Abstract
Collapsin response mediator proteins (CRMPs) are involved in signaling of axon guidance and neurite outgrowth during neural development and regeneration. Among these, CRMP2 has been identified as an important actor in neuronal polarity and axon outgrowth, these activities being correlated with the reorganization of cytoskeletal proteins. In contrast, the function of CRMP5, expressed during brain development, remains obscure. Here, we find that, in contrast to CRMP2, CRMP5 inhibits tubulin polymerization and neurite outgrowth. Knockdown of CRMP5 expression by small interfering RNA confirms its inhibitory functions. CRMP5 forms a ternary complex with MAP2 and tubulin, the latter involving residues 475-522 of CRMP5, exposed at the molecule surface. Using different truncated CRMP5 constructs, we demonstrate that inhibition of neurite outgrowth by CRMP5 is mediated by tubulin binding. When both CRMP5 and CRMP2 are overexpressed, the inhibitory effect of CRMP5 abrogates neurite outgrowth promotion induced by CRMP2, suggesting that CRMP5 acts as a dominant signal. In cultured hippocampal neurons, CRMP5 shows no effect on axon growth, whereas it inhibits dendrite outgrowth and formation, at an early developmental stage, correlated with its strong expression in neurites. At later stages, when dendrites begin to extend, CRMP5 expression is absent. However, CRMP2 is constantly expressed. Overexpression of CRMP5 with CRMP2 inhibits CRMP2-induced outgrowth both on the axonal and dendritic levels. Deficiency of CRMP5 expression enhanced the CRMP2 effect. This antagonizing effect of CRMP5 is exerted through a tubulin-based mechanism. Thus, the CRMP5 binding to tubulin modulates CRMP2 regulation of neurite outgrowth and neuronal polarity during brain development.
Introduction
Collapsin response mediator proteins (CRMPs) are cytosolic phosphoproteins, highly expressed in the developing nervous system. They act as signaling molecules involved in the regulation of microtubule polymerization, actin bundling, and endocytosis leading to neuronal differentiation and axonal growth (Arimura and Kaibuchi, 2007). Among CRMPs, CRMP2 was originally identified as the signaling molecule of the repulsive guidance cue, semaphorin 3A, inducing growth cone collapse (Goshima et al., 1995). CRMP2 has been reported to regulate neuronal polarity and axon elongation (Yoshimura et al., 2005); its overexpression induces multiple axon formation and primary axon elongation in hippocampal neurons (Inagaki et al., 2001). CRMP2 modulates axon growth through direct binding to tubulin and promoting microtubule dynamics (Fukata et al., 2002). By binding to the kinesin-1 light chain, CRMP2 regulates the transport of soluble tubulin to the distal parts of growing axons, thereby modulating axon growth (Kimura et al., 2005). Microtubule assembly can be disrupted by CRMP2 phosphorylation (Yoshimura et al., 2005). Some studies have suggested a role for CRMP2 in the etiology of Alzheimer's disease since hyperphosphorylation of CRMP2 may interfere with tubulin assembly in neurites (Cole et al., 2004; Uchida et al., 2005). In addition to its role in the transport of soluble molecules, CRMP2 can act as a cargo receptor in the transport of vesicles (Arimura et al., 2009).
Another member of the CRMP family, CRMP5, was independently identified by different approaches (Fukada et al., 2000; Inatome et al., 2000; Ricard et al., 2001). CRMP5 expression is high in developing brain, but decreases in adult brain. Our group has studied the spatiotemporal expression of CRMP5 in the cortex, hippocampus, and cerebellum, and in postmitotic neuronal precursors, suggesting a role in process extension (Ricard et al., 2001). Other studies have reported the localization of CRMP5 at the filopodia of growth cones, suggesting that it plays a role in regulating filopodial dynamics and growth cone development (Hotta et al., 2005). The association of CRMP5 with tyrosine kinase Fes/Fps or mitochondrial septin has been previously reported (Mitsui et al., 2002; Takahashi et al., 2003), although the functional significance of these interactions has not been elucidated. In addition to its expression during development, CRMP5 plays a role in neurological disorders. In adults, the reexpression of CRMP5 causes paraneoplastic neurological syndromes, as a result of cancer-induced autoimmune process. CRMP5 autoantibodies were reported as markers of small-cell lung cancer and thymoma (Ricard et al., 2001; Honnorat et al., 2009). Considering the importance of CRMP5 in development and disease, it is possible that it helps the remodeling of neuronal networks, although, to date, its function is still poorly understood.
In the present study, we show that CRMP5 forms a ternary complex with microtubule-associated protein 2 (MAP2) and tubulin, resulting in the inhibition of neurite elongation. In hippocampal neurons, CRMP5 is highly expressed in short processes at early stages of development, resulting in inhibition at the dendritic but not axon level. When both CRMP5 and CRMP2 are coexpressed, CRMP5 abrogates neurite outgrowth promotion induced by CRMP2, thereby acting as a signal modulating CRMP2 activity.
Materials and Methods
Recombinant protein production.
The cDNA encoding full-length human CRMP5 (residues 1-564) or truncated forms of the protein (1-520, 1-471) were amplified by PCR and inserted into the pT7-7 expression vector, which generated a protein with six His residues at its C terminus. The plasmid was transformed into Escherichia coli strain BL21 (DE3) (Codon-Plus; Stratagene). The bacterial culture was grown at 37°C to an OD600 of 1.8, before induction with 75 μm isopropyl β-d-thiogalactopyranoside (IPTG). After overnight incubation at 16°C, cells were harvested by centrifugation. The pellet was suspended in buffer before disruption by sonication and purification of the recombinant protein as described previously (Moradi-Améli et al., 2002).
Crude brain extract.
Postnatal day 8 (P8) mice (C57BL/6; Charles River Laboratories) were anesthetized with pentobarbital and perfused intracardially with PBS, to clear the brain of blood. After decapitation, the brains were removed and chilled on ice. All subsequent steps were performed at 4°C. Cerebral tissue was homogenized, with a Dounce homogenizer, in 25 mm HEPES, pH 7.2, 300 mm sucrose, 5 mm EDTA, 5 mm DTT, 1 mm PMSF, 1 mm orthovanadate, supplemented with protease inhibitor mixture (Complete; Roche Applied Science), and centrifuged for 30 min at 14,000 × g at 4°C. The supernatant was stored at −80°C until used. Cortex extract was prepared as above from dissected cortex.
Antibodies used, Western blot analysis, and overlay assay.
The site-specific antibody to CRMP5 (anti-CRMP5 antibody) and CRMP2 (anti-CRMP2 antibody) were produced in rabbits and purified as previously described (Ricard et al., 2001). The peptide sequence used to generate specific antisera were 505KEMGTPLADTPTRPVTRHGG524 and 454LEDGTLHVTEGS465 targeting the CRMP5 and CRMP2 C-terminal region, respectively. Other antibodies used were monoclonal anti-MAP2 antibody (clone HM-2; Sigma-Aldrich), monoclonal anti-α-tubulin (clone DM1A; Sigma-Aldrich), and anti-βIII-tubulin (clone SDL.3D10; Sigma-Aldrich) antibodies. Extracts from brain, recombinant protein, or immunoprecipitated proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with different antibodies, as described previously (Rogemond et al., 2008). Extracts from cultured hippocampal neurons, plated at a density of 1.5×106 cells/well, were obtained on days 1–6 by resuspending neurons in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 mm sodium orthovanadate, 1% Triton X-100 supplemented with protease inhibitor mixture (Complete; Roche Applied Bioscience). After centrifugation at 14,000 × g for 30 min, the supernatant containing the cytosolic fraction was subjected to SDS-PAGE and analyzed by Western blot as above.
For overlay assays, purified recombinant CRMP5 was subjected to SDS-PAGE and electrotransferred to a nitrocellulose membrane. The membrane was incubated for 1 h in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl (TBS) containing 3% (w/v) bovine serum albumin (BSA), then incubated overnight at 4°C with crude brain extract diluted 5 times in TBS containing 0.1% Tween, 1 mm DTT, 1 mm PMSF, and 3% BSA. After extensive washing with TBS containing 3% BSA, the nitrocellulose membrane was incubated for 1 h with anti-MAP2, anti-α-tubulin, or anti-βIII-tubulin antibodies. After additional extensive washing, the positive CRMP5 band was detected after an incubation for 1 h with peroxidase-conjugated anti-mouse antibody and visualized using the ECL detection system.
Immunoprecipitations and pull-down assays.
Crude brain extract was diluted six times in the immunoprecipitation (IP) buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 5 mm EGTA, 5 mm EDTA, 1 mm sodium orthovanadate, 1 mm DTT, supplemented with a mixture of protease inhibitors), mixed with the monoclonal anti-MAP2, anti-α- or anti-βIII-tubulin antibodies, and incubated overnight on a rocking platform at 4°C. Then, 150 μl of protein A-Sepharose 6MB (Sigma-Aldrich) was added to 1 ml of mixture and immunoprecipitation allowed to proceed for 1 h at 4°C. After a short spin at 14,000 × g at 4°C, the supernatant was removed, and the beads were washed six times with 1 ml of IP buffer. The beads were then suspended in sample buffer, boiled for 5 min, and subjected to SDS-PAGE and Western blotting.
For pull-down assays, cDNA fragments encoding the full-length (residues 1-564), N-terminal part ΔC472 (residues 1-471), C-terminal half ΔN244 (residues 245-564), and C-terminal part ΔN474 (residues 475-564) of human CRMP5 or full-length human CRMP2 were inserted downstream of the GST gene in the pGEX-6P-1 vector (GE Healthcare). All fragments were confirmed by DNA sequencing. E. coli BL21 (DE3) cells transformed with each plasmid were cultured to reach an OD600 of 0.6, before induction with 0.4 mm IPTG. After 4 h of incubation at 37°C, cells were harvested by centrifugation and suspended in lysis buffer before disruption by sonication. GST-fused CRMP2 or CRMP5 derivatives were purified from the cell lysate, by adsorption onto glutathione Sepharose 4B beads (GE Healthcare). To demonstrate direct interaction between CRMP5 and tubulin, GST-CRMP5 derivatives (25 μg/ml) conjugated to glutathione Sepharose 4B (50% slurry, diluted 20 times) were incubated with purified tubulin (100 μg/ml; Cytoskeleton) in PBS, 1 mm dithiothreitol, 1 mm PMSF, 20% glycerol, and 0.1% Triton X-100 for 1 h at 4°C. The beads were pulled down by centrifugation and washed thoroughly with buffer, and then subjected to SDS-PAGE and Western blotting. For the assay using mouse brain extract, the GST-CRMP5 derivatives were incubated overnight at 4°C with P8 mouse brain extract (diluted five times), instead of purified tubulin. To study the effect of CRMP5 on tubulin–CRMP2 interactions, GST-CRMP2, conjugated to glutathione Sepharose 4B, was incubated with purified tubulin for 10 min at 4°C, followed by additional incubation with increasing amounts of purified recombinant His-CRMP5 for 1 h at 4°C and treated as above.
Tubulin polymerization assay.
