Abstract
Introduction:
Colorectal cancer (CRC) is the second leading cause of cancer related death in the world after lung cancer. Early detection of CRC leads to improvement in cancer survival rate. In recent years, efforts have been made to discover a non-invasive screening marker of higher sensitivity and specificity. Fecal occult blood testing (FOBT) and genetic testing become alternative modalities to screen CRC in the population other than colonoscopy. The aim of this systematic review and meta-analysis is to determine the diagnostic accuracy, sensitivity and specificity of FOBT and genetic testing as screening tools in colorectal cancer.
Methods:
A literature search of PubMed, ScienceDirect, and Scopus was carried out. The search strategy was restricted to human subjects and studies are published in English. Data on sensitivity and specificity were extracted and pooled. Heterogeneity was assumed at significance level of p < 0.10 and was tested by chi squared. Degree of heterogeneity was quantified using the I2 statistic, and values of less than 25% is considered as homogenous. All analyses were performed using the software Meta-Disc.
Results:
A total of eleven studies were suitable for data synthesis and analysis. Five studies were analyzed for the accuracy of genetic testing, the pooled estimate for sensitivity and specificity were 71% (95% CI: 66, 75%) and 95% (95% CI: 93, 97%) respectively. Another group of studies which had been evaluated for the accuracy of FOBT, the pooled sensitivity was 31% (95% CI: 25, 38%) while the pooled specificity was 87% (95% CI: 86, 89%).
Conclusions:
FOBTs is recommended to use as population-based screening tools for colorectal cancer while genetic testing should be focusing on patients with moderate and high risk individuals.
Introduction
Colorectal cancer (CRC) is the second most common cancer in the world [1]. It is the second leading cause of cancer related death worldwide after lung cancer. The American Cancer Society found that about one in 20 people in the US will develop colorectal cancer during their lifetime [2]. Early detection is the main key to improve cancer survival rate. Screening modalities for CRC have been initiated in several countries [3,4]. Clinical practice guidelines (CPG) recommend that average risk individuals begin regular screening for CRC at 50 years of age [5]. Colonoscopy, despite being the gold standard diagnostic tool, remains invasive with risk of perforation. Because of that, the population’s uptake of colonoscopy is low [6,7,8,9,10].
On the other hand, fecal occult blood testing (FOBT) is another modality to screen CRC among the population. FOBT using either guaiac-based (gFOBT) or immunological-based (iFOBT) methods are not invasive and more preferred [11,12,13]. It detects subtle blood loss in the gastrointestinal tract as a result of cancer bleeding. gFOBT detects heme while iFOBT detect globin [14].
In the past few years, there has been a drive to discover a more sensitive and specific tool in CRC screening. Genetic testing is one of the options. It can detect if members of certain families are prone to inherit any genetic link cancer, particularly first degree relatives [15,16]. Mutations inside MLH1, MSH2, MSH6 and PMS2, the genes involved in the DNA mismatch repair (MMR), boost the risk of developing CRC, especially Lynch syndrome (LS) [17,18]. Mutations in any of these genes impair the suitable repair of DNA replication errors that accumulate in the cells, promoting their aberrant proliferation leading to cancer. MLH1 and MSH2 germline mutations account for approximately 90% of mutations in families with LS [19,20].
However, there appear to be some contention in the literature regarding its utility. Some studies on cancer genetic testing have shown a favorable sensitivity and specificity for colorectal cancer and adenoma [21,22], whilst other studies have shown the opposite [23,24]. Otherwise, it is a less invasive blood based screening, thus highly preferable and accepted among patients.
According to Clinical Practice Guidelines, moderate- and high-risk groups will be referred to do a colonoscopy, however genetic testing can be offered [5]. All individuals whose family history is suggestive of a hereditary colorectal cancer syndrome should be referred to a clinical genetics service for genetic counselling, genetic risk assessment, and consideration of genetic testing to clarify the risk. These guidelines are yet to be used locally.
The aim of this systematic review and meta-analysis is to measure the accuracy of both FOBT and genetic testing as screening tools in colorectal cancer, specifically to identify the sensitivity and specificity of these modalities and their potentials as population-based screening tools.
