Abstract
The ability of natural killer (NK) cells to lyse allogeneic targets, without the need for explicit matching or priming, makes them an attractive platform for cell-based immunotherapy. Umbilical cord blood is a practical source for generating banks of such third party NK cells for “off the shelf” cell therapy applications. NK cells are highly cytolytic, and their potent antitumor effects can be rapidly triggered by a lack of HLA expression on interacting target cells, as is the case for a majority of solid tumors, including neuroblastoma. Neuroblastoma is a leading cause of pediatric cancer-related deaths and an ideal candidate for NK cell therapy. However, the antitumor efficacy of NK cells is limited by immunosuppressive cytokines in the tumor microenvironment, such as TGFβ, which impair NK cell function and survival. To overcome this, we genetically modified NK cells to express variant TGFβ receptors which couple a mutant TGFβ dominant negative receptor to NK-specific activating domains. We hypothesized that with these engineered receptors, inhibitory TGFβ signals are effectively converted to activating signals. Modified NK cells exhibited higher cytotoxic activity against neuroblastoma in a TGFβ-rich environment in vitro and superior progression-free survival in vivo, as compared to their unmodified controls. Our results support the development of “off the shelf” gene-modified NK cells, that overcome TGFβ-mediated immune evasion, in patients with neuroblastoma and other TGFβ-secreting malignancies.
Introduction
Infusions of allogeneic effector cells are used with increasing success in cancer immunotherapy(1–4). Natural killer (NK) cells have innate immunity, rapidly lysing target cells without prior priming(5, 6), and are often used as agents of cell-based immunotherapy. Cell lysis is permitted when killer immunoglobulin-like receptors on the NK cell fail to engage with their cognate HLA molecule on the target cell. Antitumor cytolysis is triggered through the “missing self” mechanism when NK cells engage with tumor cells lacking surface HLA molecules. Although NK cells are critical for successful antitumor activity in early oncogenesis(7), their effect is reduced in advanced disease, in tumors which upregulate HLA molecules to evade NK-based surveillance, and in those which produce immunosuppressive cytokines such as IL-6, IL-10, and TGFβ(8). As such, there is a clear need to improve the therapeutic activity of NK cells against solid tumors.
Many solid tumors, including neuroblastoma (NB), evade immune control by generating a suppressive tumor microenvironment, dominated largely by the cytokine transforming growth factor beta (TGFβ)(8–10). Secretion of TGFβ is a potent immunosuppressive strategy employed by NB which inhibits immune effector cell mediated cytotoxicity and promotes a pro-tumorigenic environment(8, 9, 11, 12). This highly tumor-protective environment can only be overcome by significant immune modulation(13), which is why outcomes for advanced relapsed/refractory neuroblastoma is dismal and the development of immunotherapy strategies to overcome tumor evasion is urgently needed.
Adoptive cell therapy using NK cells has clear but limited clinical efficacy in the treatment of NB(14), and the critical role of TGFβ provides compelling rationale for neutralizing this cytokine as a means to improving the efficacy of NK cell-based therapeutics. Soluble TGFβ interacts with the TGFβRII and TGFβRI heterodimer complex on the surface of NK cells. This interaction leads to the phosphorylation of Smads 2 and 3(15), which recruits other soluble proteins and triggers a regulatory cascade leading to decreased cytolytic function and impaired expression of activating receptors(16). Specifically, TGFβ has a detrimental effect on the innate expression of NK cell activating receptors NKG2D(8, 17) and DNAM1(18). Therefore, we hypothesized that modifying the TGFβ receptor in NK cells to abrogate or subvert immunosuppressive signaling could allow NK cells to maintain therapeutic efficacy in the presence of this suppressive cytokine.
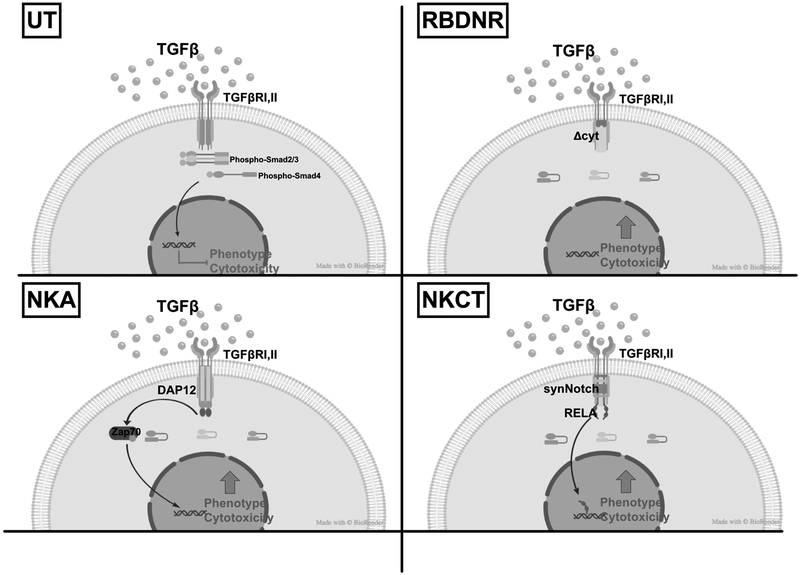
Several groups, including our own, have demonstrated that targeting the TGFβ pathway by arming immune effectors with a dominant negative receptor (DNR) can enhance effector function in a TGFβ-rich environment, with clear superiority over unmodified cells(19–23). In this report, we extended this approach by developing two novel variant TGFβ receptors that couple the TGFβ dominant negative receptor to intracellular signaling domains, mediating NK cell activation. One construct, “NKA”, contains the truncated TGFβRII domain fused to the DNAX-activation protein 12 (DAP12) NK activation motif, which initiates signaling through its single immunoreceptor tyrosine-based activation motif (ITAM), with the aim of enhancing NK cell activation(24, 25). The other construct, “NKCT” contains the truncated TGFβRII domain fused to a synthetic Notch-like receptor (“synNotch”)(26, 27) coupled to RELA, to initiate NK cell activation directly at a transcriptional level. These innovative approaches to hijack the TGFβ receptor and target TGFβ in the tumor microenvironment allows for NK cells to simultaneously (1) resist the immune suppression in the microenvironment, and (2) initiate activation to increase their ability to kill target tumor cells. (Fig. 1)
Fig. 1. TGFβ signaling in untransduced vs. RBDNR, NKA, or NKCT TGFβ-receptor modified NK cells.
Schematic depicting the effects of TGFβ binding to the receptor complex: Untransduced (UT) NK cells express the wild-type TGFβRII, which, when engaged with TGFβ in the tumor microenvironment, initiates a signaling cascade that culminates in impaired NK cell phenotype and cytotoxicity. NK cells transduced with the RBDNR, NKA, or NKCT variant TGFβ receptors alter the intracellular signaling and allow for maintained or enhanced NK cell phenotype and cytotoxicity in the setting of tumor-associated TGFβ.
Initial clinical efforts with NK cells as agents of adoptive immunotherapy have used NK cell lines(28, 29), autologous ex vivo expansion(30), or allogeneic peripheral blood(31, 32) as a cell source. In vitro studies suggest that umbilical cord blood (UCB)-derived NK cells may be more advantageous(33). With over 500 000 validated banked UCB units worldwide(34), in addition to a constant supply of fresh cells, UCB represents a practical and readily available source for generating banks of “off the shelf” cell products. The ability to select optimally mismatched donor-recipient pairs to enhance cytotoxicity contributes to the practical and functional appeal of CB as a source of cells for adoptive NK cell immunotherapy(33, 35, 36).
Here, we demonstrate robust generation of gene-modified NK cells from UCB, which resisted the suppressive effects of tumor-associated TGFβ and exhibited enhanced antitumor effects in vitro and in vivo. This strategy using allogeneic UCB-derived NK cells genetically modified to resist suppression in the tumor microenvironment is therefore a potentially new treatment modality for patients with neuroblastoma and other malignancies that are amenable to NK cell attack and utilize TGFβ secretion as a potent immune evasion mechanism.
