Abstract
The prevalence of rheumatic heart disease (RHD) in the Aboriginal population of the Australian Northern Territory is high, and Streptococcus pyogenes skin infections likely contribute to this. A promising candidate S. pyogenes “30mer” vaccine is composed of 30 pharyngitis associated type-specific antigens from the S. pyogenes M protein. Cross opsonisation experiments suggest that 30mer vaccine protection may extend to non-cognate emm types. A new “emm cluster” scheme for classifying M protein is based on the full-length coding sequence, and correlates with functional and immunological properties, and anatomical tropism. Twenty-seven years of research in the Northern Territory has yielded 1810 S. pyogenes isolates with clinical and emm type data. The primary aim was to analyse these data with reference to the emm cluster scheme and cross opsonisation information, to inform estimation of 30mer vaccine efficacy in the Northern Territory. The isolates encompass 101 emm types. Variants of cluster A-C were enriched in throat isolates, and variants of emm cluster D enriched in skin isolates. Throat isolates were enriched for 30mer vaccine cognate emm types in comparison with skin isolates of which only 25% were vaccine emm types. While cross opsonisation data indicates potential for enhancing 30mer vaccine coverage, more than one third of skin isolates were within 38 emm types untested for cross opsonisation. Emm cluster D variants, in particular emm cluster D4, were not only all non-cognate with the vaccine, but were abundant and diverse, and less likely to be cross-opsonisation positive than other emm clusters. Long term persistence of many emm types in the study area was revealed. It was concluded that the 30mer vaccine efficacy in the Northern Territory will likely require both cross protection, and additional measures to elicit immunity against variants of emm cluster D.
Author summary
The bacterium Streptococcus pyogenes causes throat and skin infections. A danger from such infections is an immune response that attacks human heart tissue, leading to rheumatic heart disease, which is difficult to treat and potentially deadly. Disadvantaged populations such as the Indigenous people in remote tropical northern Australia have high burdens of S. pyogenes skin infection, and rheumatic heart disease. An effective vaccine would be a benefit, but none is approved for clinical use. We analysed data from 1810 S. pyogenes isolates from north Australia obtained over 28 years, to determine the potential of a previously described S. pyogenes vaccine candidate to be effective in this region. Only one quarter of the isolates from skin infections had a surface antigen corresponding to any one of the 30 antigen variants in the candidate vaccine. Previous work in animals indicates potential cross-protection from the vaccine against strains with mismatched antigens. However, even if this occurs in humans, protection against skin infection strains would likely remain compromised, unless there were additional components in the vaccine. Further studies on cross-protection are critical to defining the potential of this type of vaccine in populations burdened with S. pyogenes skin infections and rheumatic heart disease.
Introduction
The Indigenous population of the Australian Northern Territory is disproportionately impacted by Streptococcus pyogenes. There is a high prevalence and incidence of skin infection (pyoderma, impetigo, skin sores) in children, for which S. pyogenes is the primary cause [1–3]. The prevalence of rheumatic heart disease (RHD) is one of the highest in the world [4–6], and there is evidence indicating a relationship to skin infections as well as pharyngitis [7, 8]. Also, the incidence of invasive S. pyogenes infections is substantially higher than in the Australian non-Aboriginal population. [9–11]. The Northern Territory Aboriginal population would benefit greatly from an effective GAS vaccine.
Analysis of S. pyogenes isolates from the Northern Territory in a series of studies has provided a consistent picture of high genetic diversity, with coexistence of multiple strains, even in small communities [12–15]. This is similar to observations in other disadvantaged populations [16–21]. There is currently no evidence that fundamentally novel lineages arise in the Northern Territory, so a current model is that social determinants leading to high skin infection prevalence facilitate on-going sustainable transmission of numerous lineages that ultimately originated outside the Northern Territory [22].
S. pyogenes emm types differ in their associations with anatomical sites. This has been described using the well-established concept of “emm patterns” [23, 24], with pattern A-C being throat associated, pattern D skin associated, and pattern E not associated with clear tropism. Previous work in the Northern Territory has shown such associations between emm pattern and site of isolation [14, 25].
More recently, the “emm cluster” scheme for classification of M protein variants was developed [21, 24, 26]. This is based on full length M protein sequences, and there is demonstrated correlation between emm cluster and host biomolecule binding specificity. The emm cluster nomenclature is an elaboration and modification of the emm pattern scheme. Phylogenetic analysis of the whole M protein sequence distinguished two main clades, X and Y. In Clade X, emm clusters E1-E6 are approximately equivalent to emm pattern E. In Clade Y, emm-clusters D1-D5 (D1-5) are approximately equivalent to emm pattern D, and emm clusters A-C1-A-C5 (A-C1-5) are approximately equivalent to emm pattern A-C. Also, some emm types within the X and Y clades are not related closely enough to other sequences to be assigned to clusters. Here, we term these collectively as “Clade Y/X. Other variants outside the X and Y clades are classified as “Outlier”.
S. pyogenes vaccine development strategies have largely been based on the M protein. A dominant hypothesis has been that immunity is emm type specific. However, it has been shown that immune sera from rabbits immunised with a 30-valent M-protein-based vaccine candidate (30mer vaccine) had significant killing activity against a large percentage of a test set of S. pyogenes strains with non-vaccine emm types [27, 28]. Furthermore, cross protection is more likely to occur within emm clusters [26]. Evidence for immunity across emm types, in particular within emm clusters, has been shown to arise from S. pyogenes impetigo infections in humans [29].
Vaccine candidates targeting conserved regions of the M protein rather than the highly variable N-terminal component have been developed. [30, 31]. These are primarily based upon peptides derived from the conserved C-repeat region. An extensively studied example is the C-region derived J8 B-cell epitope fused to non-streptococcal peptides and conjugated to diphtheria toxoid. This approach has reached human trials [32].
We aimed to use an archive of Northern Territory S. pyogenes isolates of known emm type, with corresponding data on site of isolation, epidemiology and clinical features, to understand the Northern Territory S. pyogenes population structure. Specifically, we sought to classify types according to the emm cluster classification scheme and anatomical site of isolation, to estimate the effectiveness of the candidate 30mer vaccine in this population. The long time-span over which isolates were collected (1987–2015) provided insight into strain persistence, total diversity, and diversity at time points. We identify knowledge gaps that could be prioritised in future studies directed towards the implementation of S. pyogenes vaccination in the Northern Territory.
Methods
This study utilises data from stored S. pyogenes isolates, derived from hospital and community-based settings in the Northern Territory. Emm typing data and clinical information were available for 1810 of the isolates in the collection (S1 Dataset). These comprised 1713 isolates analysed as part of a study of GAS emm diversity in the Northern Territory covering 1987–2008 (Towers et al, 2013), which included isolates from studies described in a number of publications [7, 8, 12–14, 25, 33–37]. In general, these isolates were from remote dwelling community members through testing for throat carriage, or investigating clinical syndromes consistent with S. pyogenes infection. Eighty-two isolates were emm typed as part of a study of invasive GAS infections at two Northern Territory hospitals from 2011–2013 [10] and a further 15 isolates were collected and emm typed as part of a response to a cluster of cases of acute rheumatic fever (ARF) in 2014–5 [38].
Data extracted for each isolate were emm type, date of isolation (with the exception of one isolate for which date was unavailable), anatomical site of isolation, and disease description. Isolates derived from the throat that had a disease description of “pharyngitis” were classified as “pharyngitis” isolates. Other throat derived isolates were classified as “throat carriage”. Isolates from the skin that had a collection site of “skin sore/abscess” were classified as “skin and soft tissue infections” (SSTI). These encompassed disease descriptions including pyodermas (skin sores), abscesses, cellulitis, necrotising fasciitis, and infected burns, with the majority being pyodermas. All other skin isolates had a collection site recorded as “normal skin”. Forty-one isolates were from other anatomical sites such as the urogenital tract or the ear. They were classified as “other”. Isolates in the “other” category were omitted from determination of correlation between emm cluster and anatomical site because of the low probability of useful correlations being observed. The complete data set is provided (S1 Dataset).
