Summary
Seeds are dormant and desiccated structures, filled with storage products to be used after germination. These properties are determined by the maturation program, which starts, in Arabidopsis thaliana, mid-embryogenesis, at about the same time and developmental stage in all the seeds in a fruit. The two factors, chronological and developmental time, are closely entangled during seed development, so their relative contribution to the transition to maturation is not well understood. It is also unclear whether that transition is determined autonomously by each seed or whether it depends on signals from the fruit. The onset of maturation follows the cellularization of the endosperm, and it has been proposed that there exists a causal relationship between both processes. We explored all these issues by analyzing markers for maturation in Arabidopsis mutant seeds that develop at a slower pace, or where endosperm cellularization happens too early, too late, or not at all. Our data show that the developmental stage of the embryo is the key determinant of the initiation of maturation, and that each seed makes that transition autonomously. We also found that, in contrast with previous models, endosperm cellularization is not required for the onset of maturation, suggesting that this transition is independent of the hexose/sucrose ratio in the seed. Our observations indicate that the mechanisms that control endosperm cellularization, embryo growth, and embryo maturation act independently of each other.
Keywords: seed, embryo, maturation, endosperm, Arabidopsis thaliana
Introduction
The development of organisms is characterized by the ordered progression through a set of stages, by the duration of those stages, and by the timing of the transitions between them. Development can thus be described as the interaction between two kinds of “time”: developmental time (defined by the sequence of stages) and chronological time (age) (Ebisuya and Briscoe 2018). The relationship between both types of time is distinctive for each species. Heterochronic changes, modifications in the relative timing and duration of developmental programs, have been proposed to be a significant driver of morphological evolution (Gould 1977, Geuten and Coenen 2013, Buendía-Monreal and Gillmor 2018). For instance, in snakes the faster oscillation of a developmental clock relative to the overall chronological time of development leads to a large increase in the number of vertebrae (Gomez, Özbudak et al. 2008). In the parasitic plant Rafflesia embryonic development is terminated very early, as a proembryo, followed directly by germination on its host (Nikolov, B. et al. 2014). In many plants, modifications of the vegetative phase change pathway, in particular the relative duration of the juvenile and adult stages, have been postulated to influence the variation of flowering time and leaf shape (Buendía-Monreal and Gillmor 2018). Organisms have, as a consequence, evolved timing mechanisms that coordinate age with stage, the passage of time with cell division and tissue differentiation. Some of these timers are internal, or autonomous, to the tissue in question. These include the gradual accumulation or depletion of a product, until its concentration crosses a threshold (count-up and count-down timers), and molecular oscillators based on delayed negative-feedback circuits (like the circadian clock) (Ebisuya and Briscoe 2018). These timers can be based on transcription factors, small RNAs, hormones, or metabolites (Buendía-Monreal and Gillmor 2018, Ebisuya and Briscoe 2018). Other timers depend on environmental signals, whether external to the individual (such as temperature or daylength), or emanating from a different tissue, like the non-autonomous action of a hormone or a metabolite. Finally, external and internal timers may interact to ensure proper development, like the entrainment of the oscillations of the circadian clock by daylight (Oakenfull and Davis 2017).
Embryogenesis in most eudicot plants is characterized by a specific sequence of developmental stages defined based on the shape of the embryo, rather than on the specific sequence of cell divisions (Natesh and Rau 1984). The fertilization of the female gametophyte leads to the formation of two tissues: the embryo and the endosperm. The development of the embryo is usually divided into two phases, morphogenesis and maturation (Goldberg, de Paiva et al. 1994). Morphogenesis, lasting until the late heart or torpedo stages, establishes the basic body plan of the embryo. Embryo maturation, in turn, leads to a dry, resilient, nutrient-filled seed. This latter process was a key innovation leading to the evolutionary success of the angiosperms (Linkies, Graeber et al. 2010). During early maturation, also known as “seed filling”, the embryos turn green (in 40% of plant families; (Yakovlev and Zhukova 1980)) and accumulate storage products which, in the case of Arabidopsis and related oilseed plants, are proteins (cruciferins/12S globulins and arabins/2S albumins) and oils (Baud, Dubreucq et al. 2008). Late maturation involves the loss of water (desiccation) and the establishment of a dormant state (Leprince, Pellizzaro et al. 2016). The endosperm serves as a support tissue for the embryo and, depending on the species, it can be degraded as the embryo grows, or remain as part of the mature seed (Vijayaraghavan and Prabhakar 1984).
Early maturation in Arabidopsis embryos becomes first apparent around the early heart stage. At this time chloroplast development and accumulation of chlorophyll start in the protoderm and then expand centripetally. Color becomes apparent at the late heart stage (embryo greening) (Fig. 1, Fig. S1a-c) (Mansfield and Briarty 1991, Willmann, Mehalick et al. 2011). In oilseeds like Arabidopsis, photosynthesis is necessary for the accumulation of storage lipids (Goffman, Alonso et al. 2005, Liu, Wang et al. 2017), and therefore appears to be an integral part of the seed maturation process. The onset of seed filling itself is first evidenced by the expression of the genes encoding the seed storage proteins (SSPs), enzymes involved in the synthesis of storage lipids, and proteins associated with oil bodies (oleosins). This occurs at the late heart to early torpedo stages (Fig. 1) (Guerche, Tire et al. 1990, Mansfield and Briarty 1992, Baud and Lepiniec 2009, Belmonte, Kirkbride et al. 2013, Miquel, Trigui et al. 2014).
Figure 1:
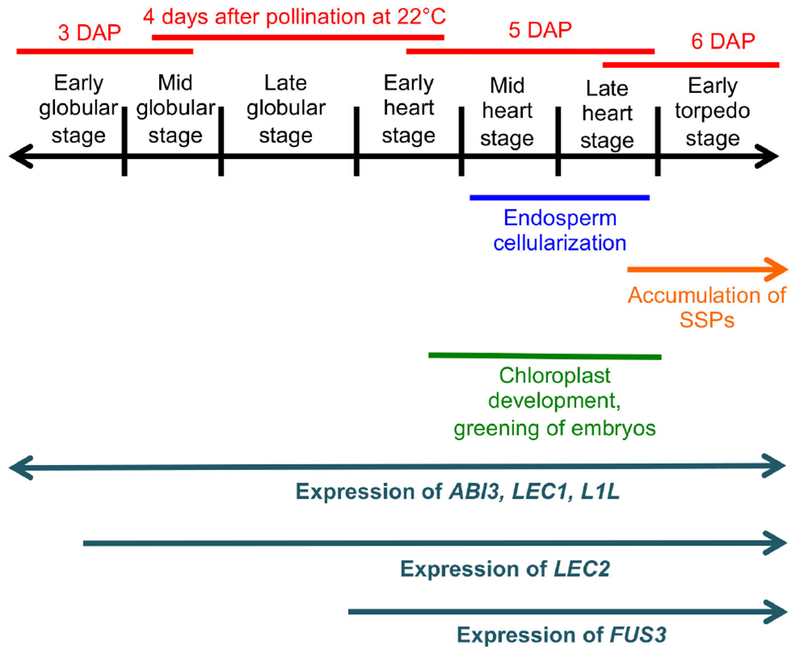
Events occurring in the seeds around the time of onset of the maturation program.
The transition between morphogenesis and early maturation involves a dramatic change in the seed transcriptome, evidencing a need for a tight regulation of its timing (Belmonte, Kirkbride et al. 2013). Although this process has been intensively studied in Arabidopsis, it is not clear whether developmental time (stage) or chronological time (age, measured in days after pollination, DAP), or a combination of both trigger maturation. It is also not well understood whether an internal timer or a signal external to the embryo drive the process. All the seeds in an Arabidopsis silique develop at similar rates. This means that in any given silique the embryos, all of which are of the same chronological age, comprise a narrow range of developmental stages (Fig. S3, 4) (Jürgens and Mayer 1994, Jenik, Jurkuta et al. 2005). A consequence of this fact is that maturation starts both at the heart/late heart stage but also at 5-6 DAP (Fig. 1, Fig. S3, 4). It has therefore been difficult to disentangle the relative contribution of these two factors to the regulation of its onset.
Research so far has focused mainly on internal factors that promote the maturation program. There is also some evidence of negative control mechanisms, which prevent premature maturation (Nodine and Bartel 2010, Willmann, Mehalick et al. 2011). The central regulators (the “master genes”) that initiate maturation are known as the LAFL genes, from their initials (LEAFY COTYLEDON1 [LEC1], LEC1-LIKE [L1L], ABSCISIC ACID INSENSITIVE3 [ABI3], FUSCA3 [FUS3], and LEC2). These genes encode transcription factors that are conserved across angiosperms (Lotan, Ohto et al. 1998, Siefers, Dang et al. 2009, Carbonero, Iglesias-Fernández et al. 2017). The LAFL genes are expressed sequentially during embryo development, with L1L, LEC1 and ABI3 starting very early, followed by LEC2 at the early/mid globular stage, and FUS3 at the late globular/early heart stage (Parcy, Valon et al. 1994, Lotan, Ohto et al. 1998, Stone, Kwong et al. 2001, Kwong, Bui et al. 2002, Kroj, Savino et al. 2003, Tsuchiya, Nambara et al. 2004) (Fig. 1, Fig. S1d-f). The LAFL proteins cooperate to turn on all the genes related to maturation by interacting with each other and with other transcriptional regulators (reviewed by (Boulard, Fatihi et al. 2017). Loss-of-function mutations in the LAFL genes lead to the reduction or elimination of SSPs and lipids. These mutants also have significant defects in later maturation processes, such as growth arrest, desiccation tolerance, and dormancy (Keith, Kraml et al. 1994, Meinke, Franzmann et al. 1994, Parcy, Valon et al. 1997, Raz, Bergervoet et al. 2001, Kroj, Savino et al. 2003, Yamamoto, Kagaya et al. 2009, Roscoe, Guilleminot et al. 2015).