Materials and Methods
Cell Sources and Cell Lines
Umbilical cord blood mononuclear cells were harvested from fresh cord blood units obtained from MD Anderson Cancer Center under approved IRB protocols (Pro00003896) by density gradient separation, and NK cells were isolated by negative selection with the EasySep Human NK Cell Isolation Kit (Stem Cell Technologies, Vancouver, Canada). Cord blood units were obtained under informed written consent and in accordance to the Declaration of Helsinki and the guidelines of the Institutional Review Board at MDACC. After 24 hours of activation with 10 ng/mL of human IL-15 (R&D Systems, Minneapolis, MN), NK cells were stimulated with K562 feeder cells, modified to express membrane-bound IL-15 and 41BBL(31, 37) (generously obtained from Baylor College of Medicine (Pro00003869)), irradiated at 200 Gy and cultured with NK cells at a 2:1 K562:NK cell ratio. NK cells were expanded in Stem Cell Growth Medium (CellGenix, Germany) supplemented with 200 IU/mL human IL-2, 15 ng/mL human IL-15, 10% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA), and 1% Glutamax (Gibco, Thermo Fisher Scientific, Waltham, MA). NK cells were isolated from 30 total cord blood donors for downstream use, and untransduced and transduced cells were generated from each individual donor line. Sample size (number of donor-derived lines) used for each experiment is specified in each figure legend. Modified and unmodified K562 cell lines were cultured with IMDM (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA), 1% Penicillin-Streptomycin, and 1% Glutamax (Gibco, Thermo Fisher Scientific, Waltham, MA). Neuroblastoma line SHSY5Y was purchased from ATCC (Manassas, VA) and grown in a 1:1 medium of DMEM and F12K medium supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA), and 1% Glutamax (Gibco, Thermo Fisher Scientific, Waltham, MA). We performed HLA and STR profiling to verify the identity and type of the SHSY5Y tumor line (Genetica Cell Line Testing, Burlington, NC). We also verified that the SHSY5Y neuroblastoma line produces high levels of TGFβ in vivo from SHSY5Y-inoculated NSG mice, and expresses low levels of MHC class I molecules (Supplementary Fig. S1). For generating the bioluminescent neuroblastoma line used in vivo, SHSY5Y was transduced with 2.5 × 106 CFU of CMV-Firefly-luciferase-puro-resistant (Cellomics Technology, Halethorpe, MD) as per manufacturer’s protocol. Bioluminescence was assessed with the Pierce Luciferase Dual Assay Kit (Thermo Fisher Scientific, Waltham, MA) and positive clones isolated by puromycin-resistance and expanded for use, and the cell line was identified as SHSY5Y-luc. Identical in vitro experiments were performed with the neuroblastoma line HTLA230, purchased from ATCC (Manassas, VA).
Generation of Plasmids and Retrovirus Production
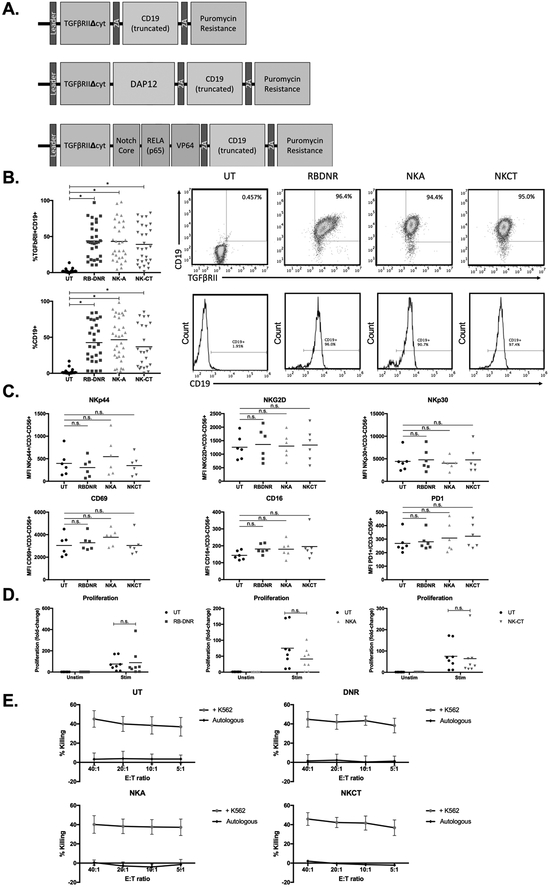
Three modified plasmids were constructed as follows (Fig. 2A): (1) RBDNR: human type II TGFβ receptor cDNA was truncated at nt597 as previously described(38) and coupled to a truncated CD19 tag and pac puromycin resistance gene via T2A sequences. (2) NKA: human type II TGFβ receptor cDNA was truncated at nt597 as previously described(38), containing extracellular and transmembrane moieties, and coupled to the transmembrane and intracellular coding region of DAP12 as derived from full-length DAP12 cDNA(39), a truncated CD19 tag and a pac puromycin resistance gene via T2A sequences. (3) NKCT: human type II TGFβ receptor cDNA was truncated at nt597 as previously described(38) and coupled to a “SynNotch” receptor(26) composed of the Notch1 minimal regulatory region fused to the DNA binding domain for RELA (p65) and a VP64 effector domain(40), coupled to a truncated CD19 tag and a pac puromycin resistance gene via T2A sequences. The RBDNR, NKA, and NKCT constructs were then individually integrated at the BamHI and NcoI sites of the retroviral vector SFG in order to generate plasmids of the same name. A control GFP-containing plasmid was generated elsewhere(41). Phoenix-ecotropic cells (ATCC, Manassas, VA) were transfected with SFG:RBDNR, SFG:NKA, and SFG:NKCT, with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) reagents used as per manufacturer’s protocol. Transient retroviral supernatant was collected 48 and 72 hours following transfection and was used to transduce the PG13 stable packaging cell line (ATCC, Manassas, VA). Transduced PG13 cells were evaluated for transduction efficiency as described below, and single cell FACS sorting was performed to isolate single clonally derived producer lines for RBDNR, NKA, and NKCT constructs. For FACS sorting, single cells that expressed high levels of CD19 and TGFβRII expression were isolated with the Becton Dickinson Influx Cell Sorter (BD Biosciences, Franklin Lakes, NJ) and selectively expanded in puromycin-containing DMEM with 10% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA) and 1% Glutamax (Gibco, Thermo Fisher Scientific, Waltham, MA). Retroviral supernatants containing RBDNR, NKA, NKC, and NKA2 constructs were harvested from sub-confluent PG13 cells, passed through a 0.45 μM filter, and stored at −80 °C until needed for transduction.
Fig. 2. Generating and characterizing TGFβ receptor-modified NK cells.
(A) Vector maps of RBDNR (top), NKA (middle), and NKCT (bottom) constructs. (B) Flow cytometry demonstrating transduction efficiency based on TGFβRII and/or CD19 positive staining. Representative flow dot plots and histograms are on the right, and summarizing data on the left. (C) The phenotype of transduced and untransduced NK cells were examined by flow cytometry, and mean fluorescent intensity values for a given surface receptor is depicted in each panel. (D) Transduced and untransduced NK cells were stained with CFSE, and stimulated with irradiated feeder cells. After 3 days, cells were harvested and assessed for CFSE dilution by flow cytometry. (E) 51Cr-labeled K562 target cells were co-cultured at various effector:target ratios with transduced or untransduced NK cells, and cytotoxicity after 5 hour co-culture was determined based on chromium content in the supernatant, calculated with spontaneous and maximum release controls. All data is representative of experiments with >8 donor lines, with * indicating significant p values <0.05.
NK Cell Transduction and Expansion
Activated NK cells were plated on retronectin-coated non-tissue culture treated plates (Takara, Japan), and transduced with RBDNR, NKA, or NKCT – containing retroviral supernatant in the presence of IL-2 (200 IU/mL). After transductions, NK cells were assessed for transduction efficiency by staining with antibodies against CD19 conjugated to allophycocyanin (BD Biosciences, Franklin Lakes, NJ) and TGFβRII conjugated to phycoerythrin (R&D Systems, Minneapolis, MN). After transduction, NK cells were expanded with additional stimulations with irradiated modified K562s, as described above, and exogenous IL-2 and IL15. To enrich for phenotypic, functional, and in vivo assays, transduced NK cells were stained with CD19 microbeads (Miltenyi Biotec, Germany), and enriched by positive immunomagnetic bead selection according the manufacturer’s protocol.