The emm type for each isolate was assigned to an emm cluster in accordance with the conversion provided by the USA Centres for Disease Control, at https://www.cdc.gov/streplab/downloads/distribution-emm-types.pdf. The emm type nomenclature was updated according to the conversion provided by the USA Centres for Disease Control at https://www.cdc.gov/streplab/downloads/emm-numeric-sort.pdf.
Categorisation of emm types according to predicted protective efficacy of the 30mer vaccine was on the basis of cross opsonisation experiments in animals [27, 28]. Emm types encompassed in the 30mer vaccine were classed as “vaccine”. Non-vaccine emm types that were more than 50% killed in both cross opsonisation studies [27, 28] were classed as “cross opsonisation-positive”. Non-vaccine emm types that were more than 50% killed in one of the two cross opsonisation studies were classed as “cross opsonisation-equivocal”. Non-vaccine emm types that were less than 50% killed in both of the crossopsonisation studies were classed as “cross opsonisation-negative”. Non-vaccine emm types that were untested in the cross opsonisation experiments were classed as “cross opsonisation unknown”. This convention is similar to that described by Williamson and co-workers [19].
For some analyses, all emm55 isolates were excluded. This is because 65 of the 81 emm55 isolates were isolated in 2005, coinciding with a large outbreak of acute post-streptococcal glomerulonephritis (APSGN) [39]. All remaining emm55 isolates were isolated in 1991 (14 isolates) or 1992 (one isolate). This indicates that emm55 causes outbreaks, which has the potential to bias analyses.
Confidence intervals for proportions were calculated using the Wilson variant of the binomial proportions method, without continuity correction, at this site: http://vassarstats.net/prop1.html. The significance of differences in proportions were assessed using the Chi squared N-1 test, accessed at https://www.medcalc.org/calc/comparison_of_proportions. p values stated in the text are modified by application of the Bonferroni correction for multiple testing.
The Simpsons Index of Diversity (D) was calculated as described by Hunter and Gaston [40], using a Microsoft Excel application developed by the authors.
This study was classed by the Human Research Ethics Committee of the Northern Territory Government Department of Health, and the Menzies School of Health Research as meeting the requirements of a negligible risk activity, and eligible for waiver of full ethical review. This is documented in a letter to P.M.G on August 30, 2018.
Results
The 1810 S. pyogenes isolates collected between 1 January 1987 and 26 February 2015 were classified into emm clusters (Table 1).
Table 1. Distribution of the 1810 isolates in this study into emm clusters.
| Emm cluster | Number of isolates | % total | Group % total |
|---|---|---|---|
| AC-1 | 1 | 0.06 | 4.81 |
| AC-2 | 14 | 0.77 | |
| AC-3 | 42 | 2.32 | |
| AC-4 | 25 | 1.38 | |
| AC-5 | 5 | 0.28 | |
| Clade_X_4 | 1 | 0.06 | 7.57 |
| Clade_Y_2 | 1 | 0.06 | |
| Clade_Y_3 | 54 | 3.0 | |
| Clade_Y_5 | 9 | 0.50 | |
| Clade_Y_6 | 13 | 0.72 | |
| Clade_Y_8 | 2 | 0.11 | |
| Clade_Y_14 | 18 | 0.99 | |
| Clade_Y_15 | 21 | 1.16 | |
| Clade_Y_16 | 9 | 0.50 | |
| Clade_Y_17 | 2 | 0.11 | |
| Clade_Y_20 | 4 | 0.22 | |
| Clade_Y_21 | 3 | 0.17 | |
| D1 | 45 | 2.49 | 33.11 |
| D2 | 88 | 4.86 | |
| D3 | 26 | 1.44 | |
| D4 | 416 | 22.99 | |
| D5 | 24 | 1.33 | |
| E1 | 83 | 4.59 | 47.73 |
| E2 | 106 | 5.86 | |
| E3 | 262 | 14.48 | |
| E4 | 169 | 9.33 | |
| E5 | 1 | 0.06 | |
| E6 | 243 | 13.42 | |
| Outlier 1 | 81 | 4.48 | 5.75 |
| Outlier 2 | 4 | 0.22 | |
| Outlier 5 | 14 | 0.77 | |
| Outlier 6 | 5 | 0.28 | |
| Unknown | 19 | 1.10 | 1.05 |
The distribution of the emm types identified in the study into emm clusters is shown in Table 2. One hundred and one emm types were identified, encompassing all defined emm clusters.
Table 2. Distribution of the emm types identified in this study into 30mer vaccine protection categories.
| Emm-cluster | Vaccine | Cross opsonisation-positive |
Cross opsonisation-equivocal | Cross opsonisation-negative | Cross opsonisation-unknown |
|---|---|---|---|---|---|
| A-C1 | 142 | ||||
| A-C2 | 197 | ||||
| A-C3 | 1 | 1–4 | |||
| A-C4 | 12 | 39, 193, 229 | |||
| A-C5 | 3 | ||||
| Clade_X | 236 | ||||
| Clade_Y | 6, 14, 18, 19, 24 | 74, 105, 122 | 57, 218, 233 | ||
| D1 | 54 | 207 | |||
| D2 | 71, 100 | ||||
| D3 | 123 | 217 | |||
| D4 | 33, 52 | 53 | 70, 80, 116 | 41, 56, 86, 91, 93, 98, 101, 108, 178, 192, 225, 230 | |
| D5 | 97 | ||||
| E1 | 4, 78 | 60 | 165 | ||
| E2 | 92 | 68, 76 | 13, 90, 106, 110, 117, 166 | ||
| E3 | 44, 49, 58, 82, 87 | 15, 25, 183 | 9, 103, 231, 144 | ||
| E4 | 2, 22, 28, 73, 77, 89, 114 | 8, 102, 109 | 124 | 88, 112, 232 | |
| E5 | 170 | ||||
| E6 | 11, 75, 81 | 48, 65, 69, 85 | 63 | 67, 99 | |
| Outlier | 95 | 55 | 241, 222 | ||
| Unknown | stG653, stGrobn | ||||
| Total emm types | 26 | 20 | 5 | 6 | 45 |
Variants of emm clusters D1-5 and E1-6 are numerically dominant, together accounting for 80.8% of the isolates. The majority (69.4%) of emm cluster D1-5 isolates are emm cluster D4, with these isolates comprising nearly one quarter of the entire collection and encompassing 18 emm types.
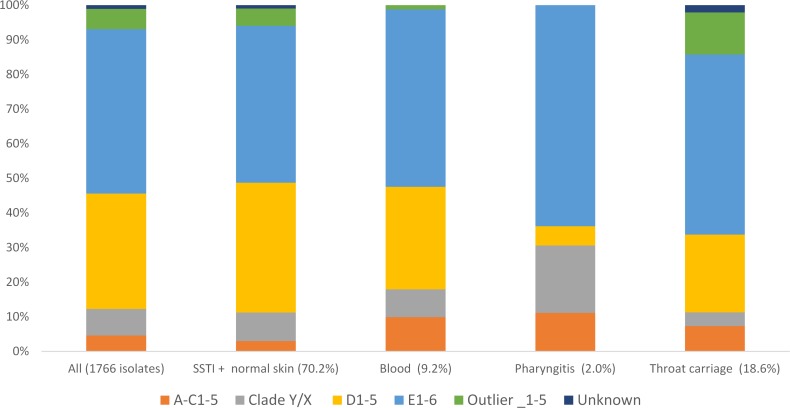
To reveal anatomic tropism, the relationship between emm cluster and anatomical site of isolation was determined (Fig 1, S2 Dataset). For this analysis, 41 isolates from anatomical locations in the “other” category for site of isolation were excluded (see Methods). Strongest statistical support (p<0.007)) was for enrichment (i.e. an elevated proportion) of emm cluster D1-5 isolates in skin isolates with respect to throat carriage isolates, enrichment of emm cluster A-C1-5 isolates in blood with respect to SSTI, and enrichment of cluster Outlier_1–5 (including emm55 isolates) in throat carriage with respect to blood. Other statistically significant but less strongly supported associations included enrichment of emm cluster D1-5 isolates in SSTI with respect to pharyngitis (p = 0.007), and emm cluster A-C1-5 in throat carriage with respect to SSTI (p = 0.028).
