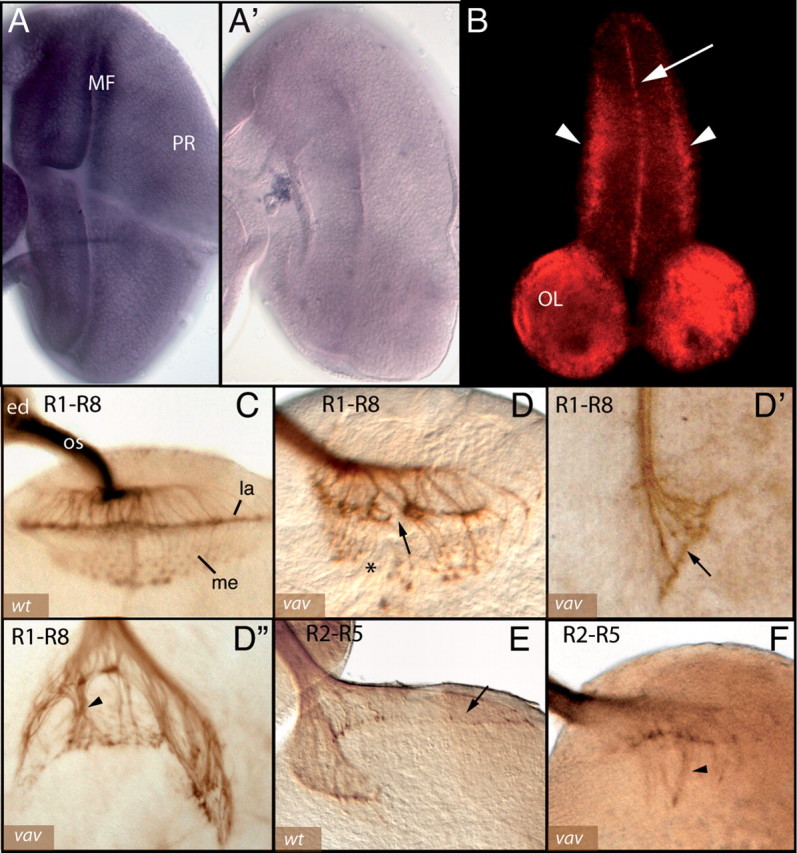
Vav is required for photoreceptor axon projections. A, B, Whole-mount in situ hybridization to detect vav expression in Drosophila third instar larval eye–brain complexes. A, A′, Anterior is toward the left. In wild-type larval eye imaginal disc (A) vav is expressed ubiquitously, including in photoreceptors (PR), with higher levels observed in the morphogenetic furrow (MF) region. Vav expression is abolished in vav mutants (A′). B, Confocal view of vav fluorescent in situ hybridization. Vav expression is observed in the larval CNS, including the optic lobes (OL), with higher levels seen in the midline (arrow) and two lateral lines (arrowheads) of the ventral nerve cord. C, D, Third instar photoreceptor axon projections are visualized with anti-Chaoptin (mAb 24B10) in eye–brain complexes. Axons project from the eye disc (ed) (upper left) into the brain through the optic stalk (os). C, In wild type (wt) the R1–R6 subset of photoreceptor axons stop in the lamina neuropil (la), where their growth cones form a continuous line of staining, while R7 and R8 continue into the medulla (me), forming a topographic array. D–D″, Various phenotypes can be observed in vav mutants: there are gaps in the lamina plexus (D, arrow), disruption of the typical medulla array pattern (D, asterisk), complete stalling of all axons in the lamina (D′, arrow) or increased numbers of axons entering the medulla (D″, arrowhead). E, F, R2–R5 photoreceptor axons are visualized selectively using a ro-tau-lacZ reporter to assess targeting to the lamina. E, In wild type, all R2–R5 axons stop in the lamina neuropil (arrow). F, In vav mutants, some R2–R5 axons display a lamina bypass phenotype (arrowhead).