Methods
Search Protocol
A systematic search was conducted (1 October 2018) on PubMed, ScienceDirect, and Scopus databases covering a period of five years (2013 to 2018) guided by Lui et al. (2013) [25]. Search was done for the relevant titles, abstracts, and keywords and adhered to the PICO strategy as stated in PRISMA checklist. The terms used for P (Problem or Population) was “colorectal cancer”, followed by terms for I (Intervention) which were “fecal occult blood test” OR “FOBT” OR “genetic testing”. The keywords used for O (Outcome) were “sensitivity” OR “specificity” OR “validity” OR “diagnostic accuracy”. For this review, there was no C (Comparison) terms in the search protocol. The respective MeSH terms for P, I, and O were also searched. Only English language literatures were searched and there was no restriction placed on location. Grey literatures and other sources were not searched.
Study Selection
Studies were first screened by the title. After the first screening was done and the studies’ titles were deemed as having appropriate research question as this review, two reviewers were randomly allocated for each study for the screening of abstracts. Following this, the next phase of the article selection involved retrieval of the full articles for further scrutinization. Two reviewers, who were different from the initial reviewers, were allocated for each study for the perusal of the full text. Data extraction was conducted for the studies which had been accepted after review of the full article. Studies were only considered for this review and included if 1) it was an original article (not a review paper or commentary) and 2) the study had included a quantitative measurement of colorectal screening tool validity. The exclusion criteria were 1) inability to access the full article; 2) full article was not in English; and 3) the study had used combination of multiple tools or assessment criteria for screening of colorectal cancer (the study had used single or with combination of FOBT, colonoscopy and genetic testing).
Assessment of Study Quality
Quality assessment for accepted studies was performed using the “Quality Assessment of Diagnostic Accuracy Studies” (QUADAS) tool. This tool was validated for the purpose of assessing study quality of studies included in diagnostic accuracy systematic review. It was developed by a consensus from nine experts in the field of diagnostics which took part in the Delphi procedure. QUADAS consists of 14 items of questions showed in Table 1 which cover bias (items 3, 4, 5, 6, 7, 10, 11, 12 and 14), variability (items 1 and 2), and the quality of reporting (items 8, 9 and 13) [26]. The validity of QUADAS by an evaluation by three reviewers rated 30 studies, with the range of agreements were very good; 85 to 91% [27].
Table 1.
Items of assessment in QUADAS.
| Items | Assessment | Questions |
|---|---|---|
| Q1 | Adequate spectrum composition | Was the spectrum of patient’s representative of the patients who will receive the test in practice? |
| Q2 | Clear description of selection criteria | Were selection criteria clearly described? |
| Q3 | Adequate reference standard | Is the reference standard likely to correctly classify the target condition? |
| Q4 | Absence of disease progression bias | Is the period between reference standard and index test short enough to be reasonably sure that the target condition did not change between the 2 tests? |
| Q5 | Absence of partial verification bias | Did the whole sample or a random selection of the sample, receive verification using a reference standard of diagnosis? |
| Q6 | Absence of differential verification bias | Did patients receive the same reference standard regardless of the index test result? |
| Q7 | Absence of incorporation bias | Was the reference standard independent of the index test (i.e. the index test did not form part of the reference standard)? |
| Q8 | Adequate description of the index test execution | Was the execution of the index test described in sufficient detail to permit replication of the test? |
| Q9 | Adequate description of the reference test execution | Was the execution of the reference standard described in sufficient detail to permit its replication? |
| Q10 | Absence of index test review bias | Were the index test results interpreted without knowledge of the results of the reference standard? |
| Q11 | Absence of reference test review bias | Were the reference standard results interpreted without knowledge of the results of the index test? |
| Q12 | Absence of clinical review bias | Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? |
| Q13 | Report of uninterpretable results | Were uninterpretable/intermediate test results reported? |
| Q14 | Description of withdrawals | Were withdrawals from the study explained? |
For the purpose of this review, item 12 was omitted because it was not relevant to the test and tool reviewed. Each study was randomly allocated to a reviewer for study quality assessment and subsequently the studies were also reviewed by another reviewer. The reviewers scored as ‘yes’, ‘no’, or ‘unknown’ for each items, and the level of agreement was stratified accordingly. Any disagreement was resolved by discussion and consensus.
Data Extraction
A standardized table with relevant headings was used to extract data, namely the study design, population, sample size, gold standard for validity measurement, sensitivity, specificity, and other measure of screening tool validity. Studies were arranged according to year of publication.
Statistical Analysis
Composite estimates of the sensitivity and specificity of each tool analyzed were tabulated in Forest Plots and compared with one another. Heterogeneity was assumed at significance level of p < 0.10 and was tested by chi squared. Degree of heterogeneity was quantified using the I2 statistic, and values of less than 25% was considered as homogenous. Arbitrary cut-off points of 50% and 75% were used to divide the studies into low, moderate, and high level of heterogeneity. All analyses were performed using the software Meta-Disc (version 1.4).
