Abstract
Purpose
The heterogeneity of tumor cells presents a major challenge to cancer diagnosis and therapy. Cutaneous T cell lymphomas (CTCL) are a group of T lymphocyte malignancies that primarily affect skin. Lack of highly specific markers for malignant lymphocytes prevents early diagnosis, while only limited treatment options are available for patients with advanced-stage CTCL. Droplet-based single-cell transcriptome analysis of CTCL skin biopsies opens avenues for dissecting patient-specific T lymphocyte heterogeneity, providing a basis for identifying specific markers for diagnosis and cure of CTCL.
Methods
Single-cell RNA-sequencing was performed by Droplet-based sequencing (10X Genomics), focusing on 14,056 CD3+ lymphocytes (448 cells from normal and 13,608 cells from CTCL skin samples) from skin biopsies of 5 patients with advanced-stage CTCL and 4 healthy donors. Protein expression of identified genes was validated in advanced-stage CTCL skin tumors by immunohistochemistry and confocal immunofluorescence microscopy.
Results
Our analysis revealed a large inter- and intra-tumor gene expression heterogeneity in the T lymphocyte subset, as well as a common gene expression signature in highly proliferating lymphocytes that was validated in multiple advanced-stage skin tumors. In addition, we established the immunological state of reactive lymphocytes and found heterogeneity in effector and exhaution programs across patient samples.
Conclusions
Single-cell analysis of CTCL skin tumor samples reveals patient-specific landscapes of malignant and reactive lymphocytes within the local microenvironment of each tumor, giving an unprecedented view of lymphocyte heterogeneity and identifying tumor-specific molecular signatures, with important implications for diagnosis and personalized disease treatment.
INTRODUCTION
Cutaneous T-cell lymphomas (CTCLs) are a heterogeneous group of malignancies characterized by chronic inflammation and accumulation of malignant T lymphocytes in the skin (1). CTCL encompasses diverse presentations including Sezary syndrome (SS) where patients present with erythroderma, lymphadenopathy, and circulating malignant T lymphocytes, as well as mycosis fungoides (MF) in which malignant cells reside primarily in the skin (2). MF is the most common form of CTCL and typically runs an indolent course with an excellent 5-year survival rate in early stages, but significantly decreased survival in advanced disease (3). In the early stages, most T cells reside in the skin and only a few circulate in peripheral blood and lymph nodes. However, a small number of patients progress, and tumor cells may involve other sites of the body with a fatal outcome (4). About 20% of patients progress to advanced-stage MF (Stages IIB to IV) (5), and the prognosis for patients with widespread CTCL manifestation beyond the skin is poor with a 5-year survival rate of only 40% (6). Large cell transformation occurs in 56–67% of advanced-stage MF patients (6) and is accompanied by clinically aggressive disease and shortened survival. Diagnosis of MF is difficult, especially in the early stages, due to absence of specific markers for malignant lymphocytes that distinguish them from nonmalignant tumor infiltrating T lymphocytes (TILs). Diagnosis is usually based on clinic-pathological correlation, and the average time-to-diagnosis is seven years (7). Delays prevent timely treatment and result in poorer clinical outcomes, while the treatment options for patients with aggressive forms of MF are limited, reflecting our poor understanding of disease pathogenesis.
Lymphocyte proliferation in CTCL is largely restricted to the skin, implying that malignant cells are dependent on their specific cutaneous microenvironment. Cytokines and other immunomodulator factors produced by malignant lymphocytes and TILs (8,9) as well as by other immune and stromal cells (10) affect cutaneous inflammation (1,8) and are important constituents of tumor local microenvironments, fostering survival, proliferation and suppression of tumor cell immunosurveillance (8,11). In this context, reactive TILs are exposed to multiple immunosuppressive pressures, including negative regulatory pathways and up-regulation of inhibitory receptors such as PD1, CTLA4, LAG3, TIM3, and TIGIT that render them dysfunctional (12–14) and unable to elaborate their full effector functions for ultimately killing tumor cells (12,13). However, the heterogeneity and immunological state of malignant and reactive lymphocytes within CTCL skin tumors remain incompletely characterized.
Recent advances in single cell transcriptome technology, including droplet-based single-cell RNA-sequencing (scRNA-seq) (15), profile gene expression across thousands of individual cells from a large heterogeneous population (16,17) such as a patient biopsy. This high-resolution analysis of cellular heterogeneity reveals individual cell functions in the context of their microenvironment and provides striking insights into the complex cellular composition of normal and diseased tissue. Here we report scRNA-seq analysis of skin tumor cells from patients with advanced-stage CTCL. This analysis provides an unprecedented view of lymphocyte heterogeneity within the skin-microenvironment of individual CTCL tumors by identifying molecular signatures that are unique for each tumor. We also established a common gene expression signature in highly proliferating lymphocytes as well as the immunological state of TILs within each tumor. Together, these data provide important implications for personalized disease management.
METHODS
Subjects and skin biopsies
Skin samples were obtained at the Comprehensive Skin Cancer Center, Columbia University Medical Center, from 10 patients with confirmed diagnoses of advanced CTCL (stage IIB-IVA; described in Supplemental Methods and Table S1) and staged according to the most recent consensus (4,5). Five patient samples were used for scRNA-seq and all ten for immunohistochemistry. Participants gave written informed consent. Human research protocols were approved by the Institutional Review Board, Columbia University. Controls included human normal skin (NS, n=8; 4 each for scRNA-seq and immunohistochemistry) and atopic dermatitis (AD, n=4; all for immunohistochemistry) obtained from The Health Sciences Tissue Bank, University of Pittsburgh. This study was conducted in accordance with the Declaration of Helsinki. Experimental procedures followed established techniques using the Chromium Single Cell 3’ Library V2 kit (10x Genomics) (18). Briefly, cell suspensions from enzymatically digested skin biopsies were loaded into the Chromium instrument (10X Genomics), and the resulting barcoded cDNAs were used to construct libraries. RNA-seq was performed on each sample (approximately 200 million reads/sample). Cell-gene unique molecular identifier counting matrices generated were analyzed using Seurat (19) to identify distinct cell populations using Louvain clustering (15). See Supplementary Methods for details. All scRNA-seq data have been deposited in the GEO database (accession GSE128531).
Multicolor immunohistochemistry
Single and dual antibody staining using tyramide signal amplification (ThermoFisher) were performed on formalin-fixed, paraffin-embedded skin samples as previously described (18). Antibodies were all purchased from Sigma. Immunohistochemistry images were obtained with an Evos FL Auto microscope (Life Technologies). Confocal images were captured on an Olympus Fluoview 1000 confocal microscope using an oil immersion 100X objective.