Fig 1. Distribution of isolates from different anatomical sites into emm clusters.
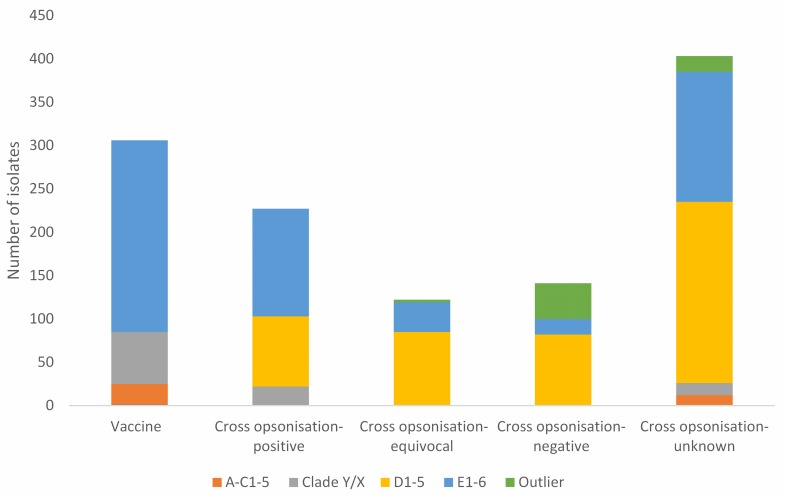
The distribution amongst vaccine classes was determined (Fig 2, S3 Dataset). Strongest statistical support (p<0.008) was for enrichment of vaccine emm type isolates in pharyngitis and throat carriage with respect to SSTI, enrichment of vaccine emm type isolates in pharyngitis with respect to normal skin, and enrichment of cross opsonisation-unknown isolates in SSTI with respect to throat carriage and also in blood with respect to throat carriage. If emm55 isolates are included, there was also strong statistical support for enrichment of cross opsonisation-negative emm type isolates in throat carriage with respect to SSTI, and in throat carriage with respect to “other”. Other less strongly supported differences can be seen in Part B of S3 Dataset. The numerically abundant emm cluster D4 isolates were specifically examined, because of their likely significance to vaccine implementation in the study area. Notably, there are 264 isolates of emm types that are emm cluster D4 and cross opsonisation-unknown. This is 14.6% of the entire collection and 44.5% of the non-vaccine, cross opsonisation-unknown emm type isolates.
Fig 2. Distribution of isolates from different anatomical sites amongst vaccine protection classes.
We then determined the diversity of the isolates on the basis of emm type, as high diversity, particularly of non-vaccine emm type strains, may complicate vaccine development. Diversity was calculated for each combination of vaccine protection class and anatomical site of isolation (Table 3).
Table 3. Diversities of isolates in different protection classes and anatomical sites.
| Vaccine | Cross opsonisation-positive |
Cross opsonisation-equivocal | Cross opsonisation-negative |
Cross opsonisation-negative, emm55 excluded |
Cross opsonisation-unknown |
||
|---|---|---|---|---|---|---|---|
| Total emm type number | 26 | 19 | 6 | 6 | 5 | 50 | |
| SSTI | Numbers of emm types | 21 | 19 | 6 | 6 | 5 | 38 |
| D | 0.93 | 0.93 | 0.78 | 0.82 | 0.80 | 0.94 | |
| Blood | Numbers of emm types | 19 | 12 | 4 | 4 | 3 | 25 |
| D | 0.90 | 0.95 | 0.65 | 0.90 | 0.83 | 0.93 | |
| Pharyngitis | Numbers of emm types | 13 | 4 | 2 | 0 | 0 | 6 |
| D | 0.95 | 1.0 | 1.0 | - | - | 1.0 | |
| Throat carriage | Numbers of emm types | 18 | 13 | 5 | 4 | 3 | 20 |
| D | 0.90 | 0.89 | 0.73 | 0.22 | 0.80 | 0.92 | |
| Normal skin | Numbers of emm types | 3 |
3 | 2 | 3 | 3 | 7 |
| D | 1.0 | 1.0 | 0.67 | 0.6 | 0.6 | 0.81 | |
| Other | Numbers of emm types | 15 | 4 | 3 | 1 | 1 | 11 |
| D | 0.97 | 0.90 | 1 | - | - | 0.99 |
It was notable that the isolates characterised as “cross opsonisation-unknown” were particularly diverse. Diversity was also determined for isolates obtained in calendar years that yielded >100 isolates (1991, 95, 97, 99, 2004, 05). The D values were 0.94 (1999, 2004, and 2005), 0.95 (1991 and 1995) and 0.96 (1997), showing that high diversity is not a consequence of the long period over which isolates were collected.
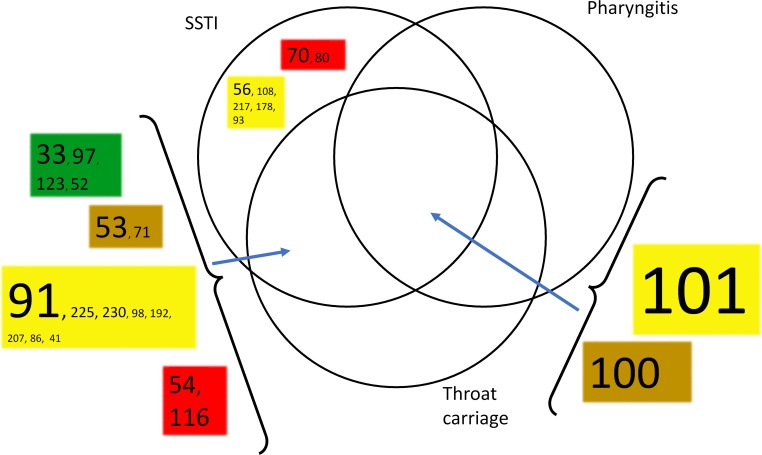
To further investigate the SSTI isolates, their distribution into vaccine coverage classes and emm clusters was determined (Fig 3, S4 Dataset). Fig 3 shows large differences between the vaccine protection classes with respect to emm cluster make-up, and the S4 Dataset shows that a large proportion of these differences are strongly supported by statistical analysis (p <0.006).
Fig 3. Distribution of SSTI isolates according to emm cluster and vaccine class.
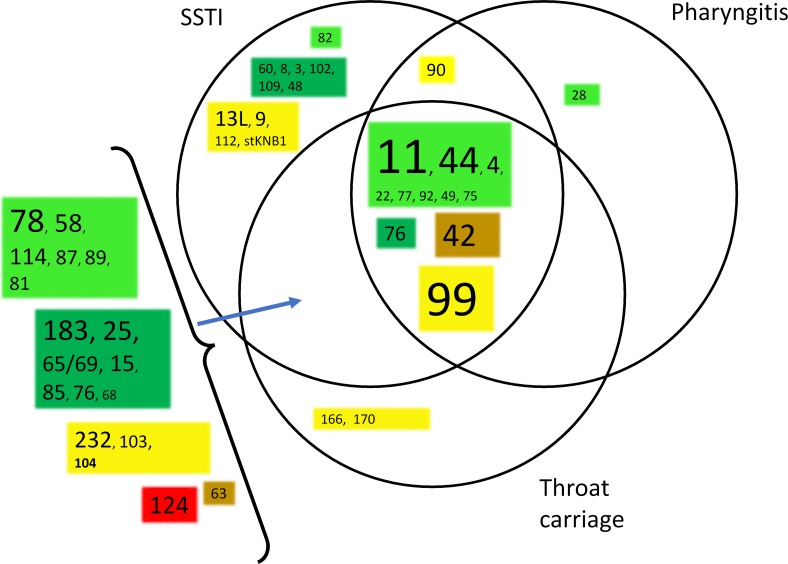
Emm type persistence was determined on the basis of the number of calendar year(s) in which each emm type was isolated. This is shown graphically in (Fig 4, S5 Dataset), with the emm types resolved on the basis of 30mer vaccine protection class. This analysis showed extensive co-existence of emm types within single calendar years, in combination with numerous examples of long term emm type persistence. There was no evidence for correlation of persistence with vaccine class. The study data set encompasses many examples of the same emm types being isolated from different anatomical sites. This is relevant to understanding strain tropism and mobility between anatomical sites. In combination with strain abundance data, this information can assist in prioritising emm types and emm clusters for further study. The relationship between number of isolates of each emm type, site of isolation, and emm cluster was determined. The results for the D1-5 and E1-6 emm clusters are shown in Fig 5 and Fig 6 and for the other emm clusters in S1 Fig. For the abundant emm clusters D1-5 and E1-6, there is extensive commonality in emm types isolated from different anatomical sites. In particular, this is seen with SSTI and throat carriage isolates. A similar analysis was performed for blood isolates (S2 Fig), and this showed that it was common for emm types found in blood to be isolated from different sites. For example, 40 of the 66 emm types (60%) were also isolated from SSTIs and throat carriage. Complete numeric emm type distributions according to anatomical sites of isolation, emm cluster, isolate numbers and year(s) of isolation are provided (S6 Dataset).
