Abstract
Fast photochemical oxidation of protein (FPOP) has become an important mass spectrometry-based protein footprinting approach. Although the hydroxyl radical (·OH) generated by photolysis of hydrogen peroxide (H2O2) is most commonly used, the pathways for its reaction with amino-acid side chains remain unclear. Here we report a systematic study of ·OH oxidative modification of 13 amino acid residues by using 18O isotopic labeling. The results differentiate three classes of residues on the basis of their oxygen uptake preference towards different oxygen sources. Histidine, arginine, tyrosine and phenylalanine residues preferentially take oxygen from H2O2. Methionine residues competitively take oxygen from H2O2 and dissolved oxygen (O2) whereas the remaining residues take oxygen exclusively from O2. Results reported in this work deepen the understanding of ·OH labeling pathway on a FPOP platform, opening new possibilities for tailoring FPOP conditions in addressing many biological questions in a profound way.
Graphical Abstract
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.
Proteins adopt different higher order structures to facilitate their unique biological functions.1 Over more than five decades, researchers have developed and applied numerous approaches to understand structure-function relationships. X-ray crystallography,2 nuclear magnetic resonance (NMR),3 mass spectrometry (MS),4 infrared spectroscopy,5 fluorescence,6 circular dichroism,7 and cryo-electron microscopy (CryoEM)8 are fulfilling in part the demanding needs in the field. One advantage of MS-based protein footprinting approaches are that the technology has improved immensely owing to advances in proteomics. Further, MS has lower sample requirements, shorter time for analysis while preserving mid-to-high resolution for proteins in solution, where biochemistry occurs.4 Prior to MS sampling, proteins are usually submitted to a labeling process that “marks” the solvent accessible surface area (SASA) of the proteins. By tracking the changes of SASA, it is possible to distinguish structure and locate sites of binding, unusual dynamics, differences between mutants and wild-type proteins, to follow fast folding, and to map epitopes to provide deep understanding of protein structure.9–10
Protein Footprinting is usually accomplished by either reversible hydrogen/deuterium exchange (HDX)11 labeling or by irreversible covalent labeling12–14. Among various covalent labeling methods, hydroxyl radical (·OH) based fast photochemical oxidation of proteins (FPOP) is among the fastest (on the timescale of sub-milliseconds), which is competitive in time with most protein conformational changes.12, 15 This feature allows “snapshots” of complex biological systems, making feasible the capture of transient intermediate states and providing new insights that previously required a combination of many sophisticated approaches.16–19 In a FPOP experiment that uses ·OH as labeling reagent, ·OH is generated through laser photolysis of hydrogen peroxide (H2O2) in a flow system, allowing ·OH to react with various amino acid residues and producing an oxygen uptake after labeling. This oxygen uptake can be located even on a specific amino-acid residue level often as a “+16” modification through a combination of post labeling protein digestion and high-resolution MS2 analysis.9, 12, 20–21 During the past 15 years, FPOP has been expanding in academics and biotechnology. It is being used to address successfully significant biological questions including epitope mapping,22–25 protein folding/unfolding,18–19, 26–27 protein aggregation,28–29 ligand-binding affinity determination,16–17 and in vivo footprinting.30–32 Development of ·OH dosimetry control33 and incorporation of a reporter peptide34 also enable precise quantification.
Despite its success, labeling pathways of ·OH in a FPOP platform remain unclear even though the reaction mechanisms between ·OH and free amino acids were extensively studied over decades by using electron paramagnetic resonance (EPR),35–39 absorption spectroscopy40–44, MS45–46, NMR47 and NMR-based hydrogen deuterium exchange.48 The outcomes allowed key reaction intermediates and reaction pathways to be identified. The principal focus has been hydroxyl radicals generated primarily by radiolysis, utilizing high-energy X-rays to ionize water molecules and subsequently produce ·OH. Turning to more complex substrates, we expect that ·OH reaction pathways for amino acid residues in a peptide or protein differ significantly owing to elimination of the free-standing carboxyl and amino group when peptide bonds form. A good example is methionine, which forms a cyclic intermediate when ·OH forms an S· with a free-standing amino acid48 but not for a methionine residue in a peptide or protein.
Chance and coworkers13, 49–52 reviewed reaction pathways between ·OH and amino-acid residues in a peptide or protein with a focus on synchrotron-based hydroxyl radical footprinting (HRF) that utilizes ·OH produced by ionization of water followed by loss of H+. Stable isotope was also introduced, where 18O-enriched water (H218O) was used to reveal the reaction pathway for phenylalanine in an HRF system.49
Although both synchrotron-based HRF and FPOP utilize ·OH as labeling reagents, they are likely to be mechanistically different. In FPOP, ·OH is from photolysis of H2O2, whose distribution in a protein solution is potentially heterogeneous owing to localized hydrogen bonding between selected amino acid residues and H2O2.53 The heterogeneity of H2O2 distribution prior to laser irradiation will induce fluctuations in local ·OH concentration during labeling and possibly alter the oxygen uptake scheme for selected residues and can be potentially utilized to tailor the labeling conditions. In synchrotron-based HRF, the ·OH, coming from water ionization, is distributed homogeneously.
In this article, we present a systematic study of ·OH labeling pathway on a FPOP platform. We distinguished the three oxygen sources for the labeling, dissolved oxygen (O2), H2O2 and water (H2O), in a set of FPOP labeling experiments. We substituted these components with 18O-enriched oxygen sources one at a time to reveal the oxygen uptake preferences across different experimental conditions owing to a 2 Da mass shift between high natural abundance 16O (99.76 %) and purposely introduced 18O. Through analyzing reactions with two different samples, 69 distinct residues were successfully resolved. Among these residues, we covered 13 different amino-acid residues and analyzed their isotope patterns as a function of different experimental conditions. The outcome allowed us to propose, on the basis of our new results and previous mechanism studies, reaction pathways for each kind of amino acid residue. We found that amino-acid residues can be clearly differentiated in three classes based on their oxygen-uptake preferences. Besides revealing the pathways for FPOP footprinting and the differences between H2O radiolysis and H2O2 photolysis, the outcome also provides a foundation for tailoring FPOP conditions in the future.
EXPERIMENTAL
2.1. Materials
Peptide retention time calibration mixture (RTC) and bovine serum albumin protein digest (BSA digest) were chosen as the test samples; they were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Composition of RTC is provided in Supporting Information. Concentrations for each of the samples was determined by UV absorption with a Thermo Scientific NanoDrop OneC (Thermo Fisher Scientific, Waltham, MA, USA). Catalase (from bovine liver), Trizma base, potassium chloride (anhydrous, ≥ 99.0%), L-methionine (≥ 99.5%), L-histidine (≥ 99.0%), hydrogen peroxide solution (containing inhibitor, 30% wt. in H2O), hydrochloric acid (36.5% - 38%) were acquired from Sigma-Aldrich (St. Louis, MO, USA). 18O-enriched oxygen gas (18O2, 99 atom % 18O, 99 % purity), 18O-enriched hydrogen peroxide (H218O2, 90 atom % 18O, 2–3 % in H2O) and 18O-enriched water (H218O, 97 atom % 18O) were obtained from Sigma-Aldrich (St. Louis, MO, USA). All chemicals were used without further purification. The solvent in the 0.5 pmol/µL RTC solution was first removed by a SpeedVac (Thermo Fisher Scientific, Waltham, MA, USA), and the residue was reconstituted with water to the concentration of 5.0 pmol/µL. Tris buffer solution (10 mM) was made by dissolving Trizma base in water. Hydrochloric acid solution was added to obtain a pH of 7.4 (measured with an Orion Star A211 pH meter, Thermo Fisher Scientific, Waltham, MA, USA). Potassium chloride was added to the tris buffer to give a concentration of 100 mM to ensure appropriate ionic strength of the buffer solution.
