Abstract
Many Ig superfamily members are expressed in the developing nervous system, but the functions of these molecules during neurogenesis are not all clear. Here, we explore the expression and function of one of members of this superfamily, protogenin (PRTG), in the developing nervous system. Expression of PRTG protein is strong in the neural tube of mouse embryos between embryonic days 7.75 and 9.5 but disappears after embryonic day 10.5 when the neural progenitor marker nestin expresses prominently. Perturbation of PRTG activity in P19 embryonal carcinoma cells and in chick embryos, by either RNA interference or a dominant-negative PRTG mutant, increases neuronal differentiation. Using yeast two-hybrid screening and an in situ binding assay, we were able to identify ERdj3 (a stress-inducible endoplasmic reticulum DnaJ homolog) as a putative PRTG ligand. Addition of purified ERdj3 protein into the P19 differentiation assay reduced neurogenesis. This effect was blocked by addition of either a neutralizing antibody against PRTG or purified PRTG ectodomain protein, indicating that the effect of ERdj3 on neurogenesis is mediated through PRTG. Forced expression of ERdj3 in the chick neural tube also impairs neuronal differentiation. Together, these results suggest that expression of PRTG defines a stage between pluripotent epiblasts and committed neural progenitors, and its signaling plays a critical role in suppressing premature neuronal differentiation during early neural development.
Introduction
Proteins of the Ig superfamily (IgSF) are characterized by the presence of conserved Ig domains in the extracellular portion and can function as cell adhesion molecules and/or as cell-surface receptors for particular ligands. Several members of IgSF are important molecules in the development of the nervous system. For example, the neural cell adhesion molecule NCAM and L1 control neuronal migration and survival, axon fasciculation, synaptic plasticity, and regeneration (Becker et al., 1998; Demyanenko et al., 1999; Polo-Parada et al., 2001; Rolf et al., 2002). Deleted in colorectal cancer (DCC) and neogenin are netrin receptors that mediate the growth of the commissural axons toward the floor plate in the embryonic spinal cord (Keino-Masu et al., 1996). The human Down's syndrome cell adhesion molecule gene (DSCAM) was originally identified as a gene associated with mental retardation (Yamakawa et al., 1998). Recent studies indicate that DSCAM is involved in neurite arborization, cell body spacing, and lamina-specific synaptic targeting in the vertebrate retina via homophilic interactions (Fuerst et al., 2008; Yamagata and Sanes, 2008). Intriguingly, DSCAM also acts as a receptor for netrin-dependent commissural axon outgrowth, turning responses and pathfinding in collaboration with DCC (Ly et al., 2008; Liu et al., 2009). Because there are 765 predicted IgSF members in the human genome and many are expressed in the developing nervous system (Lander et al., 2001), it is likely that other uncharacterized IgSF members may also influence various aspects of neurogenesis.
Protogenin (PRTG) is a recently identified IgSF member in chick and mouse embryos (Toyoda et al., 2005; Vesque et al., 2006) and belongs to the DCC/neogenin subclass of IgSF. Unlike DCC and neogenin, the expression of which starts around midgestation (Gad et al., 1997), the expression of PRTG mRNA initiates immediately after gastrulation in mouse embryos and undergoes a sharp downregulation after embryonic day 10 (E10) (Vesque et al., 2006). Recently, Wigg et al. (2008) identified a possible association between PRTG and human attention deficit hyperactivity disorder. These interesting observations prompt us to examine the functions of PRTG during neural development.
In the current study, we report that PRTG protein is present in proliferating neural precursors between mouse E7.75 and E9.5 but downregulates when the cells initiate differentiation at E10. By using ex vivo and in vivo neuronal differentiation models, we demonstrate that the major role of PRTG in the nervous system is to prevent precocious differentiation. The fact that PRTG activity is perturbed by the PRTG ectodomain and a neutralizing antibody implies that PRTG is a receptor. We further show that ERdj3 (a stress-inducible endoplasmic reticulum DnaJ homolog) may act as a secreted protein that binds to PRTG, and ERdj3 itself has a similar inhibitory effect on neuronal differentiation. These findings establish a novel role for ERdj3/PRTG signaling in neurogenesis.
Materials and Methods
Animals.
Sprague Dawley rats and C57BL/6J and BALB/cJ mice were obtained from the National Laboratory Animal Center (Taipei, Taiwan). Handling of rats and mice was according to university guidelines and was approved by the National Yang-Ming University Animal Care and Use Committee. For timed pregnancies, mice were put together in the late afternoon, and the morning on which plugs were observed was designated as E0.5. The white leg-horn eggs were purchased from a local poultry farm (Taichung, Taiwan) and incubated at 100°F until desired stages. All chicken experiments were performed according to the Institutional Animal Care and Use Committee protocol 97-97 of National Chung-Hsing University.
Materials.
Superscript reverse transcriptase II (RTase) was purchased from Invitrogen. Restriction enzymes, dATP, dCTP, dGTP, and dTTP were obtained from Roche. RNase inhibitor (RNasin) and TaqDNA polymerase were purchased from Promega. Primers were synthesized by MDBiol. All other chemicals, unless specified, were purchased from Sigma.
Vector construction.
The full-length or mutant PRTG fragments were amplified by PCR from the cDNA of mouse E9.5, rat E10.5, or chick Hamburger–Hamilton (HH) stage 10 whole embryos using high-fidelity DNA polymerase (Roche) with various appropriate primer pairs (supplemental Table S1, available at www.jneurosci.org as supplemental material) and were cut with appropriate restriction enzymes before being ligated into either pEF1/Myc–His (Invitrogen) to give pm(r)PRTGf (for mouse or rat full-length PRTG expression), pm(r)PRTGΔc (for PRTG without cytoplasmic domain), and pm(r)PRTGc (for cytoplasmic domain of PRTG) or into pCAG–EYFP to give pgPRTGf and pgPRTGΔc. The complete open reading frame of ERdj3 was amplified by PCR from E9 mouse cDNA or chick HH stage 10 whole embryos and inserted in pEF1/Myc–His to give pERdj3 or into pCAG–EYFP to give pgERdj3 and pgERdj3–KDEL. To construct the yeast bait, the rat extracellular fragment of PRTG (amino acids 40-934) was ligated into pBTM116 to give pBTM116–rPRTGe. Deletion constructs were derived from pBTM116–rPRTGe by restriction enzyme digestion and in-frame ligation. The Ig Fc region was first cloned into pEF1/Myc–His to give psFc. The extracellular domain of PRTG and ERdj3 were then cloned into psFc after PCR as pPRTGe–Fc and pERdj3–Fc. Protocols described by Chung et al. (2006) were followed to construct the microRNA mimics. DNA fragments containing PRTG or ERdj3 sequences and scramble sequences (supplemental Table S1, available at www.jneurosci.org as supplemental material) were cloned into vectors UI4–puro–SIBR or UI4–GFP–SIBR to give shPRTG(m), shERdj3(m), shPRTG(g), shERdj3(g), and shS [scrambled short-hairpin RNA (shRNA)]. All constructs were verified by restriction enzyme mapping and sequencing.
Antibodies.
