Abstract
Genetic and rare diseases (GARDs) affect more than 350 million patients worldwide and remain a significant challenge in the clinic. Hence, continuous efforts have been made to bridge the significant gap between the supply and demand of effective treatments for GARDs. Recent decades have witnessed the impressive progress in the fight against GARDs, with an improved understanding of the genetic origins of rare diseases and the rapid development in gene therapy providing a new avenue for GARD therapy. RNA-based therapeutics, such as RNA interference (RNAi), messenger RNA (mRNA) and RNA-involved genome editing technologies, demonstrate great potential as a therapy tool for treating genetic associated rare diseases. In the meantime, a variety of RNA delivery vehicles were established for boosting the widespread applications of RNA therapeutics. Among all the RNA delivery platforms which enable the systemic applications of RNAs, non-viral RNA delivery biomaterials display superior properties and a few biomaterials have been successfully exploited for achieving the RNA-based gene therapies on GARDs. In this review article, we focus on recent advances in the development of novel biomaterials for delivery of RNA-based therapeutics and highlight their applications to treat GARDs.
Keywords: RNA delivery biomaterials, genetic and rare diseases (GARDs), RNA therapeutics, RNAi, mRNA, CRISPR
1. Introduction
Genetic and rare diseases (GARDs) represent a large category of disorders which share the similarity of each affecting a small portion of the population.[1, 2] While a single rare disease may affect only a small group of people, with the definition as fewer than 200,000 patients in the United States or less than 1 in 2,000 people in the European Union, the collective impact of rare diseases can be enormous.[1–3] Approximately seven thousand rare diseases affect around 30 million individuals in the US and over 350 million patients worldwide.[3–5] Despite the great efforts that have been devoted to supporting the development of orphan drugs, many rare diseases still lack effective cures and call for new therapeutics.[3] Since a majority of GARDs stem from genetic disorders, recent progresses in gene therapies could offer more options for treating GARDs.[3, 5]
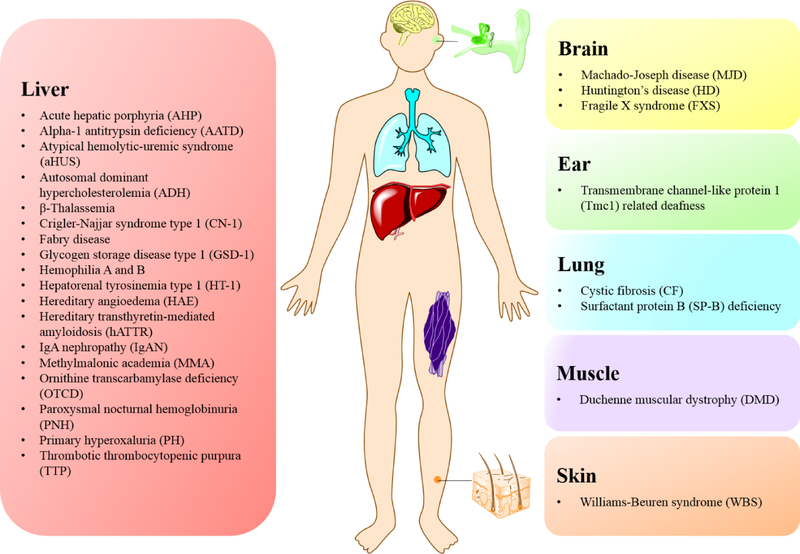
In the past several decades, RNA-based therapeutics have been explored by researchers for diverse disease applications, ranging from cancers, infections, and autoimmune diseases to genetic disorders.[6–11] RNA vaccines, for example, have exhibited encouraging results in both infection therapy and cancer immunotherapy.[6, 7, 11–15] Another worthwhile application of RNA technology is to apply RNAs as a gene therapy tool to treat GARDs, which currently lack access to effective therapeutics. To date, a variety of RNA therapeutics have been developed for GARD therapies based on the versatile family of RNA molecules, including microRNA (miRNA), small interfering RNA (siRNA), messenger RNA (mRNA), long non-coding RNA (lncRNA), and genome editing systems containing RNA components.[6, 9, 16] These different RNA-based therapeutics can be used to treat genetic diseases, primarily at the RNA level via distinct mechanisms of action. For instance, RNA interference (RNAi) technology enables the downregulation of disease related genes and protein expression; mRNA mediates the expression and supplementation of therapeutic proteins; the CRISPR gene editing system facilitates gene engineering in vivo or through cell engineering ex vivo.[6, 9, 11, 17–20] Significant research has demonstrated the feasibility of RNA technologies to treat or cure GARDs (Figure 1).[17, 21]
Figure 1.
A representation of genetic and rare diseases (GARDs) that have been explored using non-viral biomaterials based RNA therapeutics.
In terms of RNA therapeutics, RNA molecules alone have difficulty penetrating target cells, are vulnerable to degradations in biological fluids, and are incapable of accumulating in target organs following systemic administration.[18] Although some naked RNAs and chemically modified RNAs were applied for therapies based on local administration or ex vivo applications, appropriate delivery methods are still essential for both enabling systemic applications of RNA therapeutics and further maximizing their therapeutic window.[22] Several commonly used systemic RNA delivery methods include viral vectors, DNA/RNA nanotechnology and non-viral biomaterials.[20, 23, 24] Viral vectors are a type of efficient tool for gene transfection and account for many gene-therapy clinical trials.[20, 25, 26] Several viral vector mediated cellular and gene-therapy products, such as Kymriah for CAR-T immunotherapy on lymphoblastic leukemia, have already gained FDA approval.[27] Also, many viral vectors have been employed for treating GARDs, such as hemophilia, cystic fibrosis, or rare inherited retinal diseases, etc.[23, 28] In addition to viral vectors, DNA and RNA nanotechnologies have also been applied to deliver RNA therapeutics such as siRNA and miRNA, which are mainly used for cancer therapy.[24, 29–33] Non-viral RNA delivery biomaterials, such as ligand-RNA conjugates, lipid or polymer-based nanoparticles, and cell derived vehicles (exosomes), constitute another large category of RNA delivery vehicles. Different non-viral materials have been designed and optimized for accommodating efficient delivery of diverse RNA molecules as well as multi-component genome editing systems.[22, 34–36] Moreover, many of these biomaterials exhibit organ targeting properties, which make it possible for RNA therapeutics to achieve targeted effects at diseased sites.[18] In recent years, a number of non-viral delivery systems have been exploited for enabling RNA-based gene therapies on GARDs (Figure 2). Given the promising results from clinical trials and preclinical studies, biomaterials will continue to contribute to RNA therapeutics on GARDs.
Figure 2.
An illustration of representative non-viral biomaterials (outer circle) together with versatile RNA therapeutics (inner circle) that have been exploited for treating genetic and rare diseases (GARDs). [9, 37–49]
In this review, we summarize the non-viral biomaterials developed for the RNA therapeutic delivery, including siRNA, mRNA, genome editing systems with RNA, miRNA, and lncRNA. We categorize these RNA delivery systems based on the different types of RNA payloads they afford and discuss in detail their different targeted organs, such as the liver, lungs, brain, muscle etc. In addition, we highlight clinical trials and therapeutic applications of these RNA delivery biomaterials in the field of GARD therapies. Finally, we briefly conclude with the advantages and limitations of different delivery vehicles and provide our future perspectives.
2. Small interfering RNA (siRNA)
siRNA, based on the RNA interference (RNAi) technology,[50] enables efficient downregulation of specific gene expression through complementary-based mRNA cleavage.[51] A number of genetic and rare diseases (GARDs) have shown their close link to specific genetic abnormalities, where mutant or misfolded proteins are expressed as a result and lead to disease symptoms.[1] Therefore, siRNA therapy can be applied for treating GARDs by targeted silencing of the disease-causing genes.[50, 52]
siRNA is typically double-stranded RNA oligonucleotides composed of 19–25 base pairs.[22] Their gene silencing mechanism involves the interaction of siRNA with an RNA-induced silencing complex (RISC), where siRNA is separated into sense and antisense strands. The sense strand of siRNA is subsequently discarded while the antisense strand recognizes the complementary mRNA and, together with the endonuclease Argonaute 2 (AGO2), guides the cleavage of targeted sequence.[9] Through these processes, siRNA could achieve targeted gene silencing and thus mediate the downregulation of gene activity.[51]
In recent years, siRNA for treating GARDs has aroused prominent interest in both pharmaceutical industries and academia - emphasized by the US FDA approval of the first siRNA drug, Patisiran, in 2018.[53] This is also marked by a growing number of siRNA-based therapies, which are entering clinical trials.[6, 51, 54] Early applications of siRNA in treating GARDs, such as TD101 siRNA (NCT00716014) for treating an inherited skin disorder, pachyonychia congenital, emerged with therapies based on local administration, since naked and unmodified siRNA are suspected to degrade in blood circulation.[55] However, many siRNA delivery biomaterials have been successively exploited to enable the long circulation and organ targeting effects of siRNA for more systemic applications.[52] To date, various delivery biomaterials have been developed for siRNA-based therapies and some of them have demonstrated potentials in translational applications. In this section, we summarize the non-viral siRNA delivery systems that have been applied for GARDs based on their different target organs, particularly the liver, lungs, and brain (Table 1). We also review several typical delivery systems, such as lipid and lipid derived nanoparticles (LNPs) and ligand-siRNA conjugates, which enable the siRNA-based therapeutics on GARDs.
Table 1.
List of siRNA delivery systems for the treatment of genetic and rare disease (GARD)
| Name of siRNA Therapeutics or siRNA | Delivery Material | Administration Route | Indication/Disease model | Target Organ | Clinical Trial | Reference |
|---|---|---|---|---|---|---|
| Patisiran (ALN-TTR02) | LNP (MC3) | i.v. | Hereditary transthyretinmediated amyloidosis (hATTR) | liver | FDA approved (ONPATTRO™) | [56, 58, 64] |
| DCR-PH1 | LNP | i.v. | Primary hyperoxaluria type 1 (PH-1) | liver | Pre-clinical | [71] |
| Tmprss6 siRNA | Ionizable LNP | i.v. | β-Thalassemia | liver | Pre-clinical | [68, 70] |
| Givosiran (ALN-AS1) | (ESC)-GalNAc delivery platform | subcutaneous | Acute hepatic porphyria (AHP) | liver | Phase 3 (NCT03338816) | [74] |
| Fitusiran (ALN-AT3) | (ESC)-GalNAc delivery platform | subcutaneous | Hemophilia A and B | liver | Phase 3 (NCT03754790) | [84] |
| Lumasiran (ALN-GO1) | (ESC)-GalNAc delivery platform | subcutaneous | Primary hyperoxaluria type 1 (PH-1) | liver | Phase 3 (NCT03681184) | [108, 109] |
| Vutrisiran (ALNTTRsc02) | (ESC)-GalNAc delivery platform | subcutaneous | Hereditary transthyretinmediated amyloidosis (hATTR) | liver | Phase 3 (NCT03759379) | |
| Inclisiran (ALNPCSsc) | (ESC)-GalNAc delivery platform | subcutaneous | Autosomal dominant hypercholesterolemia (ADH) | liver | Phase 3 (NCT02963311) | [85] |
| Cemdisiran (ALNCC5) | (ESC)-GalNAc delivery platform | subcutaneous | IgA nephropathy (IgAN), atypical hemolytic-uremic syndrome (aHUS), paroxysmal nocturnal hemoglobinuria (PNH) | liver | Phase 2 (NCT03841448) Phase 1/2 (NCT02352493) |
[110, 111] |
| ALN-AAT02 | (ESC)-GalNAc delivery platform | subcutaneous | Alpha-1 antitrypsin deficiency (AATD) | liver | Phase 2 (NCT03767829) | [112] |
| ALN-TMP | (ESC)-GalNAc delivery platform | subcutaneous | β-Thalassemia | liver | Pre-clinical | [80] |
| ALN-F12 | (ESC)-GalNAc delivery platform | subcutaneous | Hereditary angioedema (HAE) | liver | Pre-clinical | [113] |
| Revusiran (ALNTTRsc) | (STC)-GalNAc delivery platform | subcutaneous | Hereditary transthyretinmediated amyloidosis (hATTR) | liver | Phase 2 (NCT02292186) | |
| DCR-PHXC | GalXC™ | subcutaneous | Primary hyperoxalurias (PH) | liver | Phase 2 (NCT03847909) | [87] |
| SLN124 | GalNAc-siRNA conjugation | subcutaneous | β-Thalassemia | liver | Pre-clinical | [88] |
| ARO-AAT | TRiM™ | subcutaneous | Alpha-1 antitrypsin deficiency (AATD) | liver | Phase 1 (NCT03362242) | [114] |
| ARC-AAT | DPCiv™ (Ex1) | i.v. | Alpha-1 antitrypsin deficiency (AATD) | liver | Phase 2 (NCT02900183) withdrawn | [90] |
| Lac-α-CDE (G3)/siRNA complex | Lac-α-CDE (G3) conjugate | i.v. | Familial amyloid polyneuropathy (FAP) | liver | Pre-clinical | [115, 116] |
| GUG-β-CDE (G2, DS 1.8)/siTTR complex | GUG-β-CDE (G2) conjugate | i.v. | Familial amyloid polyneuropathy (FAP) | liver | Pre-clinical | [117] |
| ENaCsiRNA | nanocomplexes | oropharyngeal | Cystic fibrosis (CF) | lung | Pre-clinical | [95, 96] |
| ARO-ENaC | TRiM™ | intratracheal/ oropharyngeal/ nebulized | Cystic fibrosis (CF) | lung | Pre-clinical | [98] |
| siMutAtax3 siRNA | RVG-9r-targeted SNALPs | i.v. | Machado-Joseph disease (MJD) | brain | Pre-clinical | [37, 105] |
| hsiRNA-loaded exosomes | exosomes | striatum | Huntington’s disease (HD) | brain | Pre-clinical | [106] |
| siRNA SNA-NCs | SNA-NCs | topical | Dominant genetic skin disorders | skin | Pre-clinical | [118] |
i.v., intravenous; LNP, lipid and lipid derived nanoparticle; MC3, DLin-MC3-DMA; ESC, Enhanced Stabilization Chemistry; STC, Standard Template Chemistry; TRiM™, Targeted RNAi Molecule Platform; DPCiv™, Dynamic Polyconjugate (DPC) i.v. drug delivery vehicle; SNALPs, Stable Nucleic Acid Lipid Particles; SNA-NCs, Spherical Nucleic Acid Nanoparticle Conjugates.