2.2. Custom-made Degassing Apparatus and Experimental Conditions
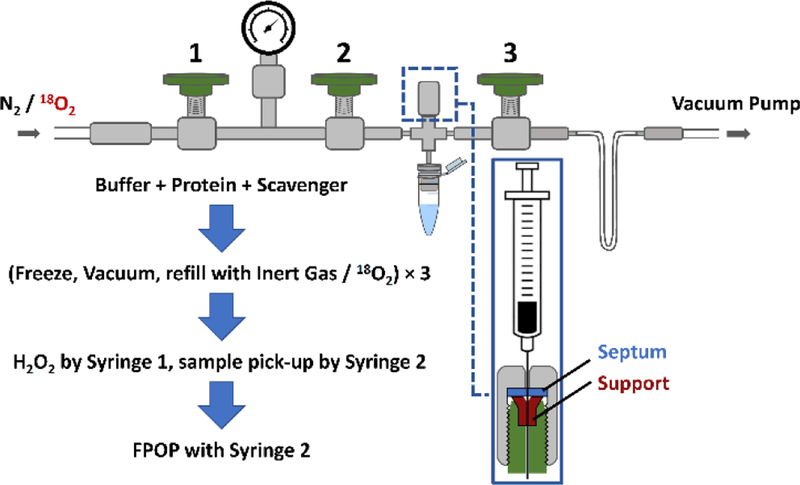
To examine systematically the oxygen source of the common oxygen addition in a FPOP experiment, a custom-designed, mini degassing apparatus was assembled. All parts of the apparatus were from Swagelok (Solon, OH, USA) and were assembled by following the manufacturer’s instructions. The design, as can be seen in Figure 1, was inspired by that of a Schlenk line where three valves with a pressure gauge were connected to both vacuum pump and nitrogen / 18O2 source through two ends. Stainless-steel tubbing was bent into a U-shape, which was submerged in liquid nitrogen (LN2) to serve as a trap for the vacuum pump. During degassing, an Eppendorf tube containing peptide sample, L-histidine scavenger in Tris buffer was connected to the apparatus and gas-sealed by an O-ring. Degassed aliquots were back-filled with either N2 or 18O2 depending on experimental conditions. Detailed operation procedures are described in Supporting Information.
Figure 1.
Schematic illustration of custom-designed degassing and sample-handling apparatus used in current study. A brief summary of the protocol steps for degassing is separated with the large blue arrows.
Experiments were executed under seven conditions. “Regular control” indicates that the sample was prepared in air whereas “vacuum control” communicates that the sample was degassed and backfilled with N2 prior to FPOP labeling. Both do not contain any 18O-enriched components. “Vacuum + 18O2” indicates that the aliquot was degassed and backfilled with 18O2. “Regular + H218O2” and “vacuum + H218O2” indicates that the hydrogen peroxide in these two conditions was 18O-enriched, with first one being prepared under air and the second one being prepared with the degassing procedure (backfilled with N2). Following the same scheme, “regular + H218O” and “vacuum + H218O” denotes that water in these two conditions were 18O-enriched and were prepared under air and then degased (back-filled with N2), respectively. Each experimental condition was given an identifying number in Table 1, and the numbers were used in all data plots.
Table 1.
Summary of experiments used for isotopic labeling
| Number | Experimental Condition |
|---|---|
| 1 | Regular Control |
| 2 | Vacuum Control |
| 3 | Vacuum + 18O2 |
| 4 | Regular + H218O2 |
| 5 | Vacuum + H218O2 |
| 6 | Regular + H218O |
| 7 | Vacuum + H218O |
2.3. FPOP Conditions
For experiments with RTC, aliquots of 50 µL with the final concentration of [RTC] = 0.2 µM, [L-histidine] = 20 µM and [H2O2] = 3.2 mM were prepared for FPOP labelling. With the BSA digest, aliquots of 50 µL containing [BSA digest] = 1 µM, [L-histidine] = 0.1 mM and [H2O2] = 16 mM were used. For each experimental condition, RTC samples were prepared in duplicate, and the BSA digest was in triplicate.
The FPOP platform was the same as previously reported.20, 28 The sample was introduced through a capillary tubing (150 µm I.D.) by a syringe pump (Pump 11 Elite, Harvard Apparatus, Holliston, MA, USA). The beam from a KrF excimer laser (EX50/250, GAM Laser, Orlando, FL, USA) with a wavelength of 248 nm and frequency of 7.2 Hz was introduced through a transparent window. The average laser energy was tuned to ~ 25 mJ/pulse and measured with a pyroelectric energy sensor (PE25-SH-V2, Ophir Optronics Solutions, North Logan, UT, USA). The flow rate was 22–25 µL/min chosen by considering the laser spot width and the operating frequency to ensure a 25% exclusion volume. Final quenching solutions containing 10 µL of 0.7 mM methionine and 0.5 µL 500 nM catalase (for RTC experiments) and 10 µL of 7 mM methionine and 1 µL 500 nM catalase (for BSA experiments) were placed at the end of capillary tube to remove any remaining radicals and consume leftover H2O2 before storing the samples and analyzing them.
2.4. LC/MS Conditions
After FPOP labeling, the samples were submitted to a Thermo Scientific Q Exactive Plus Orbitrap mass spectrometer (Waltham, MA, USA) coupled with a Thermo Scientific Dionex UltiMate 3000 RSLCnano UPLC system (Waltham, MA, USA) for analysis. Liquid chromatography (LC) separation was by a custom-packed C-18 column with bead size of 3 µm. A 10 min desalting followed by an 80 min LC gradient was used for separation, in which water with 0.1% formic acid was used as A phase and 80% water, 20% acetonitrile with 0.1% formic acid was phase B. The solvent gradient was started from 2.5% phase B, increasing to 17.5% in 30 min, 50% in 52 min and 80% in 57 min. Followed by a steady 80% B phase until 65 min, the organic phase subsequently dropped to 2.5% in 70 min and remained until the end of the gradient to re-equilibrate the column. Flow rate during the gradient was 0.4 µL/min. Fragmentation was by an HCD cell at the end of the instrument, where the top ten ions were selected for MS2. Maximum ion injection time was 100 ms and dynamic exclusion was 5 s to ensure the observation of both 16O and 18O labeled species.
