Abstract
Dopaminergic neurons develop at distinct anatomical sites to form some of the major neuromodulatory systems in the vertebrate brain. Despite their relevance in neurodegenerative diseases and the interests in reconstitutive therapies from stem cells, mechanisms of the neurogenic switch from precursor populations to dopaminergic neurons are not well understood. Here, we investigated neurogenesis of different dopaminergic and noradrenergic neuron populations in the zebrafish embryo. Birth-dating analysis by EdU (5-ethynyl-2′-deoxyuridine) incorporation revealed temporal dynamics of catecholaminergic neurogenesis. Analysis of Notch signaling mutants and stage-specific pharmacological inhibition of Notch processing revealed that dopaminergic neurons form by temporally distinct mechanisms: dopaminergic neurons of the posterior tuberculum derive directly from neural plate cells during primary neurogenesis, whereas other dopaminergic groups form in continuous or wavelike neurogenesis phases from proliferating precursor pools. Systematic analysis of Notch ligands revealed that the two zebrafish co-orthologs of mammalian Delta1, DeltaA and DeltaD, control the neurogenic switch of all early developing dopaminergic neurons in a partially redundant manner. DeltaA/D may also be involved in maintenance of dopaminergic precursor pools, as olig2 expression in ventral diencephalic dopaminergic precursors is affected in dla/dld mutants. DeltaA/D act upstream of sim1a and otpa during dopaminergic specification. However, despite the fact that both dopaminergic and corticotropin-releasing hormone neurons derive from sim1a- and otpa-expressing precursors, DeltaA/D does not act as a lineage switch between these two neuronal types. Rather, DeltaA/D limits the size of the sim1a- and otpa-expressing precursor pool from which dopaminergic neurons differentiate.
Introduction
Control of specification and differentiation of dopaminergic (DA) neurons has been intensely studied because of their prominent roles as neuromodulatory systems and the devastating effects on human health in dopamine system dysfunction and degeneration (Dauer and Przedborski, 2003; Iversen and Iversen, 2007). Although signals and transcription factors involved in development of mes-diencephalic DA neurons have been characterized in detail (Ang, 2006; Smidt and Burbach, 2007), little is known about the mechanisms of Notch-signaling mediated neurogenic lineage decisions from stem and precursor cells to DA neurons. The core components of the Notch signaling pathway comprise a family of transmembrane ligands (Delta/Serrate/Jagged) and their receptor Notch (Lewis, 1998; Louvi and Artavanis-Tsakonas, 2006). Ligand binding triggers the release of the Notch intracellular domain and its translocation to the nucleus, where it activates the expression of basic helix–loop–helix transcription factors of the suppressor of hairless HES/HER family (Fortini and Artavanis-Tsakonas, 1994), which in turn repress proneural genes and thus neuronal fate. Proneural transcription factors control general as well as neuronal type-specific aspects of differentiation (Bertrand et al., 2002).
Neurogenesis of mammalian DA neurons proceeds at defined anatomical sites in the forebrain and midbrain during extended developmental periods (Puelles and Verney, 1998; Björklund and Dunnett, 2007). Specific contributions of Notch signaling to DA neurogenesis in mammals have not been investigated so far. However, the proneural gene Neurogenin-2 (Ngn2) has been demonstrated to be required for development of mesencephalic DA neurons (Andersson et al., 2006; Kele et al., 2006). Both Mash1 and Ngn2 are expressed in DA precursors, but only Ngn2 is strictly required for DA development. Ngn2 continues to be expressed in postmitotic DA neurons, indicating a potential role during DA neuronal differentiation and subtype specification (Ang, 2006). For migrating telencephalic olfactory DA neurons, Id2 is required for dopaminergic neurogenesis in adult mice (Havrda et al., 2008). A recent study indicates that a soluble Dll4 form may contribute to rescue of DA neurons from precursor pools in an adult rat model (Androutsellis-Theotokis et al., 2009), suggesting that a better understanding of DA neurogenesis may also open new therapeutic perspectives.
The zebrafish (Danio rerio) has emerged as a model to study DA systems development (Rink and Wullimann, 2002b; Schweitzer and Driever, 2008). Given that the fish brain grows, forms new DA neurons, and regenerates throughout life (Adolf et al., 2006; Grandel et al., 2006), the understanding of mechanisms of neurogenesis may provide a paradigm for DA regeneration. Although mutational and experimental analyses have shed light on signals and transcriptional control of DA development (Guo et al., 1999; Holzschuh et al., 2003; Ryu et al., 2007; Russek-Blum et al., 2008; Löhr et al., 2009), neurogenesis is not understood in detail. In zebrafish, the proneural gene neurogenin1 (ngn1) is required for development of basal diencephalic DA neurons (Jeong et al., 2006), and its overexpression may induce ectopic DA differentiation, arguing that, similar to mammalian Ngn2, zebrafish Ngn1 may also be involved in DA differentiation.
Here, we investigated mechanisms of DA neurogenesis in zebrafish and compare them with neurogenesis of noradrenergic (NA) neurons. We applied detailed birth-dating analysis and mapped the time windows during which DA precursors are sensitive to Notch signaling to identify DA subgroup-specific mechanisms of neurogenesis during development. These experiments revealed distinct mechanisms for specific DA groups. Furthermore, we analyzed which of the Delta and Jagged family Notch ligands mediate DA neuron development and could demonstrate that two paralogous Delta ligands, DeltaA and DeltaD, mediate, in a partially redundant manner, neurogenic decisions of most early-forming DA neurons, regardless of DA subtype. Finally, we analyzed whether Notch signaling may also have effects on DA precursor pool maintenance, and on expression of DA differentiation genes and maintenance of the differentiated state. Our findings provide a basic understanding of mechanisms of neurogenesis during distinct phases of DA systems development in zebrafish.
Materials and Methods
Fish maintenance and mutant lines.
Embryos were staged at 28.5°C (Kimmel et al., 1995). To inhibit pigmentation embryos were raised in medium containing 0.2 mm phenylthiourea (Sigma-Aldrich). AB/TL wild type was used. Mutant strains were as follows: deltaA dlahi781 (Amsterdam and Hopkins, 2004), deltaD aei/dldtr233 (van Eeden et al., 1996), and mibta52b (Itoh et al., 2003). dlahi781 embryos were genotyped by PCR using the following primers: dla_1F, 5′-CGGCTCTGGAGATGCATTGG-3′, located in exon 3 3-prime of the viral insertion; dla_vir_1R, 5′-GCTAGCTTGCCAAACCTACAGGT-3′ binds to viral sequence; dla_JM_R2, 5′-CATGGGACGCCACTTACTGTTGCTCC-3′, located 5-prime to the viral insertion in exon 2 of the dla gene. dldtr233 mutant embryos carry a point mutation (T-to-A exchange) that introduces a restriction site for Tsp509I. Thus, they were genotyped by performing a PCR (primers: dld_in7_F1, 5′-ACTCGCTCGTTCATTGGATT-3′; dld_ex9_R1, 5′-TCTCCTCTGAGTCGGAATCG-3′) followed by enzymatic restriction with Tsp509I.
In situ hybridization and immunohistochemistry.
Whole-mount in situ hybridization (WISH) was performed as described previously (Hauptmann and Gerster, 1994). The following digoxigenin or fluorescein (Roche)-labeled antisense riboprobes were used: th (Holzschuh et al., 2001), sox2 (Katoh and Katoh, 2005), otpa (Ryu et al., 2007), sim1a (Eaton and Glasgow, 2006), isl1 (Korzh et al., 1993), deltaA (Haddon et al., 1998), deltaB, deltaC, deltaD, jagged1a, jagged1b, and jagged2a (Zecchin et al., 2005), dlx5a (Akimenko et al., 1994), ascl1a and ascl1b (Allende and Weinberg, 1994), ngn1 (Blader et al., 1997), nkx2.2a (Barth and Wilson, 1995), olig2 (Park et al., 2002), crh (Chandrasekar et al., 2007), and gfap (Nielsen and Jørgensen, 2003). We used exclusively th1 expression as marker for DA neurons, because the second paralog, th2, is predominantly expressed from early larval stages on (Chen et al., 2009; Filippi et al., 2010), which is later than the stages considered in the current analysis. Fluorescent whole-mount in situ hybridization (FISH) for deltaA, deltaB, deltaD combined with fluorescent immunohistochemistry for tyrosine hydroxylase (TH) was performed as described by Filippi et al. (2007).
Birth dating.
To determine when precursor cells exit the cell cycle, embryos were treated with 5-ethynyl-2′-deoxyuridine (EdU) (Buck et al., 2008; Salic and Mitchison, 2008) using the Click-iT EdU imaging kit (Invitrogen). We incubated wild-type embryos for 15–30 min in EdU: the age of EdU addition was spaced at 4 h intervals between 8 and 24 h post-fertilization (hpf), and at 6 h intervals between 24 and 60 hpf. After the treatment, EdU was removed and embryos developed until 75 hpf under standard conditions. Immunofluorescent staining with anti-TH (Alexa 555 cytoplasmic signal; red) (Ryu et al., 2007) and modified Alexa 488 covalently bound to EdU (nuclear signal; green) was performed, and embryos were analyzed using confocal microscopy.
DAPT treatments and heat shock experiments.
The γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine-t-butyl ester (DAPT) was dissolved at 10 mm in DMSO and diluted to a final concentration of 100 μm in embryo medium (Geling et al., 2002). Different clutches of wild-type embryos were incubated in DAPT solution at 28.5°C for time windows of 10–12 h, starting at time points between 15 and 60 hpf. The DAPT solution was removed by washing five times with embryo medium at 5 min intervals. All embryos were allowed to develop until 75 hpf and then fixed.
For gain-of-function experiments, Hsp70:GAL4 transgenic fish were crossed to UAS:NICD (myc-tagged) transgenic fish (Scheer and Campos-Ortega, 1999). Notch1aICD overexpression was induced by transferring the embryos into preheated medium at 40°C for 30 min at the desired time points of development. After the heat shock, embryos were incubated at 28.5°C until 72 hpf. To confirm the activation of Notch intracellular domain (NICD) expression, all embryos were analyzed by anti-Myc (Sigma-Aldrich) immunofluorescence.
Morpholinos and microinjections.
For knockdown of gene function antisense morpholinos (MOs) (Gene Tools) (Nasevicius and Ekker, 2000) were injected into embryos at one-cell stage. deltaAsplMO, deltaAmismMO, jagged2aATGMO, jagged2asplMO, and jagged2amismMO were previously reported (Zecchin et al., 2007). The sequence of the deltaBATGMO was 5′-GCA GTA CAA AGA CAG ATG CGC CAT C-3′. As a control, we used the Standard Control Oligo provided by Gene Tools. To exclude nonspecific morpholino-induced apoptosis, we coinjected the p53MO (Robu et al., 2007).
Microscopy and imaging.
Stained embryos were mounted in 100% glycerol, and light images were acquired using a Zeiss Axioskop 2 compound microscope, 10× or 20× differential interference contrast optics, and a Zeiss AxioCam MRc digital camera. Fluorescently labeled embryos were documented as confocal stacks of images using a Zeiss LSM 5 DUO upright laser-scanning confocal microscope. The figures were assembled using Adobe Photoshop CS2.
