Abstract
Platinum-based anticancer drugs are widely used as a front line drug for cancers, such as non-small-cell lung carcinoma (NSCLC) and bladder cancer. However, the efficacy is limited due to intrinsic or acquired resistance to these drugs. DNA polymerase eta (PolH, Polηbelongs to the Y-family of DNA polymerases and mediates DNA translesion synthesis, a major mechanism for DNA damage tolerance. Here, we showed that a high level of PolH is associated with cisplatin resistance in lung and bladder cancer. Consistent with this, loss of PolH markedly attenuates cisplatin resistance in both cisplatin-sensitive and -resistant lung cancer cells. Interestingly, we found that due to the presence of multiple polyadenylation sites, alternative polyadenylation (APA) produces three major PolH transcripts with various lengths of 3’untranslated region (3’UTR) (427-/2516-/6245-nt). We showed that the short PolH transcript with 427-nt 3’UTR is responsible for high expression of PolH in various cisplatin-resistant lung and bladder cancer cell lines. Importantly, loss of the short PolH transcript significantly sensitizes cancer cells to cisplatin treatment. Moreover, we found that miR619 selectively inhibits the ability of the long PolH transcript with 6245-nt 3’UTR to produce PolH protein and subsequently, PolH-dependent cell growth. Together, our data suggest that PolH expression is controlled by APA and that the short PolH transcript produced by APA can escape miR619-mediated repression and subsequently, confers PolH-mediated cisplatin resistance.
Introduction
Platinum-based anticancer drugs, such as cisplatin, are designed to inhibit DNA replication by cross-linking DNA and have been effective as frontline drugs for many types of cancers, including lung and bladder cancer. In most cases, tumors are responding to the initial treatment, but soon develop chemo-resistance, leading to an aggressive form of the diseases. Much effort has been spent to understand the mechanism of cisplatin resistance in cancer cells. One proposed mechanism is that tumor cells acquire chemoresistance via translesion DNA synthesis (TLS) [1–4]. TLS represents a major mechanism for DNA damage tolerance, which is carried out by low-fidelity TLS DNA polymerases [5]. Unlike high-fidelity DNA polymerases, TLS DNA polymerases have a flexible active site that can accommodate large distortions of DNA structures and subsequently replicate across DNA lesions [6]. As a result, TLS DNA polymerases not only allow cells to avoid double strand breaks associated with replication fork stalling, but also lead to mutagenesis by introducing incorrect nucleotides. Indeed, TLS is found to be responsible for mutations in normal and tumor cells [7] as well as in relapsed malignancies [8].
Most TLS polymerases belong to the Y-family of DNA polymerases, including PolH, PolI, PolK, and Rev1 [9], along with the B family DNA polymerase PolZ (consisting of two subunits, Rev3 and Rev7) [10]. Among them, PolH is unique in its proficiency for error-free replication across UV induced cyclobutane pyrimidine dimers (CPDs) [11]. The biological significance of PolH is highlighted by the hereditary disease Xeroderma Pigmentosum Variant (XPV), which is caused by germline mutations that inactivate PolH [12, 13]. XPV patients are highly sensitive to UV irradiation and prone to skin cancer [14], due to error-prone bypass by another TLS polymerase, possibly PolI [15–17]. Consistent with this, mice deficient in PolH recapitulate the cancer susceptibility observed in XPV patients and rapidly develop UV-induced tumors [18]. However, as a TLS polymerase, PolH can bypass a broad range of DNA lesions in an error-prone manner and induce mutagenesis. Indeed, we showed that PolH is induced by DNA damage in a p53-dependent manner [19]. Moreover, PolH was found to be highly expressed in tumors after platinum-based chemotherapy and associated with poor survival of cancer patients, including non-small-cell lung cancer (NSCLC) [3, 20, 21]. Furthermore, a recent study found that enhanced expression of PolH is associated with cisplatin resistance by ovarian cancer stem cells [1]. However, the mechanism by which PolH confers cisplatin-resistance remains to be elucidated.
Alternative polyadenylation (APA) is recognized as an important mechanism for gene regulation as more than half of human genes contain multiple polyadenylation sites (PASs) [22, 23]. APA is governed by the core cleavage and polyadenylation (C/P) machinery [24, 25], which is composed of: cleavage and polyadenylation stimulatory factor (CPSF); cleavage stimulatory factor (CSTF); cleavage factors Im and IIm (CFIm, CFIIm); and multiple RNA-binding proteins (RBPs), including nuclear poly(A) binding protein 1 (PABPN1), Symplekin, and Poly(A) polymerase (PAP). Depending on the locations of the PAS sites, the isoforms of an mRNA produced through APA can differ in their coding sequences and/or the lengths of their 3’UTRs. Recent studies have shown that APA-mediated global 3’UTR shortening plays a crucial role in cell proliferation and transformation [26–30]. For example, a truncated form of cyclin D1 transcript generated by APA was found to be highly expressed in a subset of mantle cell lymphomas and associated with poor survival [31, 32]. Additionally, distinct APA patterns have been found in cisplatin-sensitive and -resistant ovarian cancer cells [33, 34]. Nevertheless, very little is known about the role of APA in tumor development, especially the genes regulated by APA.
In the current study, we showed that high expression of PolH is associated with cisplatin resistance in lung and bladder cancer cells. We also found that loss of PolH attenuates cisplatin resistance in cancer cells via enhanced DNA damage response. Moreover, we showed that PolH transcripts with a short 3’UTR escape miR619-mediated repression and are responsible for high expression of PolH in cisplatin-resistant cells. Importantly, knockout of the short PolH transcript abrogate PolH expression and subsequently, sensitizes cancer cells to cisplatin.
Materials and Methods
Cell culture and cell line generation
WI-38, A549, H460, H1299, H358, H1975, and CL1 were cultured in RPMI-1640 medium supplement with 10% fetal bovine serum (Hyclone, Logan, UT, USA). MCF7 and RKO cells were cultured in DMEM medium supplement with 10% fetal bovine serum (Hyclone, Logan, UT, USA). CL1 cell line was generated by Dr. Wu [35]. All other cell lines were obtained from ATCC. Cells were tested negative for mycoplasma and used at below passage 20. To generate PolH-KO cell line, pSpCas9(BB)-2A-Puro vector expressing PolH guide RNA#1 (5’ GCA CAA GTT CGT GAG TCC CG 3’) and guide RNA#2 (5’ TGG AGT CAC TAG AAG TAT GT 3’)was transfected into A549, H1299, and H1975 cells. The cells were selected with puromycin and each individual clone was confirmed by sequencing for the deletion in PolH gene. To generate PolH-S-KO cell lines that contain deletion of PAS-1, two guide RNAs were used, guide RNA#1 (5’ TTA CAG ATT TCC CTG AGA AA 3’) and guide RNA #2 (5’TTT TTA ATC TTT AGC ATT TA 3’). To generate PolH-L-KO cell lines that contain deletion of PAS-3, two guide RNAs, guide RNA#3 (5’ TGT GCT TAT GCG TTA GCT AC 3’) and guide RNA #4 (5’ ACT CAG GGA TTT GTT GGC TA 3’), were used. The genotyping primers for PolH-S-KO cell lines were forward primer, 5’-GTT TCT GCC GTA TCT CAT CAA GG-3’, and reverse primer, 5’ CAT TGC AAT AAC CAG AGC 3’. The genotyping primers for PolH-L-KO cell lines were forward primer, 5’-CTG CTC ACT TGA ACT ACG GA-3’, and reverse primer, 5’-CAC TCT CAC CCC ACT TTC C-3’.
