Abstract
Down's syndrome cell adhesion molecule (DSCAM) belongs to the Down's syndrome critical region of human chromosome 21, and it encodes a cell adhesion molecule involved in dendrite morphology and neuronal wiring. Although the function of DSCAM in the adult brain is unknown, its expression pattern suggests a role in synaptic plasticity. Local mRNA translation is a key process in axonal growth, dendritogenesis, and synaptogenesis during development, and in synaptic plasticity in adulthood. Here, we report the dendritic localization of DSCAM mRNA in the adult mouse hippocampus, where it associates with CPEB1 [cytoplasmic polyadenylation element (CPE) binding protein 1], an important regulator of mRNA transport and local translation. We identified five DSCAM isoforms produced by alternative polyadenylation bearing different combinations of regulatory CPE motifs. Overexpression of DSCAM in hippocampal neurons inhibited dendritic branching. Interestingly, dendritic levels of DSCAM mRNA and protein were increased in hippocampal neurons from Ts1Cje mice, a model of Down's syndrome. Most importantly, DSCAM dendritic translation was rapidly induced by NMDA in wild-type, but not in Ts1Cje neurons. We propose that impairment of the NMDA-mediated regulation of DSCAM translation may contribute to the alterations in dendritic morphology and/or synaptic plasticity in Down's syndrome.
Introduction
Down's syndrome (DS) is the most frequent genetic cause of mental retardation (Antonarakis et al., 2004). Although it is well established that DS occurs because of trisomy of human chromosome 21 (HSA21), the precise molecular and cellular mechanisms responsible for the neurological DS phenotype remain poorly understood. In DS, dendritic branching and dendritic spine density are decreased in the cortex and hippocampus (Marin-Padilla, 1972; Suetsugu and Mehraein, 1980; Ferrer and Gullotta, 1990). In addition, dendritic spines display morphological abnormalities thought to contribute to reduced plasticity and learning potential (Marin-Padilla, 1976; Takashima et al., 1981).
Several studies have sought to identify candidate genes that alone, or in combination, may mediate the observed dendritic pathology in DS. Potential candidates include several HSA21 genes known to be involved in dendrite morphogenesis. Among them, Down's syndrome cell adhesion molecule (DSCAM) has been shown to participate in dendritic tiling and self-avoidance in the mouse visual system (Fuerst et al., 2008, 2009), two fundamental phenomena related to the formation of neural circuits. DSCAM is strongly expressed during brain development, and although it is downregulated in the adult, it persists in brain areas with high levels of synaptic plasticity, including the cortex, hippocampus, and cerebellum (Agarwala et al., 2001; Barlow et al., 2001). Nonetheless, the function of DSCAM in the adult brain remains unknown.
Local translation of mRNAs in dendritic spines is crucial for synaptic plasticity (Sutton and Schuman, 2006). It also plays an important role in dendritic morphogenesis during development and may contribute to the neuronal pathology associated to DS, as seen in fragile X mental retardation (Bassell and Warren, 2008). Several proteins regulate local mRNA translation. Among them, cytoplasmic polyadenylation element binding protein 1 (CPEB1) recognizes cytoplasmic polyadenylation elements (CPEs) located to the 3′-untranslated region (3′-UTR) of synaptic mRNAs, facilitating their transport to dendrites and regulating their polyadenylation-dependent local translation in response to synaptic activity (Richter, 2007). CPEB1 is also thought to participate in both synaptic plasticity (Alarcon et al., 2004; Berger-Sweeney et al., 2006; McEvoy et al., 2007) and dendrite morphogenesis (Bestman and Cline, 2008, 2009). Recently, a combinatorial code for the influences of CPEs on translational control was defined, whereby the number of CPEs and their positions relative to the polyadenylation hexamer modulate CPEB1-mediated translational regulation of a given mRNA (Piqué et al., 2008).
Here, we provide evidence that DSCAM mRNA is localized to dendrites and associated with CPEB1 in the adult mouse hippocampus, and describe the expression of at least five DSCAM isoforms, bearing different combinations of CPEs at their 3′-UTR. We show a role of DSCAM in dendrite branching of hippocampal neurons. Dendritic levels of both DSCAM mRNA and protein augment in Ts1Cje mice, a widely used DS model. Most importantly, we show that dendritic levels of DSCAM protein are regulated by synaptic activity at NMDA receptors (NMDARs). Such regulation is dependent on mRNA polyadenylation and protein synthesis, and it is lost in Ts1Cje mice. Together, these findings suggest a function of DSCAM in dendritogenesis and synaptic plasticity, which may be compromised in DS.
Materials and Methods
Animals.
Animals were maintained, handled, and killed in accordance with national and international laws and policies, and all protocols were approved by the University of Seville Animal Care and Use Committee. C57BL/6 mice were obtained from the Animal Production Centre (CPYA) at the University of Seville and they were used in some experiments that did not require comparison with Ts1Cje mice. Partially trisomic Ts1Cje mice were obtained from The Jackson Laboratory, and maintained on a mixed genetic background as recommended by backcrossing Ts1Cje males to C57BL/6JOlaHsd × C3H/HeNHsd F1 females (B6C3F1 hybrid females; supplied by Harlan). Littermates were used in all experiments that required comparisons to be made between wild-type and trisomic mice to ensure that the genetic background did not influence the results.
Xenopus females were obtained from the Animal Facility Service at the University Miguel Hernández (Alicante, Spain).
Synaptoneurosome preparations.
Synaptoneurosomes were isolated as described previously (Bagni et al., 2000) with some modifications. Briefly, the hippocampus from eight adult mice were homogenized in 9 ml of 320 mm sucrose and 1 mm EDTA, supplemented with protease and RNase inhibitors (10 μm leupeptin, 2 μg/ml aprotinin, 1 μm pepstatin, 20 U/ml RNaseOUT; Invitrogen). The homogenate was centrifuged at 1000 × g for 10 min at 4°C, and the resulting supernatant (total extract) was subjected to four discontinuous Percoll–sucrose gradients (3, 10, 15, and 23% Percoll). The material located at the 15–23% interface was collected, washed, and separated on an Optiprep gradient (9, 12.5, 15, 25, and 35% Optiprep), and the Optiprep solutions were prepared by diluting the corresponding volume of an Optiprep working solution (OWS) (50% Optiprep, 10 mm HEPES, pH 7.4, 65 mm sucrose; final pH adjusted to 7.4 with NaOH) in Optiprep gradient buffer (OGB) (10 mm HEPES, pH 7.4, 300 mm sucrose). Finally, the material located at interface 15–25% (synaptoneurosomes) was collected and used for RNA isolation.
RNA isolation and real-time quantitative reverse transcription-PCR.
RNA was isolated on spin columns (PureLink Micro to Midi total RNA purification system; Invitrogen) and treated on the column with DNase (Sigma-Aldrich) during the isolation procedure.
For real-time quantitative reverse transcription (RT)-PCR, RNA was reverse transcribed with random hexamer primers (Transcriptor First Strand cDNA Synthesis kit; Roche) and the cDNA samples were subjected to real-time PCR (two or three replicates) using the Brilliant SYBR Green QPCR Master Mix (Stratagene) in a Mx3000P apparatus (Stratagene). Negative reverse transcription controls in which retrotranscription was not performed were included routinely. Thermal cycling was performed over 40 cycles as follows: denaturation for 10 min at 95°C; amplification for 15 s at 95°C, 30 s at 58°C, and 10 s at 72°C. After amplification, melting curves were analyzed to confirm the identity of the PCR products. Primers sequences for DSCAM coding region (DSCAM-fw and -rv oligos), DSCAM 3′-UTR isoform 5 (DSCAM2A-fw and -rv oligos), α-Ca2+-calmodulin-dependent protein kinase II (α-CaMKII), small nuclear ribonucleoprotein N (SnrpN), and hypoxanthine-guanine phosphorybosyltransferase (HPRT) amplification were as follows: DSCAM-fw, 5′-ATGGAGGACGAGTGGTGAACAT; DSCAM-rv, 5′-GTCCTAAACTCAGGTCCCTGCT; DSCAM2A-fw, 5′-CTGATACCTCCCTATCCATTGC; DSCAM2A-rv, 5′-ATCTTCTTGTCTCTTCCCAGCA; α-CaMKII-fw, 5′-GGAGGAAACAAGAAGAACGATG; α-CaMKII-rv, 5′-TCAGCTGCTCTGTCACTTTGAT; SnrpN-fw, 5′-GGGAGAACTTGGTTTCAATGAC; SnrpN-rv, 5′-AATAGGTACACCTGCTGGCACT; HPRT-fw, 5′-GGACCTCTCGAAGTGTTGGATA; HPRT-rv, 5′-ACGTGATTCAAATCCCTGAAGT.
The efficiencies of the primers, calculated from standard curves using cDNA serial dilutions, were as follows: 97.3% for DSCAM, 83.6% for DSCAM2A, 90.4% for α-CaMKII, 89.9% for SnrpN, and 85.5% for HPRT. The r2 values of the standard curves were 0.985, 0.968, 0.991, 0.997, and 0.996, respectively. These figures were taken into account in the calculations using qBase software (Hellemans et al., 2007).
RNA-binding protein immunoprecipitation.
RNA-binding protein immunoprecipitation (RIP) was performed using the Magna RIP kit (Millipore), following the manufacturer's instructions. Total hippocampal extracts were used as the input material for CPEB1 immunoprecipitation with a polyclonal CPEB1 antibody (Abcam; ab73287), and a purified rabbit IgG (Millipore) was used in parallel as a negative control. The levels of DSCAM, SnrpN, and HPRT mRNAs in immunoprecipitated CPEB1–mRNA or IgG–mRNA complexes, as well as in the initial extracts from which the complexes were immunoprecipitated, were quantified by real-time RT-PCR.
RNA ligation poly(A) test.
RNA ligation poly(A) test (RL-PAT) was performed as described previously (Rassa et al., 2000). Briefly, total RNA from the hippocampus or hippocampal synaptoneurosomes was ligated to oligo P1 (5′-[Phos] GGTCACCTTGATCTGAAGC [AmC3]) using T4 RNA ligase (Promega) and reverse-transcribed with the specific oligo P'as (5′-GCTTCAGATCAAGGTGACCTTTTT) using the Transcriptor First-Strand cDNA Synthesis kit (Roche). The oligo P'as was used in combination with different DSCAM primers to amplify the corresponding DSCAM isoform by PCR (Platinum TAQ DNA polymerase; Invitrogen). Negative controls using genomic DNA as a template were included routinely. HPRT was used as a control in some experiments. Sequences of DSCAM and HPRT primers were as follows: s0 (for DSCAM isoforms 1 and 2), 5′-GCGGTTGTAAATACAATTTAAACGAG; s0-c (for DSCAM isoform 3), 5′-CACAGACAGCGCAAAGACTACTC; s1 (for DSCAM isoform 4), 5′-GTTAATCAGGATGAGTGCTGTCAGT; s2 (for DSCAM isoform 5), 5′-CCTGCTCTCTAACTCAGTAGGGTC; HPRT, 5′-CATGTTTCAGCAGTGTTGGCT. RL-PAT PCR products were cloned in pGEM-T Easy vector (Promega) and verified by sequencing.