Vav is therefore expressed in the appropriate place and time to play a role in photoreceptor axon targeting to the optic lobe. To test this hypothesis, photoreceptor axons of third instar larvae eye–brain complexes from wild-type and vav mutants were analyzed using the 24B10 monoclonal antibody. This antibody recognizes Chaoptin, a photoreceptor (R cell)-specific protein (Reinke et al., 1988). Axons for the eight R cells of each eye ommatidium fasciculate and project from the eye disc through the optic stalk to appropriate locations in the optic lobe, establishing a stereotypic topographic map (Clandinin and Zipursky, 2002; Mast et al., 2006). In wild-type flies, the outer R1–R6 cell axons innervate the first optic ganglion, the lamina, forming a layer of expanded growth cones seen as a smooth and uniform line. The inner R7 and R8 cell axons project through the lamina, and their growth cones elaborate a regular array in a distinct ganglion, the medulla (Fig. 4 C). Within the ganglions, R cell axons defasciculate, and each projects to a unique target. In vav mutants, while R cell axons converge in the eye disc and project into the optic lobe, they form abnormal patterns of connections in the lamina that appear discontinuous. This reflects an uneven spread of the R1–R6 axon bundles in which some regions are hyperinnervated and others lack innervation, creating gaps in the lamina (Fig. 4 D, arrow). In addition, the characteristic retinotopic pattern of the medulla is also disrupted in vav mutants (Fig. 4 D, asterisk). As it is the case for the embryonic midline guidance phenotype, photoreceptor axon targeting is similarly disrupted in the unrelated vav11837FRT19A allele, confirming the specificity of the vav gene in this process. Interestingly, mistargeting of photoreceptor axons is observed in 100% of the vav mutants analyzed (n = 60 optic lobes), although the severity of the defects is variable. We can classify the axonal phenotype using the following criteria: (1) mild, when only one or two gaps are found in the lamina, occuring in 22% of the cases analyzed; (2) strong, when more than two gaps are observed and/or too many axons seem to bypass the lamina and terminate in the medulla, seen in 58% of the cases (Fig. 4 D″); and (3) severe, found in 20% of cases, when only a few R cell axons penetrate the optic lobe and stall in the lamina instead of extending deeper in the medulla (Fig. 4 D′). The phenotype observed in the strong mutant category, where too many axons target the medulla, suggests that some of the R1–R6 axons that should stop in the lamina might be misrouted and targeted to the wrong ganglion. To assess axon targeting to the lamina in more detail, we used the ro-tau-lacZ marker that labels specifically a subset of R cell axons that stop in the lamina, R2–R5 (Garrity et al., 1999). In contrast to the wild-type situation (Fig. 4 E), in vav mutants several R2–R5 axons fail to terminate in the lamina but instead project aberrantly into the medulla (Fig. 4 F).
Thus, in addition to the role for vav in axon guidance during embryogenesis, vav is also required for photoreceptor axon guidance and targeting during larval development.
Rac proteins are Vav effectors in photoreceptors
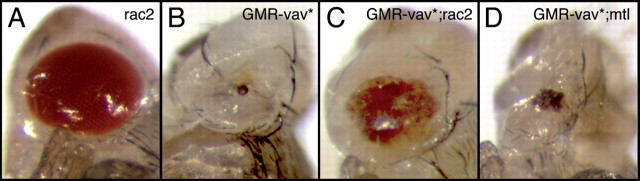
Axon guidance defects also occur in the visual system of Rac mutants (Hakeda-Suzuki et al., 2002). These defects are similar to those we observed in vav mutants larvae (data not shown), suggesting that vav could mediate its function in photoreceptor axon guidance through the activation of Rac proteins. To test this, we performed an epistasis experiment in the eye. Ectopic expression of a constitutively active form of Vav (UAS-Vav*) in the photoreceptors by using the GMR–Gal4 driver results in a severe disruption of the adult eye morphology (Fig. 5 B). We reasoned that if Vav signals through the Rac proteins in vivo, these defects should depend on Rac activity. Indeed, the eye morphology defects induced by the ectopic expression of Vav* are largely suppressed in animals homozygous for a loss-of-function mutation in either rac2 or rac1 (Fig. 5 C) (data not shown), while it is only mildly suppressed in mtl homozygous mutant flies (Fig. 5 D). These data indicate that, at least in the photoreceptors, Vav does indeed act through Rac proteins in vivo and further suggest that Rac1 and Rac2 are its preferred substrates.
Figure 5.
Differences in the ability of Rac proteins to rescue a vav gain-of-function phenotype in the eye. Adult eyes from flies with different genotypes are shown. A, GMR/+;rac2/rac2 flies do not display eye phenotype. B, GMR/vav*;rac2/+ flies showing a severe eye phenotype. C, GMR/vav*;rac2/rac2 flies display a mild eye phenotype. D, GMR/vav*;mtl/mtl flies display an eye phenotype almost similar as that in B.