Seeds have to coordinate their development with the maturation of the surrounding fruit. While there are some known examples of seeds signaling to the fruit (Seymour, Østergaard et al. 2013, Giovannoni, Nguyen et al. 2017), to our knowledge, very few signals in the reverse direction have been described. One example is the control of seed dormancy by the fruit, which senses the temperature history of the plant (Chen, MacGregor et al. 2014). The hormone abscisic acid (ABA), synthesized both in the fruit and the embryo, has been proposed to be a maternal signal that triggers early maturation (Karssen, Brinkhorst-van der Swan et al. 1983, Kanno, Jikumaru et al. 2010). However, the available evidence for this effect is contradictory (Koornneef, Hanhart et al. 1989, Parcy, Valon et al. 1994). Sugars may have an effect on late maturation, as the amount of photosynthesis in the silique wall correlates with the oil content of the seed (Hua, Li et al. 2012).
A hypothetical signal for maturation that is endogenous to the seed is a redistribution of sugars resulting from the cellularization of the endosperm. Studies in legumes led to the proposal that early seeds have a high hexose/sucrose ratio that promotes embryo cell division, while a later shift to low hexose/sucrose ratios below a certain threshold (“sugar switch”) triggers maturation and accumulation of storage products (reviewed by (Weber, Borisjuk et al. 2005)). Only about half of plants species have a cellularizing endosperm (Geeta 2003), but the sugar switch was nonetheless proposed as a general mechanism for the control of maturation (Weber, Heim et al. 1998). In Arabidopsis the endosperm develops first as a syncytium, and contains a large central vacuole (Morley-Smith, Pike et al. 2008). This vacuole has been suggested to serve as a sink for sucrose and accumulates high levels of hexoses. The endosperm starts cellularizing at the early heart stage, consuming the vacuole in the process (Mansfield and Briarty 1990, Brown, Lemmon et al. 1999) and transforming the embryo into the main sucrose sink, lowering its hexose/sucrose ratio. This infusion of sucrose is necessary for embryo growth and overall viability (Hehenberger, Kradolfer et al. 2012, Lafon-Placette and Köhler 2014). If the “sugar switch” hypothesis applies to Arabidopsis, endosperm cellularization should also be crucial for seed maturation (Wobus and Weber 1999).
In this context, one goal of our study was to answer whether chronological age and/or developmental stage trigger maturation. We also wanted to discern whether seeds control their developmental progression autonomously or whether there is a silique-wide maternal signal that determines it. Finally, we sought to investigate whether the cellularization of the endosperm plays a role in the onset of maturation. The present work only addresses the regulation of early maturation, and does not focus at all on the events of late maturation (reviewed by (Leprince, Pellizzaro et al. 2016, Shu, Liu et al. 2016). We analyzed seeds in siliques from self-pollinated plants heterozygous for recessive mutations that result in slower embryo development. In this way we were able to uncouple age and stage, and also to evaluate whether the seeds matured autonomously. Our data suggest that developmental stage, and not chronological time, is key to the transition between morphogenesis and the onset maturation. Furthermore, our observations are consistent with the seeds developing autonomously, independent of any putative fruit signal. By analyzing mutants with defective endosperm cellularization, we also showed that this process is not required for the onset of maturation. This implies that embryo maturation may be uncoupled from its growth and from the development of the endosperm.
Results
The LAFL genes are important for the proper timing of embryo greening
The development of chloroplasts and subsequent greening of the embryo are the first visible signs of its maturation. There is very little known about the control of this process. The LAFL genes are obvious candidates for positive regulators of greening, since they regulate other aspects of early embryo maturation, in particular seed filling (Meinke, Franzmann et al. 1994, Parcy, Valon et al. 1997, Kroj, Savino et al. 2003, Roscoe, Guilleminot et al. 2015). Although FUS3 and LEC1 have been shown to regulate photosynthetic genes (Yamamoto, Kagaya et al. 2010, Pelletier, Kwong et al. 2017), all lafl single and double mutant embryos contain chlorophyll at the mature green stage. The levels are in some cases reduced (fus3-3), patchy (lec2-1), or mostly limited to the cotyledons (lec1) (Meinke, Franzmann et al. 1994, Kroj, Savino et al. 2003). However, the extent to which the LAFL genes regulate the onset of greening has not been characterized in detail.
To our knowledge, there is no established protocol to analyze greening in embryos. In the past, researchers have looked at individual embryos either by excising them from the seed (e.g. (Li and Thomas 1998, Liu, Wang et al. 2017) and Fig. S1a), or by looking at chlorophyll fluorescence with a confocal microscope (e.g. (Allorent, Osorio et al. 2015). In order to assess greening in large numbers of seeds, we used differential interference contrast (DIC) microscopy to look at single, double and triple lafl mutant seeds cleared in Hoyer’s solution, and compared them to their respective wild type accessions. Although treatment with Hoyer’s eventually leaches the chlorophyll out of the seed, incubating freshly collected seeds at room temperature for 4-6 hours resulted in enough clearing to stage the embryos while still retaining most of the chlorophyll (e.g. (Willmann, Mehalick et al. 2011). This method allowed us to distinguish the greening of the embryo from that of the endosperm (Fig. S1b, c), and was robust and reproducible (Fig. S1j). It was therefore used for subsequent experiments as well. It’s worth keeping in mind that what we are assessing is greening, but that chlorophyll accumulation starts a stage earlier (Fig. S1a). The lafl alleles used have all been reported to be nulls. The only exception is fus3-3, which produces a misspliced product that leads to strong early maturation defects (Nambara, Keith et al. 1994, Lotan, Ohto et al. 1998, Stone, Kwong et al. 2001, Tiedemann, Rutten et al. 2008).
Wild type embryos turn green mostly during the late heart stage, with some showing color at the heart stage, the greening being nearly complete by early torpedo stage (Table 1, Fig. S1a-c, j) (Mansfield and Briarty 1991, Willmann, Mehalick et al. 2011).
Table 1: Embryo greening in wild type and lafl single, double, and triple mutants.
lec1-1 and lec1-2 are in Wassilewskija (Ws) background, while fus3-3, lec2-1 and abi3-3 are in Columbia (Col). Asterisks mark significant differences between the single mutant and the corresponding wild type at that stage. For the double and triple mutants asterisks mark significant differences with at least one of the single mutants (for the doubles) or one of the double mutants (for the triples) at that stage. For sample sizes and p-values for Fisher’s Exact Test for each genotype and stage see Tables S1a-d.
| Percentage of green embryos at each stage | ||||||
|---|---|---|---|---|---|---|
| Genotype | Early heart | Mid heart | Late heart | Early torpedo | Mid torpedo | Late torpedo |
| Ws | 0% | 12% | 55% | 80% | 100% | 100% |
| lec1-1 | 0% | 0%** | 0%** | 21%** | 60%** | 90%** |
| Col | 0% | 22% | 66% | 100% | 100% | 100% |
| fus3-3 | 0% | 0%** | 0%** | 11%** | 58%** | 100% |
| lec2-1 | 0% | 12% | 42%** | 76%** | 100% | 100% |
| abi3-3 | 0% | 0%** | 11%** | 50%** | 90%** | 100% |
| abi3-3 lec1-2 | 0% | 0% | 9%** | 26% | 64% | 92% |
| abi3-3 lec2-1 | 0% | 0% | 8% | 32%** | 71%** | 100% |
| fus3-3 lec1-2 | 0% | 0% | 10%** | 23% | 74%* | 94% |
| fus3-3 lec2-1 | 0% | 0% | 7%* | 29%* | 62% | 94% |
| lec1-2 lec2-1 | 0% | 0% | 16% | 38%** | 72%* | 90% |
| lec1-2 lec2-1 fus3-3 | 0% | 0% | 9% | 23% | 56%** | 84% |
| lec1-2 lec2-1 abi3-3 | 0% | 0% | 5%* | 17%** | 49%** | 88% |
: p < 0.05;
: p < 0.01
All single lafl mutants showed a delay in embryo greening (Table 1, Table S1a). The most severe were lec1-1 and fus3-3, with onset at the early torpedo stage. These are followed by abi3-3, with onset at the late heart stage. Finally, lec2-1 had the mildest phenotype, starting to green at mid heart, like the wild type, but showing lower percentages of green embryos until the mid-torpedo stage. Most of the mutant embryos are green by the late torpedo stage. Remarkably, the severity of these mutants in terms of greening largely mirrors their defects in the accumulation of SSPs and oils (Meinke, Franzmann et al. 1994, Parcy, Valon et al. 1997, Kroj, Savino et al. 2003, Roscoe, Guilleminot et al. 2015). This suggests that individual LAFL genes regulate chloroplast development to a similar degree than they do other aspects of early maturation. The double mutant embryos (abi3-3 lec1-2, abi3-3 lec2-1, fus3-3 lec1-2, fus3-3 lec2-1 and lec1-2 lec2-1) also greened considerably later than the wild type ones, but the phenotype was generally intermediate between that of the two mutant alleles, and more similar to the stronger one (Table 1, Tables S1a-c). Of the two triple mutant combinations we analyzed, lec1-2 lec2-1 fus3-3 was not different from the double mutants, while lec1-2 lec2-1 abi3-3 was only slightly stronger (Table 1, Table S1d). The distribution and amounts of chlorophyll in older multiple mutant embryos corresponded to that of the single mutants that made them up: combinations with lec1-2 had very pale or white hypocotyls, with lec2-1 patchy or white cotyledons, and with fus3-3 less chlorophyll overall (Fig. S2a-h).
In light of these results, we can conclude that embryo greening is part of the early maturation program and that it is at least partially regulated by the same transcription factors. The lack of significant additive effects in double and triple mutants shows that this process, like others regulated by the LAFL genes, is controlled by these products acting together in the same pathway (Baud, Kelemen et al. 2016, Pelletier, Kwong et al. 2017). There is spatial variation in the requirements, with LEC1 having a much more important role in the hypocotyl than in the cotyledons. Similar spatial differences are seen in the cross regulation of LAFL genes (To, Valon et al. 2006), and may underlie these disparities. Unlike SSPs and storage oils, whose accumulation is strictly dependent on the action of the LAFL factors, chloroplast development proceeds even in the absence of their activity. These results suggest that, in addition to LAFL function, additional transcription factors may participate in turning on the pathways for chlorophyll synthesis and chloroplast maturation. Potential candidates are AGL23, GOLDEN2-LIKE1 and 2, and GNC and GNC-LIKE, whose mutations lead to absent or underdeveloped chloroplasts (Fitter, Martin et al. 2002, Colombo, Masiero et al. 2008, Bastakis, Hedtke et al. 2018, Zubo, Clabaugh Blakley et al. 2018).
