Abstract
Background
Although rare in the general population, highly penetrant germline mutations in CDKN2A are responsible for 5–40% of melanoma cases reported in melanoma-prone families. We sought to determine whether MELPREDICT was generalizable to a global series of melanoma families and whether performance improvements can be achieved.
Methods
2,116 familial melanoma cases were ascertained by the international GenoMEL Consortium. We recapitulated the MELPREDICT model within our data (GenoMELPREDICT) to assess performance improvements by adding phenotypic risk factors and history of pancreatic cancer. We report areas under the curve (AUC) with 95% confidence intervals (CI) along with net reclassification indices (NRI) as performance metrics.
Results
MELPREDICT performed well (AUC=0.752; 95%CI: 0.730, 0.775), and GenoMELPREDICT performance was similar (AUC=0.748; 95% CI: 0.726, 0.771). Adding a reported history of pancreatic cancer yielded discriminatory improvement (p<0.0001) in GenoMELPREDICT (AUC=0.772; 95%CI: 0.750, 0.793; NRI=0.40). Including phenotypic risk factors did not improve performance.
Conclusion
The MELPREDICT model functioned well in a global dataset of familial melanoma cases. Adding pancreatic cancer history improved model prediction. GenoMELPREDICT is a simple tool for predicting CDKN2A mutational status among melanoma patients from melanoma-prone families and can aid in counselling these patients towards genetic testing or cancer risk counselling.
Capsule Summary
Available prediction tools for CDKN2A status were developed among small, homogeneous populations and lack generalizability. GenoMELPREDICT is a globally generalizable and simple clinical tool for predicting CDKN2A mutational status among familial melanoma patients.
GenoMELPREDICT can aid in appropriate patient management, whether that is genetic testing or cancer risk counselling.
Introduction
Inherited mutations in the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene are major risk factors for familial melanoma.[1–3] The frequency of CDKN2A mutations in melanoma-prone families varies widely (<5% to 40%) with the number of family members diagnosed with melanoma and the number of primary melanomas diagnosed within an individual.[1, 4–6] The penetrance of CDKN2A mutations in melanoma-prone families is a function of population incidence rates of melanoma and is modified by environmental factors, melanoma-associated phenotypes, and MC1R variants.[3, 7] In light of geographic variability in mutation penetrance, a standard guideline for recommending CDKN2A genetic testing has not been suitable for heterogeneous populations.[8] GenoMEL, the International Melanoma Genetics Consortium, supports a qualitative framework to identify candidate individuals for CDKN2A mutation testing based on population-based melanoma incidence rates, diagnosis of multiple primary melanomas, and a verified family history of melanoma and/or pancreatic cancer.[8] Rapid identification of familial melanoma patients with low probability of a germline mutation in CDKN2A could aid to direct patients toward risk counseling and away from inappropriate genetic testing, especially since a negative test result is unlikely to influence their risk management, and/or in fostering potential conversation about genetic testing for mutations in other known, but much rarer, high-penetrance melanoma genes.
MELPREDICT is a published logistic regression model to predict CDKN2A mutation carrier status.[9] MELPREDICT performed well (area under the curve (AUC)=0.881) among melanoma patients (n=116) belonging to melanoma-prone families in Boston, Massachusetts, USA, and similarly (AUC=0.803) among those from melanoma-prone families in Toronto, Ontario, Canada (n=143).[9] We sought to determine whether MELPREDICT was generalizable to a large series of melanoma families from 20 countries participating in GenoMEL. Further, we evaluated whether improvements in model performance can be achieved by adding personal or family history of pancreatic cancer and/or phenotypic risk factors for melanoma.
Methods
Study population
The GenoMEL consortium comprises 29 study centers from Australia, Europe, the Middle East, and North and South America. GenoMEL used a common protocol to obtain research data as previously described.[10] Written informed consent was obtained from each participant, and individual GenoMEL centers received study approval from their respective institutional review boards. Consenting participants completed a self-administered questionnaire that solicited information on phenotypic characteristics, and personal and family history of melanoma and other cancers.[10, 11]
Study sample
Our study sample reflects 2,116 melanoma patients with CDKN2A genotype. These participants were from 900 melanoma-prone families defined by the presence of three or more verified melanoma cases among blood relatives (individuals who share a common ancestor and are not related by marriage) or two verified melanoma cases in first-degree blood relatives recruited at 20 GenoMEL centers (Table 1). There were 359 reports in 122 families of a personal or family history of pancreatic cancer, and pathologic verification was available for 79 (22%) of these reports; the remainder were self-reported.