Translation assays in Xenopus oocytes.
Sequences corresponding to the 3′-UTR of the DSCAM isoforms 1, 3, 4, and 5 were amplified from mouse hippocampal cDNAs by PCR (Expand High Fidelity Plus PCR system; Roche) and cloned downstream the Firefly luciferase coding region of the pLucCassette vector (Piqué et al., 2006). The primers used for PCR amplification were as follows: BglII-3UTR-DSCAM-fw, 5′-GTTAACAGATCTCACATGGCACCTGATG (common for all isoforms); isof1-DSCAM-BamHI, 5′-TCCGGATCCAATATATTTTGGCAATTTT (for isoform 1); isof3-DSCAM-BamHI, 5′-TCCGGATCCACTCTTGTGTGCTAGAACT (for isoform 3); isof4-DSCAM-BamHI, 5′-GGATCCAAAGGCAGACCACCTTTTCTCTTT (for isoform 4); isof5-DSCAM-BamHI, 5′-GGATCCGGACTTCTTCCAATAAAACTTTAT (for isoform 5). The PCR products were first cloned into the pGEM-T Easy vector (Promega), and they were verified by sequencing. Inserts from double digested BglII-BamHI plasmids were then isolated and cloned into the BamHI site of the pLucCassette vector, and the plasmids bearing each DSCAM isoform in the correct orientation (sense) were selected.
In vitro production of luciferase.
DSCAM 3′-UTR transcripts was performed using the mMESSAGE mMACHINE kit (Ambion). To obtain the luciferase transcripts with the 3′-UTRs of similar length to those of interest but lacking the regulatory sequences, pLucpBSK plasmid was linearized with the appropriate restriction enzymes and used as a template for in vitro transcription (Piqué et al., 2006). For isoform 4, a plasmid containing the 3′-UTR of the DSCAM isoform 4 in the antisense orientation was used.
The firefly luciferase-3′-UTR transcripts and a normalizing Renilla luciferase mRNA were coinjected in stage VI Xenopus oocytes using an automatic nanoliter injector (Nanoject II; Drummond Scientific) as described previously (Piqué et al., 2006). Oocytes were incubated with or without progesterone (Sigma-Aldrich), and firefly and Renilla luciferase activities were sequentially determined using the Dual-Luciferase reporter assay kit (Promega) in a GloMax luminometer (Promega).
Hippocampal cultures and transfection.
The hippocampus from postnatal day 0 (P0) wild-type or trisomic mouse littermates was dissected out in HBSS medium (Invitrogen) and mechanically dissociated after trypsin treatment (0.2% trypsin for 10 min at 37°C) in DB1 culture medium (DMEM high glucose with l-glutamine, without sodium pyruvate; 10% fetal bovine serum; Glutamax; 08% glucose; penicillin/streptomycin). The cells were seeded on poly-l-lysine-treated coverslips (0.5 mg/ml) and cultured in Neurobasal A medium (Neurobasal A; B27 supplement; Glutamax; penicillin/streptomycin), in 24-well plates. After 48 h in culture, 0.3 mm 5-fluoro-2′-deoxyuridine (Sigma-Aldrich) and 0.8 mm uridine (Sigma-Aldrich) were added to the culture medium to inhibit glial growth.
Lipofectamine 2000 (Invitrogen) was used, following the manufacturer's instructions, for transfection of day 7 in vitro (DIV7) hippocampal neurons. A total of 0.8 μg of a plasmid expressing mouse DSCAM along with green fluorescent protein (GFP) protein (CMV-DSCAM-IRES-GFP) (Yamagata and Sanes, 2008) or a control vector expressing GFP protein (pEGFP-C1; Clontech) was transfected per well. Effects on dendrite arborization were analyzed 2 d after transfection. Three independent experiments, using different batches of neurons, were performed (six transfections per experiment and plasmid).
DSCAM antibodies.
A custom polyclonal antiserum against the synthetic peptide CGHPEPDYRWLKDNMPL, corresponding to amino acid residues 250–265 (Ig Ig3 domain) of murine DSCAM, was produced by Biomedal. This antibody immunoprecipitated a 230 kDa protein from mouse hippocampal extracts, which was more abundant (1.7-fold) in extracts from Ts1Cje than wild-type mice. As proof of specificity, the protein immunoprecipitated with this custom antiserum was detected using a commercial goat anti-human DSCAM antibody (R&D Systems), which also recognizes murine DSCAM (Fuerst et al., 2008).
Immunochemistry and image analysis.
For the immunohistochemical analyses, adult mice were first perfused with PBS and then with 4% paraformaldehyde in PBS. The brains were dissected out and postfixed overnight at 4°C with 4% paraformaldehyde in PBS. Coronal vibratome sections (30–50 μm) were permeabilized for 10 min at room temperature with 1% Triton X-100 in PBS, washed in PBS, and blocked for 1 h at room temperature in the same solution supplemented with 2% bovine serum albumin (fraction V). The rabbit polyclonal DSCAM (1:50) and mouse monoclonal MAP2 (1:1000; Millipore Bioscience Research Reagents) antibodies were diluted in the same buffer, and brain slices were incubated overnight at 4°C.
For immunocytochemistry, neurons were fixed in 4% paraformaldehyde in PBS for 20 min, and after fixation, the cells were washed three times with PBS at room temperature and they were blocked for 30 min at 37°C in PBS, 0.1% Triton X-100, and 10% fetal bovine serum. The corresponding primary antibodies were diluted in blocking buffer and the cells were incubated overnight at 4°C. The cells were then washed three times in PBS for 10 min at room temperature. DSCAM and MAP2 antibodies were used at the same dilution as above; chicken polyclonal GFP antibody (Aves GFP-1020) was used at 1:500.
Secondary antibodies conjugated to Alexa 488, Alexa 555 (Invitrogen), or Cy3 (The Jackson Laboratory) were applied for 1 h at room temperature in blocking buffer without Triton X-100 (with 3% fetal bovine serum for immunocytochemistry), and the cells or slices were washed three times in PBS for 10 min.
An Olympus Fluoview FV1000 confocal microscope (objective: PLAPON 60×; numerical aperture, 1.42) was used for immunocytochemistry image acquisition. Images were taken using fixed confocal settings, maximizing the dynamic range of pixel intensity for DSCAM labeling in dendrites when used for dendritic signal quantification. Matlab software (The MathWorks) was used to determine the mean pixel intensity for DSCAM immunofluorescence in dendrites identified using MAP2 as a marker.
Images from immunohistochemistry experiments were taken using Olympus Fluoview (objective, 10×) or Leica DM IRE2 (objective, 20×) confocal microscopes.
Morphometric analysis and quantification were performed using NIH ImageJ software. The neuron soma size was obtained by outlining the cell soma, followed by automatic calculation of pixel area in squared micrometers. Total dendritic length was calculated using the NeuronJ plug-in (Meijering et al., 2004); dendrites were identified as MAP2-positive neurites. Sholl analysis was automatically performed using the Sholl analysis plug-in (http://biology.ucsd.edu/labs/ghosh/software/).
Statistical analysis.
The quantitative data are presented as the means and SEM. The significance for comparisons was calculated using Student's t test (SigmaStat software), and statistical significance was accepted when p < 0.05. The Mann–Whitney rank sum test was used when the tests of normality or equal variance failed.
Results
DSCAM mRNA is enriched in mouse hippocampal synaptoneurosomes and it associates with the CPEB1 protein
Synaptoneurosomes are neuronal membrane fractions containing sealed presynaptic elements attached to their corresponding postsynaptic counterparts. These preparations have been used extensively to identify dendritically localized mRNAs (Bagni et al., 2000; Crispino et al., 2001; Sung et al., 2004; Di Nardo et al., 2007; Matsumoto et al., 2007; Troca-Marín et al., 2010). Hence, the localization of DSCAM mRNA was studied in total hippocampal extracts and synaptoneurosomes by real-time RT-PCR normalizing this expression against that of the HPRT gene (Di Nardo et al., 2007). Moreover, the well known dendritic transcript α-CaMKII (Mayford et al., 1996) and the soma-restricted mRNA SnrpN (Buettner et al., 2000) were used as positive and negative controls, respectively. The expression of α-CaMKII was enriched 6.6-fold in synaptoneurosomes compared with total extracts, whereas SnrpN showed no enrichment (Fig. 1A). Interestingly, DSCAM mRNA was enriched 8.1-fold in synaptoneurosomes, suggesting that it localizes in the dendrites of adult mouse hippocampal neurons.
Figure 1.
