Abstract
The utility of human pluripotent stem cell–derived kidney organoids relies implicitly on the robustness and transferability of the protocol. Here we analyze the sources of transcriptional variation in a specific kidney organoid protocol. Although individual organoids within a differentiation batch showed strong transcriptional correlation, we noted significant variation between experimental batches, particularly in genes associated with temporal maturation. Single-cell profiling revealed shifts in nephron patterning and proportions of component cells. Distinct induced pluripotent stem cell clones showed congruent transcriptional programs, with interexperimental and interclonal variation also strongly associated with nephron patterning. Epithelial cells isolated from organoids aligned with total organoids at the same day of differentiation, again implicating relative maturation as a confounder. This understanding of experimental variation facilitated an optimized analysis of organoid-based disease modeling, thereby increasing the utility of kidney organoids for personalized medicine and functional genomics.
The ability to derive induced pluripotent stem cells (iPSCs) from the somatic cells of patients1, together with directed differentiation protocols, provides a capacity to model the cell types affected by disease. This has proved effective for the validation of disease genes in cardiomyocytes and neurons, as well as for analysis of their effects on cellular function2–6. Particularly for monogenic conditions, the utility of such approaches has substantially improved with the advent of CRISPR–Cas9-mediated gene editing, facilitating the creation of isogenic corrected lines for direct comparison with mutant lines7–9.
Recently, iPSC differentiation protocols have been developed in which multiple cell types arise and self-organize in a fashion similar to the developing tissue10,11. Protocols have been reported for the generation of optic cup, cerebral cortex, intestine, and stomach organoids12–18. Unlike methods that generate a relatively homogeneous cell type, the complexity of organoids increases the likelihood of substantial variation between individual experiments. Even comparisons of patient and isogenic controls are affected by variation between cell lines, as individual iPSC clones vary in rates of cell proliferation and response to ligands that may not be associated with an inherited genetic defect. Indeed, organoid protocols also tend to involve many weeks of differentiation during which overall variation can increase.
We recently developed a stepwise protocol for generating three-dimensional (3D) kidney organoids from human iPSCs19,20. The resulting organoids contain the key epithelial cell types of the forming filtration units, the nephrons, together with collecting ducts, endothelium, pericytes, and interstitial fibroblasts. With so many distinct cell types present, the potential variation in cell type proportions raises the challenge of reproducibility, which may compromise the capacity to identify novel disease-associated transcriptional changes.
In this study, we provide a comprehensive transcriptional and morphological evaluation of our kidney organoid protocol. Applying RNA sequencing (RNA-seq) to 57 whole organoids and 8,323 organoid-derived single cells, we examined variation between organoids within a single differentiation experiment, between different differentiation experiments (batches), between iPSC clones, and between epithelial compartments isolated from kidney organoids. Our data identify a set of highly variable genes that reflect differences in organoid maturation, nephron segmentation, and off-target populations, with no greater variation between lines than between experimental batches. We demonstrate how knowledge of this gene set can be used to improve disease model analysis.
We show that even between healthy organoids, there is significant batch-to-batch variation driven by differences in the rates of organoid maturation. In disease-modeling studies, it is crucial to account for batch variation, which can confound differences between patient and control organoids and reduce the ability to identify mechanisms of disease. Careful experimental design, such as concurrent differentiations between lines, can mitigate the effects of batch-to-batch variation.
Results
Kidney organoid differentiation protocol.
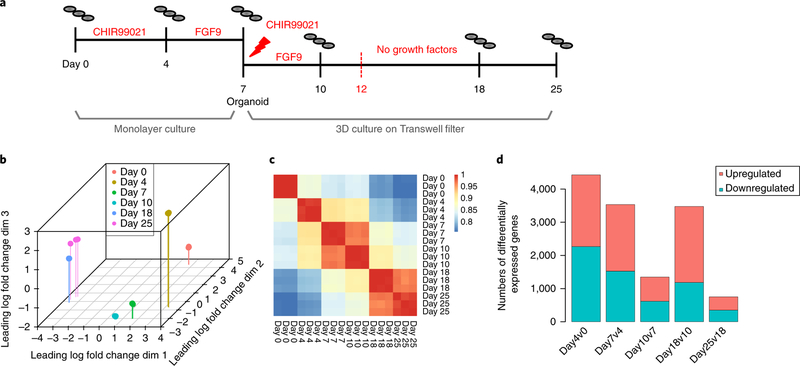
The kidney organoid protocol employed in this study (Fig. 1a) was previously described in detail19,20. For this study, each differentiation experiment began with the thawing of one vial of single-cell-adapted human iPSCs plated onto Matrigel, representing day −1. At day 0, differentiation commenced in APEL media with a specific regimen of small-molecule and recombinant proteins (Methods). Between days 0 and 7, cells were cultured as a monolayer in a six-well culture plate. Primitive streak was induced via canonical Wnt signaling (CHIR99021) and intermediate mesoderm patterning with recombinant fibroblast growth factor 9. At day 7, all cells were enzymatically dissociated and counted before individual organoids of 5 × 105 cells were pelleted for subsequent culture on Transwell filters (10–30 organoids per filter) (Fig. 1a). Organoids were grown in 3D culture from day 7 to day 25. All growth factors and inhibitors were removed on day 12. Data are referred to as originating from a vial, experiment (single differentiation), or batch. Organoids generated from different vials of the same iPSC line but simultaneously differentiated are referred to as distinct ‘experiments’, and a differentiation commenced at a different time is referred to as a distinct ‘batch’. For all transcriptional profiling, we collected either duplicates or triplicates sampled from days 0, 4, 7, 10, 18 and/or 25. At days 0 (when iPSCs were plated) and 4 (the end of CHIR99021 induction), replicates represent individual wells from the same starting vial. From day 7 onward, replicates are individual kidney organoids grown from the same starting vial.
Fig. 1. Temporal characterization of human kidney organoid differentiation.
a, Overview of differentiation protocol, noting the collection time points. On days 0 and 4, individual wells were collected from a six-well plate; from day 7 onward, independent replicates refer to individual organoids. (CHIR99021 is an aminopyrimidine derivative that is an extremely potent inhibitor of glycogen synthase kinase-3 alpha.) FGF9, fibroblast growth factor 9. b, MDS plot of all samples demonstrates a clear developmental trajectory. c, Pairwise Spearman’s ρ correlation coefficients between the samples collected across the time course (n = 15,685 genes). d, Number of significant differentially expressed genes upregulated and downregulated between consecutive time points. Significant genes were identified on the basis of TREAT statistics with absolute log fold change > 1 and FDR < 5% (two-sided).
Temporal characterization of organoid differentiation.