2.1. Liver delivery of siRNA
Lipid and lipid derived nanoparticles (LNPs)
In 2018, Patisiran (ONPATTRO™) gained its approval by the US FDA and became the first marketed RNAi-based therapy. Patisiran is a siRNA drug encapsulated within LNPs that was developed by Alnylam® Pharmaceuticals for hereditary transthyretin amyloidosis (hATTR) therapy. hATTR is an inherited genetic disease characterized by the aggregation of mutant transthyretin (TTR) proteins, which leads to disease symptoms such as heart and nerve function impairment. Patisiran, by adopting the RNAi technique, can be used to target and inhibit the TTR mRNA from producing mutant TTR proteins. Meanwhile, the use of LNPs as a well-established delivery vehicle enables the efficient delivery and accumulation of siRNA into hepatocytes, where TTR proteins are primarily produced. The combined functionality of Patisiran’s RNAi technique coupled with the use of LNPs as a delivery vehicle enables a robust reduction in the expression of the hATTR disease-linked gene, ultimately providing a therapeutic treatment for the genetic disease.[56, 57] The approval of Patisiran for hATTR therapy not only marks a significant milestone for the gene therapy based treatment of GARDs but also represents the fruit of research in the field of RNAi therapy.[53, 58]
In addition to Patisiran, a growing number of research studies reported siRNA delivery using LNPs. Many of these siRNA LNPs showed efficient delivery of siRNA to the liver. Except from the favorable physiological properties of the liver for LNPs accumulation, one potential mechanism of hepatocyte targeting effect by LNPs involves the interaction of ionizable LNPs with apolipoprotein E (apoE), a protein that facilitates the transport of molecules such as lipids and cholesterol.[59, 60] Binding on the surface of LNPs, apoE mediated the hepatocellular uptake of LNPs through multiple receptors, such as the low-density lipoprotein receptor (LDLR).[59, 60] The exceptional properties of LNPs for RNA delivery include, but are not limited to, the encapsulation and protection of RNAs from degradation during circulation, as well as the delivery of RNA payloads to the target organ following systemic administration.[18, 54, 61] Among the reported non-viral siRNA delivery platforms, LNPs represent a major intravenously (i.v.) administered siRNA delivery system for clinical applications.[22, 38] The most commonly reported siRNA LNP formulation contains five key components, which are the lipid or lipid derivatives, phospholipid, cholesterol, polyethylene glycol (PEG), and the payload siRNA.[59] In addition to physiochemical properties of the main lipid material, the combination as well as the relative ratios of all the key components also have profound effects on the delivery efficiency of LNP formulations.[59, 62] To date, many different lipid and lipid derived materials have been exploited for establishing siRNA delivery LNPs.[59, 62]
Early exploration of systemic siRNA delivery LNPs mainly focused on lipid materials such as 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane (DLin-DMA).[52] Further modification of the DLin-DMA lipid based on rational design identified DLin-KC2-DMA with an enhanced gene silencing activity.[63] Instead of generating a large library of compounds for screening, Semple and coworkers adopted a rational design strategy for the modification of the key structures, such as the head group and the linker region, within the ionizable lipid DLin-DMA.[63] Their rational design was guided by the knowledge of the relationship between the chemical structures of key groups within the lipid materials and the physicochemical properties as well as the in vivo activity of nanoparticles.[63] Semple et al. reported that DLin-KC2-DMA containing stable nucleic acid lipid particles (SNALPs) exhibited in vivo activity at siRNA doses of 0.01 mg/kg in the mouse model and was capable of silencing the TTR gene in nonhuman primates with ED50 around 0.3 mg/kg.[63] A number of lipid materials were further developed by rational design of a large library of compounds followed by in vitro and in vivo screenings, such as the DLin-MC3-DMA (MC3) used in the Patisiran LNPs formulation, C12–200, and 304O13.[38, 64, 65] For example, MC3 lipid was identified through further modulating and screening on the head group structure of DLin-KC2-DMA.[66] All these lipid materials showed improved delivery efficiency as compared with DLin-KC2-DMA LNPs.[38, 64, 65] Some other LNPs were reported with organ specificity, such as cKK-E12 lipopeptide nanoparticles, wherein the lipid tails were conjugated to peptide derivatives and achieved liver targeting siRNA delivery.[67]
In 2013, Schmidt et al. reported siRNA LNPs targeting transmembrane serine protease 6 (Tmprss6) for β-thalassemia therapy.[68] β-Thalassemia is an inherited blood genetic disorder caused by the deficient or absent synthesis of β-globin (HBB). TMPRSS6 gene encodes the matriptase-2 protein, which is expressed in hepatocytes and negatively modulates the production of hepcidin. Mutations of TMPRSS6 were found to elevate hepcidin expression and cause iron-refractory iron deficiency anemia (IRIDA).[69] By targeting and suppressing TMPRSS6 using the Tmprss6 siRNA LNPs, Schmidt et al. achieved decreased iron overload and ameliorated anemia in the mouse model of β-thalassemia intermedia.[68] In 2015, they employed the combination therapy of the Tmprss6 siRNA LNPs with Deferiprone, an oral iron chelator, for further mitigating anemia and secondary iron overload in murine β-thalassemia intermedia.[70]
In 2016, Dutta et al. explored the use of siRNA LNPs for treating primary hyperoxaluria type 1 (PH-1). PH-1 is a rare metabolic disorder characterized by the defect of a liver peroxisomal enzyme, alanine glyoxylate aminotransferase (AGT).[71] The deficiency of AGT activity could increase the production of oxalate in the liver, which leads to the formation and accumulation of calcium oxalate crystals in kidneys.[72] Current treatment for the prevention of primary manifestations of PH-1 is limited to vitamin B6 therapy, however, patients who are not pyridoxine responsive may require a combined liver/kidney transplant.[71] Dutta et al. adopted siRNA LNPs to target and downregulate hydroxyacid oxidase 1 (HAO1). HAO1 gene encodes glycolate oxidase (GO), which can transform glycolate into glyoxylate, and thus can be used as a gene silencing target for reducing oxalate levels in the liver.[73] The identified lead HAO1 siRNA-LNP showed potent suppression of GO expression in both mice and nonhuman primates. Moreover, the reduction of GO protein in the liver also resulted in reduced urine oxalate levels and calcium oxalate accumulation in the PH-1 mouse model.[71]
Although a variety of LNPs have been developed and many studies have revealed their feasibility of siRNA-based therapies, few clinical applications of siRNA LNPs for GARDs are ongoing. Many preclinical studies adopted LNPs as a liver targeting delivery system to test gene silencing activity, whereas siRNA was further developed into N-acetylgalactosamine (GalNAc)-siRNA conjugates for clinical trials.[70, 71, 74]
N-acetylgalactosamine (GalNAc) conjugates
With the development of chemical modifications on RNA molecules, modified siRNA are reported to remain intact for long durations under circulation, which provides the possibility of delivering siRNA without the shielding of nanoparticles.[19, 75, 76] Hence, siRNA delivery through ligand conjugation emerged with efficient and targeted delivery of siRNA therapeutics.
One of the well-established ligand-siRNA conjugates is that of siRNA to GalNAc, which is known as a potent ligand to the asialoglycoprotein receptor (ASGPR). The mechanism of action for the ligands interacting with ASGPR has been thoroughly investigated since the 1960s.[77] The ASGPR is highly expressed on hepatocytes and GalNAc facilitates the uptake of its payload into hepatocytes through ASGPR via clathrin-mediated endocytosis.[77] Based on this mechanism, researchers developed multivalent GalNAc-siRNA conjugates for liver targeting siRNA delivery.[77] Results showed that by conjugating the engineered ASGPR ligand to the chemically modified siRNA molecule, GalNAc-siRNA conjugates could achieve systemic stability against nucleases, which would in turn improve pharmacokinetics. Most importantly, single or multiple low-volume subcutaneous injections of these conjugates resulted in effective suppression of the target gene in the liver.[51, 77, 78] As compared with many other siRNA delivery systems which are based on repeated high-dose i.v. administrations, this subcutaneously delivered GalNAc-siRNA conjugate is thus more desirable for some therapeutic purposes. Due to these favorable properties of GalNAc-siRNA conjugates, many linker chemistries and RNA modification methods have been subsequently explored by researchers for maximizing the therapeutic window. A series of GalNAc-siRNA conjugates were established in recent years to treat a variety of GARDs with the liver as the target organ (Table 1).[64, 68, 79, 80]
Moreover, Alnylam® Pharmaceuticals developed the Enhanced Stabilization Chemistry (ESC)-GalNAc-siRNA conjugate platform, aiming to further enhance the stability, potency, and duration of siRNA molecules in the liver following subcutaneous administration.[81] As compared with their first generation ‘Standard Template Chemistry’ (STC)-based GalNAc-siRNA, in which RNA molecules were 2’-OMe and 2’-F modified and contained two terminal phosphorothioate (PS) linkages, the newly developed ESC platform features higher 2’-OMe content and four additional terminal PS linkages.[81] Many of these ESC-based GalNAc-siRNA conjugates have entered clinical trials, among which some have already reached Phase 3 clinical trials (Table 1). For example, acute hepatic porphyrias (AHP) is a rare genetic disease caused by an inherited defect in one of the enzymes in the heme biosynthesis pathway.[74, 82, 83] The elevated activity of the 5-aminolevulinic acid synthase 1 (ALAS1) enzyme, which regulates the heme pathway, can lead to the buildup of neurotoxic heme intermediates in the liver and result in disease symptoms.[82] ALN-AS1 (Givosiran), a GalNAc-conjugated RNAi therapeutic, could effectively prevent and treat induced attacks of AHP by targeting and silencing ALAS1.[74] Similarly, another GalNAc-siRNA conjugate (ALN-AT3, Fitusiran) targeting antithrombin (AT) was developed as a therapy for hemophilia A and B, inherited bleeding disorders, based on the reports that co-inheritance of prothrombotic mutations in patients results in the ameliorated clinical phenotype in hemophilia.[84] Additionally, ALN-PCSsc (Inclisiran) is a synthetic GalNAc-siRNA conjugate designed against the proprotein convertase subtilisinkexin type 9 (PCSK9). PCSK9 is known as a crucial regulator of the low-density lipoprotein receptor (LDLR), since it interacts with the LDLR and mediates the lysosomal degradation of the receptors. Therefore, the inhibition of PCSK9 offers a strategy for lowering the plasma LDL-cholesterol (LDL-C) level in patients. Inclisiran was reported to be able to significantly decrease the production of PCSK9 in liver and thus reduce the LDL-C level, which enables it to be used for the management of autosomal dominant hypercholesterolemia (ADH).[85, 86]
In addition to the GalNAc-siRNA conjugates developed based on the ESC delivery platform designed by Alnylam® Pharmaceuticals, Dicerna™ Pharmaceuticals recently initiated Phase 2 clinical trials (NCT03847909) for DCR-PHXC in treating primary hyperoxalurias (PH), and Silence Therapeutics, in year 2019, filed CTA for SLN124 for β-thalassemia and myelodysplasia syndrome.[87, 88]
Other ligand conjugates
Arrowhead Pharmaceuticals’ Targeted RNAi Molecule platform (TRiM™) is another example of using ligand conjugation strategy to mediate targeted siRNA delivery.[89] The TRiM™ platform utilizes ligand-mediated delivery and stringent bioinformatics to design tissue-specific targeting conjugates.[89] Although detailed information about TRiM™ platform is not disclosed, it is known that the platform comprises a highly potent RNA trigger as well as optimized high affinity targeting ligand and a linker for each drug candidate based on their specific applications.[89] The TRiM™ platform provides advantages such as enabling the delivery of RNAi to tissues beyond the liver and through multiple routes of administration.[89]
ARO-AAT, one of the candidates built on the TRiM™ platform, was developed as a therapy for alpha-1 antitrypsin deficiency (AATD) and has completed a Phase 1 clinical study (NCT03362242). AATD is a rare genetic liver disease caused by the production of excessive alpha-1 antitrypsin (AAT) mutant Z (Z-AAT) in the hepatocytes.[90] Accumulation of misfolded Z-AAT in the liver can lead to highly variable liver diseases with a variety of clinical presentations, such as hepatic injury and hepatocellular carcinoma (HCC).[90, 91] ARO-AAT is a liver targeting siRNA conjugate developed to suppress the production of Z-AAT protein by gene silencing with the aim to prevent accumulation of disease-related proteins in the liver, allow clearance of accumulated proteins, prevent repeated cycles of cellular damage, and reverse fibrosis associated with prior damage.[90]
Additionally, siRNA conjugations with cholesterol, aptamer, peptide and other moieties were also reported to facilitate the distribution and gene silencing effects of siRNA molecules in different organs, liver included.[92–94] These siRNA conjugates are under extensive exploration in preclinical studies.
2.2. Lung delivery of siRNA
Cystic fibrosis (CF) is a type of inherited rare disease characterized by a genetic mutation that causes mucus buildup in the lungs and pancreas. Patients with CF show symptoms such as having difficulty breathing or experiencing recurrent and persistent lung infections. To date, several publications have applied lung siRNA delivery systems for the treatment of CF.[95–98] Their common therapeutic strategy is based on the downregulation of epithelial sodium channel (ENaC). ENaC was found to be a target for CF therapy, since the upregulation of ENaC in the lungs of CF patients leads to the increased absorption of sodium and fluids from the airway, and eventually results in impaired mucociliary clearance.
In 2017, Manunta et al. reported the development of an ENaCsiRNA nanocomplex that induced effective in vitro and in vivo silencing of airway ENaC.[95, 96] This self-assembling ENaCsiRNA nanocomplex formulation was composed of a liposome (DOTMA/DOPE; L), an epithelial targeting peptide (P) and an siRNA (R).[95] Results showed that the established LPR nanocomplexes facilitated the translocation of siRNA through mucus, mediated efficient ENaCsiRNA transfections in primary CF epithelial cells, and reduced 30% of the αENaC and βENaC mRNA. These ENaCsiRNA nanocomplexes also modulated the activity of ENaC and showed effective delivery and silencing in vivo.[95, 96]
Based on ENaC inhibition mediated by siRNA, Arrowhead Pharmaceuticals developed the ARO-ENaC siRNA conjugates, by using the TRiM™ platform as the delivery technique.[98] ARO-ENaC is designed to target and suppress the production of the epithelial sodium channel alpha subunit (αENaC) in the airways of the CF lungs.[98] Preclinical studies showed that by adopting an integrin αvβ6 receptor ligand (EpL) as a targeting ligand, the ARO-ENaC siRNA conjugate can achieve the functional delivery of siRNA to the airway epithelium. ARO-ENaC thus mediated targeted silencing of αENaC mRNA in the lungs of rats, and dose-dependently improved mucociliary clearance in sheep after inhaled aerosol exposure.[98]
2.3. Brain delivery of siRNA
Although a variety of diseases require therapeutics to be delivered to the brain, brain targeted delivery has always been a formidable issue, despite the continuous efforts that have been made to help overcome the barrier. While RNAi holds great promise for treating genetic associated neurological disorders, efficient delivery of siRNA into the brain is still a remarkable obstacle to the clinical applications.[99–101] Strategies that have been applied for achieving functional siRNA delivery into the brain mainly rely on employing potent protein or peptide ligands to target the receptors, such as transferrin, insulin and LDL receptors, which are highly expressed on the blood brain barrier (BBB).[102] In recent years, some brain targeting siRNA delivery systems, including spherical nucleic acid (SNA), stable nucleic acid lipid particles (SNALPs) and targeted exosomes, have demonstrated potentials for treating neurological genetic disorders.[63, 103] Two representative studies using brain targeting siRNA delivery systems to treat rare disease are summarized as follows.
Machado-Joseph disease (MJD) is a rare neurodegenerative disease, which lacks available treatment.[104] As an autosomal dominant disorder, MJD is caused by the abnormal repetition of CAG in the ATXN3/MJD1 gene, which results in mutations in the ataxin-3 protein. In 2015, Conceiç~ao et al. successfully developed siRNA-SNALPs for the treatment of MJD, which incorporated a rabies virus glycoprotein (RVG) derived short peptide RVG-9r and encapsulated mutant ataxin-3 targeting siRNA.[37] The afforded formulation exhibited favorable features for systemic administration and in vitro efficacy to silence the mutant ataxin-3 in neuronal cells. In vivo studies further showed that RVG-9r targeted siRNA-SNALPs mediated silencing of MJD causing gene, ameliorated disease symptoms in the MJD mouse models following i.v. administration. Additionally, a safety profile of the repeated i.v. administrations of RVG-9r targeted mutant ataxin-3 targeting siRNA-SNALPs was further reported in a follow-up study.[105]
Another example of brain targeting siRNA delivery system adopted exosome vehicles for the treatment of a fatal neurological genetic disorder, Huntington’s disease (HD).[106] The cause of HD is related to the heightened CAG repeats in the huntingtin (HTT) gene.[101, 106] Many studies have demonstrated that siRNA could mediate the reduction of HTT expression in the brain.[101, 107] In 2016, Didiot et al. efficiently loaded modified small interfering RNA (hsiRNAs targeting HTT mRNA) into exosomes, which are endogenous vesicles applied as siRNA delivery biomaterials. Results showed that the hsiRNA-loaded exosomes could be uptake into mouse primary cortical neurons and, therefore, facilitate the suppression of HTT mRNA and protein. Moreover, administration of hsiRNA-loaded exosomes through unilateral infusion into mouse striatum further showed in vivo efficacy of these established exosomes, where 35% of HTT mRNA were silenced in the brain of mice.[106]
3. Messenger RNA (mRNA)
mRNA functions as the bridge between DNA and proteins in eukaryotic cells.[8, 12, 119] Endogenous mRNA carries genetic information from DNA and is translated into functional proteins.[8, 12, 119] Alternatively, by using the in vitro transcription techniques (IVTs), mRNA can be synthesized and, therefore, have become a potential new drug class for diverse therapeutic applications.[34, 39, 120, 121] When introduced into cytosol via mRNA delivery systems, the IVT mRNAs can be translated to produce desired functional proteins.[8, 12, 119]
Many genetic and rare diseases (GARDs) are characterized by protein deficiencies or malfunctions that require protein replacement therapies (PRTs).[122, 123] Hence, supplementation of exogenous proteins is a straightforward therapeutic approach.[8, 119, 120] Compared with protein drugs, mRNA can supplement both cytoplasmic and large transmembranic proteins.[8, 119, 120] Moreover, mRNA is more effective because one mRNA molecule can generate multiple copies of a protein.[8, 119, 120] Relative to plasmid DNA (pDNA), mRNA possesses temporary bioactivity and functions outside the nucleus, leading to a more controllable protein expression and avoiding the potential insertion of the genome.[8, 119, 120] Although mRNA has become a potential class of therapeutic medicines, several aspects of mRNA, including the translatability, stability, immunogenicity, and intracellular delivery have to be improved in order to achieve clinical applications.[8, 119, 120] Increased translatability and stability can be achieved though modifying and optimizing the cap structure, 5’ and 3’ untranslated regions (UTRs), coding sequences, and other structural elements.[8, 119, 120] Incorporating modified nucleosides, such as pseudouridine and 2‑thiouridine into mRNA can reduce its immune-stimulatory effects.[8, 119, 120] Ultimately, intracellular delivery remains the major challenge for widespread applications of mRNA therapeutics.