Three PRTG fragments, the extracellular domain (E peptide, rat PRTG amino acids 320–506), the cytoplasmic tail (C peptide, rat PRTG amino acids 970–1193), or the chick cytoplasmic tail (gC peptide, chick PRTG amino acids 951–1187) were fused in-frame to the His tag of pET29. The His-tagged fusion proteins were prepared in an Escherichia coli expression system and purified using a Ni2+-chelating column. Monoclonal antibody (mAb) against the E and C peptides, mouse antiserum against the gC peptide, and rabbit antiserum (W4) against the C peptide were generated by following standard protocols. Goat anti-Sox-1 (L-20, 1:100) and rabbit anti-Oct4 (H-134, 1:100) were obtained from Santa Cruz Biotechnology. Goat anti-mouse endoglin (1:100) was purchased from R & D Systems. Mouse anti-stage-specific embryonic antigen 1 (SSEA1) (1:500), anti-β-tubulin (E7, 1:10000), and anti-myc (9E10, 1:5000) antibodies were obtained from the Developmental Studies Hybridoma Bank. Mouse anti-MAP2 (1:500) was purchased from Sigma. Mouse anti-Ascl1 (1:1000) was obtained from BD Pharmingen. Mouse anti-HuC/D (16A11, 1:100) was purchased from Invitrogen. Rabbit anti-α-smooth muscle actin (ab5694, 1:500) was obtained from Abcam. Rabbit anti-cleaved caspase-3 (cCasp3) (Asp175, 1:500) was purchased from Cell Signaling Technology. Mouse anti-neuronal-specific nuclear protein (NeuN) (1:200), mouse anti-nestin (rat-401, 1:1000), rabbit anti-Sox2 (AB5603, 1:500), and FITC-labeled goat anti-mouse IgG1 antiserum were bought from Millipore Bioscience Research Reagents. Mouse anti-βIII tubulin (TuJ1, 1:4000) was obtained from Promega. Cyanine 3-labeled goat anti-mouse IgG1 antiserum was purchased from Jackson ImmunoResearch. FITC-labeled goat anti-rabbit antiserum was obtained from Zymed Laboratories. Other secondary antisera were purchased from Bethyl Laboratories.
Immunofluorescence microscopy.
Embryos were fixed for 2 h in 4% paraformaldehyde in PBS at 4°C and then processed for cryostat sectioning. Transfected cells grown on coverslips were rinsed once with PBS and then fixed in 4% paraformaldehyde for 10 min. Samples were stained with the appropriate primary and secondary antibodies and finally stained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) for 10 min. Samples were examined using a fluorescence microscope (Eclipse TE300; Nikon) equipped with a MicroMax cool CCD (Princeton Instruments) or a laser confocal microscope (Olympus FV1000). The digital images were processed using Adobe Photoshop software (Adobe Systems). Regions of chick sections with a high electroporated efficiency as revealed by EYFP (enhanced yellow fluorescence protein) or EGFP (enhanced green fluorescence protein) expression were used for the quantifications. The division between medial and lateral neural tube is indicated in Figure 3W. In each plane, the distance between ventricle and pial surface is measured. The medial part is defined as the region within three-quarters of that distance close to the ventricle. The lateral part is defined as the region within one-quarter of that distance close to the pial surface. For TuJ1 quantification, the mean of the fluorescence intensity on the electroporated region was divided by that in the corresponding region of the contralateral side (d/a and c/b). Numbers of NeuN-positive (NeuN+) cells counted manually in the electroporated regions were divided by those in the corresponding regions of the contralateral side and then normalized to the control vector. Numbers of HuC/D+ and cCasp3+ cells were also counted manually.
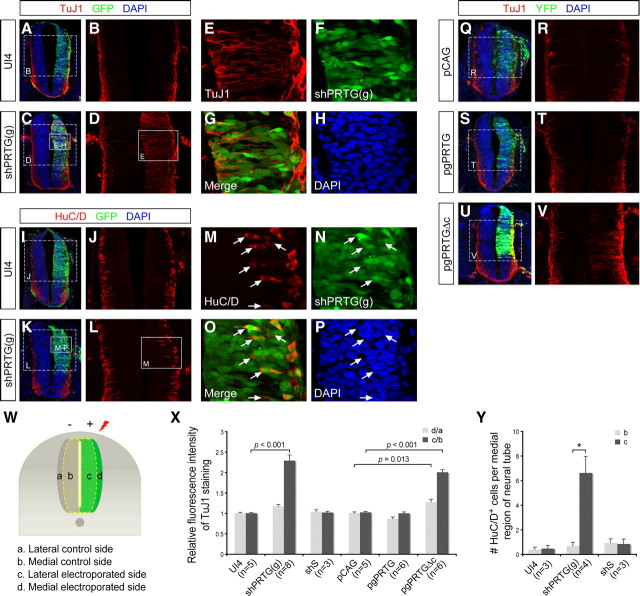
Figure 3.
PRTG represses precocious neuronal differentiation in the developing chick neural tube. A–V, Control plasmids (UI4 or pCAG) or various chick PRTG constructs were electroporated into the right side (green) of chick neural tubes at HH stage 9–10. Two days after electroporation, the chick embryos were fixed and stained with DAPI (blue) and TuJ1 antibody (A–H, Q–V, red) or anti-HuC/D antibody (I–P, red). E–H are higher magnifications of the white boxes in C and D; M–P are higher magnifications of the white boxes in K and L. W, Regions of quantification are indicated. In each plane, the distance between ventricle and pial surface is measured. The medial part is defined as within three-quarters of that distance close to the ventricle. The lateral part is defined as within one-quarter of that distance close to the pial surface. X, Quantification of the relative fluorescence intensity of TuJ1 in the chick neural tube electroporated with the indicated plasmids. Quantification was performed as described in Materials and Methods. Data are shown as the mean ± SEM. The number of chick embryo analyzed in this assay is indicated in brackets. p value is shown by one-way ANOVA and post hoc least significant difference test. Y, The number of HuC/D+ cells in the medial region is quantified and shown as the mean ± SEM (**p < 0.01; ***p < 0.001, by Student's t test).
Cell culture and transfection.
HEK293T cells were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 incubator. P19 cells were cultured in α-Minimal Essential Medium supplemented with 2.5% FBS and 7.5% newborn calf serum. To introduce the various constructs into HEK293T and P19 cells, calcium phosphate transfection mixtures (25 mm HEPES, 70 mm NaCl, 0.75 mg/ml KCl, 0.75 mm Na2HPO4, pH 7.05, 125 μm calcium chloride, and 20 μg of expression vector) were added to cells grown at 70% confluence in 10 cm culture dishes. Alternatively, cells were transfected using FuGENE 6 (Roche) or Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. The medium was changed 18 h after transfection. For the neuronal differentiation experiments (in one well of a six-well culture dish), P19 cells were transfected with 0.4 μg of pAscl1 and 1.6 μg of the indicated plasmids and then subjected to selection with 500 μg/ml G418 or 5 μg/ml puromycin at 8 h after transfection. The selection was maintained for 4 d. At 24 h after transfection, the culture medium was changed to SF21 serum-free medium (Segal et al., 1992). The secreted Fc, PRTGe–Fc, and ERdj3–Fc fusion proteins were produced in HEK293T cells and purified on a protein A/G-Sepharose column (Oncogene).
Protein extraction, immunoprecipitation, and Western blotting.
Embryonic tissue was homogenized in Laemli's lysis buffer [25 mm Tris-HCl, pH7.4, 50 mm NaCl, 0.5% sodium deoxycholate, 2% NP-40, 0.2% SDS, 50 mm NaF, and protease inhibitor cocktail (Roche)] for Western blot analysis. Transfected cells were washed once with PBS and dissolved in radioimmunoprecipitation assay lysis buffer (20 mm HEPES, pH 7.8, 150 mm NaCl, 1 mm EDTA, 0.1% Triton X-100, 50 mm NaF, 1 mm dithiothreitol, and protease inhibitor cocktail). The cell lysates were cleared by centrifugation at 13,000 × g for 10 min, and the supernatants were used for Western blotting analysis or immunoprecipitation (IP). The protein concentration was determined using a BCA kit (Pierce).