Results
Dopaminergic neurons of the zebrafish forebrain arise from distinct phases of neurogenesis
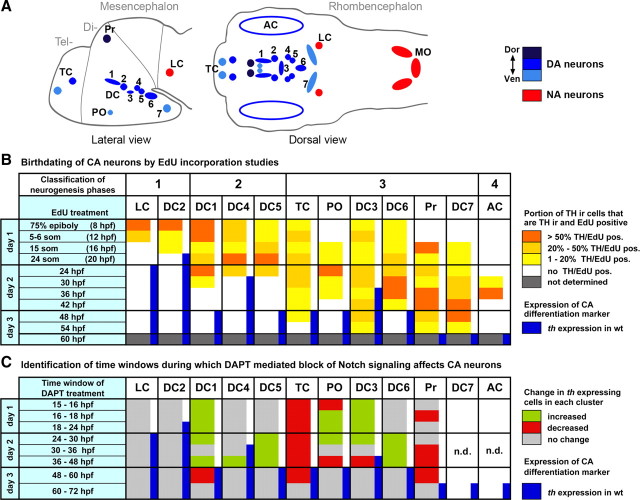
We wanted to determine at which developmental stages catecholaminergic (CA) precursors became postmitotic. Recently, BrdU incorporation experiments in zebrafish have been used to address this question for the earliest developing DA neurons (Russek-Blum et al., 2008). Here, we performed a more extensive analysis of the complete embryonic and early larval period during which DA neurons form (8–54 hpf). We used sequential pulse labeling to resolve distinct periods in which neurons of specific DA groups become postmitotic. As S-phase label, we used Click-iT EdU, which, in contrast to BrdU, can be detected without applying denaturing conditions (Warren et al., 2009) and thus provides a better preservation of the TH antigen. All EdU-labeled embryos were analyzed at 75 hpf by recording confocal stacks throughout the brain of EdU Click-iT–Alexa 488 and anti-TH immunohistochemistry double-labeled larvae (Figs. 1, 2). EdU was selectively incorporated into those proliferative zones of the brain that have been previously described (Fig. 1A–F) (Wullimann et al., 1999). Nuclei that retained a bright and uniform label were considered as having become postmitotic soon after the pulse. In contrast, other nuclei had weak and speckled EdU label, indicating multiple successive cell cycles after the EdU pulse, causing dilution of label (Chan and Gargett, 2006). A more punctuate stain could potentially also result from cells labeled only during a short time window of late S-phase when heterochromatin is replicated (Cameron and McKay, 2001); however, we consider this explanation unlikely given the long EdU pulses used in our study. For our analysis, we distinguished three distinct patterns of EdU label in the nucleus (Fig. 1G–I and legend) that correlate with nuclei (1) already postmitotic at time of pulse, (2) becoming postmitotic soon after pulse, and (3) that continued cycling after pulse. The birth-dating results are summarized in Figure 3B.
Figure 1.
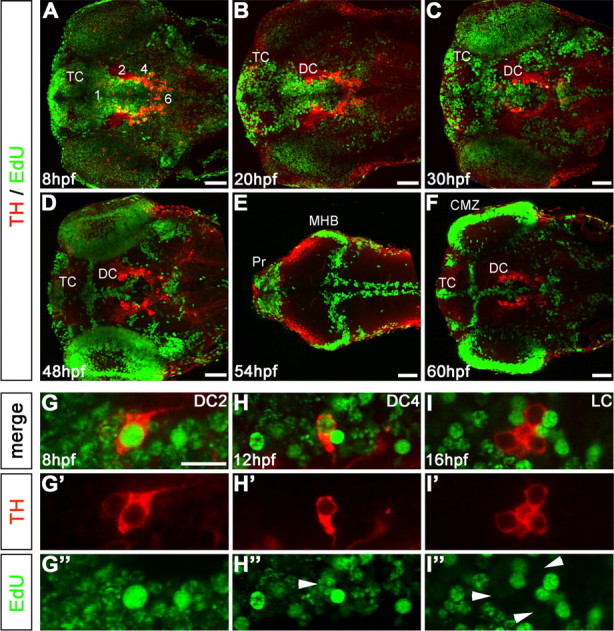
Birth dating of catecholaminergic neurons by pulsed EdU incorporation. A–F, Embryos were labeled with EdU at defined developmental stages, as indicated in panels, and analyzed at 75 hpf by TH immunohistochemistry (red) and EdU Click-iT Alexa 488 label (green). A, Pulse at 8 hpf (75% epiboly) shows most of the neural precursors were still proliferating. B, C, Pulse label at 20 and 30 hpf, Proliferative areas became more restricted. D, Pulse at 48 hpf, EdU labeling retracted to the ventricular proliferative zones (VZ). E, Pulse label at 54 hpf, Dorsal focal planes only: proliferative zones of the midbrain hindbrain boundary (MHB), rhombencephalic VZ, and many cells in the pretectum (Pr) were labeled. F, Pulse at 60 hpf, VZ of the diencephalon and telencephalon (DC and TC, respectively), and retinal ciliary marginal zone (CMZ) were labeled. G–I, EdU label evaluation. G–G″, DC2 THir cell with a bright, homogenous EdU nuclear label considered as a cell that became postmitotic soon after the incorporation of EdU. H–H″, DC4 THir cell with a spotted green nuclear label, considered as a cell that passed through several more cell cycles after incorporating EdU (white arrowhead). I–I″, LC THir cells without green-labeled nuclei (white arrowheads), considered either as cells already postmitotic at the time point of EdU incubation or as cells that passed through so many cell divisions until 75 hpf, that the EdU was not detectable anymore. Given that time windows of CA differentiation are known based on the onset of TH immunoreactivity, both options could be distinguished. A–F, Z-projections of every fifth image plane to show all relevant THir clusters in one image; dorsal views, anterior left. Scale bars: A–F, 50 μm; (in G) G–I″, 20 μm.
Figure 2.
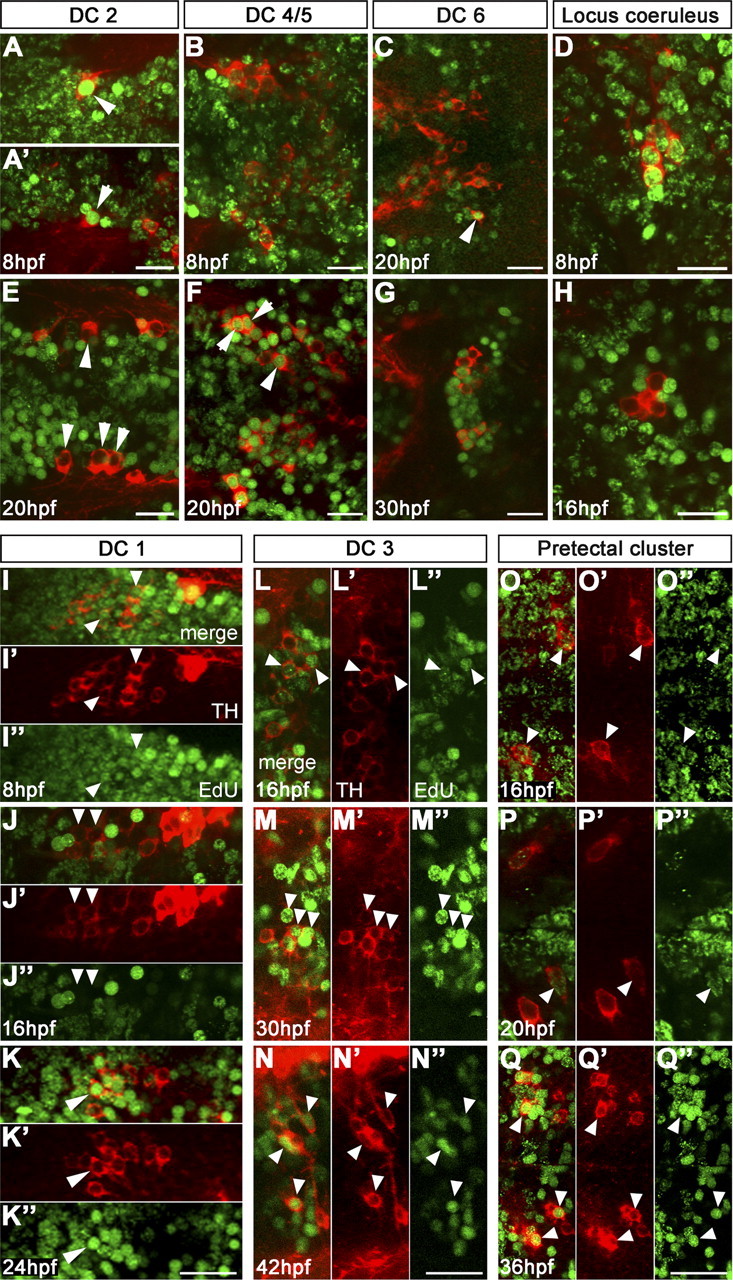
Birth dating reveals distinct developmental stages when neurons in specific DA and NA groups become postmitotic. Embryos were EdU pulse labeled as indicated in panels and analyzed at 75 hpf for EdU incorporation (green) and TH immunoreactivity (red). The arrowheads point at DA neurons. A, E, Precursors of DC2 were brightly labeled by EdU pulse at 8 hpf (different examples in A and A′), whereas after 20 hpf they were postmitotic and did not incorporate EdU anymore. B, F, Precursors of DC4/5 were faintly labeled at 8 hpf and thus continued to cycle, whereas many of them became postmitotic around 20 hpf. C, G, Few DC6 precursors left the cell cycle earlier than 30 hpf, but the vast majority became postmitotic around 30 hpf. D, H, THir cells of the LC incorporated EdU at 8 hpf; no EdU-incorporating nuclei of THir cells were detectable from pulses later than 16 hpf. I–K, Precursors of DC1 were still cycling at neural plate stage; at 16 hpf, only a few DC1 neurons were labeled, and thus early differentiating DC1 precursors were postmitotic, whereas later differentiating neurons showed EdU incorporation at 24 hpf. L–Q″, DA cells of DC3 and pretectum derived continuously over longer developmental times from proliferating precursor pools that still incorporate EdU at 36 and 42 hpf. A–Q, Projections of confocal z-stacks of up to 5 μm depth or single confocal planes; dorsal views, anterior left. A–H display merged TH and EdU detection channels. I–Q display merged channels, TH channel (′), and EdU channel (″) separately. Scale bars, 20 μm.
Figure 3.