We previously performed RNA-seq of individual organoids at days 7, 10, 18, and 25 within a single differentiation experiment19. In this study, we extended this dataset by collecting triplicate RNA samples at days 0 and 4 using the same iPSC cell line (CRL1502-C32). Organoids collected at a single time point were highly reproducible within a given differentiation experiment (Spearman’s ρ > 0.986 between all samples at each time point; Fig. 1b,c), and pairwise differential expression comparisons indicated that the largest transcriptional changes were between days 0 and 4 and the smallest were between days 18 and 25 (log fold change > 1, false discovery rate (FDR) < 0.05, Fig. 1d; Supplementary Fig. 1a, Supplementary Table 1). The top 100 upregulated genes included significantly enriched gene ontology terms for somitogenesis and anterior/posterior pattern specification between days 0 and 4, and nephron, urogenital system, and kidney development between days 10 and 18 (Supplementary Fig. 1b–f and Supplementary Table 2).
Unsupervised fuzzy clustering identified 20 synexpression clusters, which displayed correlated nonlinear expression patterns across the time course (Supplementary Figs. 2 and 3a). For each cluster, we extracted the core contributing genes (Supplementary Table 3), performed gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses (Supplementary Table 4), and identified key FANTOM5 transcription factors (see “URLs” section) within the core gene sets (Supplementary Table 5). PCR of selected core genes within seven of these clusters validated the expression patterns in independent differentiations (Supplementary Fig. 3b).
Prior knowledge of transcriptional changes in the developing mouse19 indicates that nephrons form between days 10 and 18 of the organoid time course. We observed a large-scale upregulation of genes suggestive of nephron development and vascular differentiation (2,292 upregulated genes, 1,185 downregulated genes, log fold change > 1, FDR < 0.05; Supplementary Table 1), supported by gene ontology analysis (Supplementary Fig. 1b–f). In addition, synexpression clusters 7, 10, and 12 captured genes that change within this time period (Supplementary Fig. 3a). At day 18, the most highly expressed genes included markers of podocytes, a key component of the forming glomerulus (NPHS1, NPHS2, PTPRO, MAFB, SYNPO, ITIH5, CLIC5, NPNT)21–23. Simultaneous with the upregulation of nephron markers between days 10 and 18 was the downregulation of the nephron progenitor markers LIN28A, MEOX1, CITED1, and EYA1. These genes are also known to mark the metanephric mesenchyme in humans24. On the basis of the presence of maturing nephrons, we chose day 18 for the subsequent analysis of reproducibility. The identification of these nephron- and maturity-related genes, particularly markers of podocyte differentiation, was key to our subsequent interpretation of batch variation.
Variation within and between differentiation experiments.
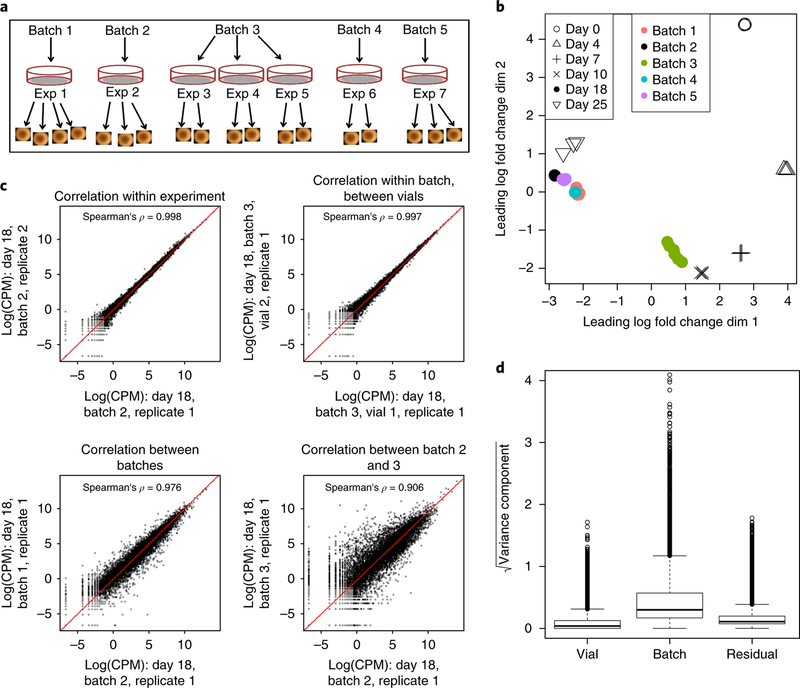
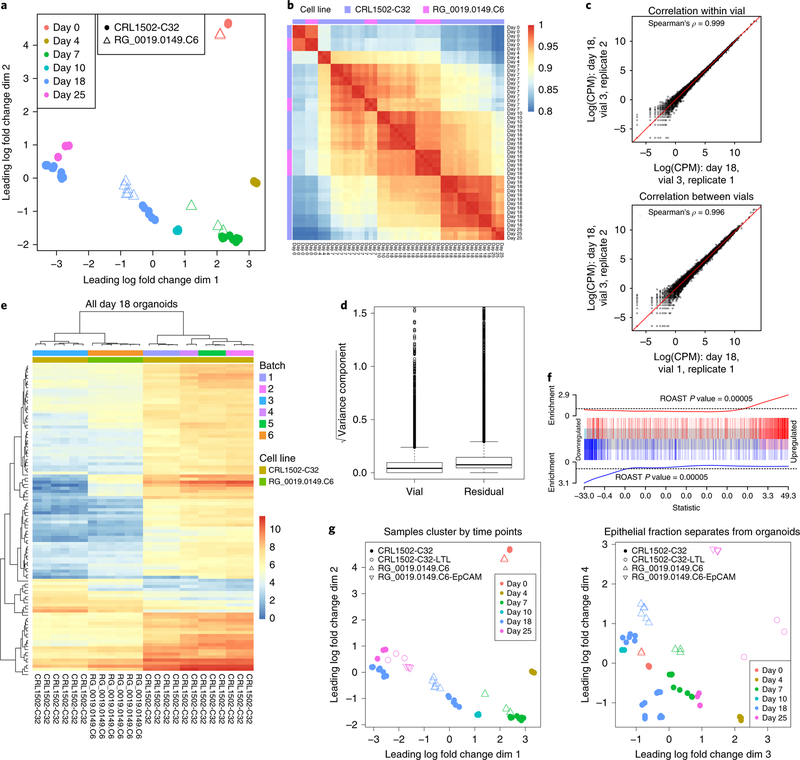
We previously identified at least nine distinct cell types in kidney organoids on the basis of immunofluorescence19. Given this cellular complexity, sources of transcriptional variation could include biological variation in component cell types or intrinsic cell line or clone heterogeneity, technical variation from vial or passage, batch variation in culture components, and variations in downstream processing procedures. Although the correlation between organoids within this initial differentiation experiment was very high (Spearman’s ρ = 0.986), we wanted to understand the level of transcriptional variability that could arise when differentiation experiments are repeated. Using the same iPSC cell line, we carried out another six organoid differentiation experiments and collected RNA from individual organoids at day 18 (Fig. 2a). Distinct differentiations were performed up to 12 months apart and hence across different reagent batches (culture media, recombinant growth factors). Organoids from experiments 3, 4, and 5 were initiated with distinct iPSC vials, but were differentiated and processed in parallel and classed as one batch. The remaining organoids from experiments 1, 2, 6, and 7 were differentiated at different times and considered to be distinct batches. In total, 18 organoids were profiled at day 18, and all samples were sequenced at the same facility using the same protocol (Methods).