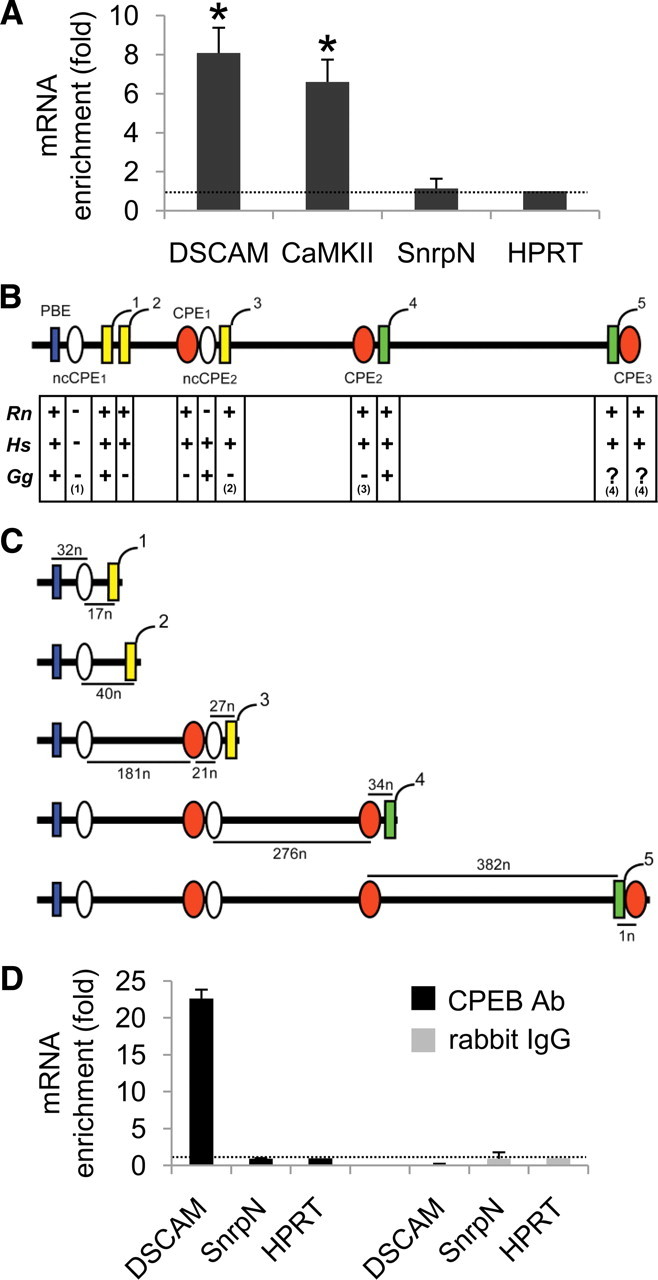
Dendritic localization of DSCAM mRNA and its association to the CPEB1 protein in the mouse hippocampus. A, DSCAM, α-CaMKII, and SnrpN mRNAs were quantified by real-time RT-PCR in synaptoneurosomes and in total extracts from the mouse hippocampus, and normalized to the levels of HPRT mRNA in the corresponding sample. The enrichment in synaptoneurosomes versus total extract is shown as the mean from four to five independent experiments (2–3 independent RT-PCR determinations per experiment). The error bars indicate SEM. Statistically significant differences when compared with HPRT are indicated as follows: *p < 0.05. B, Regulatory motifs found in the most 3′ exon of the mouse DSCAM gene are shown, including a Pumilio-binding element (PBE) (in blue), nonconsensus and consensus cytoplasmic polyadenylation elements (ncCPEs and CPEs) (in white and orange, respectively), and alternative polyadenylation hexamers (numbered from 1 to 5; AUUAAA sequence in yellow; AAUAAA sequence in green). Conservation of regulatory motifs in Rattus norvegicus (Rn), Homo sapiens (Hs), and Gallus gallus (Gg) is also shown. Notes: (1) In G. gallus, ncCPE1 is a consensus CPE (UUUUAUU sequence); (2) an additional poly(A) signal (AUUAAA) is present 33 nt downstream; (3) an additional consensus CPE [(U)15AAUUA sequence] is present 122 nt upstream the polyA4 signal; (4) sequence not available. C, Theoretical DSCAM 3′-UTR isoforms produced in mouse by alternative usage of polyadenylation hexamers 1–5. The distances between the pairs of regulatory motifs are indicated in nucleotides (n). D, Levels of DSCAM and SnrpN mRNAs in CPEB1–mRNA and rabbit IgG–mRNA (negative control) complexes immunoprecipitated from the mouse hippocampus. The mRNA enrichment in the corresponding complex compared with the mRNA levels in the extract from which they were immunoprecipitated, is shown as the mean of two independent RT-PCR determinations. The error bars indicate the SEM, and the values were normalized to HPRT expression.
The dendritic localization of mRNAs is determined by motifs collectively known as dendritic targeting elements (DTEs), usually located at the 3′-UTR. DTEs are recognized by trans-acting factors that support RNA transport and/or localization (Kindler et al., 2005). We analyzed the sequence of the mouse DSCAM 3′-UTR to identify possible DTEs that might account for the dendritic localization and regulation of DSCAM mRNA. Five putative polyadenylation signals (AAUAAA or AUUAAA hexamers) were found, all located in the 3′-most exon of DSCAM (Fig. 1B). In addition, a Pumilio binding element (PBE), two nonconsensus CPEs (ncCPEs), and three consensus CPEs were also identified (Fig. 1B). As stated before, CPE sequences serve as DTEs that are recognized by the CPEB1 protein, which is not only involved in dendritic targeting of mRNAs (Huang et al., 2003) but also in the regulation of local dendritic translation (Richter, 2007). Moreover, PBE sequences are recognized by Pumilio, which is a CPEB-interacting protein (Nakahata et al., 2001).
The alignment of the mouse DSCAM 3′-UTR with the corresponding human, rat, and chicken sequences showed that most of these regulatory motifs were conserved among vertebrates, and in particular, the PBE, consensus CPEs, and polyadenylation hexamers were conserved in mammals (Fig. 1B). In addition, alternative polyadenylation of DSCAM should theoretically produce five isoforms with different 3′-UTRs (Fig. 1C), resulting in a variety of CPE combinations as recently described for many CPEB-regulated transcripts (Piqué et al., 2008).
To determine whether DSCAM mRNA associates with CPEB1, this protein was immunoprecipitated from total hippocampus extracts in experimental conditions that preserved interactions with target mRNAs. The CPEB1-associated mRNAs were then isolated and the levels of the DSCAM, SnrpN, and HPRT transcripts (the latter used as a normalizer) were quantified by real-time RT-PCR. Accordingly, DSCAM mRNA was enriched 22-fold in mRNA–CPEB1 complexes when compared with the total extract from which these complexes were immunoprecipitated (Fig. 1D). By contrast, SnrpN showed no enrichment, and hence these results suggested that DSCAM mRNA could be a target of CPEB1 in the adult mouse hippocampus.
Alternative polyadenylation produces a repertoire of DSCAM isoforms bearing different combinations of regulatory motifs in the 3′-UTR
To characterize the regulation of DSCAM mRNA, we first assessed the functionality of the polyadenylation hexamers using the RL-PAT (Rassa et al., 2000). In this test, a 3′-amino-modified oligonucleotide (P1) (see Materials and Methods) is directly ligated to the 3′ end of the mRNA [i.e., to the poly(A) tail] and a complementary oligo (P'as) is then used to carry out reverse transcription. The cDNAs serve as templates for PCR amplification using an oligo annealing upstream the poly(A) signal and P'as as the primers. Hence, total RNA was isolated from the adult or neonatal (P0) mouse hippocampus and subjected to RL-PAT using oligonucleotides s0, s0-c, s1, or s2 to detect the DSCAM isoforms 1/2, 3, 4, and 5, respectively (Fig. 2A). Bands of the expected sizes were obtained from both adult and P0 RNA samples (Fig. 2B) and the intensity of RL-PAT products obtained with the s0 (isoforms 1/2), s0-c (isoform 3), and s2 (isoform 5) oligonucleotides were systematically higher from the neonatal than from the adult hippocampus, probably reflecting the stronger expression in P0 mice. For example, in the case of isoform 5, which could be discriminated from the other isoforms (using DSCAM2A oligos) (see Materials and Methods), the expression in neonatal hippocampus was fourfold higher than in the adult (Fig. 2C).
Figure 2.
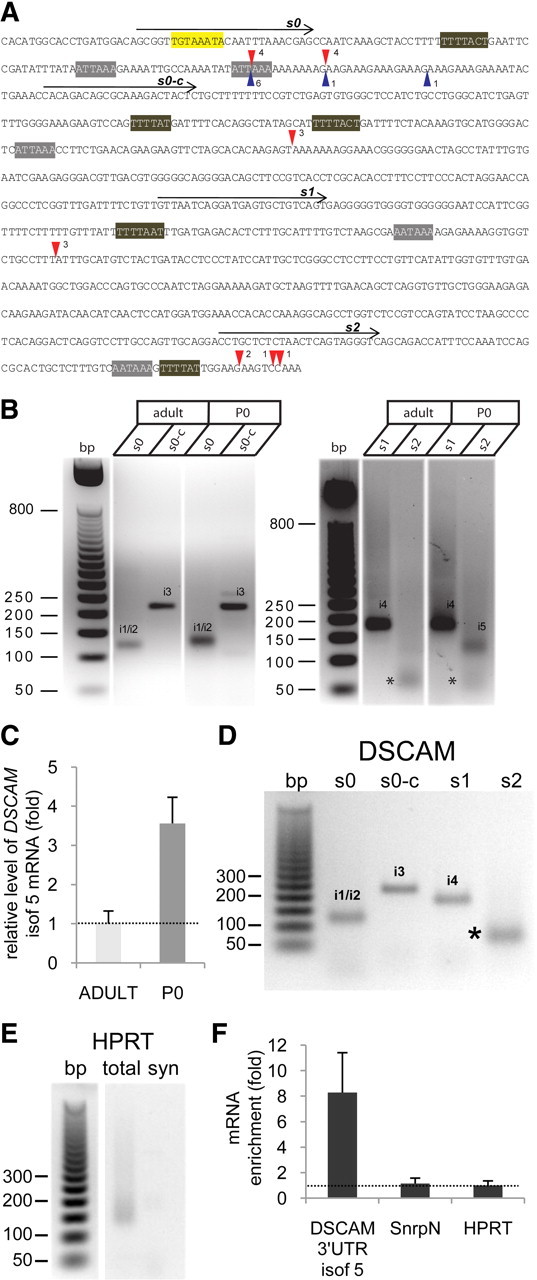
Functionality of alternative DSCAM polyadenylation signals in the mouse hippocampus. A, DSCAM 3′-UTR sequence showing the position of the oligonucleotides s0, s0-c, s1, and s2, used to identify the functional polyadenylation signals by RL-PAT. The red triangles show the cleavage positions for the corresponding polyadenylation hexamers, as deduced from sequencing of RL-PAT products; the number of sequenced clones supporting the corresponding cleavage site is also indicated. The blue triangles show the cleavage sites identified when using synaptoneurosomal RNA in the s0 RL-PAT assay. The PBE sequence (in yellow), CPE motifs (in brown), and poly(A) sites (in gray) are also marked. B, RL-PAT assay on the adult and neonatal (P0) mouse hippocampus. The estimated sizes of the PCR products were ∼120 bp (s0 primer), 220 bp (s0-c primer), 180 bp (s1 primer), and 130 bp (s2 primer). The bands marked with asterisks were identified as primer-dimer artifacts after cloning and sequencing; i1/i2, isoforms 1/2; i3, isoform 3; i4, isoform 4; i5, isoform 5. C, The mRNA levels of DSCAM isoform 5 in total extracts of the adult and neonatal (P0) mouse hippocampus were quantified by real-time RT-PCR, and normalized to the HPRT mRNA in the corresponding sample. The error bars indicate the SEM from two independent RT-PCR determinations in a representative experiment. D, RL-PAT for hippocampal synaptoneurosomes using DSCAM oligos s0, s0-c, s1, and s2 produced PCR products of a similar size to those shown in B. The band marked with an asterisk corresponded to the primer-dimer PCR artifact. E, RL-PAT for total or synaptoneurosomal (syn) hippocampal RNA using a HPRT specific oligo. F, Levels of DSCAM isoform 5 (isof 5) and of the SnrpN mRNAs quantified by real-time RT-PCR in synaptoneurosomes and in total extracts from mouse hippocampus, and normalized to HPRT mRNA in the corresponding sample. Enrichment in synaptoneurosomes versus total extract is shown for a representative experiment, in which the error bars indicate the SEM from two independent RT-PCR determinations.