Purified bovine brain tubulin (6 mg/ml; Cytoskeleton) was reconstituted in tubulin buffer (TB) containing 80 mm PIPES, pH 6.9, 2 mm MgCl2, 0.5 mm EGTA, and 5% glycerol. Tubulin, diluted twofold with purified recombinant CRMP5 equilibrated in TB, was incubated for 20 min at 4°C before starting the polymerization reaction by adding GTP (1 mm) at 37°C. Control incubations contained the TB buffer instead of CRMP5 protein. The ability of tubulin to polymerize into microtubules was monitored by the increase in absorbance at 340 nm versus time, with a Cary 100 Scan spectrophotometer (Variant) thermostated at 37°C.
Expression constructs, cell culture, and transfection.
Full-length CRMP2 or CRMP5 were amplified by PCR and inserted directionally into the pCMV2-FLAG vector (Sigma-Aldrich), which generated a protein with a FLAG tag at its N terminus, essentially as described previously (Rogemond et al., 2008). Full-length EGFP-CRMP5 or different deletion mutants were amplified by PCR introducing EcoRI and NotI restriction sites. cDNA fragments encoding full-length human CRMP5, ΔN244 CRMP5, ΔC472 CRMP5, and ΔC518 CRMP5 were digested by EcoRI and NotI and sequentially ligated into pCEFL-tagged EGFP vectors, which generated a protein with enhanced green fluorescent protein (EGFP) at its N terminus. The sequences of all constructs were confirmed by automated DNA sequencing. PC12 cells were transfected with purified plasmids containing FLAG-tagged CRMP5, EGFP, or EGFP-tagged CRMP5 derivatives as described previously (Rogemond et al., 2008). At 8 h after transfection, PC12 cells were stimulated by 100 ng/ml NGF (7S; Sigma-Aldrich), for up to 36 h. N1E-115 cells were transfected with the above plasmid constructs, or FLAG-tagged CRMP2 using Lipofectamine LTX (Invitrogen) as described previously (Rogemond et al., 2008), and fixed at 48 h after transfection.
Primary culture of hippocampal neurons.
Hippocampal neurons from embryonic day 18 (E18) mouse embryos were prepared and plated at a density of 3 × 105 cells/well as previously described (Yoshimura et al., 2005). In one set of experiments, neurons were transfected 24 h after plating with EGFP, EGFP-tagged CRMP5, or FLAG-tagged CRMP2 plasmid constructs using the Nanojuice kit (Novagen; Merck) and fixed 48 h later at 3 d of in vitro culture (DIV 3), before staining. In another set of experiments, neurons were cultured for 4 d before single transfection with the above plasmid constructs or double transfection with both EGFP-tagged CRMP5 derivatives and FLAG-tagged CRMP2 constructs using Lipofectamine LTX. Hippocampal neurons were fixed 48 h after transfection at DIV 6 before immunolabeling.
Immunocytochemistry and morphology assay.
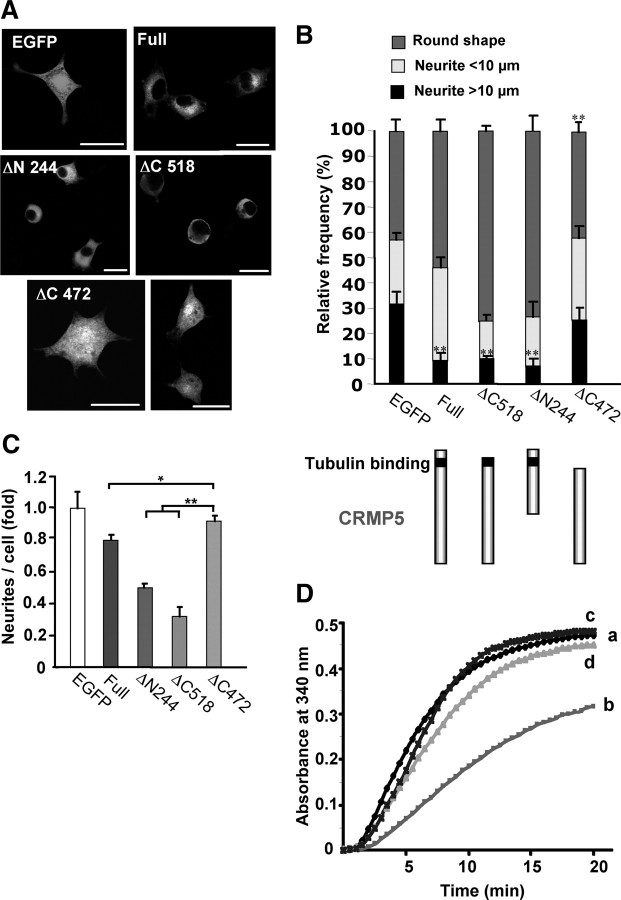
Cells were fixed in 4% paraformaldehyde (PFA) for 20 min at 4°C and blocked with PBS containing 2% BSA and 0.1% Triton X-100 before staining. PC12 cells were double-stained with polyclonal anti-FLAG (Sigma-Aldrich) and anti-rabbit Alexa Fluor 488 (Invitrogen) and with monoclonal anti-α-tubulin and anti-mouse Alexa Fluor 555 antibodies, respectively. In some studies, anti-CRMP5 antibody was used to label endogenous CRMP5. PC12 cells were observed using a laser-scanning confocal system (Leica TCF SP2 imaging platform). For morphometric analysis, PC12 cells were transfected with EGFP-CRMP5 constructs (full-length or different mutants of CRMP5 ΔN244, ΔC472, and ΔC518) and observed by green fluorescence emission without additional staining, using an Axioplan II fluorescence microscope (Carl Zeiss). The percentage of transfected PC12 cells exhibiting rounded shapes or bearing neurite-like extensions less than and greater than 10 μm was measured.
N1E-115 cells transfected with FLAG-tagged constructs, were stained using monoclonal anti-FLAG (Sigma-Aldrich), followed by anti-mouse Alexa Fluor 488 (Invitrogen) antibodies. In double-transfection experiments, FLAG-tagged CRMP2 was stained using anti-mouse Alexa Fluor 555 antibody, whereas EGFP-tagged CRMP5-transfected cells were observed by green fluorescence emission. The percentage of CRMP5-expressing cells bearing neurite length >25 μm from the cell body was measured in each transfection and compared with EGFP-expressing and/or CRMP2-expressing cells. N1E-115 cells were observed with the fluorescence microscope.
Hippocampal neurons, at either DIV 3 or DIV 6, were fixed in 4% PFA for 20 min at room temperature, and stained after single or double transfection as described above. To identify dendrites and axons in each set of experiments, labeling was performed using anti-MAP2, followed by anti-mouse Alexa Fluor 555 and anti-tau (clone H-150; Santa Cruz Biotechnology), followed by anti-rabbit Alexa Fluor 350 antibodies, respectively. The length of longest neurite was measured as that of axon. The percentage of neurons for which the second longest neurite was more than one-half the length of the longest neurite, was counted as that of multiple long neurites. The length of all remaining shorter neurites of an individual neuron, beginning from the cell body, was measured as that of dendrite. For morphometric analysis, the images of labeled neurons were captured with the fluorescence microscope, and the axons and their branching, as well as dendrites, were traced on a video screen and analyzed using analysis Image Processing 3.2 software.
Evaluation of CRMP5, and CRMP2 spatiotemporal distribution in cultured hippocampal neurons.
Hippocampal neurons from E18 mouse embryos were plated at a density of 5 × 104 cells/well as above and fixed on days 1–6 of in vitro culture (DIV 1–6). To follow CRMP5, CRMP2, and MAP2 expression at different time intervals, hippocampal neurons were double stained with anti-CRMP5 and anti-CRMP2 or anti-MAP2 followed by anti-rabbit Alexa Fluor 488 and anti-mouse Alexa Fluor 555 antibodies, respectively. Images of fluorescent neurons were captured with an Axioplan II fluorescence microscope. To determine the fluorescence intensity, all fluorescent images were digitally converted into a grayscale image before the analysis. Quantitative measurements of fluorescence intensities were obtained by averaging the intensity of different 40 × 40 square areas of pixel intensities obtained from defined area across axon and dendrites, but no cell bodies, using image processing, ImageJ 1.42q software. The grayscale intensity was corrected against the intensity point of the background. Numerical fluorescence values were graphed as fluorescence intensity for CRMP5, CRMP2, or MAP2 at DIV 1–6. For three-dimensional intensity projection, individual neurons were manually selected and the optical center of mass was used to center the measurement box (1376 × 1032 pixels). Areas for imaging contained cell bodies and networks of dendrites and axon. The luminance of an image was interpreted as the height for the plot.
Knockdown of gene expression by small interfering RNA.
Validated small interfering RNA (siRNA) against rat CRMP5 (5′-GTTTCAGCCTCTCTGGTTC-3′) sequence (Hotta et al., 2005) and control (scrambled sequence with the same percentage of CG but no sequence homology) were purchased from Invitrogen. N1E-115 cells or hippocampal neurons were transfected 24 h after plating with 100 nm vector-siRNA or scrambled siRNA (SC) using the Ribojuice kit (Novagen). N1E-115 cells were fixed 36 h after transfection and stained with phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) to label the F-actin and nuclei, respectively. Hippocampal neurons were transfected either with siRNA and EGFP and observed by green fluorescence emission or with siRNA and FLAG-CRMP2 plasmid construct and stained with anti-FLAG and anti-mouse Alexa Fluor 555 antibodies. In neurons, the transfection was performed with the simultaneous use of Ribojuice and Nanojuice kits (Novagen).
Statistical analyses.
Differences between means were compared by unpaired two-tailed Student's t test. Data are expressed as mean ± SE, with p < 0.05 considered as the level of significance. Data are the mean values of at least three individual experiments.
Results
CRMP5 binding to MAP2 and tubulin
To identify CRMP5-interacting proteins in the developing nervous system, we searched for high-molecular-weight proteins, from P8 mouse brain extract, which copurified with CRMP5 on an affinity column (Cibacron Blue agarose). Among different proteins eluted from this column and subjected to SDS-PAGE, a band migrating with an apparent molecular mass of ∼270 kDa was excised, subjected to mass spectrometry, and identified as MAP2 (data not shown). The CRMP5–MAP2 interaction was confirmed by the coimmunoprecipitation of CRMP5 by anti-MAP2 antibody in a lysate from the brain cortex of P8 mouse. Western blotting of the immunoprecipitate showed that the band migrating with an apparent mass of 66 kDa was recognized by anti-CRMP5 site-specific antibody (Fig. 1 A). The use of two other monoclonal antibodies against the N-terminal and C-terminal parts of CRMP5 further confirmed the 66 kDa band as CRMP5 (data not shown). Two forms of MAP2 can be distinguished in the neonatal stage: high-molecular-weight MAP2b and low-molecular-weight MAP2c. As a control, both forms of MAP2 were also precipitated by the anti-MAP2 antibody (Fig. 1 A). This is consistent with the colocalization of MAP2 with CRMP5 previously reported in the dendrites of embryonic cortical neurons in culture, as well as in the brain cortex of adults (Bretin et al., 2005).
Figure 1.