Fig. 2. Sources of transcriptional variation within and between experiments.
a, Diagram of organoids profiled at day 18, showing the relationship between experiment (single differentiation from a unique vial), batch (multiple experiments separated in time), and organoid. Samples refer to individual organoids within an experiment. b, MDS plot of day 18 organoid and time series samples indicating batch and day. c, Log-normalized expression for pairs of representative samples showing correlation between organoids (Spearman’s ρ, n = 15,685 genes). d, Contribution to each source of variation (vial, batch, and residual) across all 15,685 genes (center line, median; hinges, first and third quartiles; whiskers, most extreme values within 1.5× the interquartile range of the box).
Day 18 organoids within the same batch clustered tightly on a multidimensional scaling (MDS) plot, in a region between days 10 and 25 of the time course (average Spearman’s ρ across all batches, 0.956) (Fig. 2b). The highest correlation was observed between organoids differentiated at the same time (Spearman ρ = 0.997, Fig. 2c; Supplementary Fig. 4). Day 18 organoids in batch 3 were closer to day 10 organoids than to organoids from the remaining day 18 batches (Fig. 2b, Supplementary Fig. 4). For each gene, we fitted a random effects model to estimate the contribution of three variance components representing (1) batch-to-batch variability, (2) vial-to-vial variability of organoids differentiated in parallel in batch 3, and (3) ‘residual’ organoid-to-organoid variability (Supplementary Table 6). Across all genes, the largest contribution to transcriptional variability was batch-to-batch variability; vial-to-vial variability was only a small contributor, and residual variance was minimal (Fig. 2d).
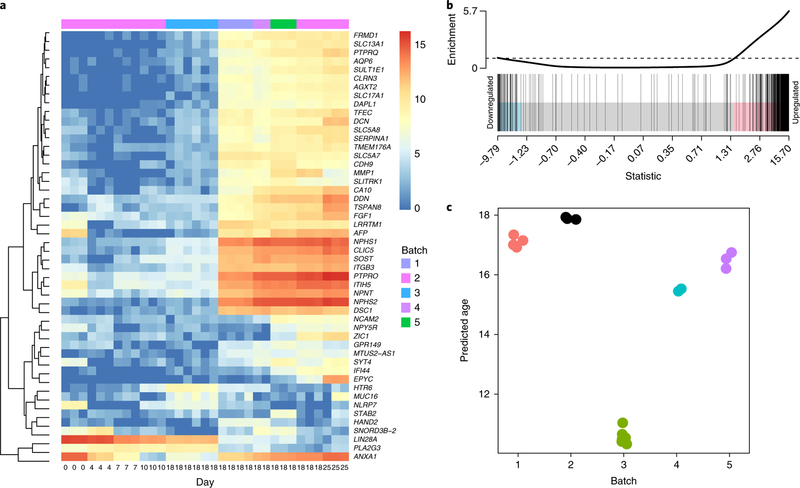
We examined genes with the greatest total variability between batches to understand how transcriptional variability arose at day 18. Many of the highly variable genes were related to nephron maturation (for example, the podocyte markers NPHS2, PTPRO, and NPHS1) (Supplementary Fig. 5). Of the top 50 most variable genes between individual day 18 organoids (Supplementary Fig. 6a), 7 and 16 genes showed synexpression patterns consistent with time-course clusters 10 and 12, respectively (Fig. 3a; Supplementary Fig. 6b), suggesting that variability may result from differences in nephron maturity. Comparison of days 10 and 25 of the original time course identified differentially expressed genes associated with nephron formation. The 500 most variable genes at day 18 were strongly enriched among nephron-related genes, with approximately 80% of the variable genes significantly upregulated between days 10 and 25 (one-sided P = 0.0005, ROAST test25, Fig. 3b).
Fig. 3. Prediction of relative organoid maturation between batches.
a, Log2-normalized expression across time points for the most variable genes identified at day 18. Note that the original time series is part of batch 2. b, ROAST analysis of the 500 most variable genes at day 18 shows strong enrichment between days 10 and 25 and suggests an association between nephron maturation and transcriptional variability (one-sided P = 0.0005). c, Prediction of organoid age based on ten genes marking the progression of differentiation.
To define a relative time scale of molecular differentiation, we constructed a multivariate linear regression for the day 7–25 time-series data using the ten genes with the greatest linear association across this time period (Supplementary Fig. 7). This framework enabled us to estimate the normalized ‘age’ of additional day 18 organoids relative to the time-series data (Fig. 3c). The results mirrored the trend in the MDS plots: batch 3 organoids had the youngest predicted ages (10.3–11 d), and organoids in batches 1, 2, 4, and 5 were closer in developmental age to day 18 (median predicted ages: batch 1, 17.1 d; batch 2, 17.9 d; batch 4, 15.5 d; batch 5, 16.5 d). This approach could be used to normalize variation between kidney organoid batches for a given iPSC line and could potentially extend to other organoid models.
Coimmunofluorescence for collecting duct (GATA-binding factor 3 and cadherin-1), distal tubule (cadherin-1), proximal tubule (Lotus tetragonolobus lectin (LTL) and protein jagged-1), and podocyte (nephrin) markers confirmed that changes between days 10 and 25 were related to nephron patterning (Supplementary Fig. 8a). This revealed evidence of a transition in nephron morphology from an initial renal vesicle to capillary loop stages across this time interval (Supplementary Fig. 8b,c). Quantitative PCR (qPCR) also confirmed the temporal pattern of core gene expression in clusters 10 and 12 (Supplementary Fig. 8d). Together, our analyses support a model in which transcriptional variation between organoids reflects significant changes in nephron morphogenesis occurring between days 10 and 25 of organoid culture.
Single-cell profiling of component cell type variation.
RNA-seq analysis suggested that organoid maturation contributed strongly to transcriptional variation. This variation may arise from expression changes within constituent cell types or from differing ratios of component cell types, including both on-target (kidney) and off-target (non-kidney) populations, in any given organoid. In addition, expression variation may result from differences in nephron segmentation between organoids, such that nephrons may be preferentially proximalized (more podocytes) or distalized (more tubular). To investigate this further, we isolated single cells from four CRL1502-C32-derived kidney organoids at day 25 (Methods). Three organoids from one differentiation were prepared and sequenced independently from the fourth organoid from a distinct differentiation.