Recently, many biomaterials have been developed for mRNA delivery, including lipids and lipid derived materials, polymers, and many others. [8, 39, 120, 124] Among them, lipids and lipid derived materials are the backbone for mRNA delivery systems.[8, 39, 124, 125] Cationic lipids, such as DOTMA (1,2-di-O-octadecenyl-3-trimethylammonium-propane), DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), zwitterionic DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), and lipofectamine have been widely studied in mRNA delivery.[39, 126] Meanwhile, ionizable lipid and lipid derived materials, which acquire positive charges at low pH and become neutral at physiological pH, have been developed for decreasing the toxicity and immunogenicity while retaining the delivery efficiency.[8] 1,2-dilinoleyloxy-3 dimethylaminopropane (DLin-DMA) and its derivatives, DLin-KC2-DMA and DLin-MC3-DMA (MC3) are representative ionizable lipid materials.[63, 127–132] In 2018, the nucleic acid drug Patisiran (ONPATTRO™) based on MC3 lipid have been approved by the US FDA.[124] Another ionizable lipid material named cKK-E12 has been investigated to deliver mRNA for multiple applications.[67, 133–135] Moreover, its derivative with unsaturated fatty chains, OF-02, and its biodegradable derivative, OF-Deg-Lin, showed efficient mRNA delivery in vivo.[136–138] The efficacy of ionizable lipid and lipid derived materials such as N,N,N-tris(2-aminoethyl) benzene-1,3,5-tricarboxamide (TT3)[40, 139], C12–200[133, 140–142], and 306Oi10[143] have also been proved for in vivo mRNA delivery. In addition, other ionizable lipids developed by Moderna Therapeutics[47, 144], Intellia Therapeutics[48], and Arcturus Therapeutics[145, 146] have been tested in preclinical studies.
Cationic polymers, such as polyamines, polypeptides, and block polymers, are another category of biomaterials for mRNA delivery. Polyethylenimine (PEI) is one of the most developed cationic polymers [39], but its toxicity has limited the therapeutic applications.[8, 124] In addition to PEI, chitosan[147–149], poly(β-amino esters) (PBAEs)[41, 150, 151], poly(glycoamidoamine) brushes (TarN3C10)[152], and N-substituted polyaspartamides (PAsp)[44, 153–157] have been used for mRNA delivery. Charge-altering releasable transporters (CARTs)[158–160], polyamidoamine (PAMAM) or polypropylenimine-based dendrimers[161, 162], and poly(lysine)-based polypeptides[163], have also been developed for in vivo mRNA delivery. Other biomaterials, including peptide complexes and inorganic materials for mRNA delivery, which mainly focus on cancer and other diseases, have been reviewed in the literature.[10, 34, 39, 121, 124]
In this section, we describe several representative mRNA delivery systems aiming at GARD therapy (Table 2), mainly based on liver and lung delivery biomaterials. Additionally, we highlight several mRNA delivery systems for other organs which may be further developed for GARD treatment.
Table 2.
List of mRNA delivery systems for the treatment of genetic and rare disease (GARD)
| Name of mRNA Therapeutics or mRNA | Delivery Material | Administration Route | Indication/Disease model | Target organ | Clinical Trial | Reference |
|---|---|---|---|---|---|---|
| hFIX mRNA | LNP (TT3) | i.v. | Hemophilia B | liver | Pre-clinical | [40] |
| hFIX mRNA | LNP (C12–200) | i.v. | Hemophilia B | liver | Pre-clinical | [168] |
| hFIX mRNA | LNP (ATX) | i.v. | Hemophilia B | liver | Pre-clinical | [145] |
| hADAMTS13 mRNA | LNP (ATX) | i.v. | Thrombotic thrombocytopenic purpura (TTP) | liver | Pre-clinical | [170] |
| hMUT mRNA | Ionizable LNP | i.v. | Methylmalonic academia (MMA) | liver | Phase 1/2 (NCT03810690) | [47] |
| FAH mRNA | Dendrimer LNP (5A2-SC8) | i.v. | Hereditary tyrosinemia type 1 (HT-1) | liver | Pre-clinical | [162] |
| hOTC mRNA | HMT (GalNAc and DOTAP) | i.v. | Ornithine transcarbamylase deficiency (OTCD) | liver | Pre-clinical | [177] |
| MRT5201 | MRT™ | i.v. | Ornithine transcarbamylase deficiency (OTCD) | liver | Phase 1/2 (NCT03767270) | [178] |
| hGLA mRNA | LNP (C12–200) | i.v. | Fabry disease | liver | Pre-clinical | [142] |
| hGLA mRNA | LNP (MC3) | i.v. | Fabry disease | liver | Pre-clinical | [129] |
| hPBGD mRNA | LNP (MC3) | i.v. | Acute intermittent porphyria (AIP) | liver | Pre-clinical | [130] |
| hUGT1A1 mRNA | Ionizable LNP | i.v. | Crigler-Najjar syndrome type 1 (CN-1) | liver | Pre-clinical | [181] |
| hSERPINA1 mRNA | Ionizable LNP | i.v. | Alpha-1 antitrypsin deficiency (AATD) | liver | Pre-clinical | [182] |
| hG6PC mRNA | Ionizable LNP | i.v. | Glycogen storage disease type 1 (GSD-1) | liver | Pre-clinical | [183] |
| hCFTR mRNA | LNP (MC3) | intranasal | Cystic fibrosis (CF) | lung | Pre-clinical | [131] |
| hCFTR mRNA | NP (chitosancoated PLGA) | i.t. or i.v. | Cystic fibrosis (CF) | lung | Pre-clinical | [149] |
| MRT5005 | MRT™ | nebulized | Cystic fibrosis (CF) | lung | Phase 1/2 (NCT03375047) | [178] |
| hCFTR mRNA | LUNAR LNP | nebulized | Cystic fibrosis (CF) | lung | Pre-clinical | [191] |
| hTE mRNA | lipofectamine 2000 | subcutaneous | Williams-Beuren syndrome (WBS) | skin | Pre-clinical | [198] |
i.v., intravenous; i.t., intratracheal; LNP, lipid and lipid derived nanoparticle; NP, nanoparticle; LUNAR, Lipid-enabled and Unlocked Nucleomonomer Agent-modified RNA; MRT™, mRNA Therapeutic Platform; HMT, Hybrid mRNA Technology; MC3, DLin-MC3-DMA.
3.1. Liver delivery of mRNA
For many GARDs, the liver is a preferred organ for mRNA-based PRTs because systemically administrated mRNA vehicles are predominantly trapped by the reticuloendothelial system (RES) in the liver. In addition, the interaction between apolipoprotein E (apoE) and biomaterials such as lipid and lipid derived materials facilitates the accumulation in the liver.[8, 18, 34, 39, 59, 120, 121, 124, 164, 165]
Hemophilia B is a blood genetic disease due to the deficiency of the blood coagulation factor IX (FIX).[166] Patients with hemophilia B are characterized by bleeding tendency, leading to recurrent bleeding episodes following trauma or spontaneous hemorrhages.[166] Repeated bleeding, especially into the joints, muscles, and soft tissues, causes chronic lesions including pain, deformities, and loss of movement.[166] Although the FIX PRT improves the outcome of hemophilia B, its short half-life, frequent intravenous (i.v.) administration method, and cost still limit the broader utilization.[167] Several lipid derived materials have been synthesized for mRNA-based PRT of Hemophilia B. For example, TT3 was developed for human FIX (hFIX) mRNA delivery.[40] After 6 hours of i.v. injection, the plasma concentration of FIX was 1740 ng/mL and the activity was 791 mIU/mL, which were well within the normal physiological values.[40] In another study, C12–200 was used to deliver hFIX mRNA to the liver of hemophilia B mice.[168] A single administration of lipid derived nanoparticles (LNPs) containing 0.5 mg/kg hFIX mRNA yielded 1800 ng/mL of FIX in the plasma.[168] The LNP formulation based on the ionizable ATX lipid developed by Arcturus Therapeutics was also prepared for delivering hFIX mRNA.[145] The therapeutic level of FIX in the plasma was maintained up to 6 days following a single i.v. administration of LNPs complexed with 2.0 mg/kg hFIX mRNA.[145]
Thrombotic thrombocytopenic purpura (TTP) is another blood genetic disease characterized by the clinical symptoms including haemolytic anaemia, thrombocytopenia, renal failure, and neurologic dysfunction.[169] One of the revealed pathogeneses of TTP is the inherited mutation of the zinc metalloproteinase gene (ADAMTS13).[169] The ATX-based mRNA delivery system provided an approach for TTP treatment.[170] One-time tail vein injection of formulated human ADAMTS13 (hADAMTS13) mRNA induced a therapeutic level of hADAMTS13 (140 ng/mL in the circulation at 24 h, and 90 ng/mL at 48 h) in ADAMTS13-deficient mice. Furthermore, therapeutic level was maintained for up to five days, which was significantly longer than the half-life (24 hours) of the recombinant human ADAMTS13 (rhADAMTS13) protein.[170]
Methylmalonic academia (MMA) is an amino acid metabolism disorder caused by the dysfunctional methylmalonyl-CoA mutase (MUT), leading to an excessive accumulation of methylmalonic acid.[171] Depending on the mutated genes, the metabolic disorder results in different clinical symptoms, including vomiting, acidosis, hyperammonemia, lethargy hypertonia, and failure to thrive.[171, 172] Although diagnosis and management have been improved, effective therapies remain elusive and patients still suffer from MMA associated complications and lifelong mortality.[171, 172] Systemic injection of human MUT (hMUT) mRNA, formulated with MC3 lipid, demonstrated a rapid (as early as 2 h after administration) and effective (maximum 90% at 24 h) reduction of plasma methylmalonic acid.[47] In addition, repeated administrations improved the growth and survival of MMA mice without obvious liver damage and inflammation.[47] Based on its mRNA platform, Moderna Therapeutics initiated a clinical trial (NCT03810690) to evaluate the mRNA-3704 in patients with MMA (Table 2).
Another amino acid metabolism disorder is hepatorenal tyrosinemia type 1 (HT-1) characterized by the mutated fumarylacetoacetate hydrolase (FAH), an important enzyme for tyrosine catabolism.[173–175] In HT-1 patients, the accumulation of tyrosine metabolites generally causes hepatic cirrhosis, renal dysfunction, neurologic crises, and cancer.[173–175] Clinical treatment of HT-1 combines a tyrosinerestricted diet and nitisinone, an inhibitor for preventing the formation of tyrosine metabolites.[175] However, long-term use of nitisinone increases the risks of developing hepatic neoplasms and liver failure.[162] In a recent report, the 5A2-SC8 dendrimer-based lipid nanoparticle (DLNP) was screened out to deliver human FAH (hFAH) mRNA in Fah knockout mice.[162] The mice were i.v. administrated with hFAH mRNA loaded DLNPs every 3 days for a month, during which hFAH expression was elevated in the liver.[162]
Ornithine transcarbamylase deficiency (OTCD) is also an amino acid metabolism disorder which is caused by the loss of ornithine transcarbamylase (OTC), a key enzyme in the urea cycle.[176] The deficiency of OTC results in hyperammonemia, giving rise to neurological injury, coma, and even death.[176] mRNA-based PRT is a promising alternative for liver transplant which is currently the only option for treating OTCD.[176] The Hybrid mRNA Technology (HMT) was developed for targeting expression of OTC in hepatocytes.[177] HMT contained two types of nanoparticles: the N-acetylgalactosamine (GalNAc) polymer micelle which allowed hepatic targeting and facilitated endosomal evasion, and the DOTAP LNP which protected mRNA from enzymatic degradation. After i.v. injection of OTC mRNA with HMT, a prolonged survival was observed in OTCD mice. Meanwhile, no obvious changes of cytokines or significant pathological lesions were detected in the blood and liver, respectively.[177] Another study for treating OTCD is MRT5201 developed by Translate Bio based on their proprietary mRNA therapeutic platform (MRT™).[178] MRT5201 aims to restore the OTC level in patients with OTCD using mRNA LNPs. Recently, a phase 1/2 clinical trial (NCT03767270) on MRT5201 has been launched (Table 2).