2.5. Data Analysis
The LC-MS2 data were first searched against a database containing corresponding peptide sequences by Byonic (Protein Metrics, San Carlos, CA, USA) to identify all 16O and 18O modifications at the peptide and amino-acid residue levels. Searching results were analyzed by Byologic (Protein Metrics, San Carlos, CA, USA), allowing three extracted ion chromatograms (EIC) representing wildtype, 16O modified (+16) and 18O modified (+18) to be processed for each peptide. Peak assignments in EIC were by MS2, and quantification was by comparing integrated EICs as described in Supporting Information. All the normalized +18O fractions were plotted against experimental conditions (denoted by a number code, Table 1) as solid bars for each resolved residue. Error bars at the top of solid bars represent the standard deviations of three and two independent runs for BSA digest and RTC samples, respectively. Peptide-specific references denoting the A + 2 contributions (of combinations of 13C, 15N, or 18O, 34S at their natural abundance) were plotted as horizontal dotted lines in the bar graph. Any normalized +18O fraction that is greater than the dashed reference line was considered to be a contribution by the purposely introduced 18O isotope.
RESULTS AND DISCUSSION
By using a mini vacuum line, we designed an approach whereby oxygen sources in the footprinting can be replaced one at a time with 18O2, H218O2 and H218O to highlight the preferred oxygen uptake pathways of 13 amino acid residues. We chose two complex samples of peptides as tests for the footprinting. The following discussion is built around the amino acids and amino-acid types according to their uptake of 18O in this pathway study.
3.1. Histidine
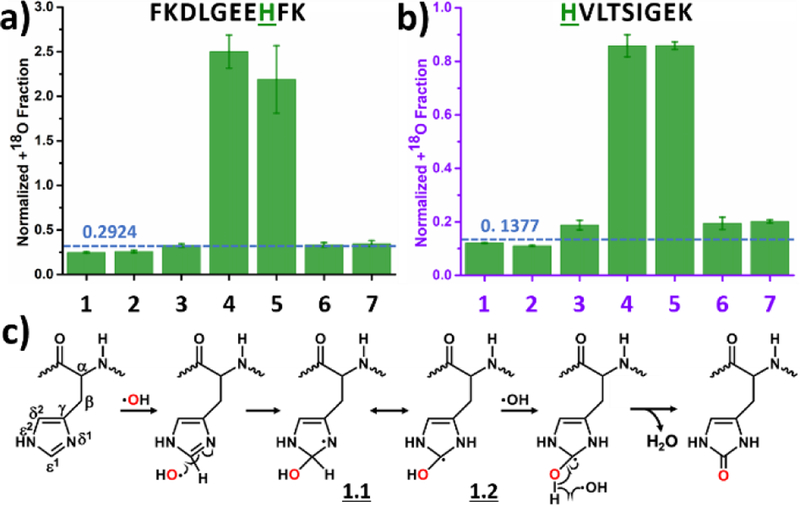
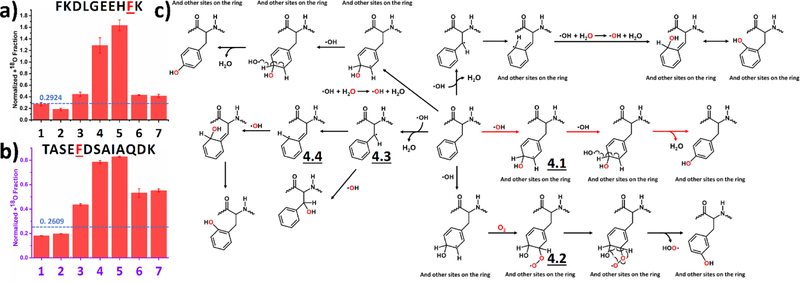
We present two different histidine residues, one from the BSA digest (Figure 2a) and one from RTC (Figure 2b) to show the remarkable singularity and simplicity of the footprinting chemistry for this amino-acid residue. For the regular control (1, Table 1 and Figure 2) and vacuum control (2) experiments, where no 18O was intentionally added into the system, experimental values for the normalized +18O fraction agrees reasonably well with the calculated value (dashed line), validating the data processing method and serving as a control for the upcoming data interpretation. From Figures 2a and 2b, the normalized +18O fractions for the experimental conditions 4 and 5 are all significantly higher than the background, suggesting the footprinting reaction involves an uptake of 18O only from H2O2, and essentially no 18O labeling by oxygen gas or by water molecules. Results for three other resolved histidines show uptake consistent with that seen in Figure 2 (see Figure S3).
Figure 2.
Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for two representative peptides from (a) BSA digest and (b) RTC containing histidine. Error bars represent standard deviations of three and two independent runs for BSA digest and RTC, respectively. Dashed horizontal line represents calculated background A + 2 fraction. (c) Proposed reaction pathway based on the results. Oxygen highlighted in red represents 18O.
Based on our observations, we can propose a reaction pathway (Figure 2c). The reaction starts by ·OH addition at the ε1 position of the side chain, resulting an OH addition and an unpaired electron at the δ1 nitrogen.35, 39, 48 The unpaired electron, now delocalized,40 is capped with a second ·OH to provide a final product (Figure 2c).13 It is also possible that ·OH to attacks the β position of the side chain by ·H abstraction,48 but this reaction route is minor (not shown in Figure 2c). In the HRF setup, whose ·OH was from synchrotron water ionization, allyl-type intermediate 1.1 and 1.2 subsequently react with O2 for the same product.13, 50
3.2. Arginine
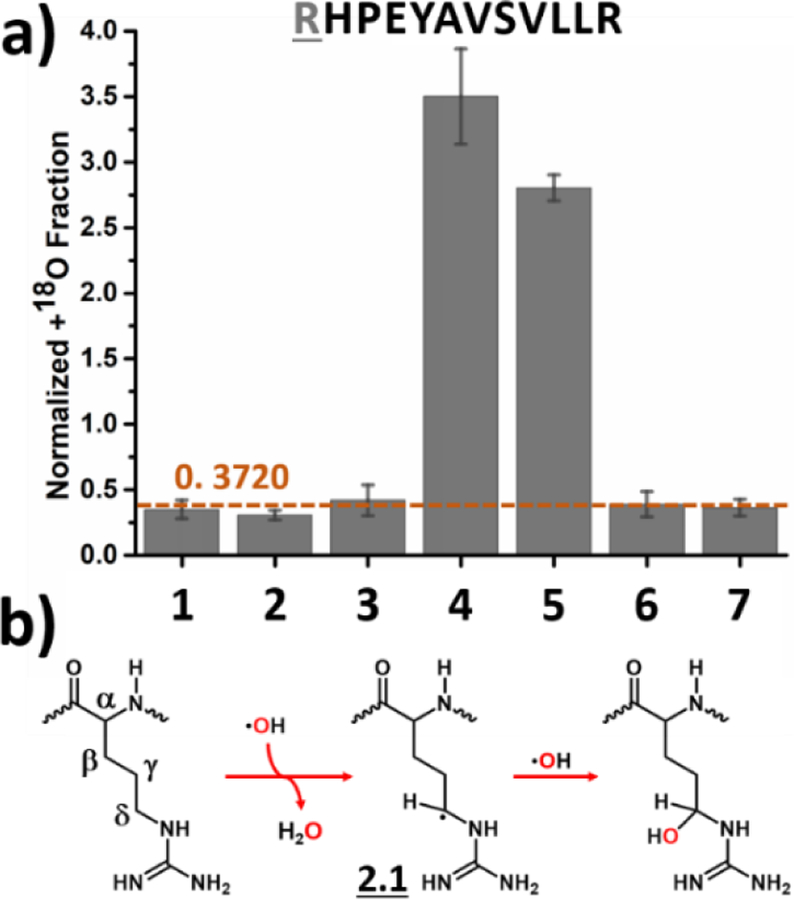
Like histidine, arginine takes oxygen exclusively from H2O2 (Figure 3a) where a specific arginine residue in a peptide from the BSA digest was analyzed under the different experimental conditions. In the proposed reaction pathway, ·OH abstracts ·H from the sidechain, yielding an arginine radical intermediate 2.1 that can be readily quenched by a second ·OH to form the final product (Figure 3b). Previous studies indicate that ·H abstraction by ·OH preferentially happens at the δ position,44 with a ratio of δ-CH2 : γ-CH2 : β-CH2 of 11 : 3 : 1.48 Under physiological conditions, the guanidyl is protonated, and its charge is delocalized over the guanidyl group. The delocalized positive charge increases the electron density on the adjacent δ-CH2 to a greater extent than on γ-CH2 and β-CH2.36 Given that ·H abstraction by ·OH is an electrophilic process, the ·OH preferentially reacts with the electron-rich δ-CH2. Higher electron density at δ-CH2 also leads to a higher reaction rate of arginine towards ·OH.13, 54 For synchrotron-based HRF, the arginine radical was quenched by O2 instead of by a second ·OH as it is when footprinting is on the current FPOP platform.13, 50
Figure 3.