Western blot analysis
Cell lysates were prepared with 2x SDS sample buffer and boiled for 5 min. Proteins were resolved in an SDS-PAGE gel (8–12%) and then transferred to nitrocellulose membranes. Membranes were subjected to blocking, washing, antibody incubation, and detection by enhanced chemiluminescence. The antibodies used were anti-PolH (Santa Cruz Biotechnology), γ-H2AX (Cell signaling), and actin (Sigma).
Northern blot analysis
Northern blot was performed as previously describe [19]. Northern blots were prepared using total RNAs isolated from various cells. To prepare PolH and GAPDH probes, cDNA fragments were amplified and then labeled with [32P]-α-dCTP using Random Primer DNA Labeling Kit (Takara). The primers to amplify the GAPDH cDNA were forward primer, 5’-TGA AGG TCG GAG TCA ACG GAT TTG GT-3’ and reverse primer, 5’-CAT GTG GGC CAT GAG GTC CAC CAC-3’. The primers to amplify to PolH cDNA were forward primer, 5’-CTT ACA TTG AAG GGT TGC CC-3’, and reverse primer, 5’-GTT GCC TGG GTT TAA CTG GA-3’.
MicoRNA transfection
miRNA control, miR619, anti-miR control, and anti-miR619 were purchased from Life Science Technology. For transfection, miRNA mimics or inhibitors were transfected at 50 nM using RNAiMax (Life Technology) according to the user’s manual.
Xenograft assay and Histological Analysis
5 × 106 cells, suspended in matrigel (1:1 ratio), were injected subcutaneously (s.c.) into 8-week-old BALB/c athymic nude mice (Charles River). Tumor growth were monitored for every 2 days for a period of up to 6 weeks. Tumor volume were calculated according to the standard formula: V = length × width × depth × 0.5236 [36]. At the endpoint, all animals were sacrificed and the tumors weighed. One half of the tumor were stored at −80°C and the other half will be fixed in formalin and embedded with paraffin. Tumors were be sectioned and H.E. stained for histopathology examination as previously described [37]. All animals and use protocols were approved by the University of California at Davis Institutional Animal Care and Use Committee.
Colony formation assay
Cells were seeded in triplicates in a six-well plate overnight, followed by treatment with or without cisplatin (2.5uM) for 18 h. After treatment, cells were washed one time with RPMI 1640 medium to remove cisplatin and then maintained in fresh medium for 2 weeks. Cells were then fixed with methanol/glacial acetic acid (7:1) and stained with 0.1% of crystal violet. To quantify the colony results, Image J software was used with the ColonyArea plugin installed as previously described [38].
Immunofluorescence Assay
The assay was performed as previously described [39]. Briefly, cells were grown on chamber slides and treated with or without cisplatin for 18h. Cells were then fixed and incubated with primary antibody overnight, followed by incubation with fluorescein isothiocyanate (FITC)- or Texas red-conjugated secondary antibodies (Jackson ImmunoResearch and Molecular Probes) for 2 h. Images of stained cells were captured using a Leica TCS-SP8 confocal laser scanning microscope (Leica, Mannheim, Germany).
RNA Isolation, RT-PCR, and Quantitative PCR (qPCR)
Total RNA was isolated with Trizol reagent as described according to user’s manual. cDNA was synthesized with Reverse Transcriptase (Promega) according to user’s manual. The PCR program used for amplification was (i) 94 °C for 5 min, (ii) 94 °C for 45 s, (iii) 60 °C for 45 s, (iv) 72 °C for 45 s and (v) 72 °C for 10 min. From steps 2–4, the cycle was repeated 22 times for actin or 30 times for PolH. The primers for actin were forward primer, 5’-CTG AAG TAC CCC ATC GAG CAC GGC A-3’, and reverse primer, 5’-GGA TAG CAC AGC CTG GAT AGC AAC G-3’. The primers for PolH were forward primer, 5’-CCA TTC GCA AAA TCC GTA GT-3’, and reverse primer, 5’-GTT GCC TGG GTT TAA CTG GA-3’. The qPCR analysis was performed in 20-μl reactions using 2X qPCR SYBR Green Mix (ABgene, Epsom, UK) along with 5 μM primers. The reactions were run on a StepOne plus (Invitrogen) using a two-step cycling program: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s. A melt curve (57–95 °C) was generated at the end of each run to verify the specificity. The common primers for all PolH transcripts were forward primer, 5’- CCA GAG TCA TTT TGG GGA GA −3’, and reverse primer, 5’-GTT GCC TGG GTT TAA CTG GA-3’. The distal primers for long PolH transcript were forward primer, 5’-CAG CCT GAG TGG TAG GGA AG-3’, and reverse primer, 5’- AAC AAT GAG GGC CAC TTG AC-3’. The short PolH transcript was calculated based on the difference between the level of PCR amplicons using common primers and the ones using distal primers. To determine the level of miR619 transcript, both miR619- and U6 spliceosomal nuclear RNA- specific cDNA was synthesized using their specific primers. The primer for miR619 was 5’-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG GGC TCA TG-3’ and the primer for U6 spliceosomal nuclear RNA was 5’-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCAG TTG AGT ATG GAA C-3’. Next, these cDNAs were used for q-PCR to examine the level of miR619 and U6 spliceosomal nuclear RNA using a specific forward primer and a common reverse primer. The specific forward primer for miR619 was 5’-ACA CTC CAG CTG GGG CTG GGA TTA CAG GCA TG-3’. The specific forward primer for U6 spliceosomal nuclear RNA was 5’- CTG CGC AAG GAT GAC ACG CA-3’. The reverse primer for both miR619 and U6 spliceosomal nuclear RNA was 5’-CTC AAC TGG TGT CGT GGA GTC GG-3’. The relative level of miR619 was calculated using the delta-delta Ct method (ddCt) by normalizing to the U6 spliceosomal nuclear RNA.
Results
Loss of PolH sensitizes cancer cells to cisplatin treatment
To determine whether PolH plays a role in cisplatin resistance, PolH expression was examined in immortalized lung fibroblasts WI-38 and six lung cancer cell lines: A549, H460, H1299, H358, H1975, and CL1. We found that the levels of PolH protein and transcript were much higher in H1299, H358, H1975, and CL1 cells than that in WI38, A549, and H460 cells (Fig. 1A-B). Interestingly, cells with higher PolH expression (H1299, H1975, CL1) were more resistant to cisplatin than the ones with lower PolH expression (A549, H460) (Fig. 1C-D). In addition, PolH expression was examined in a panel of bladder cancer cell lines (5637, TCCSUP, T24, J82, RT4, and HT1197) that exhibit various levels of resistance to cisplatin. Among them, 5637 cells are most sensitive, whereas HT1197 cells are most resistant, to cisplatin (http://www.cancerrxgene.org/). Interestingly, we found that HT1197 cells expressed the highest, whereas 5637 cells expressed the lowest, levels of PolH transcripts (Supplementary Fig. 1).