RESULTS
Single-cell transcriptome profiles from advanced-stage CTCL skin tumors and healthy control skin
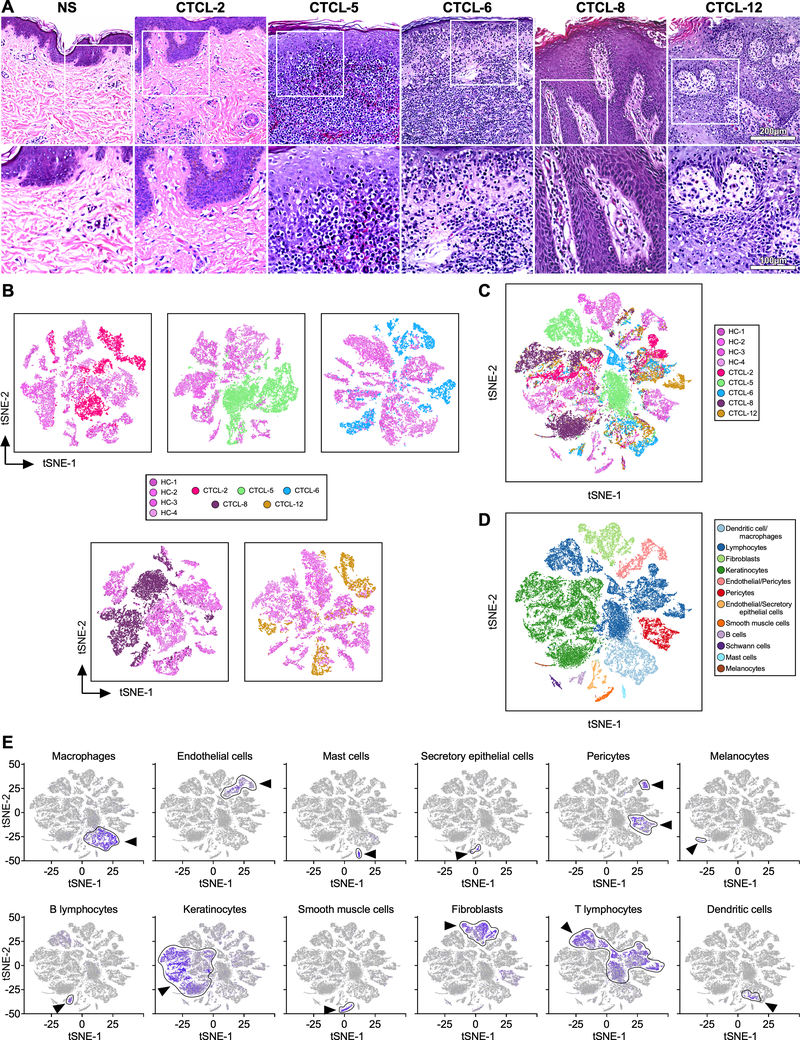
We used scRNA-seq to profile gene expression in cells obtained from the enzymatically digested skin of five advanced-stage CTCL skin tumors and four healthy control skin samples (Table S1). Fig. 1A depicts histological features of the tumors studied, as described in Table S2. A 3mm skin biopsy from each donor yielded 3,607–9,272 cells from skin tumor samples and 2,200–4,847 cells from healthy skin. After reverse transcription from each cell, we constructed cDNA libraries and performed massive parallel sequencing, obtaining an average of 47,894 mapped reads per cell and a median of 1,261 unique genes detected per cell, comparable to previous studies (18). Cells were grouped according to their expression profiles by principal components analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) dimensional reduction (20). Comparison of whole skin cell distribution from each tumor sample with the four control skin samples shows overlap in the transcriptional profiles of cells from healthy skin samples but generally no overlap between cells from the tumor samples and the healthy samples (Fig. 1B). Strikingly, the combination of five tumors and four controls show that the tumor sample profiles do not overlap with each other, thus exhibiting significant inter-tumor heterogeneity (Fig. 1C). Unsupervised graph-based Louvain clustering by Seurat (19) identified 26 clusters of cells (Fig. 1D) whose types were identified by the expression of cell-specific marker genes (18) (Fig. 1E). We found the greatest heterogeneity between tumors and controls, as well as across tumors, was at the level of lymphocytes, keratinocytes, fibroblasts and macrophages. In addition to PCA, canonical correlation analysis showed comparable results (19). Thus, scRNA-seq analysis characterizes details of the large inter-tumor cell transcriptional heterogeneity in advanced CTCL skin samples.
Fig. 1. Grouping of CTCL and normal skin populations.
Transcriptomes of 44,842 cells from four normal (14,179 cells) and five advanced-CTCL (30,663 cells) skin biopsies clustered using Seurat (19). (A) Hematoxylin and eosin staining (H&E) of skin biopsies from representative normal skin (NS) and the five tumors analyzed by scRNA-seq: top row at 200X, bottom at 400X. (B) Two-dimensional t-SNE shows dimensional reduction of reads from single cells, revealing grouping in each CTCL sample compared to all healthy control skin samples. Cells from each subject are indicated by different colors. (C) All samples in (b) are combined. (D) Distinct gene expression signatures are represented by the clustering of known markers for multiple cell types and visualized using t-SNE. Clusters belonging to each cell type are color coded. (E) Cell types in skin cell suspensions were identified by cell-specific marker as previously described (18), including AIF1 - macrophages; VWF - endothelial cells; TPSAB1 - mast cells; SCGB1B2P - secretory (glandular) cells; RGS5 - pericytes; PMEL - melanocytes; MS4A1 - B cells; KRT1 - keratinocytes; DES - smooth muscle cells; COL1A1 – fibroblasts; CD3D - T lymphocytes; and CD1C - dendritic cells. Intensity of purple color indicates the normalized level of gene expression. Cell-type specific clusters are indicated by an arrow and gates are drawn around each cluster. t-SNE, t-distributed stochastic neighbor embedding.