Fabry disease is a genetic disease due to the deficiency of lysosomal alpha galactosidase A (α-GLA).[179] The deficient α-GLA contributes to a progressive accumulation of glycolipids in many tissues and cell types, giving rise to acroparesthesia, angiokeratomas, myocardial infarction, renal failure, and other clinical symptoms.[179] Two recent studies explored the therapeutic potential of mRNA LNPs in mice with Fabry disease. DeRosa et al. encapsulated human GLA (hGLA) mRNA in C12–200-based LNPs and i.v. dosed each mouse with 1.0 mg/kg mRNA LNPs or recombinant hunam GLA (rhGLA) proteins.[142] One week after administration, hGLA proteins in the heart and kidneys of the LNP treated group was 10-fold more than the rhGLA protein treated group. Meanwhile, Fabry disease associated biomarkers, globotriaosylceramide (Gb3) and globotriaosylsphingosine (lyso-Gb3), decreased by 92% and 88% in the LNP treated group. In contrast, these biomarkers decreased by only 67% and 61% in the rhGLA protein treated group. Similar effects were observed from the study using MC3-based LNPs to deliver hGLA mRNA.[129, 144] A single i.v. administration restored the α-GLA activity with concomitant clearance of Gb3 and lyso-Gb3 in plasma and different tissues in mice with Fabry disease. Moreover, the therapeutic effects were maintained for up to six weeks.[129]
Acute intermittent porphyria (AIP), a rare metabolic disease, is correlated with hepatic deficiency of porphobilinogen deaminase (PBGD), leading to over-accumulated porphyrin precursors.[180] Patients with AIP typically experience abdominal pain, psychosis, hypertension, and tachycardia.[180] A recent preclinical study demonstrated that i.v. injection of human PBGD (hPBGD) mRNA encapsulated in MC3-based LNPs restored hepatic PBGD expression followed by a normalization of porphyrin precursor levels.[130] In AIP mice, a single dose alleviated mitochondrial dysfunction, hypertension, and pain. Importantly, the PBGD expression in the liver lasted for at least 10 days, which provided a full protection throughout the diseased period (5 to 7 days).[130]
Additionally, there are three other GARDs (Crigler-Najjar syndrome type 1, alpha-1 antitrypsin deficiency, and glycogen storage disease type 1) which are currently treated by mRNA LNPs in preclinical studies (Table 2).[181–183]
3.2. Lung delivery of mRNA
In addition to liver delivery, several lung delivery systems were recently reported in the literature.[97, 126] For delivery of mRNA to the lungs, i.v. and intratracheal (i.t.) administration were mainly investigated in the past.[126] In general, lung-targeted mRNA delivery via i.v. administration is challenging because a substantial amount of mRNA nanoparticles will be trapped in the liver and spleen.[126] Furthermore, pulmonary capillaries are non-fenestrated, hindering free exchanges of large molecules from the blood to the lung tissues.[126] However, several strategies have been developed for lung targeting mRNA delivery after systemic administration. For example, coupling pulmonary affinity ligands to nanoparticles can significantly increase mRNA delivery to the lungs.[42] Some biomaterials, interactions with serum proteins or surface receptors of pulmonary cells, may contribute to the lung specific accumulation.[150] The pathologic characteristics of certain lung diseases, such as hyper-vascularization, may also be utilized to facilitate nanoparticles reaching these areas. In contrast to i.v. administration, i.t. administration via suspension nebulization or dry powder inhalation can directly deliver mRNA to the airways of the lungs.[184] Moreover, using i.t. administration, mRNA can be delivered to a variety of cell types, such as alveolar cells, pulmonary endothelium, and immune cells.[126, 184]
Cystic fibrosis (CF) is associated with the defective cystic fibrosis transmembrane conductance regulator (CFTR), functioning as the cAMP mediated chloride channel on epithelial cells.[185, 186] Patients with CF usually experience repeated airway infections, chronic pulmonary inflammation, and respiratory distress.[187] Some small molecules that aim to restore the chloride transport function of the mutant CFTR have shown promising clinical results. However, their narrow application range, poor improvements in lung function, and side effects limit their utilization.[188–190] Recently, MC3-based LNPs were prepared to intranasally deliver human CFTR (hCFTR) mRNA to Cftr knockout mice.[131] On day 3 post-administration, chloride secretion reached 55% of that in healthy mice and the secretion maintained more than 15 days.[131] In a separate study, Haque et al. complexed hCFTR mRNA with chitosan-coated PLGA nanoparticles to relieve CF symptoms. The restored lung function was determined by increasing CFTR expression and improved forced expiratory volume (FEV).[149] Another clinical candidate, MRT5005, designed by Translate Bio to address CF through delivery of CFTR mRNA, has been studied in a clinical trial (NCT03375047, Table 2). MRT5005 will be administered by nebulization to the respiratory tract of adult subjects with CF and its safety will be evaluated in the clinical trial.[178] LUNAR-CF, developed by Arcturus Therapeutics, is another mRNA-based therapeutic for treating CF.[191] Preclinical studies indicated LUNAR nanoparticles effectively delivered mRNA into pulmonary epithelial cells and were resistant to the degradation in CF sputum during nebulization.[191]
Several mRNA delivery systems were inclined to accumulate in the lungs, and thus are propitious for treatment of pulmonary GARDs. Schrom et al. prepared lung-targeted nanoparticles by mixing mRNA with preassembled C12-(2–3-2) oligoalkylamines lipid micelles.[192, 193] Compared with other organs, the stronger luciferase signal in the lungs indicated the pulmonary specificity. Based on these results, they further i.v. delivered angiotensin-converting enzyme 2 (ACE2) mRNA, a therapeutic agent for lung fibrosis, by the lung-targeted nanoparticles. After 6 hours, obvious accumulation of ACE2 mRNA and proteins were observed in the lungs.[192, 193] Kaczmarek et al. developed a hybrid polymer–lipid nanoparticle consisting of PBAEs and lipid-PEG for lung mRNA delivery.[150] Through optimization of the polymer construction and nanoparticle formulation, they obtained a nanoparticle which showed comparable lung-specificity but two orders-of-magnitude more effective in mRNA delivery than the base nanoparticle.[194] Recently, the same group synthesized hyperbranched poly(beta amino esters) (hPBAEs) to deliver mRNA within aerosol formulation to the lungs.[151] The hPBAE nanoparticles were stable without lipid-PEG and withstood the shearing forces from the vibrating mesh nebulizer. After 24 h of nebulization, luciferase signal was observed in the lungs but not in the spleen and liver.[151] Yan et al. identified the top material named PE4K-A17–0.33C12 from a poly-(trimethylolpropane allyl ether-co-suberoyl chloride) library[195]. The PE4K-A17–0.33C12 nanoparticles formulated with 5% pluronic F127 resulted in obvious luciferase expression in the lungs after systematic administration in mice.[195] An alternative strategy for selectively delivering mRNA to the lungs is coupling nanoparticles with affinity ligands. Platelet endothelial cell adhesion molecule-1 (PECAM-1) is a typical target of endothelial cells of the lungs. Parhiz et al. developed PECAM-1 antibody modified LNPs for lung targeting delivery of mRNA.[42] Relative to non-targeted LNPs, systemic administration of PECAM-1 coupled mRNA LNPs exhibited approximately 200-fold inhibition and 25-fold elevation of luciferase expression in the liver and the lungs, respectively.[42]
3.3. Other organ delivery of mRNA
Additionally, many biomaterials are developed and applied for delivering mRNA to other organs, such as skin, eye, nasal cavity, and heart. For these organs, topical delivery is the main administration route.
Williams-Beuren syndrome (WBS) is caused by spontaneous microdeletion of about 27 genes from chromosome 7q11.23.[196, 197] Patients with WBS suffer from approximately 50 % of elastin loss which is associated with complications such as cardiovascular diseases, nervous disorders, and developmental problems.[196, 197] Lescan et al. utilized tropoelastin (TE) mRNA/lipofectamine 2000 complexes to restore the elastin level in mesenchymal stem cells isolated from a WBS patient. After 24 h of treatment, about 13-fold higher amounts of elastin was observed compared to untreated cells. [198] Furthermore, in porcine skin, over 1.2-fold increased expression of elastin was observed after the subcutaneous delivery of the complexes.[198]
Inherited retinal degeneration (IRD) is caused by progressive death of retinal photoreceptors.[199] Patients typically experience visual disturbances in childhood and gradually lose their central vision.[199] Recently, Patel et al. studied the kinetics and localization of photoreceptor expression mediated by MC3-based mRNA LNPs in the eyes of albino BALB/c mice.[132] After a subretinal injection, the photoreceptor expression persisted for 96 h (began within 4 h and reached peak expression at 24 h), and mainly distributed in the retinal pigmented epithelium (RPE).[132]
There are also some mRNA delivery systems that may be applied to GARDs in other organs. Polyplex nanomicelles composed of PEG and poly (N-(N-(2-aminoethyl)-2-aminoethyl) aspartamide) (PAsp (DET)) were designed for the delivery of the mRNA encoding neutrophilic factor into the nasal cavity of the mice with olfactory dysfunction.[156] Intranasal administration of the nanomicelles promoted the recovery of their olfactory function. Magadum et al. recently reviewed mRNA delivery systems in cardiac therapy.[200] For example, an intramyocardial injection of mRNA expressing human vascular endothelial growth factor A (VEGF-A) with RNAiMAX decreased the morbidity of mice with myocardial infarction following the repair of heart function.[201]
4. Genome editing systems containing RNA components
The advent of genome editing tools, including meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9), CRISPR from Prevotella and Francisella 1 (CRISPR-Cpf1, CRISPR-Cas12a), and many others have enabled gene engineering to potentially correct disease-causing mutations.[202–206] Among them, CRISPR-Cas9 is one of the most widely used genome editing systems for in vivo applications. The CRISPR–Cas9 system composes of the Cas9 nuclease, a single guide RNA (sgRNA) and if needed, a donor DNA template for homology-directed repair (HDR).[202, 205, 207] The Cas9 nuclease utilizes a sgRNA to recognize the target sequence and subsequently cleaves the genome at the specific site. The system can be designed to edit a large number of sequences in the genome with different sgRNAs and donor DNA templates, thus making it a powerful genome editing tool.[202, 205, 207] However, there are some challenges for in vivo applications of therapeutic genome editing tools.[205–208] Firstly, genome editing components must reach the cell nucleus to achieve therapeutic effects. Thus, extracellular and intracellular barriers need to be overcome, including avoiding enzymatic degradation in the blood, circumventing clearance by the reticuloendothelial system (RES), penetrating cell membranes, escaping endosome entrapment, and infiltrating nuclear membranes. Secondly, the complexity and large molecular weight of these components are significant challenges for delivery materials. For example, the molecular weight of Cas9 protein is about 160 kDa, and the phosphate backbones of sgRNA and DNA are highly negatively charged. These features hinder the efficiency of delivery materials. Thirdly, genome editing components should be delivered to target tissues and edit specific sequences in a controllable manner to prevent off-target events and immune responses. Therefore, an effective delivery system for genome editing tools needs to overcome these challenges in order to achieve clinical applications in vivo.
Genome editing delivery systems are generally classified into categories of physical delivery, viral delivery, and non-viral delivery. Physical delivery such as electroporation and microinjection have been widely used, which was discussed by a few review articles.[207, 209, 210] Viral delivery, such as lentivirus (LV) and adeno-associated virus (AAV), is also an effective method to deliver genome editing components.[202, 210] In addition, researchers investigated numerous non-viral materials for delivery of genome editing tools. These materials are similar to those used for mRNA delivery.[202, 205, 207–211] Targeted delivery is crucial for achieving genome editing specificity and reducing off-target effects. Although many targeting strategies have been developed for drug delivery, they have not be thoroughly applied to genome editing delivery systems. Currently, tissue targeting genome editing delivery has been limited to topical administration except the liver. Therefore, targeted delivery of genome editing tools need be addressed in order to facilitate future clinical applications.
In this section, we summarize representative non-viral materials applied to deliver genome editing components containing mRNA or/and sgRNA for the treatment of genetic and rare diseases (GARDs). (Table 3). We also review some typical non-viral materials which may be developed for delivering genome editing components to treat GARDs in the future.
Table 3.
List of CRISPR and other genome editing delivery systems for the treatment of genetic and rare disease (GARD)
| Name of the genome editing system | Delivery Material | Administration Route | Indication/Disease model | Target organ | Clinical Trial | Reference |
|---|---|---|---|---|---|---|
| Cas9 mRNA and sgRNA | LNP (LP01) | i.v. | Hereditary transthyretinmediated amyloidosis (hATTR) | liver | Pre-clinical | [48] |
| Cas9 mRNA, gRNA, and DNA template | LNP (C12–200) and AAV vector | i.v. | Hereditary tyrosinemia type 1 (HT-1) | liver | Pre-clinical | [141] |
| Cas9 mRNA and sgRNA | LNP (ZALs) | i.v. | Ai9 mouse | liver, lung, and spleen | Pre-clinical | [214] |
| Cas9 mRNA and sgRNA | LNP (TT3) | i.v. | Autosomal dominant hypercholesterolemia (ADH) | liver | Pre-clinical | [139] |
| Cas9 mRNA and enhanced sgRNA | LNP (cKK-E12) | i.v. | Autosomal dominant hypercholesterolemia (ADH) | liver | Pre-clinical | [135] |
| ZFN mRNA and DNA template | NP (chitosan-coated PLGA) and AAV vector | i.t. | Surfactant protein B (SP-B) deficiency | lung | Pre-clinical | [219] |
| SB100X-mRNA and pDNA | NP (peptide-poloxamine) | i.t. | Cystic fibrosis (CF) | lung | Pre-clinical | [45] |
| Cas9 and sgRNA | CRISPR-Gold | intracranial | Fragile X syndrome (FXS) | brain | Pre-clinical | [44] |
| Cas9 and sgRNA | LNP (8-O14B) | intracranial | Ai9 mouse | brain | Pre-clinical | [221] |
| Cas9, gRNA, and DNA template | CRISPR-Gold | intramuscular | Duchenne muscular dystrophy (DMD) | muscle | Pre-clinical | [224] |
| Cpf1 and 5’ extended crRNA | NP (PAsp(DET)) | intramuscular | Ai9 mouse | muscle | Pre-clinical | [157] |
| Cas9 and sgRNA | lipofectamine 2000 | intraaural | Transmembrane channellike protein 1(Tmc1) related deafness | ear | Pre-clinical | [226] |
i.v., intravenous; i.t., intratracheal; LNP, lipid and lipid derived nanoparticle; NP, nanoparticle.
4.1. Liver delivery of genome editing components
Transthyretin (TTR), secreted by the liver and cerebral choroid plexus, is a transport protein for retinol and thyroxine.[212, 213] The aggregation of mutated transthyretins is associated with hereditary transthyretin amyloidosis (hATTR).[212, 213] In a recent study, LP01-based lipid derived nanoparticles (LNPs) were developed for delivering Cas9 mRNA and sgRNA to edit hepatic Ttr gene in mice and rats.[48] Following a single intravenous (i.v.) injection, over 90% reduction of serum TTR was observed and the knockdown effect persisted for at least 12 months. Moreover, multiple administrations achieved cumulative editing efficacy without obvious cytokine stimulation and body weight loss.[48] The combined delivery of Cas9 mRNA and sgRNA by C12–200-based LNPs and DNA donor by AAV was used to treat hepatorenal tyrosinemia type 1 (HT-1) by correcting the fumarylacetoacetate hydrolase (Fah) gene on Fah knockout mice.[141] After a single treatment, the FAH expression in more than 6% of hepatocytes was repaired and the symptoms of hepatic damage were prevented in the following 30 days. Another material named zwitterionic amino lipid (ZAL) was synthesized in order to deliver genome editing components.[214] With i.v. injection of ZAL nanoparticles containing Cas9 mRNA and sgRNA, the stop gene sequences in Rosa26tdTomato mice were edited, leading to strong tdTO fluorescent signal in the liver, lungs and kidneys.[214]
Autosomal dominant hypercholesterolemia (ADH) is associated with mutations in the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene.[215] Patients with ADH usually suffer from coronary heart disease because of the high level of low-density lipoprotein cholesterol.[215] TT3 LNPs were used to deliver Cas9 mRNA and sgRNA to edit Pcsk9 gene in vivo.[139] After i.v. injection of Cas9 mRNA TT3 LNPs following the injection of sgRNA TT3 LNPs, the PCSK9 protein level was remarkably decreased in C57BL/6 mice.[139] In another study, Anderson et al. developed enhanced sgRNA (e-sgRNA) by incorporating chemically modified nucleotides into guide RNA sequences.[135] They formulated Cas9 mRNA and e-sgRNA targeting Pcsk9 with cKK-E12 LNPs.[134, 216] Following a single i.v. injection, hepatic PCSK9 protein was reduced by 80% and plasma cholesterol levels were reduced by about 40%.[135]
4.2. Lung delivery of genome editing components
Surfactant protein B (SP-B) deficiency is a lung genetic disease caused by mutated SP-B gene.[217] SP-B is essential for maintaining alveolar stability by interacting with lipid molecules to relieve surface tension.[217] Newborn infants with SP-B deficiency typically develop fatal respiratory failure within the first year.[217, 218] Currently, no effective treatment for SP-B deficiency is available.[218] Mahiny et al. utilized chitosan-PLGA nanoparticles to deliver ZFN mRNA and TALEN mRNA.[219] According to the efficiency of HDR, they finally chose ZFN mRNA loaded chitosan-PLGA nanoparticles to treat SP-B deficiency in mice.[219] Following intratracheal (i.t.) co-administration of nanoparticles encapsulating ZFN mRNA and AAV containing a DNA donor, SP-B expression was maintained for as long as 20 days in the lungs, which prevented the deterioration of lung function and resulted in longer lifetimes compared with negative controls.[219]
The peptide–poloxamine nanoparticle containing the Sleeping Beauty (SB) transposon system, which consists of SB transposon (pDNA) and transposase-encoding mRNA, was investigated to knock in the cystic fibrosis transmembrane conductance regulator (CFTR) gene in lung airway epithelium of cystic fibrosis (CF) mice. With the hydrophobic moiety of the peptide, the nanoparticle can overcome both the extracellular and intracellular barriers to achieve effective gene delivery in vivo.[45] After three-time i.t. administration, CFTR expression lasted for 18 weeks in CF mice and the level was similar to that of the wild type group. Meanwhile, lung pathohistological sections indicated no visible signs of inflammatory infiltrates and blood sample analysis showed a normal level of white blood cells.[45]
4.3. Brain delivery of genome editing components
Fragile X syndrome (FXS) is a familial mental disease caused by abnormal trinucleotide sequence CGG in the Fragile X mental retardation 1 (FMR1) gene.[220] The lack of effective therapeutic options causes patients with FXS to develop intellectual disabilities and autism spectrum disorders (ASDs).[220] Recently, CRISPR-Gold consisting of DNA functionalized gold nanoparticles and the PAsp (DET) polymer was developed for delivering Cas9 ribonucleoproteins (RNPs) to knock out the mGluR5 gene. The overexpressed signaling of mGluR5 gene is associated with both FXS and ASDs.[44] Through intracranial injection of 2 μL of CRISPR-Gold, both the mRNA and protein levels of mGluR5 decreased 40–50% in FXS mice. Furthermore, the treated FXS mice significantly resumed exaggerated repetitive behaviors after three weeks.[44]
The bioreducible LNP’s encapsulation of negatively charged Cre recombinase or anionic Cas9 RNPs by electrostatic assembly may lead to an approach for correcting mental disorders.[221] The disulfide bonds in the bioreducible lipids were degradable in response to the intracellular environment, which promoted intracellular delivery efficiency and reduced lipid toxicity. After six days of intracranial injection of Cre recombinase complexed with bioreducible lipid 8-O14B, visible tdTomato signal in the brain of Ai9 mice indicated an effective DNA recombination.[221]
4.4. Muscle delivery of genome editing components
Duchenne muscular dystrophy (DMD) is an inherited muscle degeneration due to the dysfunctional dystrophin protein which connects the muscle cytoskeleton to surrounding extracellular matrix to maintain muscle integrity and strength.[222, 223] Mutations result in progressive muscle degeneration and loss of mobility.[222, 223] Although life expectancy has been extended due to advanced management, effective therapies for DMD are still urgently needed.[222, 223] Lee et al. used CRISPR-Gold to correct the mutated gene in DMD mice.[224] After two weeks of intramuscular injection of CRISPR-Gold with cardiotoxin in DMD mice, 5.4% of the dystrophin gene was repaired and obvious increase of dystrophin protein was observed in the treated muscle tissues. Restored muscle function was also demonstrated by increasing limb tension.[224]
PAsp(DET) polymer was designed to in vivo deliver Cpf1 and 5′ extended crRNA.[157] The 5′ extension of crRNA improved the serum stability and enhanced the nanoparticle formulation with PAsp(DET) polymer, resulting in a higher in vivo delivery efficiency than wild type Cpf1 RNPs. Two weeks after intramuscular injection of PAsp(DET) polymer encapsulating 200 pmol Cpf1 and 200 pmol 5′ extended crRNA on Rosa26tdTomato mice, the stop sequences of tdTomato were effectively deleted, and robust tdTomato signal was observed.[157]
4.5. Ear delivery of genome editing components
TMC1 (transmembrane channel-like protein 1) is an essential element for auditory transduction in hair cells.[225] Mutations in TMC1 result in inherited hearing loss. [225] Most recently, Gao et al. applied lipofectamine 2000 to deliver Cas9 RNPs to disrupt the mutant Tmc1 for the treatment of genetic deafness.[226] The intraaural injection of Cas9 RNPs/lipofectamine 2000 complexes into the scala media of neonatal Tmc1Bth/+ mice significantly increased the survival of hair cells. Additionally, lower auditory response thresholds and improved acoustic responses were observed in injected mice.[226]
4.6. Other biomaterials for genome editing delivery
Many other distinctive delivery materials have been reported for delivering genome editing components and can possibly be developed for GARDs in the future. Ramakrishna et al. demonstrated that conjugating Cas9 and sgRNA with cell-penetrating peptides (CPPs) enhanced intracellular delivery efficiency because of the membrane-disturbed properties of CPPs.[227] Similarly, Wang et al. synthesized a membrane-penetrating cationic α-helical polypeptide which enhanced cellular internalization of Cas9 plasmid and sgRNA.[228] In addition to cellular uptake, endosomal trap is another barrier for genome editing delivery systems. Some materials, such as zeolitic imidazole frameworks (ZIFs) [229] and graphene oxide (GO)-PEG-polyethylenimine (PEI) [230] have been investigated to facilitate endosomal evasion. Furthermore, Mout et al. prepared cationic arginine gold nanoparticles (ArgNPs) to directly deliver Cas9 RNPs into the cellular cytoplasm without endocytosis.[231] For non-viral vectors, good biocompatibility enables the reduction of side effects and improves tolerability. Biodegradable lipid derived materials designed by Zhang et al. achieved efficient delivery of Cas9 mRNA in vivo without significant organic damage.[232] Timin et al. used hybrid microcarriers consisting of biocompatible SiO2 capsules and degradable-polymer coatings for CRISPR-Cas9 delivery.[233] Other methods based on nucleic acid materials, including DNA nanoclews [234] and RNA aptamer-streptavidin complexes [235], were also explored for delivering gene editing components.