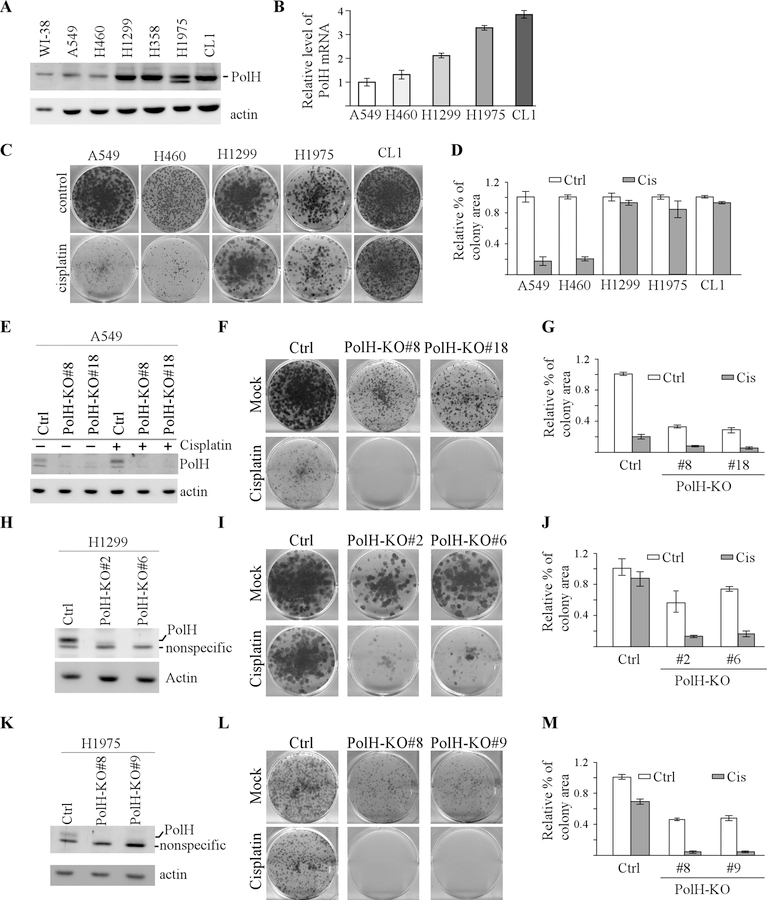
Figure 1. Loss of PolH sensitizes cancer cells to cisplatin treatment.
(A) The level of PolH and actin were examined by western blot analyzing using cell lysates from WI-38, A549, H460, H1299, H358, H1975, and CL1. (B) The level of PolH transcript was examined in A549, H460, H1299, H1975, and CL1 by performing qRT-PCR analysis. (C) Colony formation assay was performed with A549, H460, H1299, H1975, and CL1 cells treated with or without cisplatin (2.5 μM, 18h). (D) The colony formation assay in (C) was quantified using ColonyArea plugin. (E) Isogenic control and PolH-KO (clone #8 and #18) cells were mocked treated or treated with cisplatin (2.5 μM, 18h), followed by western blot analysis to examine the level of PolH and actin. (F) Colony formation assay was performed with isogenic control and PolH-KO A549 cells treated with or without cisplatin (2.5 μM, 18h). (G) The colony formation assay in (F) was quantified using ColonyArea plugin. (H and K) The level of PolH and actin was examined by western blot analysis in isogenic control and PolH-KO H1299 (F) and H1975 (H) cells. (I and L) Colony formation assay was performed with isogenic control and PolH-KO H1299 (G) and H1975 (I) cells mock-treated or treated with cisplatin (2.5 μM, 18h). (J and M) Quantification of colony formation assays in I (J) and L (M).
The above observation let us speculate that high expression of PolH confers cisplatin resistance in cancer cells. To test this, A549 cells were used to generate stable cell lines in that the PolH gene is knocked out by CRISPR-Cas9. Two representative PolH-KO clones (#8 and #18) and one isogenic control clone were chosen for further studies. Sequence analysis indicated that both PolH-KO clones had a 61-nt deletion in exon 3 of the PolH gene. We found that PolΗ was expressed and increased by cisplatin in isogenic control cells (Fig. 1E), consistent with previous report [19]. In contrast, PolH protein was undetectable in PolH-KO cells possibly due to nonsense-mediated mRNA decay (Fig. 1E). Next, colony formation assay was performed and showed that loss of PolH suppressed colony formation in A549 cells, which was further decreased by cisplatin (Fig. 1F-G). To determine whether loss of PolH enhances chemosensitivity of cisplatin-resistant lung cancer cells, H1299 and H1795 cells were used to generate PolH-KO cell lines by CRISPR-cas9, which were then confirmed by DNA sequencing. As expected, PolH protein was undetectable in PolH-KO H1299 (Fig. 1H). Importantly, we found that loss of PolH reduced number of colonies in both H1299 cell and a further reduction was observed upon cisplatin treatment (Fig. 1I-J). Consistent with this, we found that loss of PolH enhanced the sensitivity of H1975 cells to cisplatin (Fig. 1K-M). Together, these data suggest that loss of PolH markedly enhances cisplatin sensitivity of lung cancer cells and that high expression of PolH confers cisplatin resistance in lung cancer.
Loss of PolH inhibits tumor growth in vivo
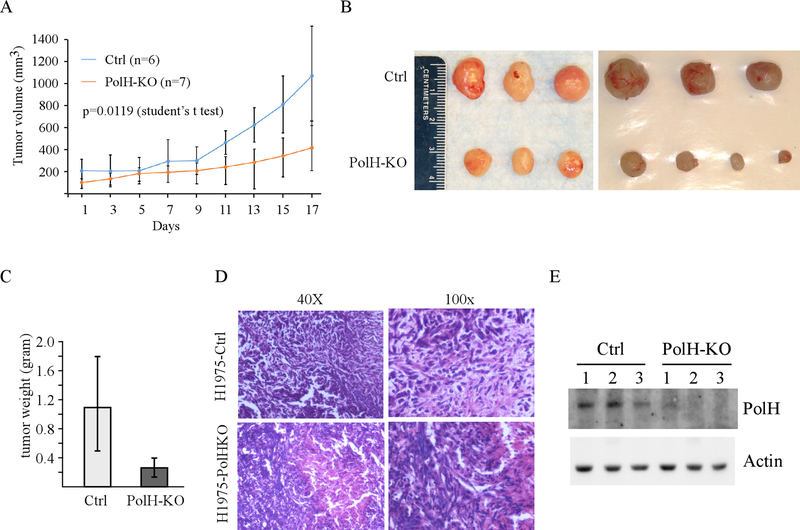
To determine whether loss of PolH inhibits tumor growth in vivo, cisplatin-resistant H1975 xenograft models were established. Briefly, both isogenic control and PolH-KO H1975 cells were inoculated subcutaneously to athymic mice and tumor growth was monitored every other day for 3 weeks. We found that PolH-KO tumors grew much slower than control tumors (Fig. 2A) (p=0.0119 by student’s t-test). Moreover, the tumor size from PolH-KO group was significantly smaller than the one from control group (Fig. 2B). Consistent with this, the average weight of PolH-KO tumors (0.29 ± 0.15 g) was significantly lower than that from control tumors (1.0 ± 0.7 g) (p<0.05 by student’s t-test) (Fig. 2C). Next, H.E. staining assay was performed and showed that control H1975 tumors and PolH-KO H1975 tumors were human origin and exhibited similar histology characteristics (Fig. 2D). Furthermore, we showed that PolH protein was detectable in the tumors from control group but remained undetectable in the tumors from PolH-KO group (Fig. 2E).