CRMP5 interacts with MAP2 and tubulin. A, Brain extract from P8 mice was immunoprecipitated by anti-MAP2 antibody, and Western blots were probed for CRMP5 and α-tubulin immunoreactivity. CRMP5 and tubulin were coimmunoprecipitated in a complex with MAP2 protein. Immunoprecipitation of MAP2 is shown as positive control. Note the absence of reactivity in the control IgG. B, Endogenous and overexpressed CRMP5 colocalized with tubulin in NGF-induced differentiated PC12 cells. Left panel, Cells transfected with FLAG-CRMP5 and immunostained with anti-α-tubulin and anti-FLAG antibodies show colocalization of CRMP5 and tubulin in the cytosol and in thin extensions. The inset is a magnification of this area. Middle panel, High magnification of one PC12 cell clearly shows colocalization of CRMP5 and tubulin. Right panel, Endogenous CRMP5 colocalized with tubulin at the cytosolic level and in thin extensions. The inset is a magnification of one PC12 cell. Scale bars, 20 μm. C, Immunoprecipitation of brain extract from P8 mice with anti-α- or anti-β-tubulin antibodies. Western blots were probed for CRMP5 immunoreactivity. CRMP5 was immunoprecipitated in a complex with α- and β-tubulin. As expected, α- and β-tubulin were also coimmunoprecipitated with anti-β- and anti-α-tubulin antibodies, respectively. Control IgG failed to precipitate any protein. D, Overlay assay using purified recombinant CRMP5 (15 μg) subjected to SDS-PAGE and transferred to nitrocellulose membrane. After an overnight incubation with brain extract from P8 mouse (+) or buffer (−), the membrane was probed for MAP2, α-tubulin, or βIII-tubulin immunoreactivity. CRMP5 interacts with MAP2, α-tubulin, or βIII-tubulin present in the brain extract. The intensity of interaction is higher for MAP2 and α-tubulin than for βIII-tubulin. E, Pull down of tubulin from mouse brain extract with GST or GST-CRMP5 (10 μg) conjugated to glutathione Sepharose 4B beads. Western blots of the pellet were probed for tubulin and MAP2 immunoreactivity. GST-CRMP5, but not GST, precipitated MAP2 and α-tubulin. Each result is representative of three to five separate experiments with similar results.
As MAP2 was the major microtubule-associated protein of brain tissue, we assessed whether tubulin was present in the immunoprecipitate formed by MAP2 and CRMP5. A band corresponding to tubulin was detected using α-tubulin antibody, thereby indicating the presence of tubulin as well as MAP2 and CRMP5 in the immunoprecipitate (Fig. 1 A). Next, we examined the expression patterns of CRMP5 and tubulin on NGF-induced differentiated PC12 cells transfected with the FLAG-CRMP5 expression vector. As the distribution of MAP2 in PC12 cells was well known (Brugg and Matus, 1988), we did not investigate its expression pattern in this study. Immunofluorescence labeling using anti-FLAG and anti-α-tubulin antibodies showed that CRMP5 was clearly colocalized with α-tubulin (Fig. 1 B, left and middle panels). However, after close examination, it seemed that this colocalization did not extend to the minor membrane extensions, since clear green staining, corresponding to CRMP5 expression, could be observed in the merge images of the transfected cells (Fig. 1 B, high magnification in left panels). Endogenous CRMP5 labeled with anti-CRMP5 antibody also showed a clear overlapping expression with tubulin (Fig. 1 B, right panels).
We then performed coimmunoprecipitation assays using α-tubulin and βIII-tubulin antibodies. CRMP5 was unambiguously coimmunoprecipitated from brain lysate by both antibodies (Fig. 1 C). To further confirm these interactions, we performed an overlay experiment in which recombinant CRMP5 protein was transferred to a nitrocellulose membrane, and then subsequently incubated in the presence or absence of brain lysate, before the detection of CRMP5-bound proteins with different antibodies. In the presence of brain lysate, CRMP5 bound strongly to MAP2 and α-tubulin (Fig. 1 D). However, anti-βIII-tubulin detected the CRMP5 protein as a faint band, indicative of a weak interaction. Reciprocally, α-tubulin and MAP2 were pulled down from lysates of brain cortex using GST-CRMP5 immobilized on glutatione-Sepharose beads, suggesting their coassembly in vivo. Figure 1 E shows that GST-CRMP5, but not GST, precipitates α-tubulin, which is present in the supernatant fractions of both GST and GST-CRMP5. Similarly, incubation of GST-CRMP5 with brain lysate led to the coprecipitation of two forms of MAP2, MAP2b and MAP2c (Fig. 1 E), albeit at lower levels than that of tubulin. Together, the above data suggest the formation of a ternary complex between CRMP5, tubulin, and MAP2. However, within this complex, the CRMP5-interacting molecule is not distinguished.
Identification of a direct interaction between the C-terminal domain of CRMP5 and tubulin
We examined whether the CRMP5-MAP2 coassembly was mediated by tubulin subunits, this hypothesis being reinforced by the fact that tubulin binding to CRMP1-4 had previously been reported (Fukata et al., 2002). An in vitro binding assay was performed using recombinant GST-CRMP5 and purified tubulin, containing both α- and β-tubulin. Both α-tubulin and β-tubulin were pulled down by GST-CRMP5 (Fig. 2 A), demonstrating a direct interaction between CRMP5 and α- and β-tubulin. However, α-tubulin showed greater binding to CRMP5 than β-tubulin. To identify the CRMP5 domain responsible for mediating interactions with tubulin, deletion constructs of CRMP5 were designed (Fig. 2 B), and purified proteins generated as GST-CRMP5 (Fig. 2 C) were used in an in vitro binding assay. After incubation of the purified tubulin with these mutated CRMP5-GST constructs (Fig. 2 D, top panel), tubulin was found to bind strongly to the full-length CRMP5 (amino acids 1-564), N-terminal-deleted mutants ΔN244 (245-564) and ΔN474 (475-564), but not to the C-terminal-deleted mutant ΔC472 (1-471). The most C-terminal 90 aa fragments of CRMP5 (475-564) coprecipitated with tubulin more strongly than the full-length form and any of the other mutants (Fig. 2 D). The same binding pattern could be observed when the above pull-down assays with GST-CRMP5 were performed using a lysate from brain cortex instead of purified tubulin (Fig. 2 D, bottom panel), thus ensuring that the identified tubulin-binding site was also valid in vivo. To determine the shortest effective fragment for tubulin binding, we generated two GST-CRMP5 additional fragments, 475–522 and 523–564, which were immobilized on beads and incubated with purified tubulin. Fragment 523-564 failed to pull down the tubulin, whereas fragment 475–522 did (Fig. 2 E). These data demonstrate that the tubulin-binding site on CRMP5 was located in a 48 aa fragment in the C-terminal region within residues 475-522. A recent study identified a tubulin-binding site within residues 480-509 of the CRMP2 sequence (Chae et al., 2009). Comparison between the CRMP5 and CRMP2 sequences within the region encompassing tubulin binding indicated that, despite a strong sequence identity between CRMP5 and CRMP2 on both sides of the identified site, only a weak sequence homology within the identified site could be observed (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Figure 2.
CRMP5 interacts directly with tubulin within C-terminal residues 475-522. A, Pull down of purified tubulin protein (40 μg) with GST or GST-CRMP5 (10 μg) conjugated with Sepharose beads. Western blots were probed by anti-α- or anti-β-tubulin antibodies. Tubulin subunits were precipitated by GST-CRMP5, but not GST, indicating a direct interaction between CRMP5 and tubulin. B, Diagram depicting CRMP5 truncation mutants. C, SDS-PAGE showing each GST-CRMP5 truncated protein confirms the correct mass for recombinant proteins. The bars on the left indicate the positions of molecular markers. The arrows on the right indicate the apparent molecular mass of each GST-tagged mutant. D, Pull down of purified tubulin (top panel) or tubulin from mouse brain extract (bottom panel) incubated with different truncated GST-CRMP5 recombinant proteins, shown in C. Western blots of the pellets, probed with an anti-α-subunit antibody, show that tubulin, purified or present in the brain extract, interacts with all truncated forms of CRMP5 except CRMP5 ΔC472. The presence of the C-terminal residues 475-564 is sufficient to ensure tubulin pull down. Tubulin is present in the supernatant fraction of all assays. E, Pull down of tubulin with the shorter fragments of recombinant GST-CRMP5. The shortest fragment, which precipitates tubulin, is encompassed by residues 475-522 of CRMP5.
To gain insight into the spatial organization of this site, the structure of CRMP5 (residues 8-483) was modeled using DHPase as a template and the structure of the C-terminal part (residues 484-552) was predicted as an “intrinsically unstructured protein” using the NORS prediction program. Within the tubulin-binding site, residues 475-478 formed the most C-terminal part of the last helix, whereas the following residues (484-522) seemed to be unstructured (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) giving the molecule a malleable and flexible structural state, and importantly facilitating its interaction with other proteins. This region seems to be surface exposed.
CRMP5 acts as an inhibitor of neurite outgrowth and formation mediated by tubulin binding
Since one of the functions reported for some CRMPs is the promotion of neurite outgrowth mediated by tubulin binding (Fukata et al., 2002), we examined the effect of CRMP5 on neurite outgrowth activity by transfecting neuroblastoma cells (N1E-115) with the FLAG-CRMP5 expression vector. In parallel, we performed transfection with CRMP2, which has been reported to promote neurite outgrowth (Suzuki et al., 2003; Rogemond et al., 2008). After immunostaining with the anti-FLAG antibody, the lengths and numbers of neurite-like extensions were compared with the control EGFP-transfected cells. CRMP2-transfected cells showed, as expected, polygonal shapes presenting longer neurites >25 μm (Fig. 3 A). The CRMP5-expressing cells (n = 206) showed mostly an overall round shape. They clearly displayed less neurite elongation (3.5-fold) and also reduced the number of neurites per cell (2-fold) compared with the control EGFP-expressing cells (**p < 0.01), as shown in Figure 3, D and E. For additional examination of CRMP5 function, we depleted the CRMP5 protein in N1E-115 cells, by siRNA interference. The effect of siRNA against CRMP5 (siCRMP5) was first confirmed by immunoblot analysis (Fig. 3 B). The immunostaining of endogenous CRMP5 in cells treated with the scrambled siRNA (SC, n = 150 cells) showed a clear green staining indicative of CRMP5 expression, whereas the absence of green staining in siCRMP5-treated cells (n = 100 cells) revealed that CRMP5 expression was totally impaired (Fig. 3 C, top panels). To evaluate neurite length, the cells were also stained by phalloidin (red). Knockdown of CRMP5 (Fig. 3 C, bottom panels; D,E) drastically increased not only the neurite elongation (3.5-fold) but also the number of neurites per cell (3-fold) compared with SC (***p < 0.005). To confirm that these effects were attributable to CRMP5 depletion, we performed rescue experiments by overexpressing EGFP-CRMP5 in siCRMP5-transfected cells showing strong level of CRMP5 expression (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). In this case, the negative effect of CRMP5 on the elongation and number of neurites was rescued by restoring the expression level of CRMP5, since the N1E-115 cells again presented an overall round shape (Fig. 3 C, right panel; D,E). Together, these data suggested that CRMP5 is involved in neurite retraction as well as neurite formation in N1E-115 cells.