Interestingly, although RL-PAT for the DSCAM isoforms 4 and 5 (oligos s1 and s2) (Fig. 2B) produced a smearing, suggesting some variability in the length of the poly(A) tails [including long poly(A) mRNAs], this was not the case in RL-PAT for isoforms 1/2 (oligo s0) and 3 (oligo s0-c) (Fig. 2B). Cloning and sequencing of RL-PAT products confirmed that the poly(A) tails of isoforms 1/2 and 3 were very short, ranging from 5 to 10 adenine residues. The poly(A)-binding protein (PABP) requires a minimum of 27 adenine residues for binding (Baer and Kornberg, 1983). As efficient mRNA translation requires an interaction between eukaryotic translational initiation factor 4G (eIF4G) and PABP (Wakiyama et al., 2000), the above results suggest that isoforms 1/2 and 3 are translationally repressed.
Sequencing permitted the exact cleavage position of each transcript to be determined (Fig. 2A, red triangles), which showed some variability for oligo s0 and s2. The first cleavage site in the case of oligo s0, 20 nt downstream of the poly(A) hexamer 1, supported the functionality of poly(A) signal 1. However, the second cleavage site 31 nt downstream the poly(A) hexamer 1, and 8 nt downstream the poly(A) hexamer 2, may be under the control of either poly(A) signal 1 or 2. Thus, the sequencing results were not conclusive regarding the functionality of poly(A) signal 2.
To determine whether the same repertoire of DSCAM isoforms was present in dendrites, RL-PAT was performed on RNA from hippocampal synaptoneurosomes and products of the appropriate sizes were obtained for oligos s0, s0-c, and s1, evidence that isoforms 1/2, 3, and 4 are present in dendrites (Fig. 2D). As a control, RL-PAT using a specific HPRT primer produced a smear with total hippocampal RNA and no amplification was obtained from synaptoneurosomal RNA (Fig. 2E), as expected for a somatic transcript.
The cleavage site for the s0 RL-PAT product was determined in eight clones from independent experiments (Fig. 2A, blue triangles), and of these, six clones corresponded to cleavage site 1 and one to cleavage site 2. Interestingly, one clone supported the functionality of poly(A) signal 2 [Fig. 2A, blue triangle located 23 nt downstream the poly(A) hexamer 2]. Finally, although RL-PAT with oligo s2 (isoform 5) produced a very faint band (Fig. 2D), as was the case in the total adult hippocampal RNA (Fig. 2B), quantitative real-time RT-PCR with DSCAM2A primers confirmed the dendritic localization of isoform 5, which was enriched approximately eightfold in synaptoneurosomes when compared with the total extract (Fig. 2F).
In summary, together, these results support the dendritic localization of all DSCAM 3′-UTR isoforms.
The combinatorial CPE code dictates the translational behavior of most DSCAM isoforms in Xenopus oocytes
As mentioned before, DSCAM isoforms produced by alternative polyadenylation displayed different combinations of PBE and CPE motifs (Fig. 1C), and DSCAM mRNA was found to be associated with the CPEB1 protein in adult hippocampus (Fig. 1D). To investigate the regulatory effect of CPEB1 protein on DSCAM translation, plasmids were generated bearing the 3′-UTR of DSCAM isoforms 1, 3, 4, or 5 downstream of the firefly luciferase coding region. As the combinatorial CPE code (Piqué et al., 2008) predicted a similar translational behavior for isoforms 1 and 2 (see below), isoform 2 was not analyzed. The luciferase-DSCAM 3′-UTR plasmids were used as templates to produce synthetic transcripts and each luciferase-DSCAM 3′-UTR reporter was coinjected with a normalizing Renilla luciferase transcript into Xenopus oocytes at stage VI. At this stage, oocytes express the molecular machinery necessary to carry out repression or cytoplasmic polyadenylation, and translational activation of CPE-containing mRNAs, triggered by progesterone (Richter, 2007). This system is therefore suitable to analyze CPEB-mediated translational regulation of mRNAs, including heterologous neuronal transcripts (Wu et al., 1998; Di Nardo et al., 2007). As the length of the 3′-UTR has a clear effect on translation (Tanguay and Gallie, 1996), a reference mRNA with a 3′-UTR of similar size, but lacking regulatory sequences, was used as a control for each luciferase-DSCAM 3′-UTR reporter (Piqué et al., 2006). As an additional control, luciferase-DSCAM 3′-UTR mRNAs containing a downstream stretch of 73 adenines (i.e., polyadenylated transcripts) were synthesized for each isoform tested, as described previously (Piqué et al., 2006). Injected oocytes were incubated with or without progesterone, and the luciferase activity from firefly and Renilla was measured sequentially.
Taking into account the CPE code (Piqué et al., 2008), isoforms 1 and 2 were not expected to be specifically regulated, isoform 3 should be repressed by CPEB, and isoforms 4 and 5 both repressed and activated by CPEB (although strongly activated in the case of isoform 4 and weakly activated in the case of isoform 5). Accordingly, the 3′-UTR of DSCAM isoform 1 produced neither repression nor activation of firefly luciferase translation (Fig. 3A). The 3′-UTR of the DSCAM isoform 3 produced a strong repression of translation as the luciferase activity was >10 times lower than in oocytes injected with a reference transcript with a similar size 3′-UTR (Fig. 3B). The 3′-UTR of DSCAM isoform 4 produced 2.5-fold repression when compared with the reference mRNA, and a 2.2-fold activation when oocytes were induced with progesterone (Fig. 3C). However, this translational activation was relatively limited and should rather be considered as a de-repression as the luciferase activity did not reach the level observed for the reference mRNA. Finally, the 3′-UTR of the DSCAM isoform 5 had no significant effect on luciferase activity, as was the case for isoform 1 (Fig. 3D). In conclusion, both CPEB-regulated and CPEB-independent DSCAM isoforms appear to exist, and with the exception of isoform 5, their translational regulation fitted the CPE code.
Figure 3.
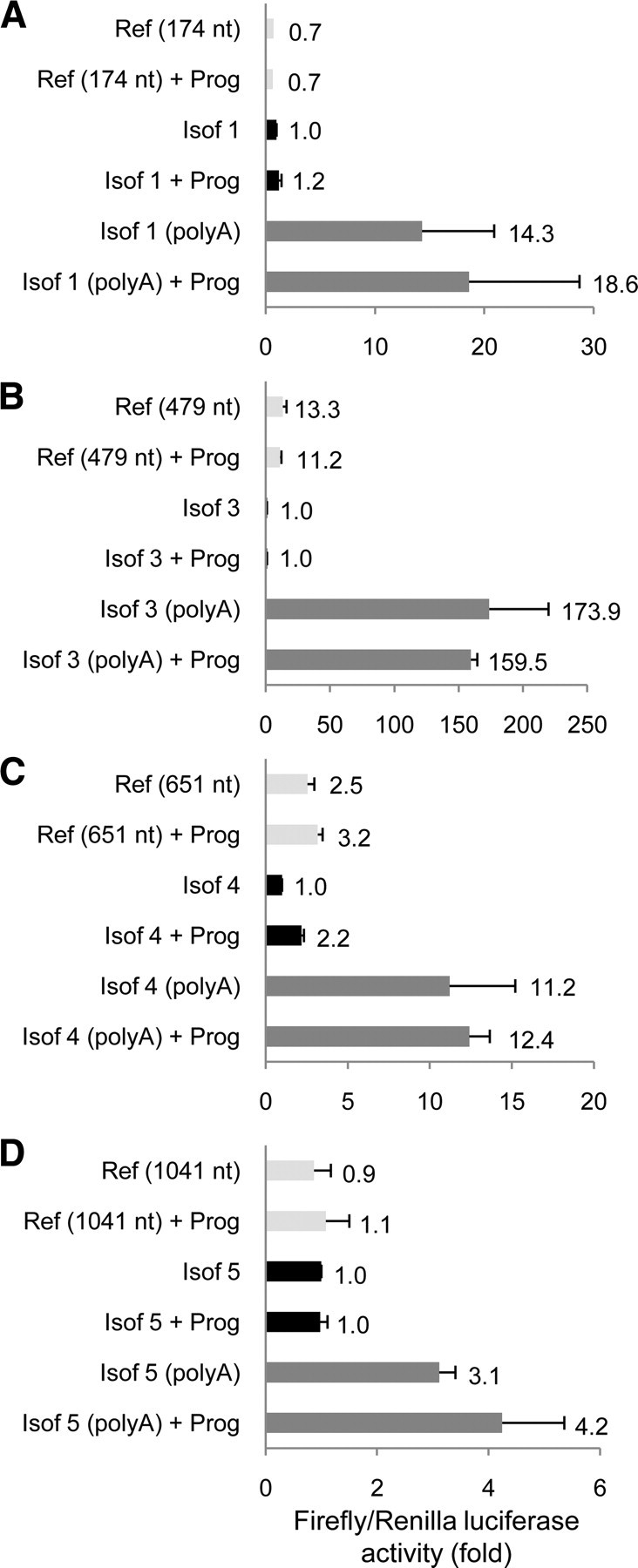
Translational regulation of DSCAM isoforms in Xenopus oocytes. Oocytes were injected with a normalizing Renilla luciferase mRNA and a synthetic firefly luciferase transcript containing the 3′-UTR of the DSCAM isoforms 1 (A), 3 (B), 4 (C), or 5 (D). For each isoform, control transcripts with an unrelated 3′-UTR [reference (Ref)] of the indicated length (in nucleotides) or the same 3′-UTR followed by a poly(A) tail of 73 nt [poly(A)] were also tested. The oocytes were incubated with or without or progesterone (Prog) and in each condition, the Renilla-normalized firefly luciferase activity is shown in arbitrary units. The mean of two independent experiments (n = 25 injected oocytes per condition) is indicated next to the corresponding error bar (SEM).
DSCAM overexpression in hippocampal neurons inhibits dendritic branching
DSCAM has been recently shown to be involved in dendrite morphogenesis in mouse retina (Fuerst et al., 2008, 2009). To assess the impact of DSCAM overexpression on dendrite morphology of hippocampal neurons, we decided to transfect DIV7 neurons with a plasmid encoding DSCAM-IRES-GFP (Yamagata and Sanes, 2008), and to analyze the dendritic phenotype 2 d later (DIV7 + 2). A dramatic negative effect in dendrite arborization was observed when compared with control GFP-transfected neurons (Fig. 4A). Accordingly, Sholl analysis, which measures the number of neurites crossing circles at various radial distances from the soma (Sholl, 1953), showed that the number of crossings at distances between 15 and 150 μm from the soma was much higher in control, GFP-transfected neurons, than in DSCAM-overexpressing neurons (Fig. 4B). Moreover, DSCAM-overexpressing neurons showed a 2.3-fold reduction of the total dendrite length (Fig. 4C). No significant differences were appreciated regarding the number of primary dendrites (Fig. 4D), which are indeed fully formed at the developmental stage selected for these experiments. Finally, a slight reduction of somatic area was measured in DSCAM-transfected neurons (Fig. 4E). These results clearly demonstrate an effect of DSCAM overexpression in dendrite branching of hippocampal neurons.