5. MicroRNA (miRNA) and long non coding RNA (LncRNA)
miRNA is small, non-coding RNA which regulates gene expression in biological systems primarily by silencing the target mRNA.[236] miRNA shares many similarities with siRNA. Specifically, both miRNA and siRNA could mediate gene silencing post-transcriptionally by targeting the mRNA. However, miRNA may regulate multiple mRNA targets whereas siRNA inhibits one specific mRNA target.[237] Studies have identified that the dysregulation of miRNA is closely associated with various disease statuses.[237, 238] Therefore, molecules that mimic the function of endogenous miRNA (miRNA mimics) as well as molecules that target endogenous miRNA (antimiRs) were widely investigated in preclinical studies.[237, 238] While many miRNA mimics and antimiRs have been focused on cancer therapies due to their biological roles and properties, several miRNA related therapies were reported for genetic and rare diseases.[237, 239] For example, MRG-201, developed by miRagen Therapeutics, is a cholesterol-conjugated miRNA duplex used for the treatment of Scleroderma and Keloids.[240] MRG-201 is a synthetic miRNA mimic of miR-29 designed to reduce the expression of collagen and other scar formation related proteins.[238, 241] MRG-201 is now in Phase 2 clinical trials (NCT03601052). Recently, brain targeted nanoparticles for miRNA delivery have been reported. In 2011, Hwang et al. reported that the rabies virus glycoprotein (RVG)-labeled non-toxic SSPEI (RVG-SSPEI) nanomaterials were able to facilitate the in vivo delivery of miR-124a to neuron cells.[242] Combining RVG-SSPEI nanoparticles with mannitol solution could further improve the delivery of miR-124a across the BBB to the brain.[242]
Although extensive preclinical studies on miRNA therapeutics have been carried out, only a small fraction of them have reached phases of clinical development.[238] Besides, only a small portion of miRNA therapeutics targets diseases other than cancer.[237] It is still of great interest to develop more functional miRNA delivery biomaterials to enable both the better understanding the roles of miRNA in vivo and miRNA-based gene therapy on GARDs.
LncRNA is another important type of non-coding RNA molecule, which is typically more than 200 bases in length.[243] While only a small proportion of lncRNA transcripts have been functionally characterized to date, lncRNA is found to be involved in many different biological processes.[244] Therefore, it is realized that mutations affecting lncRNA expression could lead to diseases, as do protein coding genes. Numerous studies in the past several years showed that altered lncRNA transcriptional levels were associated with a variety of genetic disorders ranging from monogenic to complex disorders.[245] LncRNA may thus become a therapeutic target for treating GARDs.[246]
6. Conclusions and future perspectives
In summary, we provide an overview of delivery biomaterials for RNA-based therapies, with an emphasis on genetic and rare diseases (GARDs). Recently, an increasing number of RNA molecules are emerging as promising breakthrough therapies for many diseases.[3] A variety of delivery systems have been explored and developed to enable the RNA therapeutics for treating different GARDs (Figure 3).
Figure 3.
A list of biomaterials and RNAs that have been explored for the treatment of different GARDs.
Among various delivery materials, lipid and lipid derived nanoparticles (LNPs) exhibit compelling delivery efficiency towards many different types of RNAs, ranging from small RNA molecules such as siRNA to much larger mRNA molecules or more complicated genome editing systems containing RNA components. By exploring diverse chemical scaffolds of lipids, researchers can develop LNPs with optimized physiochemical and biological properties to match a specific application. However, LNPs still have some limitations. For example, a majority of LNPs accumulate in the liver which, to some extent, limits the use of LNPs to treatment of liver-oriented diseases. Additional efforts are needed in order to further develop new LNPs and expand their applications in different organs. This may be promoted by high-throughput technologies, optimizing screening methodologies, and modulating cellular signal pathways to identify tissue specific and highly efficient delivery systems.[133, 247–249]
Ligand conjugations have achieved prominent success in delivering small RNA molecules with the development of RNA chemical modification technology. Liver targeting N-acetylgalactosamine (GalNAc) conjugation represents the lead conjugation strategy and has a growing number of RNA therapeutics in clinical trials. One of the most significant advantages of GalNAc-siRNA conjugations is that they can be administered subcutaneously, which improves the compliance of treatment. However, new ligands, especially for targeting organs other than the liver, need to be discovered. Linker chemistry can be further optimized to maximize the therapeutic window of the conjugates. Meanwhile, chemical modification of large RNA molecules is also an important area that is worth further investigation.
In addition, many other biomaterials, such as cell derived vehicles, gold nanoparticles, polymeric nanoparticles, and DNA/RNA nanotechnology, were also used as delivery systems for in vivo RNA therapeutics. Their applications in treating GARDs have not been widely explored yet. More categories of delivery systems await application for RNA-based GARD therapies.
Although many RNA delivery biomaterials have been explored for treating GARDs, a large number of rare diseases are still awaiting effective treatments. In addition to the need for deep understandings of the rare diseases and identifications of new therapeutic targets, more efforts are needed to expand the chemistry of biomaterials and enable the delivery of RNA therapeutics to different target organs and cell populations in GARD applications.
Acknowledgements
Y.D. acknowledges the support from the Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences as well as the start-up fund from the College of Pharmacy at The Ohio State University. W.Z. acknowledges the support from the Professor Sylvan G. Frank Graduate Fellowship.
Footnotes
Conflicts of interest
The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Chen B, Altman RB, Opportunities for developing therapies for rare genetic diseases: focus on gainof-function and allostery, Orphanet J Rare Dis 12(1) (2017) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].<https://rarediseases.info.nih.gov/diseases>.
- [3].Sun W, Zheng W, Simeonov A, Drug discovery and development for rare genetic disorders, Am J Med Genet A 173(9) (2017) 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lapteva L, Vatsan R, Purohit-Sheth T, Regenerative Medicine Therapies for Rare Diseases, Translational science of rare diseases 3(3–4) (2018) 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ekins S, Industrializing rare disease therapy discovery and development, Nature Biotechnology 35 (2017) 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kaczmarek JC, Kowalski PS, Anderson DG, Advances in the delivery of RNA therapeutics: from concept to clinical reality, Genome Medicine 9(1) (2017) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, Melero I, An RNA toolbox for cancer immunotherapy, Nature Reviews Drug Discovery 17 (2018) 751. [DOI] [PubMed] [Google Scholar]
- [8].Hajj KA, Whitehead KA, Tools for translation: non-viral materials for therapeutic mRNA delivery, Nature Reviews Materials 2 (2017) 17056. [Google Scholar]
- [9].Kole R, Krainer AR, Altman S, RNA therapeutics: beyond RNA interference and antisense oligonucleotides, Nat Rev Drug Discov 11(2) (2012) 125–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Meng Z, O’Keeffe-Ahern J, Lyu J, Pierucci L, Zhou D, Wang W, A new developing class of gene delivery: messenger RNA-based therapeutics, Biomater Sci 5(12) (2017) 2381–2392. [DOI] [PubMed] [Google Scholar]
- [11].Sullenger BA, Nair S, From the RNA world to the clinic, Science 352(6292) (2016) 1417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pardi N, Hogan MJ, Porter FW, Weissman D, mRNA vaccines - a new era in vaccinology, Nat Rev Drug Discov 17(4) (2018) 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bryant CE, Sutherland S, Kong B, Papadimitrious MS, Fromm PD, Hart DNJ, Dendritic cells as cancer therapeutics, Semin Cell Dev Biol 86 (2019) 77–88. [DOI] [PubMed] [Google Scholar]
- [14].Garg AD, Coulie PG, Van den Eynde BJ, Agostinis P, Integrating Next-Generation Dendritic Cell Vaccines into the Current Cancer Immunotherapy Landscape, Trends Immunol 38(8) (2017) 577–593. [DOI] [PubMed] [Google Scholar]
- [15].Kramps T, Elbers K, RNA Vaccines: Methods and Protocols, Springer; New York: 2016. [Google Scholar]
- [16].Pereira TC, Lopes-Cendes I, Emerging RNA-based drugs: siRNAs, microRNAs and derivates, Cent Nerv Syst Agents Med Chem 12(3) (2012) 217–32. [DOI] [PubMed] [Google Scholar]
- [17].Hu M, Ni Q, Yang Y, Luo J, RNAi-based Gene Therapy for Blood Genetic Diseases, (2016). [Google Scholar]
- [18].Cullis PR, Hope MJ, Lipid Nanoparticle Systems for Enabling Gene Therapies, Mol Ther 25(7) (2017) 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dowdy SF, Overcoming cellular barriers for RNA therapeutics, Nat Biotechnol 35(3) (2017) 222–229. [DOI] [PubMed] [Google Scholar]
- [20].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nature Reviews Genetics 15(8) (2014) 541. [DOI] [PubMed] [Google Scholar]
- [21].Sasaki S, Guo S, Nucleic Acid Therapies for Cystic Fibrosis, Nucleic Acid Ther 28(1) (2018) 1–9. [DOI] [PubMed] [Google Scholar]
- [22].Tam YYC, Chen S, Cullis PR, Advances in Lipid Nanoparticles for siRNA Delivery, Pharmaceutics 5(3) (2013) 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lundstrom K, Viral Vectors in Gene Therapy, Diseases (Basel, Switzerland) 6(2) (2018) 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jasinski D, Haque F, Binzel DW, Guo P, Advancement of the Emerging Field of RNA Nanotechnology, ACS Nano 11(2) (2017) 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao K, Huang L, Nonviral methods for siRNA delivery, Molecular pharmaceutics 6(3) (2009) 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Finer M, Glorioso J, A brief account of viral vectors and their promise for gene therapy, Gene Therapy 24 (2017) 1. [DOI] [PubMed] [Google Scholar]
- [27].Prasad V, Tisagenlecleucel — the first approved CAR-T-cell therapy: implications for payers and policy makers, Nature Reviews Clinical Oncology 15 (2017) 11. [DOI] [PubMed] [Google Scholar]
- [28].Spencer HT, Riley BE, Doering CB, State of the art: gene therapy of haemophilia, Haemophilia 22 Suppl 5 (2016) 66–71. [DOI] [PubMed] [Google Scholar]
- [29].Jones MR, Seeman NC, Mirkin CA, Nanomaterials. Programmable materials and the nature of the DNA bond, Science 347(6224) (2015) 1260901. [DOI] [PubMed] [Google Scholar]
- [30].Seeman NC, Nanomaterials based on DNA, Annu Rev Biochem 79 (2010) 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mirkin CA, Petrosko SH, Spherical Nucleic Acids: Adding a New Dimension to Nucleic Acids and Clinical Chemistry, Clin Chem 64(6) (2018) 971–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jiang Q, Liu S, Liu J, Wang ZG, Ding B, Rationally Designed DNA-Origami Nanomaterials for Drug Delivery In Vivo, Adv Mater (2018) e1804785. [DOI] [PubMed] [Google Scholar]
- [33].Liu Q, Wang D, Xu Z, Huang C, Zhang C, He B, Mao C, Wang G, Qian H, Targeted Delivery of Rab26 siRNA with Precisely Tailored DNA Prism for Lung Cancer Therapy, Chembiochem (2019). [DOI] [PubMed] [Google Scholar]
- [34].Islam MA, Reesor EKG, Xu Y, Zope HR, Zetter BR, Shi J, Biomaterials for mRNA delivery, Biomaterials science 3(12) (2015) 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, Lee GY, Zhou K, Zope H, Yu M, Cao W, Oswald JT, Dinarvand M, Mahmoudi M, Langer R, Kantoff PW, Farokhzad OC, Zetter BR, Shi J, Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA, Nature Biomedical Engineering 2(11) (2018) 850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miller JB, Siegwart DJ, Design of synthetic materials for intracellular delivery of RNAs: From siRNA-mediated gene silencing to CRISPR/Cas gene editing, Nano Research (2018) 1–28. [Google Scholar]
- [37].Conceicao M, Mendonca L, Nobrega C, Gomes C, Costa P, Hirai H, Moreira JN, Lima MC, Manjunath N, Pereira de Almeida L, Intravenous administration of brain-targeted stable nucleic acid lipid particles alleviates Machado-Joseph disease neurological phenotype, Biomaterials 82 (2016) 124–37. [DOI] [PubMed] [Google Scholar]
- [38].Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG, Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity, Nature Communications 5 (2014) 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li B, Zhang X, Dong Y, Nanoscale platforms for messenger RNA delivery, Wiley Interdiscip Rev Nanomed Nanobiotechnol 11(2) (2019) e1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li B, Luo X, Deng B, Wang J, McComb DW, Shi Y, Gaensler KM, Tan X, Dunn AL, Kerlin BA, Dong Y, An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo, Nano Lett 15(12) (2015) 8099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kaczmarek JC, Kauffman KJ, Fenton OS, Sadtler K, Patel AK, Heartlein MW, DeRosa F, Anderson DG, Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells, Nano Lett 18(10) (2018) 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parhiz H, Shuvaev VV, Pardi N, Khoshnejad M, Kiseleva RY, Brenner JS, Uhler T, Tuyishime S, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D, Muzykantov VR, PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake, J Control Release 291 (2018) 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li B, Zhao W, Luo X, Zhang X, Li C, Zeng C, Dong Y, Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency, Nat Biomed Eng 1(5) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY, Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours, Nature Biomedical Engineering 2(7) (2018) 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guan S, Munder A, Hedtfeld S, Braubach P, Glage S, Zhang L, Lienenklaus S, Schultze A, Hasenpusch G, Garrels W, Stanke F, Miskey C, Johler SM, Kumar Y, Tümmler B, Rudolph C, Ivics Z, Rosenecker J, Self-assembled peptide–poloxamine nanoparticles enable in vitro and in vivo genome restoration for cystic fibrosis, Nature Nanotechnology 14(3) (2019) 287–297. [DOI] [PubMed] [Google Scholar]
- [46].Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair, Nat Biomed Eng 1 (2017) 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].An D, Schneller JL, Frassetto A, Liang S, Zhu X, Park J-S, Theisen M, Hong S-J, Zhou J, Rajendran R, Systemic messenger RNA therapy as a treatment for methylmalonic acidemia, Cell reports 21(12) (2017) 3548–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Shah RR, Shah A, Ling D, Growe J, Pink M, Rohde E, Wood KM, Salomon WE, Harrington WF, Dombrowski C, Strapps WR, Chang Y, Morrissey DV, A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing, Cell Rep 22(9) (2018) 2227–2235. [DOI] [PubMed] [Google Scholar]
- [49].Weng Y, Xiao H, Zhang J, Liang X-J, Huang Y, RNAi therapeutic and its innovative biotechnological evolution, Biotechnology Advances (2019). [DOI] [PubMed] [Google Scholar]
- [50].Lares MR, Rossi JJ, Ouellet DL, RNAi and small interfering RNAs in human disease therapeutic applications, Trends in biotechnology 28(11) (2010) 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Huang Y, Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics, Molecular therapy. Nucleic acids 6 (2017) 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I, RNAi-mediated gene silencing in non-human primates, Nature 441(7089) (2006) 111–4. [DOI] [PubMed] [Google Scholar]
- [53].Ledford H, Gene-silencing technology gets first drug approval after 20-year wait, Nature 560(7718) (2018) 291–292. [DOI] [PubMed] [Google Scholar]
- [54].Zatsepin TS, Kotelevtsev YV, Koteliansky V, Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside, International journal of nanomedicine 11 (2016) 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Leachman SA, Hickerson RP, Schwartz ME, Bullough EE, Hutcherson SL, Boucher KM, Hansen CD, Eliason MJ, Srivatsa GS, Kornbrust DJ, Smith FJ, McLean WI, Milstone LM, Kaspar RL, First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder, Mol Ther 18(2) (2010) 442–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Suhr OB, Coelho T, Buades J, Pouget J, Conceicao I, Berk J, Schmidt H, Waddington-Cruz M, Campistol JM, Bettencourt BR, Vaishnaw A, Gollob J, Adams D, Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study, Orphanet J Rare Dis 10 (2015) 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DW, Gollob JA, Suhr OB, Safety and efficacy of RNAi therapy for transthyretin amyloidosis, N Engl J Med 369(9) (2013) 819–29. [DOI] [PubMed] [Google Scholar]
- [58].Wood H, FDA approves patisiran to treat hereditary transthyretin amyloidosis, Nature Reviews Neurology 14(10) (2018) 570–570. [DOI] [PubMed] [Google Scholar]
- [59].Kulkarni JA, Cullis PR, van der Meel R, Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility, Nucleic Acid Ther 28(3) (2018) 146–157. [DOI] [PubMed] [Google Scholar]
- [60].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, Fougerolles A, Maier MA, Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms, Mol Ther 18(7) (2010) 1357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Huang L, Liu Y, In vivo delivery of RNAi with lipid-based nanoparticles, Annu Rev Biomed Eng 13 (2011) 507–30. [DOI] [PubMed] [Google Scholar]
- [62].I. Sangamo Therapeutics, Deep Dive on Lipid Nanoparticle (LNP) Delivery System for Nucleic Acid Therapy. <https://www.investorvillage.com/smbd.asp?mb=1933&mn=84687&pt=msg&mid=16754759>, 2017.