Fig 4. Persistence of emm types.
Each row with colours blocks is an emm type. The top row is calendar year. A block is coloured if there was at least one of that emm type isolated in that year. Pale green: vaccine types; dark green: cross opsonisation-positive, brown: cross opsonisation-equivocal; red: cross opsonisation-negative; yellow: cross opsonisation-unknown. The Excel file from which this is derived, containing emm type numerical information, is provided as S5 Dataset.
Fig 5. Relationship between emm types within emm cluster D1-5, number of isolates, vaccine class, and anatomical sites in which emm types were identified.
The font size is proportional to the number of isolates from SSTI infections, where the number is >9. Pale green: vaccine types; dark green: cross-opsonisation- positive, brown: cross opsonisation-equivocal; red: cross opsonisation-negative; yellow: cross opsonisation-unknown. B. emm cluster E1-6.
Fig 6. Relationship between emm types within emm cluster E1-6, number of isolates, vaccine class, and anatomical sites in which emm types were identified.
The font size is proportional to the number of isolates from SSTI infections, where the number is >9. Pale green: vaccine types; dark green: cross-opsonisation- positive, brown: cross opsonisation-equivocal; red: cross opsonisation-negative; yellow: cross opsonisation-unknown.
Discussion
Here we have described emm type diversity of S. pyogenes collected over nearly three decades in the tropical north of Australia. We found that less than one third of the isolates overall, and 25.2% of the SSTI isolates, are cognate with the 30mer vaccine. In the absence of cross protection between emm types, 30mer vaccine efficacy would be likely to be poor in the Northern Territory.
The bacterial population was described in terms of the emm cluster classifications scheme, its relationship with anatomical tropism, and previously reported cross opsonisation data for a subset of known emm types [27, 28]. We show that isolates of emm types for which there are no cross-opsonisation data are abundant and diverse. This limits inference of vaccine efficacy and suggests directions for further work. The relationship between emm cluster and anatomical tropism was broadly consistent with previous reports. However, there were numerous instances of nominally skin tropic strains colonising the throat without causing signs or symptoms of pharyngitis.
The most abundant emm cluster categories in the study were E1-6 and D1-5. Emm cluster D1-5/emm pattern D strains are regarded as “skin tropic” [24, 26, 41] and our findings were consistent with this, with the proportions of cluster D1-5 in both pharyngitis and throat carriage isolates significantly less than for SSTI isolates (pharyngitis p = 0.007, throat carriage, p<0.007). Emm cluster/emm pattern E1-6 strains are regarded as non-tropic and consistent with this, no significant differences in relative prevalence at different anatomical sites were observed. Cluster A-C1-5 strains are regarded as throat tropic [24, 26]. The point estimates for the relative prevalence were higher for throat carriage and pharyngitis isolates than for non-throat isolates, but the difference was only significant for throat carriage isolates (p = 0.028), likely due to the larger number of throat carriage isolates in the study. The point estimate for the relative prevalence of Cluster A-C1-5 in blood isolates was similar to that for throat isolates and significantly higher than the relative proportion in SSTI (p<0.007). However, 12 of the 13 Cluster A-C1-5 isolates from blood were emm197 isolates isolated in connection with a cluster of invasive infections in 2012 [10], compromising the generalisability of this finding. The emm Cluster Clade Y/X isolates encompass a low frequency of throat carriage isolates as compared to pharyngitis isolates, and normal skin isolates. This is not currently understood. Given that Clade Y/X emm cluster category is composed of very diverse emm types, it is likely that the category encompasses a range of anatomical tropism conferring properties, making these results difficult to interpret. For the Outlier category, the only significant differences observed were for comparisons including emm55 isolates. Specifically, all the throat carriage isolates in the Outlier category were emm55 isolated in 2005, and likely associated with an outbreak of APSGN [39]. Therefore, the observation of elevated Outlier category emm types in throat carriage isolates may not be generalisable to the Northern Territory when there is not an emm55 outbreak in progress. In summary, tropism observed is either consistent with current models, explainable with sampling effects, or anomalous but involving a diverse emm cluster category.
The proportions of emm cluster categories derived from different anatomical sites were similar to findings from a meta-analyses of anatomical tropism on the basis of emm pattern [41]. However, previous analyses have not generally differentiated between pharyngitis and throat carriage isolates. Our study identifies a weaker tropism signal, on the basis of point estimates, for throat carriage strains, with many examples of the same emm types being recovered from SSTIs and throat carriage. This suggests that adaptation to throat carriage differs from adaptation to cause pharyngitis. Pharyngitis is rare in tropical Northern Territory, despite a high burden of skin disease and RHD [7, 42]. Thus, in this setting it appears that nominally skin tropic strains have markedly impaired capacity to cause symptomatic pharyngitis but do not experience strong barriers to colonising the throat. Open questions are the contributions of throat colonisation and SSTIs to ARF/RHD, and the extent to which throat colonisation contributes to sustainable transmission of SSTIs.
The coverage by the 30mer vaccine in the absence of cross protection is 66.7%, 40.1% and 25.2% for the pharyngitis, throat carriage and SSTI isolates respectively, with the reduced coverage of SSTI isolates in comparison to pharyngitis and throat carriage isolates strongly supported by statistical analysis (S3 Dataset) (p<0.008). The high coverage of the pharyngitis isolates is unsurprising given that the 30mer vaccine was designed with reference to strains circulating in North America, where pharyngitis is the dominant S. pyogenes infection [27]. The intermediate figure for throat carriage is consistent with our finding of weaker tropism for throat carriage, and extensive commonality of emm types from SSTI and throat carriage.
Isolates of emm types that have not been tested for cross reaction are numerous and diverse, with the proportion in SSTI isolates significantly higher than in throat carriage isolates (S3 Dataset). Extrapolation from cross opsonisation data could perhaps be used to attempt to predict 30mer vaccine efficacy. For example, from the top data row of S3 Dataset, 490 SSTI isolates are of non-vaccine emm types that have been tested for cross opsonisation. Of these, 349 (71.2%) are of cross opsonising emm types. A point estimate regarding vaccine efficacy can be made by assuming that the same proportion of isolates of “non-vaccine, cross opsonisation-unknown” emm types would show cross opsonisation. 71.2% of 415 isolates is 295 isolates. This generates an efficacy estimate regarding SSTI isolates of: (305 vaccine emm type isolates + 227 cross opsonising emm type isolates + 122 equivocal cross opsonising emm type isolates, + 295 (inferred) non-vaccine cross opsonisation-unknown isolates)/1210 total SSTI isolates = 78.4%. This could be regarded as promising. However, this estimate is potentially optimistic, because it encompasses an assumption that there will indeed be cross protection in humans against all the non-vaccine cross opsonising emm types. This remains to be demonstrated. Accordingly, while the above calculation may be useful in contributing to a conceptual framework for future research, we do not believe this defines a current robust evidence-based estimate for 30mer vaccine effectiveness in the Northern Territory.