Figure 4.
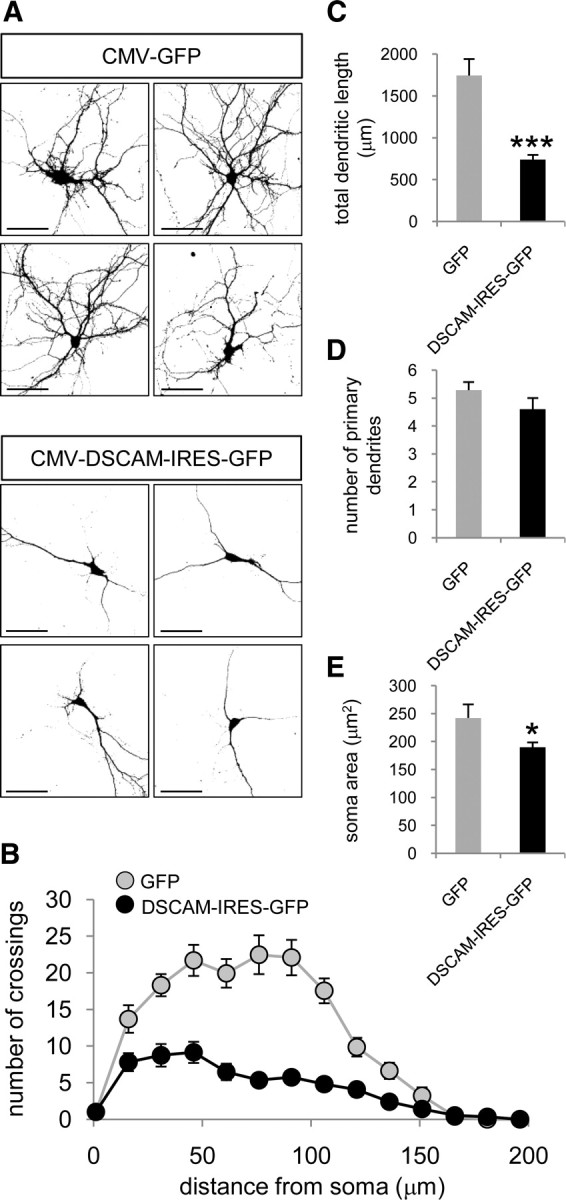
Decreased dendritic complexity in DSCAM-overexpressing hippocampal neurons. A, Representative micrographs of hippocampal neurons transfected at DIV7 with the indicated constructs. Neuronal morphology was visualized by GFP immunocytochemistry 2 d after transfection. Dendrites were identified as MAP2-positive neurites. Scale bars, 60 μm. B, Sholl analysis of the neurons transfected with GFP or DSCAM-IRES-GFP vectors. C, Total dendritic length of neurons transfected with the indicated constructs. D, Number of primary dendrites of neurons transfected with the indicated constructs. E, Somatic area of neurons transfected with the indicated plasmids. Mean values are shown (12–15 neurons); the error bars indicate the SEM, and asterisks denote statistically significant differences: *p < 0.05, ***p < 0.001.
Dendritic DSCAM mRNA and protein levels increase in the hippocampus of trisomic Ts1Cje mice
DSCAM belongs to the DS critical region, a HSA21 fragment containing 33 genes necessary in trisomy to produce the learning and memory impairment associated with the hippocampus in DS (Delabar et al., 1993; Korenberg et al., 1994; Olson et al., 2007; Belichenko et al., 2009). Hence, we quantified the levels of DSCAM mRNA and protein in the dendrites of neurons from the hippocampus of Ts1Cje mice, a widely used model of DS. These mice are trisomic for ∼81 genes, including the DS critical region genes (Sago et al., 1998). Quantitative real-time RT-PCR indicated there was a 1.5-fold increase in the level of DSCAM mRNA (as expected for a gene in trisomy) in the hippocampus of Ts1Cje mice when compared with wild-type (Fig. 5A). Interestingly, a greater increase (2.6-fold) was detected in hippocampal synaptoneurosomes (Fig. 5A).
Figure 5.
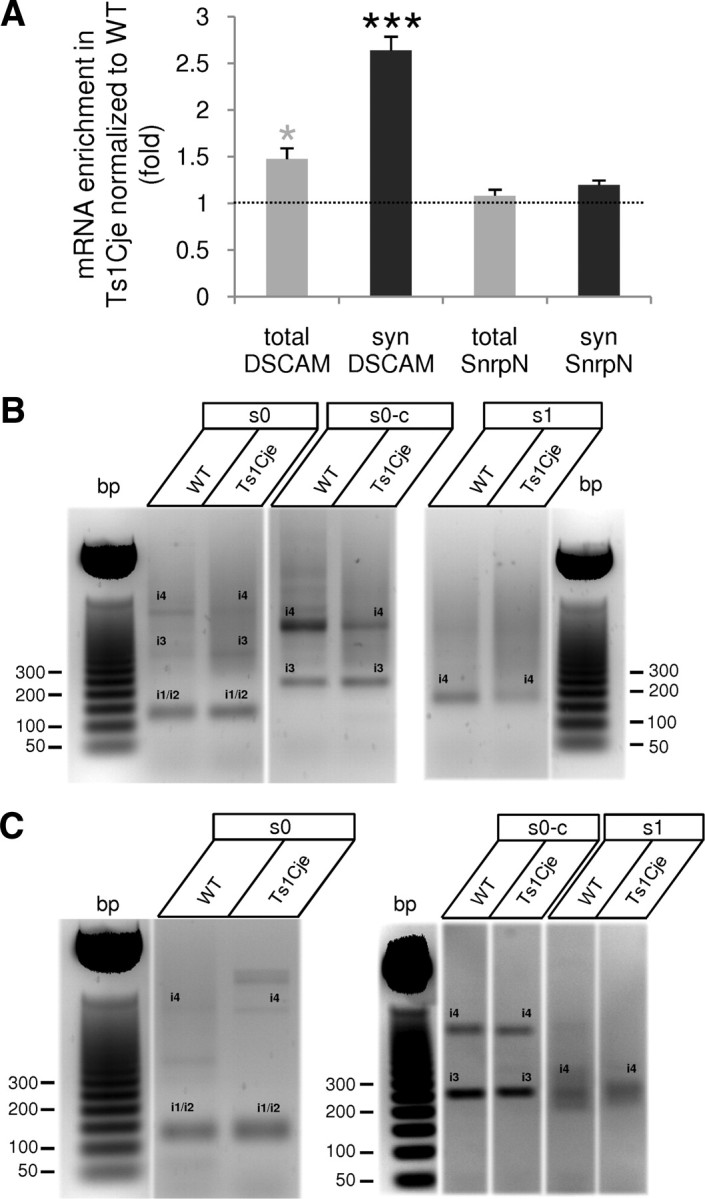
Increased dendritic level of DSCAM mRNA in the hippocampus of Ts1Cje mice. A, Increased levels of DSCAM mRNA in Ts1Cje total hippocampal extract (total) and synaptoneurosomes (syn), when compared with their wild-type (WT) littermates. The soma-restricted SnrpN mRNA was used as a control. The mRNA levels, determined by quantitative real-time RT-PCR, were normalized to HPRT mRNA, and the means from three to four independent experiments (2 independent RT-PCR determinations per experiment) are shown. The error bars indicate the SEM, and the gray and black asterisks denote statistically significant differences with respect to total SnrpN and synaptoneurosomal SnrpN values, respectively: *p < 0.05, ***p < 0.001. B, RL-PAT assay on total hippocampal RNA with oligos s0, s0-c, and s1 showing a similar pattern of DSCAM isoforms in Ts1Cje and wild-type (WT) mice. Note that, in this particular experiment, s0 oligo amplified isoforms 3 and 4 (i3 and i4, identified after cloning and sequencing) in addition to isoforms 1/2 (i1/i2); similarly, i4 was also visible when using oligo s0-c (identified by sequencing). C, RL-PAT with oligos s0, s0-c, and s1 showing a pattern of DSCAM isoform similar in wild-type (WT) and Ts1Cje hippocampal synaptoneurosomes. In this particular experiment, isoform 4 (i4) was also visible when using oligos s0 and s0-c.
RL-PAT of DSCAM isoforms 1/2, 3, and 4 using RNA from Ts1Cje adult hippocampus or hippocampal synaptoneurosomes showed a profile very similar to that of the wild-type counterpart (Fig. 5B,C), suggesting that the overall polyadenylation signal usage was not altered in trisomic mice. RL-PAT with oligo s2 (isoform 5) was not performed because of the low signal obtained when using adult tissue (see Fig. 2B,D). However, quantitative real-time RT-PCR with DSCAM2A primers let us to conclude that isoform 5 was present and overexpressed (∼1.5-fold) in Ts1Cje hippocampal synaptoneurosomes (data not shown).
To determine whether the higher dendritic DSCAM mRNA levels were correlated with higher protein expression in dendrites, a polyclonal antibody against a synthetic DSCAM peptide was raised (for details, see Materials and Methods). This antibody was used to detect the DSCAM protein in brain slices from adult wild-type and Ts1Cje mice by immunohistochemistry. DSCAM expression was detected in somatic and dendritic layers of the hippocampus (Fig. 6), and it increased in dentate gyrus, pyramidal cell layer, and, particularly, in dendritic layers of the Ts1Cje hippocampus when compared with the wild type. Together, these findings demonstrate an increase in both dendritic DSCAM mRNA and protein in DS mice.
Figure 6.