Results
Search results returned a total of 592 articles, of which 569 articles were excluded due to irrelevant title and abstracts during screening. Of these, 23 articles were eligible for review of full text. After full text article were extracted, 11 studies were included in review with six studies of FOBT suitable for meta-analysis (Figure 1).
Figure 1.
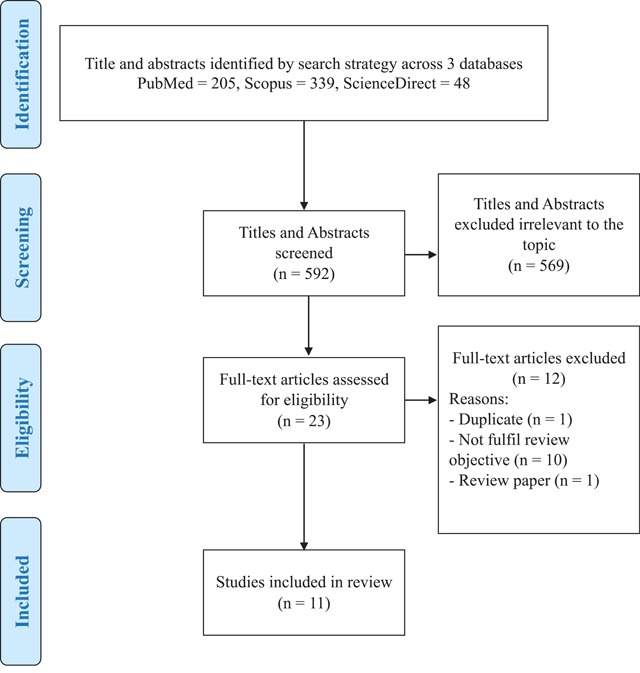
Flow Chart of Studies Selection.
Among the 11 studies included in diagnosing colorectal cancer, six studies examined the accuracy of FOBT test and another five studies examined genetic testing. For the purpose of quality assessment, we described the percentage of agreement by each QUADAS items according to the type of screening tools. The overall results of the quality assessment using QUADAS for each of the 11 studies reviewed, were presented in Table 2. An average, high percentage of agreements on both FOBT and genetic testing studies reviewed, it shows a good quality of diagnostic studies included in this review. Lower rate of agreement (FOBT 33.3%, genetic 40%) was observed on the withdrawal description among the studies.
Table 2.
Assessment of methodological quality using QUADAS (n: 11).
| No. | First Author (Year) | Screening Tool | Q1 | Q2 | Q3 | Q4 | Q5 | Q6 | Q7 | Q8 | Q9 | Q10 | Q11 | Q13 | Q14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Yeasmin F (2013) | FOBT | Y | Y | Y | Y | Y | Y | Y | Y | Y | U | Y | N | N |
| 2. | Lohsiriwat (2014) | FOBT | Y | Y | Y | Y | Y | Y | Y | Y | Y | U | U | U | N |
| 3. | Redwood (2014) | FOBT | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | U | Y |
| 4. | Elsafi (2015) | FOBT | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| 5. | Mario (2015) | FOBT | N | Y | Y | Y | N | N | Y | U | U | Y | N | N | N |
| 6. | Shapiro (2017) | FOBT | Y | Y | Y | U | Y | Y | Y | Y | Y | Y | U | U | Y |
| 7. | Dvorak (2014) | Genetic | Y | Y | Y | U | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| 8. | Amiot (2014) | Genetic | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | N |
| 9. | Johnson DH (2016) | Genetic | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | N |
| 10. | Kanth P (2016) | Genetic | Y | Y | Y | Y | Y | Y | Y | Y | Y | U | Y | Y | Y |
| 11. | Xie (2018) | Genetic | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | N |
| % of agreement ‘yes’ | FOBT | 83.3 | 100 | 100 | 83.3 | 83.3 | 83.3 | 100 | 83.3 | 83.3 | 66.7 | 50 | 16.7 | 33.3 | |
| Genetic | 100 | 100 | 100 | 80 | 100 | 100 | 100 | 100 | 100 | 80 | 100 | 100 | 40 | ||
Characteristic and Accuracy of Selected Studies
A total of eleven studies have been selected for this review. All those studies have been conducted at all regions across the world. Majority of the studies were carried out by United States of America and there were four studies done in Asia. Different study designs have been used in the selected studies, those are; prospective cohort, case-control, and cross-sectional with diverse objectives. Among the eleven studies, six studies assessed accuracy of FOBT (Table 3) and five on genetic testing (Table 4).