Single-cell transcriptome profiles reveal inter-tumor T lymphocyte heterogeneity in CTCL skin tumors
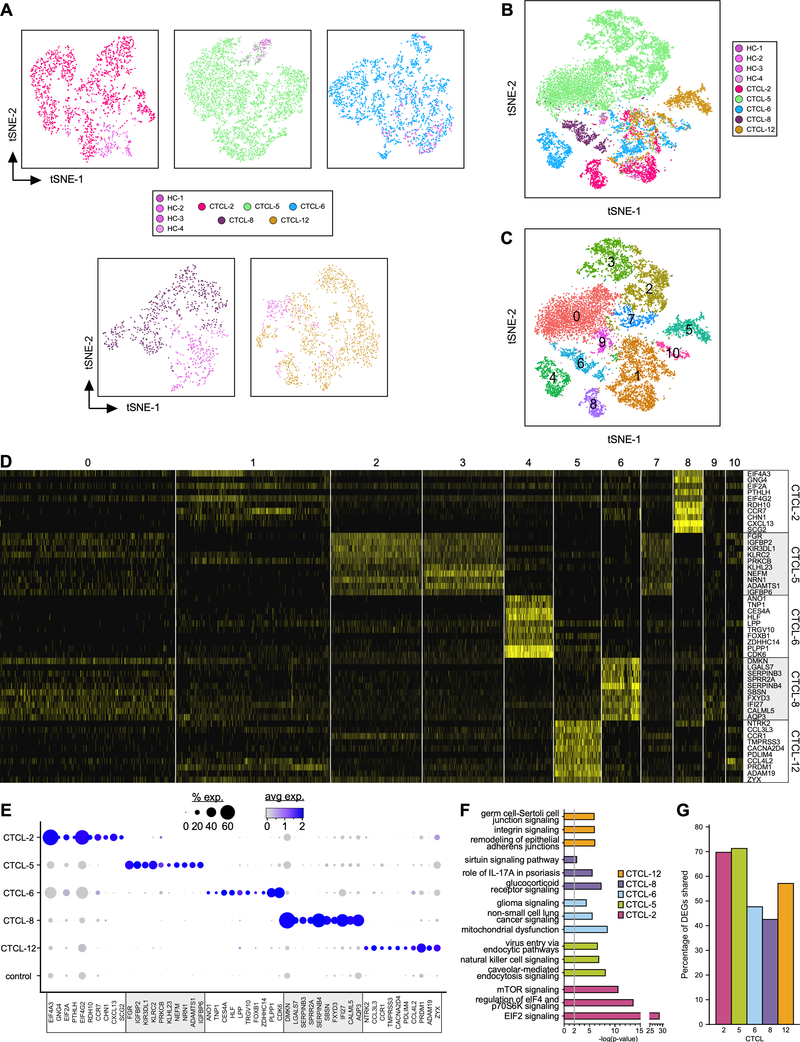
T lymphocyte transcriptional profiles of the tumors overlapped minimally with profiles of lymphocytes purified from healthy skin (Fig. 2A). Strikingly, we also observed impressive inter-tumor heterogeneity by the marginal overlap between transcriptional profile of tumor-derived lymphocytes (Fig. 2B). Comparison of the transcriptomes of each lymphocyte subset from the tumors and control skin samples identified 11 clusters (Fig. 2C). Some lymphocyte clusters were unique to individual tumors, such as cluster 8 (CTCL-2), clusters 2 and 3 (CTCL-5), cluster 4 (CTCL-6), cluster 6 (CTCL-8), and cluster 5 (CTCL-12), while clusters 1 and 10 included lymphocytes derived from all tumor and healthy skin samples.
Fig. 2. Transcriptional profiles of lymphocytes from CTCL tumors and normal skin samples.
(A-B) Transcriptomes of 14,056 cells (448 cells from normal and 13,608 cells from CTCL skin samples) expressing CD3 in groups #1, 2, 3, 13,15, 18, 19 and 20 from original t-SNE of all cells (Fig. 1E) were reanalyzed (color coded by subject) and represented as in Fig. 1B–C, revealing 11 discrete Louvain clusters using Seurat (19) (C). (D) Heat map showing examples of the most highly significant differentially expressed genes (n=10) for each cluster from (C). Cluster numbers are indicated at the top, while the cell source is indicated on the right side. Each column represents a cell. (E) Dot-plot showing the proportion of cells and the scaled average gene expression of the DE genes selected in (D). (F) Pathway analysis by Ingenuity of the most significant genes from the tumor-specific clusters. (G) Percentage of DEGs shared between tumor-specific clusters and TOX+ cells from the same tumor.
We determined the differential expression (DE) of genes in each of the unique clusters from each tumor by comparing gene expression from each cell in the cluster to that of all other cells in the dataset, using a cut-off of p<0.05 and further requiring expression of the gene from >25% of cells in the cluster. Thus, DE-identified genes are expressed either uniquely or by a large proportion of cells within each cluster compared to all other clusters. Examples of the most highly significant DE genes for each cluster are highlighted by heatmap (Fig. 2D) and the proportion of cells and the scaled average expression of these genes by all tumors and controls show strong and specific expression within individual tumors (Fig. 2E). Uniquely expressed genes included RDH10, CXCL13, SCG2 (CTCL-2), FGR, IGFBP2/P6, NEFM (CTCL-5), ANO1, TNP1, CES4A, ZDHHC14 (CTCL-6), LGALS7, SERPINB3/B4, SPRR2A (CTCL-8), NTRK2, TMPRSS3 (CTCL-12). From these and other DE genes (Table S3) we identified distinct gene expression signatures for each tumor-specific cluster, including expression of eukaryotic initiation factors (eIFs) and oncogenes (CTCL-2); NK-cell receptor and signaling molecules (CTCL-5); genes associated with tumor cell survival, proliferation, and metastasis (CTCL-6); members of the serpin, S100, and galectin families (CTCL-8), and genes associated with increased cell motility and invasiveness (CTCL-12). Ingenuity Pathway Analysis (IPA) (21) identified activation of key molecular pathways in these patient-specific tumor clusters. Highly significant examples of distinct pathways activated in each tumor were unrelated between patient samples (Fig. 2F) but followed the gene expression signatures identified above. These included eIF2, eIF4 and mTOR signaling (CTCL-2); NK-cell signaling and virus entry via endocytic pathways (CTCL-5); tumorigenic pathways common to glioma and non-small cell lung cancer (CTCL-6); pathways related to skin inflammation and skin-barrier dysfunction (CTCL-8); and pathways associated with epithelial-mesenchymal-transition (CTCL-12).
TOX (thymus high-mobility group box) is considered a marker of malignant lymphocytes in CTCL tumors (9,22,23). Strikingly, clustering of the TOX+ cells from each tumor sample (Fig. S1) revealed a large overlap of gene expression with the corresponding tumor-specific clusters identified above (Fig. 2G). Additionally, in these tumor-specific clusters we found significant but heterogeneous overexpression of genes associated with tumorigenesis, tumor-cell proliferation, and resistance to apoptosis (Fig. S2). Although the single-cell gene expression approach employed (3’ Library kit) does not allow TCR repertoire profiling, and therefore the identification of malignant lymphocytes by alpha-betaTCR clonality (24), we still could detect strong expansions of TRBC1 or TRBC2 genes in these tumor-specific clusters (Fig. S3), consistent with the occurrence of alpha-betaTCR clonality. Newly available scRNA-seq tools will allow characterization of this clonality in future work as was demonstrated in a recent study on CD4+ T cells from the peripheral blood of a Sezary patient (25). Nonetheless, we conclude from our lines of evidence that the heterogeneous but tumor-specific signatures confirmed in TOX+ cells represent patient-specific gene expression of malignant lymphocytes that may have implications for personalized therapy focusing on specific pathways.