Vav is not required for the differentiation and/or migration of lamina glial cells
Proper R cell projection patterning relies on bidirectional interactions between the photoreceptor axons and glial cells in the optic lobe. On the one hand, R cell axons provide signals to induce the differentiation and migration of the lamina glia. On the other hand, lamina glial cells produce retrograde signals that act as guidance cues for R cell axons to induce their termination within the lamina (Mast et al., 2006). For instance, in non-stop and CSN5 mutants where fewer glial cells enter the lamina, R1–R6 axons extend into the medulla (Poeck et al., 2001; Suh et al., 2002), a phenotype that resembles what we observe in vav mutants. To test whether the vav mutant phenotype could be due to defects in the formation and migration of the lamina glia, we decided to analyze the pattern of lamina glial cells in vav mutant embryos using the anti-Repo antibody as a marker (Xiong et al., 1994). In wild-type optic lobes, R cell axons target between two layers of glial cells, epithelial glia and marginal glia, at the lamina plexus (Fig. 6 A). In vav mutants, the number of glial cells and their characteristic distribution in rows surrounding the lamina plexus within the R1–R6 target region are similar to those in wild-type (Fig. 6 B). Furthermore, glial cells are found in areas in which R1–R6 axons mistarget (Fig. 6 B, arrowhead). This indicates that vav is not required for either the formation or the migration of lamina glial cells to the target area.
Figure 6.
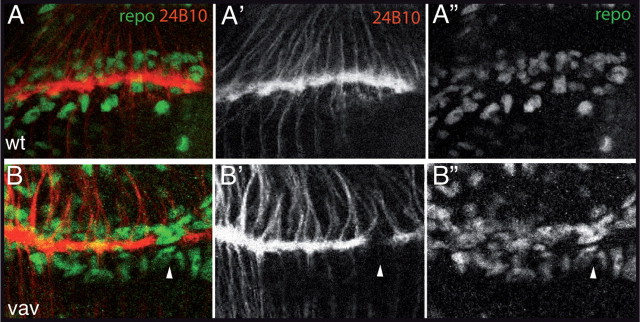
Lamina glia differentiation and migration occur normally in vav mutant optic lobes. Preparations of wild-type (wt) and vav mutant eye–brain complexes dissected from third instar larvae. Laser confocal microscopy 2 μm sections of optic lobes in which glial cells and R cell axons were visualized with anti-repo antibody (green) and mAb 24B10 (red), respectively. A, In wild-type larvae, glial cells, which have already migrated at this stage, are organized in three layers at the lamina plexus. R cell axons target between two layers of glial cells. B, In vav mutants, glia differentiation and migration are not compromised. In addition, the number of glial cells is not reduced and the lamina regions lacking axon innervation do not correlate with a lack of glial cells (arrowheads).
Vav function is required within the photoreceptor axons to control guidance
Having established that vav was not required to position glial cells in the lamina, vav could function either in R cell axons or in glial cells to dictate axon targeting. We first tried to test whether vav was required autonomously in the R cells by trying to rescue the vav mutant phenotype expressing a wild-type version of vav in the R cells by using the GMR-Gal4 driver. We performed these experiments using different UAS-vav lines and different temperatures. We could verify expression of the transgene in the R cells, as it is tagged with a hemagglutinin (HA) epitope. However, we could not rescue the phenotype in any case. There are at least two alternative explanations for these results. On the one hand, vav could be required in both axons and glia to regulate proper axon guidance. The fact that in the embryo ubiquitous expression of a vav cDNA could partially rescue the midline crossing phenotype (Fig. 3 I–K), while targeted expression of vav in either the axons or the midline glia could not (data not shown), support this idea. Alternatively, Vav might need to be tightly regulated within the cells, regulation that cannot be achieved using the GAL4-UAS system. Therefore, we used a different approach to test whether vav was required in the R cell axons or in the optic lobe target region to mediate its function in axon guidance. For this, we eliminated vav function specifically in each of these cell types by carrying out a series of clonal analyses using the MARCM technique (Lee and Luo, 1999). This system allowed us to generate and positively label vav mutant R cells or glial cells. In controls, GFP-labeled wild-type R cell axons project into appropriate locations in both the lamina and the medulla, and their growth cones expand in size when they reach their target (Fig. 7 A). However, homozygous vav mutant axons display targeting defects in the vast majority of mosaic animals (80%; n = 15 optic lobes). As in the case of the whole vav mutant larvae, many vav mutant axons in otherwise wild-type larvae do not stop at the lamina but target abnormally into the medulla (compare Fig. 7 B,C, Fig. 4 D″). In contrast, vav mutant MARCM clones affecting the lamina glia (Fig. 7 D) or the medulla (data not shown) were not associated with defects in R cell axon projections. Even when large vav mutant clones were generated in both target regions, wild-type axons were able to project normally (Fig. 7 E). Based on these observations, we conclude that vav is mainly required in R cells to regulate axon targeting.
Figure 7.