Table 3.
Characteristic and accuracy of selected fecal occult blood test studies.
| No | Author (Year) | Country | Study design | Study population | Index test | Reference test | Sample size | Sensitivity % | Specificity % | PPV % | NPV % | Accuracy % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Shapiro et al. (2017) [29] | USA | Cross-sectional | Asymptomatic patients from clinics, aged 50–75 years. | HS-gFOBT | Not mentioned | 1095 | 7.4 | 98.6 | 23.6 | 86.0* | |
| 2. | Mario et al. (2015) [30] | Brazil | Cross-sectional | Asymptomatic patients, aged ≥ 50 years. | gFOBT & flexible RSS | Colonoscopy | 102 | 30.0 | 92.4 | 30.0* | 5.0* | 86.3* |
| 3. | Elsafi et al. (2015) [31] | Saudi Arabia | Cohort | Asymptomatic patients aged 50–74 years old from 2 hospitals and confirmed CRC patients. | gFOBT | Cases 257 Control 20 |
50.0 | 77.9 | 3.5 | 99.0 | 71.8* | |
| 4. | Lohsiriwat et al. (2014) [32] | Thailand | Case-control | Histologically proven adenocarcinoma of the colon and rectum patients and individuals with normal colonoscopic findings. | FOBT | Colonoscopy | Cases 96 Control 101 |
41.0 | 97.0 | 93.0 | 63.0 | 70.0 |
| 5. | Redwood et al. (2014) [33] | USA | Cross-sectional | Asymptomatic adults aged ≥ 40 years. | gFOBT | Colonoscopy | 304 | 28.5 | 75.7 | 10.6 | 91.3 | 71.4* |
| 6. | Yeasmin et al. (2013) [34] | Bangladesh | Cross-sectional | Patients suspected to have occult bleeding. | gFOBT | Colonoscopy | 110 | 75.0 | 21.6 | 7.0 | 91.7 | 25.5* |
FOBT- Fecal occult blood test.
gFOBT- Guaiac fecal occult blood test.
HG-gFOBT- High-sensitivity guaiac fecal occult blood test.
RSS- Recto sigmoidoscopy.
PPV- Positive Predictive Value.
NPV- Negative Predictive Value.
* indirectly calculated from data.
Table 4.
Characteristic and accuracy of genetic testing.
| No | Author (Year) | Country | Study design | Study population | Index Test | Reference Test | Sample size | Sensitivity % | Specificity % | PPV % | NPV % | Accuracy % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Xie (2018) [35] | China | Case control | Patients aged >18 years old with histologically confirmed mCRC. | mSEPT9 | Colonoscopy | 123 Cases 125 Controls |
61.0 | 89.0 | 89* | 62.8* | 73.4* |
| 2. | Johnson (2016) [36] | USA | Case control | Advanced CRC patients. | BMP3 | Histology | 17 Cases 12 Controls |
76.0 | 92.0 | 92.9* | 73.3* | 82.8* |
| 3. | Kanth (2016) [37] | USA | Case control | Asymptomatic patients from 2 medical centers, aged age 45–75. | BRAF | Colonoscopy | 41 Cases 20 Controls |
94.0 | 72.0 | 79.5* | 90.9* | 83.6* |
| 4. | Dvorak (2014) [38] | Australia | Cross-sectional | Colorectal and Papillary thyroid cancer patients. | BRAF | Histology | 352 | 98.6 | 99.1 | 98.6 | 99.1 | 98.9* |
| 5. | Amiot (2014) [39] | France | Case control | Asymptomatic patients from a teaching hospital. | Wif-1 Gene | Colonoscopy | 90 Cases 157 Controls |
33.0 | 99.0 | 96.9* | 70.7* | 74.1* |
PPV- Positive Predictive Value.
NPV- Negative Predictive Value.
* indirectly calculated from data.
FOBT
A total of six studies assessed on accuracy of FOBTs. All of the studies evaluated Guaiac FOBTs. Majority of the studies were done cross-sectional, while there are two studies done as cohort and case-control. The study population were mostly asymptomatic adults aged 50 years and above. Diagnostic accuracy of FOBT ranged from 25.5% to 86.3% with sensitivity and specificity ranged from 7.4%–75.0% and 21.6%–98.6%, respectively.