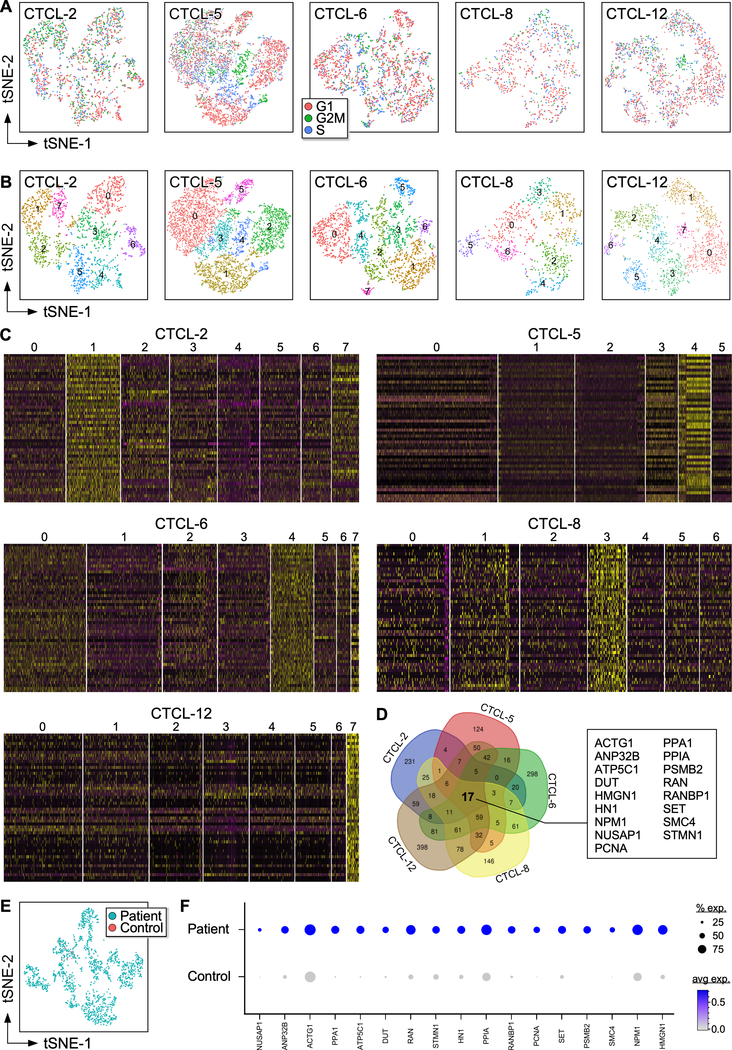
A gene expression signature identifies highly proliferating lymphocytes in advanced-stage CTCL skin tumors
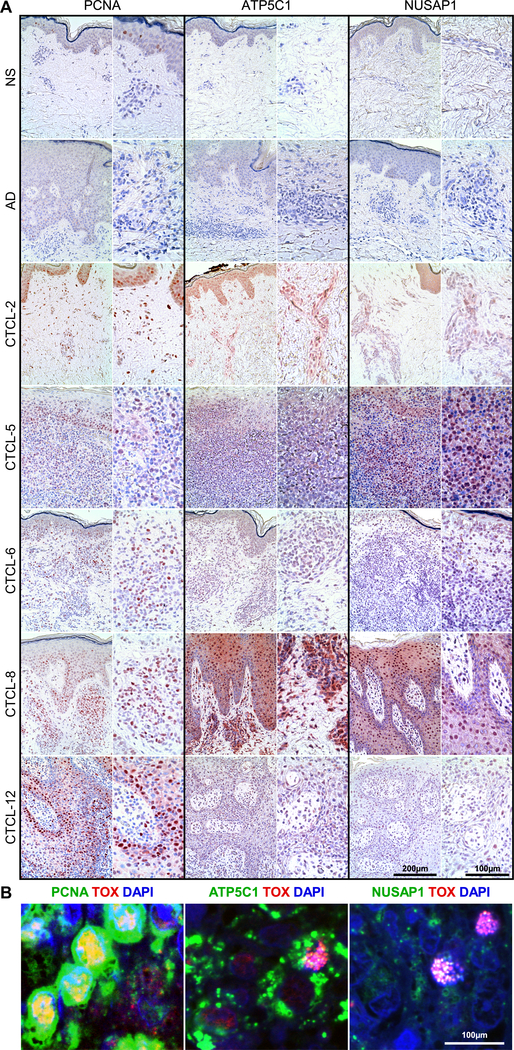
Cell cycle analysis identified actively proliferating lymphocytes by expression of G2/M and S phase genes (Fig. 3A–B). The fifty highest DE genes for each cluster of the five tumors analyzed (Fig. 3C) show specific clusters characterized by strong gene expression signatures, such as clusters 1 and 7 (CTCL-2), cluster 3 and 4 (CTCL-5), cluster 4 (CTCL-6), cluster 3 (CTCL-8), and cluster 7 (CTCL-12). Strikingly, these clusters corresponded to the highly proliferating lymphocytes identified in Figure 3A–B and highly expressed genes involved in cell cycle progression (e.g., PCNA, CDK6, CCND1, NUSAP1, CENPE, CCNA2, HMMR, CDCA8, CDK1, CENPM, CDC20, ATP5C1), proliferation (e.g., MIK67, KIAA0101, TOP2A, NPM1, IGF2, PLK1, MYC, FOS, NPM1, PRDX1, PIM2, RAN) and survival (e.g., BCL2, BIRC5, BIRC3, BCL2L12, MCTS1, TSC22) (Table S4), therefore likely representing highly proliferating malignant lymphocytes. Comparison of these clusters identified a 17-gene expression signature common to all five tumors tested (Fig. 3D). High expression by these genes was detected in all tumors while only few positive cells were found in the lymphocyte clusters of healthy controls for most genes identified (Fig. S4). Strikingly, this 17-gene expression signature was also found in TOX+ cells from all patient samples but not from controls (Fig. 3E–F). We focused on three of these common genes, PCNA, ATP5C1, and NUSAP1, that presented low expression in normal lymphocytes. These were further investigated by immunohistochemistry in the five tumors analyzed by scRNA-seq (Fig. 4A) as well as in additional samples from patients with advanced-stage CTCL (Fig. S5). Staining in tumor samples was compared to normal (NS) and atopic dermatitis (AD) skin. Results showed that apart from scant PCNA+ cells in the epidermis, NS and AD skin were negative for expression of these markers, while all tumor samples tested exhibited high numbers of positive cells both in the epidermis and in the dermis for all three markers tested. By multicolor immunofluorescence microscopy we next demonstrated that these markers co-localized with TOX (Fig. 4B). Thus, we have identified a gene expression signature of highly proliferating malignant lymphocytes that is common to all tumors tested and could be developed as a marker for the diagnosis of CTCL.
Fig. 3. Gene expression signature of proliferating lymphocytes.
(A) t-SNE analysis of CTCL and normal T lymphocytes in the cell cycle. Expression of S and G2 genes highlights proliferating cells. (B) Louvain clusters from T lymphocytes of individual CTCL tumors. (C) Heat maps of lymphocyte transcriptomes from individual tumors showing 50 examples of highly significantly DE genes in each of the clusters in (b). Cluster numbers are indicated at the top. Each column represents a cell. (D) Venn diagram showing overlap of expressed genes in highly proliferating lymphocytes from clusters 1, 7 (CTCL-2); clusters 3, 4 (CTCL-5); cluster 4 (CTCL-6); cluster 3 (CTCL-8), and cluster 7 (CTCL-12). (E) Transcriptomes of TOX+ T lymphocytes from patient tumors and healthy control skin samples. (F) Dot-plot shows the proportion of cells and the scaled average gene DE expression of the 17 common genes identified in (D).
Fig. 4. High numbers of ATP5C1+TOX+, PCNA+TOX+, and NUSAP1+TOX+ T cells accumulate in the skin tumors of patients with advanced-stage CTCL.