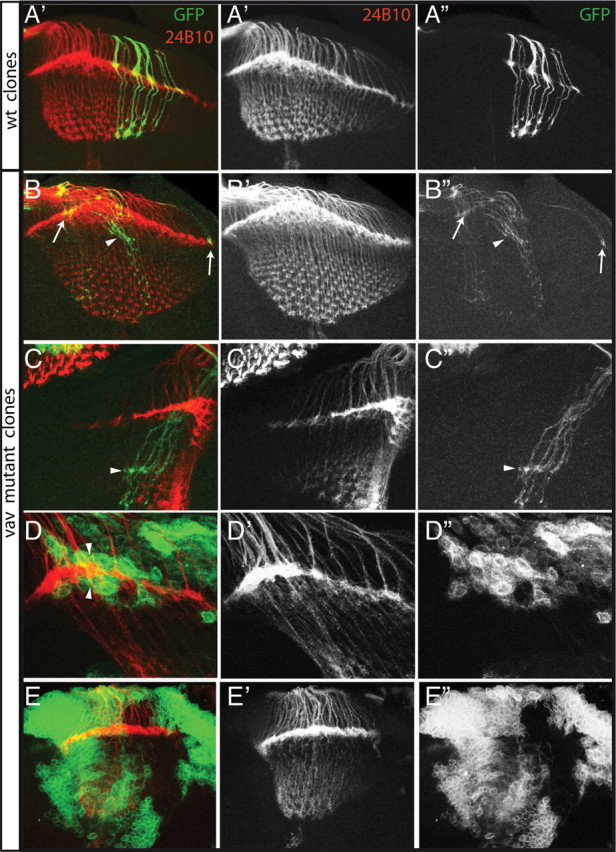
Vav function is required in R cell axons but not in the target to control axonal projections. A–E, Laser confocal microscopy of double-immunolabeled wild-type (wt) (A) and vav mutant (B–E) mosaic late third instar visual systems is shown. R cell axons were visualized with mAb 24B10 (red), and MARCM clones are visualized with GFP. Stacks of various 1 μm sections were made. A, In wild type, GFP-marked R cell axons target to appropriate locations in the lamina and medulla. B, C, While some homozygous vav mutant R cell axons target the lamina correctly (arrows), other axons (presumably R1–R6) project to a wrong layer below the lamina (arrowhead), from where some of them (presumably R7 and R8) extend deeper in the medulla. D, Wild-type R cell axons project correctly in the lamina plexus between two layers of vav mutant epithelial and marginal glial cells. E, Large clones of vav mutant cells in the medulla and the lamina do not affect R cell axon projections.
Vav is required for the correct growth of axons forming the ellipsoid body in the adult brain
The correct formation of several Drosophila adult brain structures depends on axon growth and guidance that takes place during metamorphosis. As we mentioned above, few vav mutants are able to reach adulthood and, when they do, they display strong locomotion defects. Interestingly, walking and coordination abilities are controlled by the central complex that is located between the two protocerebral hemispheres in the brains of all insects (Strauss, 2002). In addition, abnormal walking and flying behaviors have been observed in various mutants where the central complex structure is disrupted (Strauss and Heisenberg, 1993). As the four interconnected regions forming the central complex (the fan-shaped body, the ellipsoid body, the protocerebral bridge, and the paired noduli) are composed of axonal bundles, we decided to examine these structures in vav mutant animals by performing brain sectioning. The FasII antibody is a good marker to analyze the general pattern of adult brains, as it stains a subset of the axons forming the central complex structures but also other axon-rich regions outside of the central complex, such as the mushroom bodies and an asymmetrical structure important for long-term memory (Pascual et al., 2004). In vav mutant adults, all these regions seem to be present and the general aspect of the brain appeared similar to that of wild type (Fig. 8, compare A,B). However, the majority of vav mutant brains show axon growth defects in the ellipsoid body. Thus, while axonal projections constituting the ellipsoid bodies of all wild-type brains analyzed adopt a characteristic ring shape (n = 17 brains) (Fig. 8 C), 63% of vav mutant brains display ellipsoid bodies partially interrupted ventrally along the brain midline (n = 49 brains) (Fig. 8 D–E). The expressivity of the phenotype is variable, with 34% of the mutants displaying a mild phenotype in which only the external ring remains opened (Fig. 8 D), whereas 29% display a stronger phenotype with both ellipsoid body rings being opened (Fig. 8 E). The ellipsoid body precursor, a flat neuropilic structure consisting of dorsal fibers, elongates downward during metamorphosis to form the final adult ring shape (Renn et al., 1999; Boquet et al., 2000). Thus, we believe the phenotype observed in vav mutants is most likely due to ellipsoid body neurons failing to achieve complete growth. As the ellipsoid body is formed during pupation (Renn et al., 1999), these results therefore indicate that vav is not only required for axogenesis during embryonic and larval development, but is also required at later stages.