Labeling of arginine: (a) Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for a peptide from BSA digest containing arginine. Error bars represent standard deviations of three independent runs. Dashed horizontal line represents calculated background A + 2 fraction. (b) Proposed reaction pathway based on the results. Oxygen highlighted in red represents 18O and red arrows denote preferred pathway.
3.3. Tyrosine
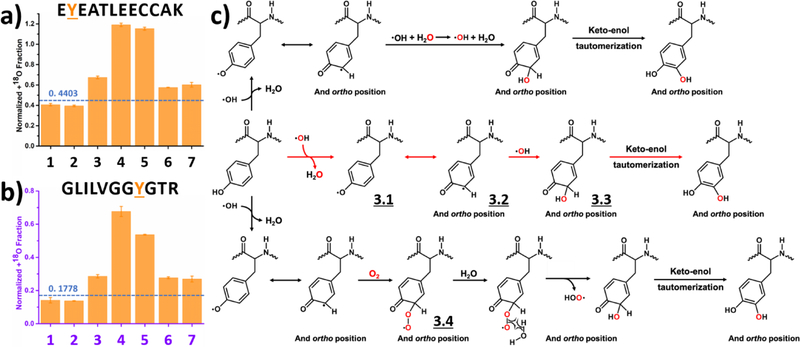
We selected two different tyrosine residues to represent a typical oxygen uptake preference, one in a peptide from the BSA digest (Figure 4a) and another from RTC (Figure 4b). Normalized +18O fractions for the last five experimental conditions are all significantly higher than the background, even when H218O2 is the main source of 18O. Tyrosine takes oxygen from all three oxygen sources (i.e., dissolved O2, H2O2 and H2O), with a preference for H2O2.
Figure 4.
Labeling of tyrosine: Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for two representative peptides from (a) BSA digest and (b) RTC. Error bars represent standard deviations of three and two independent runs for BSA digest and RTC, respectively. Dashed horizontal line represents calculated background A + 2 fraction. (c) Proposed reaction pathways based on the results. Oxygen highlighted in red represents 18O, and red arrows denote preferred pathway.
In total, we resolved 10 distinct tyrosine residues (results for the other 8 tyrosines are given in Supporting information as Figure S4). Whereas six of ten tyrosines preferentially pick up oxygen from H2O2, the favored sources of oxygen for the remaining four are unclear. Part of the uncertainty is that the modification fraction for these four residues is only a few percent, allowing background noise and interference with other chromatographic peaks to contribute. Effects due to adjacent residues may also lead to the complexity.
Based on the experimental results, we propose a reaction pathway (Figure 4c). Laser irradiation homolytically cleaves H2O2 into two ·OH that initiate all reactions by ·H abstraction, as evidenced by an NMR-based hydrogen deuterium exchange study.48 The unpaired electron in the resulting tyrosyl radical is delocalized in the aromatic ring (3.1 and 3.2),47–48 which is electron-rich, stabilizing the tyrosyl radical. Following attack of a second ·OH (red arrows in Figure 4c), the tyrosyl radical is quenched to form a single bond (3.3), and keto-enol tautomerization produces dihydroxyphenylalanine (DOPA) is the major product of the preferred reaction route.13, 45 As the electron-rich aromatic ring stabilizes the unpaired electron, the tyrosyl radical is long-lived and available for other competitive modifications. For example, dissolved O2 reacts via a minor route with the tyrosyl radical (3.4) and subsequently eliminates a HOO·, yielding the DOPA product through a keto-enol tautomerization.13 In contrast, this is the preferred reaction route in the HRF setup.13 Via another route, ·OH exchanges with solvent water, affording a 18O labeled radical.55 Given that the concentration of water is over 50 M, this minor route becomes competitive. The secondary ·OH then reacts with tyrosyl radical to give a labeled DOPA product, showing that the radical chemistry can be complex. We note that reaction pathways proposed here are the preferred routes, and that minor routes are not precluded.
3.4. Phenylalanine
We present outcomes from two different phenylalanine residues in the BSA digest (Figure 5a) and RTC (Figure 5b). Like tyrosine, phenylalanine takes oxygen from all three sources, with a preference for H2O2. An additional 13 phenylalanine residues were resolved (Figure S5); nine show clear preference of oxygen uptake form H2O2. We propose a reaction pathway that accounts for these observations (Figure 5c). It is well-accepted that ·OH predominately reacts with phenylalanine by addition to the phenyl ring to give a radical intermediate 4.113, 48, 56–57 that is quenched by a second ·OH, giving a tyrosine residue as final product (represented by red arrows in Figure 5c). The initial attack of ·OH has small preferences for different positions on the phenyl ring of ortho : meta : para = 2 : 1 : 1.5,48 and thus the reaction is not regiospecific. Because the lifetime of intermediate 4.1 is relatively long, as discussed earlier, it can be quenched by dissolved O2 (4.2), followed by a loss of HOO· to give m-tyrosine residue as resulting product. Similar with tyrosine, quenching by O2 is the dominant reaction pathway in the HRF scheme.13 The phenylalanine can also be labeled with a secondary ·OH,55 which is produced, for example, by an exchange between primary ·OH with H2O as illustrated in the top left of Figure 5c.
Figure 5.