Phenotypic and Functional Assessment of NK Cells
NK cells were harvested from 21-day or 28-day cultures, washed with FACS buffer, and incubated with human FcR Blocking Reagent for 10 minutes (Miltenyi Biotec, Germany). 21-day cultures were used for analysis of NK cell molecular signaling, whereas 28-day cultures were used for all other endpoint NK cell assays including phenotype, cytotoxicity, and in vivo applications, to allow for maximal cell expansion. Unmodified and modified NK cells, or cell lines, were stained with antibodies specific for NKp30, NKG2D, NKp44, CD16, PD1, CD56, CD3, DNAM1, CD19, TGFβRII (R&D Systems, Minneapolis, MN), HLA-ABC, or MICA/B. Antibodies were conjugated to FITC, PE, PerCP, APC, APC-Cy7, Pe-Cy7, or PerCP-Cy5.5 (BD Biosciences, Franklin Lakes, NJ unless otherwise identified). Samples were run on the Accuri C6 (BD Biosciences, Franklin Lakes, NJ) or CytoFLEX S (Beckman Coulter, Indianapolis, IN) flow cytometers and analysis conducted using Flow Jo 7.6.5 (FlowJo LLC, Ashland, OR). To assess the cytokine profile of transduced and untransduced NK cells, cell supernatant was harvested from 21/28-day NK cultures and used in the Bio-Plex Human Cytokine 17-plex Assay according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA). For examination of cellular proliferation at endpoint, NK cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) as per manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA) and co-cultured with modified K562 cells for 72 hours following assay establishment. To determine the cytolytic properties of unmodified and modified NK cells in various conditions, standard 51Cr release cytotoxicity assays were performed as described elsewhere(22). NK cells were incubated with 51Cr-labeled target cells (unmodified K562s, SHSY5Y cell lines – loaded with 10 μCi 51Cr per 10 000 cells) at 40:1, 20:1, 10:1, and 5:1 ratios for 5 hours in triplicate, and percent killing was determined by the following formula: (experimental count − spontaneous count) / (maximum count − spontaneous count) × 100%. For phenotypic and functional assessment of NK cells after exposure to TGFβ, NK cells were cultured with 10 ng/mL TGFβ (activated with 4mM HCl) added every other day. Five days following assay establishment, NK cells were isolated and examined by flow cytometry, multiplex assays, or cytotoxicity assays, as described above. Further details of NK cell culture can be found in the Supplementary Data.
Molecular Assessment of NK Cells after TGFβ Exposure
To examine the molecular effects of TGFβ, unmodified and modified NK cells (from 21-day cultures) were cultured with 10 ng/mL TGFβ (activated with 4mM HCl) at 37 °C. At 30 minutes, 1, 3, 24, 48, and 72 hours post-TGFβ addition protein was isolated for molecular assessment. Briefly, unmodified or modified NK cells were pelleted and resuspended in RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA) containing protease inhibitor and phosphatase inhibitor cocktails (Roche Diagnostics, Indianapolis, IN). Following 10 minutes of incubation at 4 °C, protein was isolated and particulate matter removed by filtration with Ultafree-CL centrifugal filter units (EMD Millipore, Burlington, MA). Protein was quantified with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA) and 30 μg of protein lysate was isolated and used in the TGFβ Signaling Pathway Magnetic Bead 6-plex Cell Signaling Multiplex Assay (EMD Millipore, Burlington, MA) as per manufacturer’s instructions. Protein expression of phospho-Akt (Ser473), phospho-ERK (Thr185/Tyr187), phospho-Smad2 (Ser465/467), phospho-Smad3 (Ser423/425) was quantitated with Luminex xMap detection, based on positive and negative quantified protein controls.
Mice and in vivo Experiments
Male and Female NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in-house in accordance with approved protocols with the Institutional Animal Care and Use Committee at Children’s National Health System. For in vivo neuroblastoma treatment experiments, 6–10 week old male and female mice were preconditioned with sublethal irradiation (300 cGy) and inoculated with 2.5 × 106 SHSY5Y-luc cells, administered subcutaneously in the dorsal flank of animals. This sublethal irradiation was performed at doses similar to that reported by other groups, which has verified successful immune depletion and immune engraftment in these models(42–45).
Animals were treated immediately following inoculation, a model commonly used in the field(43), with systemic administration of 15 × 106 unmodified or modified NK cells via tail veins. For long-term studies, animals received weekly doses of 5–10 × 106 unmodified or modified NK cells, administered systemically (5 doses in total). All mice were treated with 0.2 μg human IL-2, administered intraperitoneally every other day over the course of their cell therapy doses. The SHSY5Y neuroblastoma line was specifically chosen over the HTLA230 neuroblastoma line due its superior production of TGFβ both in vitro and in vivo in preliminary xenograft experiments (Supplementary Fig. S2). Additionally, The SHSY5Y neuroblastoma line derives from the SK-N-SH line originating from a 4-year old neuroblastoma patient and is a well-established neuroblastoma line used in the field and published in other immunotherapy studies(46–50). For examination of tumor progression, animals were imaged every other day with the IVIS Lumina 100 (Perkin Elmer, Waltham, MA), and images were scaled to the same minimum and maximum photon distribution prior to analysis. Animals were injected with 150 mg/kg Xeno-Light D-Luciferin (Perkin Elmer, Waltham, MA) 10 minutes prior to imaging with the IVIS, during which time animals were anesthetized with 2% isoflurane. Bioluminescent images were captured with 15 s exposure, with small binning and f-stop 2, and total bioluminescence was quantified by photon counts under individual murine regions of interest (photon counts). For analysis of NK cell persistence, blood was collected at designated time points from submandibular veins with Goldenrod Animal Lancets (Braintree Scientific Inc. Braintree, MA) and stored in K2EDTA-containing Microtainer tubes (BD Biosciences, Franklin Lakes, NJ) at −80 °C.
Assessment of NK Cell Persistence in vivo
Transduced NK cells were detected and quantified in the peripheral blood using digital droplet PCR (ddPCR) methods. RNA was extracted from collected blood using the Whole Blood Quick-RNA kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA). cDNA was prepared from 2000 ng of isolated RNA by performing PCR amplification with RT buffer, dNTP Mix, MultiScribe RT, RNAse inhibitor, random primers, and nuclease free water according to the High Capacity RT cDNA kit (Thermo Fisher Scientific, Waltham, MA) and samples were run with the BioRad QC200 Droplet system according to manufacturer’s protocols (Bio-Rad Laboratories Inc. Hercules, CA). For identification of NK cells, primers specific to GFP, RBDNR, NKA, and NKCT construct were used, as described in the Supplementary Data and Methods.
Statistical Analysis
All experiments were performed in duplicate or triplicate, with sample sizes indicated in each corresponding figure legend. Data was analyzed using GraphPad Prism software (GraphPad, La Jolla, CA), and across all figures the solid color bars indicate non-TGFβ-treated groups, whereas striped bars indicate TGFβ-treated groups. Comparisons between untransduced, RBDNR, NKA, and NKCT data were performed using Student’s t-test or Chi-squared tests, with p<0.05 as considered significant an denoted with a star (*) and p<0.0001 denoted with a two stars (**), unless otherwise noted. For in vivo experiments, we performed the log-rank (Mantel-Cox) test for Kaplan-Meier generated survival data, with p<0.05 as considered significant. Schematic signaling diagrams were generated using Biorender (Toronto, Canada).