The onset of maturation is seed autonomous and depends on developmental stage, not the time elapsed since fertilization
Two of the questions we wanted to explore were whether developmental stage or time since fertilization (in a constant environment) control the onset of embryonic maturation, and whether each seed autonomously regulates maturation or there is a coordinated response involving all the seeds in a silique. To answer these questions, we needed to separate age from stage, and to be able to evaluate maturation in individual seeds in a silique. We took advantage of recessive mutations that slow down embryonic development with only minor effects on patterning and morphology. Siliques of heterozygous plants were collected at specified time points after manual self-pollination. These siliques contained seeds that were all of the same age but had attained different stages, the wild type seeds being developmentally ahead of the mutant ones. This approach effectively uncoupled age and stage. The seeds were all enclosed in the same maternal tissue. Thus, if there were a maternal signal, it would reach all of them at the same time. We chose three parameters to evaluate the onset of maturation: embryo greening, the expression of the master regulatory gene FUS3 (using a FUS3p:GUS transcriptional reporter), and the expression of the SSP gene At2S3 (using a At2S3p:GFP transcriptional reporter) (Kroj, Savino et al. 2003). To ensure that our results addressed the regulation of maturation and not the idiosyncrasies of a particular mutant or biochemical process, we compared two separate mutants that affected different pathways: fusca12-U228 (fus12) and tilted1-4 (til1-4). fus12 is a null allele of the gene encoding the CSN2 subunit of the COP9 signalosome, which modulates the activity of the cellular protein degradation machinery (Schwechheimer and Isono 2010). fus12 embryos grow at a slower rate than wild type ones, likely because the COP9 complex indirectly influences the cell cycle (Betsch, Boltz et al. 2019). The fus12 allele shows low maternal transmission, resulting in 15% or fewer mutant embryos in self-pollinated heterozygous siliques (Franciosini, Moubayidin et al. 2015). til1-4 is a strong hypomorphic allele of the gene coding for DNA polymerase epsilon. Homozygous mutants show longer cell cycles and consequent retarded development (Jenik, Jurkuta et al. 2005).
fus12 is seedling lethal (Franciosini, Moubayidin et al. 2015) and til1-4 homozygotes have an extremely reduced seed set (Jenik, Jurkuta et al. 2005). Therefore, the comparisons presented below are always between homozygous mutant embryos and wild type embryos either from the same siliques or from sibling wild type plants grown at the same time. This also controls for the minor differences in growth rate we observed depending on genetic background or growth conditions. In all cases studied, wild type embryos behaved in the same manner whether developing in homozygous wild type or in heterozygous siliques.
The FUS3p:GUS reporter is expressed first in the suspensor, as early as the dermatogen stage. Then it is turned on in the embryo proper at the late globular/early heart stage, where it persists for most of embryogenesis (Kroj, Savino et al. 2003) (Figs. S1d-f, S3, S4). This reporter had the unfortunate tendency to get silenced in a fraction of the seeds whenever crossed into any mutant background. Therefore, seeds showing no GUS staining at all were not considered. Embryos with only suspensor staining were scored as “negative” for expression, while embryos that stained in both the suspensor and embryo proper were scored as “positive”. The At2S3p:GFP reporter is expressed first at the late heart or early torpedo stage, in a few cells of the hypocotyl, and then spreads out in older embryos (Willmann, Mehalick et al. 2011) (Figs. S1g-i, S3, S4). In this case we used plant lines that were homozygous for the reporter. Because we were interested in the onset of expression, embryos with just one GFP-expressing cell were counted as “positive” for expression.
For both mutants we first present the percentage of wild type and mutant embryos of different stages at each time point that expressed the marker under study (Figs. 2 and 3, panels a, d, g). fus12 and til1-4 embryos show mild root pole defects starting at the early to mid-globular stages (Jenik, Jurkuta et al. 2005, Franciosini, Moubayidin et al. 2015). To decide which embryos were mutant (those to the left of the red line in Figs. 2, 3 a, d, g), we scored for these defects and also considered the delay when compared to embryos in sibling wild type plants at that time point (Figs. S3, 4). We then collated all the wild type or mutant embryos and sorted them by age or by stage, to facilitate comparisons (Figs. 2 and 3, panels b, c, e, f, h, i).
Figure 2: Greening and expression of maturation reporters in fus12 embryos.
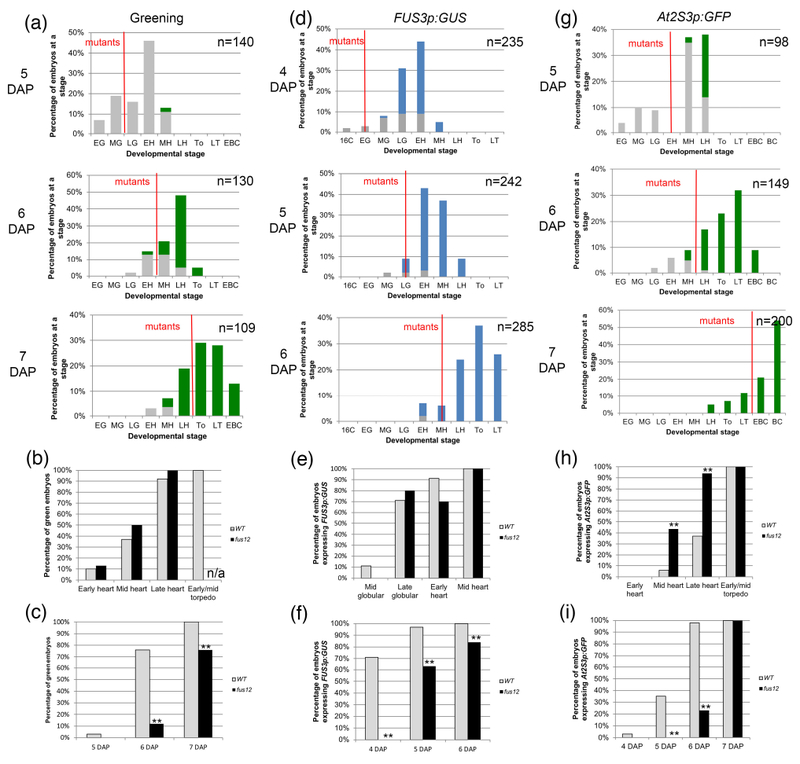
(a, d, g) Distribution of embryo stages in FUS12/fus12 siliques at different times after pollination. The embryos to the left of the red line are the fus12 embryos. A red line bisecting a bar indicates that the stage contained both wild type and mutant embryos. Stages: E; early; M: mid; L: late; 16C: 16-cell; G: globular; H: heart; T: torpedo (To: early/mid torpedo); BC: bent cotyledon. (a) Embryo greening. Gray bars denote white embryos, green bars green embryos. (b) Green embryos from the graphs in (a) sorted by developmental stage. (c) Green embryos from the graphs in (a) sorted by developmental time (DAP). (d) Expression of FUS3p:GUS. Gray bars denote GUS− embryos, blue bars GUS+ embryos. (e) GUS+ embryos from the graphs in (d) sorted by developmental stage. (f) GUS+ embryos from the graphs in (d) sorted by developmental time (DAP). (g) Expression of At2S3p:GFP. Gray bars denote GFP− embryos, green bars GFP+ embryos. (h) GFP+ embryos from the graphs in (g) sorted by developmental stage. (i) GFP+ embryos from the graphs in (g) sorted by developmental time (DAP). For (b), (c), (e), (f), (h) and (i) the sample sizes per condition and p-values from Fisher’s Exact Test between wild type and mutant are listed on Tables S2a-c (*: p < 0.05; **: p < 0.01). n/a: not applicable, the mutant embryos hadn’t reached that stage by the last time point.
Figure 3: Greening and expression of maturation reporters in til1-4 embryos.
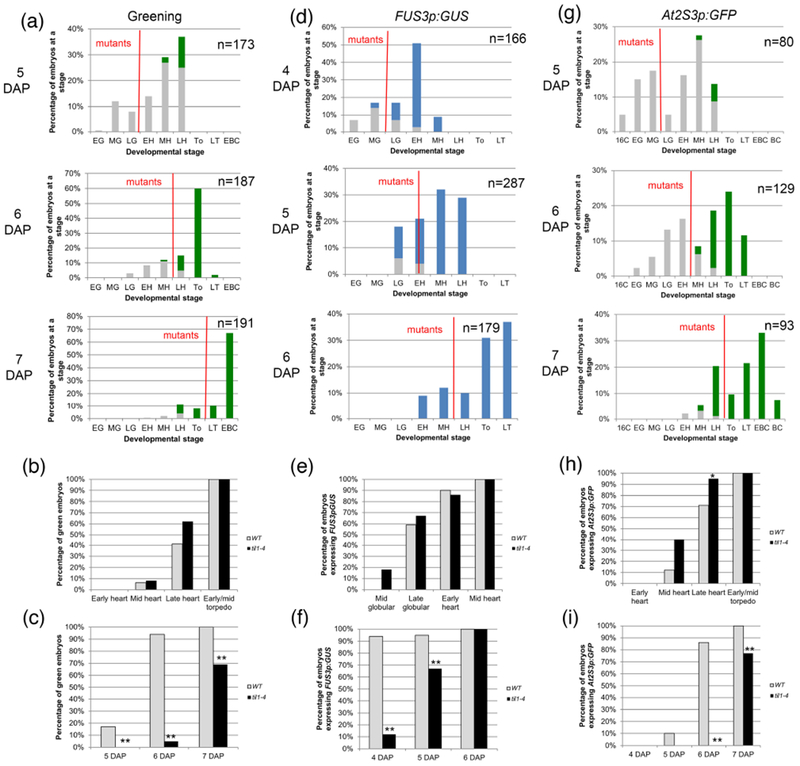
(a, d, g) Distribution of embryo stages in TIL1/til1-4 siliques at different times after pollination. The description of panels (a) to (i) is the same as in Figure 3. For (b), (c), (e), (f), (h) and (i) the sample sizes per condition and p-values from Fisher’s Exact Test between wild type and mutant are listed on Tables S2a-c.
fus12 embryos turned green following the same developmental trajectory as wild type ones, starting at the early heart stage and being mostly completed by the late heart stage (Fig. 2a, b; Table S2a). Because a wild type early heart corresponded to 5 DAP, while a fus12 early heart to 6 DAP, they greened at different ages (Fig. 2c; Table S2a). We observed the same pattern with FUS3p:GUS, with expression in the embryo proper appearing at the same stage in both wild type and fus12 (some at the late globular stage, the rest at the early heart stage) (Fig. 2d, e; Table S2b). This meant that fus12 embryos turned on the reporter a day later than wild type ones (5 vs. 4 DAP) (Fig 2f; Table S2b). For At2S3p:GFP both genotypes initiated expression at the mid heart stage, but it progressed developmentally faster in the mutant (94% vs. 37% of late heart stage embryos expressing it) (Fig. 2g, h; Table S2c). Again, this was significantly later in time for fus12 than for wild type (6 vs. 4 DAP) (Fig. 2i; Table S2c).