- [63].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ, Rational design of cationic lipids for siRNA delivery, Nature Biotechnology 28 (2010) 172. [DOI] [PubMed] [Google Scholar]
- [64].Butler JS, Chan A, Costelha S, Fishman S, Willoughby JL, Borland TD, Milstein S, Foster DJ, Goncalves P, Chen Q, Qin J, Bettencourt BR, Sah DW, Alvarez R, Rajeev KG, Manoharan M, Fitzgerald K, Meyers RE, Nochur SV, Saraiva MJ, Zimmermann TS, Preclinical evaluation of RNAi as a treatment for transthyretin-mediated amyloidosis, Amyloid 23(2) (2016) 109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG, Lipid-like materials for low-dose, in vivo gene silencing, Proceedings of the National Academy of Sciences 107(5) (2010) 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo, Angewandte Chemie International Edition 51(34) (2012) 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AKR, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG, Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates, Proceedings of the National Academy of Sciences 111(11) (2014) 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schmidt PJ, Toudjarska I, Sendamarai AK, Racie T, Milstein S, Bettencourt BR, Hettinger J, Bumcrot D, Fleming MD, An RNAi therapeutic targeting Tmprss6 decreases iron overload in Hfe(−/−) mice and ameliorates anemia and iron overload in murine beta-thalassemia intermedia, Blood 121(7) (2013) 1200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hershko C, Camaschella C, How I treat unexplained refractory iron deficiency anemia, Blood 123(3) (2014) 326–333. [DOI] [PubMed] [Google Scholar]
- [70].Schmidt PJ, Racie T, Westerman M, Fitzgerald K, Butler JS, Fleming MD, Combination therapy with a Tmprss6 RNAi-therapeutic and the oral iron chelator deferiprone additively diminishes secondary iron overload in a mouse model of beta-thalassemia intermedia, Am J Hematol 90(4) (2015) 310–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dutta C, Avitahl-Curtis N, Pursell N, Larsson Cohen M, Holmes B, Diwanji R, Zhou W, Apponi L, Koser M, Ying B, Chen D, Shui X, Saxena U, Cyr WA, Shah A, Nazef N, Wang W, Abrams M, Dudek H, Salido E, Brown BD, Lai C, Inhibition of Glycolate Oxidase With Dicer-substrate siRNA Reduces Calcium Oxalate Deposition in a Mouse Model of Primary Hyperoxaluria Type 1, Mol Ther 24(4) (2016) 770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Milliner DS, Harris PC, Cogal AG, Lieske JC, Primary Hyperoxaluria Type 1, in: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (Eds.), GeneReviews((R)), University of Washington, Seattle: University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., Seattle (WA), 1993. [Google Scholar]
- [73].Martin-Higueras C, Luis-Lima S, Salido E, Glycolate Oxidase Is a Safe and Efficient Target for Substrate Reduction Therapy in a Mouse Model of Primary Hyperoxaluria Type I, Mol Ther 24(4) (2016) 719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chan A, Liebow A, Yasuda M, Gan L, Racie T, Maier M, Kuchimanchi S, Foster D, Milstein S, Charisse K, Sehgal A, Manoharan M, Meyers R, Fitzgerald K, Simon A, Desnick RJ, Querbes W, Preclinical Development of a Subcutaneous ALAS1 RNAi Therapeutic for Treatment of Hepatic Porphyrias Using Circulating RNA Quantification, Mol Ther Nucleic Acids 4 (2015) e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Janas MM, Zlatev I, Liu J, Jiang Y, Barros SA, Sutherland JE, Davis WP, Liu J, Brown CR, Liu X, Safety evaluation of 2′-deoxy-2′-fluoro nucleotides in GalNAc-siRNA conjugates, Nucleic acids research (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sun Y, Zhao Y, Zhao X, Lee RJ, Teng L, Zhou C, Enhancing the Therapeutic Delivery of Oligonucleotides by Chemical Modification and Nanoparticle Encapsulation, Molecules 22(10) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nair JK, Willoughby JLS, Chan A, Charisse K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel’in AV, Milstein S, Taneja N, O’Shea J, Shaikh S, Zhang L, van der Sluis RJ, Jung ME, Akinc A, Hutabarat R, Kuchimanchi S, Fitzgerald K, Zimmermann T, van Berkel TJC, Maier MA, Rajeev KG, Manoharan M, Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing, Journal of the American Chemical Society 136(49) (2014) 16958–16961. [DOI] [PubMed] [Google Scholar]
- [78].Springer AD, Dowdy SF, GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics, Nucleic Acid Ther 28(3) (2018) 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Akinc A, Liu J, Qin J, Castoreno A, Schlegel M, Maier M, Fitzgerald K, Meyers R, An investigational RNAi therapeutic targeting factor XII (ALN-F12) for the treatment of hereditary Angioedema, Journal of Allergy and Clinical Immunology 137(2) (2016) AB254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fishman S, Racie T, Hettinger J, Bettencourt BR, Charisse K, Fitzgerald K, Aln-TMP: a subcutaneously administered RNAi therapeutic targeting TMPRSS6 for the treatment of β-thalassemia, Am Soc Hematology, 2013. [Google Scholar]
- [81].Janas MM, Zlatev I, Liu J, Jiang Y, Barros SA, Sutherland JE, Davis WP, Liu J, Brown CR, Liu X, Schlegel MK, Blair L, Zhang X, Das B, Tran C, Aluri K, Li J, Agarwal S, Indrakanti R, Charisse K, Nair J, Matsuda S, Rajeev KG, Zimmermann T, Sepp-Lorenzino L, Xu Y, Akinc A, Fitzgerald K, Vaishnaw AK, Smith PF, Manoharan M, Jadhav V, Wu JT, Maier MA, Safety evaluation of 2’-deoxy-2’-fluoro nucleotides in GalNAc-siRNA conjugates, Nucleic Acids Res (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sardh E, Harper P, Balwani M, Stein P, Rees D, Bissell DM, Desnick R, Parker C, Phillips J, Bonkovsky HL, Vassiliou D, Penz C, Chan-Daniels A, He Q, Querbes W, Fitzgerald K, Kim JB, Garg P, Vaishnaw A, Simon AR, Anderson KE, Phase 1 Trial of an RNA Interference Therapy for Acute Intermittent Porphyria, N Engl J Med 380(6) (2019) 549–558. [DOI] [PubMed] [Google Scholar]
- [83].Bissell DM, Wang B, Acute Hepatic Porphyria, Journal of clinical and translational hepatology 3(1) (2015) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sehgal A, Barros S, Ivanciu L, Cooley B, Qin J, Racie T, Hettinger J, Carioto M, Jiang Y, Brodsky J, Prabhala H, Zhang X, Attarwala H, Hutabarat R, Foster D, Milstein S, Charisse K, Kuchimanchi S, Maier MA, Nechev L, Kandasamy P, Kel’in AV, Nair JK, Rajeev KG, Manoharan M, Meyers R, Sorensen B, Simon AR, Dargaud Y, Negrier C, Camire RM, Akinc A, An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia, Nat Med 21(5) (2015) 492–7. [DOI] [PubMed] [Google Scholar]
- [85].Kosmas CE, Munoz Estrella A, Sourlas A, Silverio D, Hilario E, Montan PD, Guzman E, Inclisiran: A New Promising Agent in the Management of Hypercholesterolemia, Diseases 6(3) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ogura M, PCSK9 inhibition in the management of familial hypercholesterolemia, J Cardiol 71(1) (2018) 1–7. [DOI] [PubMed] [Google Scholar]
- [87].I. Dicerna Pharmaceuticals, Dicerna Files Clinical Trial Application for DCR-PHXC, the Company’s Most Advanced GalXC™ Product Candidate, for Phase 1 Study in Primary Hyperoxaluria (PH). 2017. ).
- [88].Altamura S, Muckenthaler MU, Dames S, Frauendorf C, Schubert S, Aleku M, Vadolas J, Grigoriadis G, Zügel U, SLN124, a Galnac-siRNA Conjugate Targeting TMPRSS6, for the Treatment of Iron Overload and Ineffective Erythropoiesis Such As in Beta-Thalassemia, Am Soc Hematology, 2018. [Google Scholar]
- [89].A. Pharmaceuticals, Platforms that Accelerate Drug Discovery-Targeted RNAi Molecule (TRiM™) Platform. <https://arrowheadpharma.com/science/>. [Google Scholar]
- [90].Turner AM, Stolk J, Bals R, Lickliter JD, Hamilton J, Christianson DR, Given BD, Burdon JG, Loomba R, Stoller JK, Teckman JH, Hepatic-targeted RNA interference provides robust and persistent knockdown of alpha-1 antitrypsin levels in ZZ patients, Journal of Hepatology 69(2) (2018) 378–384. [DOI] [PubMed] [Google Scholar]
- [91].Patel D, Teckman JH, Alpha-1-Antitrypsin Deficiency Liver Disease, Clin Liver Dis 22(4) (2018) 643–655. [DOI] [PubMed] [Google Scholar]
- [92].Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M, Mechanisms and optimization of in vivo delivery of lipophilic siRNAs, Nat Biotechnol 25(10) (2007) 1149–57. [DOI] [PubMed] [Google Scholar]
- [93].Lee SH, Kang YY, Jang HE, Mok H, Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics, Adv Drug Deliv Rev 104 (2016) 78–92. [DOI] [PubMed] [Google Scholar]
- [94].Kruspe S, Giangrande PH, Aptamer-siRNA Chimeras: Discovery, Progress, and Future Prospects, Biomedicines 5(3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Manunta MDI, Tagalakis AD, Attwood M, Aldossary AM, Barnes JL, Munye MM, Weng A, McAnulty RJ, Hart SL, Delivery of ENaC siRNA to epithelial cells mediated by a targeted nanocomplex: a therapeutic strategy for cystic fibrosis, Sci Rep 7(1) (2017) 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Tagalakis AD, Munye MM, Ivanova R, Chen H, Smith CM, Aldossary AM, Rosa LZ, Moulding D, Barnes JL, Kafetzis KN, Jones SA, Baines DL, Moss GWJ, O’Callaghan C, McAnulty RJ, Hart SL, Effective silencing of ENaC by siRNA delivered with epithelial-targeted nanocomplexes in human cystic fibrosis cells and in mouse lung, Thorax 73(9) (2018) 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Buxton DB, Nanomedicine for the management of lung and blood diseases, Nanomedicine 4(3) (2009) 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bush EW, Nicholas A, Li X, Kuipers I, Hamilton H, Hegge J, Zhu R, Chen B, Srivastava J, Schluep T, A Novel Targeted RNAi Molecule Delivery Platform for the Therapeutic Inhibition of ENaC in Cystic Fibrosis Lung Disease, PEDIATRIC PULMONOLOGY, WILEY; 111 RIVER ST, HOBOKEN 07030–5774, NJ USA, 2018, pp. 256–256. [Google Scholar]
- [99].Pardridge WM, shRNA and siRNA delivery to the brain, Adv Drug Deliv Rev 59(2–3) (2007) 141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zheng M, Tao W, Zou Y, Farokhzad OC, Shi B, Nanotechnology-Based Strategies for siRNA Brain Delivery for Disease Therapy, Trends Biotechnol 36(5) (2018) 562–575. [DOI] [PubMed] [Google Scholar]
- [101].Gherardini L, Bardi G, Gennaro M, Pizzorusso T, Novel siRNA delivery strategy: a new “strand” in CNS translational medicine?, Cell Mol Life Sci 71(1) (2014) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J, Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics, J Control Release 203 (2015) 1–15. [DOI] [PubMed] [Google Scholar]
- [103].Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, Daniel WL, Scott AW, Rotz MW, Meade TJ, Giljohann DA, Mirkin CA, Stegh AH, Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma, Sci Transl Med 5(209) (2013) 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Mendonca N, Franca MC Jr., Goncalves AF, Januario C, Clinical Features of Machado-Joseph Disease, Adv Exp Med Biol 1049 (2018) 255–273. [DOI] [PubMed] [Google Scholar]
- [105].Conceicao M, Mendonca L, Nobrega C, Gomes C, Costa P, Hirai H, Moreira JN, Lima MC, Manjunath N, de Almeida LP, Safety profile of the intravenous administration of brain-targeted stable nucleic acid lipid particles, Data Brief 6 (2016) 700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Didiot M-C, Hall LM, Coles AH, Haraszti RA, Godinho BMDC, Chase K, Sapp E, Ly S, Alterman JF, Hassler MR, Echeverria D, Raj L, Morrissey DV, DiFiglia M, Aronin N, Khvorova A, Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing, Molecular Therapy 24(10) (2016) 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Keiser MS, Kordasiewicz HB, McBride JL, Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia, Hum Mol Genet 25(R1) (2016) R53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Frishberg Y, van’t Hoff W, Hulton S, Haslett P, Erbe D, McGregor T, Deschênes G, A phase 1/2 trial of lumasiran (ALN-GO1), an investigational RNAi therapeutic for primary hyperoxaluria type 1, 2018. [Google Scholar]
- [109].Liebow A, Li X, Racie T, Hettinger J, Bettencourt BR, Najafian N, Haslett P, Fitzgerald K, Holmes RP, Erbe D, An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria, Journal of the American Society of Nephrology 28(2) (2017) 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Borodovsky A, Yucius K, Sprague A, Banda NK, Holers VM, Butler J, Fishman S, Vaishnaw A, Maier M, Kallanthottathil R, Aln-CC5, an investigational RNAi therapeutic targeting C5 for complement inhibition, Complement 20 (2014) 40. [Google Scholar]
- [111].Hill A, Taubel J, Bush J, Borodovsky A, Kawahata N, Mclean H, Powell C, Chaturvedi P, Warner G, Garg P, A subcutaneously administered investigational RNAi therapeutic (ALN-CC5) targeting complement C5 for treatment of PNH and complement-mediated diseases: interim Phase 1 study results, American Society of Hematology, 2015. [Google Scholar]
- [112].Schlegel MK, Janas MM, Babu IR, Blair LE, Brown CR, Castoreno A, Harbison CE, Hinkle G, Jayaraman M, Jiang Y, Improved Specificity and Therapeutic Index with ESC+ siRNA Conjugates Utilizing Seed-Pairing Destabilization via Novel Chemical Modifications.