Table 1:
Number of participants and families by ascertainment center
| GenoMEL Center | Participants* | Families† | Average number of participants per family‡ | Average number of affected members per family¶ |
|---|---|---|---|---|
| Barcelona, ES | 44 | 25 | 1.8 | 2.1 |
| Bethesda, US | 199 | 46 | 4.3 | 4.8 |
| Cesena, IT | 50 | 24 | 2.1 | 2.1 |
| Copenhagen, DK | 47 | 34 | 1.4 | 2.5 |
| Genoa, IT | 34 | 16 | 2.1 | 2.3 |
| Leeds, GB | 158 | 77 | 2.1 | 2.8 |
| Leiden, NL | 210 | 60 | 3.5 | 4.6 |
| Ljubljana, SI | 9 | 4 | 2.3 | 2.3 |
| Lund, SE | 20 | 7 | 2.9 | 4.4 |
| Montevideo, UY | 8 | 4 | 2.0 | 2.0 |
| Paris, FR | 341 | 176 | 1.9 | 2.5 |
| Philadelphia, US | 78 | 36 | 2.2 | 2.4 |
| Porto Allegre, BR | 9 | 5 | 1.8 | 2.2 |
| Queensland, AU | 96 | 21 | 4.6 | 6.2 |
| Riga, LV | 5 | 5 | 1.0 | 2.6 |
| Santiago, CL | 3 | 2 | 1.5 | 2.0 |
| São Paulo, BR | 13 | 8 | 1.6 | 2.1 |
| Stockholm, SE | 39 | 21 | 1.9 | 2.8 |
| Sydney, AU | 722 | 305 | 2.4 | 3.4 |
| Tel Aviv, IL | 21 | 18 | 1.2 | 2.0 |
| Valencia, ES | 10 | 6 | 1.7 | 2.2 |
| Total | 2116 | 900 | 2.2 | 3.1 |
Verification of melanoma was available for >99% of participants by: pathology report (74%), physician letter or clinical document verifying melanoma diagnosis (23%), cancer registry data (2%), or death certificate (<1%). Excludes affected individuals with a diagnosis of non-cutaneous melanoma or who are members of melanoma families by marriage and not ancestry.
Family members with a melanoma of the uveal tract or conjunctiva did not contribute to defining a melanoma family.
Includes only participants who contribute to prediction modeling.
Includes family members who may not contribute to prediction modeling because of missing data.
CDKN2A genotyping
Germline DNA was screened for mutations in CDKN2A (including exons 1α, 1β, 2 and 3), and mutations were classified as pathogenic (i.e. positive) or non-pathogenic (i.e. negative) as previously described.[10, 11] Eleven families had at least one member who was known to carry a mutation in another melanoma high-penetrance gene; these families were included in our analyses.
Statistical analysis
Using the MELPREDICT logistic regression model for which the probability of CDKN2A mutation carriage is defined as with L = 1.99 + [(0.92 × number of primary melanoma diagnoses) + (0.74 × number of additional family members diagnosed with melanoma) – (2.11 × ln(age at first melanoma diagnosis))], we estimated the predictive probability of CDKN2A mutation carriage among study participants, and the AUC was derived from the set of predictive probabilities.[9, 12] Using data from GenoMEL, we modeled the probability of CDKN2A mutation carriage as a function of these three variables and considered this our baseline model (GenoMELPREDICT). We used a generalized estimating equation with a logit link function and independence covariance structure with robust standard errors to account for familial clustering. We evaluated changes in baseline model performance associated with the addition of reported personal or family history of pancreatic cancer (yes, no), facial freckling (none, very few, few, some many, very many), proclivity to burn (tan with no burning, mild sun burning, sun burning with peeling, severe sun burning with blistering), proclivity to tan (very tanned, moderate tanning, mild tanning, no tanning), eye color (brown or black, blue, other), hair color (black, brown, blonde or fair, red), and skin type (very fair, fair, olive or brown or black), including all pairwise and triplet combinations of these phenotypic variables.