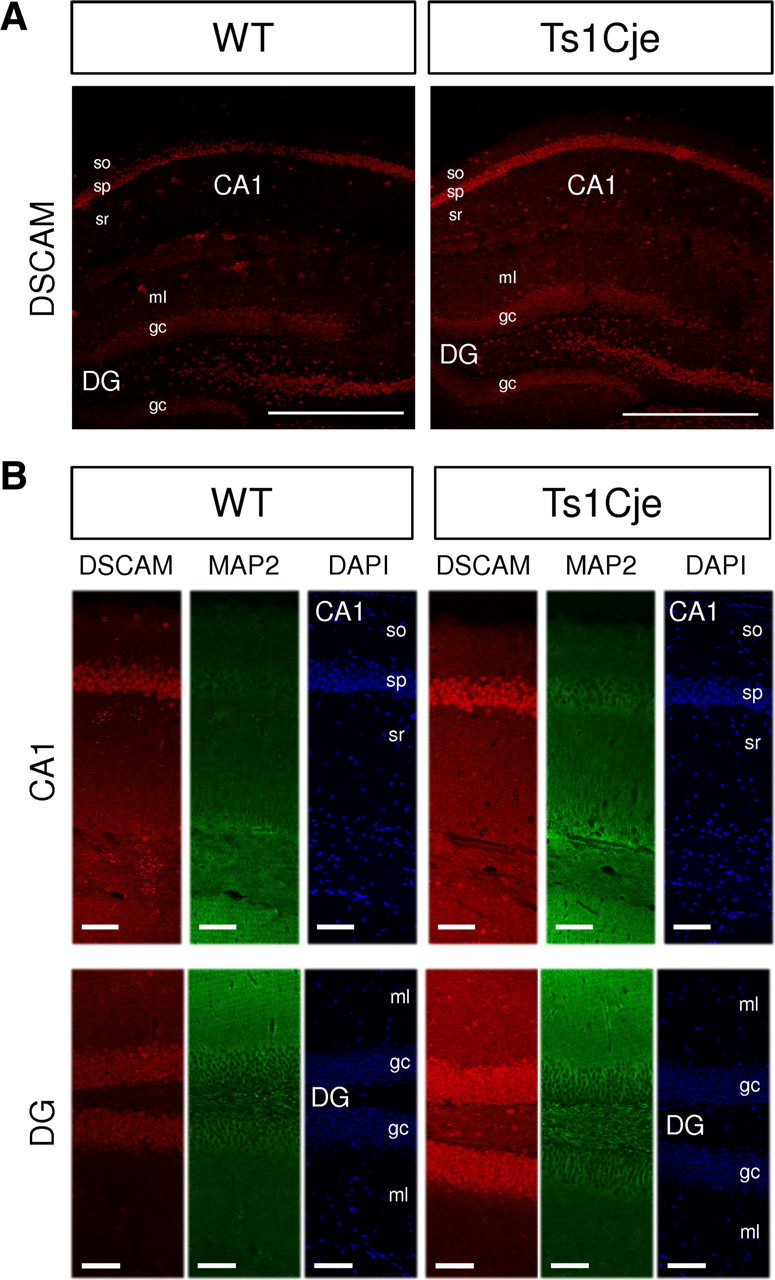
Increased dendritic level of DSCAM protein in the hippocampus of Ts1Cje mice. A, Immunohistochemical detection of DSCAM protein showing an increase in the Ts1Cje adult hippocampus compared with the wild type (WT). A confocal section is shown. Scale bars, 500 μm. so, Stratum oriens; sp, stratum pyramidale; sr, stratum radiatum; DG, dentate gyrus; ml, molecular layer; gc, granule cell layer. B, Comparison of DSCAM signal in dendritic (MAP2-positive) and somatic (DAPI-labeled) layers of wild-type (WT) and Ts1Cje hippocampus. Signal intensities were higher in dendritic layers of CA1 and DG, as well as in pyramidal cell layer (sp, stratum pyramidale) and granule cells (gc) of Ts1Cje hippocampus. A maximal projection of four consecutive confocal sections taken at equivalent locations at the hippocampus is shown in each case. Scale bars, 80 μm. A and B correspond to different experiments.
NMDAR-mediated synaptic activity regulates the levels of DSCAM protein in dendrites, and this regulation is lost in Ts1Cje neurons
In dendrites, CPEB1 is phosphorylated in response to NMDA receptor signaling, leading to cytoplasmic polyadenylation of target mRNAs coupled to translational activation (Huang et al., 2002). To investigate whether NMDAR activation also regulates the local translation of dendritic DSCAM, primary cultures of DIV12 hippocampal neurons were treated with NMDA and the amount of DSCAM protein was estimated by quantifying the fluorescent immunocytochemical dendritic (MAP2-positive) signal.
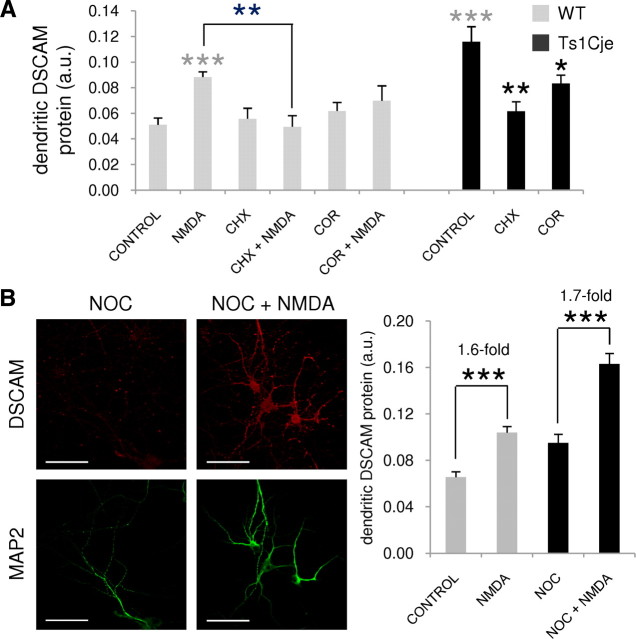
In wild-type neurons, exposure to NMDA (10 min) produced a 1.8-fold increase in dendritic DSCAM protein when compared with untreated control neurons (Fig. 7A1,A2,B). Preincubation of the cultures for 10 min with APV, a specific NMDAR antagonist, abolished this effect (Fig. 7A4,B) (p < 0.001 for Mann–Whitney rank sum test comparison between NMDA and APV plus NMDA treatments).
Figure 7.
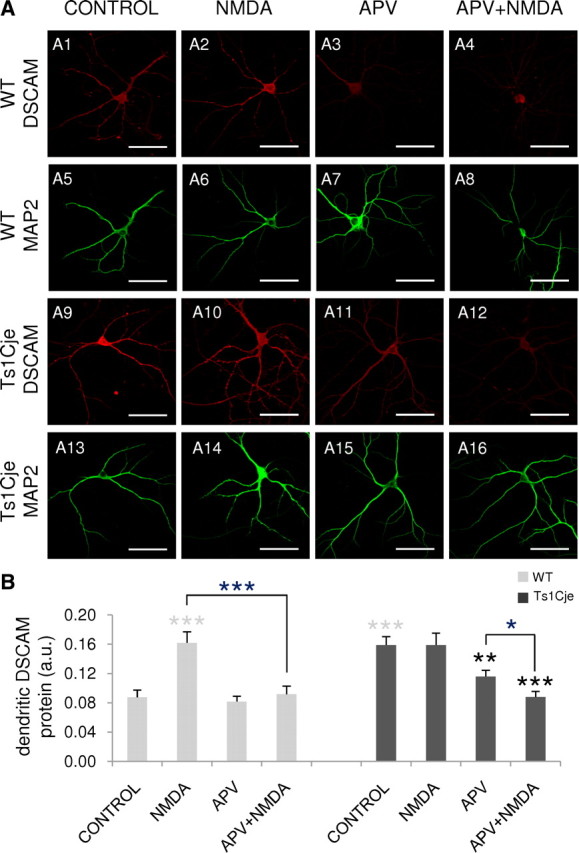
Regulation of the dendritic expression of DSCAM by NMDA. A, DSCAM immunocytochemistry with DIV12 hippocampal neurons from wild-type (WT) and Ts1Cje littermates untreated (CONTROL) and exposed to 50 μm NMDA (NMDA) or 120 μm APV (APV) for 10 min, or preincubated with 120 μm APV for 10 min before the addition of 50 μm NMDA for 10 min (APV+NMDA). The dendrites are labeled by MAP2 immunofluorescence. Scale bars, 60 μm. B, Relative DSCAM protein levels in dendrites of neurons treated as described in A [arbitrary units (a.u.)]. The mean pixel intensity for DSCAM immunofluorescence in dendrites was quantified in 12–20 images from two independent experiments. Error bars indicate the SEM. The statistically significant differences with respect to the wild-type and Ts1Cje controls are indicated with gray and black asterisks, respectively: *p < 0.05, **p < 0.01, ***p < 0.001.
Remarkably, NMDA stimulation had no effect on dendritic DSCAM protein levels in Ts1Cje hippocampal neurons (Fig. 7A10,B), in which basal conditions were in fact 1.8-fold higher (Fig. 7A9,B) than the wild-type counterparts (Fig. 7A1,B). However, a 10 min treatment of Ts1Cje neurons with APV decreased DSCAM immunolabeling in dendrites (Fig. 7A11,B). Significantly lower levels of dendritic DSCAM were found in APV plus NMDA-treated neurons (Fig. 7A12,B) when APV was applied for 20 min (a 10 min pretreatment plus an additional 10 min treatment in the presence of NMDA treatment as APV was not removed from the medium). This suggests that NMDAR signaling is necessary to sustain the relatively high levels of DSCAM protein in dendrites of DS mice.
Inhibition of protein synthesis and polyadenylation block CPEB1-dependent local translation of dendritic mRNAs (Wu et al., 1998; Wells et al., 2001; Atkins et al., 2004). To test whether mRNA polyadenylation and protein synthesis were required for the NMDA-induced increase in dendritic DSCAM protein, wild-type neurons were preincubated with cordycepin (COR) or cycloheximide (CHX), before treatment with NMDA (10 min). Preincubation with cycloheximide to inhibit translation abolished the NMDA-induced increase in dendritic DSCAM, and the presence of cordycepin, an inhibitor of polyadenylation, appeared to attenuate the NMDA-induced increase in dendritic DSCAM (Fig. 8A), although this decrease was not statistically significant. In Ts1Cje neurons, the high basal level of dendritic DSCAM protein (2.3-fold higher than wild-type in this particular experiment) diminished significantly when cells were pretreated with cycloheximide or cordycepin (Fig. 8A). Thus, mRNA polyadenylation and translation were necessary to maintain elevated DSCAM expression in the dendrites of trisomic mice.
Figure 8.
Effect of cycloheximide, cordycepin, and nocodazole on NMDA regulation of the dendritic levels of DSCAM. A, DIV12 hippocampal neurons from wild-type (WT) or Ts1Cje littermates receiving no treatment (CONTROL), 50 μm NMDA (NMDA), 1 μm cycloheximide (CHX), or 80 μm cordycepin (COR) for 10 min, or preincubated with cycloheximide or cordycepin for 10 min before addition of 50 μm NMDA for an additional 10 min period (CHX+NMDA and COR+NMDA, respectively), as indicated. Mean pixel intensity of dendritic DSCAM labeling after immunocytochemistry was quantified in six to eight images, and is shown in arbitrary units (a.u.). Error bars indicate the SEM. Statistically significant differences with respect to wild-type and Ts1Cje controls are indicated with gray and black asterisks, respectively: *p < 0.05, **p < 0.01, ***p < 0.001. B, DSCAM and MAP2 immunocytochemistry with DIV10 hippocampal neurons preincubated with 1 μg/ml nocodazole (NOC) during 6 h before addition or not of 50 μm NMDA for an additional 10 min period, as indicated. Scale bars, 60 μm. Note the punctuated MAP2 immunolabeling, probably reflecting the effect of nocodazole on the microtubule network. Mean pixel intensity of dendritic DSCAM labeling after immunocytochemistry was quantified in 20 images and is shown in arbitrary units (a.u.). Error bars indicate the SEM. Statistically significant differences are indicated with asterisks: ***p < 0.001. Note that, in this experiment, values between nontreated and nocodazole-treated neurons are not comparable, because nocodazole solution contained DMSO as vehicle.