Genetic Testing
These five articles were studies from 2014–2018 from USA, China, Australia, and France. All this study used high risk group as their population. In the other hand, two studies used BRAF as index test, and others used mSEPT9, BMP3, and Wif-1 Gene, respectively. A total of 937 patient data were pooled from the five articles. Of these, 302 patients had a confirmed diagnosis of colorectal cancer on colonoscopy and histology. Using genetic testing, 302 (32.23%) of participants were identified as true positives, 23 (2.45%) as false positives, 486 (51.87%) as true negatives and 126 (13.45%) as false negatives. The accuracy of genetic testing is as showed in Table 4. The sensitivity and specificity of genetic testing for diagnosing colorectal cancer was range 33–98 and 72–99 respectively. The diagnostic accuracy of genetic testing for diagnosing colorectal cancer was range 73.4–98.9.
Meta-Analysis
The sensitivity and specificity figures of all the studies were pooled to obtain an estimate of the diagnostic accuracy of FOBT in detecting colorectal cancer. There were six studies which had evaluated the accuracy of chemical or guaiac FOBT to detect colorectal cancer (Shapiro et al. [2017]; Mario et al. [2015]; Elsafi et al. [2015]; Lohsiriwat et al. [2014]; Redwood et al. [2014]; Yeasmin et al. [2013]). For this group, the pooled sensitivity was 31% (95% CI: 25, 38%) while the pooled specificity was 87% (95% CI: 86, 89%) (Figures 2 and 3). For analyses within the subgroups, significant heterogeneity was observed, ranging from 83.2% to 98.9% of heterogeneity as quantified by I-square test. All findings are summarized in Table 5.
Figure 2.
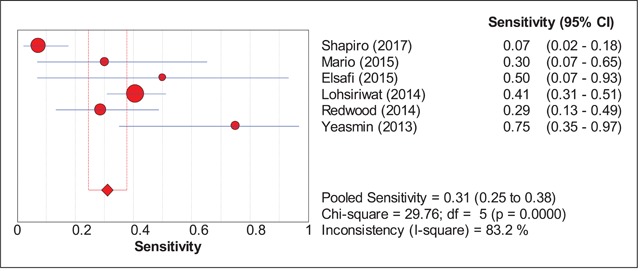
Pooled Sensitivity FOBT.
Figure 3.
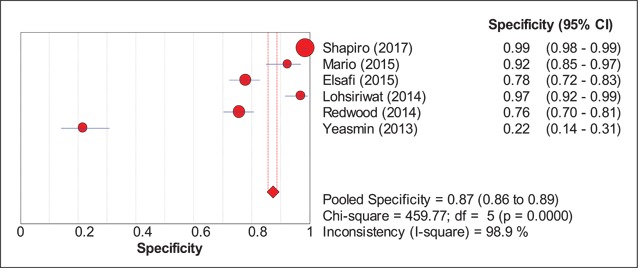
Pooled Specificity FOBT.
Table 5.
Summary Statistics for the Diagnostic Accuracy of FOBT.
| Sub-groups | Number of Studies | P-valuea | I2 (%)b | Sensitivity % (95% CI) |
Specificity % (95% CI) |
|---|---|---|---|---|---|
| FOBT | 6 | <0.001 | 83.2–98.9 | 31 (25, 38) | 87 (86, 89) |
a P-value for heterogeneity (chi-square) for both sensitivity and specificity analyses.
b I2 statistics for heterogeneity quantification for both sensitivity and specificity analyses.
Discussion
Principal Findings
This review had found 11 studies that addressed the diagnostic accuracy of genetic testing modalities and FOBT to detect colorectal cancer in the last 5 years. Chemical or guaiac FOBT has been used as a screening tool for colorectal cancer in symptomatic persons mostly due to its affordable cost. Hirai et al. (2016) reported pooled sensitivity & specificity of FOBTs for CRC detection in the proximal colon were 71.2% (95% CI 61.3–79.4%) and 93.6% (95% CI 90.7–95.7%) respectively. Both gFOBT and iFOBT showed significantly lower sensitivity but comparable specificity for the detection of proximally located CRC compared with distal CRC [28].
Moving into the era of precision medicine, genetic testing is fast gaining traction as a screening tool for those without symptoms but having strong family history or having known family members with genetic mutations associated with colorectal cancer. However, its high cost and the need for specialized service is hampering efforts to introduce the tool in resource-modest countries, including developing countries. Present study shows that genetic testing modalities had relatively better diagnostic accuracy as compared to FOBT in detecting colorectal cancer. Genetic testing was shown to be superior to FOBT in diagnostic accuracy range 73.4–98.9 versus diagnostic accuracy of FOBT range 25.5–86.3. However, most of the genetic testing studies were conducted among the high-risk group or confirmed CRC patients which may affect the outcome.