(A) Immunohistochemical stain from skin biopsies of normal skin (NS, n=4), atopic dermatitis (AD, n=4), and advanced-stage CTCL (n=5) used in scRNA-seq experiments, each at 200X (left) and 400X (right). (B) Representative examples from 3 patient samples tested of double color immunofluorescence staining for ATP5C1/TOX, PCNA/TOX, and NUSAP1/TOX, as indicated, at 1000X. DAPI stains nuclei.
Tumor-infiltrating CD8+ T lymphocytes exhibit heterogeneity on effector and exhaustion programs across patients
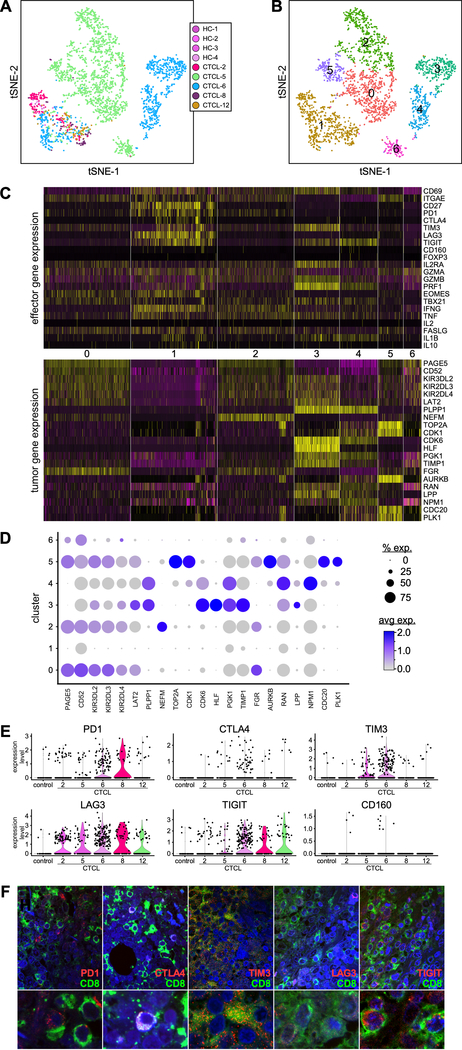
Tumor-infiltrating lymphocytes (TILs), particularly CD8+ T cells, are the major effector cell-type for fighting and killing cancer cells (12,13). To define the molecular signature of CD8+ TILs in the tumor microenvironment of advanced-stage CTCL skin tumors, we examined gene expression of effector molecules, checkpoint receptor inhibitors, and markers of T regulatory (Treg) cells in CD8+ T cells from the tumor and control skin samples. Transcriptome profiles were distinct and non-overlapping for CTCL-5 and CTCL-6 (Fig. 5A), while most CD8+ T cells from tumors and controls appear to overlap in cluster 1 (Fig. 5B). The cell composition of each CD8+ cluster is reported in Table S5. Thus, the cells in cluster 1 appeared to reflect reactive CD8+ T cells, i.e. TILs, as they did not express tumor-associated genes (Fig. 5C, lower panel, and 5D), while CD8+ T cells from the other clusters expressed genes associated with tumors likely indicating malignant lymphocytes but which are also associated with dysregulation. CD8+ TILs in cluster 1 expressed markers of skin-residency, such as CD69 and ITGAE, and of memory cells (CD27). Multiple co-inhibitory receptors were expressed by cells in this cluster, although we found variability across tumors (Fig. 5C, upper panel, and 5E), including expression of PD1, CTLA4, TIM3, LAG3, and TIGIT by most CD8+ TILs from CTCL-6; PD1 and LAG3 expression by several cells from CTCL-2, and expression by few cells from the other tumors. However, co-inhibitory receptors were also expressed by CD8+ lymphocytes from tumor-specific clusters such as PD1 and TIM3 (clusters 0, 2, and 5: CTCL-5) as well as TIM3 and TIGIT (clusters 3 and 4: CTCL-6) (Fig. 5C). Similarly, we observed variable expression of co-inhibitory receptors by CD3+CD4+ T cells across all samples and within tumor-specific clusters (Fig. S6). Multicolor immunofluorescence microscopy shows representative expression of co-inhibitory receptors by CD8+ lymphocytes in advanced CTCL tumors (Fig. 5F). No cells in any CD8+ clusters expressed Treg markers such as FOXP3 and only cells in clusters 0, 2, and 3 expressed IL2RA. Conversely, we were able to identify specific CD3+CD4+ lymphocyte clusters that co-expressed FOXP3, IL2RA, PD1, CTLA4, LAG3, and TIGIT, which likely identified Treg TILs (Fig. S6C).
Fig. 5. Expression of effector and exhaustion genes by CD8+ T cells across patient skin tumors.
(A) Transcriptomes of CD8+ T lymphocytes from individual CTCL tumors and normal skin samples (color coded by subject), revealing 7 discrete Louvain clusters (B). (C) Gene expression from the 7 discrete Louvain clusters in (b), showing DE of co-inhibitory receptors and effector molecules (upper panel) and of tumor-associated genes (lower panel). Cluster numbers are indicated in the middle. Each column represents a cell. (D) Dot-plot shows the proportion of cells and the scaled average gene DE expression of the tumor-associated genes selected in (c, lower panel). (E) Violin plots show expression of co-inhibitory receptors by CD8+ T lymphocytes from cluster 1. (F) Immunofluorescence microscopy shows co-expression of CD8 and co-inhibitory receptors, as indicated, in advanced-stage CTCL skin tumors. A representative experiment is shown at 1000X (top) and zoomed-in by 3X (bottom).
Analysis of effector molecule expression indicated that several CD8+ TILs in cluster 1 expressed granzyme A (GZMA), while only few cells expressed granzyme B (GZMB) and perforin (PRF1). CTCL-6 CD8+ T cells from clusters 3 and 4 highly expressed GZMB and PRF1, or PRF1 only, respectively. None of the other CD8+ clusters contained significant numbers of cells expressing cytolytic molecules (Fig. 5C). However, we detected a variable and modest up-regulation of FASL in most clusters, potentially providing an alternate cytolytic mechanism. CD8+ lymphocytes from clusters 1 and 2 expressed IFNγ while cells from most clusters expressed TNFα and IL1B. Interestingly, specific CD4+ clusters expressed IFNγ and TNFα as well as immunosuppressive cytokines such as IL-10, TGFß, IL-4 and IL-13 (Fig. S6C). Finally, we detected no IL-2 production by any clusters of CD8+ or CD4+ T cells.
Together these results reveal a complex landscape of CD8+, CD4+, and Treg TILs gene expression characterized by different levels of effector molecules and a variable combination of co-inhibitory receptors likely impairing an effective anti-tumor response.
DISCUSSION
Tumor cellular heterogeneity poses challenges to cancer diagnosis and treatment. Advances in single-cell gene expression profiling of patient samples open new avenues for dissecting this heterogeneity, which is a central feature of precision medicine. We employed scRNA-seq technology to profile the transcriptomes of thousands of individual cells from advanced-stage CTCL skin tumors. Our analysis revealed a large inter- and intra-tumor gene-expression heterogeneity, particularly in the T lymphocyte subset, as well as a common gene expression signature in highly proliferating lymphocytes that was validated in multiple advanced-stage skin tumors. In addition, we established the immunological state of TILs and found heterogeneity in effector and exhaution programs across patient samples. Thus, single-cell analysis provides an unprecedented view of all major cellular components simultaneously and their individual gene expression states, with important implications for diagnosis and personalized disease treatment.