Greening also had the same stage of onset (mid heart stage) and developmental progression in til1-4 embryos compared to wild type embryos (Fig. 3a, b; Table S2a). It started at 5 DAP in wild type embryos but, because of the slower growth, one day later in the mutants (Fig. 3c; Table S2a). Wild type and til1-4 expressed FUS3p:GUS at the same stages as well (onset mostly at late globular), but at day apart (Fig. 3d-f; Table S2b). Finally, the same behavior was observed for At2S3p:GFP, with onset for both genotypes at mid heart stage but a day later for the mutant (the difference at the late heart stage was barely significant, p = 0.389) (Fig. 3g-i; Table S2c).
Our data from the analysis of slower-developing embryos strongly suggest that what determines the onset of maturation (at least as defined by the proxies used in our experiments) is the developmental stage and not the time after pollination. It is also very clear from our observations that each seed begins maturation autonomously, since embryos in the same maternal environment matured at different chronological times.
The timing of endosperm cellularization does not determine the onset of maturation
The timing of endosperm cellularization is tightly correlated with that of maturation during the development of wild type seeds. The endosperm starts to cellularize during the early heart stage. At around this stage FUS3 is expressed in the embryo proper, and the embryos start accumulating chlorophyll. A couple of stages later the embryos begin to express the genes encoding or responsible for seed storage products (Fig. 1). It has been hypothesized that this correlation is not just temporal but causal: endosperm cellularization leads to a redistribution of sucrose, and this triggers the maturation process (Weber, Heim et al. 1998, Wobus and Weber 1999, Weber, Borisjuk et al. 2005).
We realized we could take advantage of the slowly developing mutant embryos to see whether the correlation held. We could compare the stage at which cellularization occurred with the stage at which maturation began in these mutants and the corresponding wild types. We observed endosperm cellularization primarily in fresh seeds cleared in Hoyer’s (Fig. S5a, b), which allows for the observation of large numbers of seeds. This method provides equivalent data to the more traditional, but laborious, analysis of sectioned and stained tissue (Fig. S5f, g). We analyzed both heterozygous and homozygous wild type siliques, and scored for mutant embryos as described above.
In fus12 seeds the endosperm cellularized at the same stage as in wild type seeds (Fig. 4a; Table S2d), confirming our previous qualitative observations (Franciosini, Moubayidin et al. 2015). Because fus12 and wild type embryos also mature at the same stage, the correlation between cellularization and maturation holds in this case. In contrast, in til1-4 endosperm cellularization occurred much earlier than in the wild type, at the mid to late globular stage (Fig. 4b; Table S2d). The data for til1-4 are, again, consistent with our previous work (Jenik, Jurkuta et al. 2005). These results indicate that endosperm cellularization is not always correlated with maturation as previously postulated.
Figure 4: Endosperm cellularization in mutants with delayed development, and greening and expression of maturation reporters in embryos from seeds with altered endosperm cellularization.
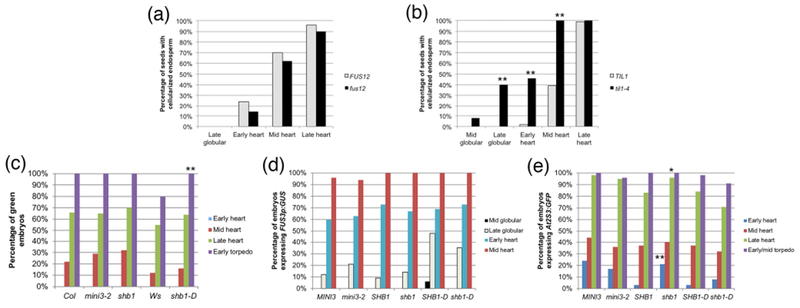
(a, b) Percentage of embryos at a certain stage in seeds with cellularized endosperm in (a) fus12 and wild type (FUS12) seeds, (b) til1-4 and wild type (TIL1) seeds. (c) Greening, (d) FUS3p:GUS expression, and (e) At2S3p:GFP expression in embryos from seeds with altered timing of endosperm cellularization. In (d) and (e) MINI3, SHB1 and SHB1-D refer to the wild type siblings of the corresponding homozygous mutants. The sample sizes per condition and p-values from Fisher’s Exact Test between wild type and mutant are listed on Tables S2d and S3a-c (*: p < 0.05; **: p < 0.01).
To further explore the connections between cellularization and maturation, and to see if we could buttress the conclusions derived from the previous experiment, we made use of mutants that are known to cellularize the endosperm either at an earlier or later embryonic stage than the wild type. In this experiment the stage of cellularization was known, and we tested whether the stage of the onset of maturation was altered. MINISEED3 (MINI3) and SHORT HYPOCOTYL UNDER BLUE1 (SHB1) encode transcription factors that act in the same pathway to regulate the timing of endosperm cellularization (Kang, Li et al. 2013). The null alleles mini3-2 and shb1 result in identical phenotypes, with endosperm cellularization starting at the late globular stage and ending at the mid heart stage, one embryonic stage earlier than wild type (Fig. S5c) (Luo, Dennis et al. 2005). shb1-D is a hypermorphic allele with the opposite phenotype: cellularization starts at the late heart stage and is completed at the early torpedo stage (one embryonic stage later than wild type) (Fig. S5d, e) (Zhou, Zhang et al. 2009).
We evaluated the same parameters as before: greening and expression of FUS3p:GUS and At2S3p:GFP. For greening, homozygous mutant plants were compared to the corresponding wild type accessions (Col for mini3-2 and shb1, Ws for shb1-D). In the case of the reporters, we compared homozygous mutant plants from the F2 generation of the cross to the reporters with segregating sibling homozygous wild type plants (labeled MINI3, SHB1, and SHB1-D, respectively, in Fig. 4c-e), to control for possible differences in genetic background. Remarkably, for all three markers the onset of maturation in all mutants was at the same embryonic developmental stage as in the wild types (Fig. 4c-e; Tables S3a-c). The only exception was a higher percentage of shb1 early heart embryos expressing At2S3p:GFP (21% vs. 3% in SHB1) (Fig. 4e; Table S3c). The data confirm the absence of correlation between the timing of endosperm cellularization and the beginning of maturation: regardless of when cellularization happens maturation starts at the same embryonic stage. Even more interestingly, in shb1-D the endosperm finishes cellularizing after the embryo has greened and At2S3p:GFP expression has been induced, further affirming our conclusions.
The onset of embryo maturation does not require a cellularized endosperm
Given the last intriguing observation, that maturation can start before endosperm cellularization is initiated, we wondered whether the onset of maturation required endosperm cellularization at all. According to the “sugar switch” model discussed earlier, maturation should depend on endosperm cellularization. If this were correct, mutants where the endosperm does not cellularize should not progress to the maturation phase.
Seeds mutant for gametophytic subunits of the Polycomb Repressive Complex 2 (PRC2) fail to cellularize their endosperm. In these seeds the embryos arrest with a late heart stage morphology, and eventually the seeds shrivel and die. These are gametophytic maternal effect mutations: heterozygous self-pollinated plants produce 50% mutant seeds (Sørensen, Chaudhury et al. 2001, Ingouff, Haseloff et al. 2005). We picked two of these mutants for our studies, fertilization independent seed2-6 (fis2-6) and fertilization independent endosperm-12 (fie-12), both null alleles with indistinguishable seed phenotypes (Guitton, Page et al. 2004, Wolff, Weinhofer et al. 2011). To make sure that the effects we saw were due to defects in endosperm cellularization and not just abnormal PRC2 complex activity, we chose a mutant in an unrelated pathway, endosperm defective1 (ede1-1). EDE1 encodes a microtubule-associated protein and mutants show aberrant or absent cytokinesis, leading to an uncellularized endosperm and embryonic arrest in about 11% of the seeds of homozygous plants (Pignocchi, Minns et al. 2009, Hehenberger, Kradolfer et al. 2012).
In fis2 and fie mutants embryo development is indistinguishable from wild type until the time of embryo cellularization (Sørensen, Chaudhury et al. 2001, Ingouff, Haseloff et al. 2005). In ede1-1 it is not possible to predict which seeds will fail to cellularize (Pignocchi, Minns et al. 2009). We concentrated our analysis, therefore, in seeds where cellularization had failed (and hence were mutant) and where the wild type (or wild type-looking) embryos in the same silique were either in the early maturation (early torpedo to early bent cotyledon) or late maturation (bent cotyledon to almost full seed) phases. Wild type embryos at these stages are all green, and all express FUS3p:GUS and At2S3p:GFP (Figs. S3, 4). Accordingly, the question was simply whether the mutant embryos expressed these markers of maturation or not.
A majority of embryos for all three mutants turned green and expressed the two reporters (see Table S3d for sample sizes). 74 to 89% of the embryos expressed FUS3p:GUS (Fig. 5a-d), indicating that the maturation-promoting factors are not only expressed but also maintained in these mutants. We observed that in fis2-6, but not in other mutant backgrounds, about a third to a half of the embryos exhibited patchy GUS staining, mainly in the cotyledons and root pole (Fig. 5b). 42 to 72% of the embryos turned green, with the lowest percentage corresponding to the earlier fie-12 embryos (Fig. 5e-h). The most dramatic difference between mutants was seen for At2S3p:GFP, where 83-93% of fis2-6 and ede1-1 embryos expressed the reporter, but only 26-38% of fie-12 embryos did (Fig. 5i-l). Expression levels of At2S3p:GFP were lower in all mutants compared to wild type embryos from the same silique (e.g. Fig. 5l), and this was much more pronounced in fie-12 embryos (Fig. 5k). It appears, therefore, that a majority of embryos can initiate the maturation program even in the absence of endosperm cellularization, even if not as robustly as in the wild type. The difference in the levels of expression of At2S3p:GFP in fie-12 compared to fis2-6 might be related to the fact that, while FIS2 is only expressed in the endosperm, FIE is also expressed in the embryo (Luo, Bilodeau et al. 2000). It has been shown that dying fie seeds accumulate little storage oils (Fatihi, Zbierzak et al. 2013). The activity of PRC2 in the embryo may therefore be important for the accumulation of storage products.