Temporal correlations of cell cycle exit, neurogenesis, and differentiation of dopaminergic and noradrenergic groups. A, Schematic drawing representing the distribution of DA (blue) and NA (red) neurons in the zebrafish larval brain. The ventral diencephalic groups are numbered from 1 to 7 according to Rink and Wullimann (2002b). Abbreviations: AC, Amacrine cells; DC1–7, diencephalic DA groups 1–7; MO, medulla oblongata; PO, preoptic group; Pr, pretectal group; TC, telencephalic groups. B, Summary of the results from birth-dating studies. EdU birth-dating results were evaluated as follows: the percentage of THir cells labeled brightly by EdU pulse at a given time point was calculated for each CA neuronal group. Cells THir but only sparsely EdU labeled were considered as continuing to cycle after pulse, and not included in calculation. The percentage of THir- and brightly EdU-labeled cells in each cluster is represented by color code. Based on EdU incorporation pattern, the DA clusters can be classified into four different types of neurogenesis: DA precursors arising mainly from (1) neural plate neurogenesis, (2) neural plate neurogenesis plus proliferative precursor pools, (3) continuous release of DA neurons from precursor populations, (4) a neurogenic wave limited to distinct time window. These classes are indicated at top of table. C, Summary of the results of DAPT-mediated inhibition of Notch signaling. For better comparison of birth-dating and Notch inhibition experiments, this table summarized results from supplemental Figure S2 (available at www.jneurosci.org as supplemental material). For quantification of results, the size of each CA cell cluster at the 75 hpf endpoint was compared for treated and control embryos for each time window of treatment (indicated at left). The color code indicates whether size of CA cluster is increased, decreased, or unchanged. n.d., Not determined, as severe developmental abnormalities interfered with analysis. The blue bars indicate th expression detectable by WISH in wild-type controls.
For a better understanding of the results of our birth-dating and neurogenesis studies, we want to briefly review development of zebrafish DA neurons. Zebrafish develop DA neurons in the diencephalon and telencephalon, but lack mesencephalic DA groups (for a schematic representation of DA and NA cells distribution, see Fig. 3A) (Holzschuh et al., 2001; Rink and Wullimann, 2002b; Filippi et al., 2010). However, ventral diencephalic DA neurons of the zebrafish posterior tuberculum provide ascending projections to the subpallium, and thus may provide functions equivalent to the mammalian mes-diencephalic DA systems (Rink and Wullimann, 2001, 2002a; Kastenhuber et al., 2010). Zebrafish DA groups have been classified by Rink and Wullimann (2002b): the earliest DA neurons start to express the terminal differentiation marker th in the posterior tuberculum (ventral diencephalon group DC2) from 18 hpf on, soon followed by group DC4 developing within the posterior tuberculum slightly more posterior (Fig. 3; blue bars in B and C indicate appearance of CA neurons) (Fig. 4A,C,E). Slightly later, DA neurons develop in the ventral thalamus (DC1), medial hypothalamus (DC3), and posterior hypothalamus (DC5/6/7) (Figs. 3A, 4G,I). DA neurons also form in the preoptic region (PO) as well as in the dorsal pretectum (Pr). Within the telencephalon, DA neurons form in the subpallium and the olfactory bulb. Finally, similar to mammals, DA amacrine cells form in the retina. Compared with mammalian DA groups, DC2 and DC4 are specified by Otp activity in zebrafish, similar to A11 DA cells in mammals (Del Giacco et al., 2006; Blechman et al., 2007; Ryu et al., 2007). The other groups correspond with their mammalian anatomical correlates in the hypothalamus (A12, A14–A15) and ventral thalamus (A13), whereas there are no direct correlates of subpallial and pretectal DA groups in mammals.
Figure 4.
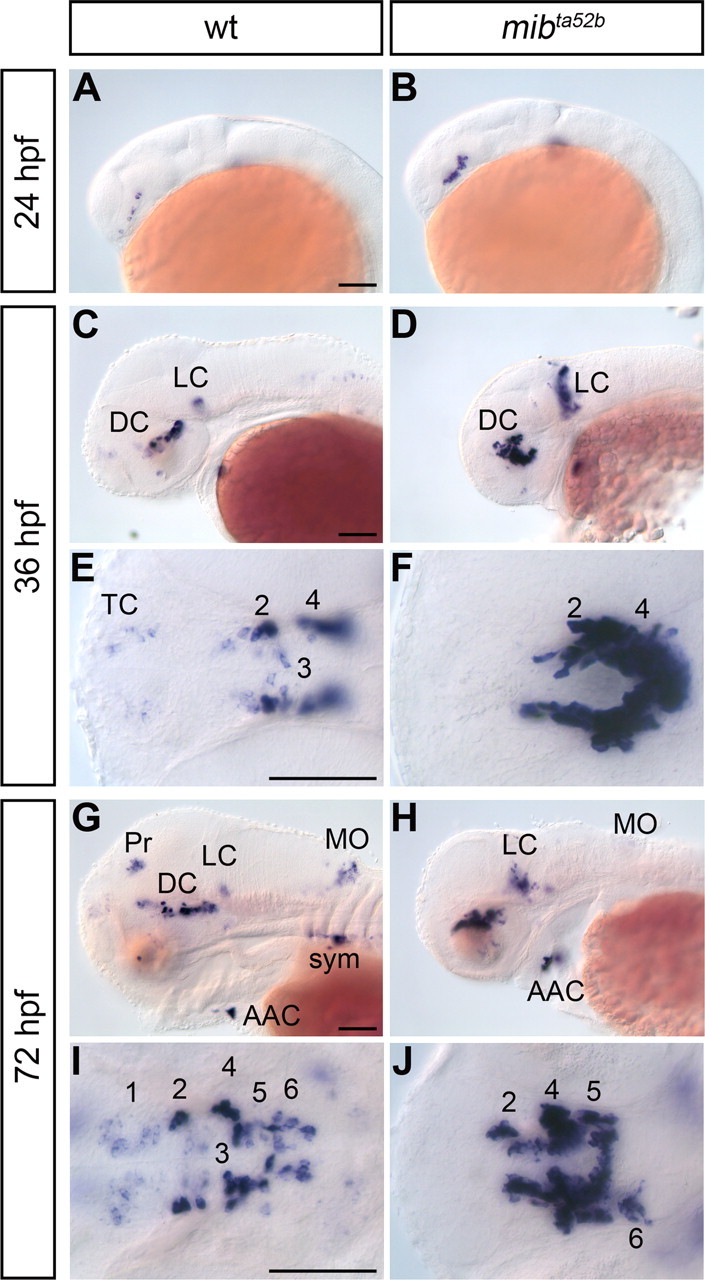
Neurogenesis of catecholaminergic neurons is affected in Notch signaling defective mind bomb mutants. A–J, Comparison of th expression in wild-type (wt) and mind bomb (mibta52b) mutants at the indicated developmental stages. Abbreviations: AAC, arch-associated neurons; DC, diencephalic dopaminergic cluster; LC, locus coeruleus; MO, medulla oblongata; Pr, pretectal cluster; sym, sympathetic neurons; TC, telencephalic cluster; 1–6: diencephalic DA clusters DC1–DC6 (see text). A–D, G, H, Lateral views. E, F, I, J, Dorsal views. Shown are minimum intensity z-projection of transmitted light image stacks to reveal all CA groups in a single image, anterior left. Scale bars, 100 μm.
Our analysis revealed that the first DA precursors that became postmitotic are those of DC2 DA neurons and of locus coeruleus (LC) NA neurons (Fig. 2A,E; D,H). These neurons were brightly labeled after EdU pulse at 8 hpf (Fig. 2A,A′,D) but clearly not labeled after EdU pulses at 16 hpf (LC) (Fig. 2H) or 20 hpf (DC2) (Fig. 2E). This suggested that DC2 DA and LC NA precursors were derived directly from neural plate cells.
The EdU labeling patterns for DC1 and DC4/5 indicated that their earliest postmitotic precursors were derived from the neural plate as well (Figs. 2I, 3B), but that also later in development precursors of these clusters exited cycling precursor pools [DC1, 24–30 hpf (Figs. 2K–K″, 3B); DC4/5, 20–24 hpf (Figs. 2F, 3B)]. Precursors of DC5 exit the cell cycle at approximately the same time as those of DC4, but they differentiate slightly later than DC4 (Fig. 3B).
For the telencephalic clusters (TCs) and medial hypothalamic DC3, it was not possible to determine a clearly delimited time window during which most of their precursors become postmitotic. We rather detected that, during all investigated intervals, several precursors of DC3 and the TC stopped cycling (Figs. 2L–N″, 3B). The same was observed for DA neurons of DC6/7, pretectum, and preoptic region (Figs. 2O–Q″, 3B). This indicated that cells were continuously added to these DA clusters, even if we observed a slight increase in the number of precursors becoming postmitotic shortly before the first differentiated TH-immunoreactive (THir) cells appeared. All precursors of the amacrine DA cells of the retina became postmitotic in a clearly delimited wave between 36 and 42 hpf (Fig. 3B).
Based on our birth-dating analysis, we conclude that, for distinct DA and NA clusters, four different temporal modes of neurogenesis can be distinguished: (1) early neurogenesis directly from neural plate cells (DC2, LC), (2) neurogenesis from neural plate and proliferating precursor pools during clearly delimited time windows (DC1/4/5), (3) continuous neurogenesis from precursors over longer developmental time (TC, DC3/6/7, Pr, PO), and (4) short wave of neurogenesis (AC) (Fig. 3B) (data not shown).
Catecholaminergic clusters are differentially affected in Notch signaling deficient mind bomb mutants
For effective activation of Notch signaling, ubiquitination of Delta by the E3 ubiquitin ligase encoded by the mind bomb (mib) gene is essential to mediate reuptake of the ligand into the signal-sending cell (Itoh et al., 2003). As Mib regulates all Notch ligands, both Deltas and Jaggeds (Koo et al., 2005), analysis of mib mutants allows to evaluate the contribution of Notch signaling. We analyzed th expression in antimorphic mibta52b (Koo et al., 2005; Zhang et al., 2007a,b) homozygous mutants between 24 and 72 hpf (Fig. 4). We observed that the earliest DA neurons (DC2) differentiating in the posterior tuberculum at 24 hpf were significantly increased in number in mibta52b mutant embryos compared with WT siblings (Fig. 4A,B). At 36 hpf, the number of th-expressing neurons in DC2 and 4 was increased in mibta52b mutants compared with WT (Fig. 4C–F). In some (2 of 14) mibta52b mutant embryos, we detected a stripe of th-expressing cells extending from the ventral prethalamus to the dorsalmost prethalamus (data not shown), which may correspond to additional DC1 and/or ectopic DA cells. At this stage, the number of LC NA neurons was also strongly increased, and they were spread along the dorsoventral axis (Fig. 4D). Conversely, whereas the earliest th-expressing telencephalic DA neurons were detectable in WT, their number was reduced or absent in mibta52b mutant embryos (Fig. 4C–F).
At 72 hpf, the strong increase in cell numbers of the posterior tubercular and hypothalamic DA groups DC2 and 4–6 in mibta52b mutant embryos persisted (Fig. 4G–J). DC1 cells appeared depleted in mibta52b embryos. Whether hypothalamic DC3 DA neurons formed in normal numbers was impossible to determine, as they appeared intermingled with DC2. We also observed that pretectal, subpallial, olfactory lobe, as well as amacrine DA cells and medulla oblongata NA cells did not form in mibta52b mutant embryos. The differential effect on DA groups appeared to correlate with their developmental time of differentiation: the number of th-expressing cells in early differentiating CA groups appeared increased, whereas late differentiating groups were depleted or absent. Neural crest-derived th-expressing cells later contributing to the carotid body did develop in approximately normal numbers, but were delayed in migration to the carotid body primordium (Fig. 4H), indicating that late differentiating NA cells form and survive in the peripheral nervous system of mibta52b mutant embryos.