Results
Variant TGFβ receptor-modified NK cells are phenotypically and functionally similar to unmodified NK cells
To examine NK cell phenotype and function following genetic modification of the TGFβ receptor, cord blood-derived NK cells(33, 34, 36) were isolated and stimulated with irradiated feeder cells and supplemented with recombinant human IL-2 and IL-15(31, 37). Four days after stimulation, NK cells were divided in to four groups: untransduced (UT), RBDNR-transduced, NKA-transduced, and NKCT-transduced NK cells as described. (Fig. 2A) Cord-blood derived NK cells were successfully transduced with RBDNR, NKA, or NKCT variant TGFβ receptors, as indicated by surface staining of TGFβRII and truncated CD19, which was included in receptor design for identification and selection (Fig. 2B, TGFβRII+CD19+: UT 1.92 ± 2.64% vs. RBDNR 43.9 ± 24.1% vs. NKA 43.2 ± 27.1% vs. NKCT 39.1 ± 26.3%, CD19+: UT 1.86 ± 3.57% vs. RBDNR 42.6 ± 27.6% vs. NKA 43.9 ± 30.2% vs. NKCT 36.9 ± 29.4%, n>30). Transduced NK cells could be enriched by performing immunomagnetic sorting with CD19 microbeads to achieve >90% enrichment (Supplementary Fig. S3.) Staining for natural cytotoxicity receptors NKp44 and NKp30 showed no significant difference in expression on transduced NK cells compared to their untransduced counterparts (NKp44: UT 27.4 ± 15.6% vs. RBDNR 25.1 ± 18.0% vs. NKA 31.9 ± 14.9% vs. NKCT 26.4 ± 18.2% p>0.05, NKp30: UT 41.1 ± 27.7% vs. RBDNR 44.2 ± 28.9% vs. NKA 41.7 ± 26.5% vs. NKCT 41.9 ± 31.4% p>0.05, n>5, Fig. 2C, Supplementary Fig. S4). Similarly, no impairment in the expression of other NK cell surface markers NKG2D, CD69, CD16, or PD1 was found (p>0.05, n>5, Fig. 2C, Supplementary Fig. S4). NK cells were labeled with CFSE and co-cultured with unlabeled modified K562s. Flow cytometric analysis of CFSE dilution over three days demonstrated no changes in NK cell proliferation following transduction with RBDNR, NKA, or NKCT receptors (fold-change compared to unstimulated; UT 75.3-fold vs. RBDNR 88.5-fold vs. NKA 41.3-fold vs. NKCT 64.2-fold, p>0.05, n>5, Fig. 2D, Supplementary Fig. S4). 51Cr-based cytotoxicity assays with untransduced and transduced NK cells showed maintained cytolysis of K562 target cells in all conditions (UT vs. RBDNR vs. NKA vs. NKCT p>0.05, n>5, Fig. 2E). Additional cytotoxicity assays with untransduced and transduced cells showed maintained cytolysis of HTLA230 neuroblastoma target cells in all conditions (UT vs. RBDNR vs. NKA vs. NKCT p>0.05, n>5, Supplementary Fig. S2). These results showed that introducing an engineered TGFβ receptor for any of the RBDNR, NKA, or NKCT constructs did not affect NK cell phenotype and function.
TGFβ receptor modification protects NK cells from downstream molecular effects of exogenous TGFβ
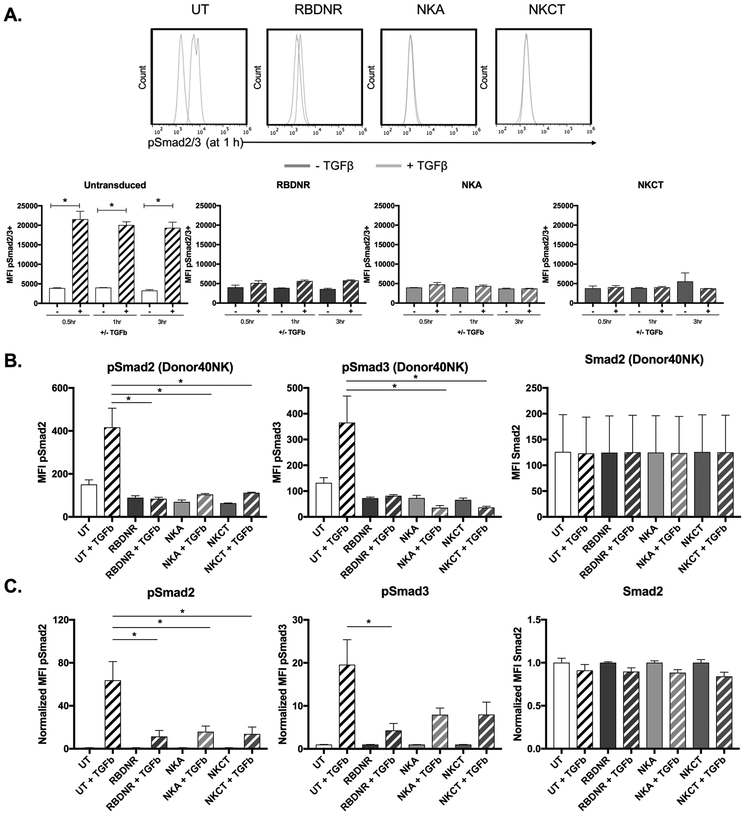
TGFβ binding initiates the phosphorylation of intracellular Smad2 and Smad3 proteins(15). To investigate the ability of RBDNR, NKA, and NKCT constructs to prevent TGFβ-mediated signaling, we co-cultured untransduced, RBDNR, NKA, and NKCT-transduced NK cells with TGFβ. Cells were harvested 0.5, 1, or 3 hours after TGFβ exposure, and either assayed by flow cytometry or lysed to isolate and characterize intracellular proteins. Flow cytometry demonstrated rapid phosphorylation (Ser465/467) of Smad2/3 when untransduced NK cells were exposed to TGFβ (pSmad2/3: UT 1.36 ± 0.95% vs. UT+TGFβ UT 73.9 ± 20.5%, p= 0.04 at 1 h, n>3, Fig. 3A), but not in NK cells transduced with either RBDNR, NKA, or NKCT receptors following TGFβ exposure (p>0.05 at 1 h, p>0.04 at 3 h, n>3, Fig. 3A). Similarly, evaluation of Smad2 (Ser465/467) and Smad3 (Ser423/425) phosphorylation from protein lysate isolated from untransduced and transduced cells after 1 hour of TGFβ exposure further demonstrated the protective effect of TGFβ receptor-modifications conferred to NK cells. Protein lysate results are shown from one representative NK line (Fig. 3B) as well as from pooled NK donor lines (pSmad2 UT+TGFβ vs. RBDNR+TGFβ p= 0.025, UT+TGFβ vs. NKCT+TGFβ p= 0.031; pSmad3 UT+TGFβ vs. RBDNR+TGFβ p= 0.037, n>5; Fig. 3C). These results demonstrated that Smad2 was only phosphorylated in UT NK cells exposed to TGFβ, while expression of the RBDNR, NKA or NKCT receptors protected from Smad2 phosphorylation.
Fig. 3. Examining the molecular effects of TGFβ-signaling.
(A) Flow cytometry was performed to examine the expression of phosphorylated Smad2/3 in transduced and untransduced NK cells after 0.5, 1, and 3 h of exposure to 10 ng/mL TGFβ. Representative histograms are on top, and summarizing data below. (B) Protein was isolated from transduced and untransduced NK cells after 1 hour of exposure to 10 ng/mL TGFβ, and was assessed for phosphorylated Smad2, phosphorylated Smad3, and Smad2 protein content by multiplex assay. Representative protein data for NK cells generated from one donor line. (C) Summarizing protein data for NK cells, where protein amounts are normalized to that of non-TGFβ conditions. All data is representative of experiments with >3 donor lines, with * indicating significant p values <0.05.