Immunoprecipitation was performed with 300 μg of protein extract and 2 μl of primary antibodies in a reaction volume of 500 μl on a rotator at 4°C overnight. The immune complex was absorbed onto 15 μl of protein A/G-Sepharose beads at 4°C for 30 min. Samples were spun down by low-speed centrifugation and washed five times with lysis buffer. For Western blotting analysis, equal amounts of either tissue or cell lysates were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membrane was blocked with 5% skimmed milk and probed with the primary antibody at 4°C overnight. After the membrane was washed and incubated with HRP-conjugated secondary antibodies at room temperature for 1 h, it was developed with an ECL kit (GE Healthcare).
In ovo electroporation.
DNA constructs were introduced into the neural tube of HH stage 9–10 chicken embryos using three 25 V/50 ms pluses. After electroporation, the embryos were incubated at 38°C. Transfection efficiency was tested by in vivo observation of GFP or YFP expression under a microscope. After 2 d (HH stage 17–20), the embryos were fixed and processed for immunostaining. At least five sections of each embryo and three embryos for each condition were analyzed.
Yeast two-hybrid analysis.
The Saccharomyces cerevisiae yeast strain L40 was grown at 30°C in YPA medium or in synthetic medium with the appropriate supplements. The pBTM116–rPRTGe bait and pACT2 preys were transformed sequentially into yeast strain L40 by the lithium acetate method. Double transformants were grown on the synthetic complete medium lacking KLUT (lysine, leucine, uracil, and tryptophan) and selected on KLUTH (lysine, leucine, uracil, tryptophan, and histidine)-depleted medium. After incubation at 30°C for 3 d, colonies on the KLUT plates were restreaked on the same media and then tested for β-galactosidase activity by the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) agarose overlay assay method.
In situ binding assay.
HEK293T cells were transfected with pPRTGf using FuGENE 6. Sixty hours after transfection, fivefold concentrated conditioned medium (CM) collected from pERdj3 transfected or nontransfected cells was added to the HEK293T cells and incubated at room temperature for 1 h. After washing once with PBS, the cells were fixed and immunofluorescence stained. Double staining of transfected PRTG and bound ERdj3 was performed using anti-PRTGc (W4) and anti-c-myc (9E10) antibodies and the appropriate secondary antisera. The fluorescence signal was quantified using NIH ImageJ software.
Results
Identification and expression of rat PRTG gene
We used suppression subtractive hybridization (Diatchenko et al., 1996) to identify genes preferentially expressed in the embryonic neural tube and obtained partial DNA fragments of PRTG as a candidate gene that was present in the embryonic neural tube but not in the postnatal day 1 (P1) rat brain (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Temporal expression of PRTG in rat brain was revealed by Northern blot analysis (Fig. 1A). Two rat PRTG transcripts, with lengths of 8.7 and 4.2 kb, are detected. Expression of PRTG is high during rat neural tube formation at E10.5 but drastically decreases in the rat brain after E12.5. Transcripts of PRTG are not detected in other non-neural tissues at E14.5, nor in any P1 or adult rat tissues (data not shown).
Figure 1.

Expression of PRTG protein in the mouse embryo. A, Expression of rat prtg in the nervous system at different stages of development as analyzed by the Northern blot. Ad, Adult; Br, brain; Nt, neural tube. B, Specificity of PRTG antibodies. Cell lysates of HEK293T cells transfected with the control vector, prPRTGf (rat full-length PRTG), prPRTGΔc (rat PRTG without cytoplasmic domain), or prPRTGc (rat cytoplasmic tail), and rat E10.5 neural tube lysate (Embryo) were subjected to Western blot using α-PRTG1 and α-PRTG2 mAbs. Addition of peptide E (E; ectodomain) or peptide C (C; cytoplasmic tail) competes out the bands recognized by the monoclonal antibodies. C, Schematic structure of PRTG. Peptide E or C for generating antibodies is marked in blue. D–H, Transverse sections through the mid-body level of mouse at E7 (D), E7.75 (E), E8.25 (F), E9.5 (G), and E10.5 (H) were stained using α-PRTG2 mAb (red). The dorsal is up in all panels. D, At E7, PRTG labeling is weakly detected in the mesoderm (me), whereas Oct4 is restricted to the ectoderm (ec; green). E, At E7.75, PRTG is detected in neuroepithelium, mesoderm, endoderm, and posterior epiblasts (arrows). The yolk sac (ys) surrounding the embryo is nonspecifically stained by the secondary antibodies. F, G, PRTG protein is present in almost all cells between E8.25 and E9.5 (red) but not in the notochord (n), differentiating cardiac cells, and some mesenchymal cells. The endocardium is labeled with anti-endoglin antibody (green). ac, Atrial chamber; dm, dermomyotome; vc, ventricular chamber; sc, spinal cord. H, PRTG is not detectable at E10.5. I, Expression of PRTG (red) and SSEA1 (green) in a transverse section of E7 and E8.25 mouse embryos. J–N, Expression of PRTG (red) and nestin (J), Ascl1 (K), TuJ1 (L), MAP2 (M), and NeuN (N) (green) in adjacent transverse sections through the thoracic level of E9.5 (top) and E10.5 (bottom) mouse embryos.
Characterization of antibodies against PRTG
We generated mAbs α-PRTG1 and α-PRTG2, which recognize the ectodomain and the cytoplasmic tail of PRTG, respectively (Fig. 1B,C). By Western blotting, two bands with apparent molecular weights of 186 and 174 kDa bind these antibodies in the lysates of HEK293T cells transfected with prPRTGf (encoding rat full-length PRTG protein). The apparent molecular weights of two detected PRTG forms are higher than expected, and this is most likely caused by glycosylation. In lysates of rat E10.5 embryos, however, only a 177 kDa band is detected by these antibodies. Binding of antibodies to these bands is blocked by the appropriate peptides, which confirms the specificity of the antibodies. Similar sizes of PRTG are detected by a rabbit polyclonal antiserum (W4) or another mAb (α-PRTG3), both of which bind the intracellular domain of PRTG (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). α-PRTG1 mAb, but not α-PRTG2 mAb, is able to label live HEK293T cells transfected with prPRTGf (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), suggesting that PRTG is localized on cell membrane. Confocal images of fixed cells labeled with α-PRTG1 show the same cell-surface staining pattern (data not shown).
Expression of PRTG in mouse embryos
We next used α-PRTG2 mAb to detect the spatial and temporal expression patterns of PRTG in the mouse embryo at the protein level. PRTG protein is first detected during the mid-streak stage (E7) in the mesoderm (Fig. 1D,I). At this stage, Oct4, a transcription factor playing an essential role in the maintenance of pluripotency in mouse embryo (Takahashi and Yamanaka, 2006), is still weakly present in the ectoderm and is mutually exclusive with PRTG expression (Fig. 1D). SSEA1, another mouse embryonic stem cell marker, is expressed in both ectodermal and mesodermal cells (Fig. 1I). At the headfold stage (E7.75), PRTG is expressed in all cells of three germ layers (Fig. 1E) and in the epiblasts of the posterior region (Fig. 1E, the region between two arrows). At the early somite stage (E8.25), PRTG is expressed abundantly in the neural tube, mesoderm, and endoderm but not in differentiated cardiac cells or endothelial cells, which can be labeled with anti-α-smooth muscle actin (α-SMA) and anti-endoglin antibodies (Fig. 1F) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). Interestingly, whereas Oct4 and SSEA1 are not detectable in the E8.25 mouse embryo (Fig. 1I) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material), PRTG continues to be expressed in the E9.5 embryo (Fig. 1G). PRTG begins to be rapidly downregulated in the E10 embryo and is not detectable after E10.5 (Fig. 1H). During the period when PRTG is highly expressed in the developing neural tube, which is between E8.5 and E9.5, the expression of nestin, Ascl1, and neuronal class III β-tubulin (detected by TuJ1) has just begun to appear (Fig. 1J–L) (supplemental Fig. S5, available at www.jneurosci.org as supplemental material). Mature neurons expressing MAP2 and NeuN do not appear until E10.5 (Fig. 1M,N) (supplemental Fig. S5, available at www.jneurosci.org as supplemental material). As illustrated by these sequential expression patterns, the expression of PRTG follows the expression of various pluripotent stem cell markers and precedes expression of neural precursor cell markers and neuronal differentiation markers; thus, expression of PRTG defines a unique period during neural development. To further confirm that PRTG is expressed in the primitive neural progenitors, we analyzed Sox2 protein expression during E7.5–E10.5 mouse embryos together with PRTG expression (supplemental Fig. S6, available at www.jneurosci.org as supplemental material). Sox2 and PRTG are both present in neuroepithelium in E7.5 mouse embryo. Costaining of Sox2 and PRTG expression in neural tube is also observed in E8.25 and E9.5 mouse embryos. Expression of Sox2 persists in the ventricular zone of the E10.5 mouse neural tube, whereas PRTG ceases to express at E10.5 neural tube. The above results support that PRTG is present in primitive neural progenitors.