Effect of inhibition and activation of Notch signaling at different stages of development
Motivated by the differential effects on early and late differentiating neuronal groups in mib mutants, we investigated potential stage-specific requirements for Notch signaling. We used the γ-secretase inhibitor DAPT to pharmacologically inhibit Notch activation (Geling et al., 2002) at defined developmental stages. We applied DAPT for overlapping time windows of 10–12 h each and analyzed th expression at 75 hpf. We chose 15 hpf as the earliest time point to start the treatment to avoid interfering with early developmental processes, but to initiate inhibition of Notch signaling before the first DA cells in the diencephalon start to express th. Compared with mibta52b mutants, DAPT-treated embryos developed a more WT-like morphology (compare Fig. 4, supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Analysis of th-expressing CA clusters at 75 hpf revealed differences in temporal requirement for Notch signaling for distinct CA clusters (supplemental Figs. S1, S2, available at www.jneurosci.org as supplemental material) (data not shown) (summarized in Fig. 3C). Because of the strong increase in th-expressing cells, it was not possible to perform absolute cell counts. Instead, the different CA clusters for each stage of treatment were classified into five different phenotypic categories ranging from depletion to very strong increase in th-expressing cells. Given that CA neurons in DC2 and LC become postmitotic during early neural plate stages, we did not significantly affect these groups in our DAPT experiments, which we started at 15 hpf. DC1 cells increased when treated with DAPT between 15 and 24 hpf and 36 and 48 hpf (supplemental Fig. S1F,L, available at www.jneurosci.org as supplemental material). Because of the strong increase in th expression in the ventral diencephalic clusters, and similar behavior of cells (data not shown), DC5/6 were analyzed together. th-expressing cells of DC5/6 were increased in embryos treated between 24 and 36 hpf (supplemental Fig. S1G–I, available at www.jneurosci.org as supplemental material), whereas treatment between 36 and 48 hpf caused an increase in cell numbers for both DC4 and DC5/6 (supplemental Fig. S1J–L, available at www.jneurosci.org as supplemental material). Conversely, we observed a depletion of DC3 th-expressing cells after 36–48 hpf DAPT treatment (supplemental Fig. S1J–L, available at www.jneurosci.org as supplemental material). For the intervals 15–24, 24–36, and 48–60 hpf, we observed slightly increased numbers of th-expressing cells in DC3 compared with WT, but this was difficult to examine because of expansion of other ventral diencephalic groups expressing th at higher levels. The DA neurons of the preoptic region were mostly depleted after inhibition of Notch signaling between 15 and 16 hpf and between 36 and 48 hpf, whereas a reduction of DA neurons in the telencephalon and in the pretectum was observed for all six time windows of DAPT treatments.
The NA neurons of the medulla oblongata were only reduced in embryos treated between 24 and 36 hpf. The peripheral NA neurons associated with the arches (AAC) and the sympathetic ganglia were not affected by any of the treatments. This suggested that they either go through neurogenesis very early or do not require Notch signaling. Our findings are at variance with previous reports on experiments performed in chicken (Tsarovina et al., 2008), which may be attributable to the fact that proliferation of peripheral precursors may play a lesser role in zebrafish at stages analyzed by us.
We also analyzed the effect of overexpression of dominant-active NICD on DA neurogenesis (supplemental Figs. S3–S5, available at www.jneurosci.org as supplemental material), using heat-shock-driven expression of NICD in transgenic zebrafish (Scheer and Campos-Ortega, 1999; Scheer et al., 2002). Supplemental Figure S6 (available at www.jneurosci.org as supplemental material) shows isl1 expression as control for effectiveness of NICD overexpression. The NICD gain-of-function experiments confirmed the results of the DAPT loss-of-function studies in that specific th-expressing clusters in the DC were found sensitive to alterations in Notch signaling during characteristic developmental stages. However, stage-specific NICD overexpression affected the formation of CA neurons to variable degrees, ranging from reduction in cell number to complete elimination of th expression (supplemental Figs. S3, S4, available at www.jneurosci.org as supplemental material). To investigate whether NICD overexpression affects DA precursors, we analyzed the expression of sim1a (Löhr et al., 2009) and otpa (Del Giacco et al., 2006; Ryu et al., 2007), which are essential for the development of DC2, 4, 5, and 6 DA neurons. We observed a downregulation of both otpa and sim1a expression in the ventral diencephalic domains of embryos heat shocked at 36 hpf (supplemental Fig. S5A,B,D,E, available at www.jneurosci.org as supplemental material). Conversely, the glial marker gfap was expressed more broadly in the rhombencephalon, which may be consistent with a delay in development, whereas the mid-hindbrain boundary domain was missing (supplemental Fig. S5C,F, available at www.jneurosci.org as supplemental material). We hypothesize that NICD overexpression already affects sim1a- and otpa-expressing precursor populations, independent of the developmental delay observed at 72 hpf. However, heat-shock-driven overexpression of NICD was accompanied by increased apoptosis (data not shown), and thus phenotypes observed at 72 hpf may constitute a combination of specific effects of Notch signaling and enhanced cell death.
deltaA and deltaD mediate dopaminergic neurogenesis in the forebrain
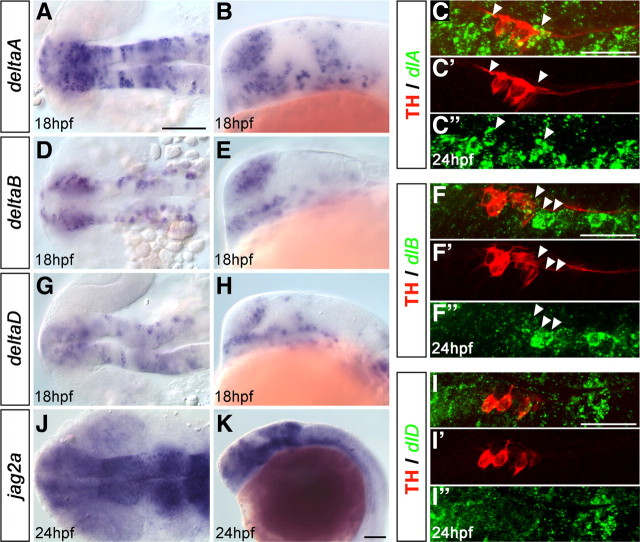
The distinct temporal modes of DA neurogenesis raised the question whether distinct Notch ligands may control neurogenesis and precursor pool maintenance for DA groups. We analyzed the expression patterns of the four delta (A–D) (Haddon et al., 1998) and three jagged genes (1a, 1b, and 2a) (Zecchin et al., 2005) during developmental stages of DA neurogenesis (Fig. 5) (supplemental Fig. S7, available at www.jneurosci.org as supplemental material). We found that four ligands, dlA, dlB, dlD, and jagged2a, are expressed in a temporospatial manner consistent with a potential role in DA neurogenesis (Fig. 5). Coexpression analysis for dlA, dlB, dlD, and TH revealed that those ligands were expressed adjacent to THir cells in DA precursor territories (Fig. 5C–C″,F–F″,I–I″). To dissect potential specific contributions of these Notch ligands, we analyzed mutants for deltaA, deltaD, and jagged2a as well as morpholino knockdown morphants for jagged2a, and deltaB (Fig. 6) (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). We found that only DeltaA loss-of-function significantly affected the th expression pattern at developmental stages ranging from 24 to 96 hpf. In dlahi781 mutants, we detected a remarkable increase in the number of DC2 DA neurons already at early stages (shown at 30 hpf in Fig. 6B,F), similar to the phenotype of mibta52b mutants. At 48 and 74 hpf, the number of th-expressing cells in DC2/4/5/6 was significantly increased, whereas it was reduced in DC1. Conversely, th expression was absent in telencephalon and pretectum (Fig. 6I,J,M,N). The peripheral (sympathetic ganglia, AAC) and medulla oblongata NA groups were not affected at 74 hpf (Fig. 6J). In dlahi781 mutants, a few more cells were detectable in prethalamic DC1 at 96 hpf compared with 74 hpf, but DC1 was still overall reduced in number compared with WT siblings (data not shown).
Figure 5.
Evaluation of Notch ligands potentially involved in diencephalic DA neurogenesis. WISH revealing expression of dlA (A, B), dlB (D, E), and dlD (G, H) at 18 hpf, and of jag2 (J, K) at 24 hpf in the ventral diencephalon. A, D, G, J, Dorsal views. B, E, H, K, Lateral views, anterior left. C, F, I, FISH for dlA (C–C″), dlB (F–F″), and dlD (I–I″) followed by fluorescent immunohistochemistry for TH in WT at 24 hpf; projected confocal z-stacks of up to 5 μm depth show THir cells of DC2 and adjacent expression of the Notch ligands (arrowheads); merged detection channels and TH channel red (′) as well as FISH channel green (″); anterior left. Scale bars: (in A) A–J, 100 μm; K, 100 μm; (in C, F, I) C–C″, F–F″, I–I″, 50 μm.
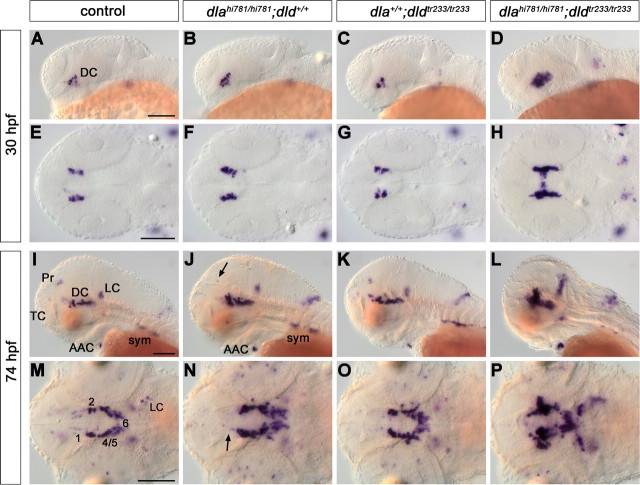
Figure 6.
deltaA and deltaD mediate diencephalic DA neurogenesis. The formation of CA neurons was analyzed in the offspring of fish carrying both dlahi781 and dldtr233 mutant alleles. The embryos were stained for th expression by WISH at 30 (A–H) and 74 (I–P) hpf, and genotyped. Controls (A, E, I, M) are homozygous WT siblings. An increase of th-expressing ventral diencephalic cells was observed in dlahi781 mutants at 30 hpf (B, F). To estimate this increase, in a separate experiment the embryos were immunostained, imaged at the confocal microscope, and DA cells counted. By 30 hpf, wild-type siblings developed on average 27.4 DA neurons (N = 8), and dlahi781 mutant embryos 40.4 neurons (N = 7). At 74 hpf, th expression in dlahi781 mutants (J, N) was differentially affected: whereas cell numbers increase in DC2/4/5/6, a reduction in DC1 (N, arrow), Pr (J, arrow), and TC was observed. The formation of AAC and sympathetic ganglia was not affected (J). The phenotype of dldtr233 mutant embryos (C, G, K, O) showed only a minor increase of th-expressing cells in the ventral diencephalon (O). Conversely, the absence of both DeltaA and DeltaD activities in dlahi781/dldtr233 double-mutant embryos (D, H, L, P) resulted in a dramatic enhancement of the th phenotype, similar to the one observed in mind bomb mutants (Fig. 4). A–D, I–L, Lateral views. E–H, M–P, Dorsal views, anterior left. E–H and M–P are z-projections of transmitted light image stacks. Scale bars, 100 μm. For abbreviations, see Figure 4.