Figure 3.

CRMP5 induces inhibition of neurite outgrowth and formation in N1E-115 cells. A, Cells were transfected with EGFP alone (top panel) or with FLAG-CRMP5 (middle panel) and FLAG-CRMP2 (bottom panel), and then labeled by anti-FLAG antibody and observed by fluorescent microscopy. The nucleus is stained blue with DAPI. Although N1E-115 cells transfected with EGFP present a polygonal shape and exhibit some neurite extensions, cells expressing CRMP5 present round cell morphologies. As reported, cells expressing CRMP2 clearly exhibit longer neurite extensions. The typical pattern of immunostaining is shown in high-magnification images in three right panels. Scale bar, 50 μm. B, Knockdown of CRMP5 expression by siCRMP5. N1E-115 cells were transfected or not with scrambled siRNA (SC) or siCRMP5, and CRMP5 expression was probed by anti-CRMP5 antibody. The GAPDH antibody is used as loading control. Note the complete depletion of CRMP5 expression after siCRMP5 treatment. C, CRMP5 depletion induces long neurite. Endogenous CRMP5 was detected by immunostaining with anti-CRMP5 antibody in SC-treated cells, whereas CRMP5 labeling was absent from siCRMP5-treated cells (top panels), detected by DAPI staining. A double staining with phalloidin, shows unambiguously the presence of longer neurite in siCRMP5-treated cells compared with SC cells (bottom panel). The CRMP5 inhibition of the neurite outgrowth is rescued by overexpression of CRMP5 in siCRMP5-treated cells (right panel). Scale bar, 50 μm. D, E, Quantification of neurite length and number after overexpression or knockdown of CRMP5. D, Comparison of neurite outgrowth between N1E-115 cells expressing EGFP (n = 77), overexpressing CRMP2 (n = 248) or CRMP5 (n = 206), or transfected with siCRMP5 (n = 100) or SC (n = 157). Overexpression of CRMP5 leads to a significant decrease in neurite outgrowth initiation when compared with controls, whereas CRMP2 promotes neurite extension; however, knockdown of CRMP5 induces an increase in neurite length when compared with cells treated with the control SC. E, Comparison of neurite numbers per cell after overexpression or knockdown of CRMP5 as for D. Overexpression or knockdown of CRMP5 generates a decrease or increase in the neurite number per cell, respectively. Data show the mean values of four individual experiments, expressed as cells bearing neurite lengths >25 μm (**p < 0.001; ***p < 0.005). Error bars indicate SEM.
To determine the impact of the tubulin-binding region of CRMP5 on its inhibitory function, we examined the neurite-like extensions, in NGF-stimulated PC12 cells expressing different EGFP-deleted constructs of CRMP5. Since the average length of neurite extension after 48 h of NGF induction was <10 μm in PC12 cells (Fontaine-Lenoir et al., 2006), this limit was chosen for the evaluation of neurite length. Confocal analysis showed that, compared with GFP-expressing cells, the cells expressing full-length CRMP5 (n = 157 cells) exhibited a lower percentage (9% instead of 30%) of neurite lengths exceeding 10 μm (**p < 0.01) but a higher percentage (36% over 27%) of extensions <10 μm (Fig. 4 A,B). Similarly, only a small percentage of the cells bearing neurites >10 μm in length could be observed among cells expressing N-terminally truncated CRMP5 (ΔN244), bearing the tubulin-binding domain (n = 95 cells), and the CRMP5 construct ΔC518 (n = 136 cells), bearing only a few additional residues responsible for tubulin binding (Fig. 4 A,B). These cells also display an overall round morphology (73–75%). In contrast, cells expressing the CRMP5 construct ΔC472, lacking the tubulin-binding domain (n = 93 cells), exhibited polygonal shape and induced neurite extensions whose lengths were comparable with EGFP-expressing cells (Fig. 4 A, bottom panels). Moreover, the number of neurite-like extensions per cell also decreased when cells expressed constructs bearing the tubulin-binding site, and increased to control levels when cells expressed the ΔC472 construct lacking this site (Fig. 4 C). It should be noted that the latter form was present in the nucleus as well as in the cytosol. These data strengthen the hypothesis that the tubulin-binding region of CRMP5 is responsible for its inhibitory function in neurite outgrowth and formation.
Figure 4.
Tubulin binding is required for the inhibition of neurite outgrowth by CRMP5. A, PC12 cells transfected by the CRMP5 EGFP-tagged constructs were stimulated 8 h later with NGF for 36 h, and then fixed and observed by confocal microscopy. Cells transfected with full-length CRMP5, or truncated mutant ΔN244 and ΔC518 constructs, bearing the tubulin-binding site, show rather round shapes and retracted extensions. However, when the tubulin-binding site is deleted (CRMP5 ΔC472), the cells fail to retract and remain spread. Scale bars, 20 μm. B, Quantification of neurite length in PC12 cells from A. The percentage of round-shaped cells or cells bearing neurite lengths greater than or less than 10 μm is shown for each construct. Below the graph is a diagram of truncated CRMP5 forms with the tubulin-binding site shown by a red box. The presence of a tubulin-binding site is necessary for neurite retraction activity. Cells expressing full-length CRMP5, ΔN244, or ΔC518 (n = 157, 95, and 136 cells, respectively) show a high percentage of round shapes (∼54–75%) and a low percentage exhibiting neurite extensions >10 μm (∼6–9%). In contrast, in CRMP5 ΔC472-expressing cells (n = 93), the percentage of round shape is lower (36%) and cells with neurite extension >10 μm are more abundant (25%). Between 19 and 36% of all cells have neurites < 10 μm. Data are the mean values of three individual experiments. **p < 0.001 versus control EGFP value. C, Quantification of neurite number per cell. Overexpression of different forms of EGFP-CRMP5 bearing the tubulin-binding site lead to a decrease in the number of neurites per cell (20–70%), whereas overexpression of CRMP5 ΔC472 has no effect (*p < 0.05; **p < 0.001). Error bars indicate SEM. D, The inhibitory effect of truncated recombinants CRMP5 ΔC521 on tubulin polymerization in vitro. Microtubule assembly was monitored at 340 nm at 37°C and recorded every 30 s. The quantity of microtubules formed is proportional to the increase in absorbance. Polymerization of purified tubulin protein (3 mg/ml) was recorded either without (a) or with 4.3 μm CRMP5 ΔC521 (b), CRMP5 ΔC472 (c), or 4 μm BSA (d) for 20 min. Data represent means for four independent observations. CRMP5 ΔC521 inhibits microtubule assembly, whereas CRMP5 ΔC472 and BSA have no effect.
We next examined the effect of CRMP5 in hippocampal neurons from E18 mouse embryos transfected with EGFP-CRMP5 on day 4 after plating, and immunostained 2 d later at DIV 6, which corresponded to stage 4 of hippocampal neuron development in culture. This stage corresponded generally to the establishment of distinct dendritic domain in cultured neurons (Dotti et al., 1988). Control EGFP neurons showed one fast-growing axon and shorter neurites considered as dendrites, although immature, at this stage (see Fig. 6 A). Double staining of the cells using antibodies to the axonal marker anti-tau1 and the somato-dendritic marker anti-MAP2 confirmed that the long neurite was the axon and the other neurites were dendrites (see Fig. 6 B). CRMP5 transfection did not significantly change axon length and branching but reduced by 20% the length of dendritic processes (n = 42 neurons). Consistent with the above data, transfection with CRMP5 ΔC518, bearing the tubulin-binding domain, reduced dendrite length by 40% (*p < 0.05), whereas the ΔC472 mutant, devoid of this domain, had no effect on dendrite length (see Fig. 6 A,C). These data suggest that the tubulin-binding region of CRMP5 is necessary for the inhibition of dendrite growth. Although CRMP2 enhanced the number of dendrites, CRMP5 showed no effect on dendrite formation at stage 4 since the number of dendrites per neuron is comparable with the control (see Fig. 6 D). The absence of effect on dendrite formation may be explained by the fact that at stage 4 the dendritic domains were established.
Figure 6.

CRMP5 counteracts CRMP2 neurite outgrowth promotion in a culture of hippocampal neurons at DIV 6 by a tubulin-based mechanism. A, E18 hippocampal neurons were cultured for 4 d, and then transfected with Lipofectamine and fixed after 2 d before antibody labeling. Transfections were performed with EGFP, EGFP-CRMP5, or FLAG-CRMP2 constructs. CRMP5 expression inhibits the length of shorter neurites (dendrites; white arrow), whereas the longest neurite (axon; white arrowhead) lengths are comparable with control EGFP. Consistent with previous results (Fukata et al., 2002), the overexpression of CRMP2 alone induces significant elongation of processes. B, Hippocampal neurons were stained with dendritic marker MAP2 (red) and axon marker tau-1 (blue) antibodies to confirm that, at DIV 6, the shorter neurites are dendrites and the longest neurite is an axon. C, Quantification of process lengths. The length of the longest neurite (axon) and all remaining shorter neurites (dendrites) of an individual neuron were measured within each treatment. Neurons expressing full-length CRMP5 (n = 42 neurons; gray bars) and CRMP5 ΔC518 (n = 38 neurons; dotted bars) show axon lengths identical with the control EGFP (n = 55 neurons), whereas they present shorter dendrite lengths. CRMP5 ΔC472 (hatched bars) has no effect on dendrite length. As reported previously (Fukata et al., 2002), CRMP2 enhances axon and dendrite lengths (n = 63 neurons). In neurons coexpressing full-length CRMP5 or CRMP5 ΔC518 and CRMP2 (n = 46 and 28 neurons, respectively), both axon and dendrite lengths decrease to the level of cells expressing CRMP5 alone, suggesting that CRMP5 negates the CRMP2 effect. In neurons coexpressing truncated CRMP5 Δ472 and CRMP2 (n = 60 neurons), axon and dendrite lengths reach those of CRMP2-expressing cells, indicating that when the tubulin-binding site is deleted, CRMP5 fails to counteract the CRMP2 activity (**p < 0.01; ***p < 0.005). D, Quantification of the number of dendrites. At DIV 6, neurons expressing CRMP5 exhibit a dendrite number identical with control EGFP, whereas neurons expressing CRMP2 show higher number of dendrite (*p < 0.05). E, Quantification of the number of secondary axons and the frequency of axon branching. The second longest neurite exhibiting no less than one-half the axon length (white bars) is considered as a secondary axon. CRMP5-expressing neurons, like EFGP-transfected neurons, do not exhibit secondary axon, whereas CRMP2 induces secondary axons. CRMP5 has no significant effect on the frequency of axon branching (gray bars), whereas CRMP2 increases axon branching. The cotransfection of both CRMP5 and CRMP2 results with an intermediate value suggests that CRMP5 does not act as a dominant signal (*p < 0.05; **p < 0.01; ***p < 0.001). Error bars indicate SEM. F, Double transfection was performed with FLAG-CRMP2 and EGFP-CRMP5 or EGFP-CRMP5 Δ472, EGFP-CRMP5 Δ518. Double-transfected cells were selected among yellow-colored cells in the merge image. Neurons coexpressing both CRMP5 (full-length or ΔC518) and CRMP2 present process lengths comparable with CRMP5 but shorter axon and dendrites lengths compared with CRMP2-expressing neurons. When the tubulin-binding site on CRMP5 is deleted (ΔC472), its coexpression with CRMP2 leads to cell morphologies similar to CRMP2, suggesting that the dominant effect of CRMP5 depends on the presence of the tubulin-binding site. Scale bars, 100 μm.