Since genetic testing has a superior diagnostic accuracy compared to FOBT, it can be suggested to be included in the CPG for screening of asymptomatic moderate- and high-risk people, provided that a prevalence study of the genetic variant responsible for CRC is available locally and its cost effectiveness analysis is done.
Analysis of Heterogeneity
Our results also showed that there was significant heterogeneity in the analysis in the subgroup. There are multiple possibilities as to why these heterogeneities came about. First, the studies were conducted in different populations and localities. The intrinsic differences between populations also extend into differences between socio-economic status, cultures, and quality of medical services. These dissimilarities may have affected the accuracy of the tools used. Since the number of studies under FOBT group (six studies) were already in single digits, it was not feasible to further sub-divide the studies into sub-groups. Doing so may improve the homogeneity between studies, but pooling accuracy estimates from two or three studies may not generate results that are generalizable to the bigger populations.
On the other hand, there were multiple genes being tested in genetic testing. Each of the five studies which had assessed genetic testing modalities had a different method of detecting the affected gene for colorectal cancer. Although these studies were classified together under “genetic testing”, there existed a measure of heterogeneity in their methods and specific marker that each study had appraised.
Strengths and Weaknesses
Our review had summarized the accuracy of genetic testing modalities and FOBT in detecting colorectal cancer from studies conducted in the last five years. This means that the results of this review are up-to-date with current evidence and the data for diagnostic accuracy of assessed tools can be incorporated into devising a health policy or guidelines in colorectal cancer prevention and management.
However, there are also a few notable weaknesses. We only searched English literatures and hence a degree of bias is expected. Pooling accuracy estimates taken only from the most recent studies may also introduce a significant bias. Apart from that, small number of studies included also made further sub-group analysis impractical and thus heterogeneity was not able to be fully explored and rectified. There were only five studies on genetic testing which studied different genes. Therefore, pool analysis for genetic testing cannot be carried out.
Lastly, we did not address other factors related to the implementation of the tools, such as the specific type of genetic testing modality, the patient clinical history, and the colorectal cancer histologic manifestations. We presume that the results were useful to suggest the level of diagnostic accuracy of the tools assessed, but is limited in robustness in view of the limitations mentioned.
Conclusion
The sensitivity and specificity of FOBTs are 31% (95% CI; 25, 38) and 87% (95% CI; 86, 89) respectively. The accuracy of genetic testing is inconclusive due to limited number of studies. For future reviews, it is recommended to omit time limitation and to include grey literatures and more evidence of FOBT and genetic testing accuracy. To propose, genetic testing should be considered as a screening tool (such as MHL1, MSH2, MSH6, Epcam, Braf v600e) for all moderate- and high-risk asymptomatic people, after genetic variant prevalence and cost effectiveness study provide further favorable evidence. It is suggested to use FOBT as population-based screening instead of opportunistic screening.
Competing Interests
There was no conflict of interest to declare. This study received no specific funding or grant from any agency, neither profit nor not-profit sector.
References
- 1.World Health Organization (WHO). Cancer; fact sheet no 297 2014. Available from: http://www.who.int/medacentre/factsheets/fs297/en.