TGFβ receptor-modified NK cells have increased expression of activation markers and maintain function in the presence of TGFβ
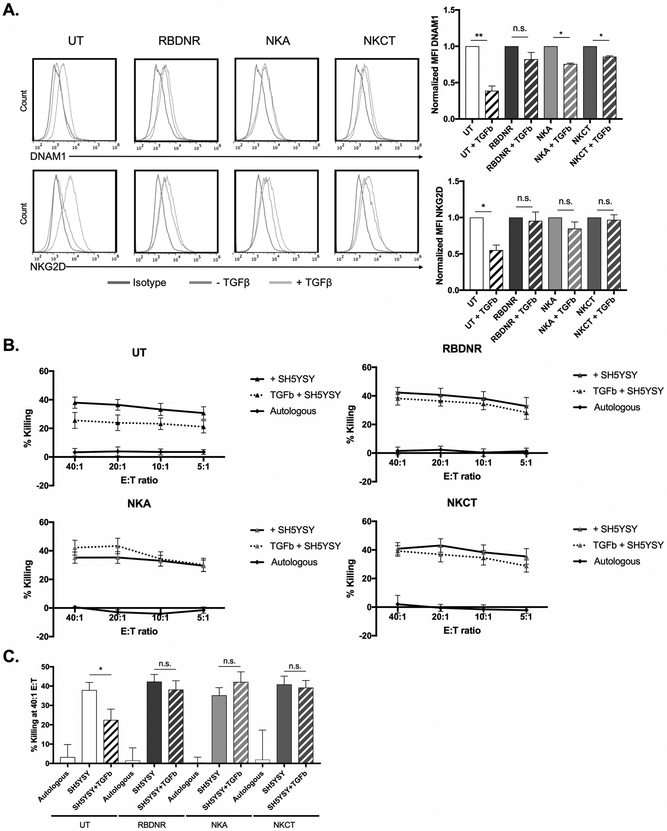
To assess whether the protection from the molecular changes occurring after TGFβ exposure translated to a phenotypic or functional advantage, untransduced and RBDNR, NKA, and NKCT-transduced NK cells were examined after 5-days in culture with TGFβ. Flow cytometry showed decreased expression of DNAX Accessory Molecule-1 (DNAM1 fold-change from non-TGFβ exposed: UT 0.39-fold, p= 0.0163, n>5, Fig. 4A) and in NKG2D (fold-change from non-TGFβ exposed: UT 0.58-fold, p= 0.04, n>5, Fig. 4A) in untransduced NK cells following exposure to TGFβ. Surface marker downregulation was not observed in RBDNR, NKA, or NKCT-transduced NK cells, which all exhibited protection from these TGFβ-mediated phenotype impairments (p>0.05, n>5, Fig. 4A). Additionally, expression of CD16 was not impaired in transduced cells following TGFβ-exposure, alluding to their potential to successfully mediate an anti-tumor effect via ADCC as well as cytolysis (Supplementary Fig. S5). Indeed, whereas untransduced NK cells showed dose-dependent cytotoxicity against SHSY5Y neuroblastoma cells (38.2 ± 4.69% killing at E:T ratio 40:1), they exhibited impaired cytolytic activity (24.6 ± 4.58% killing at E:T ratio 40:1) following pre-culture with TGFβ (Fig. 4B, 4C). Impaired cytolytic activity was not demonstrated when NK cells transduced to express the variant TGFβ-receptors (RBDNR, NKA, or NKCT) were evaluated following pre-treatment with TGFβ (Fig. 4B, 4C), suggesting their functional superiority at killing target cells in a TGFβ-rich environment. As such, we found that not only did expression of the modified TGFβ receptors protect from the molecular signaling occurring in endogenous NK cells following TGFβ exposure, but this protection translated to a protection from altered phenotype and decreased antitumor activity occurring in untransduced cells exposed to TGFβ.
Fig. 4. Examining downstream phenotypic and functional effects of TGFβ-signaling.
(A) Transduced and untransduced NK cells were exposed to TGFβ for 5 days, after which they were harvested and examined for phenotypic changes by flow cytometry. Representative histograms on the left and summarizing data on the right demonstrates changes in the expression of DNAM1 and NKG2D, with mean fluorescent intensities normalized to that of non-TGFβ conditions. (B) 51Cr-labeled SHSY5Y neuroblastoma cells were co-cultured at various effector:target ratios with transduced or untransduced NK cells, and cytotoxicity after 5 hour co-culture was determined based on chromium content in the supernatant, calculated with spontaneous and maximum release controls. (C) Cytotoxicity of NK cells against SHSY5Y neuroblastoma at a 40:1 effector:target ratio. All data is representative of experiments with >7 donor lines, with * indicating significant p values <0.05.
DAP12 and RELA-containing TGFβ receptor variant NK cells demonstrated increased expression of molecular activation markers following exposure to TGFβ
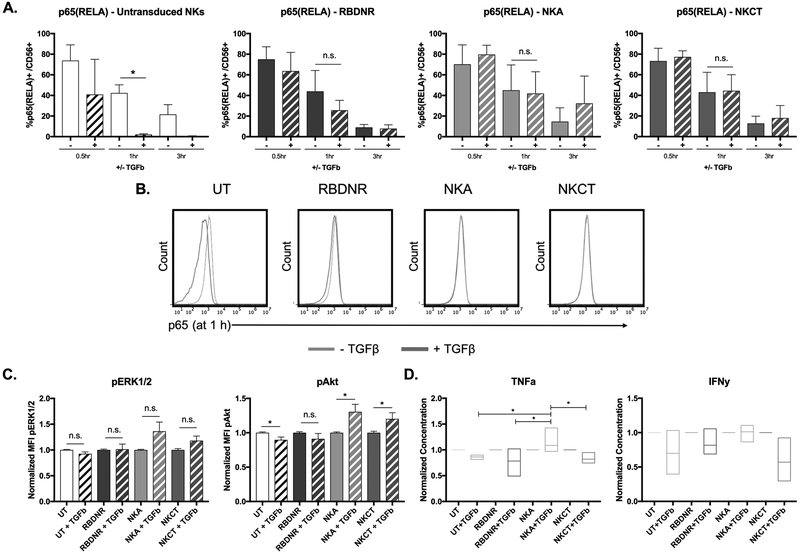
To examine the induction of NK cell activation, we co-cultured untransduced, RBDNR, NKA, and NKCT-transduced NK cells with TGFβ. Cells were harvested 0.5, 1, or 3 hours after TGFβ exposure and either lysed to isolate protein or assayed by flow cytometry. Using flow cytometry we demonstrated decreasing levels of RELA (p65) in untransduced NK cells at one and three-hours post-TGFβ exposure (UT 42.3 ± 13.7% vs. UT+TGFβ UT 2.02 ± 1.08%, p= 0.02 at 1 h UT 21.5 ± 11.5% vs. UT+TGFβ UT 0.47 ± 0.46%, p= 0.18 at 3 h, n>3, Fig. 5A, 5B). Similar trends in RELA were seen in RBDNR-transduced NK cells at one-hour post-TGFβ exposure (p= 0.31 at 1 h, p= 0.18 at 3 h, n>3, Fig. 5A, 5B) NK cells transduced with either NKA or NKCT variant TGFβ receptors demonstrated unaltered p65 expression following exposure to TGFβ (NKA p=0.92 at 1 h, p=0.61 and 3 h, n>3; NKCT p=0.96 at 1 h, p=0.75 at 3 h, n>3), suggesting that NFκB-mediated signaling persisted in these cells. Evaluation of ERK1/2 (Thr185/Tyr187) and Akt (Ser473) phosphorylation occurring in protein lysate isolated from untransduced and transduced cells after 1 hour of TGFβ exposure further showed activation in NKA and NKCT-transduced NK cells. While untransduced and RBDNR-transduced NK cells exhibited decreased or unchanged levels of Akt phosphorylation (UT vs. UT+TGFβ p= 0.0075, RBDNR vs. RBDNR+TGFβ p= 0.282, n>5; Fig. 5C), NK cells equipped with the activation-inducing TGFβ variants had increased Akt phosphorylation (NKA vs. NKA+TGFβ p= 0.0013, NKCT vs. NKCT+TGFβ p= 0.0037, n>5; Fig. 5C). In an examination of supernatant isolated from cell cultures after 12-hours of exposure to TGFβ, we found significantly increased TNFα production in NKA-transduced NK cells after cytokine exposure, as compared to either untransduced or other variant transduced NK cell groups (NKA+TGFβ vs. UT+TGFβ p= 0.039, NKA+TGFβ vs. RBDNR+TGFβ p= 0.006, NKA+TGFβ vs. NKCT+TGFβ p= 0.041; Fig. 5D). Taken together, these results suggest that NK cells transduced to express the TGFβ receptor variants, in particular the NKA modified receptor, demonstrated heightened NK activation, consistent with our observed molecular changes occurring along the NFκB and PI3K signaling pathways.
Fig. 5. Evaluation of TGFβ-signaling induced NK cell activation.
(A) Flow cytometry was performed to examine the expression of p65 (RELA) in transduced and untransduced NK cells after 0.5, 1, and 3 h of exposure to 10 ng/mL TGFβ. (B) Representative histograms for flow cytometry of p65(RELA) expression on untransduced, RBDNR, NKA, and NKCT NK cells following 1 h of exposure to 10 ng/mL TGFβ. (C) Protein was isolated from transduced and untransduced NK cells after 1 hour of exposure to 10 ng/mL TGFβ, and was assessed for phosphorylated ERK1/2 and phosphorylated Akt protein content by multiplex assay. (D) Supernatant was isolated from NK cell cultures after 12 h of exposure to 10 ng/mL TGFβ and concentration of TNFa and IFNy was quantified by multiplex assay. Summarizing protein and cytokine data is graphed, where protein amounts are normalized to that of non-TGFβ conditions. All data is representative of experiments with >3 donor lines, with * indicating significant p values <0.05.