We used the empirical method of DeLong[13] to estimate and compare (via a Wald test) paired AUCs of receiver operating characteristic (ROC) curves. For each model, AUCs and 95% confidence intervals (CI) were calculated by ten-fold cross validation to evaluate discrimination between CDKN2A mutation carriers and non-carriers, and we used one-stage cluster sampling to randomly assign all members of a family to the same fold. Optimal discrimination was determined by maximizing sensitivity and specificity. Improvement in model performance was assessed by measuring the difference between paired model AUCs and by event and non-event net classification indices (NRI).[13–15] Models incorporating phenotypic factors were performed on sample sizes that varied according to factor missingness; for each augmented model, we reran our baseline model on the corresponding reduced sample size. Multiple imputation by the fully conditional specification method was used to restore missing values.[16] All analyses were performed using SAS v.9.4 (SAS Institute, Cary, NC) and R (R Core Team; http://www.R-project.org/).
Results
CDKN2A genotype was available for 711 (33.6%) mutation carriers and 1,405 (66.4%) non-carriers belonging to 900 melanoma-prone families. CDKN2A mutations identified in GenoMEL families have been previously published.[10, 17] Results of multivariable analyses for our 3-variable baseline and 4-variable GenoMELPREDICT model that included pancreatic cancer are presented in Table 2. Age at first melanoma diagnosis, higher numbers of primary melanomas, higher numbers of family members with a melanoma diagnosis, and a personal or family history of pancreatic cancer were independently associated (p<0.0001) with CDKN2A mutation carriage.
Table 2.
Distribution of pathogenic CDKN2A mutations among GenoMEL cases and model estimates for the baseline and 4-predictor GenoMELPREDICT models.
| Variable | No. (%) with mutation | GenoMELPEtEDICT | |||
|---|---|---|---|---|---|
| OR (95% CI)* | P | OR (95% CI)* | P | ||
| Ln(age at diagnosis) Number of primary melanomas | 0.29 (0.22, 0.39) | <0.0001 | 0.28 (0.22, 0.37) | <0.0001 | |
| 1 | 378/1426 (26.5%) | 1.20(1.10, 1.31) | <0.0001 | 1.20(1.10, 1.32) | <0.0001 |
| 2 | 153/380(40.3%) | ||||
| ≥3 | 180/310(58.1%) | ||||
| Number of other family members with melanoma | |||||
| 1 | 132/669(19.7%) | 1.29(1.20, 1.38) | <0.0001 | 1.26(1.17, 1.32) | <0.0001 |
| 2 | 146/560 (26.0%) | ||||
| 3 | 9½18(28.6%) | ||||
| ≥4 | 342/569(60.1%) | ||||
| Personal or family history of pancreatic cancer | |||||
| No | 495/1757(28.2%) | 3.05(1.97,4.74) | <0.0001 | ||
| Yes | 216/359(60.2%) | ||||
Odds ratios and 95% confidence intervals were estimated from a generalized estimating equation (GEE) model using a logit link function and with adjustment for familial clustering. For reference, age at first cutaneous melanoma diagnosis is modeled as ln(age at first diagnosis) with range 2.30 (10 years old) to 4.55 (95 years old). A ln(age) of 3.0 corresponds to a 20 year old, a ln(age) of 3.5 to a 33 year old, and a ln(age) of 4.0 to a 55 year old.
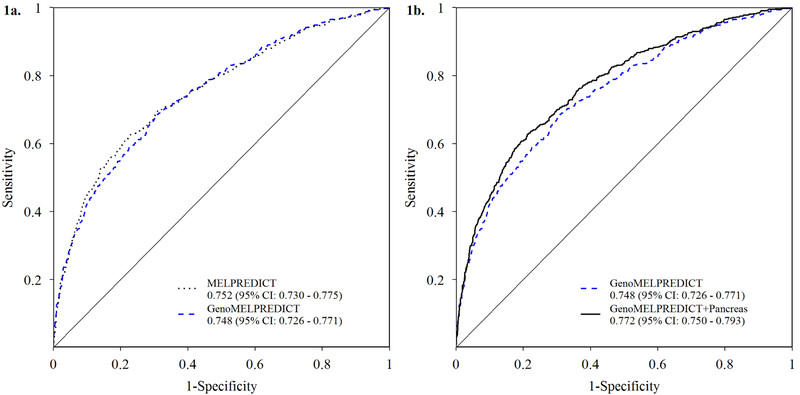
Using the published MELPREDICT model parameter coefficients to predict CDKN2A mutation carriage in the GenoMEL sample set resulted in an AUC = 0.752 (95% CI: 0.730, 0.775); the mean estimated probability of CDKN2A mutation carriage was 42.7% for mutation carriers, and 13.0% for non- carriers. De novo modeling, i.e. GenoMELPREDICT, of age at first melanoma diagnosis, number of primary melanoma diagnoses, and number of additional family members diagnosed with melanoma resulted in an AUC = 0.748 (95% CI: 0.726, 0.771). For this model, the mean estimated probability of CDKN2A mutation carriage was 46.4% for mutation carriers, and 27.2% for non-carriers. The difference in AUC values between models was not statistically significant (p = 0.21) (Figure 1a).