Figure 2. Loss of PolH inhibits tumor growth in vivo.
(A) Xenograft tumor growth in nude mice from isogenic control or PolH-KO H1975 cells (Error bars represent SEM, p=0.0119 by student t test). (B) Images of tumors excised from control and PolH-KO groups. (C) Tumor weigh distribution between isogenic control and PolH-KO groups upon termination of tumor growth experiments at day 17. (**, p<0.001, Student’s t-test). (D) Representative images of H.E-stained xenograft tumor sections from control and PolH-KO groups. (E) The level of PolH and actin were examined in isogenic control and PolH-KO tumors.
Loss of PolH leads to enhanced DNA damage response
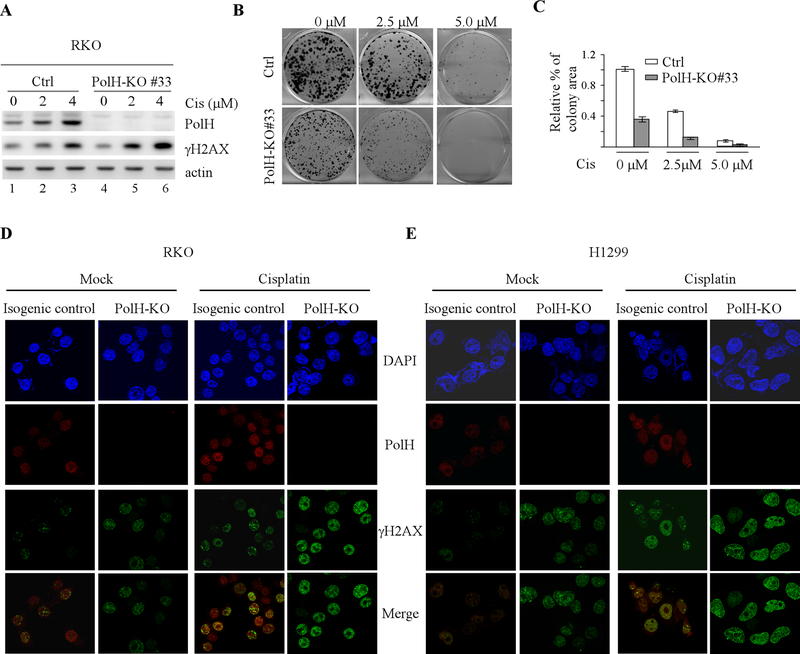
We showed previously that PolH is induced by DNA damage in a p53-dependent manner and subsequently, helps repair damaged DNA [19]. Thus, we postulate that loss of PolH enhances DNA damage response, leading to cell death. To test this, we generated RKO colon cancer cell line in that PolH was knocked out by CRISPR-Cas9. We showed that PolH was expressed in isogenic control cells but undetectable in PolH-KO cells (Fig. 3A). In addition, loss of PolH sensitized RKO cells to cisplatin treatment in a dose-dependent manner (Fig. 3B-C), consistent with the observation in lung cancer cells (Fig. 1). Moreover, we found that the level of γ-H2AX was highly increased by cisplatin in PolH-KO cells as compared to isogenic control cells (Fig. 3A, compare lanes 2–3 with 5–6, respectively). Since γ-H2AX is a marker for double strand DNA breaks [40], immunofluorescence assay was performed to measure the number of PolH and γ-H2AX foci in isogenic control and PolH-KO RKO cells. We found that PolH exhibited weak nuclear staining in mock-treated isogenic control cells and the staining intensity was markedly increased in response to cisplatin treatment (Fig. 3D and Supplemental Fig. 2A, PolH panels). By contrast, no PolH staining was observed in PolH-KO cells regardless of cisplatin treatment (Fig. 3D and Supplemental Fig. 2A, PolH panels). Interestingly, upon knockout of PolH, the number of γ-H2AX foci was increased (Fig. 3D and Supplemental Fig. 2A, mock columns, γ-H2AX panels), which was further increased by cisplatin treatment (Fig. 3D and Supplemental Fig. 2A, cisplatin columns, γ-H2AX panels). To confirm this, the effect of PolH-KO on the number of γ-H2AX foci was measured in H1299 lung cancer cells. We found that loss of PolH in H1299 lung cancer cells led to increased number of γ-H2AX foci, which was further increased by cisplatin treatment (Fig. 3E and Supplemental Fig. 2B, γ-H2AX panels). Together, these data suggest that loss of PolH leads to enhanced DNA damage response in cancer cells and subsequently, cell death.
Figure 3. Loss of PolH leads to enhanced DNA damage response.
(A) Isogenic control and PolH-KO RKO cells were treated with or without cisplatin (2.5 uM) for 18 h, followed by western blot analysis to examine the level of PolH, γ-H2AX, and actin. (B) Colony formation assay was performed using isogenic control and PolH-KO RKO cells treated with or without cisplatin for 18h. (C) Quantification of colony formation assays in B. (D-E) Immunofluorescence assay was performed with isogenic control and PolH-KO RKO (D) and H1299 (E) cells treated with or without cisplatin (2.5 uM, 8h). The PolH (red) image was obtained by anti-PolH and Texas red-conjugated secondary antibody. The γ-H2AX (green) image was obtained by anti-γ-H2AX and FITC-conjugated secondary antibody. DAPI was used for nuclear staining. The immunofluorescence images was captured using were obtained using a 63x oil objective of the Leica TCS SP8 confocal microscope.