Labeling of phenylalanine: Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for two representative peptides from (a) BSA digest and (b) RTC. Error bars represent standard deviations of three and two independent runs for BSA digest and RTC, respectively. Dashed horizontal line represents calculated background A + 2 fraction. (c) Proposed reaction pathway based on the results. Oxygen highlighted in red represents 18O, and red arrows denote preferred pathway.
Besides ·OH addition, ·OH is also able to abstract a ·H from β-CH2,48 resulting an unpaired election (4.3), which is delocalized over the phenyl ring (4.4), as depicted on the left of Figure 5c. This unpaired electron can be readily paired by the addition of a second ·OH, resulting an oxidized form of phenylalanine.
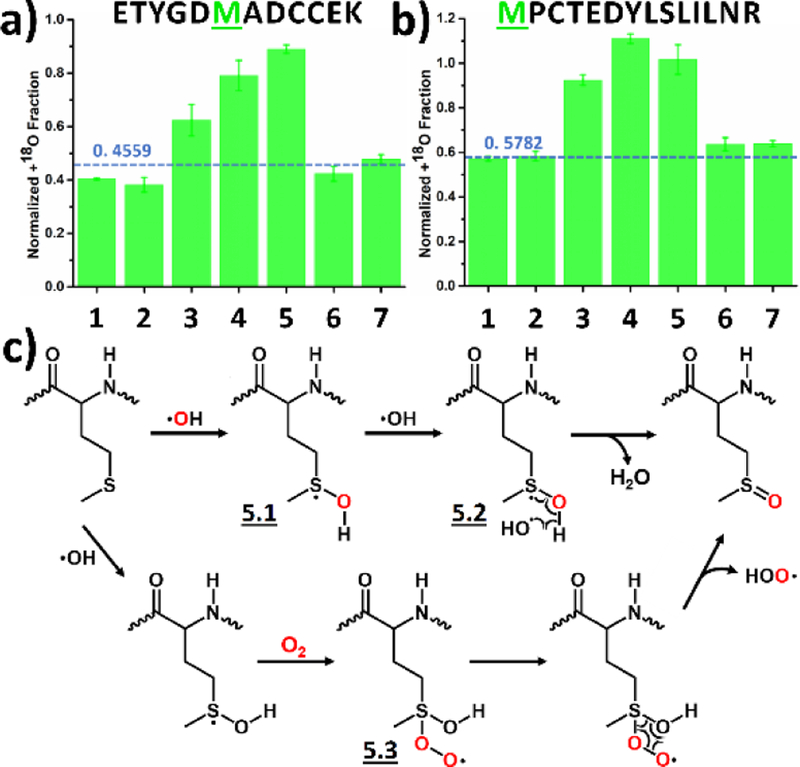
3.5. Methionine
Methionine takes oxygen from both dissolved O2 and H2O2 with comparable preferences, as shown by the comparable normalized +18O fraction between experimental conditions of vacuum + 18O2 (3), regular + H218O2 (4) and vacuum + H218O2 (5, Figure 6a and b). We explain these preferences by a reaction pathway proposed in Figure 6c. It was previously shown that ·OH reacts with methionine by adding to the sulfur atom, leaving an unpaired electron on it.48, 52, 58–59 Upon forming this intermediate 5.1, there is a comparable chance for a second ·OH (5.2) or a O2 (5.3) to react and quench the radical species. This contrasts sharply with HRF, as a distinct preference of O2 is proposed to quench intermediate 5.1.13, 52 Under both reaction routes, the final product is likely methionine sulfoxide, detected as +16 by MS2.13 Results for a third methionine residue fits well the pathway proposed above (Figure S6).
Figure 6.
Labeling of methionine: Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for two representative peptides from the BSA digest containing phenylalanine (a and b). Error bars represent standard deviations of three independent runs. Dashed horizontal line represents calculated background A + 2 fraction. (c) Proposed reaction pathway based on the results. Oxygen highlighted in red represents 18O.
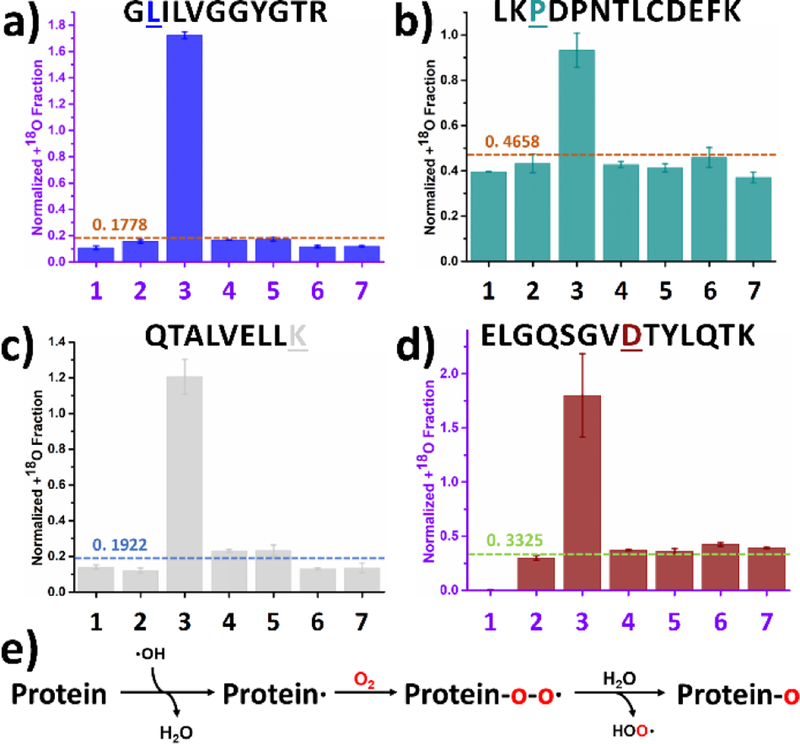
3.6. Leucine, Isoleucine, Valine and Proline
Leucine, isoleucine, valine and proline, as aliphatic amino acids, are presented together (Figure 7a and b show a representative oxidative modification of a leucine and proline residues, respectively). These aliphatic residues take oxygen nearly exclusively from O2, well aligned with HRF13 because there is unlikely any localized concentration of H2O2 in the region of these hydrophobic peptides. Mechanistically, all four residues behave similarly (Figure 7e). To begin, ·OH converts the residue into a radical by ·H abstraction. Following O2 addition to give a radical protein-O-O·, the peroxy radical is readily quenched by water, resulting an oxidized protein-O product. ·H abstraction by ·OH has been well characterized as the first step in the oxidative modification of aliphatic amino acids,13, 48, 56, 60 with a preferred abstraction occurring at the branch point (γ-CH for leucine, β-CH for isoleucine and β-CH for valine) because tertiary radical > secondary radical > primary radical in stability. The cyclic nature of proline complicates the picture. As previously reported, ·H abstraction for proline takes place at δ-CH2, γ-CH2 and β-CH2 in the ratio of 2.7 : 1.4 : 1.48 The higher reactivity of δ-CH2 is explained as an inductive effect of the adjacent peptide bond. Further evidence supporting this pathway is given as normalized +18O fraction plots for additional aliphatic residues (Figures S7–10).