The breakdown of the SSTI isolates on the basis of emm cluster and vaccine protection class, and the diversity determinations add detail to this picture (Fig 3, S4 Dataset). What is striking is the strong relationship between 30mer vaccine protection class and emm cluster composition. Significant differences in frequencies may be expected when comparing the “vaccine” class with other classes, because the vaccine contains no emm Cluster D1-5 antigens. However, we also observed strongly supported significant differences (p<0.008) regarding emm types that gave different cross opsonisation results i.e cross opsonisation-positive, -intermediate and -negative. It is noteworthy that the relative contributions of isolates that are emm Cluster E1-6 becomes less as the cross opsonisation results become more negative. The inverse is seen with emm cluster D1-5. This is consistent with correlation of emm cluster with cross protection [26], and indicates that the cross opsonisation data is biologically and potentially clinically meaningful. It was also noted that within the cross opsonisation-negative category there were four emm cluster D1-5 emm types (emm54, emm116, emm70 and emm80) and only a single emm cluster E1-6 emm type (emm124). This emphasises that emm cluster E1-6 isolates contribute very little to the cross opsonisation-negative isolates in terms of number or diversity. The substantial proportion of Outlier isolates in this category were emm55, many of which were associated with an APSGN outbreak [39]. Removing these from consideration emphasises the dominance of emm cluster D1-5 in the cross opsonisation-negative category.
The overall picture is that the emm cluster E1-6 isolates are overwhelmingly of emm types that are either cognate with the 30mer vaccine, tested positive in cross opsonisation assays, or are of emm types untested for cross opsonisation. In contrast, the emm cluster D1-5 isolates (which are primarily emm cluster D4), are all non-cognate with the 30mer vaccine, and are less likely to be of emm types that tested positive in cross opsonisation assays. Also, just over half the isolates of cross opsonisation-unknown emm types are emm cluster D1-5. The other emm cluster categories are of less concern. In particular there are no emm cluster A-C1-5 or Clade Y/X isolates in the cross opsonisation equivocal or negative categories, and very few that are untested. Outlier isolates are either cross opsonisation-negative (APSGN associated emm55) or a small number of isolates of untested emm types. In summary, these results lead to the prediction that emm clusters D1-5, in particular emm cluster D4, are particularly problematic with respect to 30mer vaccine efficacy against SSTIs in the Northern Territory. It is relevant that Frost and co-workers found evidence that eliciting an immune response against emm cluster D4 strains with a non-cognate emm type is inherently difficult [29].
The “blood” isolates in this study represent isolates from invasive infections, and so are of inherent interest. These are diverse, with the 162 blood isolates encompassing 66 emm types, 57 of which have been found in this study at other anatomical sites. The distribution between emm clusters and vaccine protection categories are broadly similar to SSTI isolates. Emm81 is the most abundant emm type in the blood isolates (emm cluster E6, 14 isolates, 8.6% of total). Emm81 is a “vaccine” emm type and was isolated in nine separate years, indicating persistence rather than particularly high prevalence at any time point. Emm81 has been associated with invasive infections in several studies outside Australia [43–45]. Of the nine emm types found only in blood, seven are represented by only 1–2 isolates. Of greater interest are the remaining two emm types, emm197 (emm cluster A-C2) (12 isolates) and emm113 (E3) (10 isolates). Other studies have not identified these as dominant invasive strains, but emm197 has been identified in New Zealand associated with ARF [46] and emm113 has been classified as “invasive”[47]. Both are non-vaccine emm types with no cross opsonisation information, and in the Northern Territory were isolated in the same geographical region in 2012 (emm197) and 2011, 2012, 2013 (emm113), suggesting outbreaks [10]. These emm types are candidates for future study and surveillance.
Isolates from sites other than the skin, throat or blood are highly diverse (S1 Dataset) with the 41 isolates encompassing 34 emm types, and no emm types are represented by more than two isolates. This is unsurprising giving the diversity of sites and the long period over which isolates were collected.
It is likely that the 30mer vaccine will not provide good protection against APSGN in the NT. Of three APSGN outbreaks in the Northern Territory, emm55 S. pyogenes is associated with the largest, in 2005 [39]. Emm55 is non-cognate with the vaccine and cross opsonisation-negative. A recent review indicates that poor 30mer vaccine coverage against APSGN associated strains is predicted in Australia, and also in South America, where emm55 appears to be an important cause of APSGN outbreaks [48]. Inclusion of an emm55 antigen in any S. pyogenes vaccine targeted to remote regions with disadvantaged populations may be justifiable.
The population structure of S. pyogenes in the Northern Territory has now been clearly established as encompassing high diversity and long term emm type persistence, resulting in high diversity at any time point and a high proportion of cluster D1-5 and E1-6 strains and isolates. This is similar to other studies based on populations with high burdens of skin infections and ARF/RHD [16–18, 20]. In particular the picture is remarkably similar to what was observed in New Zealand by Williamson and co-workers [19], where high emm type diversity, correlation of ARF burden with pyoderma burden and cluster D1-5 prevalence, and poor 30mer vaccine coverage of skin-derived isolates, were all observed.
This analysis defines specific emm types that could be prioritised for further study, in addition to the blood-associated invasive emm types mentioned above. For example, Emm cluster D1-5 types, for which there are no cross opsonisation data, and are relatively abundant and persistent in this study setting were emm101 (emm cluster D4), emm91 (D4), and emm56 (D4). Emm101 and emm91 were also of high relative prevalence in pyoderma isolates from New Zealand [19]. S6 Dataset is a potentially useful resource for prioritising further work.
The major limitation of this study is that the isolates were not collected systematically but were a convenience sample. However, the high diversity at multiple time points, in combination with evidence for extensive strain persistence, suggests that the isolates do provide a useful picture of S. pyogenes diversity in the Northern Territory over a period of decades. Another limitation is the small number of pharyngitis isolates, although this is in large part a consequence of the low prevalence of pharyngitis in the study area.
We conclude that the effectiveness of the 30mer vaccine in the study area would be dependent on emm type cross protection in humans, which cannot currently be estimated with any certainty. If there is cross protection in accordance with cross opsonisation data, then the 30mer vaccine can be reasonably predicted to provide good protection against all emm clusters except D1-5, and possibly Outlier, which together comprise 53.5% of the sample. Therefore, a potential strategy would be to combine the 30mer vaccine with a vaccine(s) specifically targeting emm cluster D1-5, and possibly emm55. It is of interest that a 26-valent precursor of the 30mer vaccine contains the emm33 and emm101 antigens [49]. These are both emm cluster D4. Both emm types are relatively abundant in our collection, with emm101 being the most abundant emm type (74 isolates), and emm33,represented by 32 isolates. Therefore, testing the 26mer vaccine, or a combination of the 30mer and 26mer, in the Northern Territory may be worth consideration. However, in the absence of cross protection, obtaining useful efficacy in the Northern Territory using an emm type specific vaccine may be difficult to achieve, in particular because coverage of the numerous emm Cluster E1-6 strains will be less, and obtaining coverage of the numerous and diverse emm cluster D1-5 may be impractical. Accordingly, determination of the correspondence between cross opsonisation data and clinical efficacy is critical for defining future directions.
Supporting information
The checklist used is that designed for observational studies.
(DOCX)
(XLSX)
Part A: Distribution of isolates on the basis of anatomical site of origin. These values were used to construct Fig 1. In Fig 1, the numbers from SSTI and normal skin were combined. 95% CI intervals were calculated, without correction for multiple testing (see Methods). No 95% CI intervals were calculated for the proportions in the bottom row, as these are in large part a reflection of sampling activity, rather than any biological properties of the isolates. Part B: Results of N-1 Chi-suared tests. This experiment addressed differences between isolates from different anatomical sites regarding their distribution into emm clusters. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of SSTI isolates that are emm cluster A-C1-5 (3% of 1210 isolates), compared with the percentage of blood isolates that are emm cluster A-C1-5 9.9% of 162 isolates). Because of potential for bias, the analysis was also performed with emm55 isolates omitted (starred data points) (see text). We took a conservative approach to assessing the credibility of the differences in proportions. We used the 70 cells in the table to apply a Bonferroni correction. The usual cut-off for significance is P = 0.05. 0.05/70 = 0.00071. Cells coloured red have values <0.0001, which equates to a Bonferroni corrected p value of <0.007, which we regard as strongly supporting the significance of the difference in proportion. Cells coloured orange have p values from 0.0001–0.0007, which equates to Bonferroni corrected p values of 0.007–0.05. We regard this as significant, but less strongly supporting the difference in proportion. The orientations of significant differences are shown using single letters reflecting the first letters of the anatomical sites. A “-”symbol designates where the comparison is of two 0% values.