The increase in th-expressing neurons in dlahi781 mutants was less dramatic compared with that in mibta52b mutants, suggesting that other ligands might contribute to DA neurogenesis. To evaluate the potential redundancy of the paralogs deltaA and deltaD, we analyzed th expression in the offspring of fish carrying both dlahi781 and dldtr233 mutant alleles. Compared with dlahi781 mutants, the DA phenotype of dldtr233 single mutant embryos was rather mild and not significantly different from WT siblings (Fig. 6C,G,K,O). However, in dlahi781/dldtr233 double mutants, the increase in the number of th-expressing neurons was strongly enhanced at all stages analyzed (Fig. 6D,H,L,P). DA phenotype and general morphology in double mutants strongly resembled those of mibta52b mutants. This indicates that both dla and dld mediate DA neurogenesis, although dla appears to have a major role in this process and to be able to compensate for the absence of dld.
Loss of DeltaA/DeltaD signaling causes loss of dopaminergic precursor pools
The increased number of DA neurons in dlahi781 mutants could be caused by excessive neurogenesis from a defined precursor pool or by extra rounds of proliferation in precursor pools inhibited to enter neurogenesis. In the latter case, the expression of proliferation markers in dlahi781 mutants should be upregulated compared with WT siblings. Analysis of the proliferation markers pcna (Wullimann et al., 1999) at 30, 48, and 72 hpf, and mcm5 (Ryu et al., 2005) at 24 hpf did not reveal an obvious increase in proliferation in the ventral diencephalon of dlahi781 mutants (supplemental Fig. S9A,B, available at www.jneurosci.org as supplemental material) (data not shown). To further investigate whether the observed increase in the number of DC2 DA neurons in dlahi781 mutant embryos was attributable to additional rounds of proliferation, we performed experiments of EdU incorporation at 8 and 24 hpf, and analyzed TH expression at 72 hpf. When the pulse was performed at 8 hpf, almost all DC2 neurons were EdU labeled in control siblings (Fig. 3B) and dlahi781 larvae (supplemental Fig. S10A,B, available at www.jneurosci.org as supplemental material), indicating that DC2 precursors were still cycling at that stage. When the pulse was performed at 24 hpf [i.e., when all DC2 precursors should have exited the cell cycle according to our analysis (Fig. 3B)], we did not observe any EdU-positive DC2 neuron, neither in control siblings (supplemental Fig. S10C, available at www.jneurosci.org as supplemental material) nor in dlahi781 mutants (supplemental Fig. S10D, available at www.jneurosci.org as supplemental material). This strongly suggests that the supernumerary DC2 neurons in dlahi781 mutant embryos do not derive from additional rounds of precursor cell divisions.
The reduction of th expression in late emerging DA cell groups, like in the pretectum, would support the hypothesis that dlA is involved in the maintenance of primary precursor pools. In this case, too many neurons may have formed prematurely, and precursor pools may already have been depleted when late emerging clusters should form. Thus, changes should be detected in expression of markers for neural stem and precursor cells. sox2 is a transcription factor involved in neural stem cell maintenance, and is expressed in neural stem cell and precursor pools in zebrafish (Adolf et al., 2006). dlahi781 mutants did not reveal a significant reduction of sox2 expression at 48 hpf when compared with WT siblings (supplemental Fig. S9C,D, available at www.jneurosci.org as supplemental material), and thus stem cell/precursor pools in general do not appear to be significantly reduced.
However, loss of DeltaA may cause depletion of selected precursor pools, including those that specifically contribute to DA neurons. The only currently known factors expressed in diencephalic DA precursors are olig2 (Borodovsky et al., 2009) and ngn1 (Jeong et al., 2006).
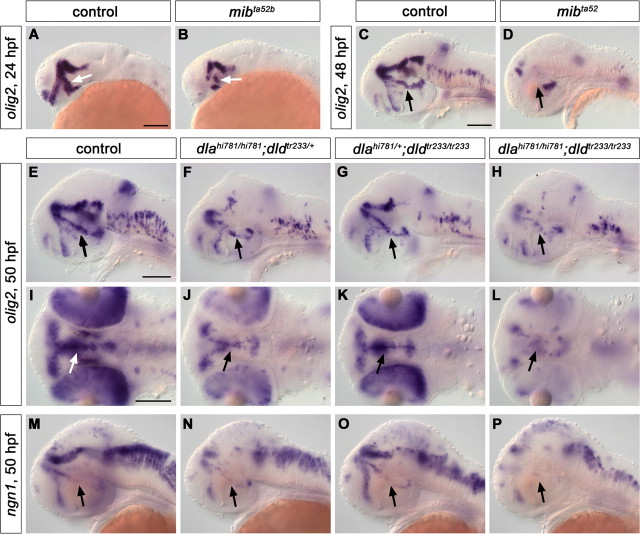
When we analyzed olig2 expression in mibta52b mutants, we observed that it was severely reduced at both 24 and 48 hpf (Fig. 7A–D), arguing that precursors for DA neurons may indeed be lost because of precocious differentiation and absence of precursor maintaining signals. Analysis of dlahi781/dldtr233 double mutants also revealed a significant reduction of olig2-expressing cells at 50 hpf (Fig. 7H,L), to which the loss of DeltaA contributed the most (Fig. 7F,J), as the lack of DeltaD activity only marginally affected olig2 expression (Fig. 7G,K). Such reduction, however, was observed in dlahi781 mutant embryos only during the second day of development, as olig2 expression appeared still comparable with control siblings at 30 hpf (data not shown).
Figure 7.
mibta52b and dlahi781/dldtr233 mutant embryos display a reduction of olig2- and ngn1-expressing precursor cells. The expression of the precursor marker olig2 was analyzed by WISH in mibta52b (A–D) and dlahi781/dldtr233 (E–L) double mutant embryos. olig2 was dramatically downregulated in mibta52b mutants compared with WT siblings at both 24 (A, B) and 48 (C, D) hpf. A remarkable reduction of olig2 expression was also observed at 50 hpf in dlahi781/dldtr233 double-mutant embryos compared with WT siblings (H, L), although less pronounced than in mibta52b mutants. The analysis of olig2 expression in single-mutant embryos revealed that lack of DeltaA activity (F, J) contributed more than lack of DeltaD (G, K) to this reduction. Similarly, ngn1 expression was dramatically reduced in the posterior tuberculum of dlahi781/dldtr233 double mutant embryos (P) compared with WT siblings (M) at 50 hpf, although it appeared only slightly reduced in dlahi781 (N) and rather normal in dldtr233 (O) single mutants. All the arrows point at the posterior tubercular area. A–H, M–P, Lateral views. I–L, Dorsal views, anterior left. Scale bars, 100 μm.
ngn1 expression has been shown to increase in mib mutants or after complete Notch inhibition at early stages (Bae et al., 2005; Cau et al., 2008). We analyzed ngn1 expression in mibta52b mutant embryos at 5 h intervals starting at 16 hpf (shortly before the first TH-expressing cells appear) until 37 hpf. We observed that, although ngn1 expression at early stages was higher in mibta52b mutants compared with control siblings, it decreased over time, and by 37 hpf no ngn1 expression domain could be detected in the posterior tuberculum of mibta52b mutants anymore (data not shown), indicating premature neuronal differentiation and concomitant depletion of precursor pools. When we analyzed ngn1 expression in dlahi781 mutants, we could not observe any significant change in its expression pattern at the stages analyzed up to 34 hpf (supplemental Fig. S9E,F, available at www.jneurosci.org as supplemental material) (data not shown). As for olig2, a general downregulation of ngn1 expression was detectable only during the second day of development in dlahi781mutants (Fig. 7M,N), although it did not appear to involve significantly the posterior tubercular domain (Fig. 7N, arrow). Although no significant change in ngn1 expression was observed in dldtr233 single-mutant embryos (Fig. 7O), the absence of both DeltaA and DeltaD activities resulted in a phenotype similar to that of mibta52b mutants, with almost no ngn1-expressing precursors left in the posterior tubercular area (Fig. 7P).
DeltaA acts upstream of sim1 and otp
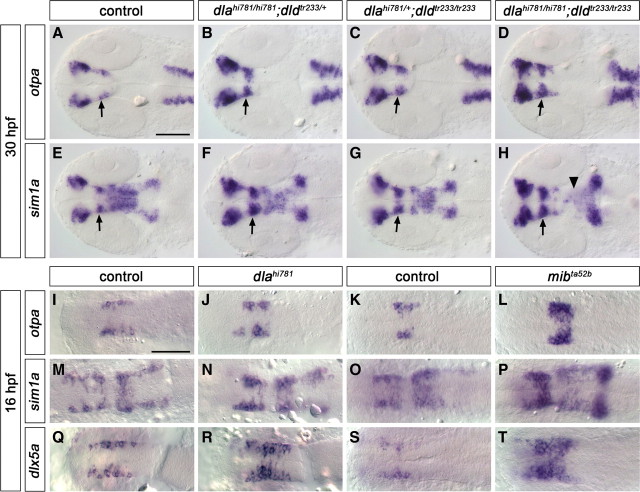
The earliest expressed transcription factors specifically required for ventral diencephalic DA neuron specification and differentiation are encoded by sim1 and otp genes. We investigated whether sim1 and otp may act as patterning genes already before the neurogenic switch, or whether they may be active predominantly after the initiation of neurogenesis. In dlahi781 and dldtr233 single mutants, analyzed at 30 hpf, both otpa and sim1a expression patterns were expanded in all their diencephalic domains (Fig. 8A–C; E–G, respectively). This expansion was even broader in dlahi781/dldtr233 double mutants (Fig. 8D,H), similar to what we observed for mibta52b mutants at 24 hpf (supplemental Fig. S11A–H, available at www.jneurosci.org as supplemental material). Conversely, a decrease of sim1a expression was detected in the mesencephalon of mibta52b (supplemental Fig. S11F, arrowhead, available at www.jneurosci.org as supplemental material) and dlahi781/dldtr233 double mutants (Fig. 8H, arrowhead).