Figure 7.
Labeling of leucine, proline, lysine, and aspartic acid: Normalized +18O fraction determined under number-coded experimental conditions (Table 1) for representative peptides containing (a) leucine from RTC, (b) proline from BSA digest, (c) lysine from BSA digest and (d) aspartic acid from RTC. Error bars represent standard deviations of three and two independent runs for BSA digest and RTC, respectively. Dashed horizontal line represents calculated background A + 2 fraction. (e) Proposed reaction pathway based on the results. Oxygen highlighted in red represents 18O.
3.7. Lysine, Aspartic Acid, Glutamic Acid and Glutamine
The charged residues of lysine, aspartic acid and glutamic acid take oxygen exclusively from O2 (Figure 7c and d) as does the polar, uncharged glutamine (reaction pathways in Figure 7e). The lysine, amine group is positively charged at physiological pH, leading to a lower electron density at ε-CH2. As a result, ·OH preferentially reacts with β-CH2, γ-CH2 and δ-CH2.37, 48 For aspartic acid, glutamic acid and glutamine, a previous NMR-based hydrogen deuterium exchange study revealed that the carboxylate and carboxamide groups deactivate the ·H abstraction at physiological pH.48 Such deactivation lowers the rate constant between ·OH and these residues.13 Moreover, oxygen uptake by these residues is not the dominant reaction pathway upon ·OH labeling as evidenced in previous studies. Other possible reaction pathways are not the primary focus of this work and will not be discussed in detail.13 Supporting evidence from other normalized +18O fraction plots for another lysine, two aspartic acids and several glutamic acid and glutamine residues are in Figure S11–S14.
3.8. Three Classes of Residues in FPOP Platform
We can group the thirteen resolved amino-acid residues into three different classes based on their reactivity and preferences towards different oxygen sources. Class 1 residues (histidine, arginine, tyrosine, and phenylalanine) preferentially take oxygen from H2O2, most likely by addition of ·OH to a previously formed radical. Among these four residues, we can identify three sub-classes based on their distinct reaction pathways.
Class 1a residues are histidine and arginine that take oxygen solely from H2O2. The rationalization for their behavior begins with an interaction of H2O2 via hydrogen bonding with histidine, arginine as well as with tyrosine, cysteine, threonine, glutamine, aspartic acid, lysine, methionine, tyrosine and tryptophan.53 This interaction gives rise to a high local concentration of H2O2 with the protein in the vicinity of these amino-acid residues, allowing photolysis to produces a high local concentration of ·OH where the first ·OH adds onto histidine and abstracts the ·H from arginine, and another ·OH, in close proximity, reacts with the radical intermediate. Such reaction pathways contrast significantly with synchrotron-based HRF,13, 50 where no local fluctuation of ·OH concentration is expected since the radical precursor is water. The two radical intermediates for histidine and arginine have shorter radical lifetimes that minimize the side reactions, causing these two residues to be less likely to uptake oxygen from dissolved O2 and water.
Class 1b residue is tyrosine. Because H2O2 hydrogen bonds to tyrosine as well, the reaction involves H abstraction to give a protein free radical that then is capped by reaction with another ·OH nearby. The unpaired electron of tyrosyl radical is delocalized into the electron-rich aromatic ring, stabilizing it and prolonging its radical lifetime. The longer lifetime makes it possible for tyrosine to react along other reaction routes, for which tyrosine also takes oxygen from dissolved O2 and from water through an exchange process as discussed earlier. We can also view the preference as experimental evidence to support the hydrogen bonding between H2O2 and these amino acid residues.
Class 1c residue is phenylalanine. The oxygen uptake by phenylalanine is similar to that of tyrosine because ·OH addition and abstraction are electrophilic processes, and both aromatic rings are electron-rich, particularly tyrosine. They differ probably because H2O2 does not hydrogen-bound with phenylalanine. Even if the local ·OH concentration is not elevated owing to a lack of a pre-formed H2O2 – phenylalanine complex, the electron density of the phenyl ring still attracts the ·OH radicals. Moreover, ·OH reacts with phenylalanine via multiple pathways, including ·OH addition of the phenyl ring and ·OH abstraction at β-CH2, both favoring a path having a clear preference for taking oxygen from H2O2. Furthermore, the long radical lifetime of the aryl radical, side reaction of taking oxygen from dissolved O2 and from water through an exchange process are weakly competitive facilitated.
Class 2 residue is methionine. Although methionine can be hydrogen bounded to H2O2 prior to laser irradiation, which leads to a high local ·OH concentration after photolysis, methionine has a high reactivity towards with both ·OH or O2 after adding the first ·OH, permitting us to classify methionine differently than class 1 residues.
Class 3 residues include leucine, isoleucine, valine, proline, lysine, aspartic acid, glutamic acid and glutamine. These residues take oxygen nearly exclusively from O2 following similar pathways for each residue (Figure 7e). Although some of these residues (lysine, aspartic acid and glutamine) hydrogen bond to H2O2 to give a high local ·OH concentration, they are sufficiently stable to show high specificity towards O2. Similar preference for these residues was proposed in the HRF system.13
CONCLUSION
To serve a need for elucidating reaction pathways for FPOP, we developed a simple but effective platform for 18O isotopic labeling on a under different reaction conditions and used it to reveal the oxidation pathways for 13 amino-acid residues. There are three classes of residues clearly differentiated based on their choice of oxygen during ·OH labeling. Class 1a and 1b residues provide experimental evidence in supporting hydrogen bonding between H2O2 and selected residues. Although our first focus is +16 modifications, this approach can be applied to the several other modifications that occur with hydroxyl radicals.
This is the first systematic study of FPOP labeling pathways, and the results elevate the fundamental understanding of ·OH-based FPOP chemistry. This foundation allows us to tailor the FPOP conditions to address specific biological questions. For example, the dissolved O2 can be replaced with another free radical (e.g., nitrogen monoxide (NO), a stable radical species with profound biological significance61) to test its reactivity with protein free radicals. NO should be inserted into proteins via class 3 residues but also participate in selected reaction routes of class 1 and 2 residues that require O2 to complete. Another exciting possibility is footprinting by FPOP in anaerobic conditions, where the application of HRF may be limited due to its demand in dissolved O2 during labeling. FPOP brings additional oxygen source (H2O2) into the system and label the protein in a different mechanism (class 1 and 2 residues), making it possible to label selected residues even under reduced oxygen conditions.
Moreover, understanding pathways may enable residue-specific detection of selected biochemical processes based on different reaction mechanisms. By time-dependent and rapid introduction of an 18O source following the initial perturbation, it may be possible to follow protein dynamics even in a residue-specific manner. Deeper mechanistic understandings of ·OH labeling opens new possibilities for the FPOP platform, providing opportunities not clear prior to this fundamental work.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institute of Health NIGMS Grant 5P41GM103422 and instrumentation was supplied by 1S10OD016298-01A1 (to MLG.). Authors are grateful to Protein Metrics for software support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Explanation of degassing apparatus operation, data analysis procedures with detailed example for peak assignment of extracted ion chromatogram and corresponding MS2, the remaining normalized +18O fraction plots for tyrosine, histidine, phenylalanine, methionine, leucine, isoleucine, valine, proline, lysine, aspartic acid, glutamic acid and glutamine residues together with detailed reaction pathways for some of the residues can be found in supporting information..