Large numbers of malignant and non-malignant reactive lymphocytes are often found infiltrating CTCL skin tumors. However, specific markers have been lacking for identifying the malignant lymphocytes since the tumor cells cannot be reliably isolated from the lesional skin of patients. This limitation prevents characterizing the transcriptional profile and heterogeneity of malignant lymphocytes or distinguishing them from benign reactive lymphocytes that might block tumor growth. Further, it delays the diagnosis of CTCL and complicates development of effective treatments. Characterizing single-cell transcriptomes overcomes these problems while providing an unbiased and comprehensive map of rare lymphocyte populations and cell states within each tumor sample. We found that the transcriptional profiles of lymphocytes isolated from healthy control skin were similar to each other, while those from tumor skin appeared mostly distinct from the healthy skin profiles and overlap only partially with each other. This variation reflects both the different subtypes of the samples studied as well as any tumor-specific expression unique to individual patients. Significantly, we could identify at least one unique lymphocyte cluster for each CTCL tumor sample analyzed, and their malignant phenotype was confirmed by the overlap in gene expression with cells from the same tumor expressing TOX, a marker previously shown to identify malignant lymphocytes in CTCL. Thus, we have demonstrated a novel basis for identifying tumor cell heterogeneity that may be developed for personalized therapies.
The unique transcriptional pattern of tumor-specific lymphocyte clusters observed for each tumor sample indicates activation of specific tumor-associated signaling pathways. Some expressed genes had not been previously associated with CTCL while the expression of others that had been linked to CTCL varied across patient samples from high level expression by many cells in some tumors to little or no expression in others. For example, the cluster unique for CTCL-2, a sample from a stage IVA Sezary syndrome patient, overexpressed chemokine genes such as CXCL13, CCR7 and CCR4 that confer enhanced migratory ability to memory Sezary cells (26). Further, multiple eIF proteins were up-regulated in this tumor sample, showing that the eIF2 and eIF4 signaling pathways are activated as well as the mTOR signaling cascade, a major regulator of eIF4 and ribosomal protein S6 kinase (27). Interestingly, deregulation or altered expression of eIFs leads to translational reprogramming and promotes several oncogenic processes, including tumor cell survival, proliferation, neo-vascularization, and metastasis (27). In contrast, lymphocytes of the two clusters unique to CTCL-5 (representing a CD8+ aggressive cytotoxic CTCL) expressed genes involved in NK cell-signaling as well as several NK-cell receptors, including killer-cell immunoglobulin-like receptors and CLEC12A that negatively regulate NK-mediated cytotoxicity against tumor cells (28,29). We also detected up-regulation of several genes involved with virus entry via endocytic pathways, which is intriguing in view of the potential role for persistent viral infections in the etiology of CTCL (30). A third pattern characteristic of cells from the CTCL-6-specific cluster was expression of genes involved with tumor cell survival, proliferation, and metastasis, some common to tumorigenic pathways specific to glioma and non-small cell lung cancer (e.g.: CDK6, KRAS, PA2G4, PIK3R1, RB1, RRAS2, RXRA, TFDP1).
Parallel to up-regulation of genes associated with carcinogenesis and metastasis such as TPT1 (31) and MALAT1 (32), cells from the CTCL-8-specific cluster expressed the cysteine-protease inhibitors SERPINB3 and SERPINB4, which are expressed by various tumors and involved in inactivating granzyme M, an enzyme that kills tumor cells (33). Moreover, SERPINB3/B4 promotes tumor-cell transformation, migration, and drug resistance (33) and contributes to inflammation and barrier dysfunction in inflammatory skin diseases (34). Indeed, we found that cells in this cluster up-regulated expression of genes from the psoriasis-like pathway as well as those associated with skin-barrier dysfunction, which is a striking match to the histopathological characterization of the CTCL-8 sample (Table S3). Previous studies have shown that malignant T lymphocytes drive the morphological and histopathological changes observed in MF skin lesions, including keratinocyte hyperproliferation and compromised skin barrier function (35). These changes lead to downregulation of keratinocyte differentiation markers and increased intercellular distance, enhancing skin permeability and contributing to the increased susceptibility to skin infections in CTCL patients, particularly in advanced-stage disease (36). The cluster unique to CTCL-12, from a patient with MF and large-cell transformation, overexpressed genes associated with epithelial-mesenchymal-transition (EMT). This process allows tumor cells to acquire migratory, invasive, and stem-like properties, promoting tumor infiltration and metastasis (37,38). Although originally associated with epithelial-derived tumors, EMT is also implicated in hematological cancers (39). During ETM, cancer cells change morphology by disruption of intercellular junctions, loss of cell polarity, reorganization of the cytoskeleton, and increased cell motility necessary for invasion (37,38). In particular, we observed up-regulation of genes associated with the remodeling of adherens junctions, changes in cell adhesion, activation of small GTPases of the Rho family such as RAC1, CDC42, and RHOA, and reorganization of the actin cytoskeleton (37,38,40) (Table S3). We also found up-regulation of TMPRSS3, a type II transmembrane serine protease that contributes to EMT in other human cancers via activation of the ERK1/2 signaling pathway (41).
While characterizing this vast inter-tumor heterogeneity in gene expression that may be essential for tailoring personalized medicine, we also searched for gene expression common to all tumors that could be used to guide diagnosis, design new medications that treat all CTCL tumor subtypes, and monitor treatment efficacy. We found that highly proliferating T lymphocytes in each tumor expressed a very defined and strong gene expression signature involving cell cycle progression, proliferation, resistance to apoptosis, and metabolic processes. Strikingly, we found that these signatures had 17 genes in common that we found to be also expressed by TOX+ cells in all tumors. We validated the protein co-expression of three of them (PCNA, ATP5C1, NUSPA1) with TOX in multiple patients with advanced-stage CTCL. Thus, these results strongly indicate that both the common and heterogeneous patterns of gene expression can be exploited for diagnosis and treatment of CTCL.
Expression of checkpoint inhibitory receptors renders CD8+ TILs incapable of mounting an efficient anti-tumor response, as manifested by impaired degranulation and reduced proinflammatory cytokine production (12,13). Thus, recent cancer immunotherapies have focused on enhancing CD8+ T cell antitumor responses by targeting the inhibitory receptors (12), yielding major clinical benefits (42). However, only a subset of patients exhibited clear long-term responses, while most patients with different types of tumors failed to respond. This failure likely results from the expression of multiple inhibitory receptors on TILs that may synergistically modulate antitumor responses by different pathways. Preclinical and clinical (43) studies have indicated that full antitumor immunity may require several inhibitory receptors to be blocked. Consistent with these studies, our analysis demonstrates that co-inhibitory receptors are simultaneously but heterogeneously expressed on both CD8+ and CD4+ T lymphocytes. A particularly striking example is the overexpression of TIGIT, LAG3 and TIM3 by CD8+ T cells in CTCL-6, and LAG3 in CTCL-2, indicating a strong patient-specific signature that may be exploited for individualized targeting. Although co-inhibitory receptors appeared to be expressed mostly by reactive TILs, we also observed their expression by T lymphocytes expressing tumor-associated genes. In some cases, these cells were clearly identifiable as malignant, while in others they may instead represent exhausted T lymphocytes acquiring a malignant phenotype or nonconventional Tregs such as CD4+ Tregs lacking FOXP3 and/or IL2RA expression.