- [113].Liu J, Qin J, Borodovsky A, Racie T, Castoreno A, Schlegel M, Maier M, Zimmerman T, Fitzgerald K, Butler J, Akinc A, An Investigational RNAi Therapeutic Targeting Factor XII (ALN-F12) for the Treatment of Hereditary Angioedema, Rna (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wooddell C, Blomenkamp K, Chen H, Griffin J, Zhu R, Chu Q, Hamilton H, Hegge J, Christianson D, Li Z, ARO-AAT, a subcutaneous RNAi-based therapeutic for alpha-1 antitrypsin-related liver disease, demonstrates liver exposure-response and efficacy in preclinical studies, Journal of Hepatology 68 (2018) S82. [Google Scholar]
- [115].Hayashi Y, Mori Y, Higashi T, Motoyama K, Jono H, Sah DW, Ando Y, Arima H, Systemic delivery of transthyretin siRNA mediated by lactosylated dendrimer/alpha-cyclodextrin conjugates into hepatocyte for familial amyloidotic polyneuropathy therapy, Amyloid 19 Suppl 1 (2012) 47–9. [DOI] [PubMed] [Google Scholar]
- [116].Hayashi Y, Mori Y, Yamashita S, Motoyama K, Higashi T, Jono H, Ando Y, Arima H, Potential use of lactosylated dendrimer (G3)/alpha-cyclodextrin conjugates as hepatocyte-specific siRNA carriers for the treatment of familial amyloidotic polyneuropathy, Mol Pharm 9(6) (2012) 1645–53. [DOI] [PubMed] [Google Scholar]
- [117].Anno T, Higashi T, Hayashi Y, Motoyama K, Jono H, Ando Y, Arima H, Potential use of glucuronylglucosyl-beta-cyclodextrin/dendrimer conjugate (G2) as a siRNA carrier for the treatment of familial amyloidotic polyneuropathy, J Drug Target 22(10) (2014) 883–90. [DOI] [PubMed] [Google Scholar]
- [118].Zheng D, Giljohann DA, Chen DL, Massich MD, Wang X-Q, Iordanov H, Mirkin CA, Paller AS, Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation, Proceedings of the National Academy of Sciences 109(30) (2012) 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sahin U, Karikó K, Türeci Ö, mRNA-based therapeutics—developing a new class of drugs, Nature reviews Drug discovery 13(10) (2014) 759. [DOI] [PubMed] [Google Scholar]
- [120].Xiong Q, Lee GY, Ding J, Li W, Shi J, Biomedical applications of mRNA nanomedicine, Nano Research 11(10) (2018) 5281–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Guan S, Rosenecker J, Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems, Gene Ther 24(3) (2017) 133–143. [DOI] [PubMed] [Google Scholar]
- [122].Gorzelany JA, de Souza MP, Protein replacement therapies for rare diseases: A breeze for regulatory approval?, Science translational medicine 5(178) (2013) 178fs10–178fs10. [DOI] [PubMed] [Google Scholar]
- [123].Crunkhorn S, Regulatory watch: Enhanced chance of success for protein replacement therapies, Nature Publishing Group, 2013. [DOI] [PubMed] [Google Scholar]
- [124].Kowalski PS, Rudra A, Miao L, Anderson DG, Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery, Molecular Therapy (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Granot Y, Peer D, Delivering the right message: Challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—An innate immune system standpoint, Seminars in immunology, Elsevier, 2017, pp. 68–77. [DOI] [PubMed] [Google Scholar]
- [126].Sahu I, Haque AA, Weidensee B, Weinmann P, Kormann MS, Recent developments in mRNA-based Protein supplementation therapy to target Lung diseases, Molecular Therapy (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, Rajeev KG, Hafez IM, Akinc A, Maier MA, Tracy MA, Cullis PR, Madden TD, Manoharan M, Hope MJ, Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo, Angew Chem Int Ed Engl 51(34) (2012) 8529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, Tam YK, Ansell SM, Kumar V, Qin J, Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics, Molecular Therapy 21(8) (2013) 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zhu X, Yin L, Theisen M, Zhuo J, Siddiqui S, Levy B, Presnyak V, Frassetto A, Milton J, Salerno T, Systemic mRNA Therapy for the Treatment of Fabry Disease: Preclinical Studies in Wild-Type Mice, Fabry Mouse Model, and Wild-Type Non-human Primates, The American Journal of Human Genetics (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Jiang L, Berraondo P, Jericó D, Guey LT, Sampedro A, Frassetto A, Benenato KE, Burke K, Santamaría E, Alegre M, Systemic messenger RNA as an etiological treatment for acute intermittent porphyria, Nature medicine 24(12) (2018) 1899. [DOI] [PubMed] [Google Scholar]
- [131].Robinson E, MacDonald KD, Slaughter K, McKinney M, Patel S, Sun C, Sahay G, Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis, Mol Ther 26(8) (2018) 2034–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Patel S, Ryals RC, Weller KK, Pennesi ME, Sahay G, Lipid nanoparticles for delivery of messenger RNA to the back of the eye, Journal of Controlled Release (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG, Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs, Nano letters 15(11) (2015) 7300–7306. [DOI] [PubMed] [Google Scholar]
- [134].Oberli MA, Reichmuth AM, Dorkin JR, Mitchell MJ, Fenton OS, Jaklenec A, Anderson DG, Langer R, Blankschtein D, Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy, Nano letters 17(3) (2016) 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yin H, Song C-Q, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing, Nature biotechnology 35(12) (2017) 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG, Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery, Advanced materials 28(15) (2016) 2939–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Fenton OS, Kauffman KJ, Kaczmarek JC, McClellan RL, Jhunjhunwala S, Tibbitt MW, Zeng MD, Appel EA, Dorkin JR, Mir FF, Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes, Advanced Materials 29(33) (2017) 1606944. [DOI] [PubMed] [Google Scholar]
- [138].Fenton OS, Kauffman KJ, McClellan RL, Kaczmarek JC, Zeng MD, Andresen JL, Rhym LH, Heartlein MW, DeRosa F, Anderson DG, Customizable Lipid Nanoparticle Materials for the Delivery of siRNAs and mRNAs, Angewandte Chemie 130(41) (2018) 13770–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, Wang Q, Guo Y, Dong Y, Tan X, A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo, Cell Res 27(3) (2017) 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].DeRosa F, Guild B, Karve S, Smith L, Love K, Dorkin JR, Kauffman KJ, Zhang J, Yahalom B, Anderson DG, Heartlein MW, Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system, Gene Ther 23(10) (2016) 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao G, Weng Z, Dong Y, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG, Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo, Nat Biotechnol 34(3) (2016) 328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].DeRosa F, Smith L, Shen Y, Huang Y, Pan J, Xie H, Yahalom B, Heartlein MW, Improved Efficacy in a Fabry Disease Model Using a Systemic mRNA Liver Depot System as Compared to Enzyme Replacement Therapy, Molecular Therapy (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Hajj KA, Ball RL, Deluty SB, Singh SR, Strelkova D, Knapp CM, Whitehead KA, Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH, Small 15(6) (2019) e1805097. [DOI] [PubMed] [Google Scholar]
- [144].Sabnis S, Kumarasinghe ES, Salerno T, Mihai C, Ketova T, Senn JJ, Lynn A, Bulychev A, McFadyen I, Chan J, A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates, Molecular Therapy 26(6) (2018) 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Ramaswamy S, Tonnu N, Tachikawa K, Limphong P, Vega JB, Karmali PP, Chivukula P, Verma IM, Systemic delivery of factor IX messenger RNA for protein replacement therapy, Proc Natl Acad Sci U S A 114(10) (2017) E1941–e1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Liu-Chen S, Connolly B, Cheng L, Subramanian RR, Han Z, mRNA treatment produces sustained expression of enzymatically active human ADAMTS13 in mice, Sci Rep 8(1) (2018) 7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lallana E, Rios de la Rosa JM, Tirella A, Pelliccia M, Gennari A, Stratford IJ, Puri S, Ashford M, Tirelli N, Chitosan/hyaluronic acid nanoparticles: rational design revisited for RNA delivery, Molecular pharmaceutics 14(7) (2017) 2422–2436. [DOI] [PubMed] [Google Scholar]
- [148].Fernández Fernández E, Santos-Carballal B, Weber W-M, Goycoolea FM, Chitosan as a nonviral co-transfection system in a cystic fibrosis cell line, International Journal of Pharmaceutics 502(1) (2016) 1–9. [DOI] [PubMed] [Google Scholar]
- [149].Haque AA, Dewerth A, Antony JS, Riethmüller J, Schweizer GR, Weinmann P, Latifi N, Yasar H, Pedemonte N, Sondo E, Chemically modified h CFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis, Scientific reports 8(1) (2018) 16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Kaczmarek JC, Patel AK, Kauffman KJ, Fenton OS, Webber MJ, Heartlein MW, DeRosa F, Anderson DG, Polymer–lipid nanoparticles for systemic delivery of mRNA to the lungs, Angewandte Chemie International Edition 55(44) (2016) 13808–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Patel AK, Kaczmarek JC, Bose S, Kauffman KJ, Mir F, Heartlein MW, DeRosa F, Langer R, Anderson DG, Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium, Adv Mater 31(8) (2019) e1805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Dong Y, Dorkin JR, Wang W, Chang PH, Webber MJ, Tang BC, Yang J, Abutbul-Ionita I, Danino D, DeRosa F, Poly (glycoamidoamine) brushes formulated nanomaterials for systemic siRNA and mRNA delivery in vivo, Nano letters 16(2) (2016) 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Aini H, Itaka K, Fujisawa A, Uchida H, Uchida S, Fukushima S, Kataoka K, Saito T, Chung U.-i., Ohba S, Messenger RNA delivery of a cartilage-anabolic transcription factor as a diseasemodifying strategy for osteoarthritis treatment, Scientific reports 6 (2016) 18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Matsui A, Uchida S, Ishii T, Itaka K, Kataoka K, Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases, Scientific reports 5 (2015) 15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Uchida S, Kinoh H, Ishii T, Matsui A, Tockary TA, Takeda KM, Uchida H, Osada K, Itaka K, Kataoka K, Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety, Biomaterials 82 (2016) 221–228. [DOI] [PubMed] [Google Scholar]
- [156].Baba M, Itaka K, Kondo K, Yamasoba T, Kataoka K, Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles, Journal of controlled release 201 (2015) 41–48. [DOI] [PubMed] [Google Scholar]
- [157].Park HM, Liu H, Wu J, Chong A, Mackley V, Fellmann C, Rao A, Jiang F, Chu H, Murthy N, Lee K, Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo, Nat Commun 9(1) (2018) 3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].McKinlay CJ, Vargas JR, Blake TR, Hardy JW, Kanada M, Contag CH, Wender PA, Waymouth RM, Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals, Proceedings of the National Academy of Sciences 114(4) (2017) E448–E456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].McKinlay CJ, Benner NL, Haabeth OA, Waymouth RM, Wender PA, Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters, Proceedings of the National Academy of Sciences 115(26) (2018) E5859–E5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Haabeth OA, Blake TR, McKinlay CJ, Waymouth RM, Wender PA, Levy R, mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice, Proceedings of the National Academy of Sciences 115(39) (2018) E9153–E9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Abedi-Gaballu F, Dehghan G, Ghaffari M, Yekta R, Abbaspour-Ravasjani S, Baradaran B, Dolatabadi JEN, Hamblin MR, PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy, Applied materials today 12 (2018) 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Cheng Q, Wei T, Jia Y, Farbiak L, Zhou K, Zhang S, Wei Y, Zhu H, Siegwart DJ, Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH mRNA to Normalize Liver Function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I, Adv Mater (2018) e1805308. [DOI] [PubMed] [Google Scholar]
- [163].Chen Q, Qi R, Chen X, Yang X, Wu S, Xiao H, Dong W, A targeted and stable polymeric nanoformulation enhances systemic delivery of mRNA to tumors, Molecular Therapy 25(1) (2017) 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Trepotec Z, Lichtenegger E, Plank C, Aneja MK, Rudolph C, Delivery of mRNA therapeutics for treatment of hepatic diseases, Molecular Therapy (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kauffman KJ, Webber MJ, Anderson DG, Materials for non-viral intracellular delivery of messenger RNA therapeutics, J Control Release 240 (2016) 227–234. [DOI] [PubMed] [Google Scholar]
- [166].Peyvandi F, Garagiola I, Young G, The past and future of haemophilia: diagnosis, treatments, and its complications, The Lancet 388(10040) (2016) 187–197. [DOI] [PubMed] [Google Scholar]
- [167].Nazeef M, Sheehan JP, New developments in the management of moderate-to-severe hemophilia B, Journal of blood Medicine 7 (2016) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].DeRosa F, Guild B, Karve S, Smith L, Love K, Dorkin J, Kauffman K, Zhang J, Yahalom B, Anderson D, Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system, Gene therapy 23(10) (2016) 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, Yang AY, Siemieniak DR, Stark KR, Gruppo R, Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura, Nature 413(6855) (2001) 488. [DOI] [PubMed] [Google Scholar]
- [170].Liu-Chen S, Connolly B, Cheng L, Subramanian RR, Han Z, mRNA treatment produces sustained expression of enzymatically active human ADAMTS13 in mice, Scientific reports 8(1) (2018) 7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Baumgartner MR, Hörster F, Dionisi-Vici C, Haliloglu G, Karall D, Chapman KA, Huemer M, Hochuli M, Assoun M, Ballhausen D, Proposed guidelines for the diagnosis and management of methylmalonic and propionic acidemia, Orphanet journal of rare diseases 9(1) (2014) 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Fraser JL, Venditti CP, Methylmalonic and propionic acidemias: clinical management update, Current opinion in pediatrics 28(6) (2016) 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Mitchell G, Larochelle J, Lambert M, Michaud J, Grenier A, Ogier H, Gauthier M, Lacroix J, Vanasse M, Larbrisseau A, Neurologic crises in hereditary tyrosinemia, New England Journal of Medicine 322(7) (1990) 432–437. [DOI] [PubMed] [Google Scholar]
- [174].Grompe M, St.-Louis M, Demers SI, Al-Dhalimy M, Leclerc B, Tanguay RM, A single mutation of the fumarylacetoacetate hydrolase gene in French Canadians with hereditary tyrosinemia type I, New England Journal of Medicine 331(6) (1994) 353–357. [DOI] [PubMed] [Google Scholar]
- [175].Chinsky JM, Singh R, Ficicioglu C, Van Karnebeek CD, Grompe M, Mitchell G, Waisbren SE, Gucsavas-Calikoglu M, Wasserstein MP, Coakley K, Diagnosis and treatment of tyrosinemia type I: a US and Canadian consensus group review and recommendations, Genetics in Medicine 19(12) (2017) 1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Rowe PC, Newman SL, Brusilow SW, Natural history of symptomatic partial ornithine transcarbamylase deficiency, New England Journal of Medicine 314(9) (1986) 541–547. [DOI] [PubMed] [Google Scholar]
- [177].Prieve MG, Harvie P, Monahan SD, Roy D, Li AG, Blevins TL, Paschal AE, Waldheim M, Bell EC, Galperin A, Targeted mRNA therapy for ornithine transcarbamylase deficiency, Molecular Therapy 26(3) (2018) 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].TranslateBio, Our Progress-Pipeline: present and future focus. <https://translate.bio/pipeline/>.