PolH expression is regulated by alternative polyadenylation
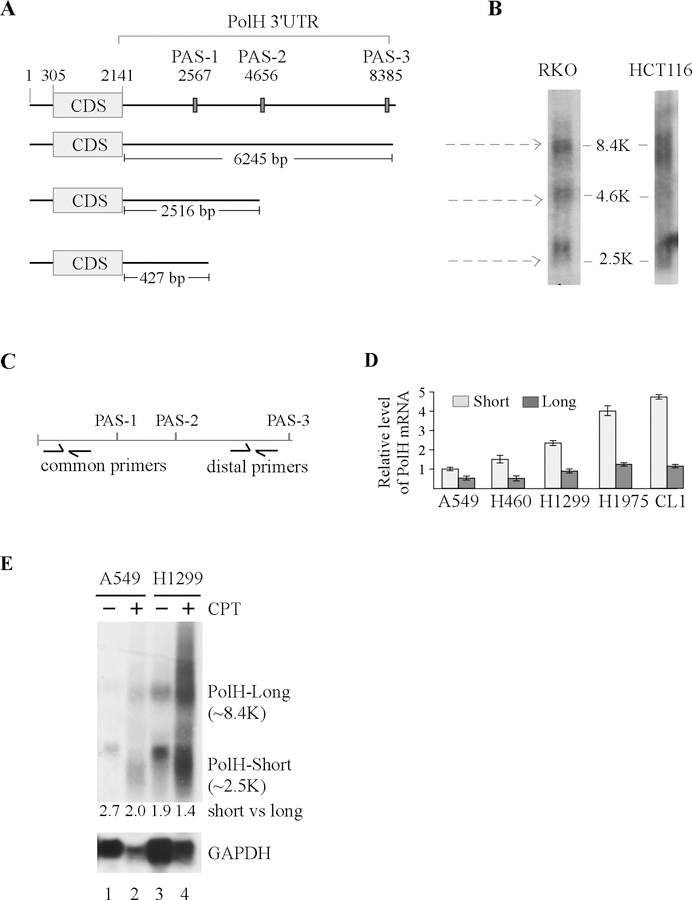
Since PolH transcript is highly expressed in cisplatin-resistant lung and bladder cancer cells (Fig. 1B and supplementary Fig. 1), it is necessary to determine the mechanism by which PolH expression is elevated in cisplatin-resistant cells, which may open a new revenue for developing novel therapeutic strategy to improve the efficacy of cisplatin. Recent studies showed that selective APA is found to regulate expression of oncogenes and tumor suppressor genes, such as IGF2BP1 and Pten [26, 41, 42]. To this end, we searched for polyadenylation sites (PAS) in PolH 3’UTR and found three PAS: PAS-1 at nt 2567; PAS-2 at nt 4656; and PAS-3 at nt 8385 (Fig. 4A). Consistent with this, a distinct group of PolH mRNA isoforms differentially expressed in cells and tissues has been annotated in the APA database that (http://mosas.sysu.edu.cn/utr). To verify this, Northern blot analysis was performed and showed that in RKO cells, three PolH transcripts were expressed from PAS-1/−2/−3 sites whereas in HCT116 cells, two transcripts were expressed from PAS-1/−3 sites (Fig. 4B). These data suggest the PAS-1 and PAS-3 are the major sites used in the cells. To determine whether APA contributes to increased expression of PolH in lung cancer cells, the relative abundance of short vs. long PolH transcripts was examined by qRT-PCR using the 2-ΔΔCt method [43]. The long PolH transcript was defined as the one cleaved at the PAS-3 site and calculated as the PCR amplicon using distal primers (Fig. 4C). The short PolH transcripts were defined as the ones cleaved at the PAS-1/−2 sites and calculated based on the difference between the level of PCR amplicons using common primers and the ones using distal primers (Fig. 4C). We found that the level of short PolH transcripts was much higher than that of long PolH transcript in all five lung cell lines (Fig. 4D). We also found that the level of short PolH transcripts was much higher in cisplatin-resistant cells (H1299, H1975, CL1) than that in cisplatin-sensitive cells (A549, H460) (Fig. 4D). Similarly, the level of short PolH transcripts was much higher in cisplatin-resistant HT1197 bladder cancer cells than that in cisplatin-sensitive 5637 cells (Supplementary Fig. 3). To further confirm that PolH transcript is subjected to APA regulation in lung cancer cells, Norther blot analysis was performed using total RNAs from A549 and H1299 cells treated with or without camptothecin. We found that in the absence of camptothecin, PolH was expressed as two major transcripts in both A549 and H1299 cells and the level of short transcript was much higher than that of the long transcript (Fig. 4E, lane 1 and 3), consistent with the data obtained from qRT-PCR (Fig. 4D). Interestingly, in response to the camptothecin treatment, both short and long PolH transcripts were increased and the ratio of short vs long transcripts was slightly decreased (Fig. 4E, compare lanes 2 and 4 with 1 and 3, respectively). Together, these data suggest that the short PolH transcripts, cleaved at PAS-1/−3 site, are responsible for increased expression of PolH in lung and bladder cancer cells.
Figure 4. PolH expression is regulated by alternative polyadenylation.
(A) Schematic diagrams of the PolH transcripts along with the location of three polyadenylation sites. (B) Northern blots were prepared using total RNAs isolated from RKO and HCT116 cells and then probed with PolH cDNA. (C) Schematic diagrams illustrating the location of common primers to amplify all PolH transcripts or distal primers to amplify the long PolH transcript. (D) The relative level of short and long PolH transcripts was examined in A549, H460, H1299, H1975, and CL1 cells. The level of short PolH transcripts is calculated by subtracting the PCR amplicons using distal primers from the ones using common primers. (E) Northern blots were prepared with total RNAs purified from A549 or H1299 cells treated with or without camptothecin for 24 h, and then probed with cDNAs derived from the PolH and GAPDH genes, respectively.