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interests.
REFERENCES
- 1.Brändén C-I; Tooze J, Introduction to protein structure Taylor & Francis: 1999. [Google Scholar]
- 2.Drenth J, Principles of protein X-ray crystallography Springer Science & Business Media: 2007. [Google Scholar]
- 3.Cavanagh J; Fairbrother WJ; Palmer AG III; Skelton NJ, Protein NMR spectroscopy: principles and practice Elsevier: 1995. [Google Scholar]
- 4.Domon B; Aebersold R, Mass Spectrometry and Protein Analysis. Science 2006, 312 (5771), 212–217. [DOI] [PubMed] [Google Scholar]
- 5.KONG J; YU S, Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochimica et Biophysica Sinica 2007, 39 (8), 549–559. [DOI] [PubMed] [Google Scholar]
- 6.Eftink MR, Fluorescence Techniques for Studying Protein Structure. In Methods of Biochemical Analysis, Suelter CH, Ed. John Wiley & Sons, Inc.: 1991. [DOI] [PubMed] [Google Scholar]
- 7.Whitmore L; Wallace BA, Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89 (5), 392–400. [DOI] [PubMed] [Google Scholar]
- 8.Adrian M; Dubochet J; Lepault J; McDowall AW, Cryo-electron microscopy of viruses. Nature 1984, 308 (5954), 32–36. [DOI] [PubMed] [Google Scholar]
- 9.Li KS; Shi L; Gross ML, Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Accounts of Chemical Research 2018, 51 (3), 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L; Gross ML, Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry. Protein and Peptide Letters 2019, 26 (1), 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z; Smith DL, Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Science 1993, 2 (4), 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hambly DM; Gross ML, Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom 2005, 16 (12), 2057–63. [DOI] [PubMed] [Google Scholar]
- 13.Xu G; Chance MR Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev 2007, 107, 3514–3543. [DOI] [PubMed] [Google Scholar]
- 14.Mendoza VL; Vachet RW, Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrometry Reviews 2009, 28 (5), 785–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gau BC; Sharp JS; Rempel DL; Gross ML, Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Analytical Chemistry 2009, 81 (16), 6563–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu XR; Zhang MM; Rempel DL; Gross ML, Protein-Ligand Interaction by Ligand Titration, Fast Photochemical Oxidation of Proteins and Mass Spectrometry: LITPOMS. Journal of The American Society for Mass Spectrometry 2019, 30 (2), 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XR; Zhang MM; Rempel DL; Gross ML, A Single Approach Reveals the Composite Conformational Changes, Order of Binding, and Affinities for Calcium Binding to Calmodulin. Analytical Chemistry 2019. [DOI] [PMC free article] [PubMed]
- 18.Jiawei Chen DLR, and Michael L Gross, Temperature Jump and Fast Photochemical Oxidation Probe Submillisecond Protein Folding. J. Am. Chem. Soc 2010, 132, 15502–15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J; Rempel DL; Gau BC; Gross ML, Fast Photochemical Oxidation of Proteins and Mass Spectrometry Follow Submillisecond Protein Folding at the Amino-Acid Level. Journal of the American Chemical Society 2012, 134 (45), 18724–18731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B; Cheng M; Rempel D; Gross ML, Implementing fast photochemical oxidation of proteins (FPOP) as a footprinting approach to solve diverse problems in structural biology. Methods 2018, 144, 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maleknia SD; Downard KM, Advances in radical probe mass spectrometry for protein footprinting in chemical biology applications. Chemical Society Reviews 2014, 43 (10), 3244–3258. [DOI] [PubMed] [Google Scholar]
- 22.Jones LM; B. Sperry J; A. Carroll J; Gross ML, Fast Photochemical Oxidation of Proteins for Epitope Mapping. Analytical Chemistry 2011, 83 (20), 7657–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y; Chen G; Wei H; Huang RY-C; Mo J; Rempel DL; Tymiak AA; Gross ML, Fast Photochemical Oxidation of Proteins (FPOP) Maps the Epitope of EGFR Binding to Adnectin. Journal of The American Society for Mass Spectrometry 2014, 25 (12), 2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin M; Krawitz D; Callahan MD; Deperalta G; Wecksler AT, Characterization of ELISA Antibody-Antigen Interaction using Footprinting-Mass Spectrometry and Negative Staining Transmission Electron Microscopy. Journal of The American Society for Mass Spectrometry 2018, 29 (5), 961–971. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y; Wecksler AT; Molina P; Deperalta G; Gross ML, Mapping the Binding Interface of VEGF and a Monoclonal Antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and Mass Spectrometry. Journal of The American Society for Mass Spectrometry 2017, 28 (5), 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calabrese AN; Ault JR; Radford SE; Ashcroft AE, Using hydroxyl radical footprinting to explore the free energy landscape of protein folding. Methods 2015, 89, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinkel F; Gsponer J, Determination of Protein Folding Intermediate Structures Consistent with Data from Oxidative Footprinting Mass Spectrometry. Journal of Molecular Biology 2016, 428 (2, Part A), 365–371. [DOI] [PubMed] [Google Scholar]
- 28.Li KS; Rempel DL; Gross ML, Conformational-Sensitive Fast Photochemical Oxidation of Proteins and Mass Spectrometry Characterize Amyloid Beta 1–42 Aggregation. Journal of the American Chemical Society 2016, 138 (37), 12090–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng Y; Capri J; Waring A; Valentine JS; Whitelegge J, Exposure of Solvent-Inaccessible Regions in the Amyloidogenic Protein Human SOD1 Determined by Hydroxyl Radical Footprinting. Journal of The American Society for Mass Spectrometry 2019, 30 (2), 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espino JA; Mali VS; Jones LM, In Cell Footprinting Coupled with Mass Spectrometry for the Structural Analysis of Proteins in Live Cells. Analytical Chemistry 2015, 87 (15), 7971–7978. [DOI] [PubMed] [Google Scholar]
- 31.Rinas A; Mali VS; Espino JA; Jones LM, Development of a Microflow System for In-Cell Footprinting Coupled with Mass Spectrometry. Analytical Chemistry 2016, 88 (20), 10052–10058. [DOI] [PubMed] [Google Scholar]
- 32.Espino JA; Jones LM, Illuminating Biological Interactions with in Vivo Protein Footprinting. Analytical Chemistry 2019, 91 (10), 6577–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie B; Sharp JS, Hydroxyl Radical Dosimetry for High Flux Hydroxyl Radical Protein Footprinting Applications Using a Simple Optical Detection Method. Analytical Chemistry 2015, 87 (21), 10719–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu B; Mackness BC; Rempel DL; Zhang H; Cui W; Matthews CR; Zitzewitz JA; Gross ML, Incorporation of a Reporter Peptide in FPOP Compensates for Adventitious Scavengers and Permits Time-Dependent Measurements. J Am Soc Mass Spectrom 2017, 28 (2), 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samunl A; Neta P, Electron spin resonance study of the reaction of hydroxyl radicals with pyrrole, imidazole, and related compounds. The Journal of Physical Chemistry 1973, 77 (13), 1629–1635. [Google Scholar]
- 36.Mossoba MM; Rosenthal I; Riesz P, Electron spin resonance of spin-trapped radicals of amines and polyamines. Hydroxyl radical reactions in aqueous solutions and γ-radiolysis in the solid state. Canadian Journal of Chemistry 1982, 60 (12), 1493–1500. [Google Scholar]
- 37.L. Hawkins C; J. Davies M, EPR studies on the selectivity of hydroxyl radical attack on amino acids and peptides. Journal of the Chemical Society, Perkin Transactions 2 1998, (12), 2617–2622.