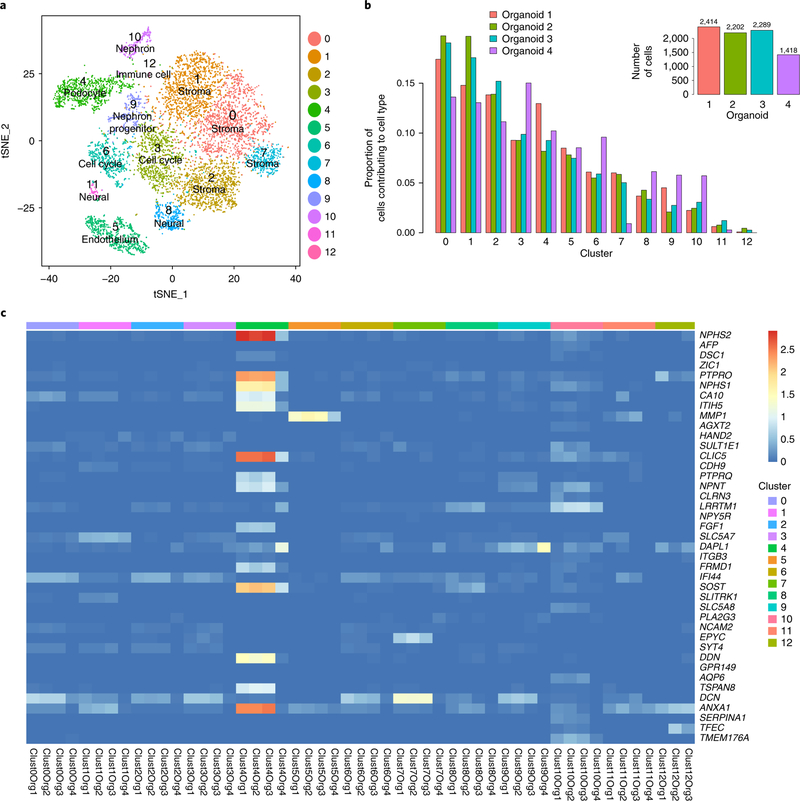
After filtering for poor-quality cells and genes with low expression, clustering of all remaining 8,323 cells from these organoids26,27 generated 13 clusters that we labeled using gene ontology term analysis and comparison to mouse and human profiling data (Fig. 4a; Supplementary Figs. 9 and 10, Supplementary Table 7, Methods). Although cells from most clusters were present in all organoids, these differed in relative proportions, most notably between batches (Fig. 4b). Organoid 4 was underrepresented for stromal populations (clusters 0, 1, 2, 7); was overrepresented for clusters 3 and 6 (cell-cycle-related), 8 (neural), 9 (nephron progenitor), and 10 (nephron); and contained no cluster 12 (immune) cells. The proportion of cells assigned to the podocyte cluster varied between all organoids, irrespective of batch. While this may reflect relative podocyte maturation, it may also reflect variation in the isolation of these tightly interdigitated cells. Differences in cell type proportion may also be a result of the dissociation methods used; however, it is notable that the three organoids from the same batch were most similar.
Fig. 4. Single-cell profiling reveals heterogeneity in component cell types among four day 25 organoids.
a, Graph-based clustering identifies 13 clusters, including anticipated and off-target cell types. The t-distributed stochastic neighbor embedding (tSNE) plot consists of 8,361 cells from n = 4 biologically independent organoids. b, Proportions of cells per cell type in each organoid. Inset bar graph, total number of cells contributed from each organoid. c, Average log-normalized expression for top variable genes for each organoid in each cluster. Most of the variable genes are expressed in cluster 4, the podocytes.
The majority of the most variable genes identified by bulk RNA-seq were restricted to the podocyte cluster, with only a few genes (MMP1, endothelium; DCN, stroma) selectively expressed in other cell types (Fig. 4c). The proportion of cells expressing highly variable cell-type-specific genes, such as NPHS2, PTPRO, and MMP1, also differed across the four organoids (Supplementary Fig. 11). A greater proportion of cells in organoids 1, 2, and 3 had high expression of NPHS2 and PTPRO in the podocyte cluster and MMP1 in the endothelial cluster, while fewer cells in organoid 4 expressed these genes at high levels (Supplementary Fig. 11). With regard to key kidney marker genes, organoid 4 had more cells expressing PAX2 (clusters 4 (podocyte), 9 (nephron progenitor), and 10 (nephron)) but much reduced MAFB expression in the podocyte cluster (Supplementary Figs. 10 and 11). Notably, organoid 4 did not show a relative depletion of cells in cluster 4 (podocytes), but the cells in this cluster still showed a distinct podocyte expression pattern. This may reflect variation in relative age, more distal nephron patterning, or a combination of both between batches. Importantly, single-cell analysis once again showed that organoids from the same batch were the most similar.
Correlation between organoids from distinct iPSC clones.
All the data presented thus far were generated using iPSC clone CRL1502-C3228,29. To examine variation between iPSC lines, we generated kidney organoids from a healthy female human iPSC line (RG_0019.0149.C6). Hereinafter we refer to the lines as CRL and RG for simplicity. The resulting organoids displayed similar brightfield morphology (Supplementary Fig. 12). RNA-seq was performed on two day 18 organoids from each of three separate but simultaneous differentiation experiments (six organoids in total); triplicate day 0 and 7 cultures were profiled from each differentiation experiment. Organoids generated from the two lines were strikingly concordant, exhibiting greater correlation and clustering by differentiation duration and not cell line (Fig. 5a,b).
Fig. 5. Transcriptional variation between iPSC lines, and temporal concordance between total organoid and enriched nephron epithelium.
a, MDS plot of organoids generated from two cell lines. b, Spearman’s correlations between all organoids. c, Log-normalized expression for pairs of RG_0019.0149. C6 organoids. Spearman’s ρ was used to calculate correlation coefficients across 15,685 genes. d, Contribution of two variance components in a random effects model estimated for each gene (n = 14,870 genes). Center line, median; hinges, first and third quartiles; whiskers, most extreme values within 1.5× the interquartile range of the box. e, Hierarchical clustering of all day 18 organoids based on the top 100 differentially expressed genes between day 18 and day 10 in the original time course data. The color scale represents the expression values as log(CPM). f, Barcode plot showing enrichment of genes differentially expressed between CRL1502-C32 day 18 batch 3 organoids (n = 6) and RG_0019.0149.C6 day 18 organoids (n = 6) when compared to day 18 (n = 3) versus day 10 (n = 3) CRL1502-C32 organoids. g, MDS plots of day 25 enriched nephron epithelium samples from the two cell lines, labeled CRL1502-C32-LTL and RG_0019.0149.C6-EpCAM, compared to all other total organoid samples.