The short PolH transcript with 427-nt 3’UTR is primarily responsible for elevated PolH expression and subsequently, PolH-mediated cisplatin resistance
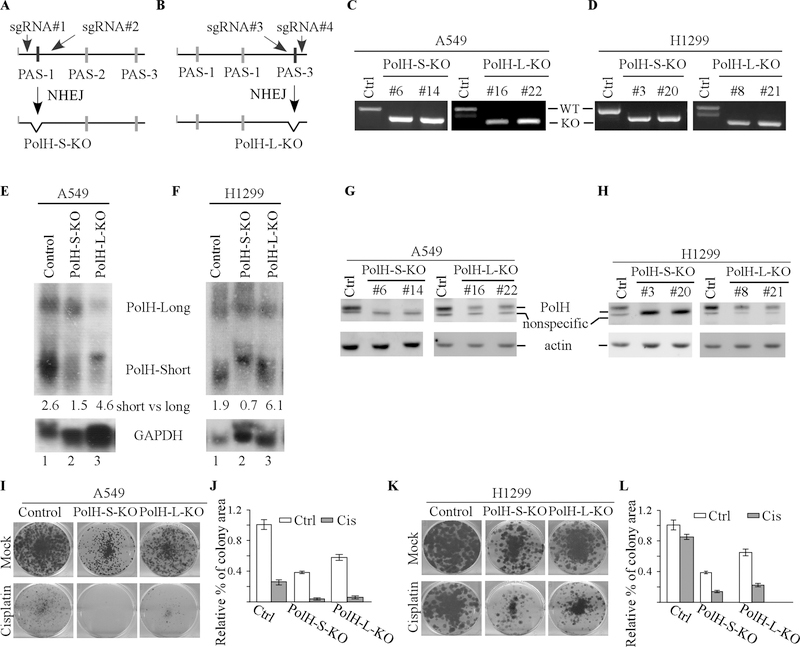
To characterize whether APA-mediated PolH expression plays a role in cisplatin resistance, we generated multiple A549 and H1299 cell lines in which the PAS-1 or PAS-3 site in the PolH gene was deleted by CRISPR-cas9 (Fig. 5A-B). Sequencing analysis indicated that a region of 97-nt containing the PAS-1 site was deleted in PolH-S-KO cells (Fig. 5C-D, left panels) whereas a region of 113-nt containing the PAS-3 site was deleted in PolH-L-KO cells (Fig.5C-D, right panels). Next, Norther blot was performed to examine the level of short and long PolH transcripts in isogenic control, PolH-S-KO and PolH-L-KO cells treated with camptothecin. We found that compared to that in isogenic control cells, the level of short PolH transcript was reduced in PolH-S-KO (Fig. 5E-F, compare lane 1 with 2), whereas the level of long PolH transcript was reduced in PolH-L-KO (Fig. 5E-F, compare lane 1 with 3). Importantly, we found that the level of PolH protein was almost undetectable in PolH-S-KO, but only reduced in PolH-L-KO, A549 and H1299 cells (Fig. 5G-H). Since PolH transcripts cleaved at PAS-2/−3 sites were still intact in PolH-S-KO cells, these data suggest that the short PolH transcript cleaved at the PAS-1 site is responsible for the majority of PolH protein expressed in A549 and H1299 cells. Moreover, we found that compared to that by isogenic control cells, the number of colonies by PolH-S-KO and PolH-L-KO A549/H1299 cells was reduced, which was further decreased by treatment of cisplatin (Fig. 5I-L). We would like to note that the number of colonies by PolH-S-KO A549/H1299 cells, in which PolH was undetectable, was much lower than that by PolH-L-KO A549/H1299 cells, in which PolH was reduced (Fig. 5I-L).
Figure 5. The short PolH transcript with 427-nt 3’UTR is primarily responsible for elevated PolH expression and subsequently, PolH-mediated cisplatin resistance.
(A-B) Schematic diagrams illustrating the strategy to use CRISPR-Cas9 technology to generate PolH-S-KO (A) and PolH-L-KO (B) cell lines. (C-D) Verification of PolH-S-KO and PolH-L-KO A549 (C) and H1299 (D) cells by genotyping. (E-F) Northern blots were prepared with total RNAs purified from isogenic control, PolH-S-KO, and PolH-L-KO A549 (E) or H1299 (F) cells treated with camptothecin for 24 h, and then probed with cDNAs derived from the PolH and GAPDH genes, respectively. (G-H )The levels of PolH and actin were examined in PolH-S-KO and PolH-L-KO A549 (G) and H1299 (H). (I-J) Colony formation assay was performed with PolH-S-KO and PolH-L-KO A549 (I) treated with or without cisplatin (2.5 μM, 18 h), and then quantified using ColonyArea software plugin (J). (K-L) Colony formation assay was performed with PolH-S-KO and PolH-L-KO H1299 (K) treated with or without cisplatin (2.5 μM, 18 h) and then quantified using ColonyArea software plugin (L).
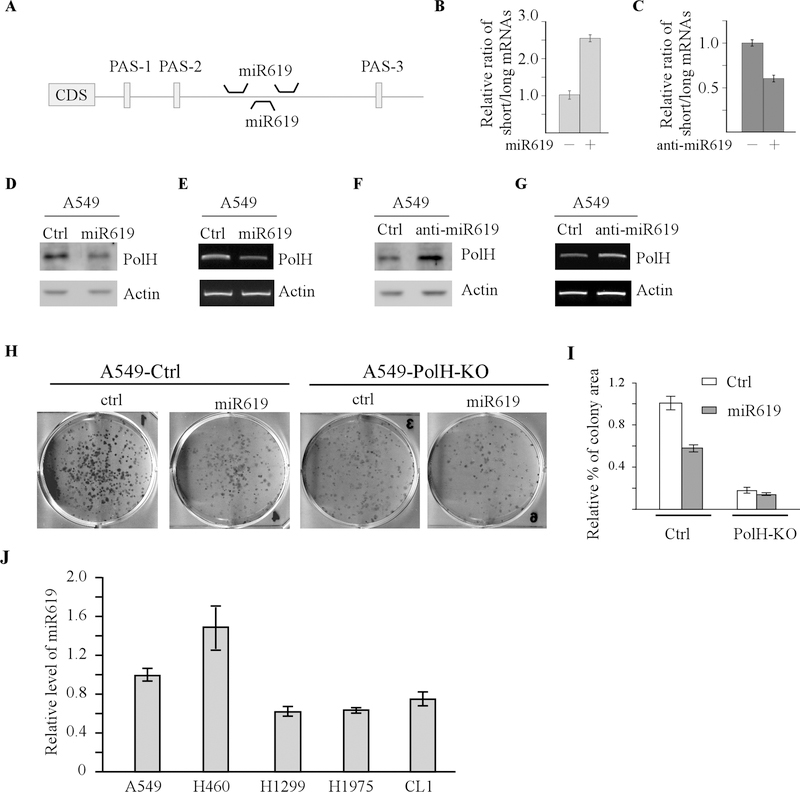
miR619 represses PolH expression by inhibiting the long PolH transcript cleaved at PAS-3 site
miRs are known to repress mRNA stability and/or translation through binding to a seed sequence, generally located in the 3’UTRs of an mRNA [44]. Interestingly, miRNAs are also involved in APA-mediated gene expression by selectively inhibiting mRNAs with long 3’UTR. Consequently, mRNA isoforms with a short 3’UTR often escape miRNA regulation. In this regard, we searched the miRNA database for a miRNA that can regulate various PolH mRNA isoforms. We found three potential binding sites for miR619 in PolH 3’UTR, which are located between PAS-2 and PAS-3 sites (Fig. 6A). Thus, miR619 would only regulate the transcript cleaved at PAS-3 site (Fig. 5B). To test this, the relative abundance of short vs. long PolH transcripts was examined in A549 cells transfected with a control, miR619, or anti-miR619. We found that miR619 increased, whereas anti-miR619 decreased, the ratio of short vs. long transcripts in A549 cells (Fig. 6B-C). Similar results were obtained in H1299, RKO, and MCF7 cells (Supplementary Fig. 4A-B). Moreover, we showed that the levels of PolH protein and transcript were reduced by miR619 (Fig. 6D-E). By contrast, anti-miR619 was able to increase PolH proteins and transcripts (Fig. 6F-G). Next, to determine whether miR619 plays a role in PolH-mediated cell proliferation, isogenic control and PolH-KO A549 cells were transiently transfected with a control miRNA or miR619, followed by colony formation assay. We found that miR619 inhibited colony formation in isogenic control A549 cells (Fig. 6H-I). By contrast, in PolH-KO cells, miR619 had very little effect on colony formation (Fig. 6H-I), suggesting that the growth inhibition mediated by miR619 is PolH-dependent. Consistent with this, we showed that miR619 was capable of reducing the number of colonies formed by isogenic control cells but not by PolH-KO H1299 cells (Supplementary Fig. 4C). Finally, we examine the level of miR619 in various lung cancer cells and found that the level of miR619 is much higher in cisplatin sensitive lung cancer cells (A549 and H460) as compared to cisplatin resistant lung cancer cells (H1299, H1975, and CL1). Together, these data suggest that miR619-mediated PolH repression sensitizes lung cancer cells to cisplatin treatment.