- 38.McCormick ML; Gaut JP; Lin T-S; Britigan BE; Buettner GR; Heinecke JW, Electron Paramagnetic Resonance Detection of Free Tyrosyl Radical Generated by Myeloperoxidase, Lactoperoxidase, and Horseradish Peroxidase. Journal of Biological Chemistry 1998, 273 (48), 32030–32037. [DOI] [PubMed] [Google Scholar]
- 39.Lassmann G; Eriksson LA; Himo F; Lendzian F; Lubitz W, Electronic Structure of a Transient Histidine Radical in Liquid Aqueous Solution: EPR Continuous-Flow Studies and Density Functional Calculations. The Journal of Physical Chemistry A 1999, 103 (9), 1283–1290. [Google Scholar]
- 40.Rao PS; Simic M; Hayon E, Pulse radiolysis study of imidazole and histidine in water. The Journal of Physical Chemistry 1975, 79 (13), 1260–1263. [Google Scholar]
- 41.Hiller KO; Masloch B; Goebl M; Asmus KD, Mechanism of the hydroxyl radical induced oxidation of methionine in aqueous solution. Journal of the American Chemical Society 1981, 103 (10), 2734–2743. [Google Scholar]
- 42.Solar S; Solar W; Getoff N, Reactivity of hydroxyl with tyrosine in aqueous solution studied by pulse radiolysis. The Journal of Physical Chemistry 1984, 88 (10), 2091–2095. [Google Scholar]
- 43.Torreggiani MTA, A pulse radiolysis study of carnosine in aqueous solution. International Journal of Radiation Biology 1998, 74 (3), 333–340. [DOI] [PubMed] [Google Scholar]
- 44.Ito T; Morimoto S; Fujita S.-i.; Nishimoto S.-i., Radical intermediates generated in the reactions of l-arginine with hydroxyl radical and sulfate radical anion: A pulse radiolysis study. Radiation Physics and Chemistry 2009, 78 (4), 256–260. [Google Scholar]
- 45.Karam LR; Dizdaroglu M; Simic MG, OH Radical-induced Products of Tyrosine Peptides. International Journal of Radiation Biology and Related Studies in Physics, Chemistry and Medicine 1984, 46 (6), 715–724. [DOI] [PubMed] [Google Scholar]
- 46.Goshe MB; Anderson VE, Hydroxyl Radical-Induced Hydrogen/Deuterium Exchange in Amino Acid Carbon-Hydrogen Bonds. Radiation Research 1999, 151 (1), 50–58. [PubMed] [Google Scholar]
- 47.Margolis SA; Coxon B; Gajewski E; Dizdaroglu M, Structure of a hydroxyl radical induced cross-link of thymine and tyrosine. Biochemistry 1988, 27 (17), 6353–6359. [DOI] [PubMed] [Google Scholar]
- 48.Nukuna BN; Goshe MB; Anderson VE, Sites of Hydroxyl Radical Reaction with Amino Acids Identified by 2H NMR Detection of Induced 1H/2H Exchange. Journal of the American Chemical Society 2001, 123 (6), 1208–1214. [DOI] [PubMed] [Google Scholar]
- 49.Maleknia SD; Brenowitz M; Chance MR, Millisecond Radiolytic Modification of Peptides by Synchrotron X-rays Identified by Mass Spectrometry. Analytical Chemistry 1999, 71 (18), 3965–3973. [DOI] [PubMed] [Google Scholar]
- 50.Xu G; Takamoto K; Chance MR, Radiolytic Modification of Basic Amino Acid Residues in Peptides: Probes for Examining Protein–Protein Interactions. Analytical Chemistry 2003, 75 (24), 6995–7007. [DOI] [PubMed] [Google Scholar]
- 51.Xu G; Chance MR, Radiolytic Modification of Acidic Amino Acid Residues in Peptides: Probes for Examining Protein–Protein Interactions. Analytical Chemistry 2004, 76 (5), 1213–1221. [DOI] [PubMed] [Google Scholar]
- 52.Xu G; Chance MR, Radiolytic Modification of Sulfur-Containing Amino Acid Residues in Model Peptides: Fundamental Studies for Protein Footprinting. Analytical Chemistry 2005, 77 (8), 2437–2449. [DOI] [PubMed] [Google Scholar]
- 53.Karmakar T; Balasubramanian S, Elucidating the interaction of H2O2 with polar amino acids – Quantum chemical calculations. Chemical Physics Letters 2014, 613, 5–9. [Google Scholar]
- 54.Buxton GV; Greenstock CL; Helman WP; Ross AB, Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (⋅ OH/⋅ O− in aqueous solution. Journal of physical and chemical reference data 1988, 17 (2), 513–886. [Google Scholar]
- 55.Vassilev P; Louwerse MJ; Baerends EJ, Hydroxyl Radical and Hydroxide Ion in Liquid Water: A Comparative Electron Density Functional Theory Study. The Journal of Physical Chemistry B 2005, 109 (49), 23605–23610. [DOI] [PubMed] [Google Scholar]
- 56.Garrison WM, Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chemical Reviews 1987, 87 (2), 381–398. [Google Scholar]
- 57.Balakrishnan I; Reddy MP, Mechanism of reaction of hydroxyl radicals with benzene in the .gamma. radiolysis of the aerated aqueous benzene system. The Journal of Physical Chemistry 1970, 74 (4), 850–855. [Google Scholar]
- 58.Schoeneich C; Aced A; Asmus KD, Mechanism of oxidation of aliphatic thioethers to sulfoxides by hydroxyl radicals. The importance of molecular oxygen. Journal of the American Chemical Society 1993, 115 (24), 11376–11383. [Google Scholar]
- 59.Barata-Vallejo S; Ferreri C; Postigo A; Chatgilialoglu C, Radiation Chemical Studies of Methionine in Aqueous Solution: Understanding the Role of Molecular Oxygen. Chemical Research in Toxicology 2010, 23 (1), 258–263. [DOI] [PubMed] [Google Scholar]
- 60.Davies MJ, The oxidative environment and protein damage. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2005, 1703 (2), 93–109. [DOI] [PubMed] [Google Scholar]
- 61.Vincent SR, Nitric oxide: A radical neurotransmitter in the central nervous system. Progress in Neurobiology 1994, 42 (1), 129–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.