This second line was differentiated in a single batch, but it was possible to examine gene-wise variation arising from distinct vials and residual (organoid-to-organoid plus unknown) variation at day 18. Organoids grown from the same vials of the RG cell line were highly correlated, as were those grown from different vials (Fig. 5c). Random effects analysis confirmed that vial contribution was small across all genes, with residual variation contributing only marginally higher transcriptional variability (Fig. 5d, Supplementary Table 8). The most variable genes appeared to be driven by differences between vials 2 and 3 (Supplementary Fig. 13) and were overrepresented for gene ontology terms involving the Golgi lumen, locomotory behavior, and epidermal cell differentiation, whereas variable genes with high residual contributions were overrepresented in signaling receptor activity (Supplementary Fig. 14, Supplementary Table 9).
The differences between day 18 organoids from the distinct iPSC lines were again associated with organoid maturation. Unsupervised clustering of all day 18 organoids using the top 100 maturity-related genes (differentially expressed between days 10 and 18 in the original time course) indicated that RG day 18 organoids were older than CRL day 18 organoids from batch 3, but younger than those from the other batches (Fig. 5e). Genes differentially expressed between RG and CRL batch 3 day 18 organoids (generated with the same replication design) were highly enriched for maturity-related genes (Fig. 5f, ROAST P = 0.0005).
Epithelial populations cluster with age-matched organoids.
Single-cell profiling demonstrated that transcriptional variation between individual experiments is largely driven by the diversity of cell types present. To simplify the pool of cells for analysis, we enriched for the epithelial cell adhesion molecule (EpCAM+) epithelial fraction of day 25 RG organoids and the LTL+ proximal tubular epithelium fraction from CRL organoids by using magnetic-activated cell sorting (MACS). This required organoids within a given differentiation to be pooled to yield enough RNA for sequencing. Hence, replicates represent sorted fractions from distinct batches. Despite this, the epithelium-enriched profiles clustered alongside the global organoid profile from the same differentiation time point, illustrating the strong contribution of epithelial cell types to the global profile (Fig. 5g). Total organoid and epithelium-enriched samples also separated along the lower dimensions, as anticipated (Fig. 5g).
Plotting the log-normalized expression of epithelial and stromal markers confirmed high expression of epithelium-enriched profiles (Supplementary Fig. 15a) and depleted expression of interstitial markers in both epithelium-enriched populations (Supplementary Fig. 15b). The LTL+ fraction showed distinctly higher expression of protein jagged-1, consistent with the selective enrichment of the proximal tubular epithelium (LTL+) versus all epithelium (EpCAM+). Although sorting for specific cell types will not overcome inherent differences in nephron maturation between batches, it reduces the complexity of the transcriptional profile and potentially improves the ability to detect cell-type-specific transcriptional changes. Such an approach is likely to assist in disease modeling.
Application to disease modeling.
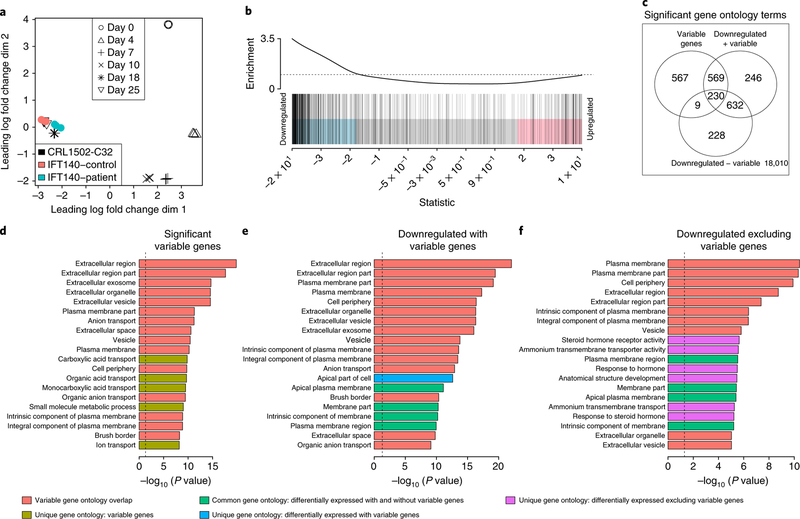
Our modeling of the sources of variability suggests that transcriptional profiling of organoids to identify disease-related changes will be challenging. We applied our approach to compare a patient iPSC line and a genetically repaired isogenic control line generated using CRISPR–Cas9-mediated gene editing. The line was derived from a person with juvenile nephronophthisis in association with retinitis pigmentosa and skeletal anomalies (Mainzer–Saldino syndrome30) resulting from distinct point mutations in both copies of IFT140, a component of the retrograde transport machinery of primary cilia. EpCAM-enriched epithelial fractions were isolated from pooled organoids at day 25, with triplicates from both patient and gene-corrected lines generated from distinct differentiation experiments. Combining these with the time-series data revealed evidence of differences in maturation, with patient samples closer to day 18 organoids and control samples closer to day 25 organoids (Fig. 6a).
Fig. 6. Transcriptional analysis of a disease model improves with accounting for highly variable genes.
a, MDS plot showing patient samples, control samples, and time series data. IFT140, intraflagellar transport protein 140 homolog. b, The previously identified 570 highly variable genes were significantly enriched in the genes downregulated between patient and control samples (ROAST one-sided P = 0.0085). Moderated t-tests were used to test for differential expression between patient and control samples (two-sided). FDR < 1% and absolute log fold change > 0.7 were used to identify significant differentially expressed genes. c, Overlap of significant gene ontology categories for the three different gene ontology analyses. d, Gene ontology categories enriched for significant, highly variable genes. e, Gene ontology categories enriched for significantly downregulated genes, including highly variable genes. f, Gene ontology categories enriched for significantly downregulated genes, excluding highly variable genes. For all gene ontology analyses, a modified hypergeometric test was used to determine statistical significance, considering gene length bias. P values are one-sided.