TIGIT expression by CD4+ T cells from the peripheral blood of patients with advanced-stage SS was recently associated with reduced IFNγ and IL-2 production (44). We found that TIGIT is also highly expressed by both CD8+ and CD4+ TILs from advanced-stage CTCL skin tumors, and in parallel with expression of other co-inhibitory receptors, most notably TIM3, PD1, LAG3, and to a lesser extent CTLA4. Since TIGIT can foster an immunosuppressive tumor microenvironment by promoting Treg function and maintenance as well as by inhibiting cytotoxic T cell activity (45), we confirmed TIGIT expression by Tregs in the samples tested and found a correlation with lack of GZMB and perforin expression in cluster 1 CD8+ TILs that characterizes exhaustion. Interestingly, we found that CTCL-6-specific CD8+TIGIT+ lymphocytes from cluster 3 expressed GZMB and perforin as well as tumor-associated genes, likely representing cytotoxic malignant lymphocytes. CD8+ TILs showed no IL-2 expression while most cells expressed TNFα and IFNγ, which was associated with EOMES expression, consistent with an exhausted CD8+ T cell phenotype (46). Such cells are defective in IFNγ production and cytotoxicity but continue expressing IFNγ and granzyme mRNAs (46), consistent with our findings. Likewise, we observed that CD4+TILs also expressed IFNγ and TNFα as well as TGFß, which is likely produced in combination with IL-10 by FOXP3+ cytotoxic Tregs (47). Furthermore, variability observed in TGFß, IL-10, IL-4 and IL-13 cytokine production from CD4+ T cells of the CTCL-12-specific and CTCL-8-specific clusters likely reflects FOXP3−Tregs or malignant lymphocytes, or CD4+FOXP3+ malignant lymphocytes with suppressive activity (48), respectively. We conclude that multiple co-inhibitor receptors are expressed by malignant and reactive lymphocytes in advanced CTCL skin tumors, conferring to the latter a dysfunctional phenotype. Understanding the heterogeneity in co-inhibitory receptor expression may be essential for developing patient-specific therapy and for guiding checkpoint inhibitor blocking in order to permit effective killing of tumor cells by TILs.
In conclusion, single-cell transcriptome profiling provides novel insights into CTCL disease heterogeneity by revealing patient-specific landscapes of malignant and reactive lymphocytes. Recent reports applying single-cell RNA-seq to peripheral blood of Sezary patients (25,49) also demonstrated heterogeneity in the malignant T-cell population and, in addition, genetic heterogeneity within the same patient over time has been observed (50). Although studies with small subsets of patients need to be expanded to confirm and extend general trends, we nonetheless demonstrate the ability to detect gene expression patterns among single skin tumor cells that can provide a framework for improving CTCL diagnosis and treatment, thus realizing the goal of precision medicine.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Advances in single-cell gene expression profiling of patient samples open new avenues for dissecting tumor cell heterogeneity, which is a central feature of precision medicine. We employed scRNA-seq technology to profile the transcriptomes of thousands of individual cells from advanced-stage CTCL skin tumors. Our analysis revealed a large inter- and intra-tumor gene-expression heterogeneity, particularly in the T lymphocyte subset, as well as a common gene expression signature in highly proliferating lymphocytes that was validated in multiple advanced-stage skin tumors. In addition, we established the immunological state of tumor infiltrating lymphocytes and found heterogeneity in effector and exhaution programs across patient samples. Thus, single-cell analysis provides an unprecedented view of all major cellular components simultaneously and their individual gene expression states. New developments in single-cell transcriptome profiling are highly relevant for discovering clinically relevant biomarkers of disease and for tailoring patient-specific treatment.
ACKNOWLEDGEMENTS
We thank Christina Morse for technical support for immunofluorescence microscopy. This work was supported by National Institutes of Health /National Cancer Institute grant R21 CA209107–02 to P.F.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. The New England journal of medicine 2004;350(19):1978–88. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127(20):2375–90 doi 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol 2010;28(31):4730–9 doi 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 4.Scarisbrick JJ, Kim YH, Whittaker SJ, Wood GS, Vermeer MH, Prince HM, et al. Prognostic factors, prognostic indices and staging in mycosis fungoides and Sezary syndrome: where are we now? Br J Dermatol 2014;170(6):1226–36 doi 10.1111/bjd.12909. [DOI] [PubMed] [Google Scholar]
- 5.Scarisbrick JJ, Prince HM, Vermeer MH, Quaglino P, Horwitz S, Porcu P, et al. Cutaneous Lymphoma International Consortium Study of Outcome in Advanced Stages of Mycosis Fungoides and Sezary Syndrome: Effect of Specific Prognostic Markers on Survival and Development of a Prognostic Model. J Clin Oncol 2015. doi 10.1200/JCO.2015.61.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arulogun SO, Prince HM, Ng J, Lade S, Ryan GF, Blewitt O, et al. Long-term outcomes of patients with advanced-stage cutaneous T-cell lymphoma and large cell transformation. Blood 2008;112(8):3082–7 doi 10.1182/blood-2008-05-154609. [DOI] [PubMed] [Google Scholar]
- 7.Scarisbrick JJ, Quaglino P, Prince HM, Papadavid E, Hodak E, Bagot M, et al. The PROCLIPI international registry of early stage Mycosis Fungoides identifies substantial diagnostic delay in most patients. Br J Dermatol 2018. doi 10.1111/bjd.17258. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 2008;454(7203):436–44. [DOI] [PubMed] [Google Scholar]
- 9.Geskin LJ, Viragova S, Stolz DB, Fuschiotti P. Interleukin-13 is over-expressed in cutaneous T-cell lymphoma cells and regulates their proliferation. Blood 2015. doi blood-2014-07-590398 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komohara Y, Takeya M. CAFs and TAMs: maestros of the tumour microenvironment. J Pathol 2017;241(3):313–5 doi 10.1002/path.4824. [DOI] [PubMed] [Google Scholar]