- 2.American Cancer Society. Colorectal Cancer Facts & Figures 2011–2013; 2013. [Google Scholar]
- 3.Sung JJ, Ng SC, Chan FK, et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2015; 64(1): 121–32. DOI: 10.1136/gutjnl-2013-306503 [DOI] [PubMed] [Google Scholar]
- 4.Cairns SR, Scholefield JH, Steele RJ, et al. Guidelines for colorectal cancer screening and surveillance in moderate- and high-risk groups (update from 2002). Gut. 2010; 59(5): 666–89. DOI: 10.1136/gut.2009.179804 [DOI] [PubMed] [Google Scholar]
- 5.Ministry of Health. Clinical Guidelines Practice: Management of Colorectal Cancer Malaysia Health Technology Assessment Section (MaHTAS) 2017. [Google Scholar]
- 6.Von Karsa L, Patnick J, Segnan N, et al. European guidelines for quality assurance in colorectal cancer screening and diagnosis: Overview and introduction to the full supplement publication. Endoscopy. 2013; 45(1): 51–9. DOI: 10.1055/s-0032-1325997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitlock EP, Lin JS, Liles E, Beil TL and Fu R. Screening for colorectal cancer: A targeted, updated systematic review for the U.S. Preventive Services Task Force. Annals of internal medicine. 2008; 149(9): 638–58. DOI: 10.7326/0003-4819-149-9-200811040-00245 [DOI] [PubMed] [Google Scholar]
- 8.Qaseem A, Denberg TD, Hopkins RH, et al. Screening for colorectal cancer: A guidance statement from the American College of Physicians. Annals of internal medicine. 2012; 156(5): 378–86. DOI: 10.7326/0003-4819-156-5-201203060-00010 [DOI] [PubMed] [Google Scholar]
- 9.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA and Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. The American journal of gastroenterology. 2009; 104(3): 739–50. DOI: 10.1038/ajg.2009.104 [DOI] [PubMed] [Google Scholar]
- 10.Lin JS, Piper MA, Perdue LA, et al. Screening for Colorectal Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. Jama. 2016; 315(23): 2576–94. DOI: 10.1001/jama.2016.3332 [DOI] [PubMed] [Google Scholar]
- 11.Zhu MM, Xu XT, Nie F, Tong JL, Xiao SD and Ran ZH. Comparison of immunochemical and guaiac-based fecal occult blood test in screening and surveillance for advanced colorectal neoplasms: A meta-analysis. Journal of digestive diseases. 2010; 11(3): 148–60. DOI: 10.1111/j.1751-2980.2010.00430.x [DOI] [PubMed] [Google Scholar]
- 12.Hewitson P, Glasziou P, Irwig L, Towler B and Watson E. Screening for colorectal cancer using the fecal occult blood test, Hemoccult. The Cochrane database of systematic reviews. 2007; 1: Cd001216 DOI: 10.1002/14651858.CD001216.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ministry of Health. Health Technology Assessment Section (MaHTAS) MDD, Ministry of Health Malaysia. Immunochemical Faecal Occult Blood Test (IFOBT) For Colorectal Cancer (CRC) Screening. MOH/P/PAK/233. 12(TR). 2011. [Google Scholar]
- 14.Fuzi SAM, Sabirin J and Hussain S. Health Technology Assessment Report- Immunochemical Fecal Occult Blood Test (IFOBT) For Colorectal Cancer (CRC) Screening; 2012. [Google Scholar]
- 15.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA: A cancer journal for clinicians. 2008; 58(3): 130–60. DOI: 10.3322/CA.2007.0018 [DOI] [PubMed] [Google Scholar]
- 16.Taylor DP, Burt RW, Williams MS, Haug PJ and Cannon-Albright LA. Population-based family history—specific risks for colorectal cancer: A constellation approach. Gastroenterology. 2010; 138(3): 877–85. DOI: 10.1053/j.gastro.2009.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on Colorectal Cancer. Diseases of the colon and rectum. 2014; 57(8): 1025–48. DOI: 10.1097/DCR.000000000000000 [DOI] [PubMed] [Google Scholar]
- 18.Vasen HF, Watson P, Mecklin JP and Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999; 116(6): 1453–6. DOI: 10.1016/S0016-5085(99)70510-X [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Bigas MA, Boland CR, Hamilton SR, et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: Meeting highlights and Bethesda guidelines. Journal of the National Cancer Institute. 1997; 89(23): 1758–62. DOI: 10.1093/jnci/89.23.1758 [DOI] [PubMed] [Google Scholar]
- 20.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. Journal of the National Cancer Institute. 2004; 96(4): 261–8. DOI: 10.1093/jnci/djh034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieuwenhuis MH, Vogt S, Jones N, et al. Evidence for accelerated colorectal adenoma-carcinoma progression in MUTYH-associated polyposis? Gut. 2012; 61(5): 734–8. DOI: 10.1136/gut.2010.229104 [DOI] [PubMed] [Google Scholar]
- 22.Kievit W, de Bruin JH, Adang EM, et al. Current clinical selection strategies for identification of hereditary non-polyposis colorectal cancer families are inadequate: A meta-analysis. Clin Genet. 2004; 65(4): 308–16. DOI: 10.1111/j.1399-0004.2004.00220.x [DOI] [PubMed] [Google Scholar]