An initial analysis found 1,244 downregulated and 1,097 upregulated genes between patient and gene-corrected samples (FDR < 0.05). Of the 1,000 genes previously identified as most variable from our healthy organoids at day 18, 570 were among the differentially expressed genes in the disease study, 44% (249/570) and 29% (167/570) of which were downregulated at an FDR < 5% and < 1%, respectively. Indeed, the set of 570 highly variable genes were significantly enriched among the downregulated genes (Fig. 6b, ROAST P = 0.0085). We found that removing the highly variable genes reduced the number of significant gene ontology terms from 1,677 to 1,099 (Fig. 6c). We identified significant gene ontology terms for (1) the 167 highly variable downregulated (FDR < 1%) genes (Fig. 6d), (2) all of the patient downregulated genes, including these variable genes (Fig. 6e), and (3) the patient downregulated genes after removal of the highly variable genes (Fig. 6f). The removal of the highly variable genes from the patient differentially expressed gene list reduced the prominence of gene ontology terms associated with transporters and extracellular matrix and increased the association of the disease state with gene ontology terms such as ‘apical part of the cell’ and plasma membrane region (Fig. 6f). Such gene ontology terms are more biologically relevant to the ciliopathic disease present in this patient. We conclude that removing highly variable genes removed noise in the data, thus highlighting more biologically relevant pathways.
Discussion
The prospect of using organoids to model development or disease relies on the accuracy of the model and its reproducibility within and between lines. Kidney organoids represent arguably the most complex human organoid generated to date. With this increased cellular complexity comes greater variation in the relative proportions of cell types between experiments. Our transcriptional analysis showed a remarkable degree of congruence between the profiles of organoids generated across considerable technical and biological variation. Both whole-organoid and single-cell approaches indicated that batch-to-batch variation was the greatest driver of overall variability, with primary contributions from nephron maturation and nephron patterning, together with shifts in the relative abundance of on-target and off-target cell types.
While the robustness of the kidney organoid protocol suggests that disease modeling is feasible, our analysis of variability also highlights challenges in comparing patient and control cell lines. To assist with normalization, we provide a list of genes that are most variable between batches at a single time point, as well as a set of ten genes to generate an estimate of relative kidney organoid maturity. Although it is possible to exclude the most variable genes from any transcriptional comparison, this may also remove information of relevance to the phenotype of a particular line. In addition, as a post-hoc analysis, the ability to estimate differentiation stage does not ensure that lines will all reach the same level of maturity in a given differentiation. While this does not provide a solution if disease lines repeatedly show differences in relative maturation state from controls, we have successfully identified disease-related transcriptional changes between a patient and gene-corrected isogenic control line by removing the 570 most variable genes identified in this study30.
Another approach to reducing technical variation is to select for specific cell types within the organoid. Even though the epithelial cells sorted from organoid cultures were enriched for epithelial and depleted for stromal marker expression, they still aligned most closely with their source organoid tissues matched for differentiation stage. Therefore, the analysis of selected cellular components is likely to improve comparative organoid phenotyping.
Our study demonstrates that directed differentiation of human iPSCs to kidney organoids is robust, reproducible, and transferable between stem cell lines. As the greatest source of variation is differences in technical parameters, rather than the cell line, cellular compartment, or individual organoid, perhaps the most important implication of this work is that researchers must carry out any comparisons between patient and control lines concurrently while controlling as many technical variables as possible.
URLs.
FANTOM5 transcription factors, http://fantom.gsc.riken.jp/5/sstar/Browse_Transcription_Factors_hg19; TopGene Suite, https://toppgene.cchmc.org; Oshlack github repository, https://github.com/Oshlack/OrganoidVarAnalysis.
Methods
Ethical approval and consent to participate.
All research complied with the relevant ethical regulations and Australian legislation pertaining to stem cell research. Ethical approval for the derivation of the human iPSC line RG_0019.0149.C6 was approved as a subproposal of HREC_14_QRBW_34 by the Human Research Ethics Committee (HREC) of the Royal Womens and Childrens Hospital. iPSC reprogramming and gene correction of patient skin fibroblasts was conducted with approval from the HRECs of the Lady Cilento Children’s Hospital (no. HREC/15/QRCH/126), the University of Queensland (Medical Research and Ethics Committee approval no. 2014000453), and the Royal Brisbane and Women’s Hospital (HREC/14/QRBW/34), including research governance approval at all sites. Written informed consent was obtained from all participants or legal guardians as appropriate.
Human iPSC derivation and directed differentiation to kidney organoids.
Human iPSCs were generated using Sendai reprogramming as previously described. CRL1502-C32 is a female human iPSC cell line derived from ATCC CRL-1502 fetal fibroblasts28. RG_0019.0149.C6 is a female human iPSC cell line derived from fresh skin fibroblasts collected from an adult. The protocol for directed differentiation to kidney organoid has been described previously19,20,30.
Single-cell sample preparation and sequencing.
The four kidney organoids were harvested into ice-cold PBS. Three organoids (batch 1) were digested over 15 min at 37 °C in trypsin-EDTA (Thermo Fisher Scientific); the fourth organoid (batch 2) was digested over 15 min on ice in Liberase (Sigma-Aldrich); both batches were stored on ice in APEL media (Stem Cell Technologies). Organoids from batch 1 were run in parallel on a chromium 10x Single Cell Chip (10x Genomics); batch 2 was run at a later date. Libraries were prepared using Chromium Single Cell Library Kit V2 (10x Genomics) and sequenced on an Illumina HiSeq using 100 base pair (bp) paired-end reads in two runs (batches 1 and 2) at the Australian Genome Research Facility.
Brightfield imaging and immunofluorescence imaging of cultures.
Brightfield images were taken using the Nikon TS-1000 inverted microscope. For the kidney organoid, antibody staining was performed as described previously20. The following antibodies and dilutions were used: mouse anti-E-cadherin (1:300; BD Biosciences); goat anti-GATA-3 (1:300; R&D Systems); sheep anti-nephrin (1:300; R&D Systems); biotinylated LTL (1:300; Vector Laboratories); and rabbit anti-Jagged1 (1:300; Abcam). Confocal imaging was performed using a ZEISS LSM780 scanning confocal microscope, with a ZEISS Plan-Apochromat 25×/0.8-NA (numerical aperture) multi-immersion objective. Confocal stacks were taken at 1.5-μm Z spacing and exported to the Imaris software (Bitplane) for 3D reconstruction and surface rendering. All other image processing was performed in Fiji31. All immunofluorescence analyses were successfully repeated more than three times; representative images are shown.
QPCR of organoids.
Total RNA was extracted from cells with the Purelink RNA Mini Kit (Thermo Fisher Scientific), and complementary DNA was synthesized from 100 ng of total RNA using GoScript reverse transcriptase (Promega). Quantitative reverse transcription PCR (qRT–PCR) analyses were performed with the GoTaq qPCR Master Mix (Promega) by a PRISM 7500 96 real-time PCR System (Applied Biosystems). All absolute data were first normalized to glyceraldehyde 3-phosphate dehydrogenase and then normalized to control samples (2−ΔΔCt method). The sequences of primers used for qRT–PCR are as listed in Supplementary Table 10.
Isolation of EpCAM-enriched cellular fractions.