(DOCX)
Part A. Distribution of the 1810 isolates included in this study into 30mer vaccine protection classes. These data were used to assemble Fig 2. Starred figures are with emm55 isolates omitted. 95% CI’s (see Methods) are provided for the distributions of isolates from different anatomical sites across 30mer protection classes. 95% CI’s are not provided for the distribution of isolates of different 30mer protection classes across sites of isolation, as this is impacted greatly by specimen collection activity, compromising the value of statistical analysis. Part B. This experiment addressed differences between isolates from different anatomical sites regarding their distribution into 30mer vaccine protection classes. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of SSTI isolates that are 30mer vaccine emm types (25.2% of 1210), vs the percentage of blood isolates that are 30mer vaccine emm types (34.6% of 162 isolates). Starred numbers were calculated with emm55 isolates omitted. As with S2 Data Set, a Bonferroni correction was applied. In this instance, 76 tests for significance were performed. Cells coloured red have values <0.0001, which equates to a Bonferroni corrected P -value of <0.0076, which we regard as strongly supporting significance. Cells coloured orange have P values from 0.0001–0.0006, which equates to Bonferroni corrected P values of 0.008–0.05. We regard this as significant, but less strongly supporting the difference in proportion. Starred values are with emm55 isolates omitted. The orientations of significant differences are indicated in the data cells, with the single letters representing the first letters of site of isolation.
(DOCX)
Part A. Relationship between 30mer vaccine protection class and emm cluster for SSTI isolates. Starred numbers are with emm55 isolates omitted. 95% CI’s are included for the percent values calculated with reference to the “vertical totals”; the number of isolates in each 30mer vaccine protection class. These are the values used for the analysis in Part B below, and also which were visualised in Fig 3. The percent values with reference to anatomical site (horizontal totals) will be in large part a function of specimen collection activity, so have not been subjected to statistical analysis. Part B. Results of N-1 Chi-squared tests. This experiment addressed differences between SSTI isolates of different emm cluster categories their distribution into 30mer vaccine protection classes. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of “vaccine emm type” isolates that are also “emm cluster A-C1-5” isolates (7.9% of 305 isolates) vs percentage of “cross-opsonisation positive” isolates that are also “emm cluster A-C1-5” isolates (0% of 227 isolates). Starred numbers were calculated with emm55 isolates omitted. As with S2 Data Set and S3 Data Set, a Bonferroni correction was applied. In this instance, 59 tests for significance were performed. Cells coloured red have p values <0.0001, which equates to a Bonferroni corrected P -value of <0.006, which we regard as strongly supporting significance. Cells coloured orange have P values from 0.0001–0.0008, which equates to Bonferroni corrected p values of ~0.006–0.05. We regard this as significant, but less strongly supporting the difference in proportion. Starred values are with emm55 isolates omitted. The orientations of significant differences are indicated in the data cells, with the single letters either designating “vaccine” (v), or the first letter of the cross-opsonisation descriptor. A “-”symbol represents where both percentages to be compared were zero. Isolates of emm types of unknown emm cluster were omitted because the numbers were too small to be meaningful.
(DOCX)
Emm types and emm clusters are shown in the left two columns.
(XLSX)
“Other” anatomical sites are excluded, although are taken into account in the “Years Isolated” column.
(DOCX)
This encompasses emm clusters not included in Fig 5 and Fig 6. The emm clusters are indicated on each part of the figure. The font size is proportional to the number of isolates from SSTI infections, when the number of isolates is >9. Pale green: vaccine types; dark green: cross opsonisation-positive, brown: cross opsonisation-equivocal; red: cross opsonisation-negative; yellow: cross opsonisation-unknown.
(PPTX)
This indicates other sites that the emm types were isolated from.
(PPTX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
SYCT is a NHMRC Career Development Fellow (#1145033). The study received support from NHMRC project grant #1098319 (SCYT, PMG). NHMRC web site: https://nhmrc.gov.au/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Andrews RM, McCarthy J, Carapetis JR, Currie BJ. Skin disorders, including pyoderma, scabies, and tinea infections. Pediatric clinics of North America. 2009;56(6):1421–40. Epub 2009/12/08. 10.1016/j.pcl.2009.09.002 . [DOI] [PubMed] [Google Scholar]
- 2.Bowen AC, Tong SY, Andrews RM, O'Meara IM, McDonald MI, Chatfield MD, et al. Short-course oral co-trimoxazole versus intramuscular benzathine benzylpenicillin for impetigo in a highly endemic region: an open-label, randomised, controlled, non-inferiority trial. Lancet. 2014;384(9960):2132–40. Epub 2014/08/31. 10.1016/S0140-6736(14)60841-2 . [DOI] [PubMed] [Google Scholar]
- 3.Bowen AC, Tong SY, Chatfield MD, Carapetis JR. The microbiology of impetigo in indigenous children: associations between Streptococcus pyogenes, Staphylococcus aureus, scabies, and nasal carriage. BMC Infect Dis. 2014;14:727 Epub 2015/01/01. 10.1186/s12879-014-0727-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carapetis JR, McDonald M, Wilson NJ. Acute rheumatic fever. Lancet. 2005;366(9480):155–68. Epub 2005/07/12. 10.1016/S0140-6736(05)66874-2 . [DOI] [PubMed] [Google Scholar]
- 5.Parnaby MG, Carapetis JR. Rheumatic fever in indigenous Australian children. J Paediatr Child Health. 2010;46(9):527–33. Epub 2010/09/22. 10.1111/j.1440-1754.2010.01841.x . [DOI] [PubMed] [Google Scholar]
- 6.Carapetis JR, Currie BJ. Clinical epidemiology of rheumatic fever and rheumatic heart disease in tropical Australia. Advances in experimental medicine and biology. 1997;418:233–6. Epub 1997/01/01. 10.1007/978-1-4899-1825-3_56 . [DOI] [PubMed] [Google Scholar]
- 7.McDonald M, Brown A, Edwards T, Hope A, Amu M, Morey F, et al. Apparent contrasting rates of pharyngitis and pyoderma in regions where rheumatic heart disease is highly prevalent. Heart, lung & circulation. 2007;16(4):254–9. Epub 2007/06/22. 10.1016/j.hlc.2007.02.087 . [DOI] [PubMed] [Google Scholar]
- 8.McDonald MI, Towers RJ, Andrews RM, Benger N, Currie BJ, Carapetis JR. Low rates of streptococcal pharyngitis and high rates of pyoderma in Australian aboriginal communities where acute rheumatic fever is hyperendemic. Clin Infect Dis. 2006;43(6):683–9. Epub 2006/08/17. 10.1086/506938 . [DOI] [PubMed] [Google Scholar]
- 9.Whitehead BD, Smith HV, Nourse C. Invasive group A streptococcal disease in children in Queensland. Epidemiology and infection. 2011;139(4):623–8. Epub 2010/07/09. 10.1017/S0950268810001378 . [DOI] [PubMed] [Google Scholar]
- 10.Boyd R, Patel M, Currie BJ, Holt DC, Harris T, Krause V. High burden of invasive group A streptococcal disease in the Northern Territory of Australia. Epidemiology and infection. 2016;144(5):1018–27. 10.1017/S0950268815002010 . [DOI] [PubMed] [Google Scholar]
- 11.Gear RJ, Carter JC, Carapetis JR, Baird R, Davis JS. Changes in the clinical and epidemiological features of group A streptococcal bacteraemia in Australia's Northern Territory. Trop Med Int Health. 2015;20(1):40–7. Epub 2014/10/31. 10.1111/tmi.12405 . [DOI] [PubMed] [Google Scholar]
- 12.Richardson LJ, Towers RJ, Cheng AC, Currie BJ, Carapetis JR, Giffard PM, et al. Diversity of emm sequence types in group A beta-haemolytic streptococci in two remote Northern Territory Indigenous communities: Implications for vaccine development. Vaccine. 2010. Epub 2010/08/06. S0264-410X(10)00744-9 [pii] 10.1016/j.vaccine.2010.05.046 . [DOI] [PubMed] [Google Scholar]