To determine whether the tubulin–CRMP5 interaction exerted an influence on tubulin polymerization, we performed an in vitro assay to evaluate the capacity of purified tubulin to polymerize into microtubules, in the presence or absence of recombinant CRMP5 protein. A truncated form of CRMP5 (ΔC521) was overexpressed since, as indicated above, this is the most active form in neurite retraction. Purified control tubulin started to polymerize in the presence of GTP after a time lag of ≈60 s, with a steep initial slope during the first 7 min, followed by an increase up to a variation of optical density (ΔOD) of 0.47 after 15 min (Fig. 4 D). The addition of truncated recombinant CRMP5 before the polymerization step delayed tubulin polymerization, since the initial rate of microtubule formation was slower and the ΔOD was 34% lower than the control level. Moreover, even after prolonged incubation, the OD did not reach a plateau level (Fig. 4 D). To ascertain the specific effect of the CRMP5 tubulin-binding site, a polymerization assay was performed in the presence of CRMP5 ΔC472, which showed very similar tubulin polymerization to control tubulin. A similar pattern of microtubule formation was observed in the presence of bovine serum albumin, as a control, with a plateau reached at a ΔOD of 0.45 (Fig. 4 D).
CRMP5 inhibition of neurite outgrowth is not counteracted by CRMP2 expression
To evaluate the effect of CRMP5 when CRMP2 was present, we cotransfected N1E-115 cells with both GFP-CRMP5 and FLAG-CRMP2 expression vectors, cells coexpressing both proteins being identified by their yellow color (Fig. 5 A). A small percentage of CRMP5-expressing cells (4%) showed neurite lengths >25 μm (**p < 0.01), whereas this percentage was higher (20.4%) for CRMP2-expressing cells (Fig. 5 B). Remarkably, cells coexpressing both CRMP2 and CRMP5 (n = 79) presented short neurite-like extensions, since only 5.3% displayed neurite lengths >25 μm (Fig. 5 A, top panels; B). Strikingly, 70% of double-transfected cells exhibited rather round shapes. These data suggest that CRMP5 acts as a dominant signal to counteract the neurite outgrowth promotion activity of CRMP2. The double transfection of cells with CRMP2 and truncated forms of CRMP5 harboring the tubulin-binding site resulted in a low percentage of cells exhibiting long neurites, whereas double transfection with CRMP2 plus CRMP5 ΔC472, devoid of the tubulin-binding site, resulted in a high percentage of cells (24%) displaying extensions longer than 25 μm (***p < 0.005) (Fig. 5 A, bottom panels; B). These results suggest that the abrogation of neurite outgrowth function of CRMP2 by CRMP5 is also dependent on the tubulin-binding capacity of CRMP5.
Figure 5.

CRMP5 neurite outgrowth inhibition abrogates the CRMP2 promotion function in N1E-115 cells. A, Cells were cotransfected with GFP-CRMP5 and FLAG-CRMP2 constructs, labeled with anti-FLAG antibody (red), and observed with fluorescent microscopy. Only the cells transfected with both vectors are yellow in the merge image. Note that the cells transfected with CRMP2 alone, which remained red, exhibited some neurite-like extensions. Cells cotransfected with full-length CRMP5, ΔN244, or ΔC518 and CRMP2 clearly show a round morphology. Cells double transfected with CRMP5 ΔC472 and CRMP2 show a polygonal shape. Scale bars, 50 μm. B, Quantification of cells from A exhibiting neurite lengths >25 μm. CRMP5 decreases, but CRMP2 increases the percentage of cells bearing longer neurite lengths. In the presence of both CRMPs, the cells (n = 79) show rather a low percentage of longer neurites, indicating the dominant effect of CRMP5 when bearing the tubulin-binding site. However, when CRMP5 ΔC472 and CRMP2 are overexpressed, the percentage of cells with longer neurite lengths reaches the value of CRMPP2 expression alone, suggesting that the antagonizing effect of CRMP5 is exerted through a tubulin-based mechanism (**p < 0.001; ***p < 0.0005). Error bars indicate SEM.
We next examined the effect of CRMP5 relative to CRMP2 in hippocampal neurons transfected with EGFP-CRMP5 and/or FLAG-CRMP2, and immunostained at DIV 6, as mentioned above. In accordance with previous reports (Fukata et al., 2002), hippocampal neurons expressing CRMP2 showed a significant increase in both axon length and the frequency of axonal branching with the cells sometimes displaying secondary axons (Fig. 6 A,C,E). Detailed dendrite length measurements showed that CRMP2 increased the length of dendrites 1.4-fold compared with control (Fig. 6 C). Strikingly, when CRMP2 and full-length CRMP5 were coexpressed (n = 46 neurons), axon elongation and dendrite length decreased to the level of CRMP5 expression alone (***p < 0.001), strengthening the hypothesis that CRMP5 abrogates CRMP2 promotion of neurite outgrowth in a dominant manner (Fig. 6 C,F). However, after close examination, the dominant effects of CRMP5 were more pronounced in dendrites than in axons. In fact, although there was less axon branching and fewer secondary axons in double-transfected compared with CRMP2-expressing neurons, they reached a level situated somewhere between CRMP2 and CRMP5 expression alone (Fig. 6 E). To elucidate whether the tubulin-binding domain of CRMP5 was involved in the inhibition of CRMP2 function, hippocampal neurons were cotransfected with CRMP2 and different truncated forms of CRMP5 (Fig. 6 F). Importantly, neurons cotransfected with CRMP2 and ΔC518 CRMP5, containing the tubulin-binding domain, reduced dendrite and axon length to the level of CRMP5 expression alone, indicating that this form abrogated the CRMP2 promotional outgrowth of axon and dendrites. On the contrary, neurons cotransfected with CRMP2 and CRMP5 ΔC472, lacking the tubulin-binding domain (n = 60 neurons), showed significant increases in axon length and dendrite outgrowth (***p < 0.0005) approximately equal to the level of CRMP2 expression alone (Fig. 6 A,C,F), indicating that when the tubulin-binding domain was missing, CRMP5 was unable to counteract CRMP2 function. These data suggest that CRMP5 abrogates the neurite outgrowth promotional activity of CRMP2 at both the dendrite and axon levels, and this effect seems to be mediated by the tubulin-binding capacity of CRMP5 in hippocampal neurons.
The role of microtubule in the extension of neuronal process has been previously well established (Yamada et al., 1970). More recently, the interaction of tubulin with CRMP2 was reported to promote the axon growth (Fukata et al., 2002). Therefore, to examine the molecular mechanisms that generate the antagonistic effect of CRMP5 on CRMP2, in vitro binding assays were performed to elucidate whether CRMP5 had an effect on the tubulin–CRMP2 interaction. This interaction was assessed using recombinant GST-CRMP2 and purified tubulin in the presence of increasing concentrations of purified full-length or truncated forms of CRMP5, either ΔC472 or a newly designed one ΔC521, generated as His-CRMP5 (Fig. 7 A). As expected, purified tubulin was pulled down by GST-CRMP2 (Fig. 7 B). When GST-CRMP2 and purified tubulin were subsequently incubated with either full-length or CRMP5 ΔC521, the binding of tubulin to CRMP2 decreased with increasing amounts of CRMP5 (Fig. 7 B), indicating that CRMP5 impaired the tubulin–CRMP2 interaction. The antagonizing effect of ΔC521 was more pronounced than the full-length CRMP5 (Fig. 7 B), probably because of the greater exposure of the tubulin-binding site because of the truncation of the C terminus in the former. This effect is specific for CRMP5, since the presence of large amounts of bovine serum albumin during the incubation did not influence the tubulin-CRMP2 interaction (data not shown). However, even at high concentrations, CRMP5 loss-of-function mutant ΔC472 lacking the tubulin-binding site could not counteract CRMP2 binding, since CRMP2 still bound to tubulin (Fig. 7 B). These data suggest that direct tubulin binding to CRMP5 impairs the binding of tubulin to CRMP2. Nonetheless, as CRMP proteins can form homocomplex and heterocomplex between isoforms (Wang and Strittmatter, 1997; Deo et al., 2004; Stenmark et al., 2007), the hypothesis that a direct CRMP5 binding to CRMP2 may slow down its ability to process neurite outgrowth cannot be totally excluded. To check this hypothesis, we performed in vitro binding assays, in the absence of tubulin, using recombinant GST-CRMP2 and various concentrations of either full-length, ΔC521, or ΔC472 forms of CRMP5. The control GST did not bind to CRMP5. Different concentrations of all CRMP5 isoforms, including ΔC472, were pulled down by GST-CRMP2 (Fig. 7 C), indicating that these forms interacted with CRMP2. Nevertheless, the fact that CRMP5 ΔC472, which did not counteract CRMP2 promotional activity still bound to CRMP2, indicates that the direct CRMP5/CRMP2 interaction does not correlate with CRMP5 inhibitory effect. Together, the above data suggest that, although CRMP5/CRMP2 oligomerization does occur, however only the direct CRMP5 binding to tubulin by preventing tubulin/CRMP2 interaction can antagonize CRMP2 promotional effect.
Figure 7.

CRMP5 antagonizes the tubulin–CRMP2 interaction. A, SDS-PAGE showing each His-CRMP5 truncated protein and GST-CRMP2 protein confirms the correct mass for recombinant proteins. The bars on the left indicate the positions of molecular marker bands. B, Purified tubulin incubated with GST-CRMP2 recombinant protein or GST, pulled down in the presence of different concentrations (0–1.3 μm) of either full-length or truncated CRMP5 (ΔC521 or ΔC472), shown in A. Western blots of the pellets, probed with an anti-α-subunit antibody, shows that CRMP2 precipitates the tubulin, in the absence or the presence of low concentrations (<0.5 μm) of all forms of CRMP5. Higher concentrations of full-length or ΔC521 CRMP5 decrease or prevent the tubulin pull down by GST-CRMP2, whereas CRMP5 ΔC472 lacking the tubulin-binding site did not prevent it. Tubulin is present in the supernatant fraction of all assays. Control GST, incubated with the highest concentration of all CRMP5 forms, failed to precipitate the tubulin. C, GST-CRMP2 or GST was pulled down in the absence or presence of different concentrations (0–1.3 μm) of either full-length, or truncated His-tagged CRMP5 (ΔC521 or ΔC472), shown in A. Western blots of the pellets, probed with an anti-His antibody (CRMP5 detection), shows that CRMP2 precipitates all different forms of CRMP5. Control GST failed to precipitate different forms of CRMP5.