Figure 1. Receiver operator characteristic (ROC) curves for GenoMELPREDICT models.
Comparison of the ROC curves derived from the (Figure 1a) 3-variable baseline GenoMELPREDICT model and MELPREDICT as reported by Niendorf et al., 2006; and (Figure 1b) 3-variable baseline GenoMELPREDICT model and the 4-variable GenoMELPREDICT model including any reported personal or family history of pancreatic cancer. Legend results are cross-validated areas under the curve (AUC) and 95% confidence intervals (CI) for GenoMELPREDICT models and AUC and 95% CI for MELPREDICT.
Adding phenotypic risk factors did not result in performance improvements of the 3-variable baseline GenoMELPREDICT model (data not tabulated and available upon request). However, including personal or family history of pancreatic cancer to the 3-variable baseline model significantly (p < 0.0001) augmented its discriminatory performance, yielding an AUC=0.772 (95%CI: 0.750, 0.793) (Figure 1b). The mean estimated probability of CDKN2A mutation carriage was 48.4% for mutation carriers and 26.2% for non-carriers. The NRI was 0.404, with noted improvement (79.6%) for reclassification of non-carriers, but at the expense of reclassification of carriers (−39.2%). Adding phenotypic variables to the 4-predictor model that included personal or family history of pancreatic cancer did not result in further model improvement (data not tabulated and available upon request). Selecting a predicted probability cutoff of 35% for this four variable model, which was similar to the theoretical best cutoff based on Youden’s index (34.4%), resulted in a sensitivity of 61%, specificity of 79%, positive predictive value of 60%, and a negative predictive value of 80%. A range of model metrics for the baseline and 4-predictor GenoMELPREDICT models is available upon request. Consistent with results using observed phenotypic data, adding imputed phenotypic variables did not result in performance improvement of either the baseline or 4-predictor GenoMELPREDICT models (data not tabulated and available upon request).
In subgroup analyses, the AUCs for the 3- and 4-predictor GenoMELPREDICT models were somewhat higher among Australian participants [0.809 (0.773, 0.844) for both], and similar or slightly higher among participants living in Northern European countries [0.760 (0.718, 0.803) and 0.775 (0.734, 0.816), respectively]. Model performance was lower among participants from Southern European and South American countries [0.625 (0.535, 0.714) and 0.635 (0.548, 0.722), respectively].
Models that excluded families with individuals who carried a mutation in other known melanoma high penetrance genes, or excluded families without a verified report of personal or family history of pancreatic cancer were consistent with our main results. In models excluding melanoma-prone families from Sydney, which comprised one-third of all data used in our analysis, AUCs for the baseline (0.772; 95% CI: 0.747, 0.797) and 4-variable (0.784; 95% CI: 0.760, 0.808) GenoMELPREDICT models were slightly higher compared to models using all available GenoMEL data. After excluding participants from the Bethesda and Queensland centers, both of which contributed higher numbers of affected members with CDKN2A genotype data per family (4.3 and 4.6 respectively), model AUCs were slightly lower than those calculated from all available GenoMEL data (0.708; 95% CI: 0.681, 0.734 for baseline; and 0.740; 95% CI: 0.714, 0.765 for the 4-variable model).
Discussion
We show that the published MELPREDICT model used to predict CDKN2A mutational status is generalizable to the global community of melanoma-prone families represented in GenoMEL. We also provide evidence that adding personal and family history of pancreatic cancer to the model, a variable that can be collected with very little additional associated cost, leads to some improvement in the ability to predict CDKN2A mutational status, and we call this augmented model GenoMELPREDICT. Predictive performance of GenoMELPREDICT is comparable to other clinical tools used to predict BRCA1 and BRCA2 mutational status among breast cancer patients.[18–20]
The diverse global sample of familial melanoma cases recruited by GenoMEL allowed us to detect a broader spectrum of CDKN2A mutations compared to the limited number (18 variants) reported by the original MELPREDICT developers.[9] A total of 85 unique, putatively pathogenic mutations were identified among GenoMEL cases, allowing for a more representative appraisal of GenoMELPREDICT’s performance.