Figure 5: Greening and expression of maturation reporters in embryos from seeds with no endosperm cellularization.
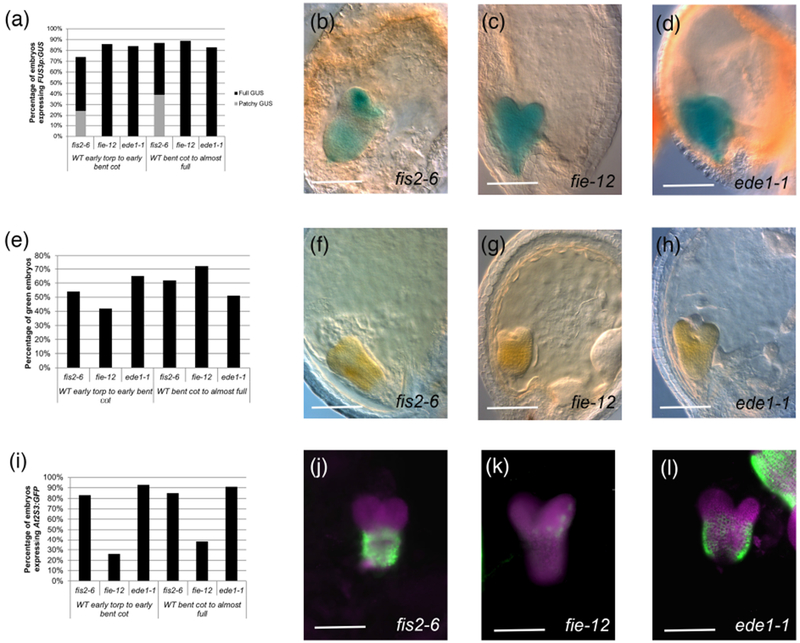
(a) Percentage of embryos expressing FUS3p:GUS. (b-d): Examples of FUS3p:GUS expression in embryos of (b) fis2-6 (mature green stage; patchy GUS), (c) fie-12 (bent cotyledon stage), (d) ede1-1 (mature green stage). (e) Percentage of green embryos in seeds with failed endosperm cellularization. (f-h): Examples of green embryos in (f) fis2-6 (early bent cotyledon stage), (g) fie-12 (early bent cotyledon stage), (h) ede1-1 (late heart stage). (i) Percentage of embryos expressing At2S3p:GFP. (j-l): Examples of At2SS3p:GFP expression in embryos of (j) fis2-6 (mid torpedo stage), (k) fie-12 (bent cotyledon stage), (l) ede1-1 (early bent cotyledon stage, with part of a wild type embryo from the same silique next to it). Magenta represents chlorophyll autofluorescence. The sample sizes per condition are listed on Table S3d. Scale bars: 100 μm.
Embryo maturation is initiated at the proper stage even if the supply of sucrose is reduced
Reductions in the provision of sucrose to the embryo, by mutating sucrose transporters, lead to slower embryo growth and reduced seed filling in a variety of species (Baud, Wuillème et al. 2005, Chen, Lin et al. 2015, Sosso, Luo et al. 2015). An unexamined question is whether the timing of maturation depends on the supply of sucrose.
To address this issue, we analyzed a triple mutant for the sucrose transporters SWEET11, 12 and 15 (sweet11;12;15). Of all the SWEET transporters, these are the ones expressed at high levels in the seed, primarily in the seed coat and endosperm. sweet11;12;15 mutant seeds have slowly developing embryos (two days slower than the wild type Col), and result in incompletely filled seeds, with storage oil concentration reduced by 71% (Chen, Lin et al. 2015). Moreover, SWEET11;12;15 function is required predominantly in the seed coat, because only the maternal genotype is important for the defects seen in the mutant embryos (Chen, Lin et al. 2015).
We repeated in sweet11;12;15 plants the studies that we had carried out in the previous mutants. Greening was examined in self-pollinated sweet11;12;15 plants. For FUS3p:GUS and At2S3p:GFP we pollinated sweet11;12;15 homozygous plants with pollen homozygous for the reporters and then collected seeds at 4-7 DAP. Because of the maternal effect, the seeds produced should have a mutant phenotype. Comparisons were made to Col (greening) or to wild type plants homozygotes for the reporter (also in Col background).
In spite of being delayed by two days compared to the wild type, the onset of FUS3p:GUS expression and greening occurred at the same stage in sweet11;12;15 embryos as in the wild type (Fig. 6a, b; Table S4). FUS3p:GUS expression then rose at a slightly higher rate, and greening developed at a slower rate than in the wild type (Fig. 6a, b; Table S4). Endosperm cellularization in sweet11;12;15 seeds occurred at the same embryonic stage as in the wild type, although with a somewhat different dynamic (Fig 6d; Table S4). In this particular case, the correlation between endosperm cellularization and onset of maturation is not different from the wild type situation. In contrast, the expression of At2S3p:GFP was significantly delayed, both in term of stages (Fig. 6c; Table S4) and DAP (mid-heart Col 5 DAP, late heart sweet11;12;15 7 DAP) . The accumulation of storage products, then, is much more sensitive to the supply of sugars than the onset of maturation itself.
Figure 6: Analyses of sweet11;12;15 embryos.
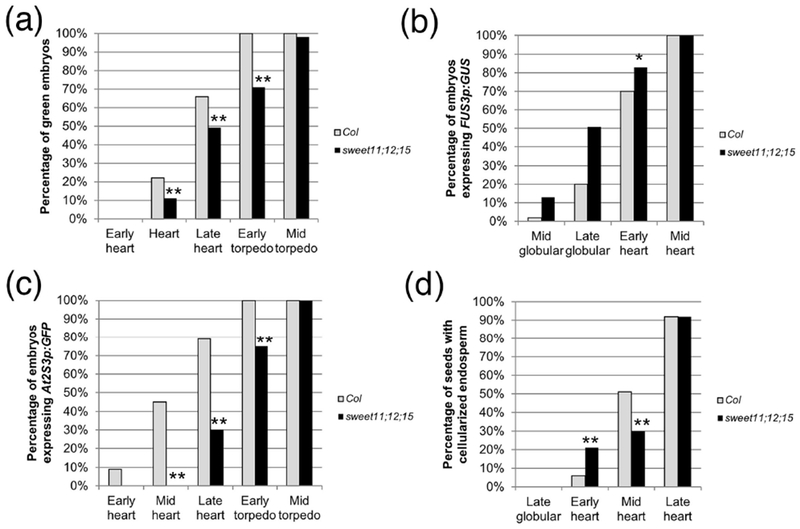
(a) Greening, (b) FUS3p:GUS expression, (c) At2S3p:GFP expression, and (d) endosperm cellularization in sweet11;12;15 embryos. The sample sizes per condition and p-values from Fisher’s Exact Test between wild type and mutant are listed on Table S4 (*: p < 0.05; **: p < 0.01).
Discussion
In this work we looked at the relative influence of developmental versus chronological time in the regulation of the onset of embryonic maturation, whether seeds start maturation autonomously, and whether the “sugar switch” model applies to Arabidopsis. Several of these issues had been difficult to address because the development of all the embryos in an Arabidopsis silique proceeds at a similar speed, obscuring the contributions of stage and age to the process.
The first conclusion from our experiments analyzing the expression of a master positive regulator (FUS3p:GUS), greening, and expression of a SSP gene (At2S3p:GFP) in mutant backgrounds that have delayed development (fus12, til1-4) is that each seed develops independently from the others in the silique, and from the maternal tissue. The only maternal signal for maturation that has been proposed to date is ABA, but the “early” peak of ABA in the seed is at 9-10 DAP (Karssen, Brinkhorst-van der Swan et al. 1983, Kanno, Jikumaru et al. 2010), much too late to start the process at 5-6 DAP. Our experiments strongly suggest that there are no signals produced by the fruit that are responsible for triggering maturation. An alternative explanation is that a signal may exist, but that the embryos will not perceive it or respond to it until they have reached the appropriate stage. Given the current paucity of evidence for fruit-derived signals, we favor the former interpretation.
The second conclusion from our studies of delayed mutants is that the onset of maturation is dependent on the developmental stage of the embryo and not the number of days since fertilization. Something similar had been observed in apetala2 mutants, but those results were not straightforward to interpret, since the mutation affected not only the embryo but also the endosperm and the seed coat (Ohto, Floyd et al. 2009). Time and stage can be uncoupled in Arabidopsis embryos, and the results imply the existence of some kind of internal timing mechanism linked to stage progression for triggering maturation.
Our data suggest that the early heart to heart stage is the key moment when the maturation program is initiated. This is the first stage at which all the gene products described as important for the expression of maturation genes are present at the same time in the embryo proper. While LEC1, L1L, ABI3, bZIP10 and 53, and NF-YC2 are expressed from early in development (Parcy, Valon et al. 1994, Lotan, Ohto et al. 1998, Kwong, Bui et al. 2002, Belmonte, Kirkbride et al. 2013), LEC2 doesn’t appear in the embryo proper at detectable levels until the early- to mid-globular stages (Stone, Kwong et al. 2001, Kroj, Savino et al. 2003) (but possibly earlier, see (Hofmann, Schon et al. 2019)), and FUS3 not until the late globular to early heart stages (Kroj, Savino et al. 2003, Tsuchiya, Nambara et al. 2004) (but see (Roscoe, Vaissayre et al. 2019) for a suggested earlier onset). The key, then, is likely to be in the regulation of the LAFL genes, of which not much is known. The expression of FUS3, but not ABI3, depends partly on LEC2 for its initiation (To, Valon et al. 2006). Both FUS3 and LEC2 are repressed at early stages, indirectly, by microRNAs. Embryos mutant for DICER-LIKE1 (DCL1), that have significantly reduced microRNA levels, show precocious expression of these genes and of maturation markers (Nodine and Bartel 2010, Willmann, Mehalick et al. 2011). miR156, in particular, has been proposed to serve as a count-down timer for embryo differentiation, in a similar fashion to its action during vegetative phase change (Nodine and Bartel 2010). However, more data is necessary to support this hypothesis. It is also likely that chromatin plays a role in regulating expression. For instance, the nucleosome remodelers CHR5 and PICKLE modulate the levels of ABI3, LEC1 and FUS3 in an antagonistic manner (Shen, Devic et al. 2015). There is also likely to be a stage-dependent layer of post-transcriptional and/or post-translational regulation, such as the phosphorylation of ABI3 or FUS3, or the stabilization of the latter by ABA (Guerriero, Martin et al. 2009, Lu, dela Paz et al. 2010, Tsai and Gazzarrini 2012).