Figure 8.
deltaA and deltaD affect expression of DA precursor and differentiation genes. The expression of transcription factors involved in DA and neuroendocrine precursor development and differentiation was analyzed at 30 hpf (A–H) or 16 hpf (I–T) in the offspring of fish carrying dlahi781, dldtr233, or mibta52b mutant alleles as indicated in the panels. All diencephalic expression domains of both otpa (A–D) and sim1a (E–H) were enlarged in dlahi781 (B, F), dldtr233 (C, G), and more severely in dlahi781/dldtr233 double mutants (D, H) compared with control siblings (A, E). In contrast, in dlahi781/dldtr233 double mutants, sim1a expression was reduced in the mesencephalon (H, arrowhead). All arrows point at the posterior tubercular area. A higher number of cells expressing otpa (I–L), sim1a (M–P), and dlx5a (Q–T) was detectable already at 16 hpf in dlahi781 (I, J, M, N, Q, R) and mibta52b (K, L, O, P, S, T) mutants. All images are dorsal views, anterior left. A–H are z-projections of transmitted light image stacks. Scale bars, 100 μm.
The first DA neurons appear in the ventral diencephalon around 18 hpf. Since otpa and sim1a are both expressed much earlier than 18 hpf, we analyzed their expression patterns at 14 somite stage and found that more cells express otpa and sim1a within the same rostrocaudal domain in dlahi781 (Fig. 8I,J,M,N) as well as in mibta52b mutants (Fig. 8K,L,O,P) at this stage. We also analyzed the expression of other transcription factors expressed in ventral diencephalon DA precursor cells, such as nkx2.2a (DC3) and dlx5a (DC1/3/7) (A. Filippi and W. Driever, unpublished data). We found that the number of dlx5a-positive cells was significantly increased in dlahi781 (Fig. 8Q,R) and mibta52b mutants (Fig. 8S,T), but not the expression pattern of nkx2.2a (data not shown).
These data reveal that Notch signaling acts upstream of sim1 and otp in ventral diencephalic DA development, and that DeltaA and DeltaD appear to be the ligands controlling Notch activity in this process. The rostrocaudal extent of sim1 and otp expression in mibta52b or dlahi781/dldtr233 mutant embryos is not affected, clearly showing that patterning is normal. Rather, it appears that a larger number of cells within the mediolateral extent of the neuroepithelium initiates sim1 and otp expression, arguing that a larger than normal portion of precursors enters neurogenesis.
Interestingly, sim1 and otp do specify not only DA neurons but also several neuroendocrine cell types in the preoptic region and posterior tuberculum/hypothalamus. Specifically, DA and corticotropin-releasing hormone (CRH) neurons are cospecified by sim1 and otp in the posterior tuberculum (Löhr et al., 2009). Therefore, we asked whether Notch signaling may act as a lineage switch to assign Sim1- and Otp-expressing precursors to either the DA or CRH lineage. We found that, in mibta52b mutants, thus in the absence of functional Notch signaling, CRH neurons were strongly increased in number in most brain areas, except for the posterior tuberculum (Fig. 9A–D) (for anatomical location of CRH neurons, see Chandrasekar et al., 2007). In deltaA and deltaD mutants, in a similar fashion, posterior tubercular CRH neurons behaved differently from other CRH groups (Fig. 9E–L). With respect to CRH groups in other areas of the brain, the dlahi781/dldtr233 double-mutant phenotype was similar to mib (Fig. 9, compare K, L, with C, D), although less pronounced. In contrast, no significant increase of the posterior tubercular CRH neurons could be detected in dlahi781 (Fig. 9G,H), dldtr233 (Fig. 9I,J), and dlahi781/dldtr233 mutant embryos (Fig. 9K,L). Thus, it appears that the mechanisms that mediate the increase in number of CRH neurons in dlahi781/dldtr233 double mutants in other regions of the brain do not apply to those in the posterior tubercular area (i.e., those cospecified with DA neurons by sim1 and otp). These neurons appear spatially disorganized in both mibta52b and dlahi781/dldtr233 double mutants, but not significantly increased in number. Therefore, we conclude that Delta/Notch signaling in otp- and sim1-expressing precursors does not establish a switch between DA and CRH differentiation. We further conclude that DeltaA/DeltaD signal specifically affects the prospective DA neurons among otp- and sim1-expressing precursors, whereas other mechanisms may mediate the neurogenic lineage switch for the CRH precursor subpopulation. Thus, Notch ligands may control the size of the sim1a/otpa-positive precursor pool and neurogenesis of DA neurons in the posterior tuberculum, whereas different neuronal populations deriving from the same precursor pool may be specified by Notch-independent mechanisms.
Figure 9.
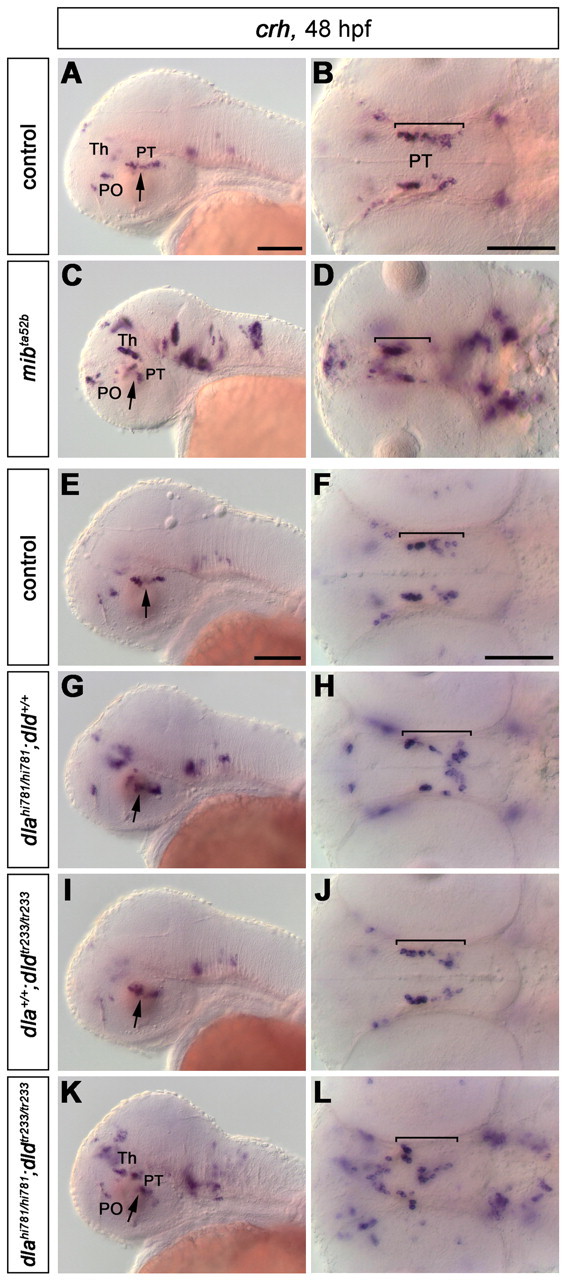
Development of crh-expressing neurons in mibta52b and dlahi781/dldtr233 mutant embryos. Analysis of crh expression in mibta52b mutants (A–D) and in the offspring of fish carrying dlahi781 and dldtr233 mutant alleles (E–L) at 48 hpf. The number of crh-expressing cells in most anatomical regions is significantly increased in mibta52b mutants (C, D; compare with A, B), and in dlahi781/dldtr233 double mutants (K, L; compare with E, F), whereas no significant change is observed in dlahi781 (G, H) or dldtr233 (I, J) single-mutant embryos compared with WT siblings (E, F). However, the number of crh-expressing neurons in the posterior tubercular area (arrows) is relatively unchanged in all embryos analyzed, indicating that dla and dld do not mediate a cell fate switch between the otpa/sim1a-dependent crh-expressing cells and DA neurons. Abbreviations: PO, Preoptic area; PT, posterior tuberculum; Th, thalamus. A, C, E, G, I, K, Lateral views; all arrows point at the posterior tubercular area. B, D, F, H, J, L, Dorsal views; the posterior tubercular domain is indicated by brackets. Anterior is always left. B, D, F, H, J, and L are z-projections of transmitted light image stacks. Scale bars, 100 μm.
Discussion
DA neurons form at distinct anatomical sites and during distinct developmental phases; therefore, we aimed at identifying shared and specific mechanistic aspects of DA neurogenesis in zebrafish. Although the developmental times for appearance of DA clusters expressing terminal differentiation markers have been described, very little is known about (1) the origin of the neural population from which DC DA clusters derive, (2) when DA precursor cells become postmitotic, (3) when Delta/Notch mediated signaling sets the neurogenic switch, (4) which Notch ligands mediate DA neurogenesis, (5) which precursor transcription factors may be affected in their expression by Notch signaling, and (6) whether Notch signaling may have functions also in differentiating or mature DA neurons.
Our birth-dating analysis revealed that catecholaminergic neurons become postmitotic during distinct phases of CNS development. NA precursors of the LC and DA precursors of DC2 are among the first to exit the cell cycle already before 16 and 20 hpf, respectively. Birth dating and stage-specific pharmacological (DAPT) suppression of Notch signaling both showed that DC4 cells are born later than those of DC2, indicating that DC2 and DC4 may arise from distinct phases of neurogenesis. Similar to DC4, some precursors of DC1 and DC5 become postmitotic early during neural plate stages, but between 20 and 30 hpf precursors for those DA clusters continued to be released from proliferating precursor pools. A recent study (Russek-Blum et al., 2008) analyzed incorporation of BrdU into proliferating DA precursors and concluded that DA precursors of DC2 and DC3–6 become postmitotic between 10 and 16 hpf. However, analysis of transcriptional mechanisms controlling DA development in the ventral diencephalon indicates that DC2 and 4–6 are specified by mechanisms clearly distinct from DC3 (Ryu et al., 2007). In addition, our results from birth-dating and Notch inhibition studies clearly suggested that precursor cells for DC3 and DC6 are released from precursor pools also later than 16 hpf in development.
Birth-dating studies together with loss- and gain-of-function analyses of Notch signaling indicated that, based on temporal and mechanistic criteria, there are four distinct modes of neurogenesis for CA neurons in the embryo CNS. (1) At neural plate stages, when primary neurogenesis of sensory and motor neurons has been described (Hartenstein, 1989; Kimmel and Westerfield, 1990), precursor cells for DC2 and the LC stop cycling and enter DA and NA differentiation, respectively. (2) Precursor cells contributing to ventral thalamic DC1 and posterior tubercular/hypothalamic DC4 and 5 initially derive from neural plate cells, but also later appear to be added continuously from proliferating neuroepithelial cells within a limited time window. Therefore, a relatively fixed number of neurons is reached early during the second day of development. (3) The third type of neurogenesis appears to constitute a continuous release of precursors from proliferating precursor pools. We showed that this mechanism holds for the telencephalic groups, the pretectal and preoptic clusters, as well as for the hypothalamic groups DC3/6/7. (4) The retinal amacrine DA cells derive from their precursor pool in a brief developmental time window during a short pulse of neurogenesis, which is consistent with the waves of neurogenesis previously described for retinal development (Vetter and Brown, 2001).