Figure 6. miR619 represses PolH expression by inhibiting the long PolH transcript cleaved at PAS-4 site.
(A) Schematic diagrams illustrating the binding sites of miR619 in the 3’UTR of PolH mRNA. (B) A549 cells were transfected with miRNA control or miR619 for 3 days, followed by qRT-PCR to examine the level of total and long PolH transcript. The level of short PolH transcripts is determined by subtracting the PCR amplicons of long PolH transcripts from the ones of total PolH transcript. The ratio of short vs long PolH transcript is calculated by dividing amplicons of the short PolH transcript by the ones of long PolH transcript. (C) The experiments were performed the same as in (B) except that anti-miR control and anti-miR619 were used. (D) The levels of PolH and actin proteins were examined by western blot analysis using lysates from A549 cells transfected with control miRNA or miR619 for 3 days. (E) A549 cells transfected with control miRNA or miR619 for 3 days, followed by RT-PCR analysis to examine the levels of PolH and actin transcripts. (F) The levels of PolH and actin proteins were examined by western blot analysis using lysates from A549 cells transfected with anti-miRNA control or anti-miR619 for 3 days. (G) A549 cells were transfected with anti-miRNA control or anti-miR619 for 3 days, followed by RT-PCR analysis to measure the levels of PolH and actin transcripts. (H-I) Colony formation assay was performed with isogenic control and PolH-KO A549 cells transfected with control miRNA or miR619, ant then quantified using ColonyArea software plugin (I). (J) The level of miR619 was examined by qRT-PCR in various lung cancer cell lines.
Discussion
Platinum-based anticancer drugs are designed to inhibit DNA replication by cross-linking DNA and have been effective as frontline drugs for many types of tumors. In most cases, tumors respond to the initial treatment, but ultimately develop resistance, leading to an aggressive form of the diseases. As a key mediator of TLS, PolH is induced by DNA damage agents to replicate damaged genomes and thus, may help cancer cells evade chemotherapy. However, it remains to be elucidated whether PolH confers cisplatin resistance in NSCLC. To address this, we examined PolH expression in a set of lung cancer cells that exhibit various degrees of resistance to cisplatin. Interestingly, we showed that high levels of PolH are associated with cisplatin resistance in lung cancer cells (Fig. 1A-D). Moreover, we found that loss of PolH sensitizes not only cisplatin-sensitive (A549) but also cisplatin-resistant (H1299 and H1975) lung cancer cells to cisplatin treatment (Fig. 1F-G, 1H-J and 1L-M). Furthermore, we showed that loss of PolH suppresses tumor growth in mice bearing H1975 xenografts (Fig. 2). Together, these data suggest that targeting PolH may have dual anticancer effects: inhibiting tumor growth and reducing chemoresistance. In support of this notion, high levels of PolH were found to be associated with poor prognosis in NSCLC patients receiving platinum-based chemotherapy [3, 45]. In contrast, low levels of PolH were found to be associated with good prognosis in head and neck cancer patients receiving cisplatin treatment [21]. Thus, further investigations are warranted to test the clinical significance of using PolH inhibitors to overcome chemoresistance.
Recent studies suggest that bypass of cisplatin-induced bulky adducts may require two TLS DNA polymerase [46, 47]. The first TLS polymerase inserts a nucleotide opposite to a lesion (insertion step), followed by a second TLS to extend DNA synthesis. In vitro studies indicate that PolH is involved in the insertion step due to its large and flexible active sites whereas the B family DNA polymerase Pol ζ is involved for the extension step [48]. Indeed, Polζ, composed of catalytic Rev3 and accessary Rev7, can bypass cisplatin-induced adducts and mediates cisplatin resistance in multiple cancer cell lines [49]. Similarly, Rev3 sensitizes lung tumors to chemotherapy [50]. Thus, future studies are needed to examine whether PolH collaborates with Pol ζ to mediate cisplatin resistance in vivo and most importantly, whether targeting these TLS pathways would enhance the anticancer effects of cisplatin.
As a critical enzyme for maintaining genome integrity, PolH is known to be tightly regulated by multiple mechanisms. For example, PolH is transcriptionally up-regulated by tumor suppressor p53 [19] . In addition, PolH mRNA is stabilized by RNA-binding protein PCBP1 [51]. Furthermore, PolH protein can be targeted by E3 ligases, including Pirh2 and Mdm2 [52, 53]. However, whether PolH expression is regulated through APA has never been examined. In the current study, we found that PolH is expressed as multiple transcripts with various lengths of 3’UTRs due to the presence of multiple polyadenylation sites (Fig. 4A). Notably, we found that the PolH transcript with a short 3’UTR is responsible for high levels of PolH protein and associated with cisplatin resistance in lung and bladder cancer cells (Fig. 4D-E and Supplemental Fig. 2). Importantly, knockout of the short PolH transcript by deleting PAS-1 site abrogates PolH expression and subsequently, sensitizes cancer cells to cisplatin treatment (Fig. 5). These observations are consistent with the observation that loss of PolH sensitizes tumor cells to cisplatin treatment (Fig. 1).
To identify a factor that regulates PolH expression via APA, we found that miR619 regulates PolH expression by selectively suppressing the long PolH transcript. In support of this notion, the ratio of short vs. long PolH transcripts is increased by miR619 but decreased by anti-miR619 (Fig. 6B-C). Moreover, we showed that levels of PolH protein and transcript were decreased by miR619 but increased by anti-miR619 (Fig. 6D-G). Furthermore, we showed that ectopic expression of miR619 inhibits cell proliferation in a PolH-dependent manner (Fig. 6H-I). Finally, we showed that the level of miR619 is higher in cisplatin sensitive lung cancer cells than that in cisplatin resistance lung cancer cells (Fig. 6J). Together, these data reveal a novel mechanism by which PolH expression is regulated by miR619 via APA, which play a role in PolH-mediated cisplatin resistance. In addition to miRNAs, RNA-binding proteins are also involved in APA [25, 54]. Previously, we showed that PCBP1 stabilizes PolH transcript [51]. Interestingly, the binding site of PCBP1 in PolH mRNA is close to PAS-1. Thus, it would be interesting to examine whether PCBP1 can regulate PolH expression via APA. Additionally, it would be important to determine how the core cleavage and polyadenylation (C/P) machinery contributes to the production of the PolH transcript with a short 3’UTR.
In summary, our study suggests that PolH expression is regulated by APA and that a short PolH transcript generated from selective APA is responsible for increased PolH expression and subsequently, PolH-mediated cisplatin resistant.