Characterization of PRTG function in P19 cells
Based on the expression pattern shown in Figure 1, we reasoned that PRTG is expressed in early committed cells, and, when these cells differentiate further, expression of PRTG is turned off. To examine this possibility, we used pluripotent P19 embryonal carcinoma cells, which can undergo neuronal differentiation during induction with all trans-retinoic acid (RA) (Jones-Villeneuve et al., 1983), to study the activity of PRTG. Both PRTG mRNA and protein were detected in P19 cells (Fig. 2A,B, lane 1). During RA treatment, PRTG increases significantly during the first day, then dramatically decreases on the second day, and is undetectable after the fourth day of induction (Fig. 2A,B). Interestingly, the peak of PRTG protein expression is between the peaks of the expression of Oct4 and Sox1 (Fig. 2B,C). Sox1 is one of the earliest transcription factors in neural progenitor cells (Pevny et al., 1998).
Figure 2.
PRTG is expressed in early committed cells, and blockage of PRTG activity promotes neuronal differentiation in P19 cells. A, B, Differentiation of P19 cells was first induced by retinoic acid for 4 d (Day 1–4), and then the cells were cultured in serum-free medium for another 4 d (Day 5–8). A, Expression of prtg mRNA was analyzed by the Northern blot. MAP2, A neuronal marker; cyclophilin, RNA loading control. B, Expression of PRTG, Oct4, Sox1, and MAP2 in differentiating P19 cells was analyzed by Western blot. A nonspecific band with molecular weight 40 kDa recognized by α-PRTG2 was used as protein loading control (Ctrl). C, Quantification of protein amounts from the Western blots shown in B. Relative protein expression levels are normalized to the highest protein expression amount of each protein. D, Lysates of HEK293T cells transfected with shPRTG(m), control vector (UI4), or shS together with pmPRTGf were subjected to Western blot (WB) by anti-myc and anti-β-tubulin antibodies. E, Lysates of HEK293T cells transfected with pmPRTGf(myc), prPRTGf(HA), and prPRTGΔc(HA) were immunoprecipitated by anti-HA or anti-myc antiserum and then subjected to Western blot by anti-myc or anti-HA antiserum. F, Experimental procedure for P19 cell neuronal differentiation analysis. P19 cells were transfected with 0.4 μg of pAscl1 and 1.6 μg of the indicated plasmids, subjected to selection with G418 or puromycin, and then cultured in SF21 serum-free medium for 4 d. G, Differentiated neuronal cells were detected by TuJ1 staining. Scale bar, 50 μm. H, The percentage of cells labeled by TuJ1 in one whole well of the six-well plate was counted. Data are then normalized relative to the percentage of the control (pEF1A or UI4) and shown as the mean ± SEM (n = 3; **p < 0.01, by Student's t test).
To elucidate roles of PRTG during neuronal differentiation, we performed gain-of-function and loss-of-function experiments in P19 cells. For the loss-of-function experiment, we prepared a microRNA-based shRNA expression vector, shPRTG(m), which contains two tandem repeats of three different sequences listed in supplemental Table S1 (available at www.jneurosci.org as supplemental material) and knocks down PRTG efficiently (Fig. 2D) (supplemental Fig. S7A,B, available at www.jneurosci.org as supplemental material). Because RA treatment of P19 cells leads to both neuronal and glial differentiation (Jones-Villeneuve et al., 1982), we adopted another neuronal differentiation paradigm by ectopic expression of the proneural gene Ascl1 into P19 cells to focus on roles of PRTG in neuronal differentiation process and to shorten the differentiation period to 4 d (Farah et al., 2000). In addition, we induced a minimal level of neuronal differentiation by choosing a suboptimal Ascl1 expression level to better observe the role of PRTG in neurogenesis. Only one-fifth of Ascl1 expression construct used in Farah's protocol were transfected into P19 cells with pmPRTGf or shPRTG(m). Transfected cells were selected with G418 or puromycin and cultured in a serum-free medium (SF21) for 4 d (for experimental procedures, see Fig. 2F). By scoring the numbers of TuJ1-positive cells, we found that overexpression of PRTG does not change the percentage of neurons induced. However, knockdown of PRTG does increase the number of neurons to 247% of the control (Fig. 2G,H). Knockdown of PRTG by another two different shRNA constructs [shPRTG(1659) and shPRTG(323), containing four repeats of nucleotides 1659-1680 or nucleotides 323-344, respectively (listed in Table S1, available at www.jneurosci.org as supplemental material)] also promotes neurogenesis (data not shown). Intriguingly, expression of Oct4 protein is decreased in cells in which endogenous PRTG activity has been perturbed by shPRTG(m) (supplemental Fig. S7C, lane 6). It should be noted that knockdown of PRTG alone without Ascl1 in the P19 cells induced very few neurons (∼1 in 10,000 cells) (data not shown).
PRTG is a transmembrane protein, and therefore we suspected that PRTG might serve as a receptor and affect cellular process through its cytoplasmic tail. We first assessed whether PRTG can form dimers or multimers, which would indicate the feasibility of using a truncated PRTG (PRTGΔc) that contains no cytoplasmic tail as a dominant-negative mutant (Amaya et al., 1991; Hemmati-Brivanlou and Melton, 1992). To do this, myc-tagged mouse PRTG and hemagglutinin (HA)-tagged rat PRTG were coexpressed in HEK293T cells and were subjected to IP and Western blot analysis. As expected, these two PRTG proteins were coprecipitated (Fig. 2E, top panels, lanes 4 and 9). Furthermore, PRTGΔc could also be coprecipitated with full-length PRTG protein (Fig. 2E, top panels, lanes 5 and 10), which suggests that it would be feasible to use PRTGΔc to interfere with the activity of endogenous PRTG. When PRTGΔc expression construct was transfected into P19 cells with the Ascl1 expression construct, the percentage of TuJ1-labeled cells increased to 202% of the cells transfected with the control vector (Fig. 2G,H), which is similar to the phenotype in PRTG knocked down cells. It was also noted that transfection of PRTGΔc into P19 cells also decreased Oct4 protein expression (supplemental Fig. S7C, lane 3, available at www.jneurosci.org as supplemental material), similar to what occurred in the shPRTG(m) experiment, demonstrating that PRTGΔc acts as a dominant-negative mutant. These findings indicate that perturbation of PRTG activity by either shPRTG(m) or PRTGΔc promotes neuronal differentiation and suggest that endogenous PRTG may act as a gatekeeper to prevent neuronal differentiation.