The only metabolic signal proposed, so far, to trigger embryo maturation is a switch in the hexose/sucrose ratio from high to low, following the cellularization of the endosperm, a hypothesis based mostly on research on legumes (Wobus and Weber 1999, Weber, Borisjuk et al. 2005). The work of several labs on oilseeds, however, offered contrasting data. Tomlinson and colleagues (Tomlinson, McHugh et al. 2004) concluded that, in tobacco seeds, altering the sugar ratios does not have an effect on the accumulation of storage oils, and that a low hexose-to-sucrose ratio is not required for the onset of seed filling. Morley-Smith and others (Morley-Smith, Pike et al. 2008), on their part, inferred that the embryo in Brassica napus seeds is not subjected to the changing ratios of hexose-to-sucrose that occur in the endosperm but that it may receive a more constant concentration of sucrose via the suspensor or the micropylar endosperm. These observations questioned the notion of a change in sucrose concentrations as a trigger for seed filling. In Arabidopsis, endosperm cellularization was hypothesized to redistribute the flow of sucrose from the vacuole to the embryo, to promote its growth (Hehenberger, Kradolfer et al. 2012, Lafon-Placette and Köhler 2014). However, its connection to maturation, which starts immediately after cellularization, hadn’t been explored.
Our experiments show that endosperm cellularization is not required for the onset of maturation. In til1-4, shb1 and mini3-2 the endosperm cellularizes one or two stages early, but maturation does not begin precociously. The opposite is true in shb1-D, cellularization is late, but maturation remains on schedule. Even more conclusive are the observations on mutants where the endosperm fails to cellularize (fis2-6, fie-12, ede1-1). These seeds (when measured whole) maintain a high hexose-to-sucrose ratio (Hehenberger, Kradolfer et al. 2012). The sugar-switch model of (Wobus and Weber 1999) would predict that the transition to maturation shouldn’t happen in these mutant embryos. However, in the vast majority of embryos we observe greening, and expression of At2S3p:GFP (and of FUS3p:GUS, which actually precedes endosperm cellularization). Our data therefore imply that, in Arabidopsis, the onset of maturation is independent of the hexose-to-sucrose ratio of the whole seed. Our observations then, appear to confirm the previous studies in oilseeds that suggested that the “sugar switch” model does not apply to these plants. They also indicate that, because endosperm cellularization and the onset of maturation can be uncoupled, they are likely to be controlled by independent genetic programs. It must be noted that in Arabidopsis the endosperm shows expression of many maturation-related genes, but their regulation is likely to be independent from that in the embryo and will need further study (Belmonte, Kirkbride et al. 2013).
Endosperm cellularization is likely to be required for embryonic cell division and growth, since all the mutants that fail to cellularize the endosperm show embryo arrest (Sørensen, Chaudhury et al. 2001, Ingouff, Haseloff et al. 2005). Whether this is due to reduced sugar levels (Hehenberger, Kradolfer et al. 2012) or elevated turgor pressure (Beauzamy, Fourquin et al. 2016) is not clear. However, culturing fis2 or sweet11;12;15 embryos in high sucrose can partially rescue their growth defects (Hehenberger, Kradolfer et al. 2012, Chen, Lin et al. 2015). A corollary of the observations that fie-12, fis2-6 and ede1-1 embryos stop growing but nevertheless begin maturing is that embryo growth and maturation are regulated by separate genetic pathways. The final size of the embryo (and seed) appears to be influenced by the timing of endosperm cellularization, by the proliferation and expansion of the seed coat, and by interactions between all these tissues and the embryo (Orozco-Arroyo, Paolo et al. 2015). The timing of maturation, on the other hand, appears to be regulated mostly by the LAFL genes (Boulard, Fatihi et al. 2017).
Our study should not be interpreted to mean that endosperm cellularization is not important for the accumulation of storage products. The levels of expression of At2S3p:GFP in the mutants that don’t cellularize never reaches the levels of the wild type. The mechanism by which the endosperm influences the final content of storage products is unknown, but it could be related to the provision of sucrose via the SWEET (or other) transporters. We observe a definite delay in the expression of At2S3p:GFP in sweet11;12;15, and the mature seeds of this mutant show a 34% reduction in seed weight and a 71% reduction in fatty acid content (Chen, Lin et al. 2015). Further studies are needed to elucidate the connections between endosperm and maturation.
Experimental Procedures
Plant material and growth conditions
Plants were grown at 22°C with 16 hours of light in a growth chamber (Conviron). Seeds were planted directly in soil (Fafard-2, SunGro Horticulture) supplemented with Osmocote Plus 15-9-12 fertilizer. The only exception were desiccation-intolerant mutants, which were grown by taking immature homozygous mutant seeds, germinating them on plates with sterile Murashige-Skoog (MS) medium (4.4 g/L MS salts, 1X Gamborg’s vitamins, 0.5 g/L MES, 10 g/L sucrose, and 7.5 g/L tissue culture agar, pH 5.7) (all reagents from Sigma unless specified otherwise) and then transplanting them to soil. For the time series, unopened flowers were emasculated and then pollinated two days later. Siliques were collected 3 to 7 days after pollination, depending on the experiment.
All the mutants and reporters used here have been described before: lec1-1 (Meinke 1992), lec1-2 (West, Matsudaira Yee et al. 1994), fus3-3 (Keith, Kraml et al. 1994), lec2-1 (Meinke, Franzmann et al. 1994), abi3-3 (Nambara, Naito et al. 1992), til1-4 (Jenik, Jurkuta et al. 2005), fus12-U228/csn2 (Miséra, Müller et al. 1994), mini3-2 (Luo, Dennis et al. 2005), shb1 and shb1-D (Kang and Ni 2006), fie-12 (Wolff, Weinhofer et al. 2011), fis2-6 (Guitton, Page et al. 2004), ede1-1 (Pignocchi, Minns et al. 2009), sweet11;12;15 (Chen, Lin et al. 2015), and FUS3p:GUS and At2S3:GFP (Kroj, Savino et al. 2003). til1-4, and the reporter genes are in a Col background, while fus12 is in a Landsberg erecta (Ler) background. Seeds were obtained from the following sources: our stocks (fus12, til1-4), John Harada (U. of California-Davis) (lec1-1, lec1-2, lec2-1, fus3-3, abi3-3, double and triple mutants), Min Ni (U. of Minnesota) (shb1-D), Claudia Köhler (Uppsala BioCenter) (fie-12), Li-Qing Chen (U. of Illinois at Urbana-Champaign) (sweet11;12;15), François Parcy (Biosciences and Biotechnology Institute of Grenoble) (FUS3p:GUS and At2S3p:GFP), and the Arabidopsis Biological Resource Center (mini3-2, shb1, ede1-1, fis2-6, Col and Ws-2). Plants were genotyped based on the phenotype of the developing seeds. The exceptions were mini3-2, shb1 and shb1-D, which were PCR-genotyped using the primers described by (Zhou, Zhang et al. 2009), and the triple LAFL mutants (lec1-2 lec2-1 fus3-3/abi3-3). In the case of fus3-3, PCR was done with primers 5’catctatggtttcttctaacgatctc3’ and 5’ttggacggttttcacgtttggaccttcaagtctag3’ and the product digested with XbaI. For abi3-3, amplification was carried out with primers 5’gggcctccggcttttgtccgctcgg3’ and 5’ccacgtcagcaggtggtaccagatc3’ and the product cut with HinfI.
Histochemistry and microscopy
All our observations were carried out on a Leica DMRB microscope equipped with ProgRes MFcool and ProgRes C5 cameras (Jenoptik). Images were acquired with the ProgRes software and processed as necessary (overall brightness, contrast). Graphs were generated in Microsoft Excel. Figures were assembled using Adobe Photoshop CC 2018.
For the clearing of whole seeds, siliques were opened with fine tweezers and the seeds placed on a slide with Hoyer’s solution (Anderson 1954). Hoyer’s was made by dissolving 50 g of chloral hydrate and 3.75 g of gum arabic into 15 ml of water and 2.5 ml of glycerol. The resulting solution was diluted 2 parts Hoyer’s to 1 part water before using. Cleared seeds were observed using Differential Interference Contrast (DIC) optics. Embryos were staged according to (Jürgens and Mayer 1994), except for the torpedo stages, which we define as: early torpedo embryos are those that first show the cotyledons parallel to each other, mid-torpedo embryos have a length that is about half of that of the seed, late torpedo embryos are about as long as the seed.
GUS staining of whole seeds involved slitting the siliques open, fixing in cold 90% acetone for 20 minutes, rinsing with 100 mM phosphate buffer pH 7, and incubating in GUS staining solution (100 mM phosphate buffer, pH 7, 1 mM EDTA, 1% Tween 20, 2.5 mM potassium ferrocyanide/ferricyanide, and 1 mg/ml X-Glc [Gold Biotechnology]) at 37°C for 4 hours. Stained tissue was rinsed with water and the seeds mounted in Hoyer’s and cleared overnight at 4°C.
To analyze GFP expression, embryos were dissected out of the seeds with needles directly onto a slide with a drop of 5% sucrose (Fisher Scientific) and observed immediately. For GFP and chlorophyll fluorescence, the excitation/emission wavelengths were 480/535 nm and 560/645 nm, respectively.
For the preparation of sectioned and stained tissues, siliques were cut into three pieces and fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol) overnight at 4°C. The fixative was vacuum infiltrated for 15 minutes. The fixed tissue was dehydrated through an ethanol and ethanol:CitriSolv (Fisher Scientific) series and embedded in Paraplast Plus (McCormick Scientific). 7 μm sections were cut with a Leica RM2155 microtome and mounted on albuminized slides. The mounted sections were deparaffinized in CitriSolv, rehydrated, and stained for 10 minutes in 0.5% Celestine Blue (dissolved in 5% ferric ammonium sulfate). The stained slides were rinsed in water, dehydrated with an ethanol:CitriSolv series, and mounted with Omnimount (Electron Microscopy Sciences).