In wild-type neurons, exposure to NMDA also produced an increase of DSCAM protein in the cell body (Fig. 7, compare A1, A2). In fact, quantification of the fluorescent immunocytochemical DSCAM signal in somata showed an increase of 1.5-fold in NMDA-treated neurons (data not shown). Nocodazole inhibits the protein transport from the neuronal body to the dendrites, by affecting the amount of polymerized tubulin (Dotti and Banker, 1991; Cid-Arregui et al., 1995). Accordingly, it has been shown that activity-induced dendritic local translation of BDNF and TrkB mRNAs also occurs (even if at lower levels) in nocodazole-treated hippocampal neurons (Tongiorgi et al., 1997). Hippocampal neurons were preincubated with nocodazole for 6 h as described by Tongiorgi et al. (1997), before the NMDA treatment, to evaluate a possible contribution of DSCAM protein transport from the soma to the dendrites. In these experiments, the NMDA-induced dendritic DSCAM increase was similar in control and nocodazole-treated neurons (Fig. 8B), suggesting that it was attributable to local translation of dendritic DSCAM mRNA, and not to the transport of DSCAM protein from the neuronal body.
Together, these results demonstrate that DSCAM protein levels in dendrites of hippocampal wild-type neurons are regulated by NMDAR-mediated synaptic activity and that this regulation, which requires polyadenylation and protein synthesis, and is independent of the microtubule-based protein transport from the soma, is lost in Ts1Cje neurons.
Discussion
DSCAM encodes a neural cell adhesion molecule belonging to the Ig superfamily that mediates homophilic binding (Yamakawa et al., 1998; Agarwala et al., 2000). Most studies have focused on Drosophila DSCAM1, a gene alternatively spliced to produce >38,000 possible protein isoforms (Schmucker et al., 2000), providing the basis for self-avoidance and tiling (Millard and Zipursky, 2008). Although the mammalian DSCAM gene is unable to generate such wide molecular diversity, it has been demonstrated to mediate homophilic repulsion, promoting dendrite self-avoidance in the developing mouse retina (Fuerst et al., 2008, 2009). Moreover, roles of DSCAM in both de novo and learning-related synaptogenesis have been shown using diverse biological models (Yamagata and Sanes, 2008; Li et al., 2009), and the interaction of DSCAM with PSD-95 and MAGI scaffolding proteins seems to be necessary for its synaptic function (Yamagata and Sanes, 2010).
Two DSCAM isoforms have been described in humans (Yamakawa et al., 1998), a full-length isoform and a soluble isoform lacking the transmembrane domain, and they may also exist in the mouse (Agarwala et al., 2001). Here, we provide evidence that there is an additional complexity of DSCAM isoforms, alternative polyadenylation producing at least five isoforms bearing different 3′-UTRs. Moreover, for the first time, we report the dendritic localization of DSCAM mRNA and its association to CPEB1 in the mouse hippocampus. CPEB1 mediates the transport of mRNAs to dendrites (Huang et al., 2003), thereby regulating local dendritic translation after glutamatergic synaptic activity (Huang et al., 2002). In addition to its role in synaptic plasticity (Alarcon et al., 2004; Berger-Sweeney et al., 2006; McEvoy et al., 2007), CPEB1 has also been shown to regulate dendrite morphogenesis and neuronal circuit assembly (Bestman and Cline, 2008, 2009). Thus, CPEB1-dependent local dendritic translation of DSCAM mRNA might play an important role in dendrite morphogenesis in different brain areas, including the hippocampus, and it might also influence synaptic plasticity in adulthood.
Although all DSCAM 3′-UTR isoforms appear to be transported to dendrites, we identified different translational behavior of these isoforms in Xenopus oocytes. The CPE code predicts the translational regulation of CPE-containing mRNAs (Piqué et al., 2008). It has been established that repression requires at least two CPEs located anywhere in the 3′-UTR and that the distance between the CPEs dictates the degree of repression. The optimal distance for repression is 10–12 nt, whereas at a distance >50 nt they become nonfunctional (Piqué et al., 2008). Thus, DSCAM isoforms 3, 4, and 5 were predicted to be repressed.
Conversely, polyadenylation-mediated translational activation requires a consensus CPE or a nonconsensus CPE together with a PBE (separated in this case by 15–30 nt). The CPE must be <100 nt from the polyadenylation hexamer (optimal distance, 25 nt), but not overlapping (Piqué et al., 2008). In this way, DSCAM isoforms 4 and 5 were predicted to be cytoplasmically polyadenylated and activated (strongly in the case of isoform 4 and weakly in the case of isoform 5).
Finally, it was predicted that DSCAM isoforms 1 and 2 would not be regulated by CPEB1, given that the distance between the PBE and ncCPE1 was >30 nt. Furthermore, ncCPE1 is not conserved in mammals, indicating that these isoforms are not regulated by CPEB1 in rat or human.
Experiments with luciferase-DSCAM 3′-UTR reporters in Xenopus oocytes confirmed the predicted CPE-mediated translational control, although the DSCAM isoform 4 was de-repressed rather than activated, and paradoxically isoform 5 was not regulated. As previously reported for other mRNAs (Piqué et al., 2008), modification of CPE-mediated translational regulation by additional regulatory elements present in the 3′-UTR of DSCAM isoforms 4 and 5 may explain this behavior. In any case, DSCAM translational regulation is likely to be more complex in the mouse hippocampus than in the heterologous molecular context of Xenopus oocytes. Indeed, DSCAM mRNA has been found to be selectively associated with fragile X mental retardation protein (FMRP)–mRNP complexes in the mouse brain (Brown et al., 2001). FMRP is another key regulator of transport and local translation of synaptic mRNAs, and the absence of FMRP provokes fragile X syndrome, a cause of mental retardation (Bassell and Warren, 2008). Thus, DSCAM provides an interesting molecular link between Down's and fragile X syndromes, emphasizing the potential significance of local translation in the context of DS mental retardation.
DSCAM is located in the critical region of HSA21. Increased DSCAM protein levels in the brains of DS patients have been reported (Saito et al., 2000). We observed that overexpression of DSCAM in hippocampal neurons produced a dramatic inhibition of dendrite branching, in agreement with the dendritic phenotype observed in DS patients (Marin-Padilla, 1972).
The 1.5-fold increase in DSCAM expression in the Ts1Cje hippocampus demonstrated here might well be expected for a trisomic gene. However, in Ts1Cje hippocampal synaptoneurosomes, DSCAM mRNA was 2.6-fold more abundant than in wild-type counterparts. This suggests that the accumulation of dendritic DSCAM mRNA in the Ts1Cje hippocampus was not merely because of gene overexpression produced by the trisomy and that additional deficits in mRNA transport, docking, and/or stability may occur in these trisomic mice.
As expected, an increase in the dendritic levels of DSCAM mRNA was correlated with an increase in the amount of dendritic DSCAM protein. Most importantly, the results presented here demonstrate that dendritic levels of DSCAM protein are regulated by NMDAR-mediated synaptic activity and that this regulation is lost in trisomic hippocampal neurons. Dendritic levels of DSCAM protein increased rapidly in hippocampal neurons treated with NMDA in a manner dependent on protein synthesis and polyadenylation. Moreover, it occurred at the same extent in nocodazole-treated neurons, precluding the possibility that it was attributable to DSCAM protein transport along microtubules from the soma to dendrites. In conjunction with the observed enrichment of DSCAM mRNA in synaptoneurosomes and its association with immunoprecipitated CPEB1 protein in hippocampus, we propose that DSCAM is a mRNA locally translated in dendrites, probably under the regulation of CPEB1 in the mouse hippocampus (Fig. 9).
Figure 9.
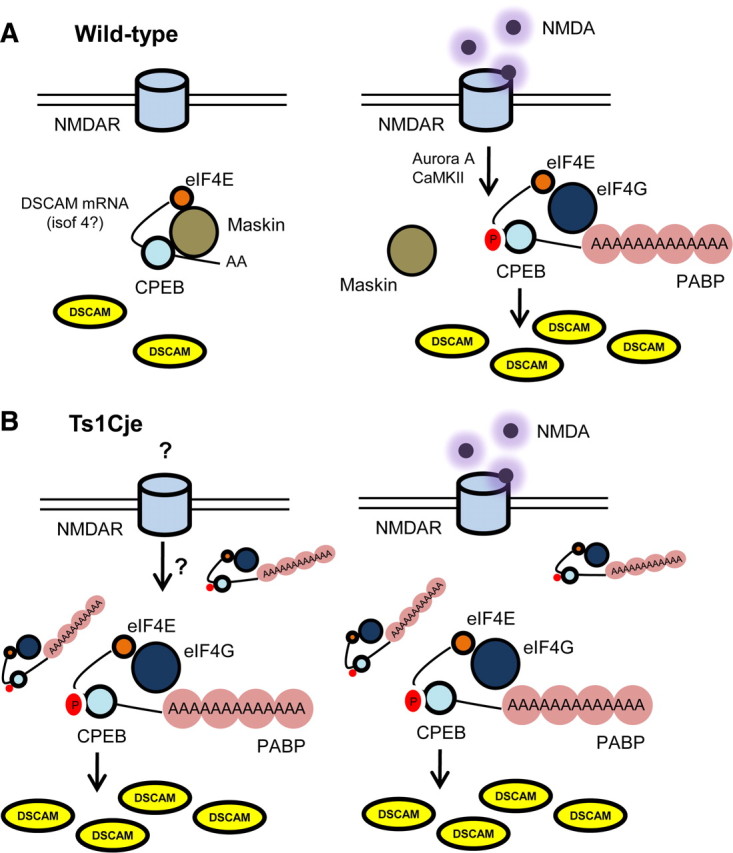
Hypothetical model describing the local NMDA-regulated dendritic translation of DSCAM in wild type and Ts1Cje mice. A, In wild-type hippocampal neurons, synaptic activity mediated by NMDA activates kinases (Aurora A or CaMKII) capable of phosphorylating CPEB1 protein. CPEB1 phosphorylation disrupts the inhibitory Maskin–eIF4E interaction, promoting polyadenylation of DSCAM mRNA. PABP associates to DSCAM poly(A) tail and interacts with eIF4G, stabilizing the eIF4E–eIF4G interaction, which is essential for the initiation of translation. B, In trisomic hippocampal neurons, dendritic DSCAM mRNA levels and translation are constitutively increased, regardless of the presence of NMDA. However, DSCAM translation requires both NMDA receptor activity and polyadenylation as it is blocked by APV and cordycepin. Thus, Ts1Cje neurons are unable to modulate DSCAM translation in response to NMDAR-mediated synaptic activity. This impairment may contribute to DS neuropathology.