We found that Notch signaling differentially affects the development of DA neurons in the forebrain. Although we detected a strong increase in DA neurons of the posterior tuberculum and medial and caudal hypothalamus in mibta52b mutants, most telencephalic and pretectal DA neurons did not form by 72 hpf. This loss of late differentiating DA neurons could be explained either by a depletion of precursor pools because of excessive premature differentiation, or by increased apoptosis. For mibta52b mutants, we also cannot exclude indirect effects on neurogenesis, as mibta52b mutant embryos start to develop gross abnormalities from 72 hpf on, including impairment of somitogenesis, neural crest and vasculature development, and apoptosis in the brain (Schier et al., 1996; Itoh et al., 2003). However, in our experiments using stage-specific pharmacological inhibition of Notch by treating embryos with the γ-secretase inhibitor DAPT (Dovey et al., 2001), we basically phenocopied mibta52b, revealing that the loss of these clusters reflects a stage-specific requirement for Notch signaling rather than being a secondary effect.
To understand which of the different Notch ligands may be involved in DA neurogenesis, we performed coexpression analysis as well as mutant, double mutant, and morpholino knockdown analyses. Our data indicate that deltaA and deltaD are the ligands involved in the neurogenic selection of DA neurons in the zebrafish forebrain, as the lack of both DeltaA and DeltaD activities results in an enhanced DA phenotype, similar to that observed in the complete absence of Notch signaling. deltaA appears to play the major role in DA neurogenesis, as it is able to compensate for the absence of deltaD, whereas deltaD can only partially compensate loss of deltaA. dla and dld are both orthologs of mammalian Dll1. Although the role of Dll1 in DA development has not been functionally analyzed, it has been shown to be expressed in regions of mes-diencephalic DA neurogenesis, and its expression, together with Hes5, depends on Ngn2 activity (Kele et al., 2006). Thus, conserved Delta orthologs may control DA neurogenesis in all vertebrates. A second Delta-related gene, Dlk1, has been shown to be involved in DA neurogenesis (Bauer et al., 2008), but its function is not clear.
Although Notch signaling may restrict the number of differentiating DA neurons forming from precursor pools early in development, prolonged loss of Notch signaling may differentially affect late differentiating DA neurons. Our birth-dating and loss-of-function experiments suggested that Notch signaling may be required for maintenance of precursor pools for late differentiating neurons. We focused our analysis of precursor populations on dlahi781 mutants, which prominently affect CA neurogenesis. mibta52b mutant or DAPT-treated embryos are less suitable for this analysis, as they show much broader defects in neurogenesis and make it more difficult to distinguish secondary effects. We were not able to detect a loss of sox2-expressing precursor and stem cells, or changes in pattern of pcna- or mcm5-expressing proliferating cells in dlahi781 mutants. However, DeltaA signaling may only affect specific DA precursor pools rather than general neural stem and precursor cells. Therefore, we analyzed the expression of olig2, which has been demonstrated to be expressed in precursors of ventral diencephalic DA neurons and required for their development (Borodovsky et al., 2009). Our data revealed a severe loss of olig2-expressing precursors in both mibta52b and dlahi781/dldtr233 mutants, demonstrating that DeltaA/DeltaD are required for maintaining DA precursor pools in the early zebrafish embryo (Fig. 9). A similar reduction in ngn1-expressing precursors in the posterior tuberculum further supports the finding that DeltaA/DeltaD signaling is required for maintaining olig2/ngn1-positive DA precursors in this territory.
In addition to its roles in precursor pool maintenance and the neurogenic switch, Notch signaling may also be involved in later specification and differentiation processes of DA clusters. Similar dual roles of Notch in precursor pool maintenance and neuronal specification have been previously shown for spinal cord neurogenesis (Appel et al., 2001), telencephalic precursor pools (Yun et al., 2002), and neural stem cells (Hitoshi et al., 2002). Although we were not able to test this hypothesis for telencephalic or pretectal DA groups because of our lack of knowledge on specific factors involved in their specification and early differentiation, we investigated whether Notch signaling may contribute to DA specification and differentiation in the posterior tuberculum and hypothalamus. otpa (Ryu et al., 2007) and sim1a (Löhr et al., 2009) are expressed in DA precursors of DC2/4/5/6 as well as in differentiated neurons, and are required for specification of these groups. In mibta52b and dlahi781/dldtr233 mutants, the number of sim1a- and otpa-expressing cells was increased. The fact that sim1 and otpa domains did not expand rostrocaudally reveals that Notch signaling does not have an effect on developmental patterning at this stage. Rather, as revealed by the lateral to medial expansion of the domains, it appears that more cells from the more medially located precursor domains enter DA neurogenesis in absence of Notch signaling. Our data show that Notch signaling acts upstream of sim1a and otpa, indicating that both genes may not act during neural pattern formation but are involved in differentiation processes after the neurogenic selection of neural precursors. Surprisingly, Notch signaling gain-of-function by NICD overexpression appeared to repress th as well as both sim1a and otpa expression, even at developmental stages when their expression domains have already been well established. Although gain-of-function experiments may activate mechanisms not normally engaged in a specific differentiation process, this opens the possibility that Notch signaling may also contribute to control of transcription factors involved in specification and differentiation of dopaminergic neurons.
Given that Notch signaling has previously been shown to be involved in the switch between two distinct lineages (Spana and Doe, 1996; Shin et al., 2007), we investigated whether Notch signaling may be involved in selecting DA versus CRH neurons, which both derive from sim1a- and otpa-expressing precursors. Complete loss of Notch signaling led to expansion of DA neurons but did not significantly affect CRH neurons in the posterior tuberculum, arguing against a role for Notch signaling in a DA–CRH lineage decision from a common precursor pool. Surprisingly, DeltaA/D act selectively during DA but not CRH neurogenesis, despite the fact that both neural types depend on DeltaA/D to determine the number of sim1a- and otpa-expressing precursors. These findings indicate neural subtype specificity for distinct Notch ligands within the same multilineage precursor population (Fig. 10). A survey of the effect of loss of other Notch ligands on posterior tubercular CRH neurons did not identify a single ligand with a prominent neurogenic effect (data not shown). Thus, a combination of Notch ligands other than DeltaA/D may be involved, or, alternatively, the lineage decision between DA and CRH neurons from sim1/otpa precursors may be mediated by other signaling pathways or mechanisms.
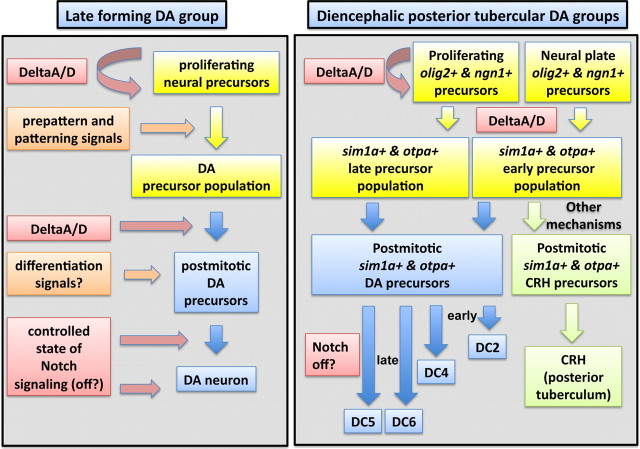
Figure 10.
Model of Delta/Notch signaling during dopaminergic development in zebrafish. This study revealed shared and distinct aspects of the earliest differentiating posterior tubercular DA neurons and of other late differentiating DA groups. The left part of the figure presents a model summarizing how Notch signaling may act during neurogenesis of late differentiating neurons that derive from proliferating precursor pools. The patterning and differentiation signals are by and large unknown so far. DeltaA/D appears to be the prominent Notch signal acting to maintain DA precursor pools. DeltaA/D are also involved in the neurogenic switch selecting DA neurons. Our Notch ICD overexpression experiments indicate that Notch signaling should be off during late phases of DA development to enable stable differentiation. The right part of the figure summarizes the effects of Notch signaling we observed for diencephalic posterior tubercular DA neurons, many of which derive already during primary neurogenesis at neural plate stages. DeltaA/D are required to maintain olig2- and ngn1-expressing DA precursors and to regulate the size of the sim1a/otpa-expressing precursor population. These otpa- and sim1a-expressing ventral diencephalic precursors differentiate into DA and CRH neurons. Among the otpa- and sim1a-expressing precursors, DA neurons are selected in temporal sequence, starting with DC2 and DC4 neurons already during late neural plate stages (which may not depend on DeltaA/D for precursor maintenance), and then later DC4, DC5, and DC6 also from proliferating precursor pools. CRH neurons appear to be selected from the same precursor pool, but by unknown mechanisms: these could be other signaling events, or several partially redundant acting Notch ligands other than DeltaA/D, as our survey of Notch ligands did not reveal strong effects on these CRH neurons for any single Notch ligand (data not shown).
Our data raise the question whether there is a conserved mechanism of neurogenesis for all DA neurons or whether there are different mechanisms that converge with other transcriptional input to one DA neurotransmitter phenotype. Although transcriptional input from local prepattern would suggest that multiple transcriptional modules might converge on specification of the DA neurotransmitter phenotype (Ohyama et al., 2005; Smidt and Burbach, 2007; Löhr et al., 2009), our observations on dla/dld in zebrafish and previous reports on Dll1 in mammals (Kele et al., 2006) suggest a unifying mechanism for DA neurogenesis. This mechanism appears to apply both to DA systems established by primary neurogenesis from neural plate cells as well as to neural stem and precursor cell-based mechanisms that control continuous expansion of later developing DA systems. Interestingly, primary neurogenesis provides DC2 and DC4 neurons, which are part of the evolutionary ancient diencephalospinal system that contributes to somatomotor control at hindbrain and spinal levels as well as neuroendocrine and sympathetic control (Takada et al., 1988; Björklund and Dunnett, 2007; Löhr et al., 2009). Therefore, the rapid development of the zebrafish larvae and the intricate contribution of diencephalospinal DA connectivity to somatomotor control may have made this system coevolve with primary neurogenesis mechanisms that derive neurons very early directly from neural plate cells.
Footnotes
This work was supported by the European Union Collaborative Project mdDANEURODEV-222999, Deutsche Forschungsgemeinschaft (DFG) Graduierten Kolleg 1104, DFG Sonderforschungsbereich 505, 592, and 780, and the Excellence Initiative of the German Research Foundation EXC 294. We are grateful to Jochen Holzschuh and Stephanie Eckerle for sharing the protocol for the Click-iT EdU cell proliferating assay and for helpful advice, and to Olga Kolesow for technical assistance. We thank Martina Jänicke, Bernhard Saeger, Annette Neubüser, Daria Onichtchouk, and Soojin Ryu for scientific discussion, and Verdon Taylor, Jochen Holzschuh, Heiko Löhr, and Jörn Schweitzer for comments on this manuscript. We thank Julian Lewis for kindly providing the mib mutant fish line, Patrick Blader for the dla mutants, and Matthias Hammerschmidt for providing the heat-shock Gal4 and NICD lines. We thank Natascia Tiso and Francesco Argenton for sharing the delta and jagged Morpholinos.