Kidney organoids were transferred to a drop of trypsin-EDTA, minced with a surgical blade, transferred to a 15-ml tube with 3 ml of trypsin-EDTA, and incubated at 37 °C for 10 min while the disintegrating organoid pieces were gently pipetted every 3 min. Next, 8 ml of DMEM + 10% fetal bovine serum was added and cells were pelleted by centrifugation (250g for 5 min). Then, the pellet was resuspended in 2 ml of epithelial cell culture medium (Renal Epithelial cell growth medium; Banksia Scientific). The remaining cell clumps were removed with a 40-µm cell strainer (BD Biosciences), and cells were counted. Cells (107) were repelleted by centrifugation (250g for 5 min), resuspended in 200 µl of MACS buffer (46.7 ml PBS+; 3.3 ml of 7.5% BSA; 200 µl 0.5 M EDTA), mixed with 20 µl of CD326 microbeads (Miltenyi Biotec), and refrigerated to 4 °C. After 30 min, 5 ml of MACS buffer was added and cells were pelleted by centrifugation, resuspended in 500 µl MACS buffer, and applied to a prerinsed MS column in the MACS separator. After three washings with MACS buffer, cells were collected with 1 ml of MACS buffer after the column was removed from the MACS separator. This routinely yielded approximately 3 × 105 EpCAM+ cells per organoid and live cell rates > 75%. The isolation of epithelial cellular fractions from patient organoids has been described previously30.
Isolation of LTL-enriched cellular fractions.
Pooled iPSC-derived kidney organoids were dissociated by incubation with TrypLE Select Enzyme (Thermo Fisher Scientific) for 12 min at 37 °C, with gentle pipetting every 2 min to aid dissociation. The cell solution was passed through a series of cell strainers with sequentially smaller mesh sizes, ranging from 100 to 40 µM (Corning) with extensive washing using sterile chilled MACS buffer (Dulbecco’s PBS, 2 nM EDTA, 0.5% BSA) to yield a single-cell suspension. Cells were counted, centrifuged at 300g for 5 min, and resuspended in 100 µl of MACS buffer with 1 µl of biotin-conjugated LTL primary antibody (Vector Laboratories); they were incubated on ice for 30 min. Cells were rinsed with MACS buffer and centrifuged twice before being resuspended in 300 µl of MACS buffer plus 100 µl of streptavidin microbeads (Miltenyi Biotec) for 30 min on ice. Cells were rinsed with MACS buffer and centrifuged before being resuspended in 500 µl of MACS buffer and passed through a MACS MS column according to the manufacturer’s protocol (Miltenyi Biotec). The LTL+ fraction was eluted from the column; the sorted cell population was counted and stored at −80 °C until RNA extraction was performed.
Bulk RNA sequencing, data acquisition, alignment, and quality control.
The RNA from the kidney organoid differentiations was extracted using the RNeasy Micro Kit (QIAGEN), and the sequencing libraries were prepared using the standard Illumina protocols. For the epithelial fractions, these were obtained from pooled organoids from the same differentiation. Most of the samples were sequenced at the Institute for Molecular Bioscience in Brisbane, Australia. The LTL-enriched and patient samples were sequenced at the Translational Genomics Unit, Murdoch Children’s Research Institute, Melbourne, Australia. The STAR aligner (version 2.4.0h1)32 was used to map the 75-bp single-end reads to the human reference genome (hg19) in the two-pass mapping mode. Uniquely mapped reads were summarized across genes with featureCounts (version 1.4.6)33 using GENCODE release 19 comprehensive annotation. Subsequent analyses of the count data were performed in the R statistical programming language with the Bioconductor34 packages edgeR35, limma36, RUVSeq37, and Mfuzz38, the annotation package org. Hs.eg.db, and the R package lme439. The Nature Research Reporting Summary has further details on the software used in the study. Highly expressed genes were defined as having at least one count per million (CPM) in at least two or three samples and were retained for statistical analysis. The threshold for selecting highly expressed genes was determined by the minimum group sample size in the dataset being analyzed. In addition, genes encoding ribosomal protein, mitochondrial genes, pseudogenes, and genes without annotation (Entrez Gene identification) were removed before trimmed mean of M-values normalization40 and statistical analysis. For all datasets, MDS plots were used to visualize the greatest sources of variation as part of quality control. The MDS plots were based on the top 500 most variable genes, specifying pairwise distance metrics in the limma package.
Statistical analysis.
Differential expression analysis of time-series data.
The time-series RNA-seq data consisted of three replicates at each of six time points: days 0, 4, 7, 10, 18, and 25. The samples for days 7, 10, 18, and 25 were obtained and sequenced together, with additional data for the earlier time points (days 0 and 4) generated at a later date. To account for possible batch effects, we carried out differential expression analysis using RUVSeq in conjunction with edgeR. RUVSeq was performed using empirical control genes, identified as the 5,000 genes that varied the least across the time-course data, based on an F-statistic. Genes that were differentially expressed at consecutive time points were identified as those that had an adjusted FDR < 5%. Genes significantly differentially expressed with an absolute log fold change of at least 1 and an FDR < 5% were identified by a TREAT analysis41 with robust variance estimation42. Gene ontology testing was done with the goana function in the limma package, adjusting for gene length bias43, and with the web-based ToppGene suite (see “URLs”)44.
Fuzzy clustering.
Genes that displayed similar patterns of expression across the time-course data were clustered by fuzzy c-means clustering. Clustering was limited to genes that showed evidence of differential expression across the time course based on an F-statistic, and with an absolute log fold change of at least 1 between at least one comparison. For this analysis, each time point was compared to the remaining time points. This identified 7,682 genes to use as input for the Mfuzz algorithm. The counts were transformed to log(CPM), adding a small offset of 0.25 proportional to the library sizes. Each of the three replicates were averaged per time point, and the data were standardized so that each gene had a mean of 0 and s.d. of 1 to ensure that genes with similar changes in expression were close in Euclidean space. The soft clustering approach assigned each gene gradual degrees of membership, ranging from 0 to 1, to each of the 20 clusters. We identified a core set of genes for each cluster by specifying a cutoff on the membership score of 0.5, with the number of core genes per cluster ranging from 70 to 299. Gene ontology and KEGG pathway analysis on the core genes was done with the goana and kegga functions in the limma package, adjusting for gene length bias43; the analysis was further explored with the ToppGene suite.
Random effects modeling.
To study the different variance components in comparisons across experiments and batches, we fitted a multilevel random effects model to the day 18 organoid data for each cell line separately, using the lmer function in the lme4 package. The data were transformed to log(CPM), with a small prior count of 0.5 added before model fitting; variance components for batch, vial, and residual were extracted for the CRL1502-C32 organoids, and vial and residual components were extracted for the RG_0019.0149.C6 organoids. Genes were ranked according to total variation, which we obtained by summing the variance components. For each gene, we obtained the greatest contributor to the variability by calculating the proportions of each variance component to the total variation.