Figure 8.
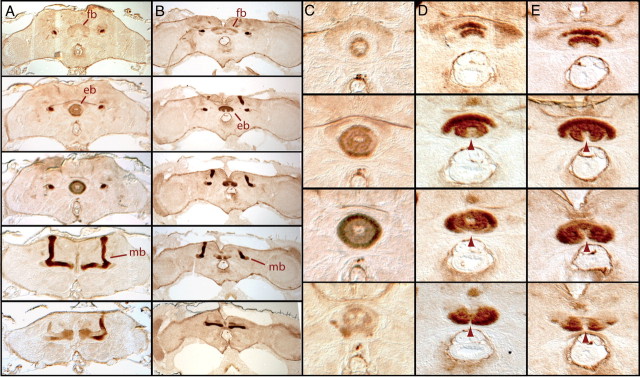
Central brain defects in the adult vav mutant. Serial 7 μm paraffin frontal sections of wild-type and vav mutant adult brains were stained with the axon marker FasII to highlight different neuropilar structures. Dorsal is above. A, In wild-type larvae, central complex regions such as the fan-shaped body (fb) and the ellipsoid body (eb) are stained with FasII, as well as the mushroom bodies (mb). B, The main brain structures are present in vav mutant brains. C, Higher magnification of wild-type ellipsoid body showing the characteristic ring-like shape of this structure. D, E, Ellipsoid bodies from vav mutants show axonal growth arrest with various expressivities (arrowheads), resulting in ventrally opened structures along the midline. In the milder cases (D) only the outside ring of the ellipsoid body is opened, whereas in stronger cases (E) axons forming both the outside and inside rings are affected.
Discussion
Vav members are key regulators of the Rho GTPases and the Rac proteins in particular. However, although many studies have implicated the Rac proteins in controlling several aspects of axon growth and guidance during development (Linseman and Loucks, 2008), our understanding of vav function in these processes is far more primitive. This is quite surprising given that all vav members are expressed in neural tissues in mammals. Recently, analysis of postnatal vav2/vav3-deficient mice has revealed abnormal retinogeniculate projections (Cowan et al., 2005). In this study, we demonstrate that in Drosophila vav is required for axon growth and guidance at embryonic, larval, and pupal stages. Hence, our data strengthen the role of vav in multiple aspects of axogenesis during development.
Vav is used reiteratively during Drosophila development to regulate distinct aspects of axogenesis
During the formation of the embryonic central nervous system of Drosophila, the neurons send out axons that project either ipsilaterally or contralaterally to form the complex axonal lattice. A small number of neurons project ipsilaterally as they receive repulsive signals from the midline glia and never cross the midline, while most neurons project contralaterally, cross the midline, and form the commissures (Schmid et al., 1999). In vav2/vav3 mutant mouse brains, ipsilateral but not contralateral projections are affected (Cowan et al., 2005). These results are consistent with our data showing that Drosophila vav is required to regulate proper ipsilateral axon projection, as in vav mutant embryos the most medial longitudinal axons occasionally cross the midline when they should not. These fascicles are particularly sensitive to perturbations in axon guidance mechanism and cross the midline whenever repulsive signaling is altered, suggesting that Vav might participate in the regulation of repulsive signaling from the midline.
We also demonstrated that vav is required during subsequent larval development in regulating photoreceptor axon targeting to the optic lobe. This is again consistent with the finding that vav2/vav3 mutant mice display abnormal projections of axons connecting the retinal cells to the brain (Cowan et al., 2005), suggesting that the role of vav in mediating axon guidance decisions is conserved between species. Interestingly, vav function in photoreceptors (R cells) seems to be more important than in the embryonic CNS. Indeed, we found that in 100% of the larvae, R cell axons projected aberrantly to the lamina and the medulla target regions, while only 14% of vav mutant embryos displayed guidance defects. Finally, we showed that later, during metamorphosis, vav is required once more for the correct formation of the ellipsoid body, one of the central brain structures. The ellipsoid body, in a majority of vav mutant adult brains, does not close properly and remains ventrally opened, most likely reflecting defects in the growth of the axons forming the ellipsoid body rather than guidance errors. Interestingly, the ellipsoid body has been involved in the control of locomotion, and vav mutant adults display strong locomotion defects. Opened ellipsoid bodies have also been found in ciboulot mutants (Boquet et al., 2000b). However these mutants do not display locomotion defects, suggesting that a disruption of the ellipsoid body alone is not sufficient to produce the locomotion phenotype observed in vav mutants. This implies that vav might also be required to regulate other aspects of axogenesis, in addition to the ones we have identified.