CRMP2 outgrowth promotion is enhanced in CRMP5-deficient neurons
A few days after axon has began its formation and rapid growth, remaining processes elongate and acquire the characteristic of dendrite (Dotti et al., 1988). As significant dendrite elongation begins between days 3 and 5, to assess the function of endogenous CRMP5 on dendrite outgrowth, at an early stage of development, we knocked down the CRMP5 expression in hippocampal neurons, by siCRMP5 on day 1 after plating, and then neurons were examined 2 d after transfection at DIV 3. First, the effect of CRMP5 expression was examined at early stage of development in hippocampal neurons from E18 mouse embryo transfected with EGFP-CRMP5. Compared with control EGFP expression (n = 52 neurons), the expression of EGFP-CRMP5 (n = 37 neurons) significantly reduced the lengths (**p < 0.01) and numbers (***p < 0.001) of neurites per cell considered as primary dendrites. Then, we studied the consequence of CRMP5 depletion by its knockdown with siCRMP5. The knockdown was confirmed by immunoblot analysis (Fig. 8 A). In CRMP5-depleted neurons (n = 45), the length and number of primary dendrites were unambiguously higher (**p < 0.01) than the control SC neurons (n = 25), indicating that in vivo, CRMP5 exerted an inhibitory effect on dendrite outgrowth and initiation at an early stage of the development (Fig. 8 A,B). A double staining of the neurons with anti-tau1 and anti-MAP2 antibodies confirmed that the long neurite was the axon and the other neurites were dendrites (Fig. 8 D). The reexpression of CRMP5 in neurons treated with siCRMP5 confirmed that the negative effect of CRMP5 was rescued by the expression of CRMP5 (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). As observed for neurons at DIV 6, CRMP5 overexpression, at DIV 3, did not have any significant effect on axon length (Fig. 8 B). However, endogenous depletion of CRMP5 slightly, but not significantly, increased the axon length compared with SC-expressing neurons (Fig. 8 B). Because CRMP5 overexpression antagonized the neurite outgrowth promotion induced by CRMP2 expression, we asked whether the knockdown of endogenous CRMP5 had an effect on the CRMP2 induced outgrowth. Remarkably, in neurons knocked down for CRMP5 expression but overexpressing the CRMP2 construct (n = 28 neurons), the length and numbers of primary dendrites were higher (**p < 0.01) than in neurons expressing CRMP2 and SC (n = 32 neurons), indicating an antagonizing effect of endogenous CRMP5 at the dendritic level (Fig. 8 C,E). The negative effect of CRMP5 on dendrite growth and formation was rescued by restoring CRMP5 expression (Fig. 8 E). These results strongly support the inhibitory effect of endogenous CRMP5 on both the outgrowth and formation of primary dendrite counterbalancing the outgrowth promotion activity of CRMP2. Compared with cultured hippocampal neurons at DIV 3, CRMP2 expression in the presence of SC (n = 40 neurons) increased by 1.3-fold the axon length. Interestingly, knockdown of CRMP5 in hippocampal neurons expressing CRMP2 (n = 37 neurons) increased even more (1.5-fold; ***p < 0.005) axon length and enhanced (3-fold) the number of multiple secondary axons (**p < 0.01), with neurons presenting complicated multibranched axon trees (Fig. 8 C,F). The overexpression of CRMP5 in CRMP2 plus siCRMP5-transfected neurons brought down the axon length to that of the control (Fig. 8 F), strengthening the argument that endogenous CRMP5 controlled the axon growth effect of CRMP2. However, it should be noticed that CRMP5 did not exert an inhibitory effect on axon growth since axon length reached the level of control. Together, our data clearly confirmed the role of endogenous CRMP5 at early developmental stages, in preventing dendrite growth and formation and in the regulation of CRMP2 promotional growth.
Figure 8.

Effect of CRMP5 depletion on dendrite and axon outgrowth in cultured hippocampal neurons at DIV 3. A, E18 hippocampal neurons were cultured for 1 d, and then transfected with EGFP or EGFP-CRMP5 or cotransfected with EGFP and SC or siCRMP5. The knockdown of CRMP5 expression by siCRMP5 in cultured neurons is shown by Western blots probed with anti-CRMP5 antibody. Overexpression of EGFP-CRMP5 at DIV 3 inhibits neurite growth (arrow), whereas the length of the longest neurite, the axon (arrowhead) is comparable with the control EGFP. CRMP5 depletion considerably enhances the length and the number of short neurites, without effect on axon length, whereas the control neurons transfected with SC show phenotypes comparable with EGFP-transfected neurons. The bottom panels show dendrites and soma under high magnification. Scale bars, 100 μm. B, Quantification of process lengths and numbers shown in A. At DIV 3, neurons expressing CRMP5 (n = 37) show axonal lengths identical with the control EGFP (n = 52 neurons), whereas neurons depleted of CRMP5 (n = 45) show a very slight increase in axon length (red bars) compared with SC-treated neurons (n = 62). However, neurite lengths (yellow bars) and numbers (green bars) are decreased with CRMP5 expression, but increased with CRMP5 depletion when compared with SC (**p < 0.01; ***p < 0.001). C, CRMP2 activity is enhanced by CRMP5 depletion. E18 hippocampal neurons were cultured for 1 d, and then cotransfected with FLAG-CRMP2 plus siCRMP5 or SC, as control, for 2 d before immunostaining with anti-FLAG antibody. Depletion of CRMP5 allows an enhanced effect of CRMP2 on dendrite length and number as well as on axon growth and branching, indicating that endogenous CRMP5 restricts CRMP2 functions. Dendrites are shown under high magnification in bottom right panels. Scale bars, 100 μm. D, Immunostaining of cultured hippocampal neurons at DIV 3 with dendritic marker MAP2 and axon marker tau-1 antibodies. E, Quantification of process lengths and numbers shown in C. At DIV 3, neurons cotransfected with FLAG-CRMP2 and siCRMP5 (n = 28 neurons) show an increase in the promotional activity of CRMP2 on neurite length (yellow bars) and numbers (green bars) when compared with neurons cotransfected with CRMP2 and SC (n = 31 neurons). However, when CRMP5 is overexpressed in the above cotransfected neurons (n = 15), the CRMP2 effect is completely abolished, leading to an inhibition of neurite growth and number similar that of CRMP5 expression indicated in B. F, Quantification of axon length and multiple axon, shown in C. CRMP5 knockdown in cultured neurons at DIV 3 expressing FLAG-CRMP2 amplifies the CRMP2 effect on axon growth when compared with control SC, whereas CRMP5 overexpression abolishes this effect (**p < 0.01). In addition, as previously described (Fukata et al., 2002), we observed that CRMP2 induces a multiple axon formation (n = 40 neurons), but the knockdown of CRMP5 (n = 37 neurons) leads to a significant increase (3-fold; **p < 0.01) in neurons bearing multiple axons, indicating that endogenous CRMP5 may exert an effect on delimiting CRMP2 promotion of axon growth. Data are the mean values of three individual experiments. Error bars indicate SEM.
CRMP5 expression is not persistent in hippocampal neurons
Because CRMP5 inhibited dendrite outgrowth and modulated CRMP2 activity, we tested whether these effects were correlated with the CRMP5 level of expression in dendrites by studying the spatiotemporal distribution of CRMP5. The subcellular distribution of CRMP5 had previously been studied, in cultures of hippocampal neurons by Hotta et al. (2005), but only at DIV 1 and 7, with a focus on the growth cone accumulation of CRMP5. Therefore, we studied the endogenous expression of CRMP5, in cultured hippocampal neurons at DIV 1–6, and compared it with endogenous CRMP2 expression and MAP2 by double staining using different combination of anti-CRMP5, anti-CRMP2, and anti-MAP2 antibodies (Fig. 9 A,B). Green fluorescence intensities representing CRMP5 expression, determined from selected neurons, were shown as a three-dimensional intensity projection (Fig. 9 A, left panels). Fluorescence intensities, indicative of CRMP5, CRMP2, and MAP2 expression, were determined from defined region on dendrites and axon using imaging software (ImageJ) and graphed as fluorescence values (Fig. 9 D). The precise length of dendrites and axon at each day of culture was determined, specifically in the absence of CRMP2 and CRMP5 expression, using CellTracker Green, CMFDA, 5-chloromethyl fluorescein diacetate (supplemental Fig. S5, available at www.jneurosci.org as supplemental material). Minor CRMP5 and MAP2 expressions could be detected only in soma after 1 and 2 d of culture (Fig. 9 A), whereas CRMP2 expression in neurites was clearly detected since day 2 of culture (38 ± 12 AU) (Fig. 9 B,D). At DIV 3, one of the immature neurite, showing high CRMP2 expression (46.8 ± 9.6 AU) elongated rising 332 μm (±50 μm) in length to become the future axon (Fig. 9 B,D), at the same time a strong expression of endogenous CRMP5 was detected in the soma and primary dendrites (37.1 ± 6.9 AU) presenting a colocalization with MAP2 and CRMP2 (Fig. 9 A,B). In contrast to axon, the dendrite length did not change at DIV 3 (30 ± 6.7 μm instead of 28 ± 6.5 μm at DIV 2) (Fig. 9 D). Accordingly, the MAP2 expression in dendrite, considered as an indication of dendrite outgrowth, remained low (Fig. 9 D, middle panel). This suggested that high CRMP5 expression during axonogenesis maintained the primary dendrites inhibited. CRMP5 was also distributed in the axon, but to a lesser extent (10.5 ± 2.6 AU) (Fig. 9 D), whereas CRMP2 expression was almost equivalent in axon and primary dendrites. During following stages of neuronal development, corresponding to dendrite outgrowth (Fig. 9 D, top panel), a decrease in CRMP5 expression was observed in dendrites (17.7 ± 4.6 AU) at DIV 4, followed by a very low expression level at DIV 5 (2.1 ± 1 AU), at which an absence of CRMP5 staining was noticed in dendrites (Fig. 9 A,B; supplemental Fig. S5, available at www.jneurosci.org as supplemental material), whereas the CRMP2 expression remained almost constant (Fig. 9 D, bottom panel). On the opposite, a strong labeling was observed with MAP2, indicating the dendrite outgrowth, whose length reached at DIV 5 50 ± 10 μm (Fig. 9 D, top panel), which perfectly matched with that previously reported (Dotti et al., 1988). It should be noted that MAP2 expression increased consistently in dendrites from DIV 1 to 6 (from 1.6 ± 1.3 to 37.5 ± 6 AU) (Fig. 9 D). In accordance with a previous report (Hotta et al., 2005), at DIV 6, a wide redistribution of CRMP5 was observed within hippocampal neurons, including soma, dendrites, and axon. Although CRMP5 expression was very low in axon at the beginning, it became very high (38.7 ± 10.7 AU) at this stage of development (Fig. 9 A,D), suggesting a role for CRMP5 in controlling the axon elongation at later stages of development. At DIV 6, CRMP5 was also observed de novo, at the dendritic level, however, to a lesser extent than at DIV 3 (14.1 ± 3.3 AU) (Fig. 9 D), whereas CRMP2 expression in dendrites and axon remained at high level, suggesting that, in the absence of CRMP5, CRMP2 promoted dendrite outgrowth. The expression level of both CRMP5 and CRMP2 in hippocampal neurons was further examined by Western blot analysis of a lysate of neurons from each day of culture. Figure 9 C showed that the band migrating at 66 kDa recognized by anti-CRMP5 antibody presented high intensity at DIV 3, its intensity progressively decreased to reach a very low level at DIV 5, but at DIV 6 this band showed again very high intensity. The higher intensity at DIV 6 compared with DIV 3 might be explained by the fact that, as mentioned above, the CRMP5 expression was increased at the axon level at DIV 6. The band recognized by anti-CRMP2 antibody, although exhibiting low intensity at DIV 1–2, showed similar high intensity from DIV 3 onward, with the highest intensity reached at DIV 5 at which multiple isoforms of CRMP2, previously described (Rogemond et al., 2008), could be visualized (Fig. 9 C). These results, in perfect agreement with the above data on fluorescent quantification of CRMP5 and CRMP2 expression, demonstrate the transient expression of CRMP5. Together, our data demonstrated that a correlation might exist between CRMP5 expression and its inhibitory effect on dendrite growth during axonogenesis; subsequently, its absence of expression at DIV 5 of neuronal development allowed dendrites to grow, suggesting a role for CRMP5 in maintaining them in a quiescent state at early stages of development, thus regulating the establishment of neuronal polarity.