Comparing highly variable genes to the time-series data.
To formally test whether the highly variable genes arise because of the varying maturity of the organoids, we tested for differential expression between day 25 (n = 3) and day 10 (n = 3) organoids to identify a set of maturity-related genes. We used RUVSeq in conjunction with edgeR to identify differentially expressed genes between days 25 and 10, and tested whether the highly variable genes were changing as a set between these two time points by using the ROAST gene set test25 in the limma package.
Comparison of CRL1502-C32 and RG_0019.0149.C6 day 18 organoids.
To obtain a set of genes that were differentially expressed between these two cell lines, we compared the RG_0019.0149.C6 day 18 organoids (n = 6) with the CRL1502-C32 organoids (n = 6) from batch 3. A list of differentially expressed genes was obtained using voom45 and TREAT with a log fold change > 1 and an FDR < 5%. The upregulated and downregulated genes were tested as distinct gene sets for enrichment with ROAST in differentially expressed genes between day 18 (n = 3) and day 10 (n = 3) CRL1502-C32 organoids from the original time-series data (independent biological replicates from batch 2).
Single-cell data analysis.
Cell Ranger (version 1.3.1; 10x Genomics) was used to process and aggregate raw data into gene-level counts for each cell in each organoid. CellrangerRkit version 1.1.0 was used to read the data into the R programming language. Quality control was performed separately on batch 1 and batch 2 organoids. Poor-quality cells were defined as those that had more than 95% zeros across the cells, and were discarded. Additional quality control was performed; we checked the proportions of reads assigned to ribosomal and mitochondrial genes, as well as cell diversity. Genes with low expression were defined as genes that had zeros in more than (total number of cells − 20) cells, which allowed for a minimum cluster size of 20 cells. In addition, mitochondrial and ribosomal genes, as well as genes without gene annotation, were filtered out. After quality control, there were 6,942 cells with expression measurements for 15,245 genes for batch 1 organoids, and 1,419 cells and 13,710 genes for the fourth batch 2 organoid. To identify clusters in the data, we used the alignment method in the Seurat package (version 2.0.0)27. The two batches were merged using canonical correlation analysis based on 2,662 highly variable genes and 20 canonical correlation vectors. Thirteen clusters were identified using the 20 canonical correlation vectors with the resolution parameter set to 0.8. Marker genes for the 13 clusters were defined using the ROCtest in the Seurat package, which allowed for cell types to be assigned to each cluster. In addition, differential expression analysis between each cluster versus the remaining clusters was performed in the edgeR package; genes that had a log fold change > 1 and an FDR < 5% were identified using TREAT. The upregulated genes from this analysis further assisted with assignment of cell types to the clusters.
Supplementary Material
Acknowledgements
We thank A. Christ and G. Baillie at the Institute for Molecular Bioscience, The University of Queensland, for sequencing services. We acknowledge A. Mallett and S. Alexander for assistance in ethics applications and patient recruitment. We thank D. Vukcevic and G.K. Smyth for valuable discussion regarding random effects modeling, and J. Maksimovic for initial analysis and mapping of the patient RNA-seq data. This study was funded by the National Institute of Diabetes and Digestive and Kidney Diseases (grant no. DK107344) and National Health and Medical Research Council of Australia (NHMRC) (grant nos. GNT1041277, GNT1100970, GNT1098654). The Murdoch Children’s Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. M.H.L. is an NHMRC Senior Principal Research Fellow. A.O. is an NHMRC Career Development Fellow (grant no. GNT1126157). T.A.F. is an NHMRC Postgraduate Scholarship (grant no. GNT1114409) and Royal Australian College of Physicians Jacquot Award Recipient (grant no. APP1114409).
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41592-018-0253-2.
Competing interests
M.H.L. and M.T. hold intellectual property around the kidney organoid differentiation protocol. M.H.L. holds contract research agreements with Organovo Holdings. All other authors declare that they have no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-018-0253-2.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability. The scripts for analyzing all the bulk RNA-seq and single-cell RNA-seq data are available from the Oshlack github repository (see “URLs”).
References
- 1.Takahashi K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bellin M et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J 32, 3161–3175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim C et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 494, 105–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phelan DG et al. ALPK3-deficient cardiomyocytes generated from patient-derived induced pluripotent stem cells and mutant human embryonic stem cells display abnormal calcium handling and establish that ALPK3 deficiency underlies familial cardiomyopathy. Eur. Heart J 37, 2586–2590 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A & Vaccarino FM Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol 13, 265–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aksoy I et al. Personalized genome sequencing coupled with iPSC technology identifies GTDC1 as a gene involved in neurodevelopmental disorders. Hum. Mol. Genet 26, 367–382 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Jang Y-Y & Ye Z Gene correction in patient-specific iPSCs for therapy development and disease modeling. Hum. Genet 135, 1041–1058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paquet D et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Howden SE, Thomson JA & Little MH Simultaneous reprogramming and gene editing of human fibroblasts. Nat. Protoc 13, 875–898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ader M & Tanaka EM Modeling human development in 3D culture. Curr. Opin. Cell Biol 31, 23–28 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Huch M & Koo B-K Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Suga H et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature 480, 57–62 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Eiraku M et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Spence JR et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano T et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadoshima T et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl Acad. Sci. USA 110, 20284–20289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCracken KW et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Takasato M, Er PX, Chiu HS & Little MH Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc 11, 1681–1692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavenstädt H, Kriz W & Kretzler M Cell biology of the glomerular podocyte. Physiol. Rev 83, 253–307 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Brunskill EW, Georgas K, Rumballe B, Little MH & Potter SS Defining the molecular character of the developing and adult kidney podocyte. PLoS One 6, e24640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindström NO et al. Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J. Am. Soc. Nephrol 29, 806–824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs JA et al. Integration-free induced pluripotent stem cells model genetic and neural developmental features of Down syndrome etiology. Stem Cells 31, 467–478 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Yu J et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes TA et al. Patient-iPSC-derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am. J. Hum. Genet 102, 816–831 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Huber W et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Risso D, Ngai J, Speed TP & Dudoit S Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Futschik ME & Carlisle B Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol 3, 965–988 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Bates D, Mächler M, Bolker B & Walker S Fitting linear mixed-effects models using lme4. J. Stat. Softw 67, 1–48 (2015). [Google Scholar]
- 40.Robinson MD & Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy DJ & Smyth GK Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25, 765–771 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phipson B, Lee S, Majewski IJ, Alexander WS & Smyth GK Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat 10, 946–963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young MD, Wakefield MJ, Smyth GK & Oshlack A Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Bardes EE, Aronow BJ & Jegga AG ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law CW, Chen Y, Shi W & Smyth GK voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.