Which isoforms are responsible for the NMDA-induced increase in dendritic DSCAM? Although all DSCAM isoforms were detected in synaptoneurosomes and they were presumably located in dendrites, only isoform 4 was activated by progesterone in Xenopus oocytes. Furthermore, although isoforms 1/2, 3, and 5 appeared to be more abundant in the neonatal hippocampus (as deduced by intensity of RL-PAT products), isoform 4 was expressed at similar levels in P0 and adult tissues. These findings might suggest that the DSCAM isoform 4 may play a predominant role in adult neuronal physiology (i.e., synaptic plasticity), whereas other isoforms may mediate developmental processes (i.e., dendritogenesis). Finally, the RL-PAT product for isoform 4 (oligo s1) appeared highly polyadenylated in the total adult hippocampal extracts but not in synaptoneurosomes, suggesting that isoform 4 should be repressed in dendrites [having a short poly(A) tail] and thus it may be susceptible to NMDA activation. In any case, as the inhibitory effect of cordycepin was limited other isoforms may participate in the activity-regulated increase of dendritic DSCAM protein in a polyadenylation-independent manner.
Interestingly, Ts1Cje neurons exhibited a basal level of dendritic DSCAM protein similar to that found in NMDA-activated wild-type neurons. Although addition of NMDA did not further increase dendritic DSCAM labeling in Ts1Cje neurons, NMDAR signaling was necessary to maintain DSCAM protein above the basal wild-type levels. Polyadenylation and protein synthesis were also required. It is interesting to note that the NMDA-regulated pool of dendritic DSCAM protein in Ts1Cje neurons has a rapid turnover, as short APV or cycloheximide treatments (10 min) were sufficient to markedly reduce the amount of dendritic DSCAM protein. Alternatively, a more complex regulatory process as ectodomain shedding could explain the rapid loss of DSCAM labeling. The release of the extracellular domain through limited proteolysis is indeed a frequent mechanism to regulate the function of transmembrane proteins (Arribas and Borroto, 2002).
In conclusion, the regulation of local dendritic translation of DSCAM mediated by NMDA signaling is lost in trisomic neurons (Fig. 9). In support of this idea, Ts1Cje are hypersensitive to the channel blocker MK-801 [(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate], suggesting dysregulation of NMDAR signaling (Siddiqui et al., 2008). We hypothesize that impairment of NMDA regulation of local dendritic mRNA translation, such as that of DSCAM, may contribute to deficits in dendrite morphology and/or synaptic plasticity in DS. Additional studies are warranted to fully address this question.
Footnotes
This work was supported by the Ministerio de Ciencia e Innovación (Instituto de Salud Carlos III, Spain, Grants PI050600 and PI081401), Fondation Jérôme Lejeune (France), Fundación Ramón Areces (Spain), and Junta de Andalucía Grant P09-CTS-4610. We are very grateful to Dr. Raúl Méndez and Dr. María Piqué for providing the luciferase vectors; Dr. Joshua Sanes for the CMV-DSCAM-IRES-GFP vector; Dr. Antonio Leal, Dr. Javier Salguero, and Leonardo Gómez Sánchez for help with the image analysis; and Dr. Miriam Echevarría for the use of the oocyte injector. We also thank Itziar Benito for expert technical assistance in mouse handling and management with the surgical procedures in Xenopus, as well as Dr. Francisco Tejedor for his critical comments on this manuscript.
References
- Agarwala KL, Nakamura S, Tsutsumi Y, Yamakawa K. Down syndrome cell adhesion molecule DSCAM mediates homophilic intercellular adhesion. Brain Res Mol Brain Res. 2000;79:118–126. doi: 10.1016/s0169-328x(00)00108-x. [DOI] [PubMed] [Google Scholar]
- Agarwala KL, Ganesh S, Suzuki T, Akagi T, Kaneko K, Amano K, Tsutsumi Y, Yamaguchi K, Hashikawa T, Yamakawa K. Dscam is associated with axonal and dendritic features of neuronal cells. J Neurosci Res. 2001;66:337–346. doi: 10.1002/jnr.1226. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of Schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Arribas J, Borroto A. Protein ectodomain shedding. Chem Rev. 2002;102:4627–4638. doi: 10.1021/cr010202t. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Mannucci L, Dotti CG, Amaldi F. Chemical stimulation of synaptosomes modulates alpha-Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J Neurosci. 2000;20:RC76, 1–6. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow GM, Micales B, Lyons GE, Korenberg JR. Down syndrome cell adhesion molecule is conserved in mouse and highly expressed in the adult mouse brain. Cytogenet Cell Genet. 2001;94:155–162. doi: 10.1159/000048808. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belichenko NP, Belichenko PV, Kleschevnikov AM, Salehi A, Reeves RH, Mobley WC. The “Down syndrome critical region” is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of Down syndrome. J Neurosci. 2009;29:5938–5948. doi: 10.1523/JNEUROSCI.1547-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The relationship between dendritic branch dynamics and CPEB-labeled RNP granules captured in vivo. Front Neural Circuits. 2009;3:10. doi: 10.3389/neuro.04.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Buettner VL, LeBon JM, Gao C, Riggs AD, Singer-Sam J. Use of terminal transferase-dependent antisense RNA amplification to determine the transcription start site of the Snrpn gene in individual neurons. Nucleic Acids Res. 2000;28:E25. doi: 10.1093/nar/28.7.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid-Arregui A, Parton RG, Simons K, Dotti CG. Nocodazole-dependent transport, and brefeldin A-sensitive processing and sorting, of newly synthesized membrane proteins in cultured neurons. J Neurosci. 1995;15:4259–4269. doi: 10.1523/JNEUROSCI.15-06-04259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino M, Capano CP, Aiello A, Iannetti E, Cupello A, Giuditta A. Messenger RNAs in synaptosomal fractions from rat brain. Brain Res Mol Brain Res. 2001;97:171–176. doi: 10.1016/s0169-328x(01)00321-7. [DOI] [PubMed] [Google Scholar]
- Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, Prieur M, Noel B, Sinet PM. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- Di Nardo AA, Nedelec S, Trembleau A, Volovitch M, Prochiantz A, Montesinos ML. Dendritic localization and activity-dependent translation of Engrailed1 transcription factor. Mol Cell Neurosci. 2007;35:230–236. doi: 10.1016/j.mcn.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Banker G. Intracellular organization of hippocampal neurons during the development of neuronal polarity. J Cell Sci Suppl. 1991;15:75–84. doi: 10.1242/jcs.1991.supplement_15.11. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Gullotta F. Down's syndrome and Alzheimer's disease: dendritic spine counts in the hippocampus. Acta Neuropathol. 1990;79:680–685. doi: 10.1007/BF00294247. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Huang BS, Vishwasrao H, Sutedja N, Chen W, Jin I, Hawkins RD, Bailey CH, Kandel ER. Dscam mediates remodeling of glutamate receptors in Aplysia during de novo and learning-related synapse formation. Neuron. 2009;61:527–540. doi: 10.1016/j.neuron.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M. Structural abnormalities of the cerebral cortex in human chromosomal aberrations: a Golgi study. Brain Res. 1972;44:625–629. doi: 10.1016/0006-8993(72)90324-1. [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M. Pyramidal cell abnormalities in the motor cortex of a child with Down's syndrome. A Golgi study. J Comp Neurol. 1976;167:63–81. doi: 10.1002/cne.901670105. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Setou M, Inokuchi K. Transcriptome analysis reveals the population of dendritic RNAs and their redistribution by neural activity. Neurosci Res. 2007;57:411–423. doi: 10.1016/j.neures.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci U S A. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy M, Cao G, Montero Llopis P, Kundel M, Jones K, Hofler C, Shin C, Wells DG. Cytoplasmic polyadenylation element binding protein 1-mediated mRNA translation in Purkinje neurons is required for cerebellar long-term depression and motor coordination. J Neurosci. 2007;27:6400–6411. doi: 10.1523/JNEUROSCI.5211-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Millard SS, Zipursky SL. Dscam-mediated repulsion controls tiling and self-avoidance. Curr Opin Neurobiol. 2008;18:84–89. doi: 10.1016/j.conb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Olson LE, Roper RJ, Sengstaken CL, Peterson EA, Aquino V, Galdzicki Z, Siarey R, Pletnikov M, Moran TH, Reeves RH. Trisomy for the Down syndrome “critical region” is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet. 2007;16:774–782. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- Piqué M, López JM, Méndez R. Cytoplasmic mRNA polyadenylation and translation assays. Methods Mol Biol. 2006;322:183–198. doi: 10.1007/978-1-59745-000-3_13. [DOI] [PubMed] [Google Scholar]
- Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274:438–449. doi: 10.1006/viro.2000.0494. [DOI] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, Epstein CJ, Huang TT. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci U S A. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Oka A, Mizuguchi M, Motonaga K, Mori Y, Becker LE, Arima K, Miyauchi J, Takashima S. The developmental and aging changes of Down's syndrome cell adhesion molecule expression in normal and Down's syndrome brains. Acta Neuropathol. 2000;100:654–664. doi: 10.1007/s004010000230. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Lacroix T, Stasko MR, Scott-McKean JJ, Costa AC, Gardiner KJ. Molecular responses of the Ts65Dn and Ts1Cje mouse models of Down syndrome to MK-801. Genes Brain Behav. 2008;7:810–820. doi: 10.1111/j.1601-183X.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu M, Mehraein P. Spine distribution along the apical dendrites of the pyramidal neurons in Down's syndrome. A quantitative Golgi study. Acta Neuropathol. 1980;50:207–210. doi: 10.1007/BF00688755. [DOI] [PubMed] [Google Scholar]
- Sung YJ, Weiler IJ, Greenough WT, Denman RB. Selectively enriched mRNAs in rat synaptoneurosomes. Brain Res Mol Brain Res. 2004;126:81–87. doi: 10.1016/j.molbrainres.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Takashima S, Becker LE, Armstrong DL, Chan F. Abnormal neuronal development in the visual cortex of the human fetus and infant with down's syndrome. A quantitative and qualitative Golgi study. Brain Res. 1981;225:1–21. doi: 10.1016/0006-8993(81)90314-0. [DOI] [PubMed] [Google Scholar]
- Tanguay RL, Gallie DR. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troca-Marín JA, Alves-Sampaio A, Tejedor FJ, Montesinos ML. Local translation of dendritic RhoA revealed by an improved synaptoneurosome preparation. Mol Cell Neurosci. 2010;43:308–314. doi: 10.1016/j.mcn.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol. 2000;10:1147–1150. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci. 2010;30:3579–3588. doi: 10.1523/JNEUROSCI.6319-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K, Huot YK, Haendelt MA, Hubert R, Chen XN, Lyons GE, Korenberg JR. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7:227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]