Perturbation of PRTG activity causes precocious neuronal differentiation in the developing chick neural tube
Our findings in P19 cells demonstrate that PRTG suppresses neurogenesis. To further examine this possibility in vivo, we used the chick neural tube as our model. The shPRTG(g) construct (containing a GFP expression gene) was verified as able to knockdown chick PRTG efficiently in both HEK293T cells and chick embryos (supplemental Fig. S8A–C, available at www.jneurosci.org as supplemental material). It has been shown previously that endogenous PRTG mRNA is expressed at a higher level in the chick neural tube during the period of HH stage 9–18 (Toyoda et al., 2005). We delivered shPRTG(g) into one side of the chick neural tube at HH stage 9–10 via in ovo electroporation. Two days after electroporation, a marked expansion of TuJ1 staining in the medial part of the shPRTG(g)-electroporated side was noted with a fluorescence intensity increase of 229 ± 13% relative to the medial control side (Fig. 3A–H,W,X). Moreover, the number of cells expressing another neuronal marker, HuC/D, was also significantly increased in the medial region of shPRTG(g)-electroporated side (Fig. 3I–P,W,Y). These ectopic HuC/D-positive cells also lost expression of Sox2, although Sox2-positive cells did not drastically reduce in the electroporated side (supplemental Fig. S9, available at www.jneurosci.org as supplemental material). More NeuN-positive cells could be detected in the lateral region of the shPRTG(g)-electroporated side than on the control side, although the increase was not as strong as with the TuJ1 staining (supplemental Fig. S10B,F, available at www.jneurosci.org as supplemental material). One possible reason for this is the fact that NeuN is a late neuronal differentiation marker (Fig. 1N) and requires a longer period to exhibit the full extent of its expression. Expression of an shS did not affect neuronal differentiation in the chick neural tube (Fig. 3X,Y) (supplemental Fig. S10F, available at www.jneurosci.org as supplemental material). When taken together, these results suggest that PRTG maintains cells in an undifferentiated state during neural development.
We also introduced pCAG–gPRTGf and pCAG–gPRTGΔc into the chick neural tube at HH stage 9–10 via in ovo electroporation. Exogenous PRTG protein and PRTGΔc protein were expressed as expected when the cells were analyzed at the HH stage 17–20 (supplemental Fig. S8D,E, available at www.jneurosci.org as supplemental material). EYFP expression in each vector facilitates tracing of the electroporated cells. Ectopic expression of PRTG leads to a somewhat lower number of TuJ1-positive cells on the electroporated side than on the control side, although this was not statistically significant (Fig. 3S,T,X). However, the numbers of NeuN-positive neurons were reduced to 82.6 ± 3.5% on the electroporated side overexpressing PRTG when compared with the control side (supplemental Fig. S10D,F, available at www.jneurosci.org as supplemental material). More striking results were observed in embryos electroporated with pgPRTGΔc; the level of the TuJ1 intensity was found to be 100 ± 7% more within the medial part of neural tube on the electroporated side compared with the control side (Fig. 3U,V,X). Moreover, 41.6 ± 5.4% more NeuN-positive cells could be observed on the pgPRTGΔc-electroporated side than on the contralateral side (supplemental Fig. S10F, available at www.jneurosci.org as supplemental material). These results provide evidence that perturbation of PRTG function overrides the normal route of neurogenesis in chick embryos and prompts cells to differentiate prematurely. When expression of cHes1 and cHes5 was examined in chick embryos by in situ hybridization, there was no difference of expression levels of these two genes between the electroporated side and the control side, suggesting that perturbation of PRTG signaling does not drastically affect progenitor cell status as revealed by cHes1 and cHes5 expression (supplemental Fig. S11, available at www.jneurosci.org as supplemental material).
ERdj3 as a ligand for PRTG
The above results show that disturbing PRTG function using either shPRTG or PRTGΔc promotes neurogenesis in both P19 cells and the chick neural tube. However, overexpression of full-length PRTG did not decrease the percentage of neurons in P19 cells significantly. We reasoned that this might be attributable to the fact that PRTG acts as a receptor, and there is an already sufficient amount of endogenous PRTG receptor in undifferentiated P19 cells relative to a limited amount of its ligand present outside the cells; in these circumstances, any extra PRTG would not elicit any additional effect. In this context, an elucidation of functions of PRTG would be immensely helped if its ligand(s) can be identified.
To explore the possibility that PRTG acts as a receptor for an unknown ligand(s), we conducted the P19 cell differentiation assay in the presence of either α-PRTG1 mAb, which binds to the PRTG ectodomain and may serve as a neutralizing antibody, or PRTGe–Fc protein, which is a secreted Fc-tagged PRTG ectodomain protein and may compete with the endogenous PRTG for the ligand(s) (for experimental procedures, see Fig. 4A). Addition of α-PRTG1 mAb or PRTGe–Fc protein to the culture medium increases the number of TuJ1-positive neurons in Ascl1-transfected P19 cells by 34 and 139%, respectively (Fig. 4B). A control mAb (α-PRTG3) and the secreted Fc-only protein do not exhibit any such effect on neurogenesis. These results suggest that PRTG serves as a receptor and is activated by a ligand or ligands present in P19 culture medium. These results also support further the idea that perturbation of PRTG activity promotes neuronal differentiation.
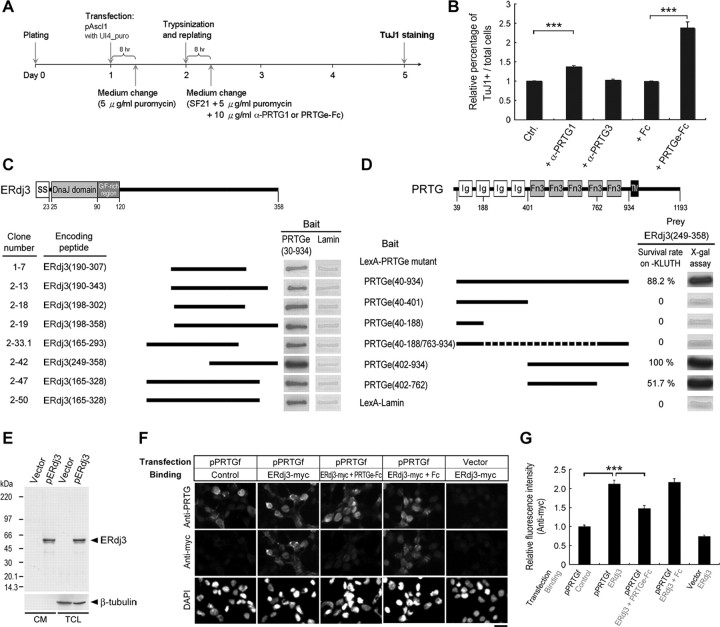
Figure 4.