- 13.McDonald MI, Towers RJ, Andrews R, Benger N, Fagan P, Currie BJ, et al. The dynamic nature of group A streptococcal epidemiology in tropical communities with high rates of rheumatic heart disease. Epidemiology and infection. 2008;136(4):529–39. Epub 2007/06/02. 10.1017/S0950268807008655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald MI, Towers RJ, Fagan P, Carapetis JR, Currie BJ. Molecular typing of Streptococcus pyogenes from remote Aboriginal communities where rheumatic fever is common and pyoderma is the predominant streptococcal infection. Epidemiology and infection. 2007;135(8):1398–405. Epub 2007/02/20. 10.1017/S0950268807008023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowen AC, Harris T, Holt DC, Giffard PM, Carapetis JR, Campbell PT, et al. Whole genome sequencing reveals extensive community-level transmission of group A Streptococcus in remote communities. Epidemiology and infection. 2016;144(9):1991–8. 10.1017/S095026881500326X . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakota V, Fry AM, Lietman TM, Facklam RR, Li Z, Beall B. Genetically diverse group A streptococci from children in far-western Nepal share high genetic relatedness with isolates from other countries. Journal of clinical microbiology. 2006;44(6):2160–6. Epub 2006/06/08. 44/6/2160 [pii] 10.1128/JCM.02456-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Athey TB, Teatero S, Sieswerda LE, Gubbay JB, Marchand-Austin A, Li A, et al. High Incidence of Invasive Group A Streptococcus Disease Caused by Strains of Uncommon emm Types in Thunder Bay, Ontario, Canada. Journal of clinical microbiology. 2016;54(1):83–92. Epub 2015/10/23. 10.1128/JCM.02201-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdem G, Mizumoto C, Esaki D, Abe L, Reddy V, Effler PV. Streptococcal emm types in Hawaii: a region with high incidence of acute rheumatic fever. Pediatr Infect Dis J. 2009;28(1):13–6. Epub 2008/12/06. 10.1097/INF.0b013e31818128ce [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson DA, Smeesters PR, Steer AC, Morgan J, Davies M, Carter P, et al. Comparative M-protein analysis of Streptococcus pyogenes from pharyngitis and skin infections in New Zealand: Implications for vaccine development. BMC Infect Dis. 2016;16(1):561 10.1186/s12879-016-1891-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9(10):611–6. Epub 2009/09/26. S1473-3099(09)70178-1 [pii] 10.1016/S1473-3099(09)70178-1 . [DOI] [PubMed] [Google Scholar]
- 21.Baroux N, D'Ortenzio E, Amedeo N, Baker C, Ali Alsuwayyid B, Dupont-Rouzeyrol M, et al. The emm-cluster typing system for Group A Streptococcus identifies epidemiologic similarities across the Pacific region. Clin Infect Dis. 2014;59(7):e84–92. Epub 2014/06/27. 10.1093/cid/ciu490 . [DOI] [PubMed] [Google Scholar]
- 22.Towers RJ, Carapetis JR, Currie BJ, Davies MR, Walker MJ, Dougan G, et al. Extensive diversity of Streptococcus pyogenes in a remote human population reflects global-scale transmission rather than localised diversification. PLoS One. 2013;8(9):e73851 Epub 2013/09/26. 10.1371/journal.pone.0073851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingshead SK, Readdy TL, Yung DL, Bessen DE. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol Microbiol. 1993;8(4):707–17. Epub 1993/05/01. . [DOI] [PubMed] [Google Scholar]
- 24.Bessen DE, McShan WM, Nguyen SV, Shetty A, Agrawal S, Tettelin H. Molecular epidemiology and genomics of group A Streptococcus. Infect Genet Evol. 2015;33:393–418. Epub 2014/12/03. 10.1016/j.meegid.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessen DE, Carapetis JR, Beall B, Katz R, Hibble M, Currie BJ, et al. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J Infect Dis. 2000;182(4):1109–16. Epub 2000/09/09. JID000584 [pii] 10.1086/315842. 10.1086/315842 . [DOI] [PubMed] [Google Scholar]
- 26.Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210(8):1325–38. Epub 2014/05/07. 10.1093/infdis/jiu260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29(46):8175–8. Epub 2011/09/17. 10.1016/j.vaccine.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dale JB, Penfound TA, Tamboura B, Sow SO, Nataro JP, Tapia M, et al. Potential coverage of a multivalent M protein-based group A streptococcal vaccine. Vaccine. 2013;31(12):1576–81. Epub 2013/02/05. 10.1016/j.vaccine.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frost HR, Laho D, Sanderson-Smith ML, Licciardi P, Donath S, Curtis N, et al. Immune Cross-Opsonization Within emm Clusters Following Group A Streptococcus Skin Infection: Broadening the Scope of Type-Specific Immunity. Clin Infect Dis. 2017;65(9):1523–31. Epub 2017/10/12. 10.1093/cid/cix599 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale JB, Batzloff MR, Cleary PP, Courtney HS, Good MF, Grandi G, et al. Current Approaches to Group A Streptococcal Vaccine Development. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK)2016. [PubMed] [Google Scholar]
- 31.Good MF, Pandey M, Batzloff MR, Tyrrell GJ. Strategic development of the conserved region of the M protein and other candidates as vaccines to prevent infection with group A streptococci. Expert Rev Vaccines. 2015;14(11):1459–70. Epub 2015/10/21. 10.1586/14760584.2015.1081817 . [DOI] [PubMed] [Google Scholar]
- 32.Sekuloski S, Batzloff MR, Griffin P, Parsonage W, Elliott S, Hartas J, et al. Evaluation of safety and immunogenicity of a group A streptococcus vaccine candidate (MJ8VAX) in a randomized clinical trial. PLoS One. 2018;13(7):e0198658 Epub 2018/07/03. 10.1371/journal.pone.0198658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carapetis J, Gardiner D, Currie B, Mathews JD. Multiple strains of Streptococcus pyogenes in skin sores of aboriginal Australians. Journal of clinical microbiology. 1995;33(6):1471–2. Epub 1995/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGregor KF, Bilek N, Bennett A, Kalia A, Beall B, Carapetis JR, et al. Group A streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources. J Infect Dis. 2004;189(4):717–23. Epub 2004/02/10. 10.1086/381452 . [DOI] [PubMed] [Google Scholar]
- 35.McDonald M, Towers R, Fagan P, McKinnon M, Benger N, Andrews R, et al. Recovering streptococci from the throat, a practical alternative to direct plating in remote tropical communities. Journal of clinical microbiology. 2006;44(2):547–52. Epub 2006/02/04. 10.1128/JCM.44.2.547-552.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald MI, Benger N, Brown A, Currie BJ, Carapetis JR. Practical challenges of conducting research into rheumatic fever in remote Aboriginal communities. The Medical journal of Australia. 2006;184(10):511–3. Epub 2006/05/25. . [DOI] [PubMed] [Google Scholar]
- 37.Richardson LJ, Tong SY, Towers RJ, Huygens F, McGregor K, Fagan PK, et al. Preliminary validation of a novel high-resolution melt-based typing method based on the multilocus sequence typing scheme of Streptococcus pyogenes. Clin Microbiol Infect. 2011;17(9):1426–34. Epub 2010/11/26. 10.1111/j.1469-0691.2010.03433.x . [DOI] [PubMed] [Google Scholar]
- 38.Francis JR, Gargan C, Remenyi B, Ralph AP, Draper A, Holt D, et al. A cluster of acute rheumatic fever cases among Aboriginal Australians in a remote community with high baseline incidence. Australian and New Zealand journal of public health. 2019. Epub 2019/04/18. 10.1111/1753-6405.12893 . [DOI] [PubMed] [Google Scholar]
- 39.Marshall CS, Cheng AC, Markey PG, Towers RJ, Richardson LJ, Fagan PK, et al. Acute post-streptococcal glomerulonephritis in the Northern Territory of Australia: a review of 16 years data and comparison with the literature. The American journal of tropical medicine and hygiene. 2011;85(4):703–10. Epub 2011/10/07. 10.4269/ajtmh.2011.11-0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of clinical microbiology. 1988;26(11):2465–6. Epub 1988/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bessen DE, Kumar N, Hall GS, Riley DR, Luo F, Lizano S, et al. Whole-genome association study on tissue tropism phenotypes in group A Streptococcus. J Bacteriol. 2011;193(23):6651–63. Epub 2011/09/29. 10.1128/JB.05263-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald M, Currie BJ, Carapetis JR. Acute rheumatic fever: a chink in the chain that links the heart to the throat? Lancet Infect Dis. 2004;4(4):240–5. Epub 2004/03/31. 10.1016/S1473-3099(04)00975-2 . [DOI] [PubMed] [Google Scholar]
- 43.Rantala S, Vahakuopus S, Siljander T, Vuopio J, Huhtala H, Vuento R, et al. Streptococcus pyogenes bacteraemia, emm types and superantigen profiles. Eur J Clin Microbiol Infect Dis. 2012;31(5):859–65. 10.1007/s10096-011-1385-9 . [DOI] [PubMed] [Google Scholar]