In summary, our results show that vav is required reiteratively throughout life to regulate different axogenesis events, including axon growth and guidance.
Vav requirements within axons during axon guidance
During larval development, R cell axon targeting to the optic lobe is controlled, on the one hand, by some genes that are acting within the axons themselves, and on the other hand, by some genes that are sending signals to the axons from the glia to guide them (Mast et al., 2006). Our MARCM experiments clearly demonstrate a role for vav within the R cell axons to regulate their projections. This is also the case in mammals, where Vav2 is highly expressed in the growth cones of cultured neurons where it is required to control guidance (Cowan et al., 2005).
Interestingly, in Drosophila, the GEF Trio has been shown to activate Rac, which in turn activates Pak, which is recruited to the membrane by Dock (Garrity et al., 1996; Hing et al., 1999; Newsome et al., 2000; Hakeda-Suzuki et al., 2002). These proteins participate in a signal transduction pathway that plays an essential role during photoreceptor axon guidance. We have shown here that Vav also acts via Rac in photoreceptors and that vav and trio interact genetically. Thus, in this context, it is tempting to speculate that like Trio, Vav could also contribute to the precise spatial control of Pak activity. In this scenario, the combination of signals via Vav, Trio, and Dock would allow growth cones to integrate multiple guidance signals.
Vav function in the axons could be to regulate the intracellular trafficking of guidance receptors through the activation of Rac. In mammals, for instance, vav2 has been proposed to be required in axons downstream of ephrin signaling for proper axon guidance (Cowan et al., 2005). In this case, when ephrins bind their Eph receptors, Vav becomes transiently activated upon phosphorylation and promotes local Rac-dependent endocytosis of the ephrin/Eph complex, a key event in axonal repulsion. In Drosophila however, mutations in Eph surprisingly show no obvious axon guidance defects in the photoreceptor axons targeting to the optic lobe nor in the embryonic CNS (Boyle et al., 2006). This suggests that in Drosophila, vav would need to be acting downstream of other guidance signals besides Eph.
In conclusion, we propose that Vav, after being activated by signaling receptors, could be required to stimulate Rac proteins to participate in the regulation of axon growth and guidance during development.
Vav and other GEFs
The Drosophila genome contains 22 GEFs. At least nine of them are expressed in the CNS (Hu et al., 2005), five of which are thought to be Rac activators. Why are there several Rac GEFs acting in the nervous system? A possible explanation is that the different GEFs might be activated in response to distinct guidance cues, thus triggering Rac-dependent specific cellular responses. For instance, beside its function in longitudinal axon growth, Trio has been involved in promoting commissure formation through its interaction with the attractive Netrin receptor Frazzled (Forsthoefel et al., 2005). Furthermore, another GEF, Sos, has been proposed to mediate Rac activation downstream of the Robo receptor to control axon repulsion at the midline (Yang and Bashaw, 2006). In this scenario, Vav, Sos, and Trio could coexist and be activated in response to different guidance molecules to control distinct aspects of axon guidance during the formation of the CNS.
In another scenario, different set of GEFs could also act redundantly to activate Rac proteins to a certain level, or at precise time points or in specific subcellular locations, allowing a unique cellular response. In fact, we have shown that the loss of both vav and sos function enhances dramatically the individual midline guidance phenotypes, suggesting that vav and sos can act redundantly in a common pathway. Similarly, the phenotype of the vav;trio double mutant in the nervous system, both at the midline and along the longitudinal axons, is more severe than the single mutants. In addition, while mutations in either vav or trio do not show any obvious defects outside the nervous system despite their widespread expression, elimination of both results in gross morphological defects. This indicates that both genes can act redundantly in vivo in different tissues and suggests that vav and trio are the main regulators of Rac activity.
A final explanation for the existence of different rac GEFs is that they could preferentially activate a particular Rac. There are three highly related rac genes in Drosophila, rac1, rac2, and mtl, and it has been suggested that Rac1 and Rac2 are preferred substrates of Trio (Hakeda-Suzuki et al., 2002). By performing a similar epistasis analysis, we have shown here that in photoreceptor cells Vav activates preferentially Rac1 and Rac2. The fact that vav and trio show similar substrate specificities could explain why we found these two genes to be redundant during embryogenesis.
In conclusion, although the vav family has been mainly implicated in the hematopoietic system and immune response, new roles are beginning to emerge for these genes. The fact that vav is required for axon growth and guidance at different stages of development suggests that it could be playing a multiplicity of functions in response to diverse signals. The existence of various protein–protein interaction domains in Vav represents a means of integrating Rac activities. Our results also suggest that vav function must overlap with that of other Rac modulators. Having isolated mutations in the Drosophila vav gene will help elucidate not only the role of this GEF during neural development but also the molecular mechanisms underlying general remodeling of the embryonic and adult nervous systems.