ERdj3 is a putative ligand of PRTG. A, Experimental procedure for P19 cell differentiation assay used in the following analyses. B, Relative percentages of TuJ1+ neurons induced from Ascl1-transfected P19 cells in the presence of 10 μg/ml α-PRTG1 mAb, α-PRTG3 mAb, PRTGe–Fc, or Fc are shown (n = 3; ***p < 0.001, by Student's t test). C, Schematic representations of eight ERdj3 clones identified by yeast two-hybrid screening are shown, and their intensities of interaction with the PRTG ectodomain are displayed as β-galactosidase activity in an X-gal assay. D, Various prtg deletion mutants were examined for their ability to interact with C-terminal ERdj3. E, ERdj3 is a secreted protein. CM and total cell lysate (TCL) of HEK293T cells transfected with pERdj3 or the control vector were subjected to Western blotting using anti-myc antibody. β-tubulin, A cytosolic protein marker. F, ERdj3 binds PRTG. HEK293T cells were transfected with pPRTGf or control vector. Cells were then treated with ERdj3-containing CM in the presence or absence of PRTGe–Fc. PRTG expression was detected by rabbit anti-PRTG serum (W4; top row). Binding of ERdj3 on the cells was detected by anti-myc mAb (middle row). Cell nuclei were visualized with DAPI (bottom row). Binding of ERdj3 to PRTG-expressing cells was partially inhibited by 40 μg/ml PRTGe–Fc (the third column). Scale bar, 20 μm. G, Quantification of ERdj3 binding to PRTG-expressing cells. The results are normalized relative to fluorescence intensity of the cells transfected with pPRTGf in the presence of control CM. Results are shown as the mean ± SEM from a total of 50 cells (***p < 0.001, by Student's t test).
To search for the ligand(s) of PRTG, we used a yeast two-hybrid approach. PRTG ectodomain (PRTGe) was used as a bait to screen a mouse E7 embryo cDNA library. After analyzing 5.2 million clones, 54 partial DNA fragments were found to produce peptides that interacted with PRTGe. Among these fragments, eight clones contained DNA sequences corresponding to the C-terminal domain of ERdj3 (Fig. 4C), a protein with a signal sequence and reported previously to be an ER-resident cochaperone (Yu et al., 2000). To confirm the association of PRTG with ERdj3 and to delineate the interaction domains involved in the interaction of PRTG with ERdj3, a series of PRTG deletion mutants were constructed, and their interaction with the C-terminal region of ERdj3 was evaluated by yeast two-hybrid assay. As shown in Figure 4D, the fibronectin type III (FnIII) motifs of the PRTG protein, especially the first three FnIII motifs, are critical for binding of PRTG with ERdj3.
Interaction of PRTG and ERdj3 in mammalian cells
To determine whether ERdj3 is released from cells, HEK293T cells were transfected with pERdj3 that produces a C-terminal myc–His-tagged ERdj3 protein. When CM was subjected to Western blot analysis using an anti-myc mAb, ERdj3 was detected in the CM (Fig. 4E). At least 50% of the total ERdj3 protein was secreted into the CM by the transfected HEK293T cells. The presence of this protein in the CM is not attributable to the rupture of dying cells, because the cytosolic protein marker β-tubulin was not detected in the CM (Fig. 4E). Identical secretion patterns were detected in STO cells (supplemental Fig. S12, available at www.jneurosci.org as supplemental material); specifically ERdj3 or ERdj3-Fc, but not EGFP, was present in CM, thus excluding the possibility that the release of ERdj3 is an artifact caused by a high expression level.
We next used an in situ binding assay to determine whether ERdj3 protein binds PRTG. Cells transfected with pPRTGf were incubated with the CM containing ERdj3 for 1 h before fixation and then double labeled with antibodies against PRTG and myc. ERdj3 was found to attach to cells displaying PRTG on the cell surface (Fig. 4F, the second column) but not to those cells without PRTG expression (Fig. 4F, the fifth column). Moreover, this binding is abolished by adding 40 μg/ml PRTGe–Fc during the incubation (Fig. 4F, the third column, G). These findings, together with results of the yeast two-hybrid assay, support the hypothesis that ERdj3 is a putative ligand of PRTG.
ERdj3 inhibits neuronal differentiation by activation of the PRTG receptor
To examine whether ERdj3 interacts with PRTG to regulate neural development, we detected the expression of ERdj3 by in situ hybridization in the mouse embryos from E7 to E10 when PRTG is expressed. Between E7 and E7.5, ERdj3 mRNA is in all cells of the three germ layers and was present in extra-embryonic tissues (supplemental Fig. S13, available at www.jneurosci.org as supplemental material). During the period of E8–E9.5, ERdj3 is expressed in the neural tube, mesoderm, and endoderm. Compared with PRTG expression, the area in which ERdj3 is expressed encompasses regions in which PRTG expresses but also extends more widely to include, for example, the developing heart in which PRTG is not detected. At E10.5, expression of ERdj3 in the neural tube is downregulated but still persists in the brachial arches, heart, posterior somites, and limbs. Together, these results show that the expression pattern of ERdj3 coincides with that of PRTG from E7 to E9.5 in mouse embryos, raising the possibility that ERdj3 functions together with PRTG to maintain cells in an undifferentiated state. To examine this possibility, we used the P19 cell differentiation assay. Overexpression of ERdj3–Fc protein in P19 cells decreases the numbers of TuJ1-positive neurons by 36 ± 0.5% (Fig. 5A,B). To verify that this phenomenon is attributable to the secreted form of ERdj3 and that it does not act through ER-associated ERdj3, we purified ERdj3–Fc by protein A/G column and applied purified ERdj3–Fc to the culture medium of P19 cells transfected with Ascl1. The purified ERdj3–Fc reduced significantly the number of TuJ1-positive cells (Fig. 5C, bar 2). Importantly, this reduction in neurogenesis by purified ERdj3–Fc can be reversed by the addition of α-PRTG1 mAb or PRTGe–Fc (Fig. 5C, bars 3, 5), demonstrating further that the effect of ERdj3–Fc is mediated through PRTG.
Figure 5.
ERdj3 inhibits neuronal differentiation through PRTG in P19 cell differentiation assay. A, P19 cells were transfected with indicated plasmids and pAscl1. Differentiated neurons were detected by TuJ1 staining 4 d after transfection. Scale bar, 100 μm. B, Quantitative results of A are shown as the mean ± SEM (n = 3; ***p < 0.001, by Student's t test). C, P19 cells were induced to differentiation and were cultured with purified 5 μg/ml ERdj3–Fc in the presence or absence of 10 μg/ml α-PRTG1 mAb, α-PRTG3 mAb, PRTGe–Fc, or Fc proteins. Quantitative results of TuJ1+ neurons are shown as the mean ± SEM (n = 3; *p < 0.05; **p < 0.01; #p = 0.053, by Student's t test).
ERdj3 suppresses neuronal differentiation in the developing chick neural tube
We further tested the role of ERdj3 in vivo using chick neural tube. Overexpression and knockdown of chick ERdj3 was done using two constructs, pgERdj3 and shERdj3(g), which were verified in HEK293T cells (supplemental Fig. S14A, available at www.jneurosci.org as supplemental material). Overexpression of chick ERdj3 in the chick neural tube reduced TuJ1 staining (Fig. 6A,B) (for images of the control vector, see Fig. 3Q,R). Moreover, the numbers of NeuN+ cells in the electroporated side versus in the contralateral side were quantified and then normalized against the control vector. The number of NeuN-positive neurons on the pgERdj3-electroporated side decreased to 88.3 ± 3.4% of the contralateral side (Fig. 6C,D) (p < 0.05, by Student's t test). To eliminate the possibility that this effect is attributable to ERdj3 activity in the ER, an ER-retained ERdj3 protein (supplemental Fig. S14B, available at www.jneurosci.org as supplemental material), ERdj3–KDEL (lysine, aspartic acid, glutamic acid, leucine), was ectopically expressed in the chick neural tube. No decrease of TuJ1 staining or HuC/D-positive neurons was observed on the pgERdj3–KDEL-electroporated side compared with the contralateral side (supplemental Fig. S15, available at www.jneurosci.org as supplemental material). These results indicate that secreted ERdj3 inhibits neuronal differentiation. Conversely, knockdown of ERdj3 leads to a thinner chick neural tube compared with the control side (Fig. 6E–H) (for the image of the control vector, see Fig. 3A). The reduction in neural tube volume on the shERdj3-electroprated side is attributable to apoptosis, because many EGFP-positive cells are also labeled with antibody against activated caspase-3 (Fig. 6F, arrows, I). Use of another shRNA construct, shERdj3(799), which contains eight repeats of nucleotides 799-820 of ERdj3(g) (listed in supplemental Table S1, available at www.jneurosci.org as supplemental material), also increased the numbers of apoptotic cells on the electroporated side (data not shown), suggesting that this is not an off-target effect. Thus, rather than the expected increase in neurogenesis, there were fewer TuJ1-stained cells and fewer NeuN-positive cells on the shERdj3-electroporated side (Fig. 6G–H′), which may be because ERdj3 knockdown cells had undergone apoptosis and cell death. The fact that ERdj3 plays a role in regulating cell survival makes it difficult to use the knockdown approach to precisely answer the question of whether ERdj3 directly regulates neurogenesis.