Supplementary Material
Table S1: Sample sizes and statistical tests for lafl mutants (corresponding to Table 1).
Table S2: Sample sizes and statistical tests for mutants with delayed development (corresponding to Figures 2–4).
Table S3: Sample sizes and statistical tests for mutants with abnormal endosperm cellularization (corresponding to Figures 4 and 5).
Table S4: Sample sizes and statistical tests for sweet11;12;15 mutants (corresponding to Figure 6).
Figure S1: Greening and expression of maturation reporters in wild type embryos.
Figure S2: Distribution of chlorophyll in older lafl double and triple mutants.
Figure S3: Greening and expression of maturation reporters in FUS12 wild type backgrounds at different times after pollination.
Figure S4: Greening and expression of maturation reporters in TIL1 wild type backgrounds at different times after pollination.
Figure S5: Endosperm cellularization.
Acknowledgements
We would like to thank Jim Engleman for taking care of our plants, Juan José Ripoll for comments on the manuscript, and Li-Qing Chen, John Harada, Claudia Köhler, Min Ni and François Parcy for providing seeds. This work was supported by Franklin & Marshall College and by a grant from the National Institute of General Medical Sciences of the National Institutes of Health to P.D.J. (1R15GM113186-01). The authors declare no conflict of interest.
Footnotes
Data statement: supporting data can be accessed as Supplementary Information on The Plant Journal website.
References
- Allorent G, Osorio S, Vu JL, Falconet D, Jouhet J, Kuntz M, Fernie AR, Lerbs-Mache S, Macherel D, Courtois F and Finazzi G (2015). “Adjustments of embryonic photosynthetic activity modulate seed fitness in Arabidopsis thaliana.” New Phytologist 205: 707–719. [DOI] [PubMed] [Google Scholar]
- Anderson LE (1954). “Hoyer’s solution as a rapid permanent mounting medium for bryophytes.” The Bryologist 57(3): 242–244. [Google Scholar]
- Bastakis E, Hedtke B, Klermund C, Grimm B and Schwechheimer C (2018). “LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis.” The Plant Cell 30: 582–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C and Leipiniec L (2008). Storage Reserve Accumulation in Arabidopsis: Metabolic and Developmental Control of Seed Filling The Arabidopsis Book. Rockville, MD, American Society of Plant Biology; 6: e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S and Lepiniec L (2009). “Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis.” Plant Physiology and Biochemistry 47: 448–455. [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L. c. and Rochat C (2005). “The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis.” The Plant Journal 43: 824–836. [DOI] [PubMed] [Google Scholar]
- Baud S. b., Kelemen Z, Thévenin J, Boulard C. l., Blanchet S, To A, Payre M, Berger N, Effroy-Cuzzi D, Franco-Zorrilla JM, Godoy M, Solano R, Thevenon E, Parcy F. o., Lepiniec L. c. and Dubreucq B (2016). “Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed.” Plant Physiology 171: 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauzamy L. n., Fourquin C, Dubrulle N, Boursiac Y, Boudaoud A and Ingram G (2016). “Endosperm turgor pressure decreases during early Arabidopsis seed development.” Development 143: 3295–3299. [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB and Harada JJ (2013). “Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed.” Proceedings National Academy of Sciences USA 110: E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsch L, Boltz V, Brioudes F, Pontier G, Girard V, Savarin J, Wipperman B, Chambrier P, Tissot N, Benhamed M, Mollereau B, Raynaud C, Bendahmane M and Szécsi J (2019). “TCTP and CSN4 control cell cycle progression and development by regulating CULLIN1 neddylation in plants and animals.” PLoS Genetics 15(1): e1007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard C, Fatihi A, Lepiniec L and Dubreucq B (2017). “Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits.” Biochimica et Biophysica Acta - Gene Regulatory Mechanisms 1860(10): 1069–1078. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H and Olsen O-A (1999). “Development of endosperm in Arabidopsis thaliana.” Sex Plant Reproduction 12: 32–42. [Google Scholar]
- Buendía-Monreal M and Gillmor CS (2018). “The times are a-changin’: heterochrony in plant development and evolution.” Frontiers in Plant Science 9: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero P, Iglesias-Fernández R and Vicente-Carbajosa J (2017). “The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds.” Journal of Experimental Botany 68(4): 871–880. [DOI] [PubMed] [Google Scholar]
- Chen L-Q, Lin IW, Qu X-Q, Sosso D, McFarlane HE, Londoño A, Samuels AL and Frommer WB (2015). “A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo.” The Plant Cell 27: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, MacGregor DR, Dave A, Florance H, Moore K, Paszkiewicz K, Smirnoff N, Graham IA and Penfield S (2014). “Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year.” Proceeding of the National Academy of Sciences USA 111: 18787–18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM and Colombo L (2008). “AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis.” The Plant Journal 54(6): 1037–1048. [DOI] [PubMed] [Google Scholar]
- Ebisuya M and Briscoe J (2018). “What does time mean in development?” Development 145: dev164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A, Zbierzak AM and Dörmann P (2013). “Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds.” Plant Physiology 163: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW and Langdale JA (2002). “GLK gene pairs regulate chloroplast development in diverse plant species.” The Plant Journal 31(6): 713–727. [DOI] [PubMed] [Google Scholar]
- Franciosini A, Moubayidin L, Du K, Matari NH, Boccaccini A, Butera S, Vittorioso P, Sabatini S, Jenik PD, Costantino P and Serino G (2015). “The COP9 SIGNALOSOME is required for postembryonic meristem maintenance in Arabidopsis thaliana.” Molecular Plant 8: 1623–1634. [DOI] [PubMed] [Google Scholar]
- Geeta R (2003). “The origin and maintenance of nuclear endosperms: viewing development through a phylogenetic lens.” Proceedings of the Royal Society, London B 270: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K and Coenen H (2013). “Heterochronic genes in plant evolution and development.” Frontiers in Plant Science 4: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ, Nguyen C, Ampofo B, Zhong S and Fei Z (2017). “The epigenome and transcriptional dynamics of fruit ripening.” Anuual Review of Plant Biology 68: 61–84. [DOI] [PubMed] [Google Scholar]
- Goffman FD, Alonso AP, Schwender J, Shachar-Hill Y and Ohlrogge JB (2005). “Light enables a very high efficiency of carbon storage in developing embryos of rapeseed.” Plant Physiology 138: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G and Yadegari R (1994). “Plant embryogenesis: zygote to seed.” Science 266: 605–614. [DOI] [PubMed] [Google Scholar]
- Gomez C, Özbudak EM, Wunderlich J, Baumann D, Lewis J and Pourquié O (2008). “Control of segment number in vertebrate embryos.” Nature 454: 335–339. [DOI] [PubMed] [Google Scholar]
- Gould SJ (1977). Ontogeny and phylogeny. Cambridge, London, Harvard Univeristy Press. [Google Scholar]
- Guerche P, Tire C, Grossi de Sa F, De Clercq A, Van Montagu M and Krebbers E (1990). “Differential expression of the Arabidopsis 2S albumin genes and the effect of incresing gene family size.” The Plant Cell 2: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G, Martin N, Golovko A, Sundström JF, Rask L and Ezcurra I (2009). “The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth.” New Phytologist 184: 552–565. [DOI] [PubMed] [Google Scholar]
- Guitton A-E, Page DR, Chambrier P, Lionnet C, Faure J-E, Grossniklaus U and Berger F (2004). “Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana.” Development 131: 2971–2981. [DOI] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D and Köhler C (2012). “Endosperm cellularization defines an important developmental transition for embryo development.” Development 139: 2031–2039. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Schon MA and Nodine MD (2019). “The embryonic transcriptome of Arabidopsis thaliana.” Plant Reproduction 32: 77–91. [DOI] [PubMed] [Google Scholar]
- Hua W, Li R-J, Zhan G-M, Liu J, Li J, Wang X-F, Liu G-H and Wang H-Z (2012). “Maternal control of seed oil content in Brassica napus: the role of silique wall photosynthesis.” The Plant Journal 69: 432–444. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Haseloff J and Berger F (2005). “Polycomb group genes control developmental timing of endosperm.” The Plant Journal 42(5): 663–674. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Jurkuta REJ and Barton MK (2005). “Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon.” Plant Cell 17: 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G and Mayer U (1994). Arabidopsis Embryos: Color Atlas of Development. J. Bard. London, Wolfe Publishing: 7–21. [Google Scholar]
- Kang X, Li W, Zhou Y and Ni M (2013). “A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement.” Public Library of Science Genetics 9(3): e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X and Ni M (2006). “Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 Contains SPX and EXS Domains and Acts in Cryptochrome Signaling.” The Plant Cell 18: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y and Seo M (2010). “Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone Interactions.” Plant and Cell Physiology 51: 1988–2001. [DOI] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Brrklend AE and Koornneef M (1983). “Indcution of dormancy during seed development by endogenous abscicic acid: studies on abscic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh.” Planta 157: 158–165. [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG and McCourt P (1994). “fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis.” The Plant Cell 6: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HWM and Karssen CM (1989). “In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana.” Plant Physiology 90: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J and Parcy F (2003). “Regulation of storage protein gene expression in Arabidopsis.” Development 130(24): 6065–6073. [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB and Harada JJ (2002). “LEAFY COTYLEDON1-LIKE Defines a Class of Regulators Essential for Embryo Development.” The Plant Cell 15(1): 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C and Köhler C (2014). “Embryo and endosperm, partners in seed development.” Current Opinion in Plant Biology 17: 64–69. [DOI] [PubMed] [Google Scholar]
- Leprince O, Pellizzaro A, Berriri S and Buitink J (2016). “Late seed maturation: drying without dying.” Journal of Experimental Botany 68(4): 827–841. [DOI] [PubMed] [Google Scholar]