- 23.Kastrinos F, Balmana J and Syngal S. Prediction models in Lynch syndrome. Fam Cancer. 2013; 12(2): 217–28. DOI: 10.1007/s10689-013-9632-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith GL, Edwards RT and Gray J. Cancer genetics services: A systematic review of the economic evidence and issues. British journal of cancer. 2004; 90(9): 1697–703. DOI: 10.1038/sj.bjc.6601792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Yao Z, Li C, et al. A step-by-step guide to the systematic review and meta-analysis of diagnostic and prognostic test accuracy evaluations. British journal of cancer. 2013; 108(11): 2299–303. DOI: 10.1038/bjc.2013.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiting P, Rutjes AWS, Reitsma JB, Bossuyt PMM and Kleijnen J. The development of QUADAS: A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Medical Research Methodology. 2003; 3(1): 25 DOI: 10.1186/1471-2288-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting PF, Weswood ME, Rutjes AWS, Reitsma JB, Bossuyt PNM and Kleijnen J. Evaluation of QUADAS: A tool for the quality assessment of diagnostic accuracy studies. BMC medical research methodology. 2006; 6: 9 DOI: 10.1186/1471-2288-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirai HW, Tsoi KK, Chan JY, et al. Systematic review with meta-analysis: Fecal occult blood tests show lower colorectal cancer detection rates in the proximal colon in colonoscopy-verified diagnostic studies. Alimentary pharmacology & therapeutics. 2016; 43(7): 755–64. DOI: 10.1111/apt.13556 [DOI] [PubMed] [Google Scholar]
- 29.Shapiro JA, Bobo JK, Church TR, et al. A Comparison of Fecal Immunochemical and High-Sensitivity Guaiac Tests for Colorectal Cancer Screening. The American journal of gastroenterology. 2017; 112(11): 1728–35. DOI: 10.1038/ajg.2017.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jucá MJ, Assuncão PRT and Hasten-Reiter Júnior HN. Fecal occult blood test and flexible rectosigmoidoscopy: Tools for the screening of colorectal neoplasms in asymptomatic patients. Journal of Coloproctology. 2015; 35(1): 35–41. DOI: 10.1016/j.jcol.2015.01.002 [DOI] [Google Scholar]
- 31.Elsafi SH, Alqahtani NI, Zakary NY and Al Zahrani EM. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clinical and Experimental Gastroenterology. 2015; 8: 279–84. DOI: 10.2147/CEG.S86419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohsiriwat V. Accuracy of self-checked fecal occult blood testing for colorectal cancer in Thai patients. Asian Pacific Journal of Cancer Prevention. 2014; 15(18): 7981–4. DOI: 10.7314/APJCP.2014.15.18.7981 [DOI] [PubMed] [Google Scholar]
- 33.Redwood D, Provost E, Asay E, et al. Comparison of fecal occult blood tests for colorectal cancer screening in an Alaska native population with high prevalence of Helicobacter pylori infection, 2008–2012. Preventing Chronic Disease. 2014; 11(4). DOI: 10.5888/pcd11.130281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeasmin F, Ali M, Rahman M, Sultana T, Rahman MQ and Ahmed A. A comparative study of chemical and immunological method of fecal occult blood test in the diagnosis of occult lower gastrointestinal bleeding. Bangladesh Medical Research Council Bulletin. 2014; 39(2): 52–6. DOI: 10.3329/bmrcb.v39i2.19641 [DOI] [PubMed] [Google Scholar]
- 35.Xie L, Jiang X, Li Q, et al. Diagnostic value of methylated Septin9 for colorectal cancer detection. Frontiers in Oncology. 2018. July; 8 DOI: 10.3389/fonc.2018.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson DH, Taylor WR, Aboelsoud MM, et al. DNA methylation and mutation of small colonic neoplasms in Ulcerative Colitis and Crohn’s Colitis: Implications for surveillance. Inflammatory Bowel Diseases. 2016; 22(7): 1559–67. DOI: 10.1097/MIB.0000000000000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanth P, Bronner MP, Boucher KM, et al. Gene signature in sessile serrated polyps identifies colon cancer subtype. Cancer Prevention Research. 2016; 9(6): 456–65. DOI: 10.1158/1940-6207.CAPR-15-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dvorak K, Aggeler B, Palting J, McKelvie P, Ruszkiewicz A and Waring P. Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: Impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma. Pathology. 2014; 46(6): 509–17. DOI: 10.1097/PAT.0000000000000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amiot A, Mansour H, Baumgaertner I, et al. The detection of the methylated Wif-1 gene is more accurate than a fecal occult blood test for colorectal cancer screening. PLoS ONE. 2014; 9(7). DOI: 10.1371/journal.pone.0099233 [DOI] [PMC free article] [PubMed] [Google Scholar]