Repeat dosing with TGFβ receptor-modified NK cells enhances survival and tumor eradication in a xenograft model of TGFβ-secreting neuroblastoma
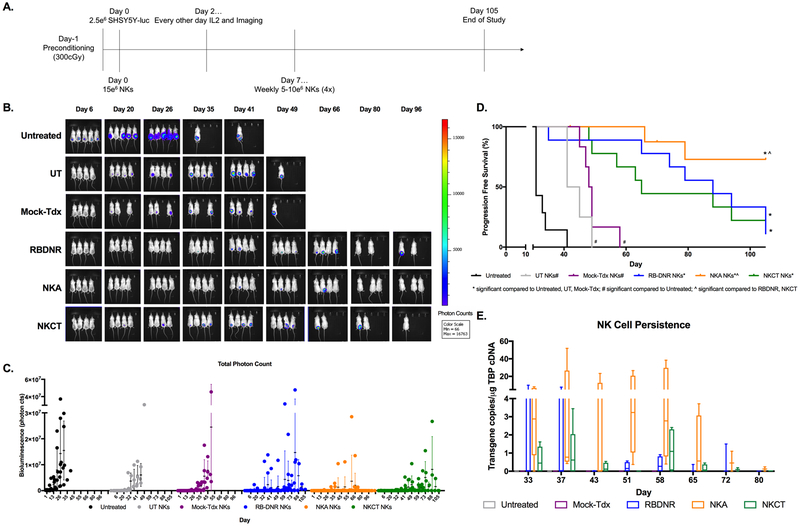
We established a xenograft model of human neuroblastoma using SHSY5Y human neuroblastoma cells(51), inoculated subcutaneously in pre-conditioned immunodeficient animals. Animals were randomly assigned to six treatment groups: untreated, untransduced NK cells (UT), mock GFP-transduced NK cells (Mock-Tdx), RBDNR-transduced NK cells (RBDNR), NKA-transduced NK cells (NKA), and NKCT-transduced NK cells (NKCT). Following inoculation, animals were treated systemically (43) with 15 × 106 NK cells, and monitored during alternate day intraperitoneal IL-2 administration for the duration of the study. Repeated doses of untransduced or transduced NK cells were subsequently given on Days 0, 7, 14, 21, and 28 following tumor inoculation (Fig. 6A), which mirrors desired clinical dosing regimens. Tumor growth was monitored every other day by quantifying bioluminescence (total photon counts) of animals imaged with the IVIS system, using a normalized photon scale(52, 53). Rapid tumor progression was seen in untreated animals, who had a median survival of 31 days (Fig. 6B, 6C, Supplementary Fig. S6). Animals infused with untransduced or mock-transduced NK cells showed delayed tumor progression compared to untreated animals; however these animals eventually succumbed to tumor progression (UT median survival = 43 days, Mock-tdx median survival = 48.5 days, Fig. 6B, 6C). In contrast, infusion of RBDNR or NKCT-transduced NK cells led to improved tumor control and prolonged survival (RBDNR median survival = 88 days, NKCT median survival = 65 days; survival untreated vs. RBDNR p<0.0001, untreated vs. NKCT p<0.0001; Fig. 6D). Animals treated with NKA-transduced NK cells exhibited superior protection from tumor progression (Fig. 6B, 6C, Supplementary Fig. S6) and significantly enhanced survival (progression free survival = 72.9%, survival untreated vs. NKA p<0.0001, UT vs. NKA p= 0.0001, RBDNR vs. NKA p= 0.0333, NKCT vs. NKA p= 0.0313; Fig. 6D). In an assessment to determine the immune populations in the peripheral blood of mice using flow cytometry, we showed that NK cells represented a very minor (<1%) population of the total lymphoid compartment (Supplementary Fig. S7), and as such, the more sensitive ddPCR assay was used to identify the presence of unmodified or modified NK cells peripherally. Therefore, peripheral blood was isolated weekly following the final therapeutic dose of NK cells on day 28, and RNA was extracted from the blood in order to evaluate the presence of the NK cell transgene (GFP or TGFβ variant receptor) by quantitative ddPCR assay. At 5 and 9 days following the final infusion, modified NK cells were identified in circulation. Over the next six weeks, there was some evidence of RBDNR and NKCT-transduced NK cells persisting, although in progressively dwindling numbers as time continued and tumors progressed (Fig. 6E, Supplementary Table 1). NKA-transduced NK cells, however, persisted in higher frequencies than either RBDNR or NKCT-transduced NK cells (Fig. 6E, Supplementary Table 1). Analysis of the TBP transgene in all samples ensured a sufficient quantity and quality of DNA, and was used to normalize all results.
Fig. 6. Long-term tumor-free survival with repeat doses of NK cell treatment in vivo.
(A) Schematic for our in vivo neuroblastoma model: immunodeficient mice were preconditioned, inoculated with luciferase-positive SHSY5Y, treated with systemically delivered transduced or untransduced NK cells on a weekly basis for 5 weeks, and received adjuvant IL2. (B) Tumor growth was monitored by evaluation bioluminescence of animals, which was (C) quantified by total photon counts taken at the same scale. (D) The effect of treatment with transduced or untransduced NK cells on animal survival over the length of the study. (E) Untransduced or transduced NK cells were identified using ddPCR methods to identify transgene copies in systemic blood isolated at weekly intervals following the last NK treatment. Tumor bioluminescence was qualitatively identified according to the heat map color scale, in vivo results are representative with n=5–9 animals/experimental group, ^ indicates significant p values <0.05 compared to RBDNR and NKCT animals, * indicates significant p values <0.05 compared to untreated, UT and Mock-tdx animals, and # indicates significant p values <0.05 compared to untreated animals only.
Taken together, these data indicate that NK cells modified to express novel variants of a TGFβ-receptor protect cells from the inhibitory effects of neuroblastoma-associated TGFβ and demonstrate superior antitumor efficacy in vivo. Further, the enhanced persistence of NKA-transduced NK cells and the significant improvement in progression-free survival in mice administered NKA-transduced NK cells over the RBDNR and NKCT transduced NK cell products suggests that coupling the TGFβ-receptor modification to the NK-specific signaling motif DAP12 confers additional therapeutic advantages and prolonged NK cell persistence in vivo. (Supplementary Fig. S6).
Discussion
In this study we genetically engineered NK cells with novel TGFβ receptors to counter any suppressive TGFβ-mediated signaling and investigated if we could switch the negative TGFβ signal into an activating signal. We demonstrated that phosphorylation of Smad2 and Smad3 occurred as early as 30 minutes after TGFβ-exposure in unmodified NK cells, but was blocked in RBDNR-, NKA-, and NKCT-transduced NK cells. The signaling cascade initiated by the phosphorylation of Smad2/3 led to impaired expression of surface receptors(54) and consequent impairment of antitumor cytolytic function. We found that not only were cord-blood modified NK cells resistant to the inhibitory effects of tumor-associated TGFβ – they also showed superior antitumor efficacy in a TGFβ-rich tumor setting, specifically when transduced with the NKA receptor. The strategy of rendering cell therapy products resistant to inhibitory TGFβ has been explored in a number of malignancies(19, 23). However, by fully inactivating the negative TGFβ pathway and converting the inhibitory signal to an ancillary signal, we created a novel and potent NK cell-specific therapeutic which could be used as an allogeneic “off the shelf” cellular therapy for the treatment of patients with neuroblastoma.