Figure 6.

ERdj3 suppresses neuronal differentiation in the chick neural tube. Overexpression or knockdown of chick ERdj3 constructs were electroporated into the right side (green) of chick neural tubes at HH stage 9–10. Two days after electroporation, the chick embryos were fixed and stained with DAPI (blue) and TuJ1 (A, B, E, E′, red), anti-NeuN (C, D, F, F′, red), or anti-cCasp3 (G, H, red) antibodies. White boxed regions in A, C, and E are enlarged in B, D, and F, respectively. A, B, Overexpression of ERdj3 leads to fewer TuJ1+ cells (bracket in B) in the chick HH stage 17–20 embryo. C, D, Overexpression of ERdj3 reduces NeuN+ cells in the chick HH stage 17–20 embryo. There are more NeuN+ cells on the control side (arrowheads in D). Moreover, the NeuN+ cell on the electroporated side is an untransfected cell (NeuN+, GFP-negative cell; arrow in D). E–H, Knockdown of endogenous ERdj3 produces thinner chick neural tube at HH stage 17–20. E, F, More cCasp3-positive cells are detected in the shERdj3(g)-electroporated side (arrows in F). I, Quantitative results of the number of cCasp3+/GFP+ cells on the electroporated side shown in F. Data are shown as the mean ± SEM quantified from at least three embryos and five sections for each embryo (***p < 0.001, by Student's t test).
Discussion
During embryogenesis, the proper balance between cell proliferation and differentiation is fundamental to the growth and functioning of the nervous system. The switch from pluripotent epiblasts to neural progenitor cells appears to be the first regulatory point. Here, we report that the expression of PRTG, an IgSF receptor, can delineate this developmental transition stage. Furthermore, the interaction between PRTG and its ligand ERdj3 suppresses neuronal differentiation, which represents a novel regulatory pathway as neuronal cell fate is acquired between E7.75 and E9.5 in mouse embryos.
Expression of PRTG defines a developmental transition period between E7.75 and E9.5
Expression of PRTG initiates at E7 mouse embryo, at the time when residual expression of Oct4, a pluripotent epiblast marker, is present in ectoderm. Intriguingly, although there is a temporal overlap of PRTG and Oct4 expression, PRTG and Oct4 are expressed in regions that are mutually exclusive. Later on, expression of PRTG is prominent in almost all cells of mouse embryos from E7.75 to E9.5 and then quickly diminishes when nestin, a neural progenitor cell marker, just begins to express conspicuously. The peak expression of PRTG falls between Oct4 and nestin (Fig. 7). More specifically, PRTG is expressed in cells whose cell fates are in a committed but undifferentiated status during the period between E7.75 and E9.5 in the mouse embryo. Note that, when cells enter the differentiation mode, they decrease PRTG expression, as showed by the results that differentiated neuronal, cardiac, and endothelial cells labeled by MAP2, endoglin, or α-SMA do not express PRTG. Up to now, the characteristics of cells that are labeled by PRTG are not well known, and how these cells become multipotent progenitor cells along particular lineages and undergo further differentiation is not clearly understood either. Reagents that recognize PRTG may be used as molecular tools to help answer these two questions.
Figure 7.
Summary of PRTG expression and the potential function of ERdj3/PRTG signaling in the mouse embryo. Schematic representations of the protein amounts from the immunohistochemical staining in Figure 1 are shown in different colors. The intensity of the color indicates the protein levels. PRTG expression is between marker proteins for epiblasts and marker proteins for neural progenitor cells and thus depicts a distinctive transition stage during mouse neural development. By extrapolating experimental results obtained from P19 cells and the chick neural tube, it is likely that ERdj3/PRTG signaling plays a critical role in the suppression of premature neuronal differentiation in early mouse embryos. Whether ERdj3/PRTG signaling acts at the generation of neural progenitor cells and/or the differentiation of neural progenitor cells into neurons is currently unclear. ERdj3 may have an additional role associated with supporting survival of neuroectoderm.
ERdj3/PRTG signaling prevents precocious neuronal differentiation
To elucidate the functions of PRTG signaling in neural development, we performed loss-of-function and gain-of-function experiments in P19 cells and in the developing chick neural tube. The reason that P19 cells were adopted as an ex vivo model to explore the functions of PRTG is attributable to the fact that temporal expression of PRTG in these cells during RA treatment recapitulates the expression pattern that occurs in mouse embryos. We found that perturbation of PRTG activity by four different reagents, RNA interference, a dominant-negative mutant (PRTGΔc), a neutralizing antibody (α-PRTG1 mAb), and the purified PRTG ectodomain protein (PRTGe–Fc), promotes more neurons in the P19 model and in the chick neural tube model.
In our initial gain-of-function experiments, we found that overexpression of PRTG in P19 differentiation assay does not decrease neurogenesis, although the same treatment does produce fewer neurons in the chick neural tube. We rationalized that these results may be attributable to the facts that PRTG acts as a receptor and that there are already sufficient PRTG present in P19 cells and in the chick neural tube; thus, an additional increase in PRTG proteins, if they are not activated by the limited ligand(s) present, is futile. The yeast two-hybrid assay was then used to identify a ligand for PRTG. It was found that the C-terminal fragment of ERdj3 interacts with first three FnIII regions of PRTG. Moreover, secreted ERdj3 binds to cells that possess PRTG on the cell surface, and this process is blocked by purified PRTG ectodomain protein. We suggest that the dissociation constant for the PRTG/ERdj3 complex is likely to be high, because repeated attempts to immunoprecipitate the PRTG and ERdj3 complex did not succeed (data not shown).
Overexpression of ERdj3 or exogenously added purified ERdj3 decreases neuronal formation in the P19 cell differentiation assay. This effect is presumably mediated through PRTG, because it is eliminated by either the PRTG neutralizing antibody or PRTG ectodomain protein. The results for whole-mount in situ hybridization from E7.5 to E9.5 show that ERdj3 is expressed in domains that also exhibit PRTG expression, which further support the possibility that ERdj3 can interact with PRTG in vivo. Together, these results suggest that ERdj3/PRTG signaling may set up a barrier for neuronal differentiation during E7.75–E9.5 neural development in mouse embryos.