MelaPRO[21] and CM-Score[22] are two other published algorithms for CDKN2A mutation prediction among melanoma prone families. MelaPRO incorporates melanoma risk among unaffected family members, uses a Bayesian approach to predict carrier status, and incorporated penetrance estimates for areas of high and low baseline incidence, and one derived from the population-based Genes, Environment, and Melanoma Study.[23] MelaPRO was tested on a patient sample drawn from the same ascertainment center used by Niendorf et al. to test the MELPREDICT algorithm, and it outperformed (n=195; AUC=0.86) MELPREDICT on prediction of carrier status among the same homogeneous familial cohort. The CM-Score algorithm is a multivariate logistic regression model developed among a training cohort of 1,227 Dutch melanoma-prone families and incorporates five clinical features (number of family members with melanoma and with multiple primary melanomas, median age at diagnosis, and presence of pancreatic cancer or upper airway cancer in a family member) to predict germline CDKN2A mutational status. CM-Score was validated in a combined Swedish and Dutch cohort of 421 melanoma-prone families. CM-Score demonstrated excellent performance characteristics among a homogeneous group of Northern Europeans (AUC=0.94; 95%CI: 0.90, 0.98), possibly due to the high incidence of specific founder mutations in this population.[22]
We opted to assess MELPREDICT rather than MelaPRO or CM-Score. CM-Score was developed among a cohort of Swedish and Dutch melanoma-prone families with a high incidence of specific founder mutations, reducing generalizability. Due to the increased incidence of upper airway cancers observed among carriers of these Swedish and Dutch founder mutations, the CM-Score algorithm incorporates any history of such cancers and may be inappropriate for a heterogeneous population of familial melanoma kindreds.[22] In our dataset, there were 295 reports of a personal or family history of laryngeal, pharyngeal, and oral cavity cancers within 97 families; pathologic verification was available for 30 (10%) of these reports. MelaPRO requires users to specify CDKN2A penetrance associated with the population under study, which involves more complex assessments of the source populations from which individual cases arise; this aspect may potentially limit MelaPRO’s utility in clinical practice. Because the GenoMEL consortium includes melanoma-prone families from around the world and simultaneous modeling of multiple CDKN2A penetrances was not feasible, our preference was to evaluate generalizability and enhancement of MELPREDICT.
The 3- and 4-predictor GenoMELPREDICT models perform best among participants living in Australia. This likely reflects the large influence of these individuals, who comprise nearly 40% of our analytic sample, on overall model estimates. Conversely, 3- and 4-predictor GenoMELPREDICT models perform poorest among participants living in Southern European and South American sites. This likely reflects our working definition of a “melanoma-prone family,” which minimally is two verified melanoma cases in a first-degree blood relation. This definition may be too strict for populations that experience lower incidence of melanoma for which a definition of two or more verified melanoma cases among blood relatives may be better suited. Of the 900 families who had at least one member who contributed to GenoMELPREDICT modeling, the Southern European and South American sites had, as expected, a lower mean number of affected members per family (2.1) compared to that for the Northern European (3.3) or Australian (3.6) sites.
We have reported on limitations of the GenoMEL study that include differences in amount of data collected across centers, possible misclassification of CDKN2A mutations, lack of centralized pathology review for reported cases of melanoma, and non-population-based ascertainment and sampling of families at some centers based on known mutation status or number of familial melanoma cases.[10, 17] Although pathological verification of reported personal or familial cases of pancreatic cancer was low (22%) in GenoMEL, the positive predictive value and sensitivity of self-report of family history for this cancer are both reported to surpass 70%.[24]
GenoMELPREDICT is an effective predictor of CDKN2A mutational status, and statistical performance improvement was made by adding any reported personal or family history of pancreatic cancer. However only 5% to 10% of melanomas can be attributed to high penetrance germline genetics, and thus only a small proportion of patients diagnosed with melanoma will benefit from genetic testing for CDKN2A.[25] Despite controversy regarding the genetic testing of individuals in melanoma-prone families,[26] there is burgeoning commercial availability of such tests. We have previously published in this journal the challenges in developing a single encompassing worldwide recommendation to best guide health professionals with respect to which patients should be considered for CDKN2A genetic testing.[8] In Table 3, we republish our candidacy criteria for consideration of genetic testing.[8] Complementing these criteria, GenoMELPREDICT may serve as a quick and robust tool, applicable worldwide, for directing patients away from unnecessary genetic testing, especially in the event of a low carrier probability estimate. Moreover, guidance considering the management of patients belonging to melanoma-prone families in the context of genetic testing is available in a Continuing Medical Education article published in this journal.[26] A user-friendly web-based interface to calculate the probability of carriage of a CDKN2A mutation is available at www.genomel.org.