Figure 9.

Transient spatiotemporal expression of CRMP5 compared with CRMP2 expression in cultured hippocampal neurons. A, E18 hippocampal neurons were cultured without any transfection, from DIV 1 to 6, and then fixed and immunostained with anti-CRMP5 (green) and anti-MAP2 (red) to follow the distribution of endogenous CRMP5 and the dendritic marker, respectively. Representative images of neurons are presented for each DIV. The images in left panels represent three-dimensional intensity projections of CRMP5 at the indicated time. At DIV 1 and 2, CRMP5 and MAP2 staining are restricted to the soma (white arrowhead). At DIV 3, CRMP5 expression is clearly detected in dendrites, axon, and soma, the merge image shows a clear colocalization of CRMP5 and MAP2 in dendrites. This colocalization is no longer observed at DIV 5, where merge image shows longer dendrites stained red with anti-MAP2. The inset is a magnification of soma and dendrites. At DIV 6, CRMP5 staining is predominantly observed at the axon level, where a clear green staining can be observed at the merge image. CRMP5 is de novo expressed in dendrites. Scale bars, 50 μm. B, E18 hippocampal neurons were cultured as in A and immunostained with anti-CRMP5 (green) and anti-CRMP2 (red) antibodies. Representative images of neurons are presented for each day as merge images showing CRMP5 and CRMP2 double staining. The inset shows the CRMP2 immunolabeling in red. At DIV 1 and 2, CRMP5 and CRMP2 staining overlapped at the soma. In contrast to CRMP5, a CRMP2 expression is detected at DIV 2 in neurites, which are clearly stained red in the merge image; note the presence of the longer neurite, which will become the future axon. From DIV 3 to 6, high CRMP2 staining is persistently and clearly observed in dendrites, axon, and soma. As observed in A, CRMP5 shows a peak of expression at DIV 3 and DIV 6, where it is unambiguously colocalized with CRMP2 in axon and dendrites stained yellow. The decrease of CRMP5 expression is confirmed by clear red staining of axon and dendrites with anti-CRMP2 at DIV 4 and 5, as observed in merged images. Scale bars, 50 μm. C, Expression of CRMP5 and CRMP2 in a culture of hippocampal neurons by Western blot analysis on DIV 1 through DIV 6. E18 hippocampal neurons in culture were collected each day. After lysis, the cells were subjected to SDS-PAGE and Western blot probed for CRMP5 and CRMP2 immunoreactivity. Antibody to GAPDH is used as a control of the amount of protein loaded. The total expression of CRMP5 protein in neurons shows unambiguously two high-intensity bands at DIV 3 and DIV 6 with a decrease in intensity in DIV 4 and 5. However, the total CRMP2 expression is similarly high from DIV 3 through DIV 6. Note the presence of multiple isoforms of CRMP2 at DIV 5 with slightly higher intensity. D, Quantification of CRMP5 and CRMP2 fluorescence in dendrites and axon in relation to their lengths. The top panel shows the length of extensions from hippocampal neurons cultured as in A at DIV 1 to DIV 6. The precise length of neurites, axon, or dendrites is determined on hippocampal culture stained with CMFDA (supplemental data 4, available at www.jneurosci.org as supplemental material). The middle and bottom panels show the fluorescence intensity from A and B determined after conversion of each fluorescent image into a gray scale. Quantitative measurements are obtained by averaging the intensity of fluorescence within a box size of 40 × 40 pixels drawn on different regions of the dendrites and axon (n = 10 measurement boxes within each neuron), using ImageJ software. Average fluorescence intensities obtained from these boxes in a number of neurons are plotted in arbitrary units (a.u.). On the dendritic level, CRMP5 show very low expression at DIV 1 and 2 (n = 22 and 19 neurons, respectively), and CRMP2 exhibits higher expression at DIV 1 and 2 (n = 11 and 13 neurons, respectively). A peak of CRMP5 expression at DIV 3 (n = 29 neurons) can be observed, concomitant with axonal growth, whereas CRMP2 expression does not change (n = 16 neurons). Compared with DIV 3, CRMP5 expression decreases by 2.2-fold at DIV 4 (n = 26 neurons; p < 0.05) and 18-fold at DIV 5 (n = 22 neurons; p < 0.0005), where dendrites begin their growth and show high MAP2 expression. On the opposite, CRMP2 expression slightly increases on DIV 4 and 5 (n = 13 neurons). CRMP5 expression increases by sixfold at DIV 6 compared with DIV 5 (n = 22 neurons; p < 0.001), whereas CRMP2 expression does not significantly change (n = 13 neurons). On the axon level, CRMP5 expression decreases from DIV 3 to DIV 5, followed by significant reexpression at DIV 6 (3.3-fold compared with DIV 3; p < 0.01). CRMP2 expression is persistent from DIV 2 to DIV 6, with a slight increase of expression at DIV 5. Error bars indicate SEM.
Discussion
In this study, we provide evidence for a novel role of CRMP5 in inhibiting neurite outgrowth through binding to tubulin and MAP2 during brain development. Overexpression of CRMP2 was reported to promote neurite elongation and axon induction (Arimura and Kaibuchi, 2007), whereas CRMP3 was described as enhancing dendrite branching in mouse hippocampus (Quach et al., 2008). A change in cell shape after CRMP5 overexpression has previously been reported (Suzuki et al., 2003). Although CRMP5 is distributed throughout axon and dendrites (Bretin et al., 2005; Hotta et al., 2005), growth inhibition is observed at the dendritic level, but not in the axon in hippocampal neurons. CRMP5 also inhibits dendrite initiation, although this inhibition occurs only at an early developmental stage (stage 3) concomitant with its strong expression in dendrites. Knockdown of CRMP5 unambiguously confirms its inhibitory function. Nevertheless, CRMP5 deficiency results in greater neurite numbers and lengths in N1E-115 cells than in neurons. In neurons, other negative signaling molecules may partially compensate for CRMP5 function.
CRMP associations with tubulin have been reported for CRMP1-4 (Fukata et al., 2002), whereas a CRMP5–tubulin interaction has not yet been identified. We demonstrate that CRMP5 interacts with tubulin and MAP2 in mouse brain cortex during development, forming a ternary complex, within which CRMP5 shows direct binding to α- and β-tubulin. The colocalization of CRMP5 and tubulin at the cytosolic level supported their interaction. The tubulin-binding domain is mapped to a C-terminal fragment of CRMP5 (residues 475-522). The C-terminal part of CRMP5 fell into the category of “intrinsically unstructured proteins,” characterized by a flexible and unfolded state, favoring interactions with other partners. Consequently, the tubulin-interacting domain, located within this region, would be easily available for binding to tubulin. Previous reports differ on the tubulin-binding properties of CRMP2. A central domain (residues 323-381) was reported to be involved in CRMP2 activity on microtubule assembly (Fukata et al., 2002). As this site is located in a hidden region in the x-ray structure (Stenmark et al., 2007), CRMP2 may have to undergo some conformational change to expose it. In contrast, a recent study identified CRMP2 residues 480-509 as being responsible for such activity (Chae et al., 2009). The latter site resembles the identified CRMP5 tubulin-binding site. However, despite the strong sequence homology between CRMPs and the surface exposure of both sites, they show a low sequence homology.
Distinct pathways are involved in different CRMP-mediated neurite outgrowth functions through actin-binding (Alabed et al., 2007) or tubulin-binding mechanisms (Fukata et al., 2002). We demonstrate that overexpression of CRMP5 bearing the tubulin-binding site inhibits neurite outgrowth, whereas the expression of CRMP5 ΔC472, devoid of tubulin-binding site, failed to inhibit it. Similarly, CRMP5 tubulin-binding capacity is required for inhibition of dendrite outgrowth in neurons. Moreover, we find that recombinant CRMP5 inhibits tubulin polymerization in an in vitro microtubule polymerization assay, whereas CRMP5 ΔC472 does not. Similar mechanism inhibits neurite formation, since CRMP5 ΔC472 expression failed to reduce the number of neurites, whereas different forms of CRMP5 bearing the tubulin-binding site did. The inhibition is more pronounced for ΔC518 and ΔN244 than for full-length forms. This may be explained by a greater accessibility of the tubulin-binding site in the C-terminally deleted form and the induction of a conformational change in the N-terminally deleted form leading to a better exposure of this site.
MAP2 plays an essential role in modulating the growth (Brugg and Matus 1988) and the initiation of neuronal processes by maintaining the linkage or mediating cross talk between microtubules and other cytoskeletal elements. The presence of MAP2 denotes a difference in the way that tubulin interacts with CRMP5 and CRMP2, since MAP2 interactions with other CRMPs were not found in previous reports (Fukata et al., 2002). Notwithstanding whether MAP2 binds to CRMP5 through direct interaction or indirectly via tubulin, its presence within the complex implies that it is a determinant factor in the inhibitory role of CRMP5 notably at the dendritic level. Several observations support this theory. First, in vivo, CRMP5 forms a complex with MAP2 and tubulin. Second, consistent with our observations, a colocalization of CRMP5 and MAP2 in dendrites was previously reported (Bretin et al., 2005). Third, in axon, devoid of MAP2, CRMP5 does not exert inhibitory activity. Therefore, we can assume that by forming a complex with tubulin heterodimers and MAP2, CRMP5 locks the microtubular structure and prevents additional microtubule polymerization, thereby inhibiting neurite outgrowth and formation.