Use of the synthetic Notch receptor into the NKCT receptor is a strategy conceptualized and first applied in the setting of chimeric antigen receptor generation for T cells(26, 27). This strategy employs logic gating, requiring the cell to receive a primary signal in order to trigger a secondary signal through a “SynNotch” receptor. The “SynNotch” receptor contains a core regulatory Notch domain, coupled to an intracellular transcriptional domain that cleaves and engages with nuclear promoters to initiate a given transcriptional change. The NKCT receptor used here contains the extracellular TGFβ dominant negative receptor coupled to a Notch and RELA-linked domain; engagement of TGFβ with this receptor would trigger cleavage of the “SynNotch” motif leading to increased transcription of RELA (p65) and consequent increase in NK cell activation. Our in vitro experiments with the NKCT construct validated this strategy for activating NK cells. However, the potential advantage of this construct was not borne out in vivo, as systemic treatment with NKCT-modified NK cells achieved antitumor efficacy and progression-free survival no better than achieved by RBDNR-modified NK cells that only block TGFβ-mediated signaling. The size of the construct might have been a limiting factor, impairing cleavage and translocation of the large intracellular signaling portion of this receptor. Additionally, because the construct bypassed a natural signaling cascade instead of leading directly to transcriptional activation, it is possible that in the TGFβ-rich environment NKCT-transduced NK cells could be chronically activated causing NK cell dysfunction and apoptosis. Alternatively chronic activation could have generated a negative feedback loop from inhibitory cytokines(55).
In contrast, the NKA receptor (containing DAP12 fused to the dominant negative receptor facilitating NK-specific intracellular signaling) led to improved activity in vivo. In unmodified NK cells, DAP12 associates with natural activating and cytotoxicity receptors such as NKG2C and NKp44. Once dimerized, the ITAM-containing cytoplasmic domain can readily dock with Zap70 and Syk proteins. Global cell activation is the resultant effect of DAP12-activation, which signals through the PI3K/ERK and Akt pathways(25, 56–58). By incorporating the transmembrane and ITAM-containing domains of DAP12 in the NKA construct, TGFβ binding with the engineered receptor triggered activation of DAP12 signaling and enhanced the NK cell activity. The antitumor efficacy of the NKA construct was superior to that obtained with NK cells engineered only to block TGFβ signaling, as in the RBDNR-engineered cells. Furthermore, this additional modification conferred a distinct survival advantage, with NKA-transduced cells persisting up to 7 weeks following their final infusion in treated animals. This in turn led to a superior antitumor effect and a survival advantage in these mice. Further assessment of activation markers expressed by NK cells isolated ex vivo from treated animals would allow a greater depth of understanding into the in vivo mechanism, and will be an important component of larger scale efforts as this approach is translated to the clinic. Although the enhanced PI3K/Akt signaling found in vitro indicates successful propagation of DAP12 mediated activation, it does not specifically address the mechanism through which the TGFβRII-DAP12 linked receptor is forming a dimer or tetramer, and the resultant signaling cascade. As such, it would be essential for future studies to further elucidate this signaling mechanism as well as examine other downstream molecular targets to ensure that enhanced Akt activity would not lead to artificially enhanced NK cell exhaustion.
Topfer et al. have also incorporated DAP12 signaling into a prostate stem cell antigen (PSCA)-specific CAR construct. Preliminary results confirmed the benefit of the DAP12 construct over non-DAP12-containing CAR cells(39), however this effort was conducted with the NK cell line YTS, which, although similar to endogenous NK cells in phenotype, lacks the KIR expression resident to primary NK cells. Our efforts genetically modifying primary NK cells derived from cord blood sources represents a clinical relevant application, where interaction between inhibitory KIRs on NK cells with MHC I variants can have a large influence on the resultant activity (cytotoxicity or suppression) of NK cells used for cell therapy. Our modification of NK cells with a combination of enhanced cell activity through DAP12 and ameliorated TGFβ blockade represents a novel and promising cell therapy approach for neuroblastoma and other malignancies.
One drawback of using CD19 expression to identify transduced cells is that selective downregulation of either the TGFβ modified receptor or the CD19 tag could occur. By using immunomagnetic beads to selectively enrich our cell populations, we minimized the likelihood of this happening (Supplementary Fig. S3). While engineered NK cells might downregulate the modified TGFβ receptor over time, our in vivo studies identified gene modified NK cells with biological activity beyond four weeks suggesting that the cell constructs were stable and could exert long-term antitumor effects.
This report demonstrates preclinical efficacy of a novel mechanism to convert a customarily inhibitory signal, TGFβ, into an activating pathway for NK cells – by doing so, the TGFβ-rich tumor microenvironment is transformed to enhance NK-cell mediated cytotoxicity of tumors. By generating NK cell products from over 30 umbilical cord blood units, and through in vitro and in vivo testing in a human xenograft model of neuroblastoma, this report supports translation to clinical applications. Further preclinical work is being pursued to identify the potential mechanisms of escape that could be faced clinically. For example, examining the function of these variant TGFβ receptors in a humanized model would be of considerable future interest since humanized neuroblastoma models would provide the opportunity to examine interactions with other immune components (e.g. myeloid derived suppressor cells) that may also play a role in promoting NK cell dysfunction in the neuroblastoma setting. Further, ex vivo profiling of immune subsets over time would allow for further in depth analysis of the interactions between NK cells and other immune effectors, and could help determine whether the gene engineered NK cells are capable of eliciting enhanced cytotoxicity through supporting ADCC in addition to tumor-targeted cytotoxicity. Although many neuroblastomas have decreased or absent levels of MHC I, rendering them attractive targets for NK-mediated cytolysis, it would also be of considerable interest in further studies to examine the efficacy of this NK-based immunotherapy in a tumor that has upregulated MHC I expression as a method of tumor escape. In such a setting however, combining NK cell therapy with other immunomodulatory agents (small molecule or epigenetic) may represent an attractive therapeutic avenue. Finally, another priority in further preclinical testing and in initial clinical readouts would be to determine the extent of NK cell migration to tumor-draining lymph nodes and other biological niches following repeat NK cell dosing. While preliminary efforts revealed that CCR2 expression is impaired in NK cells following exposure to TGFβ, and modification with the dominant negative receptor (and variants) may protect from this decline, further probing of the complete effect on NK cells migration is the subject of future study.
In summary, cord blood-derived NK cells modified to avoid the inhibitory effects of TGFβ represent an efficient way to harness fast-acting innate immune cells for therapy. Furthermore, our development of novel variant TGFβ receptors, composed of the dominant negative receptor coupled to intracellular signaling domains initiating NK cell activation, represents a unique approach to transform a classical tumor inhibitory mechanism into a therapeutic weapon. Our preclinical results support translational research to establish allogeneic, cord blood-derived, gene-modified NK cells to treat patients with neuroblastoma and other malignancies that use TGFβ secretion as a potent immune evasion mechanism.
Supplementary Material
Translational Relevance.
The relatively limited treatment options for patients with high-risk neuroblastoma emphasizes the need for enhanced, more specific therapies. Third party natural killer cells represent a cell therapeutic with the potential to successfully eradicate tumors, however their value is limited by the immunosuppressive tumor microenvironment, which produces cytokines such as TGFβ to render NK cells dysfunctional. Our approach targeting TGFβ in the microenvironment using genetically modified receptors that convert inhibitory signals into activating ones is novel, and it allows for NK cells to simultaneously resist the immune suppression and achieve enhanced activation leading to superior in vitro and in vivo anti-tumor efficacy. With this body of work, we have generated robust preclinical data which justifies scale-up and translation of this novel cell therapy platform into the clinic, thus providing a novel therapeutic option for patients suffering from neuroblastoma and other TGFβ-secreting malignancies.
Acknowledgements:
The authors would like to gratefully acknowledge the Institute for Biomedical Sciences at The George Washington University, where RAB is a doctoral candidate. This work is supported by funding to Dr. Catherine Bollard from the Department of Defense (Award W81XWH-15-1-0334) and the NIH National Center for Advancing Translational Sciences (Awards UL1TR000075 and KL2TR000076).
Abbreviations:
- ADCC
antibody-dependent cellular cytotoxicity
- CAR
chimeric antigen receptor
- DAP12
DNAX-activation protein 12
- DNAM1
DNAX accessory molecule-1
- DNR
dominant negative receptor
- ELISA
Enzyme-linked immunosorbent assay
- GFP
green fluorescent protein
- HLA
human leukocyte antigen
- ITAM
immunoreceptor tyrosine-based activation motif
- KIR
killer-cell immunoglobulin-like receptor
- MHC
major histocompatibility complex
- NCR
natural cytotoxicity receptor
- NK cell
natural killer cell
- NKA
modified TGFβ activating receptor
- NKCT
modified TGFβ activating receptor
- NB
neuroblastoma
- NKG2D
transmembrane protein belonging to NKG2 family of C-type lectin-like receptors
- PD-1
programmed cell death protein-1
- RBDNR
modified TGFβ dominant negative receptor
- RELA
Rel-like domain-containing protein/p65
- TGFβ
transforming growth factor beta
- TNFα
tumor necrosis factor alpha
- UCB
umbilical cord blood
- UT
untransduced
Footnotes
The authors declare no potential conflicts of interest.