Supplementary Material
Significance:
A short PolH transcript produced by alternative polyadenylation escapes repression by miR619 and confers resistance to cisplatin.
Acknowledgements
This work is supported in part by National Institutes of Health R01 grants (CA195828, CA224433, and CA081237).
Footnotes
Competing interests: No competing interests declared
References
- 1.Srivastava AK, et al. , Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc Natl Acad Sci U S A, 2015. 112(14): p. 4411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie K, et al. , Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci U S A, 2010. 107(48): p. 20792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceppi P, et al. , Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res, 2009. 15(3): p. 1039–45. [DOI] [PubMed] [Google Scholar]
- 4.Waters LS, et al. , Eukaryotic Translesion Polymerases and Their Roles and Regulation in DNA Damage Tolerance. Microbiology and Molecular Biology Reviews, 2009. 73(1): p. 134-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters LS, et al. , Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev, 2009. 73(1): p. 134–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash S, Johnson RE, and Prakash L, Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem, 2005. 74: p. 317–53. [DOI] [PubMed] [Google Scholar]
- 7.Sale JE, Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol, 2013. 5(3): p. a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons DW, et al. , An integrated genomic analysis of human glioblastoma multiforme. Science, 2008. 321(5897): p. 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sale JE, Lehmann AR, and Woodgate R, Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol, 2012. 13(3): p. 141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan GN, et al. , DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res, 2008. 18(1): p. 174–83. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RE, Prakash S, and Prakash L, Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science, 1999. 283(5404): p. 1001–4. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RE, et al. , hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 1999. 285(5425): p. 263–5. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, et al. , The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature, 1999. 399(6737): p. 700–4. [DOI] [PubMed] [Google Scholar]
- 14.Limoli CL, et al. , UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci U S A, 2002. 99(1): p. 233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. , Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res, 2007. 67(7): p. 3018–26. [DOI] [PubMed] [Google Scholar]
- 16.Stary A, et al. , Role of DNA polymerase eta in the UV mutation spectrum in human cells. J Biol Chem, 2003. 278(21): p. 18767–75. [DOI] [PubMed] [Google Scholar]
- 17.Gueranger Q, et al. , Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst), 2008. 7(9): p. 1551–62. [DOI] [PubMed] [Google Scholar]
- 18.Lin Q, et al. , Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res, 2006. 66(1): p. 87–94. [DOI] [PubMed] [Google Scholar]
- 19.Liu G and Chen X, DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol, 2006. 26(4): p. 1398–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teng KY, et al. , DNA polymerase eta protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J Transl Med, 2010. 8: p. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, et al. , Expression of DNA translesion synthesis polymerase eta in head and neck squamous cell cancer predicts resistance to gemcitabine and cisplatin-based chemotherapy. PLoS One, 2013. 8(12): p. e83978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian B and Manley JL, Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci, 2013. 38(6): p. 312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, et al. , A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res, 2005. 33(1): p. 201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Giammartino DC, Nishida K, and Manley JL, Mechanisms and consequences of alternative polyadenylation. Mol Cell, 2011. 43(6): p. 853–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng D and Tian B, RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv Exp Med Biol, 2014. 825: p. 97–127. [DOI] [PubMed] [Google Scholar]
- 26.Mayr C and Bartel DP, Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell, 2009. 138(4): p. 673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangone M, et al. , The landscape of C. elegans 3’UTRs. Science, 2010. 329(5990): p. 432–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elkon R, et al. , E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol, 2012. 13(7): p. R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris AR, et al. , Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res, 2012. 18(19): p. 5256–66. [DOI] [PubMed] [Google Scholar]
- 30.Sandberg R, et al. , Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science, 2008. 320(5883): p. 1643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen RW, et al. , Truncation in CCND1 mRNA alters miR-16–1 regulation in mantle cell lymphoma. Blood, 2008. 112(3): p. 822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiestner A, et al. , Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood, 2007. 109(11): p. 4599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klinck R, et al. , Multiple alternative splicing markers for ovarian cancer. Cancer Res, 2008. 68(3): p. 657–63. [DOI] [PubMed] [Google Scholar]
- 34.Zhu J, Chen Z, and Yong L, Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol, 2018. 148(2): p. 368–374. [DOI] [PubMed] [Google Scholar]
- 35.Chu YW, et al. , Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol, 1997. 17(3): p. 353–60. [DOI] [PubMed] [Google Scholar]
- 36.Janik P, Briand P, and Hartmann NR, The effect of estrone-progesterone treatment on cell proliferation kinetics of hormone-dependent GR mouse mammary tumors. Cancer Res, 1975. 35(12): p. 3698–704. [PubMed] [Google Scholar]
- 37.Zhang J, et al. , Mice deficient in Rbm38, a target of the p53 family, are susceptible to accelerated aging and spontaneous tumors. Proc Natl Acad Sci U S A, 2014. 111(52): p. 18637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzman C, et al. , ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One, 2014. 9(3): p. e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, et al. , Establishment of a dog model for the p53 family pathway and identification of a novel isoform of p21 cyclin-dependent kinase inhibitor. Mol Cancer Res, 2009. 7(1): p. 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothkamm K, et al. , DNA damage foci: Meaning and significance. Environ Mol Mutagen, 2015. 56(6): p. 491–504. [DOI] [PubMed] [Google Scholar]
- 41.Thivierge C, et al. , Alternative polyadenylation confers Pten mRNAs stability and resistance to microRNAs. Nucleic Acids Res, 2018. 46(19): p. 10340–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, et al. , Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases. Nucleic Acids Res, 2017. 45(15): p. 8930–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ and Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 2001. 25(4): p. 402–8. [DOI] [PubMed] [Google Scholar]
- 44.Bartel DP, MicroRNAs: target recognition and regulatory functions. Cell, 2009. 136(2): p. 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valk K, et al. , Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology, 2010. 79(3–4): p. 283–92. [DOI] [PubMed] [Google Scholar]
- 46.Shachar S, et al. , Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J, 2009. 28(4): p. 383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livneh Z, Ziv O, and Shachar S, Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle, 2010. 9(4): p. 729–35. [DOI] [PubMed] [Google Scholar]
- 48.Lee YS, Gregory MT, and Yang W, Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci U S A, 2014. 111(8): p. 2954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma S, et al. , DNA polymerase zeta is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol Pharmacol, 2012. 81(6): p. 778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doles J, et al. , Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A, 2010. 107(48): p. 20786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren C, et al. , DNA polymerase eta is regulated by poly(rC)-binding protein 1 via mRNA stability. Biochem J, 2014. 464(3): p. 377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung YS, Qian Y, and Chen X, DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst), 2012. 11(2): p. 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung YS, Liu G, and Chen X, Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol Cell Biol, 2010. 30(4): p. 1041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erson-Bensan AE, Alternative polyadenylation and RNA-binding proteins. J Mol Endocrinol, 2016. 57(2): p. F29–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.