This leaves the question as to how PRTG/ERdj3 signaling keeps early committed neural progenitors in an undifferentiated state. Close examination of the perturbation experiments in the chick neural tube using shPRTG(g) and pPRTGΔc suggests that this is likely to be a cell-autonomous effect among PRTG-expressing cells. For example, all TuJ1- and HuC/D-positive cells detected in the medial chick neural tubes had been electroporated with shPRTG(g) or pPRTGΔc, namely, they were always GFP- or YFP-positive electroporated cells. Conversely, GFP- or YFP-negative cells in the medial neural tube are no more likely to be TuJ1 or HuC/D positive on the electroporated side than on the contralateral side. Thus, secreted ERdj3 may act on PRTG-expressing cells to prevent premature differentiation during the period between neuroectoderm and neural progenitors (Fig. 7). It is likely that PRTG may merely act as a gatekeeper for neurogenesis in the mouse embryo, because only a marginal effect on neurogenesis can be observed when P19 cells are transfected with shPRTG(m) and pPRTGΔc in the absence of AsclI. The details of the mechanisms involved in ERdj3/PRTG signaling, however, still need additional investigation.
ERdj3 may have dual functions during neural development
When an ERdj3 shRNA constructs were electroporated in the chick neural tube, we were surprised to observe, on the electroporated side, the thinness of the chick neural tube and the presence of activated caspase 3-positive apoptotic cells. This was not observed when PRTG was knocked down (Fig. 3C and data not shown). This implies that ERdj3, in addition to preventing precocious neuronal differentiation, also maintains cell survival (Fig. 7). A recent report also supports the idea that ERdj3 may maintain cell survival. Cell death is triggered in human gastric cells treated with a nonsteroidal anti-inflammatory drug, celecoxib. A study has shown that upregulation of ERdj3 inhibits celecoxib-induced apoptosis (Tsutsumi et al., 2006). One possibility is that the ERdj3-mediated survival effect during development may be delivered through a receptor other than PRTG; this is because disturbing PRTG function does not lead to cell death. Alternatively, instead of being a ligand, ERdj3 may regulate cell survival through its role as an Hsp40/DnaJ chaperone in the ER. It is possible that ERdj3 may be important for the proper folding of cellular proteins during neural development. Therefore, knockdown of ERdj3 leads to cellular death.
ERdj3 was originally identified as a cochaperone (Yu et al., 2000; Shen and Hendershot, 2005). However, the characteristics of the ERdj3 primary amino acid sequence reveal that it may have dual roles. ERdj3, unlike other ERdjs, has no predicted transmembrane helix and no ER retrieval signal at the C terminus; this favors a tendency to enter the secretory pathway (Ohtsuka and Hata, 2000). Our observation that ERdj3, but not ERdj3–KDEL, is present in the CM of HEK293T and STO cells corroborates with this prediction. Furthermore, our finding that the C-terminal motif of ERdj3 binds to a specific protein and has effects other than one involving in protein folding is not without precedent. A study has shown that the SANT2 domain in the C-terminal region of ERdj1 binds to α1-antichymotrypsin and antagonizes its inhibitory activity (Kroczynska et al., 2004). In this context, ERdj3 is unique among ERdj family members, in both its cellular localization and its biological functions. It acts as a chaperone activator when it resides in the ER but also acts as a ligand for PRTG protein when it is secreted into the extracellular milieu.
In summary, our results show that the generation of neurons from neuroepithelium is, at least in part, regulated by PRTG protein. Interestingly, expression of PRTG is also present in both mesodermal and endodermal cells (Fig. 1) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). In light of this, it is plausible that the mechanism that commits pluripotent epiblasts to neural differentiation has parallels in the mesoderm and endoderm. This possibility suggests that PRTG might potentially be a key mediator at the molecular level that acts before the onset of circulation to coordinate the rate of proliferation and the time of differentiation between the three primary germ layers.
Footnotes
This research was supported by National Health Research Institutes Grant NHRI-GT-EX89S732C, National Science Council Grant NSC92-2320-B-010-069, the Yen Tjing Ling Medical Foundation, and the Ministry of Education (Aim for the Top University Plan) (M.-J. F.). We thank H.-M. Shih for guidance during the yeast two-hybrid screening, D. L. Turner for the expression vector (pAscl1) and two shRNA vectors (UI4-puro and UI4-GFP), N. Nakatsuji for the pCAG–EYFP–CAG vector, H.-M. Lai for preparing tissue culture reagents, the Developmental Studies Hybridoma Bank for providing monoclonal antibodies, and C.-M. Chen, L.-S. Kao, and C.-Y. Kuan for discussion and critical reading of this manuscript.
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Becker T, Bernhardt RR, Reinhard E, Wullimann MF, Tongiorgi E, Schachner M. Readiness of zebrafish brain neurons to regenerate a spinal axon correlates with differential expression of specific cell recognition molecules. J Neurosci. 1998;18:5789–5803. doi: 10.1523/JNEUROSCI.18-15-05789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Tsai AY, Maness PF. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knock-out mice. J Neurosci. 1999;19:4907–4920. doi: 10.1523/JNEUROSCI.19-12-04907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad JM, Keeling SL, Wilks AF, Tan SS, Cooper HM. The expression patterns of guidance receptors, DCC and Neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev Biol. 1997;192:258–273. doi: 10.1006/dbio.1997.8756. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. A truncated activin receptor inhibits mesoderm induction and formation of axial structures in Xenopus embryos. Nature. 1992;359:609–614. doi: 10.1038/359609a0. [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, Rudnicki MA, Harris JF, McBurney MW. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol Cell Biol. 1983;3:2271–2279. doi: 10.1128/mcb.3.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kroczynska B, Evangelista CM, Samant SS, Elguindi EC, Blond SY. The SANT2 domain of the murine tumor cell DnaJ-like protein 1 human homologue interacts with alpha1-antichymotrypsin and kinetically interferes with its serpin inhibitory activity. J Biol Chem. 2004;279:11432–11443. doi: 10.1074/jbc.M310903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci U S A. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- Polo-Parada L, Bose CM, Landmesser LT. Alterations in transmission, vesicle dynamics, and transmitter release machinery at NCAM-deficient neuromuscular junctions. Neuron. 2001;32:815–828. doi: 10.1016/s0896-6273(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Rolf B, Bastmeyer M, Schachner M, Bartsch U. Pathfinding errors of corticospinal axons in neural cell adhesion molecule-deficient mice. J Neurosci. 2002;22:8357–8362. doi: 10.1523/JNEUROSCI.22-19-08357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal RA, Takahashi H, McKay RD. Changes in neurotrophin responsiveness during the development of cerebellar granule neurons. Neuron. 1992;9:1041–1052. doi: 10.1016/0896-6273(92)90064-k. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Toyoda R, Nakamura H, Watanabe Y. Identification of protogenin, a novel immunoglobulin superfamily gene expressed during early chick embryogenesis. Gene Expr Patterns. 2005;5:778–785. doi: 10.1016/j.modgep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Tsutsumi S, Namba T, Tanaka KI, Arai Y, Ishihara T, Aburaya M, Mima S, Hoshino T, Mizushima T. Celecoxib upregulates endoplasmic reticulum chaperones that inhibit celecoxib-induced apoptosis in human gastric cells. Oncogene. 2006;25:1018–1029. doi: 10.1038/sj.onc.1209139. [DOI] [PubMed] [Google Scholar]
- Vesque C, Anselme I, Couvé E, Charnay P, Schneider-Maunoury S. Cloning of vertebrate Protogenin (Prtg) and comparative expression analysis during axis elongation. Dev Dyn. 2006;235:2836–2844. doi: 10.1002/dvdy.20898. [DOI] [PubMed] [Google Scholar]
- Wigg KG, Feng Y, Crosbie J, Tannock R, Kennedy JL, Ickowicz A, Malone M, Schachar R, Barr CL. Association of ADHD and the Protogenin gene in the chromosome 15q21.3 reading disabilities linkage region. Genes Brain Behav. 2008;7:877–886. doi: 10.1111/j.1601-183X.2008.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yamakawa K, Huot YK, Haendelt MA, Hubert R, Chen XN, Lyons GE, Korenberg JR. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7:227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]