References
- Adolf B, Chapouton P, Lam CS, Topp S, Tannhäuser B, Strähle U, Götz M, Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol. 2006;295:278–293. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Weinberg ES. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev Biol. 1994;166:509–530. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 2004;77:3–20. doi: 10.1016/s0091-679x(04)77001-6. [DOI] [PubMed] [Google Scholar]
- Andersson E, Jensen JB, Parmar M, Guillemot F, Björklund A. Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development. 2006;133:507–516. doi: 10.1242/dev.02224. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Rueger MA, Park DM, Mkhikian H, Korb E, Poser SW, Walbridge S, Munasinghe J, Koretsky AP, Lonser RR, McKay RD. Targeting neural precursors in the adult brain rescues injured dopamine neurons. Proc Natl Acad Sci U S A. 2009;106:13570–13575. doi: 10.1073/pnas.0905125106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Appel B, Givan LA, Eisen JS. Delta-Notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Shimizu T, Hibi M. Patterning of proneuronal and inter-proneuronal domains by hairy- and enhancer of split-related genes in zebrafish neuroectoderm. Development. 2005;132:1375–1385. doi: 10.1242/dev.01710. [DOI] [PubMed] [Google Scholar]
- Barth KA, Wilson SW. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development. 1995;121:1755–1768. doi: 10.1242/dev.121.6.1755. [DOI] [PubMed] [Google Scholar]
- Bauer M, Szulc J, Meyer M, Jensen CH, Terki TA, Meixner A, Kinkl N, Gasser T, Aebischer P, Ueffing M. Delta-like 1 participates in the specification of ventral midbrain progenitor derived dopaminergic neurons. J Neurochem. 2008;104:1101–1115. doi: 10.1111/j.1471-4159.2007.05037.x. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Björklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, Strähle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- Blechman J, Borodovsky N, Eisenberg M, Nabel-Rosen H, Grimm J, Levkowitz G. Specification of hypothalamic neurons by dual regulation of the homeodomain protein Orthopedia. Development. 2007;134:4417–4426. doi: 10.1242/dev.011262. [DOI] [PubMed] [Google Scholar]
- Borodovsky N, Ponomaryov T, Frenkel S, Levkowitz G. Neural protein Olig2 acts upstream of the transcriptional regulator Sim1 to specify diencephalic dopaminergic neurons. Dev Dyn. 2009;238:826–834. doi: 10.1002/dvdy.21894. [DOI] [PubMed] [Google Scholar]
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. Biotechniques. 2008;44:927–929. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- Cameron HA, McKay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol. 2001;435:406–417. doi: 10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- Cau E, Quillien A, Blader P. Notch resolves mixed neural identities in the zebrafish epiphysis. Development. 2008;135:2391–2401. doi: 10.1242/dev.013482. [DOI] [PubMed] [Google Scholar]
- Chan RW, Gargett CE. Identification of label-retaining cells in mouse endometrium. Stem Cells. 2006;24:1529–1538. doi: 10.1634/stemcells.2005-0411. [DOI] [PubMed] [Google Scholar]
- Chandrasekar G, Lauter G, Hauptmann G. Distribution of corticotropin-releasing hormone in the developing zebrafish brain. J Comp Neurol. 2007;505:337–351. doi: 10.1002/cne.21496. [DOI] [PubMed] [Google Scholar]
- Chen YC, Priyadarshini M, Panula P. Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochem Cell Biol. 2009;132:375–381. doi: 10.1007/s00418-009-0619-8. [DOI] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Del Giacco L, Sordino P, Pistocchi A, Andreakis N, Tarallo R, Di Benedetto B, Cotelli F. Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev Biol. 2006;6:50. doi: 10.1186/1471-213X-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Eaton JL, Glasgow E. The zebrafish bHLH PAS transcriptional regulator, single-minded 1 (sim1), is required for isotocin cell development. Dev Dyn. 2006;235:2071–2082. doi: 10.1002/dvdy.20848. [DOI] [PubMed] [Google Scholar]
- Filippi A, Dürr K, Ryu S, Willaredt M, Holzschuh J, Driever W. Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development. BMC Dev Biol. 2007;7:135. doi: 10.1186/1471-213X-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi A, Mahler J, Schweitzer J, Driever W. Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J Comp Neurol. 2010;518:423–438. doi: 10.1002/cne.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandel H, Kaslin J, Ganz J, Wenzel I, Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295:263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev Biol. 1999;208:473–487. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–4644. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3:399–411. doi: 10.1016/0896-6273(89)90200-6. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Havrda MC, Harris BT, Mantani A, Ward NM, Paolella BR, Cuzon VC, Yeh HH, Israel MA. Id2 is required for specification of dopaminergic neurons during adult olfactory neurogenesis. J Neurosci. 2008;28:14074–14086. doi: 10.1523/JNEUROSCI.3188-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschuh J, Ryu S, Aberger F, Driever W. Dopamine transporter expression distinguishes dopaminergic neurons from other catecholaminergic neurons in the developing zebrafish embryo. Mech Dev. 2001;101:237–243. doi: 10.1016/s0925-4773(01)00287-8. [DOI] [PubMed] [Google Scholar]
- Holzschuh J, Hauptmann G, Driever W. Genetic analysis of the roles of Hh, FGF8, and nodal signaling during catecholaminergic system development in the zebrafish brain. J Neurosci. 2003;23:5507–5519. doi: 10.1523/JNEUROSCI.23-13-05507.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Iversen SD, Iversen LL. Dopamine: 50 years in perspective. Trends Neurosci. 2007;30:188–193. doi: 10.1016/j.tins.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Jeong JY, Einhorn Z, Mercurio S, Lee S, Lau B, Mione M, Wilson SW, Guo S. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc Natl Acad Sci U S A. 2006;103:5143–5148. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber E, Kratochwil CF, Ryu S, Schweitzer J, Driever W. Genetic dissection of dopaminergic and noradrenergic contributions to catecholaminergic tracts in early larval zebrafish. J Comp Neurol. 2010;518:439–458. doi: 10.1002/cne.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative genomics on SOX2 orthologs. Oncol Rep. 2005;14:797–800. [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Westerfield M. Primary neurons of the zebrafish. In: Edelman GM, Gal WR, Cowan WM, editors. Signals and senses: local and global order in perceptual maps. New York: Wiley; 1990. pp. 561–588. [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, Lee J, Chitnis AB, Kim CH, Kong YY. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–425. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Löhr H, Ryu S, Driever W. Zebrafish diencephalic A11-related dopaminergic neurons share a conserved transcriptional network with neuroendocrine cell lineages. Development. 2009;136:1007–1017. doi: 10.1242/dev.033878. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Jørgensen AL. Structural and functional characterization of the zebrafish gene for glial fibrillary acidic protein, GFAP. Gene. 2003;310:123–132. doi: 10.1016/s0378-1119(03)00526-2. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ellis P, Kimura S, Placzek M. Directed differentiation of neural cells to hypothalamic dopaminergic neurons. Development. 2005;132:5185–5197. doi: 10.1242/dev.02094. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Puelles L, Verney C. Early neuromeric distribution of tyrosine-hydroxylase-immunoreactive neurons in human embryos. J Comp Neurol. 1998;394:283–308. doi: 10.1002/(sici)1096-9861(19980511)394:3<283::aid-cne2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal diencephalon (posterior tuberculum) Brain Res. 2001;889:316–330. doi: 10.1016/s0006-8993(00)03174-7. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Connections of the ventral telencephalon and tyrosine hydroxylase distribution in the zebrafish brain (Danio rerio) lead to identification of an ascending dopaminergic system in a teleost. Brain Res Bull. 2002a;57:385–387. doi: 10.1016/s0361-9230(01)00696-7. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain Res Dev Brain Res. 2002b;137:89–100. doi: 10.1016/s0165-3806(02)00354-1. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russek-Blum N, Gutnick A, Nabel-Rosen H, Blechman J, Staudt N, Dorsky RI, Houart C, Levkowitz G. Dopaminergic neuronal cluster size is determined during early forebrain patterning. Development. 2008;135:3401–3413. doi: 10.1242/dev.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci U S A. 2005;102:18467–18472. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Mahler J, Acampora D, Holzschuh J, Erhardt S, Omodei D, Simeone A, Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr Biol. 2007;17:873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev. 2002;112:9–14. doi: 10.1016/s0925-4773(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Harvey M, Malicki J, Solnica-Krezel L, Stainier DY, Zwartkruis F, Abdelilah S, Stemple DL, Rangini Z, Yang H, Driever W. Mutations affecting the development of the embryonic zebrafish brain. Development. 1996;123:165–178. doi: 10.1242/dev.123.1.165. [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Driever W. Development of the dopamine systems in zebrafish. In: Pasterkamp RJ, Smidt MP, Burbach JPH, editors. Development and engineering of dopamine neurons. New York: Springer Science+Business Media; 2008. pp. 1–14. [Google Scholar]
- Shin J, Poling J, Park HC, Appel B. Notch signaling regulates neural precursor allocation and binary neuronal fate decisions in zebrafish. Development. 2007;134:1911–1920. doi: 10.1242/dev.001602. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Takada M, Li ZK, Hattori T. Single thalamic dopaminergic neurons project to both the neocortex and spinal cord. Brain Res. 1988;455:346–352. doi: 10.1016/0006-8993(88)90093-5. [DOI] [PubMed] [Google Scholar]
- Tsarovina K, Schellenberger J, Schneider C, Rohrer H. Progenitor cell maintenance and neurogenesis in sympathetic ganglia involves Notch signaling. Mol Cell Neurosci. 2008;37:20–31. doi: 10.1016/j.mcn.2007.08.010. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Allende ML, Weinberg ES, Nüsslein-Volhard C. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Warren M, Puskarczyk K, Chapman SC. Chick embryo proliferation studies using EdU labeling. Dev Dyn. 2009;238:944–949. doi: 10.1002/dvdy.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullimann MF, Puelles L, Wicht H. Early postembryonic neural development in the zebrafish: a 3-D reconstruction of forebrain proliferation zones shows their relation to prosomeres. Eur J Morphol. 1999;37:117–121. doi: 10.1076/ejom.37.2.117.4739. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zecchin E, Conigliaro A, Tiso N, Argenton F, Bortolussi M. Expression analysis of jagged genes in zebrafish embryos. Dev Dyn. 2005;233:638–645. doi: 10.1002/dvdy.20366. [DOI] [PubMed] [Google Scholar]
- Zecchin E, Filippi A, Biemar F, Tiso N, Pauls S, Ellertsdottir E, Gnügge L, Bortolussi M, Driever W, Argenton F. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev Biol. 2007;301:192–204. doi: 10.1016/j.ydbio.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Q, Jiang YJ. Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific delta substrates. J Mol Biol. 2007a;366:1115–1128. doi: 10.1016/j.jmb.2006.11.096. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Q, Lim CH, Qiu X, Jiang YJ. The characterization of zebrafish antimorphic mib alleles reveals that Mib and Mind bomb-2 (Mib2) function redundantly. Dev Biol. 2007b;305:14–27. doi: 10.1016/j.ydbio.2007.01.034. [DOI] [PubMed] [Google Scholar]