Table 3.
Candidacy for consideration of genetic testing
| Low melanoma incidence area/population | Moderate to high melanoma incidence area/population |
|---|---|
|
|
|
|
This table refers to pathologically confirmed invasive melanoma. Table reprinted from Leachman et al., J Am Acad Dermatol 2009.
Acknowledgments
We acknowledge the contributions of the participants, their families and the many clinicians, geneticists, genetic counsellors and allied health professionals involved in their management. This work was performed in participation with members of the following study centers:
Leeds: Linda Whitaker, Paul Affleck, Jennifer H. Barrett, Jane Harrison, Mark M. Iles, Juliette Randerson-Moor, John C. Taylor, Kairen Kukalizch, Susan Leake, Birute Karpavicius, Sue Haynes, Tricia Mack, May Chan, and Yvonne Taylor.
Barcelona: Paula Aguilera, Llúcia Alós, Celia Badenas, Alicia Barreiro, Neus Calbet, Cristina Carrera, Carlos Conill, Mireia Domínguez, Daniel Gabriel, Pablo Iglesias, Josep Malvehy, M. Eugenia Moliner, Javiera Pérez, Ramon Pigem, Miriam Potrony, Joan Anton Puig Butille, Ramon Rull, Marcelo Sánchez, Gemma Tell-Martí, Sergi Vidal-Sicart, and Oriol Yelamos.
Valencia: Zaida García-Casado, Celia Requena, José Bañuls, Virtudes Soriano, José Antonio López-Guerrero, Manuel Moragón, Vicente Oliver. Samples for CDKN2A analysis were obtained from the Biobank of the Instituto Valenciano de Oncología.
NCI at Cesena, Italy: Paola Minghetti, Laura Fontaine, Katie Beebe, and Giorgio Landi
Genoa: Giovanna Bianchi-Scarrà, Lorenza Pastorino, Virginia Andreotti, Claudia Martinuzzi, Bruna Dalmasso, Giulia Ciccarese, Francesco Spagnolo, and Paola Queirolo.
Latvia: Kristine Azarjana, Simona Donina, Olita Heisele, Baiba Štreinerte, Aija Ozola and Ludmila Engele.
Sydney: Caroline Watts, Gayathri St. George, Robyn Dalziell and Kate McBride who assisted with recruitment of study participants; Leo Raudonikis who assisted with data management; and Chantelle Agha-Hamilton and Svetlana Pianova who assisted with biospecimen management.
Uruguay: Virginia Barquet, Javiera Pérez, Miguel Martínez, Jimena Núñez, and Malena Scarone.
São Paulo: Dirce Maria Carraro, Alexandre Leon Ribeiro de Ávila, Luciana Facure Moredo, Bianca Costa Soares de Sá, Maria Isabel Waddington Achatz, and João Duprat.
Porto Alegre: Renan Rangel Bonamigo and Maria Carolina Widholzer Rey.
Leiden: Coby Out-Luiting, Clasine van der Drift, Leny van Mourik, Wilma Bergman, Femke de Snoo, Jeanet ter Huurne, and Frans van Nieuwpoort.
Paris: wishes to thank The French familial melanoma study group: P. Andry-Benzaquen, B. Bachollet, F. Bérard, P. Berthet, F. Boitier, V. Bonadona, JL. Bonafé, JM. Bonnetblanc, F. Cambazard, O. Caron, F. Caux, J. Chevrant-Breton, A. Chompret (deceased), S. Dalle, L. Demange (deceased), O. Dereure, MX. Doré, MS. Doutre, C. Dugast (deceased), E. Maubec, L. Faivre, F. Grange, Ph. Humbert, P. Joly, D. Kerob, B. Labeille, C. Lasset, MT. Leccia, G. Lenoir, D. Leroux, J. Levang, D. Lipsker, S. Mansard, L. Martin, T. Martin-Denavit, C. Mateus, JL. Michel, P. Morel, L. Olivier-Faivre, JL. Perrot, N. Poulalhon, C. Robert, S. Ronger-Savle, B. Sassolas, P. Souteyrand, D. Stoppa-Lyonnet, L. Thomas, P. Vabres, L. Vincent-Fetita, and E. Wierzbicka. We also thank Hamida Mohamdi for managing the French MELARISK database.