- Li Z and Thomas TL (1998). “PEI1, an Embryo-Specific Zinc Finger Protein Gene Required for Heart-Stage Embryo Formation in Arabidopsis.” The Plant Cell 10: 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Graeber K, Knight C and Leubner-Metzger G (2010). “The evolution of seeds.” New Phytologist 186: 817–831. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang X, Ren K, Li K, Wei M, Wang W and Sheng X (2017). “Light deprivation-induced inhibition of chloroplast biogenesis does not arrest embryo morphogenesis but strongly reduces the accumulation of storage reserves during embryo maturation in Arabidopsis.” Frontiers in Plant Science 8: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M.-a., Matsudaira Yee K, West MAL, Lo R, Wong RW, Yamagishi K, Fischer RL, Goldberg RB and Harada JJ (1998). “Arabidopsis LEAFY COTYLEDON1 Is Sufficient to Induce Embryo Development in Vegetative Cells.” Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lu QS, dela Paz J, Pathmanathan A, Chiu RS, Tsai AY-L and Gazzarrini S (2010). “The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis.” The Plant Journal 64: 100–113. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ and Chaudhury A (2000). “Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds.” Proceedings of the National Academy of Sciences USA 97(19): 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ and Chaudhury A (2005). “MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis.” Proceedings of the National Academy of Sciences, USA 102(48): 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG and Briarty LG (1990). “Endosperm cellularization in Arabidopsis thaliana L.” Arabidopsis Information Service 27: 65–72. [Google Scholar]
- Mansfield SG and Briarty LG (1991). “Early embryogenesis in Arabidopsis thaliana. II. The developing embryo.” Canadian Journal of Botany 69: 461–476. [Google Scholar]
- Mansfield SG and Briarty LG (1992). “Cotyledon cell development in Arabidopsis thaliana during reserve deposition.” Canadian Journal of Botany 70: 151–164. [Google Scholar]
- Meinke DW (1992). “A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons.” Science 258(5088): 1647–1650. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle T and Yeung EC (1994). “leafy cotyledon mutants of Arabidopsis.” The Plant Cell 6: 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Trigui G, d’Andréa S, Kelemen Z, Baud S. b., Berger A, Deruyffelaere C, Trubuil A, Lepiniec L. c. and Dubreucq B (2014). “Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds.” Plant Physiology 164: 1866–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miséra S, Müller AJ, Weiland-Heidecker U and Jürgens G (1994). “The FUSCA genes of Arabidopsis: negative regulators of light responses.” Molecular and General Genetics 244(3): 242–252. [DOI] [PubMed] [Google Scholar]
- Morley-Smith ER, Pike MJ, Findlay K, Köckenberger W, Hill LM, Smith AM and Rawsthorne S (2008). “The transport of sugars to developing embryos is not via the bulk endosperm in oilseed rape seeds.” Plant Physiology 147: 2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P and Naito S (1994). “Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene.” Plant and Cell Physiology 35(3): 509–513. [PubMed] [Google Scholar]
- Nambara E, Naito S and McCourt P (1992). “A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele.” The Plant Cell 2(4): 435–441. [Google Scholar]
- Natesh S and Rau MA (1984). The embryo Embryology of Angiosperms. Johri BM. Berlin, New York, Springer-Verlag: 377–443. [Google Scholar]
- Nikolov LA, T PB, Manickam S, Endress PK, Kramer EM and Davis CC (2014). “Holoparasitic Rafflesiaceae possess the most reduced endophytes and yet give rise to the world’s largest flowers.” Annals of Botany 114: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD and Bartel DP (2010). “MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis.” Genes & Development 24: 2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull RJ and Davis SJ (2017). “Shining a light on the Arabidopsis circadian clock.” Plant, Cell & Environment 40(11): 2571–2585. [DOI] [PubMed] [Google Scholar]
- Ohto M. a., Floyd SK, Fischer RL, Goldberg RB and Harada JJ (2009). “Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis.” Sex Plant Reproduction 22: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Arroyo G, Paolo D, Ezquer I and Colombo L (2015). “Networks controlling seed size in Arabidopsis.” Plant Reproduction 28: 17–32. [DOI] [PubMed] [Google Scholar]
- Parcy F. o., Valon C, Kohara A, Miséra S and Giraudat J. r. m. (1997). “The ABSCISIC ACID-lNSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development.” The Plant Cell 9: 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F. o., Valon C, Raynal M, Gaubier-Comella P, Delseny M and Giraudat J (1994). “Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid.” The Plant Cell 6: 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JM, Kwong RW, Park S, Leb BH, Badena R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB and Harada JJ (2017). “LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development.” Proceedings of the National Academies of Science USA 114(32): E6710–E6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, Lloyd CW, Doonan JH and Hills MJ (2009). “ENDOSPERM DEFECTIVE1 Is a novel microtubule-associated protein essential for seed development in Arabidopsis.” The Plant Cell 21: 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JHW and Koornneef M (2001). “Sequential steps for developmental arrest in Arabidopsis seeds.” Development 128: 243–252. [DOI] [PubMed] [Google Scholar]
- Roscoe TJ, Vaissayre V, Paszkiewicz G, Clavijo F, Kelemen Z, Michaud C, Lepiniec L, Dubreucq B, Zhou D-X and Devic M (2019). “Regulation of FUSCA3 expression during seed development in Arabidopsis.” Plant & Cell Physiology 60: 476–487. [DOI] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule J-J, Berger F and Devic M (2015). “Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis.” Plant and Cell Physiology 56(6): 1215–1228. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C and Isono E (2010). “The COP9 signalosome and its role in plant development.” European Journal of Cell Biology 89: 157–162. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S and Martin C (2013). “Fruit development and ripening.” Annual Review of Plant Biology 64: 219–241. [DOI] [PubMed] [Google Scholar]
- Shen Y, Devic M, Lepiniec L and Zhou D-X (2015). “Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes.” Plant Biotechnology Journal 13: 811–820. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu X.-d., Xie Q and He Z.-h. (2016). “Two faces of one seed: hormonal regulation of dormancy and germination.” Molecular Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G and Holt BF (2009). “Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity.” Plant Physiology 149(2): 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MB, Chaudhury AM, Robert H, Bancharel E and Berger F (2001). “Polycomb group genes control pattern formation in plant seed.” Current Biology 11: 277–281. [DOI] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li Q-B, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, Rogowsky PM, Ross-Ibarra J, Yang B and Frommer WB (2015). “Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport.” Nature Genetics 47(12): 1489–1493. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Matsudaira Yee K, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB and Harada JJ (2001). “LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development.” Proceedings of the National Academies of Sciences USA 98(20): 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann J, Rutten T, Monke G, Vorwieger A, Rolletschek H, Meissner D, Milkowski C, Petereck S, Mock H and Zank T (2008). “Dissection of a complex seed phenotype: Novel insights of FUSCA3 regulated developmental processes.” Developmental Biology 317(1): 1–12. [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J and Parcy F (2006). “A network of local and redundant gene regulation governs Arabidopsis seed maturation.” The Plant Cell 18(7): 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson KL, McHugh S, Labbe H, Grainger JL, James LE, Pomeroy KM, Mullin JW, Miller SS, Dennis DT and Miki BLA (2004). “Evidence that the hexose-to-sucrose ratio does not control the switch to storage product accumulation in oilseeds: analysis of tobacco seed development and effects of overexpressing apoplastic invertase.” Journal of Experimental Botany 55: 2291–2303. [DOI] [PubMed] [Google Scholar]
- Tsai AY-L and Gazzarrini S (2012). “AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis.” The Plant Journal 69: 809–821. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nambara E, Naito S and McCourt P (2004). “The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis.” The Plant Journal 37: 73–81. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan MR and Prabhakar K (1984). The endosperm Embryology of Angiosperms. Johri BM. Berlin, Heidelberg, New York, Tokyo, Springer-Verlag. [Google Scholar]
- Weber H, Borisjuk L and Wobus U (2005). “Molecular physiology legume seed development.” Annual Review of Plant Biology 56: 253–279. [DOI] [PubMed] [Google Scholar]
- Weber H, Heim U, Golombek S, Borisjuk L and Wobus U (1998). “Assimilate uptake and the regulation of seed development.” Seed Science Research 8: 331–345. [Google Scholar]
- West MAL, Matsudaira Yee K, Danao J, Zimmerman JL, Fischer RL, Goldberg RB and Harada JJ (1994). “LEAFY COTYLEDONl 1s an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis.” The Plant Cell 6(1731–1745). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann MR, Mehalick AJ, Packer RL and Jenik PD (2011). “MicroRNAs regulate the timing of embryo maturation in Arabidopsis.” Plant Physiology 155: 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus U and Weber H (1999). “Sugars as signal molecules in plant seed development.” Biological Chemistry 380: 937–944. [DOI] [PubMed] [Google Scholar]
- Wobus U and Weber H (1999). “Sugars as Signal Molecules in Plant Seed Development.” Biological Chemistry 380: 937–944. [DOI] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MTA, Spillane C, Nordborg M, Rehmsmeier M and Köhler C (2011). “High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm.” Public Library of Science Genetics 7(6): e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev MS and Zhukova GY (1980). “Chlorophyll in embryos of angiosperm seeds, a review.” Botaniska Notiser 133: 323–336. [Google Scholar]
- Yamamoto A, Kagaya A, Toyoshima R, Kagaya M, Takeda S and Hattori T (2009). “Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors.” The Plant Journal 58: 843–856. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S and Hattori T (2010). “Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis.” Plant and Cell Physiology 51: 2031–2046. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X and Ni M (2009). “SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development.” The Plant Cell 21: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Clabaugh Blakley I, Franco-Zorrilla JM, Yamburenko MV, Solano R, Kieber JJ, Loraine AE and Schaller GE (2018). “Coordination of chloroplast development through the action of the GNC and GLK transcription factor families.” Plant Physiology 178: 130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Sample sizes and statistical tests for lafl mutants (corresponding to Table 1).
Table S2: Sample sizes and statistical tests for mutants with delayed development (corresponding to Figures 2–4).
Table S3: Sample sizes and statistical tests for mutants with abnormal endosperm cellularization (corresponding to Figures 4 and 5).
Table S4: Sample sizes and statistical tests for sweet11;12;15 mutants (corresponding to Figure 6).
Figure S1: Greening and expression of maturation reporters in wild type embryos.
Figure S2: Distribution of chlorophyll in older lafl double and triple mutants.
Figure S3: Greening and expression of maturation reporters in FUS12 wild type backgrounds at different times after pollination.
Figure S4: Greening and expression of maturation reporters in TIL1 wild type backgrounds at different times after pollination.
Figure S5: Endosperm cellularization.