- [179].Zarate YA, Hopkin RJ, Fabry’s disease, The Lancet 372(9647) (2008) 1427–1435. [DOI] [PubMed] [Google Scholar]
- [180].Anderson KE, Bloomer JR, Bonkovsky HL, Kushner JP, Pierach CA, Pimstone NR, Desnick RJ, Recommendations for the diagnosis and treatment of the acute porphyrias, Annals of internal medicine 142(6) (2005) 439–450. [DOI] [PubMed] [Google Scholar]
- [181].Apgar JF, Tang JP, Singh P, Balasubramanian N, Burke J, Hodges MR, Lasaro MA, Lin L, Miliard BL, Moore K, Jun LS, Sobolov S, Wilkins AK, Gao X, Quantitative Systems Pharmacology Model of hUGT1A1-modRNA Encoding for the UGT1A1 Enzyme to Treat Crigler-Najjar Syndrome Type 1, CPT Pharmacometrics Syst Pharmacol 7(6) (2018) 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Connolly B, Isaacs C, Cheng L, Asrani KH, Subramanian RR, SERPINA1 mRNA as a Treatment for Alpha-1 Antitrypsin Deficiency, J Nucleic Acids 2018 (2018) 8247935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Roseman DS, Khan T, Rajas F, Jun LS, Asrani KH, Isaacs C, Farelli JD, Subramanian RR, G6PC mRNA Therapy Positively Regulates Fasting Blood Glucose and Decreases Liver Abnormalities in a Mouse Model of Glycogen Storage Disease 1a, Mol Ther 26(3) (2018) 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Azarmi S, Roa WH, Löbenberg R, Targeted delivery of nanoparticles for the treatment of lung diseases, Advanced drug delivery reviews 60(8) (2008) 863–875. [DOI] [PubMed] [Google Scholar]
- [185].Riordan JR, Rommens JM, Kerem B.-s., Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA, Science 245(4922) (1989) 1066–1073. [DOI] [PubMed] [Google Scholar]
- [186].Chu C-S, Trapnell BC, Curristin S, Cutting GR, Crystal RG, Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA, Nature genetics 3(2) (1993) 151. [DOI] [PubMed] [Google Scholar]
- [187].Stoltz DA, Meyerholz DK, Welsh MJ, Origins of cystic fibrosis lung disease, New England Journal of Medicine 372(4) (2015) 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Ren CL, Morgan RL, Oermann C, Resnick HE, Brady C, Campbell A, DeNagel R, Guill M, Hoag J, Lipton A, Cystic fibrosis foundation pulmonary guidelines. Use of cystic fibrosis transmembrane conductance regulator modulator therapy in patients with cystic fibrosis, Annals of the American Thoracic Society 15(3) (2018) 271–280. [DOI] [PubMed] [Google Scholar]
- [189].Jennings MT, Dezube R, Paranjape S, West NE, Hong G, Braun A, Grant J, Merlo CA, Lechtzin N, An observational study of outcomes and tolerances in patients with cystic fibrosis initiated on lumacaftor/ivacaftor, Annals of the American Thoracic Society 14(11) (2017) 1662–1666. [DOI] [PubMed] [Google Scholar]
- [190].Hagemeijer MC, Siegwart DJ, Strug LJ, Cebotaru L, Torres MJ, Sofoluwe A, Beekman JM, Translational research to enable personalized treatment of cystic fibrosis, J Cyst Fibros 17(2s) (2018) S46–s51. [DOI] [PubMed] [Google Scholar]
- [191].A. Therapeutics, Pipeline.
- [192].Jarzębińska A, Pasewald T, Lambrecht J, Mykhaylyk O, Kümmerling L, Beck P, Hasenpusch G, Rudolph C, Plank C, Dohmen C, A Single Methylene Group in Oligoalkylamine‐Based Cationic Polymers and Lipids Promotes Enhanced mRNA Delivery, Angewandte Chemie International Edition 55(33) (2016) 9591–9595. [DOI] [PubMed] [Google Scholar]
- [193].Schrom E, Huber M, Aneja M, Dohmen C, Emrich D, Geiger J, Hasenpusch G, Herrmann-Janson A, Kretzschmann V, Mykhailyk O, Translation of angiotensin-converting enzyme 2 upon liver-and lung-targeted delivery of optimized chemically modified mRNA, Molecular Therapy-Nucleic Acids 7 (2017) 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Kaczmarek JC, Kauffman KJ, Fenton OS, Sadtler K, Patel AK, Heartlein MW, DeRosa F, Anderson DG, Optimization of a Degradable Polymer–Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells, Nano letters 18(10) (2018) 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Yan Y, Xiong H, Zhang X, Cheng Q, Siegwart DJ, Systemic mRNA Delivery to the Lungs by Functional Polyester-based Carriers, Biomacromolecules 18(12) (2017) 4307–4315. [DOI] [PubMed] [Google Scholar]
- [196].Urbán Z, Riazi S, Seidl TL, Katahira J, Smoot LB, Chitayat D, Boyd CD, Hinek A, Connection between elastin haploinsufficiency and increased cell proliferation in patients with supravalvular aortic stenosis and Williams-Beuren syndrome, The American Journal of Human Genetics 71(1) (2002) 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Pober BR, Johnson M, Urban Z, Mechanisms and treatment of cardiovascular disease in Williams-Beuren syndrome, The Journal of clinical investigation 118(5) (2008) 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Lescan M, Perl RM, Golombek S, Pilz M, Hann L, Yasmin M, Behring A, Keller T, Nolte A, Gruhn F, De Novo Synthesis of Elastin by Exogenous Delivery of Synthetic Modified mRNA into Skin and Elastin-Deficient Cells, Molecular Therapy-Nucleic Acids 11 (2018) 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Sahel J-A, Marazova K, Audo I, Clinical characteristics and current therapies for inherited retinal degenerations, Cold Spring Harbor perspectives in medicine 5(2) (2015) a017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Magadum A, Kaur K, Zangi L, mRNA-Based Protein Replacement Therapy for the Heart, Molecular Therapy (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Zangi L, Lui KO, Von Gise A, Ma Q, Ebina W, Ptaszek LM, Später D, Xu H, Tabebordbar M, Gorbatov R, Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction, Nature biotechnology 31(10) (2013) 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Yin H, Kauffman KJ, Anderson DG, Delivery technologies for genome editing, Nat Rev Drug Discov 16(6) (2017) 387–399. [DOI] [PubMed] [Google Scholar]
- [203].Oude Blenke E, Evers MJ, Mastrobattista E, van der Oost J, CRISPR-Cas9 gene editing: Delivery aspects and therapeutic potential, J Control Release 244(Pt B) (2016) 139–148. [DOI] [PubMed] [Google Scholar]
- [204].Wang M, Glass ZA, Xu Q, Non-viral delivery of genome-editing nucleases for gene therapy, Gene Ther 24(3) (2017) 144–150. [DOI] [PubMed] [Google Scholar]
- [205].Wang H-X, Li M, Lee CM, Chakraborty S, Kim H-W, Bao G, Leong KW, CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery, Chemical reviews 117(15) (2017) 9874–9906. [DOI] [PubMed] [Google Scholar]
- [206].Zhang H-X, Zhang Y, Yin H, Genome editing with mRNA encoding ZFN, TALEN and Cas9, Molecular Therapy (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Glass Z, Lee M, Li Y, Xu Q, Engineering the delivery system for CRISPR-based genome editing, Trends in biotechnology 36(2) (2018) 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Li L, Hu S, Chen X, Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities, Biomaterials 171 (2018) 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Lino CA, Harper JC, Carney JP, Timlin JA, Delivering CRISPR: a review of the challenges and approaches, Drug delivery 25(1) (2018) 1234–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Liu C, Zhang L, Liu H, Cheng K, Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications, Journal of Controlled Release 266 (2017) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Eoh J, Gu L, Biomaterials as vectors for the delivery of CRISPR–Cas9, Biomaterials science (2019). [DOI] [PubMed] [Google Scholar]
- [212].Gertz MA, Dispenzieri A, Sher T, Pathophysiology and treatment of cardiac amyloidosis, Nat Rev Cardiol 12(2) (2015) 91–102. [DOI] [PubMed] [Google Scholar]
- [213].Plante-Bordeneuve V, Said G, Familial amyloid polyneuropathy, Lancet Neurol 10(12) (2011) 1086–97. [DOI] [PubMed] [Google Scholar]
- [214].Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ, Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA, Angewandte Chemie (International ed. in English) 56(4) (2017) 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C, Mutations in PCSK9 cause autosomal dominant hypercholesterolemia, Nat Genet 34(2) (2003) 154–6. [DOI] [PubMed] [Google Scholar]
- [216].Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates, Proceedings of the National Academy of Sciences 111(11) (2014) 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Whitsett JA, Weaver TE, Hydrophobic surfactant proteins in lung function and disease, N Engl J Med 347(26) (2002) 2141–8. [DOI] [PubMed] [Google Scholar]
- [218].Hamvas A, Cole FS, Nogee LM, Genetic disorders of surfactant proteins, Neonatology 91(4) (2007) 311–7. [DOI] [PubMed] [Google Scholar]
- [219].Mahiny AJ, Dewerth A, Mays LE, Alkhaled M, Mothes B, Malaeksefat E, Loretz B, Rottenberger J, Brosch DM, Reautschnig P, Surapolchai P, Zeyer F, Schams A, Carevic M, Bakele M, Griese M, Schwab M, Nurnberg B, Beer-Hammer S, Handgretinger R, Hartl D, Lehr CM, Kormann MS, In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency, Nat Biotechnol 33(6) (2015) 584–6. [DOI] [PubMed] [Google Scholar]
- [220].Persico AM, Napolioni V, Autism genetics, Behavioural brain research 251 (2013) 95–112. [DOI] [PubMed] [Google Scholar]
- [221].Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q, Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles, Proc Natl Acad Sci U S A 113(11) (2016) 2868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management, The Lancet Neurology 9(1) (2010) 77–93. [DOI] [PubMed] [Google Scholar]
- [223].Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care, The Lancet Neurology 9(2) (2010) 177–189. [DOI] [PubMed] [Google Scholar]
- [224].Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair, Nature biomedical engineering 1(11) (2017) 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Pan B, Géléoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, Holt JR, TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear, Neuron 79(3) (2013) 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Gao X, Tao Y, Lamas V, Huang M, Yeh W-H, Pan B, Hu Y-J, Hu JH, Thompson DB, Shu Y, Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents, Nature 553(7687) (2018) 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Ramakrishna S, Dad A-BK, Beloor J, Gopalappa R, Lee S-K, Kim H, Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA, Genome research 24(6) (2014) 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Wang H-X, Song Z, Lao Y-H, Xu X, Gong J, Cheng D, Chakraborty S, Park JS, Li M, Huang D, Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide, Proceedings of the National Academy of Sciences 115(19) (2018) 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, Li M, Khashab NM, Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework, Journal of the American Chemical Society 140(1) (2017) 143–146. [DOI] [PubMed] [Google Scholar]
- [230].Yue H, Zhou X, Cheng M, Xing D, Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing, Nanoscale 10(3) (2018) 1063–1071. [DOI] [PubMed] [Google Scholar]
- [231].Mout R, Ray M, Yesilbag Tonga G, Lee Y-W, Tay T, Sasaki K, Rotello VM, Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing, ACS nano 11(3) (2017) 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Zhang X, Li B, Luo X, Zhao W, Jiang J, Zhang C, Gao M, Chen X, Dong Y, Biodegradable amino-ester nanomaterials for Cas9 mRNA delivery in vitro and in vivo, ACS applied materials & interfaces 9(30) (2017) 25481–25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Timin AS, Muslimov AR, Lepik KV, Epifanovskaya OS, Shakirova AI, Mock U, Riecken K, Okilova MV, Sergeev VS, Afanasyev BV, Efficient gene editing via non-viral delivery of CRISPR–Cas9 system using polymeric and hybrid microcarriers, Nanomedicine: Nanotechnology, Biology and Medicine 14(1) (2018) 97–108. [DOI] [PubMed] [Google Scholar]
- [234].Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z, Self‐assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing, Angewandte Chemie International Edition 54(41) (2015) 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K, Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing, Nat Commun 8(1) (2017) 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Ranganathan K, Sivasankar V, MicroRNAs - Biology and clinical applications, J Oral Maxillofac Pathol 18(2) (2014) 229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Lam JK, Chow MY, Zhang Y, Leung SW, siRNA Versus miRNA as Therapeutics for Gene Silencing, Mol Ther Nucleic Acids 4 (2015) e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Rupaimoole R, Slack FJ, MicroRNA therapeutics: towards a new era for the management of cancer and other diseases, Nature Reviews Drug Discovery 16 (2017) 203. [DOI] [PubMed] [Google Scholar]
- [239].Ban JJ, Chung JY, Lee M, Im W, Kim M, MicroRNA-27a reduces mutant hutingtin aggregation in an in vitro model of Huntington’s disease, Biochem Biophys Res Commun 488(2) (2017) 316–321. [DOI] [PubMed] [Google Scholar]
- [240].BOULDER C, miRagen Therapeutics Announces Initiation of Phase 2 Clinical Trial of MRG-201, GLOBE NEWSWIRE, 2018. [Google Scholar]
- [241].Lampi MC, Reinhart-King CA, Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials, Science Translational Medicine 10(422) (2018) eaao0475. [DOI] [PubMed] [Google Scholar]
- [242].Hwang DW, Son S, Jang J, Youn H, Lee S, Lee D, Lee YS, Jeong JM, Kim WJ, Lee DS, A brain-targeted rabies virus glycoprotein-disulfide linked PEI nanocarrier for delivery of neurogenic microRNA, Biomaterials 32(21) (2011) 4968–75. [DOI] [PubMed] [Google Scholar]
- [243].Khorkova O, Hsiao J, Wahlestedt C, Basic biology and therapeutic implications of lncRNA, Adv Drug Deliv Rev 87 (2015) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Bonetti A, Carninci P, From bench to bedside: The long journey of long non-coding RNAs, Current Opinion in Systems Biology 3 (2017) 119–124. [Google Scholar]
- [245].Kung JT, Colognori D, Lee JT, Long noncoding RNAs: past, present, and future, Genetics 193(3) (2013) 651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Pastori C, Wahlestedt C, Involvement of long noncoding RNAs in diseases affecting the central nervous system, RNA Biol 9(6) (2012) 860–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Ball RL, Hajj KA, Vizelman J, Bajaj P, Whitehead KA, Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA, Nano Lett 18(6) (2018) 3814–3822. [DOI] [PubMed] [Google Scholar]
- [248].Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, Gamboa Castro M, Anderson SE, Rudoltz TG, Lando GN, Mummilal Tiwari P, Kirschman JL, Willett N, Jang YC, Santangelo PJ, Bryksin AV, Dahlman JE, High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing, Proc Natl Acad Sci U S A 115(42) (2018) E9944–e9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, Mihai C, Almarsson O, Sahay G, Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA, Nano Lett 17(9) (2017) 5711–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]