Queensland: Nicholas Martin, Grant Montgomery, David Whiteman, Stuart MacGregor, David Duffy and Michael Gattas, along with Judith Symmons and Harry Beeby who assisted with data management.
Stockholm: wishes to thank Diana Lindén, R.N. for excellent work collecting and entering data into the study data base and Rainer Tuominen for screening of CDKN2A.
Tel Aviv: wishes to acknowledge Yael Laitman.
Lund: wishes to thank Anita Zander, R.N. for invaluable help with the data from the Lund Melanoma Study Group and acknowledge Kari Nielsen, Anna Måsbäck, Katja Harbst, Goran Jonsson and Åke Borg.
The University of Utah (Salt Lake City): acknowledges the use of the Genetic Counseling Shared Resource supported by National Institutes of Health grant P30CA042014 awarded to the Hunstman Cancer Institute.
The University of Pennsylvania: acknowledges the contributions of Patricia Van Belle, Althea Ruffin, Jillian Knorr and Wenting Zhou.
Gustave Roussy: wishes to thank Christophe Blondel for technical assistance in CDKN2A genotyping and to acknowledge the work of Gustave Roussy Biobank (BB-0033–00074) in providing DNA resources.
Funding sources
This work was supported by: the European Commission under the 6th and 7th Framework Programme [LSH-CT-2006–018702]; Cancer Research UK Programme Awards (C588/A4994 and C588/ A10589); a Cancer Research UK Project Grant (C8216/A6129); the US National Institutes of Health [R01-CA83115, R01CA5558–01A2 (MTL), 5R25-CA147832–04 (NJT)]; the intramural Research Program of the NIH, National Cancer Institute (NCI), Division of Cancer Epidemiology and Genetics; the National Health and Medical Research Council of Australia [NHMRC 107359, 402761, 633004, 566946, 211172]; the Cancer Council New South Wales (project grant 77/00, 06/10); the Cancer Institute New South Wales [CINSW 05/TPG/1–01, 10/TPG/1–02]; the Cancer Council Victoria and the Cancer Council Queensland (project grant 371); CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior); FAPESP (Fundação para o Amparo da Pesquisa do Estado de São Paulo) – SP, Brazil # 2007/04313–2; the National Health and Medical Research Council of Australia and the NCI (CA88363); the Cancer Research Foundations of Radiumhemmet and the Swedish Cancer Society; the Paulsson Trust, Lund University; grant support from the Swedish Cancer Society and European Research Council Advanced Grant (ERC-2011–294576); the research at the Melanoma Unit in Barcelona is partially funded by Spanish Fondo de Investigaciones Sanitarias grants PI15/00716 and PI15/00956; CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain, co-financed by European Development Regional Fund “A way to achieve Europe” ERDF; AGAUR 2014_SGR_603 of the Catalan Government, Spain; European Commission under the 6th Framework Programme, Contract No. LSHC-CT-2006–018702 (GenoMEL) and by the European Commission under the 7th Framework Programme, Diagnoptics; The National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115); a grant from “Fundació La Marató de TV3” 201331–30, Catalonia, Spain; a grant from “Fundación Científica de la Asociación Española Contra el Cáncer” GCB15152978SOEN, Spain, and CERCA Programme / Generalitat de Catalunya. Part of the work was carried out at the Esther Koplowitz Center, Barcelona; the Italian Association for Cancer research (AIRC) n. 15460 and 5 × 1000 to PG, Italian Ministry of Health-Ricerca Finalizzata 2016 (5 × 1000 funds to IRCCS San Martino-IST, Genoa); the Programme Hospitalier de Recherche Clinique (PHRC-AOM-07–195) awarded to M-FA and FD; grant support from Institut National du Cancer (INCA) was attributed to B B-deP. for coordination of Melanoma Oncogenetics in France; the Comisión Honoraria de Lucha Contra el Cáncer, CSIC, Fundación Manuel Pérez, Montevideo, Uruguay; the work of Nelleke A. Gruis was supported in part by the Dutch Cancer Society (UL 2012–5489); Francisco Cuellar is supported by a scholarship awarded by CONACYT, Mexico (152256/158706); Anne Cust is the recipient of Career Development Fellowships from the NHMRC (1147843) and Cancer Institute NSW (15/CDF/1–14).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors state no conflicts of interest.
Statement of IRB
Individual GenoMEL centers received study approval by their respective institutional review boards.