Supplementary Data
Detailed experimental procedures, tables, and supplementary figures can be found in the Supplementary Data which is available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References:
- 1.Rosenberg SA, Restifo NP, Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Restifo NP, Dudley ME, Rosenberg SA, Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol 12, 269–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tey SK, Bollard CM, Heslop HE, Adoptive T-cell transfer in cancer immunotherapy. Immunol Cell Biol 84, 281–289 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Kalos M, June CH, Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 39, 49–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herberman RB, Nunn ME, Holden HT, Lavrin DH, Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer 16, 230–239 (1975). [DOI] [PubMed] [Google Scholar]
- 6.Kiessling R, Klein E, Wigzell H, “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol 5, 112–117 (1975). [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Lanier LL, Natural killer cells and cancer. Advances in cancer research 90, 127–156 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Bottino C et al. , Natural killer cells and neuroblastoma: tumor recognition, escape mechanisms, and possible novel immunotherapeutic approaches. Front Immunol 5, 56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HM, Kim KS, Kim J, A comparative study of the effects of inhibitory cytokines on human natural killer cells and the mechanistic features of transforming growth factor-beta. Cell Immunol 290, 52–61 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Rouce RH et al. , The TGF-beta/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia 30, 800–811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Pang Y, Moses HL, TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 31, 220–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen PS et al. , Induction of transforming growth factor beta 1 and its receptors during all-trans-retinoic acid (RA) treatment of RA-responsive human neuroblastoma cell lines. Cancer Res 55, 2380–2386 (1995). [PubMed] [Google Scholar]
- 13.Tarek N et al. , Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest 122, 3260–3270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanold J et al. , NK cell immunotherapy for high-risk neuroblastoma relapse after haploidentical HSCT. Pediatric blood & cancer 59, 739–742 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Heldin CH, Miyazono K, ten Dijke P, TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Rook AH et al. , Effects of transforming growth factor beta on the functions of natural killer cells: depressed cytolytic activity and blunting of interferon responsiveness. J Immunol 136, 3916–3920 (1986). [PubMed] [Google Scholar]
- 17.Crane CA et al. , TGF-beta downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro Oncol 12, 7–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpa S et al. , Transforming growth factor beta regulates differentiation and proliferation of human neuroblastoma. Exp Cell Res 229, 147–154 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Yvon ES et al. , Cord blood natural killer cells expressing a dominant negative TGF-beta receptor: Implications for adoptive immunotherapy for glioblastoma. Cytotherapy 19, 408–418 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Brand T, MacLellan WR, Schneider MD, A dominant-negative receptor for type beta transforming growth factors created by deletion of the kinase domain. J Biol Chem 268, 11500–11503 (1993). [PubMed] [Google Scholar]
- 21.Lacuesta K et al. , Assessing the safety of cytotoxic T lymphocytes transduced with a dominant negative transforming growth factor-beta receptor. J Immunother 29, 250–260 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Foster AE et al. , Antitumor activity of EBV-specific T lymphocytes transduced with a dominant negative TGF-beta receptor. J Immunother 31, 500–505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kloss CC et al. , Dominant-Negative TGF-beta Receptor Enhances PSMA-Targeted Human CAR T Cell Proliferation And Augments Prostate Cancer Eradication. Mol Ther, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McVicar DW et al. , DAP12-mediated signal transduction in natural killer cells. A dominant role for the Syk protein-tyrosine kinase. J Biol Chem 273, 32934–32942 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Turnbull IR, Colonna M, Activating and inhibitory functions of DAP12. Nat Rev Immunol 7, 155–161 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Morsut L et al. , Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 164, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roybal KT et al. , Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell 167, 419–432 e416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam YK, Martinson JA, Doligosa K, Klingemann HG, Ex vivo expansion of the highly cytotoxic human natural killer-92 cell-line under current good manufacturing practice conditions for clinical adoptive cellular immunotherapy. Cytotherapy 5, 259–272 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Arai S et al. , Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: a phase I trial. Cytotherapy 10, 625–632 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa E et al. , Autologous natural killer cell therapy for human recurrent malignant glioma. Anticancer Res 24, 1861–1871 (2004). [PubMed] [Google Scholar]
- 31.Fujisaki H et al. , Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res 69, 4010–4017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klingemann HG, Martinson J, Ex vivo expansion of natural killer cells for clinical applications. Cytotherapy 6, 15–22 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Lin SJ, Kuo ML, Cytotoxic function of umbilical cord blood natural killer cells: relevance to adoptive immunotherapy. Pediatr Hematol Oncol 28, 640–646 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Gluckman E, Milestones in umbilical cord blood transplantation. Blood reviews 25, 255–259 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Ruggeri L et al. , Natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Int J Hematol 81, 13–17 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Shah N et al. , Antigen presenting cell-mediated expansion of human umbilical cord blood yields log-scale expansion of natural killer cells with anti-myeloma activity. PLoS One 8, e76781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho D, Campana D, Expansion and activation of natural killer cells for cancer immunotherapy. Korean J Lab Med 29, 89–96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bollard CM et al. , Adapting a transforming growth factor beta-related tumor protection strategy to enhance antitumor immunity. Blood 99, 3179–3187 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Topfer K et al. , DAP12-based activating chimeric antigen receptor for NK cell tumor immunotherapy. J Immunol 194, 3201–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Zalatan JG et al. , Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner HJ et al. , A strategy for treatment of Epstein-Barr virus-positive Hodgkin’s disease by targeting interleukin 12 to the tumor environment using tumor antigen-specific T cells. Cancer Gene Ther 11, 81–91 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Saland E et al. , A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J 5, e297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu E et al. , Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 32, 520–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltinger C, Debatin KM, Murine models for experimental therapy of pediatric solid tumors with poor prognosis. Int J Cancer 92, 313–318 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Shultz LD et al. , Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174, 6477–6489 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Barry WE et al. , Activated Natural Killer Cells in Combination with Anti-GD2 Antibody Dinutuximab Improve Survival of Mice after Surgical Resection of Primary Neuroblastoma. Clin Cancer Res, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen R, Houston J, Chan WK, Finkelstein D, Dyer MA, The role of interleukin-2, all-trans retinoic acid, and natural killer cells: surveillance mechanisms in anti-GD2 antibody therapy in neuroblastoma. Cancer Immunol Immunother 67, 615–626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richman SA et al. , High-Affinity GD2-Specific CAR T Cells Induce Fatal Encephalitis in a Preclinical Neuroblastoma Model. Cancer Immunol Res 6, 36–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waetzig V et al. , Retinoic acid-induced survival effects in SH-SY5Y neuroblastoma cells. J Cell Biochem, (2018). [DOI] [PubMed] [Google Scholar]
- 50.Maser T et al. , Tolcapone induces oxidative stress leading to apoptosis and inhibition of tumor growth in Neuroblastoma. Cancer Med 6, 1341–1352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovalevich J, Langford D, Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol 1078, 9–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim E, Modi KD, Kim J, In vivo bioluminescent imaging of mammary tumors using IVIS spectrum. J Vis Exp, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JB et al. , Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PLoS One 5, e9364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran HC et al. , TGFbetaR1 Blockade with Galunisertib (LY2157299) Enhances Anti-Neuroblastoma Activity of the Anti-GD2 Antibody Dinutuximab (ch14.18) with Natural Killer Cells. Clin Cancer Res 23, 804–813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schleinitz N, Vely F, Harle JR, Vivier E, Natural killer cells in human autoimmune diseases. Immunology 131, 451–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanier LL, Corliss B, Wu J, Phillips JH, Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8, 693–701 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL, NKp44 triggers NK cell activation through DAP12 association that is not influenced by a putative cytoplasmic inhibitory sequence. J Immunol 172, 899–906 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Winter JN, Jefferson LS, Kimball SR, ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am J Physiol Cell Physiol 300, C1172–1180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.