Antagonistic roles of CRMP5 and CRMP2
The study of the combinatorial functions of both CRMP5 and CRMP2 led to the important finding that the neurite outgrowth-promoting function of CRMP2 is totally abrogated by neurite outgrowth inhibition induced by CRMP5, indicating that CRMP5 acts as the dominant signal. In cultured hippocampal neurons, dendrite elongation and initiation induced by CRMP2 expression is considerably enhanced by a CRMP5 deficiency, strengthening the fact that, in vivo, endogenous CRMP5 restricts CRMP2 function. CRMP2 expression results in axon induction and the formation of multiple axons, as reported previously (Inagaki et al., 2001). Abrogation of the CRMP2 outgrowth promotion is observed not only at the dendritic level but also at that of the axon. Nevertheless, CRMP5 expression does not exert inhibitory activity at the axon level since the axon length never falls below the level of controls, but it abolishes CRMP2 induction of axon growth. A CRMP5 deficiency in neurons results also in increased CRMP2-induced axon elongation and the number of multiple axons, suggesting that, in vivo, CRMP5 may modulate CRMP2 induction of both axon growth and multiple axon formation.
Different mechanisms may govern the antagonizing function of CRMP5. First, by binding to CRMP2, CRMP5 may counteract its ability to process neurite outgrowths. However, despite the direct interaction between CRMP5 and CRMP2 observed in this study and reported by others (Fukada et al., 2000), the fact that the loss-of-function CRMP5 ΔC472 still binds to CRMP2 without influencing its activity does not support this interpretation. Second, CRMP5 and CRMP2 bind competitively to tubulin. Indeed, in contrast to the full-length and CRMP5 ΔC518 bearing the tubulin-binding site, the CRMP5 ΔC472 failed to abrogate the CRMP2 promotion of axon and dendrite outgrowth in hippocampal neurons, suggesting that CRMP5 antagonistic activity is tubulin-binding dependent. In addition, only recombinant CRMP5 bearing the tubulin-binding site, but not CRMP5 ΔC472, can disrupt the in vitro binding of CRMP2 to tubulin. By binding to tubulin heterodimers, CRMP2 transports them, via kinesin-1 motor proteins, to the plus-ends of microtubules to promote neurite extension (Kimura et al., 2005). Thus, by binding to microtubules, CRMP5 impairs CRMP2 transport of tubulin, preventing it to promote microtubule assembly, which causes the modification of the microtubule networks.
CRMP5 in neuronal polarity
As CRMP2 plays a role in the establishment and maintenance of neuronal polarity by inducing axonal growth (Yoshimura et al., 2005), endogenous CRMP5, by restricting CRMP2 activity, may intervene in the regulation of neuronal polarity. In this study, using different approaches, we find that the CRMP2 expression is almost constant during neuronal development, whereas CRMP5 presents two peaks of expression at DIV 3 and 6. Neuronal polarization occurs from stage 2 to 3, at which one of the immature neurite begins to extend rapidly and becomes axon. Other neurites remain quiescent for a few days until the axon begins to growth, and then they elongate and acquire, at stage 4 (DIV 5), the characteristics of dendrites (Dotti et al., 1988). To date, it is not known why neurites other than the axon remain quiescent. Here, several lines of inquiry have helped to elucidate this point. First, we find that strong CRMP5 expression in dendrites occurs at DIV 3, corresponding to axon growth and quiescent neurite. Second, CRMP5 expression progressively diminishes and almost stops at DIV 5, when dendrites begin to extend. Third, CRMP5 acts as an inhibitor of dendrite elongation and formation with no effect on axon growth. Fourth, CRMP5 restricts the CRMP2 outgrowth promotional function. Considering the continuous expression of CRMP2, it can be assumed that CRMP5, by counteracting CRMP2, maintains neurites in a quiescent state at the early stage of the development, allowing the axon to continue its outgrowth, whereas its lack of expression thereafter allows dendrites to elongate and to reach their mature state, probably because of the CRMP2-induced promotional outgrowth. A slight CRMP5 reexpression can be observed in dendrites, at a later stage of development (DIV 6), which may help control mature dendrite length. This is consistent with our previous observations that, at the postnatal stage, CRMP5 is present in the external granular layer of cerebellum (Ricard et al., 2001) and CRMP5 is expressed in soma and dendrites of adult brain (Bretin et al., 2005). In parallel, high CRMP5 expression in the axon, at a late developmental stage, may constrain CRMP2-induced growth, delimiting axonal length. Whereas many intracellular and extracellular molecules seem to determine neuronal polarity (Arimura and Kaibuchi, 2007), our observations suggest that CRMP5 can be considered as one of the molecules involved in regulating neuronal polarity.
In conclusion, the modulation of CRMP5 expression in neurons is a critical factor in regulating neurite outgrowth and neuronal polarity, ensuring the establishment of normal neuronal communication during brain development.
Footnotes
We thank Dr. Lionel Ballut for helpful discussions.
References
- Alabed YZ, Pool M, Ong Tone S, Fournier AE. Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J Neurosci. 2007;27:1702–1711. doi: 10.1523/JNEUROSCI.5055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nature. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Arimura N, Kimura T, Nakamuta S, Taya S, Funahashi Y, Hattori A, Shimada A, Ménager C, Kawabata S, Fujii K, Iwamatsu A, Segal RA, Fukuda M, Kaibuchi K. Anterograde transport of TrkB in axons is mediated by direct interaction with Slp1 and Rab27. Dev Cell. 2009;16:657–686. doi: 10.1016/j.devcel.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Bretin S, Reibel S, Charrier E, Maus-Moatti M, Auvergnon N, Thevenoux A, Glowinski J, Rogemond V, Prémont J, Honnorat J, Gauchy C. Differential expression of CRMP1, CRMP2A, CRMP2B, and CRMP5 in axons or dendrites of distinct neurons in the mouse brain. J Comp Neurol. 2005;486:1–17. doi: 10.1002/cne.20465. [DOI] [PubMed] [Google Scholar]
- Brugg B, Matus A. PC12 cells express juvenile microtubule-associated proteins during nerve growth factor induced neurite outgrowth. J Cell Biol. 1988;107:643–650. doi: 10.1083/jcb.107.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YC, Lee S, Heo K, Ha SH, Jung Y, Kim JH, Ihara Y, Suh PG, Ryu SH. Collapsin response mediator protein-2 regulates neurite formation by modulating tubulin GTPase activity. Cell Signal. 2009;12:1818–1826. doi: 10.1016/j.cellsig.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Cole AR, Knebel A, Morrice NA, Robertson LA, Irving AJ, Connolly CN, Sutherland C. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem. 2004;279:50176–50180. doi: 10.1074/jbc.C400412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Schmidt EF, Elhabazi A, Togashi H, Burley SK, Strittmatter SM. Structural bases for CRMP function in plexin-dependent semaphorin3A signaling. EMBO J. 2004;23:9–22. doi: 10.1038/sj.emboj.7600021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Lenoir V, Chambraud B, Fellous A, David S, Duchossoy Y, Baulieu EE, Robel P. Microtubule-associated protein 2 (MAP2) is a neurosteroid receptor. Proc Natl Acad Sci U S A. 2006;103:4711–4716. doi: 10.1073/pnas.0600113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada M, Watakabe I, Yuasa-Kawada J, Kawachi H, Kuroiwa A, Matsuda Y, Noda M. Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J Biol Chem. 2000;275:37957–37965. doi: 10.1074/jbc.M003277200. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Ménager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Honnorat J, Cartalat-Carel S, Ricard D, Camdessanché JP, Carpentier AF, Rogemond V, Chapuis F, Aguera M, Decullier E, Duchemin AM, Graus F, Antoine JC. Onco-neural antibodies and tumor type determine survival and neurological symptoms in paraneaplastic neurological syndromes with Hu or CV2/CRMP5 antibodies. J Neurol Neurosurg Psychiatry. 2009;80:412–416. doi: 10.1136/jnnp.2007.138016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A, Inatome R, Yuasa-Kawada J, Qin Q, Yamamura H, Yanagi S. Critical role of collapsin response mediator protein-associated molecule CRAM for filopodia and growth cone development in neurons. Mol Biol Cell. 2005;16:32–39. doi: 10.1091/mbc.E04-08-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Chihara K, Arimura N, Ménager C, Kawano Y, Matsuo N, Nishimura T, Amano M, Kaibuchi K. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001;4:781–782. doi: 10.1038/90476. [DOI] [PubMed] [Google Scholar]
- Inatome R, Tsujimura T, Hitomi T, Mitsui N, Hermann P, Kuroda S, Yamamura H, Yanagi S. Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J Biol Chem. 2000;275:27291–27302. doi: 10.1074/jbc.M910126199. [DOI] [PubMed] [Google Scholar]
- Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Mitsui N, Inatome R, Takahashi S, Goshima Y, Yamamura H, Yanagi S. Involvement of Fes/Fps tyrosine kinase in semaphorin3A signaling. EMBO J. 2002;21:3274–3285. doi: 10.1093/emboj/cdf328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi-Améli M, Lorca T, Ficheux D, di Pietro A, Gillet G. Interaction between the antiapoptotic protein Nr-13 and cytochrome c. Antagonistic effect of the BH3 domain of Bax. Biochemistry. 2002;41:8540–8550. doi: 10.1021/bi0110286. [DOI] [PubMed] [Google Scholar]
- Quach TT, Massicotte G, Belin MF, Honnorat J, Glasper ER, Devries AC, Jakeman LB, Baudry M, Duchemin AM, Kolattukudy PE. CRMP3 is required for hippocampal CA1 dendritic organisation and plasticity. FASEB J. 2008;22:401–409. doi: 10.1096/fj.07-9012com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard D, Rogemond V, Charrier E, Aguera M, Bagnard D, Belin MF, Thomasset N, Honnorat J. Isolation and expression pattern of human Unc-33-like phosphoprotein 6/collapsin response mediator protein 5 (Ulip6/CRMP5): coexistence with Ulip2/CRMP2 in Sema3a-sensitive oligodendrocytes. J Neurosci. 2001;21:7203–7214. doi: 10.1523/JNEUROSCI.21-18-07203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogemond V, Auger C, Giraudon P, Becchi M, Auvergnon N, Belin MF, Honnorat J, Moradi-Améli M. Processing and nuclear localization of CRMP2 during brain development induce neurite outgrowth inhibition. J Biol Chem. 2008;283:14751–14761. doi: 10.1074/jbc.M708480200. [DOI] [PubMed] [Google Scholar]
- Stenmark P, Ogg D, Flodin S, Flores A, Kotenyova T, Nyman T, Nordlund P, Kursula P. The structure of human collapsing response mediator protein 2, a regulator of axonal growth. J Neurochem. 2007;101:906–917. doi: 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakagomi S, Namikawa K, Kiryu-Seo S, Inagaki N, Kaibuchi K, Aizawa H, Kikuchi K, Kiyama H. Collapsin response mediator protein-2 accelerates axon regeneration of nerve-injured motor neurons of rat. J Neurochem. 2003;86:1042–1050. doi: 10.1046/j.1471-4159.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Inatome R, Yamamura H, Yanagi S. Isolation and expression of a novel mitochondrial septin that interacts with CRMP/CRAM in the developing neurones. Genes Cells. 2003;8:81–93. doi: 10.1046/j.1365-2443.2003.00617.x. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, Ihara Y, Mikoshiba K, Kolattukudy P, Honnorat J, Goshima Y. Semaphorin3A signaling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem. 1997;69:2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Spooner BS, Wessells NK. Axon growth: roles of microfilaments and microtubules. Proc Natl Acad Sci U S A. 1970;66:1206–1212. doi: 10.1073/pnas.66.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]