- 11.Guenova E, Watanabe R, Teague JE, Desimone JA, Jiang Y, Dowlatshahi M, et al. TH2 cytokines from malignant cells suppress TH1 responses and enforce a global TH2 bias in leukemic cutaneous T-cell lymphoma. Clin Cancer Res 2013;19(14):3755–63 doi 1078-0432.CCR-12-3488 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crespo J, Sun H, Welling TH, Tian Z, Zou W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol 2013;25(2):214–21 doi 10.1016/j.coi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15(8):486–99 doi 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016;44(5):989–1004 doi 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015;161(5):1202–14 doi 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schelker M, Feau S, Du J, Ranu N, Klipp E, MacBeath G, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nature communications 2017;8(1):2032 doi 10.1038/s41467-017-02289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017;355(6332) doi 10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabib T, Morse C, Wang T, Chen W, Lafyatis R. SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin. The Journal of investigative dermatology 2018;138(4):802–10 doi 10.1016/j.jid.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33(5):495–502 doi 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushati N, Smith J, Briscoe J, Watkins C. An intuitive graphical visualization technique for the interrogation of transcriptome data. Nucleic Acids Res 2011;39(17):7380–9 doi 10.1093/nar/gkr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30(4):523–30 doi 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, et al. Molecular markers of early-stage mycosis fungoides. The Journal of investigative dermatology 2012;132(6):1698–706 doi jid201213 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Dulmage B, Geskin L, Guitart J, Akilov OE. The biomarker landscape in mycosis fungoides and Sezary syndrome. Exp Dermatol 2017;26(8):668–76 doi 10.1111/exd.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Masson A, O’Malley JT, Elco CP, Garcia SS, Divito SJ, Lowry EL, et al. High-throughput sequencing of the T cell receptor beta gene identifies aggressive early-stage mycosis fungoides. Science translational medicine 2018;10(440) doi 10.1126/scitranslmed.aar5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borcherding N, Voigt AP, Liu V, Link BK, Zhang W, Jabbari A. Single-cell profiling of cutaneous T-cell lymphoma reveals underlying heterogeneity associated with disease progression. Clin Cancer Res 2019. doi 10.1158/1078-0432.CCR-18-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picchio MC, Scala E, Pomponi D, Caprini E, Frontani M, Angelucci I, et al. CXCL13 is highly produced by Sezary cells and enhances their migratory ability via a synergistic mechanism involving CCL19 and CCL21 chemokines. Cancer Res 2008;68(17):7137–46 doi 10.1158/0008-5472.CAN-08-0602. [DOI] [PubMed] [Google Scholar]
- 27.de la Parra C, Walters BA, Geter P, Schneider RJ. Translation initiation factors and their relevance in cancer. Curr Opin Genet Dev 2018;48:82–8 doi 10.1016/j.gde.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wechsler J, Bagot M, Nikolova M, Parolini S, Martin-Garcia N, Boumsell L, et al. Killer cell immunoglobulin-like receptor expression delineates in situ Sezary syndrome lymphocytes. J Pathol 2003;199(1):77–83 doi 10.1002/path.1251. [DOI] [PubMed] [Google Scholar]
- 29.Benson DM Jr., Caligiuri MA. Killer immunoglobulin-like receptors and tumor immunity. Cancer Immunol Res 2014;2(2):99–104 doi 10.1158/2326-6066.CIR-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litvinov IV, Shtreis A, Kobayashi K, Glassman S, Tsang M, Woetmann A, et al. Investigating potential exogenous tumor initiating and promoting factors for Cutaneous T-Cell Lymphomas (CTCL), a rare skin malignancy. Oncoimmunology 2016;5(7):e1175799 doi 10.1080/2162402X.2016.1175799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucibello M, Gambacurta A, Zonfrillo M, Pierimarchi P, Serafino A, Rasi G, et al. TCTP is a critical survival factor that protects cancer cells from oxidative stress-induced cell-death. Exp Cell Res 2011;317(17):2479–89 doi 10.1016/j.yexcr.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Kim SH, Yang WI, Kim SJ, Yoon SO. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget 2017;8(19):31305–17 doi 10.18632/oncotarget.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y, Sheshadri N, Zong WX. SERPINB3 and B4: From biochemistry to biology. Semin Cell Dev Biol 2017;62:170–7 doi 10.1016/j.semcdb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izuhara K, Yamaguchi Y, Ohta S, Nunomura S, Nanri Y, Azuma Y, et al. Squamous Cell Carcinoma Antigen 2 (SCCA2, SERPINB4): An Emerging Biomarker for Skin Inflammatory Diseases. Int J Mol Sci 2018;19(4) doi 10.3390/ijms19041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thode C, Woetmann A, Wandall HH, Carlsson MC, Qvortrup K, Kauczok CS, et al. Malignant T cells secrete galectins and induce epidermal hyperproliferation and disorganized stratification in a skin model of cutaneous T-cell lymphoma. The Journal of investigative dermatology 2015;135(1):238–46 doi 10.1038/jid.2014.284. [DOI] [PubMed] [Google Scholar]
- 36.Willerslev-Olsen A, Krejsgaard T, Lindahl LM, Bonefeld CM, Wasik MA, Koralov SB, et al. Bacterial toxins fuel disease progression in cutaneous T-cell lymphoma. Toxins (Basel) 2013;5(8):1402–21 doi 10.3390/toxins5081402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Silva-Diz V, Lorenzo-Sanz L, Bernat-Peguera A, Lopez-Cerda M, Munoz P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin Cancer Biol 2018. doi 10.1016/j.semcancer.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006;66(17):8319–26 doi 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 39.Chen SC, Liao TT, Yang MH. Emerging roles of epithelial-mesenchymal transition in hematological malignancies. J Biomed Sci 2018;25(1):37 doi 10.1186/s12929-018-0440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeichi M Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol 2014;15(6):397–410 doi 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 41.Wang JY, Jin X, Li XF. Knockdown of TMPRSS3, a Transmembrane Serine Protease, Inhibits Proliferation, Migration, and Invasion in Human Nasopharyngeal Carcinoma Cells. Oncology research 2018;26(1):95–101 doi 10.3727/096504017X14920318811695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27(4):450–61 doi 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. The New England journal of medicine 2017;377(14):1345–56 doi 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jariwala N, Benoit B, Kossenkov AV, Oetjen LK, Whelan TM, Cornejo CM, et al. TIGIT and Helios Are Highly Expressed on CD4(+) T Cells in Sezary Syndrome Patients. The Journal of investigative dermatology 2017;137(1):257–60 doi 10.1016/j.jid.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunological reviews 2017;276(1):112–20 doi 10.1111/imr.12518. [DOI] [PubMed] [Google Scholar]
- 46.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007;27(4):670–84 doi 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Akhmetzyanova I, Zelinskyy G, Littwitz-Salomon E, Malyshkina A, Dietze KK, Streeck H, et al. CD137 Agonist Therapy Can Reprogram Regulatory T Cells into Cytotoxic CD4+ T Cells with Antitumor Activity. J Immunol 2016;196(1):484–92 doi 10.4049/jimmunol.1403039. [DOI] [PubMed] [Google Scholar]
- 48.Heid JB, Schmidt A, Oberle N, Goerdt S, Krammer PH, Suri-Payer E, et al. FOXP3+CD25- tumor cells with regulatory function in Sezary syndrome. The Journal of investigative dermatology 2009;129(12):2875–85 doi 10.1038/jid.2009.175. [DOI] [PubMed] [Google Scholar]
- 49.Buus TB, Willerslev-Olsen A, Fredholm S, Blumel E, Nastasi C, Gluud M, et al. Single-cell heterogeneity in Sezary syndrome. Blood Adv 2018;2(16):2115–26 doi 10.1182/bloodadvances.2018022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology 2017;6(5):e1306618 doi 10.1080/2162402X.2017.1306618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.