- 44.Chiang-Ni C, Wu AB, Liu CC, Chen KT, Lin YS, Chuang WJ, et al. Emergence of uncommon emm types of Streptococcus pyogenes among adult patients in southern Taiwan. Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi. 2011;44(6):424–9. 10.1016/j.jmii.2011.04.005 . [DOI] [PubMed] [Google Scholar]
- 45.Paveenkittiporn W, Nozawa T, Dejsirilert S, Nakagawa I, Hamada S. Prevalent emm types and superantigen gene patterns of group A Streptococcus in Thailand. Epidemiology and infection. 2016;144(4):864–9. 10.1017/S0950268815001880 . [DOI] [PubMed] [Google Scholar]
- 46.Williamson DA, Smeesters PR, Steer AC, Steemson JD, Ng AC, Proft T, et al. M-Protein Analysis of Streptococcus pyogenes Isolates Associated with Acute Rheumatic Fever in New Zealand. Journal of clinical microbiology. 2015;53(11):3618–20. 10.1128/JCM.02129-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plainvert C, Dinis M, Ravins M, Hanski E, Touak G, Dmytruk N, et al. Molecular epidemiology of sil locus in clinical Streptococcus pyogenes strains. Journal of clinical microbiology. 2014;52(6):2003–10. 10.1128/JCM.00290-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worthing KA, Lacey JA, Price DJ, McIntyre L, Steer AC, Tong SYC, et al. Systematic Review of Group A Streptococcal emm Types Associated with Acute Post-Streptococcal Glomerulonephritis. The American journal of tropical medicine and hygiene. 2019;100(5):1066–70. Epub 2019/03/28. 10.4269/ajtmh.18-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNeil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, et al. Safety and immunogenicity of 26-valent group a streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41(8):1114–22. Epub 2005/09/16. 10.1086/444458 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The checklist used is that designed for observational studies.
(DOCX)
(XLSX)
Part A: Distribution of isolates on the basis of anatomical site of origin. These values were used to construct Fig 1. In Fig 1, the numbers from SSTI and normal skin were combined. 95% CI intervals were calculated, without correction for multiple testing (see Methods). No 95% CI intervals were calculated for the proportions in the bottom row, as these are in large part a reflection of sampling activity, rather than any biological properties of the isolates. Part B: Results of N-1 Chi-suared tests. This experiment addressed differences between isolates from different anatomical sites regarding their distribution into emm clusters. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of SSTI isolates that are emm cluster A-C1-5 (3% of 1210 isolates), compared with the percentage of blood isolates that are emm cluster A-C1-5 9.9% of 162 isolates). Because of potential for bias, the analysis was also performed with emm55 isolates omitted (starred data points) (see text). We took a conservative approach to assessing the credibility of the differences in proportions. We used the 70 cells in the table to apply a Bonferroni correction. The usual cut-off for significance is P = 0.05. 0.05/70 = 0.00071. Cells coloured red have values <0.0001, which equates to a Bonferroni corrected p value of <0.007, which we regard as strongly supporting the significance of the difference in proportion. Cells coloured orange have p values from 0.0001–0.0007, which equates to Bonferroni corrected p values of 0.007–0.05. We regard this as significant, but less strongly supporting the difference in proportion. The orientations of significant differences are shown using single letters reflecting the first letters of the anatomical sites. A “-”symbol designates where the comparison is of two 0% values.
(DOCX)
Part A. Distribution of the 1810 isolates included in this study into 30mer vaccine protection classes. These data were used to assemble Fig 2. Starred figures are with emm55 isolates omitted. 95% CI’s (see Methods) are provided for the distributions of isolates from different anatomical sites across 30mer protection classes. 95% CI’s are not provided for the distribution of isolates of different 30mer protection classes across sites of isolation, as this is impacted greatly by specimen collection activity, compromising the value of statistical analysis. Part B. This experiment addressed differences between isolates from different anatomical sites regarding their distribution into 30mer vaccine protection classes. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of SSTI isolates that are 30mer vaccine emm types (25.2% of 1210), vs the percentage of blood isolates that are 30mer vaccine emm types (34.6% of 162 isolates). Starred numbers were calculated with emm55 isolates omitted. As with S2 Data Set, a Bonferroni correction was applied. In this instance, 76 tests for significance were performed. Cells coloured red have values <0.0001, which equates to a Bonferroni corrected P -value of <0.0076, which we regard as strongly supporting significance. Cells coloured orange have P values from 0.0001–0.0006, which equates to Bonferroni corrected P values of 0.008–0.05. We regard this as significant, but less strongly supporting the difference in proportion. Starred values are with emm55 isolates omitted. The orientations of significant differences are indicated in the data cells, with the single letters representing the first letters of site of isolation.
(DOCX)
Part A. Relationship between 30mer vaccine protection class and emm cluster for SSTI isolates. Starred numbers are with emm55 isolates omitted. 95% CI’s are included for the percent values calculated with reference to the “vertical totals”; the number of isolates in each 30mer vaccine protection class. These are the values used for the analysis in Part B below, and also which were visualised in Fig 3. The percent values with reference to anatomical site (horizontal totals) will be in large part a function of specimen collection activity, so have not been subjected to statistical analysis. Part B. Results of N-1 Chi-squared tests. This experiment addressed differences between SSTI isolates of different emm cluster categories their distribution into 30mer vaccine protection classes. For example, the top left data square is derived from an N-1 Chi-squared test on the percentage of “vaccine emm type” isolates that are also “emm cluster A-C1-5” isolates (7.9% of 305 isolates) vs percentage of “cross-opsonisation positive” isolates that are also “emm cluster A-C1-5” isolates (0% of 227 isolates). Starred numbers were calculated with emm55 isolates omitted. As with S2 Data Set and S3 Data Set, a Bonferroni correction was applied. In this instance, 59 tests for significance were performed. Cells coloured red have p values <0.0001, which equates to a Bonferroni corrected P -value of <0.006, which we regard as strongly supporting significance. Cells coloured orange have P values from 0.0001–0.0008, which equates to Bonferroni corrected p values of ~0.006–0.05. We regard this as significant, but less strongly supporting the difference in proportion. Starred values are with emm55 isolates omitted. The orientations of significant differences are indicated in the data cells, with the single letters either designating “vaccine” (v), or the first letter of the cross-opsonisation descriptor. A “-”symbol represents where both percentages to be compared were zero. Isolates of emm types of unknown emm cluster were omitted because the numbers were too small to be meaningful.
(DOCX)
Emm types and emm clusters are shown in the left two columns.
(XLSX)
“Other” anatomical sites are excluded, although are taken into account in the “Years Isolated” column.
(DOCX)
This encompasses emm clusters not included in Fig 5 and Fig 6. The emm clusters are indicated on each part of the figure. The font size is proportional to the number of isolates from SSTI infections, when the number of isolates is >9. Pale green: vaccine types; dark green: cross opsonisation-positive, brown: cross opsonisation-equivocal; red: cross opsonisation-negative; yellow: cross opsonisation-unknown.
(PPTX)
This indicates other sites that the emm types were isolated from.
(PPTX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.