References
- 1.Kefford RF, Newton Bishop JA, Bergman W, et al. Counseling and DNA testing for individuals perceived to be genetically predisposed to melanoma: A consensus statement of the Melanoma Genetics Consortium. J Clin Oncol 1999;17(10):3245–51. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AM, Tucker MA. Genetic epidemiology of cutaneous melanoma: a global perspective. Arch Dermatol 2001;137(11):1493–6. [DOI] [PubMed] [Google Scholar]
- 3.Bishop DT, Demenais F, Goldstein AM, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 2002;94(12):894–903. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein AM, Tucker MA. Screening for CDKN2A mutations in hereditary melanoma. J Natl Cancer Inst 1997;89(10):676–8. [DOI] [PubMed] [Google Scholar]
- 5.Monzon J, Liu L, Brill H, et al. CDKN2A mutations in multiple primary melanomas. N Engl J Med 1998;338(13):879–87. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007;44(2):99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demenais F, Mohamdi H, Chaudru V, et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J Natl Cancer Inst 2010;102(20):1568–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leachman SA, Carucci J, Kohlmann W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009;61(4):677 e1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niendorf KB, Goggins W, Yang G, et al. MELPREDICT: a logistic regression model to estimate CDKN2A carrier probability. J Med Genet 2006;43(6):501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor NJ, Handorf EA, Mitra N, et al. Phenotypic and Histopathological Tumor Characteristics According to CDKN2A Mutation Status among Affected Members of Melanoma Families. J Invest Dermatol 2016;136(5):1066–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harland M, Goldstein AM, Kukalizch K, et al. A comparison of CDKN2A mutation detection within the Melanoma Genetics Consortium (GenoMEL). Eur J Cancer 2008;44(9):1269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143(1):29–36. [DOI] [PubMed] [Google Scholar]
- 13.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–45. [PubMed] [Google Scholar]
- 14.Pencina MJ, D’Agostino RB, Massaro JM. Understanding increments in model performance metrics. Lifetime Data Anal 2013;19(2):202–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leening MJ, Steyerberg EW, Van Calster B, et al. Net reclassification improvement and integrated discrimination improvement require calibrated models: relevance from a marker and model perspective. Stat Med 2014;33(19):3415–8. [DOI] [PubMed] [Google Scholar]
- 16.Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 17.Taylor NJ, Mitra N, Goldstein AM, et al. Germline Variation at CDKN2A and Associations with Nevus Phenotypes among Members of Melanoma Families. J Invest Dermatol 2017;137(12):2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindor NM, Lindor RA, Apicella C, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of LAMBDA, BRCAPRO, Myriad II, and modified Couch models. Fam Cancer 2007;6(4):473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindor NM, Johnson KJ, Harvey H, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Fam Cancer 2010;9(4):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer C, Kuchenbacker K, Engel C, et al. Evaluating the performance of the breast cancer genetic risk models BOADICEA, IBIS, BRCAPRO and Claus for predicting BRCA1/2 mutation carrier probabilities: a study based on 7352 families from the German Hereditary Breast and Ovarian Cancer Consortium. J Med Genet 2013;50(6):360–7. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Niendorf KB, Patel D, et al. Estimating CDKN2A carrier probability and personalizing cancer risk assessments in hereditary melanoma using MelaPRO. Cancer Res 2010;70(2):552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potjer TP, Helgadottir H, Leenheer M, et al. CM-Score: a validated scoring system to predict CDKN2A germline mutations in melanoma families from Northern Europe. J Med Genet 2018; 101136/jmedgenet-2017-105205. [DOI] [PubMed] [Google Scholar]
- 23.Begg CB, Orlow I, Hummer AJ, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 2005;97(20):1507–15. [DOI] [PubMed] [Google Scholar]
- 24.Fiederling J, Shams AZ, Haug U. Validity of self-reported family history of cancer: A systematic literature review on selected cancers. Int J Cancer 2016;139(7):1449–60. [DOI] [PubMed] [Google Scholar]
- 25.Florell SR, Boucher KM, Garibotti G, et al. Population-based analysis of prognostic factors and survival in familial melanoma. J Clin Oncol 2005;23(28):7168–77. [DOI] [PubMed] [Google Scholar]
- 26.Soura E, Eliades PJ, Shannon K, et al. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J Am Acad Dermatol 2016;74(3):395–407; quiz 408–10. [DOI] [PMC free article] [PubMed] [Google Scholar]