Footnotes
Research in our laboratories is funded by the Spanish Ministerio de Ciencia y Tecnología (MCYT) (BFU2007-64715/BMC to M.D.M.-B., CSD-2007-00008), the European Molecular Biology Organization Young Investigator Program, and the Junta de Andalucía (CVI-280 and PO6-CVI1592). M.M. was supported by postdoctoral fellowships from the MCYT and the Juan de la Cierva Program. The institutional support from the Junta de Andalucía to the Centro Andaluz de Biología del Desarrollo is acknowledged. We thank Dr. C. Klämbt, Dr. T. Suzuki, Dr. B. Dickson, Dr. L. Yang, Dr. B. Altenhein, and the Bloomington Stock Center for fly stocks and reagents. We are grateful to members of Dr. A. González-Reyes's and M. D. Martín-Bermudo's laboratories, Dr. X. Bustelo, Dr. B. Hassan, Dr. A. Hidalgo, and Dr. C. Sharpe for critically reading the manuscript. Special thanks to Dr. A. Pascual for helping us analyze the adult brain phenotype and for useful comments on this manuscript. M.M. performed experiments and analyzed data. M.M and M.D.M.-B. conceived, designed, and wrote this paper. F.F.A. provided technical support and generated stocks. D.A. contributed to the initial analysis of the photoreceptor axonal phenotype.
References
- Aoki K, Nakamura T, Fujikawa K, Matsuda M. Local phosphatidylinositol 3,4,5-trisphosphate accumulation recruits Vav2 and Vav3 to activate Rac1/Cdc42 and initiate neurite outgrowth in nerve growth factor-stimulated PC12 cells. Mol Biol Cell. 2005;16:2207–2217. doi: 10.1091/mbc.E04-10-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- Betz R, Sandhoff K, Fischer KD, van Echten-Deckert G. Detection and identification of Vav1 protein in primary cultured murine cerebellar neurons and in neuroblastoma cells (SH-SY5Y and Neuro-2a) Neurosci Lett. 2003;339:37–40. doi: 10.1016/s0304-3940(02)01457-x. [DOI] [PubMed] [Google Scholar]
- Boquet I, Hitier R, Dumas M, Chaminade M, Préat T. Central brain postembryonic development in Drosophila: implication of genes expressed at the interhemispheric junction. J Neurobiol. 2000a;42:33–48. [PubMed] [Google Scholar]
- Boquet I, Boujemaa R, Carlier MF, Préat T. Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell. 2000b;102:797–808. doi: 10.1016/s0092-8674(00)00068-4. [DOI] [PubMed] [Google Scholar]
- Boyle M, Nighorn A, Thomas JB. Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development. 2006;133:1845–1854. doi: 10.1242/dev.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bustelo XR. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet N, Prieto M, Fabre C, Noren NK, Privat A. Distribution of p120 catenin during rat brain development: potential role in regulation of cadherin-mediated adhesion and actin cytoskeleton organization. Mol Cell Neurosci. 2003;22:467–486. doi: 10.1016/s1044-7431(03)00030-7. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Making connections in the fly visual system. Neuron. 2002;35:827–841. doi: 10.1016/s0896-6273(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Couceiro JR, Martín-Bermudo MD, Bustelo XR. Phylogenetic conservation of the regulatory and functional properties of the Vav oncoprotein family. Exp Cell Res. 2005;308:364–380. doi: 10.1016/j.yexcr.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Crowner D, Madden K, Goeke S, Giniger E. Lola regulates midline crossing of CNS axons in Drosophila . Development. 2002;129:1317–1325. doi: 10.1242/dev.129.6.1317. [DOI] [PubMed] [Google Scholar]
- Dekel I, Russek N, Jones T, Mortin MA, Katzav S. Identification of the Drosophila melanogaster homologue of the mammalian signal transducer protein, Vav. FEBS Lett. 2000;472:99–104. doi: 10.1016/s0014-5793(00)01413-7. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, Seeger MA. The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila . Development. 2005;132:1983–1994. doi: 10.1242/dev.01736. [DOI] [PubMed] [Google Scholar]
- Fritz JL, VanBerkum MF. Calmodulin and Son of sevenless dependent signaling pathways regulate midline crossing of axons in the Drosophila CNS. Development. 2000;127:1991–2000. doi: 10.1242/dev.127.9.1991. [DOI] [PubMed] [Google Scholar]
- Fujikawa K, Miletic AV, Alt FW, Faccio R, Brown T, Hoog J, Fredericks J, Nishi S, Mildiner S, Moores SL, Brugge J, Rosen FS, Swat W. Vav1/2/3-null mice define an essential role for Vav family proteins in lymphocyte development and activation but a differential requirement in MAPK signaling in T and B cells. J Exp Med. 2003;198:1595–1608. doi: 10.1084/jem.20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity PA, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky SL. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, Zinn K, Zipursky SL. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–717. doi: 10.1016/s0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila . Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Hornstein I, Mortin MA, Katzav S. DroVav, the Drosophila melanogaster homologue of the mammalian Vav proteins, serves as a signal transducer protein in the Rac and DER pathways. Oncogene. 2003;22:6774–6784. doi: 10.1038/sj.onc.1207027. [DOI] [PubMed] [Google Scholar]
- Hu H, Li M, Labrador JP, McEwen J, Lai EC, Goodman CS, Bashaw GJ. Cross GTPase-activating protein (CrossGAP)/Vilse links the Roundabout receptor to Rac to regulate midline repulsion. Proc Natl Acad Sci U S A. 2005;102:4613–4618. doi: 10.1073/pnas.0409325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Linseman DA, Loucks FA. Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front Biosci. 2008;13:657–676. doi: 10.2741/2710. [DOI] [PubMed] [Google Scholar]
- Mast JD, Prakash S, Chen PL, Clandinin TR. The mechanisms and molecules that connect photoreceptor axons to their targets in Drosophila . Semin Cell Dev Biol. 2006;17:42–49. doi: 10.1016/j.semcdb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Movilla N, Bustelo XR. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Ohnishi H, Okazawa H, Murata Y, Kusakari S, Hayashi Y, Miyashita M, Itoh H, Oldenborg PA, Furuya N, Matozaki T. CD47 promotes neuronal development through Src- and FRG/Vav2-mediated activation of Rac and Cdc42. J Neurosci. 2006;26:12397–12407. doi: 10.1523/JNEUROSCI.3981-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson BJ. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila . Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Noordermeer JN, Kopczynski CC, Fetter RD, Bland KS, Chen WY, Goodman CS. Wrapper, a novel member of the Ig superfamily, is expressed by midline glia and is required for them to ensheath commissural axons in Drosophila . Neuron. 1998;21:991–1001. doi: 10.1016/s0896-6273(00)80618-2. [DOI] [PubMed] [Google Scholar]
- Pascual A, Huang KL, Neveu J, Préat Neuroanatomy: brain asymmetry and long-term memory. Nature. 2004;427:605–606. doi: 10.1038/427605a. [DOI] [PubMed] [Google Scholar]
- Perrimon N. Clonal analysis of dominant female-sterile, germline-dependent mutations in Drosophila melanogaster . Genetics. 1984;108:927–939. doi: 10.1093/genetics/108.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila . Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- Reinke R, Krantz DE, Yen D, Zipursky SL. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell. 1988;52:291–301. doi: 10.1016/0092-8674(88)90518-1. [DOI] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH. Genetic analysis of the Drosophila ellipsoid body neutrophil: organization and development of the central complex. J Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Midkiff BR, Kedar VP, Maness PF. Adhesion molecule L1 stimulates neuronal migration through Vav2-Pak1 signaling. Neuroreport. 2004;15:2791–2794. [PubMed] [Google Scholar]
- Seeger M, Tear G, Ferres-Marco D, Goodman CS. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron. 1993;10:409–426. doi: 10.1016/0896-6273(93)90330-t. [DOI] [PubMed] [Google Scholar]
- Stemerdink C, Jacobs JR. Argos and Spitz group genes function to regulate midline glial cell number in Drosophila embryos. Development. 1997;124:3787–3796. doi: 10.1242/dev.124.19.3787. [DOI] [PubMed] [Google Scholar]
- Strauss R. The central complex and the genetic dissection of locomotor behavior. Curr Opin Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci. 1993;13:1852–1861. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GS, Poeck B, Chouard T, Oron E, Segal D, Chamovitz DA, Zipursky SL. Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron. 2002;33:35–46. doi: 10.1016/s0896-6273(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Turner M, Billadeau DD. VAV proteins as signal integrators for multi-subunit immune-recognition receptors. Nat Rev Immunol. 2002;2:476–486. doi: 10.1038/nri840. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila . Nat Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Yang L, Bashaw GJ. Son of sevenless directly links the Robo receptor to rac activation to control axon repulsion at the midline. Neuron. 2006;52:595–607. doi: 10.1016/j.neuron.2006.09.039. [DOI] [PubMed] [Google Scholar]